1. Introduction
Polyketide synthases (PKSs) are a large family of multifunctional enzymes related to fatty acid synthases (Hopwood, 1997; Hopwood and Sherman, 1990; Staunton and Wiessman, 2001; Castoe et al., 2007). PKSs catalyze the biosynthesis of a very diverse group of compounds, mainly produced by bacteria, fungi, and plants. Several bacterial and fungal polyketides have antibiotic and mycotoxic properties, e.g. erythromycin, rifamycin, and actinorhodin. In higher plants, polyketide compounds play a role in flower pigmentation, pathogen defense, light-response, as well as symbiotic plant-pathogen interactions (Schroder et al., 1998; Winkel-Shirley, 2002). Polyketides are also of great interest for their therapeutic properties as antitumor and immunosuppressives. Animal pks genes were first discovered in sea urchins and in the genomes of chickens and fish (Calestani et al., 2003; Castoe et al., 2007). Polyketide compounds have been isolated from other echinoderms and marine molluscs (reviewed in Blunt et al., 2011), and in a few cases, biosynthesis has been also confirmed by isotopic studies (Cutignano et al., 2009). The pks genes have been identified in marine sponge metagenomes as well, but in this case they appear to belong to the symbiotic bacterial genomes (reviewed in Hochmuth and Piel, 2009).
Sea urchins (Strongylocentrotus purpuratus) have two pks genes, pks1 and pks2 (Castoe et al., 2007). The expression pattern and function of pks1 were previously described by Calestani et al. (2003). Pks1 is exclusively expressed in larval pigment cells and their precursors, and is required for the biosynthesis of the pigment, a naphthoquinone called echinochrome (Griffiths, 1965). Echinochrome, purified from adult immune cells (coelomocytes), has been shown to have antibiotic properties against several marine and non-marine bacteria (Service and Wardlaw, 1984). The onset of pks1 expression occurs in the early blastula stage, between 15 and 18 hours post fertilization, and this is restricted to secondary mesenchyme cell (SMC) precursors at the vegetal pole of the embryo. At this stage, SMC precursors surround the primary mesenchyme cell (PMC) precursors, which will later produce the larval skeleton (spicules; Wilt, 1999; Ettensohn et al., 1997). Pks1 expression continues throughout development, to the pluteus stage in pigment cells embedded in the aboral ectoderm (Calestani et al., 2003).
The sea urchin pks2 was identified by Castoe et al. (2007) using a phylogenomic approach. PKS2 is closely related to bacterial PKSs (e.g., from Nocardia farcinica, Clostridium acetobutylicum, and Microbulbifer degradans). The function of PKS2 and of the closely related bacterial PKSs is still not known. PKS2 and PKS1 conserved domain sequences differ to some extent, being KS-AT-DH-ER-KR-PP-TE and KS-AT-DH-ERPP, respectively (Castoe et al., 2007). PKS2 has two additional catalytic domains, KR and TE. The presence of a KR domain suggests that PKS2 might synthesize a polyketide with further reduction of cheto-groups, beyond that seen in the echinochrome pigment. The TE domain is usually involved in the termination of synthesis, sometime producing intramolecular cyclization (reviewed in Keating and Walsh, 1999).
In this work we characterized both the spatial and temporal expression patterns of pks2 to build the foundation for future functional and regulatory studies.
2. Results and Discussion
The pks2 sequence utilized in this study was initially derived from the genetic sequence database at the National Center for Biotechnical Information (NCBI; GenBank ID:XM_791684.2). This mRNA reference sequence is supported by EST sequence data, including PMC specific cDNA library ESTs (GenBank ID: CX688952, CD322502, CD338698, DN580373, DN782682, DN782682, CX698309) and by sea urchin embryo transcriptome sequence data (SpBase ID number: SPU_028395.2 at http://sugp.caltech.edu/SpBase/; Samanta et al., 2006; Cameron et al., 2009). We experimentally verified the mRNA sequence predictions by sequencing the pks2 cDNA (GenBank ID: XXXXXXXX and YYYYYYYY; 1 to 3 X coverage of 7,227 bp out of 7,821bp). The pks2 sequence we obtained confirmed the mRNA sequence predictions in the regions of interest, showing a 99% nucleotide identity with XM_791684.2, and SPU_028395.2. Moreover, the full-length mRNA sequence, including 5′- and 3′-UTRs, has been recently published (GenBank ID: JN680355; identified by mRNA-seq analysis of a sea urchin transcriptome; Tu et al., unpublished). This latest sequence also showed a 99% nucleotide sequence identity with our cDNA sequence. The aminoacid sequences are identical.
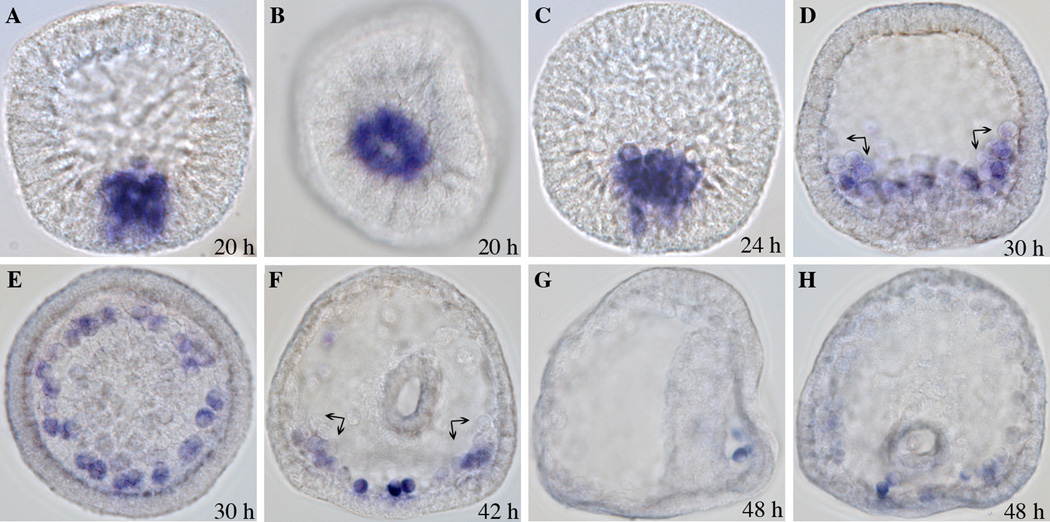
We analyzed pks2 temporal expression by real-time quantitative PCR (QPCR). The pks2 transcript levels were measured throughout development, beginning with unfertilized eggs and ending at the prism stage (50 h post fertilization; Fig. 1). Pks2 is zygotically expressed, appearing between 6 and 15 h of development. Expression peaks at the hatched blastula stage (21 h), with an average of 1273 molecules per embryo, and then steadily decreases to an average of 169 molecules per embryo at prism stage (50 h). The pks2 spatial expression pattern was analyzed by whole mount in situ hybridization (WMISH). Based on the QPCR time-course data described earlier, WMISH were performed at the following developmental stages: 17 h (not shown), 20 h, 24 h, 30 h, 42 h, and 48 h (Fig. 2). The expression of pks2 was observed in PMCs precursors at blastula stage (17–20 h; Fig. 2 A, B). Expression continues to be restricted to PMCs during mesenchyme blastula (24 h), when PMCs have ingressed into the blastocoel (Fig. 2 C). Pks2 appear to be expressed at the same level in all PMCs during these developmental stages (Fig. 2 A, B, C). At the early gastrula stage (30 h), PMCs arrange in a ring around the invaginating archenteron. At the same time, a subset of PMCs forms two aggregates (clusters) in the position corresponding to the future ventrolateral region of the embryo. The cells forming the ventrolateral clusters are responsible for the production of the first skeletogenic elements, starting at mid-gastrula (reviewed in Wilt, 1999). Our data show that, at the early gastrula stage, pks2 is still expressed in all PMCs (Fig. 2 D, E). By the late gastrula stage (42 h), pks2 expression is restricted to the oral side in a sub-group of cells of the subequatorial ring and ventrolateral clusters, which, at this point, are already producing the larval skeleton (Fig. 2 F). At the early prism stage (48 h) pks2 transcripts are limited to 3–4 PMCs localized in the ring on the oral side (Fig. 2 G, H).
Figure 1. Temporal expression of pks2.
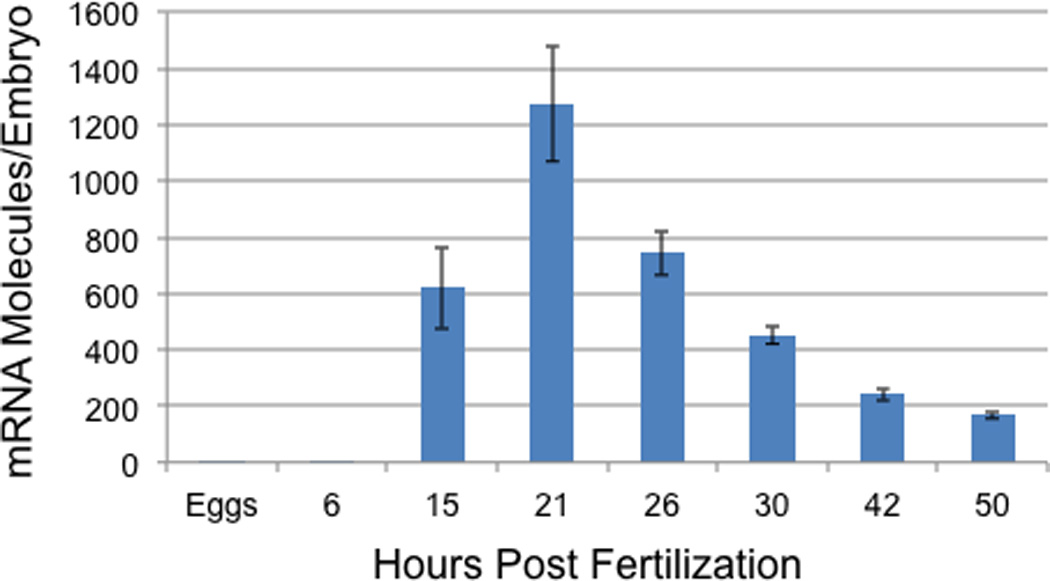
Transcript levels were measured from unfertilized eggs to the prism stage (50 h) by real-time QPCR. Data are the average number of mRNA molecules per embryo (± SE) derived from four replicate experiments (embryos produced from four sets of parents).
Figure 2. Spatial expression of pks2.
WMISH was performed on embryos at different stages of development using a RNA digoxygenin-labeled probe. Tiles are labeled with the post-fertilization time points. (A, B) Hatched blastula; (C) Mesenchyme blastula; (D, E) Early gastrula; (F) Late gastrula. (G, H) Early prism. (A, C, D, G) Side view; (B, E, F, H) Vegetal view. Expression of pks2 is specific to skeletogenic cells (PMCs) and their precursors. The black arrows indicate the ventrolateral clusters. By the prism stage, gene expression is limited to just a few PMC descendants.
Pks2 is co-expressed with other PMC differentiation genes, some of which have been shown to be required for skeletogenesis (Killian et al, 2010; Materna et al., 2010; Amore and Davidson, 2006; Livingston et al, 2006). For example, pks2 is transcriptionally activated at about the same time as cyclophilin, ficolin, and the spicule matrix protein coding gene sm50 (Materna et al., 2010). The function of pks 2 is still unknown; nevertheless, our data do not strongly support a pks2 role in skeletogenesis, because transcript levels decrease during the time PMCs begin production of the spicules. The polyketide compound synthesized by PKS2 possibly has a role in the immune defense of the developing embryo. This hypothesis is supported by the chemical nature of polyketides, which, in large part, have antibiotic properties (Hopwood, 1997; Hopwood and Sherman, 1990; Staunton and Wiessman, 2001; Blunt et al., 2011). Similarly, the sea urchin echinochrome, synthesized by PKS1 in pigment cells, has been shown to have antibiotic properties (Service and Wardlaw, 1984). Previous studies suggest that pigment cells might be immune cells in the sea urchin larva. The morphology and behavior of pigment cells are, to some extent, similar to that of macrophages (Gibson and Burke, 1987). In fact, pigment cells appear to respond to bacterial immune challenge by migrating toward the site of infection and phagocytizing bacteria (Hibino et al., 2006). In this context, the putative polyketide compound synthesized by PKS2 in skeletogenic cells may provide a first line of defense during the hatched blastula and gastrula stages, preceding the biosynthesis of echinochrome, which begins to accumulate in pigment cells during early gastrula (Chaffe and Mazia, 1963). Subsequently, from the late gastrula to the prism stage, the skeletogenic cells that still express pks2 on the oral side of the embryo might complement the immune defense role of pigment cells, which are localized on the aboral side.
Future studies of pks2 function, as well as of the chemical structure and function of the polyketide produced, will provide new insight into its role during sea urchin development, and will add to the knowledge of the biological functions of animal polyketides.
3. Experimental Procedures
3.1. Embryo culture
Gametes were collected from adult sea urchins (Strongylocentrotus purpuratus). The eggs were fertilized in filtered seawater containing penicillin 20U/ml and streptomycin 50µg/ml, and then incubated at 15°C for the duration of development.
3.2. Sequencing
We amplified by PCR four overlapping pks2 fragments from cDNA of 24 hours p.f. embryos (Table S1; supplementary data). The sequences of the PCR primers were derived from the genetic sequence database at the National Center for Biotechnical Information (NCBI; mRNA Reference Sequence; GenBank ID: XM_791684.2) and are listed in Table S1 (Supplementary data). We designed 21 sequencing primers and sequenced the four overlapping pks2 fragments (Table S2; supplementary data). The sequencing service was provided by the Nevada Genomics Center (University of Nevada, Reno, NV).
3.3. Real-time quantitative PCR
Approximately 150 embryos were collected at different developmental time points (0, 6, 15, 21, 26, 30, 42, and 50 h post fertilization). RNA was isolated using an RNeasy kit (Qiagen Valencia, CA), then DNase treated with DNAse-Free (ABI/Ambion, Austin, TX), and purified using a Qiagen RNeasy kit following the manufacturer’s procedures. The cDNA was synthesized using a High-Capacity cDNA Reverse Transcriptase Archive kit (Applied Biosystems, Foster City, CA).
A time course was generated by real-time QPCR for four cDNA batches produced from four sets of parents. The QPCR primer set was specific for pks2 (there is no significant nucleotide sequence similarity between pks1 and pks2). The forward and reverse QPCR primer sequences were AGCAACATCTGGAGGAATGG and CATTCGAAACTGGTGGTGTG, respectively. The QPCR reactions were prepared in triplicate (per time point), using SYBR Green (ABI/Ambion, Austin, TX). Ubiquitin was used as internal standard. The pks2 transcripts per embryo were estimated based on the number of ubiquitin transcripts per embryo quantified by Materna et al. (2010). For the 50 h developmental time point, we used the number of ubiquitin transcripts per embryo described for the 48 h stage (Materna et al, 2010).
3.4. Whole-mount in situ hybridization
The whole-mount in situ hybridization was performed on embryos produced from two sets of parents, as previously described (Minokawa et al., 2004). The DNA template used to synthesize the WMISH probe was PCR amplified from cDNA prepared from 21 h embryos, during the stage of highest expression of pks2. The WMISH probe was designed to target the 5' coding region within the KS domain, bridging the first and second exons. The following primers were used to amplify probes 910 nucleotides in size: pks2 F-T7 5'-ATATTAATACGACTCACTATAGGGAAATCACCGGCAAGAAACAC-3' pks2 R-SP6 5'-ATATATTTAGGTGACACTATAGAATAACTTGGCAGGGCAATACC-3' The pks2 forward primer included the T7 promoter sequence and the pks2 reverse primer included the SP6 promoter sequence.
The PCR product was purified using a QIAquick PCR purification kit (Qiagen, Valencia, CA) and subsequently gel purified using a MinElute Gel Extraction kit (Qiagen, Valencia, CA). The digoxigenin- labeled sense and antisense RNA probes were produced by transcription using either T7 or SP6 RNA polymerases, following the manufacturer’s procedures (Roche Diagnostics, Indianapolis, IN).
Highlights.
-
>
We describe the polyketide synthase2 (pks2) expression pattern in sea urchin embryos
-
>
pks2 expression is restricted to skeletogenic cells and their precursors
-
>
pks2 is first detected during the blastula stage
-
>
The transcript level peaks at hatched blastula
-
>
By prism stage pks2 expression is limited to 3–4 skeletogenic cells on the oral side
Supplementary Material
Acknowledgements
The project described was supported by the NIH award number R15HD060008 from the Eunice Kennedy Shriver National Institute Of Child Health & Human Development.
Abbreviations
- AT
acyl transferase domain
- DH
dehydratase domain
- ER
enoyl reductase domain
- KR
ketoreductase domain
- KS
ketoacyl synthase domain
- PP
phosphopantetheine attachment site
- TE
thioesterase domain
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Amore G, Davidson EH. cis-Regulatory control of cyclophilin, a member of the ETS-DRI skeletogenic gene battery in the sea urchin embryo. Dev. Biol. 2006;293:555–564. doi: 10.1016/j.ydbio.2006.02.024. [DOI] [PubMed] [Google Scholar]
- Blunt JW, Copp BR, Munro MH, Northcote PT, Prinsep MR. Marine natural products. Nat. Prod. Rep. 2011;28:196–268. doi: 10.1039/c005001f. [DOI] [PubMed] [Google Scholar]
- Calestani C, Rast JP, Davidson EH. Isolation of pigment cell specific genes in the sea urchin embryo by differential macroarray screening. Development. 2003;130:4587–4596. doi: 10.1242/dev.00647. [DOI] [PubMed] [Google Scholar]
- Cameron RA, Samanta M, Yuan A, He D, Davidson E. SpBase: the sea urchin genome database and web site. Nucleic Acids Res. 2009;37:D750–D754. doi: 10.1093/nar/gkn887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castoe TA, Stephens T, Noonan BP, Calestani C. A novel group of type I polyketide synthases (PKS) in animals and the complex phylogenomics of PKSs. Gene. 2007;392:47–58. doi: 10.1016/j.gene.2006.11.005. [DOI] [PubMed] [Google Scholar]
- Chaffee RR, Mazia D. Echinochrome synthesis in hybrid sea urchin embryos. Dev. Biol. 1963;7:502–512. doi: 10.1016/0012-1606(63)90138-6. [DOI] [PubMed] [Google Scholar]
- Cutignano A, Cimino G, Villani G, Fontana A. Origin of the C3-unit in placidenes: further insights into taxa divergence of polypropionate biosynthesis in marine molluscs and fungi. Tetrahedron. 2009;65:8161–8164. [Google Scholar]
- Ettensohn CA, Guss KA, Hodor PG, Malinda KM. The morphogenesis of the skeletal system of the sea urchin embryo. In: Collier JR, editor. Progress in Developmental Biology, Vol. VII, Reproductive Biology of Invertebrates. New Delhi: Oxford/IBH Publishing; 1997. pp. 225–265. [Google Scholar]
- Gibson AW, Burke RD. Migratory and invasive behavior of pigment cells in normal and animalized sea urchin embryos. Exp. Cell. Res. 1987;173:546–557. doi: 10.1016/0014-4827(87)90294-1. [DOI] [PubMed] [Google Scholar]
- Griffiths M. A study of the synthesis of naphthaquinone pigments by the larvae of two species of sea urchins and their reciprocal hybrids. Dev. Biol. 1965;11:433–447. doi: 10.1016/0012-1606(65)90049-7. [DOI] [PubMed] [Google Scholar]
- Hibino T, Loza-Coll M, Messier C, Majeske AJ, Cohen AH, Terwilliger DP, Buckley KM, Brockton V, Nair SV, Berney K, Fugmann SD, Anderson MK, Pancer Z, Cameron RA, Smith LC, Rast JP. The immune gene repertoire encoded in the purple sea urchin genome. Dev. Biol. 2006;300:349–365. doi: 10.1016/j.ydbio.2006.08.065. [DOI] [PubMed] [Google Scholar]
- Hochmuth T, Piel J. Polyketide synthases of bacterial symbionts in sponges--evolutionbased applications in natural products research. Phytochemistry. 2009;70:1841–1849. doi: 10.1016/j.phytochem.2009.04.010. [DOI] [PubMed] [Google Scholar]
- Hopwood DA. Genetic Contributions to Understanding Polyketide Synthases. Chem. Rev. 1997;97:2465–2498. doi: 10.1021/cr960034i. [DOI] [PubMed] [Google Scholar]
- Hopwood DA, Sherman DH. Molecular genetics of polyketides and its comparison to fatty acid biosynthesis. Annu. Rev. Genet. 1990;24:37–66. doi: 10.1146/annurev.ge.24.120190.000345. [DOI] [PubMed] [Google Scholar]
- Keating TA, Walsh CT. Initiation, elongation, and termination strategies in polyketide and polypeptide antibiotic biosynthesis. Curr. Opin. Chem. Biol. 1999;3:598–606. doi: 10.1016/s1367-5931(99)00015-0. [DOI] [PubMed] [Google Scholar]
- Killian CE, Croker L, Wilt FH. SpSM30 gene family expression patterns in embryonic and adult biomineralized tissues of the sea urchin, Strongylocentrotus purpuratus. Gene Expr. Patterns. 2010;10:135–139. doi: 10.1016/j.gep.2010.01.002. [DOI] [PubMed] [Google Scholar]
- Livingston BT, Killian CE, Wilt F, Cameron A, Landrum MJ, Ermolaeva O, Sapojnikov V, Maglott DR, Buchanan AM, Ettensohn CA. A genome-wide analysis of biomineralization-related proteins in the sea urchin Strongylocentrotus purpuratus. Dev. Biol. 2006;300:335–348. doi: 10.1016/j.ydbio.2006.07.047. [DOI] [PubMed] [Google Scholar]
- Materna SC, Nam J, Davidson EH. High accuracy, high-resolution prevalence measurement for the majority of locally expressed regulatory genes in early sea urchin development. Gene Expr Patterns. 2010;10:177–184. doi: 10.1016/j.gep.2010.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Minokawa T, Rast JP, Arenas-Mena C, Franco CB, Davidson EH. Expression patterns of four different regulatory genes that function during sea urchin development. Gene Expr. Patterns. 2004;4:449–456. doi: 10.1016/j.modgep.2004.01.009. [DOI] [PubMed] [Google Scholar]
- Samanta MP, Tongprasit W, Istrail S, Cameron RA, Tu Q, Davidson EH, Stolc V. The transcriptome of the sea urchin embryo. Science. 2006;314:960–962. doi: 10.1126/science.1131898. [DOI] [PubMed] [Google Scholar]
- Schroder J, Raiber S, Berger T, Schmidt A, Schmidt J, Soares-Sello AM, Bardshiri E, Strack D, Simpson TJ, Veit M, Schroder G. Plant polyketide synthases: a chalcone synthase-type enzyme which performs a condensation reaction with methylmalonyl-CoA in the biosynthesis of C-methylated chalcones. Biochemistry. 1998;37:8417–8425. doi: 10.1021/bi980204g. [DOI] [PubMed] [Google Scholar]
- Service M, Wardlaw AC. Echinochrome-A as a bactericidal substance in the coelomic fluid of Echinus esculentus (L.) Comp. Biochem. 1984;79 B:161–165. [Google Scholar]
- Staunton J, Weissman KJ. Polyketide biosynthesis: a millennium review. Nat. Prod. Rep. 2001;18:380–416. doi: 10.1039/a909079g. [DOI] [PubMed] [Google Scholar]
- Wilt FH. Matrix and mineral in the sea urchin larval skeleton. J. Struct. Biol. 1999;126:216–226. doi: 10.1006/jsbi.1999.4105. [DOI] [PubMed] [Google Scholar]
- Winkel-Shirley B. Biosynthesis of flavonoids and effects of stress. Curr. Opin. Plant Biol. 2002;5:218–223. doi: 10.1016/s1369-5266(02)00256-x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.