Abstract
Staphylococcus epidermidis is the most important member of the coagulase-negative staphylococci and one of the most abundant colonizers of human skin. While for a long time regarded as innocuous, it has been identified as the most frequent cause of device-related infections occurring in the hospital setting and is therefore now recognized as an important opportunistic pathogen. S. epidermidis produces a series of molecules that provide protection from host defenses. Specifically, many proteins and exopolymers, such as the exopolysaccharide PIA, contribute to biofilm formation and inhibit phagocytosis and the activity of human antimicrobial peptides. Furthermore, recent research has identified a family of pro-inflammatory peptides in S. epidermidis, the phenol-soluble modulins (PSMs), which have multiple functions in immune evasion and biofilm development, and may be cytolytic. However, in accordance with the relatively benign relationship that S. epidermidis has with its host, production of aggressive members of the PSM family is kept at a low level. Interestingly, in contrast to Staphylococcus aureus with its large arsenal of toxins developed for causing infection in the human host, most if not all “virulence factors” of S. epidermidis appear to have original functions in the commensal lifestyle of this bacterium.
Keywords: Staphylococcus epidermidis, biofilm, phenol-soluble modulins, polysaccharide intercellular adhesin, device-related infections, hospital-associated infections
1. Introduction
Coagulase-negative staphylococci (CNS) are a prominent part of the normal flora of human skin. Certain CNS species prefer specific niches, while others occur almost everywhere on our body [1,2]. In contrast to Staphylococcus aureus, CNS colonize every human being. This includes Staphylococcus epidermidis, which is the most frequently encountered CNS species on the human body and a common colonizer of the axillae, head, and nares [1,3]. Colonization with S. epidermidis may play an important role for the maintenance of a healthy skin flora, by competition with potentially harmful microorganisms such as in particular S. aureus. On the other hand, S. epidermidis is now also being recognized as an important opportunistic pathogen that can cause significant problems when breaching the epithelial barrier, especially during biofilm-associated infection of indwelling medical devices. Here, I will present molecular factors that are involved in the success of S. epidermidis as a colonizer and pathogen, with a specific focus on the interaction with S. aureus and the different “approaches” of these two related bacteria to evade human immune defenses during colonization and infection.
2. Diseases caused by S. epidermidis and other coagulase-negative staphylococci
Most diseases caused by S. epidermidis and other CNS are of a chronic character and occur as device-related infections (such as intravascular catheter or prosthetic joint infections) and their complications [4]. S. lugdunensis represents a certain exception among the CNS, as it is more often involved in severe infections that resemble those caused by S. aureus [5–7]. In CNS device-related infections, complete removal of the infected device and prolonged antibiotic therapy are often necessary. CNS bloodstream infections originating from intravascular catheter infections are estimated to reach 250,000 cases per year in the U.S., with a mortality rate of 1–25%. These infections represent a significant burden for the public health system, with a cost of $25,000 per episode and an excess hospital stay of > 7 days [8,9]. More severe manifestations, such as prosthetic valve endocarditis (PVE), are rare compared to device-related infections. However, CNS are among the most frequent causes of PVE (15–40%) [10]. Of note, neonates represent a particularly high-risk group for CNS infections [11], with 31% of all neonatal infections and 73% of neonatal bacteremias attributable to CNS [12].
3. Antibiotic resistance
Resistance to methicillin is widespread among hospital isolates of CNS and in particular, S. epidermidis (methicillin-resistant S. epidermidis, MRSE), ranging globally from 75–90% [13]. It is due to the presence of the mecA gene, which codes for a penicillin binding protein, PBP2a, with decreased affinity to methicillin [14]. Heteroresistance, which means that only one in 104 to 108 cells shows a high level of methicillin resistance, occurs in CNS similar to S. aureus [15]. Resistance to aminoglycosides and macrolides, and to a lesser extent, tetracycline, chloramphenicol, and clindamycin is also frequently observed among hospital-associated S. epidermidis strains [4]. Intermediate resistance to vancomycin (VISE, vancomycin intermediate resistant S. epidermidis) is on the rise [16]; however, spread of vancomycin high-level resistant strains has not been reported. Among the newer antibiotics, resistance is still very rare to linezolid and streptogramins, and not reported for daptomycin or tigecycline [4]. Of note, non-specific resistance (tolerance) by biofilm formation is of great concern in CNS infections and will be discussed below.
4. Virulence factors of S. epidermidis
S. epidermidis is by far the best studied member of the CNS in terms of the knowledge that we have on molecular mechanisms of virulence (Tab. 1). For the purpose of the present review, the definition of `virulence factors” will be broad, comprising genes and proteins that facilitate the establishment of infection and persistence of the organism in the human body. It will become clear that most of these factors also have important roles in the commensal life of S. epidermidis as an innocuous inhabitant of the human skin and may thus not be regarded as “virulence factors” sensu stricto.
Tab. 1.
Virulence factors of S. epidermidis
| Virulence factor | Gene | Function |
|---|---|---|
| Biofilm formation | ||
| Primary attachment | ||
| AtlE | atlE | Bifunctional autolysin/adhesin |
| Aae | aae | Bifunctional autolysin/adhesin |
| Teichoic acids | (multiple biosynthetic genes) | Attachment (shown in S. aureus) |
| SdrF | sdrF | Binds to collagen |
| SdrG (Fbe) | sdrG | Binds to fibrinogen |
| SdrH | sdrH | Putative binding function only |
| Ebp | ebp | Binds to elastin |
| Embp | embp | Binds to fibronectin |
| AtlE/Aae | atlE/aae | Bind to multiple matrix proteins |
| Aap | aap | Binds to corneocytes |
| Intercellular aggregation | ||
| PNAG/PIA | icaADBC | Intercellular polysaccharide adhesin |
| Bap | bap | Intercellular protein adhesin |
| Aap | aap | Intercellular protein adhesin precursor (requires proteolytic processing for activity) |
| Embp | embp | Intercellular protein adhesin |
| Teichoic acids | Component of biofilm matrix | |
| Protective exopolymers | ||
| PNAG/PIA | icaADBC | Protects from IgG, AMPs, phagocytosis, complement |
| PGA | capABCD | Protects from AMPs, phagocytosis |
| Resistance to AMPs | ||
| SepA protease | sepA | AMP degradation |
| Dlt, MprF, VraFG |
dltABCD
mprF vraFG |
In analogy to S. aureus: D-alanylation of teichoic acids (Dlt), lysylation of phospholipids (MprF), putative AMP export (VraFG) |
| Aps system | apsR (graR), apsS (graS), apsX | AMP sensor, regulator of AMP resistance mechanisms |
| Toxins | ||
| Enterotoxins | ||
| SEC3, SE1L | sec3, selL | Pathogenicity island-located enterotoxins |
| Phenol-soluble modulins (PSMs) | ||
| PSMα | psm α | Cytolysin (moderate activity), pro-inflammatory |
| PSMβ1, PSMβ2 | psmβ1, psmβ2 | Biofilm-structuring surfactants, pro-inflammatory |
| PSMδ | psm δ | Cytolysin (strong activity), pro-inflammatory |
| PSMε | psm ε | Cytolysin (moderate activity), pro-inflammatory |
| δ-toxin | hld | Cytolysin (moderate activity), pro-inflammatory |
| PSM-mec | psm-mec | Cytolysin (moderate activity), pro-inflammatory |
| Exoenzymes | ||
| Proteases | ||
| Cysteine protease (SspB, Ecp) | sspB | Unknown, tissue damage? |
| Metalloprotease/elastase (SepA) | sepA | Tissue damage?, AMP resistance |
| Glutamylendopeptidase (GluSE, SspA, Esp) | sspA | Degradation of fibrinogen and complement factor C5, competition with S. aureus (by proteolysis of biofilm matrix proteins?) |
| Lipases GehC, GehD | gehC, gehD | unknown |
| Others | ||
| Fatty acid modifying enzyme unidentified (FAME) | Detoxification of bactericidal fatty acids |
4.1 Biofilms
S. epidermidis, like many other CNS, is an excellent biofilm former, and most S. epidermidis infections involve biofilms. Biofilms are sticky, surface-attached agglomerations of bacteria that are embedded in an extracellular matrix and provide protection from antibiotics and mechanisms of host defense [17,18]. For many antibiotics, MIC values against bacteria in biofilms can be higher by several logs compared to those against planktonic (free-floating) bacteria.
The mechanisms underlying the protective features of biofilms are multiple and may be different for a specific antibiotic or host defense mechanism. First, the matrix represents a mechanical barrier that is hardly penetrable for immune cells. However, there are also reports suggesting that, for example, neutrophils penetrate deeply into bacterial biofilms [19], likely through the matrix-free channels that are characteristic components of the three-dimensional biofilm structure. Nevertheless, phagocyte activity is commonly regarded as severely hampered by biofilm formation. For S. aureus, this has recently been shown clearly by Thurlow et al. [20]. Furthermore, limited diffusion through the extracellular matrix contributes to biofilm tolerance towards some antibiotics [21]. This is the case for example for ciprofloxaxcin and biofilms of Pseudomonas aeruginosa [22]. In contrast, other antibiotics have been shown to diffuse easily into the biofilm matrix, such as rifampin, vancomycin, and daptomycin in S. epidermidis biofilms [23,24]. Second, biofilms are to some extent physiologically “dormant” - showing reduced activity of many active cell processes such as cell division, protein synthesis, or DNA replication [21]. This specific physiological state of biofilms significantly decreases the efficacies of many antibiotics that target those processes, including for example the large number of protein synthesis inhibitors and the cell wall-active beta-lactam antibiotics. Third, biofilms may comprise an increased number of so-called “persister” cells, which show increased tolerance to antibiotics [25]. Recently, it was shown that S. epidermidis biofilms contain a high number of persister cells upon antibiotic exposure [26]. Finally, biofilms may show over-production of proteins or other biopolymers that contribute to resistance or tolerance [27].
4.1.1 S. epidermidis biofilms protect from mechanisms of innate host defense
Many of the protective mechanisms of biofilms toward host defenses, in particular those of innate host defense, have been analyzed in S. epidermidis. Mechanisms of innate host defense allow the human body to respond swiftly to an infection. They are triggered by the recognition of invariant structures on the bacterial surface, such as peptidoglycan, lipopeptides, lipoteichoic acids, or even some species-specific molecules such as the staphylococcal phenol-soluble modulins (PSMs) discussed below. Innate host defense comprises mainly the activity of phagocytes, antimicrobial peptides (AMPs), and the complement system.
It has been suggested early that the main biological purpose of the S. epidermidis “slime” substance, now called extracellular biofilm matrix, is to prevent ingestion of bacteria by phagocytes. Beginning in the 1980s, several reports were published providing evidence for that function [28–30]. Recent research has given more mechanistic insight, showing that S. epidermidis biofilms protect from neutrophil-dependent killing by complement inactivation via prevention of the deposition of C3b and immunoglobulin G (IgG) [31]. Furthermore, the extracellular matrix decreases the activity of antimicrobial peptides (AMPs), likely mostly by hindering them from reaching their predominant target, the cytoplasmic membrane [32].
4.1.2 Molecular mechanisms contributing to biofilm development
Biofilm development proceeds in 3 steps: initial adhesion, intercellular aggregation and accumulation, and final detachment [33]. Initial adhesion may occur to abiotic surfaces, such as the uncovered plastic surface of indwelling medical devices, or to biotic surfaces, such as tissues or human matrix protein-covered devices. Intercellular aggregation is accomplished by a series of matrix proteins and non-proteinaceous polymeric substances. In addition, more recent research has indicated a key role of surfactants in biofilm detachment [34–36]. Specific molecules involved in S. epidermidis adhesion, aggregation, biofilm structuring, and detachment will be presented in the following. First, proteins and polymers involved in adhesion and/or accumulation will be discussed. Many of these have been implicated with both adhesion and accumulation. Finally, recent work on biofilm structuring and detachment will be presented.
MSCRAMMs
Several MSCRAMMs (microbial surface components recognizing adhesive matrix molecules) [37] play a role in tethering S. epidermidis cells to tissue or matrix protein-covered device surfaces. This may or may not ultimately lead to biofilm formation, but in any case is a crucial step for S. epidermidis persistence in or on the human body during colonization and infection. Similar to S. aureus, S. epidermidis has several MSCRAMMS for that purpose, with specificities for a series of different human matrix proteins [38]. This functional redundancy underscores the importance that the establishment of tissue adhesion has for S. epidermidis physiology. For example, MSCRAMMs of S. epidermidis accomplish binding to fibrinogen (SdrG/Fbe) [39,40], fibronectin (Embp) [41], vitronectin (AtlE, Aae) [42,43], and collagen (GehD) [44]. The fibrinogen and fibronectin binding proteins represent MSCRAMMs sensu stricto, with sortase-catalyzed covalent anchoring to peptidoglycan [45] and primary function as adhesins, while AtlE and Aae are non-covalently surface bound autolysins with adhesive in addition to their primary cell wall turnover functions [46]. Finally, GehD is a lipase that also appears to have an unrelated function as binding protein [44].
PIA
The exopolysaccharide polysaccharide intercellular adhesin (PIA), also named poly-N-acetylglucosamine (PNAG) was identified as the main constituent of the S. epidermidis extracellular matrix substance (“slime”) in 1996 by the group of Dietrich Mack in Hamburg, Germany [47]. While minor uncertainties persist about partial N-succinylation and yet not further identified variants of PIA, the main PIA exopolysaccharide is a linear N-acetylglucosamine polymer with partially N-deacetylated amine groups. It has a chain length of more than 130 residues in average. Notably, the sugar moieties are linked in β1–6 linkage, which contrasts one of the most abundant poly-glucosamines in nature, chitin, which has β1–4 linkages [47] (Fig. 1).
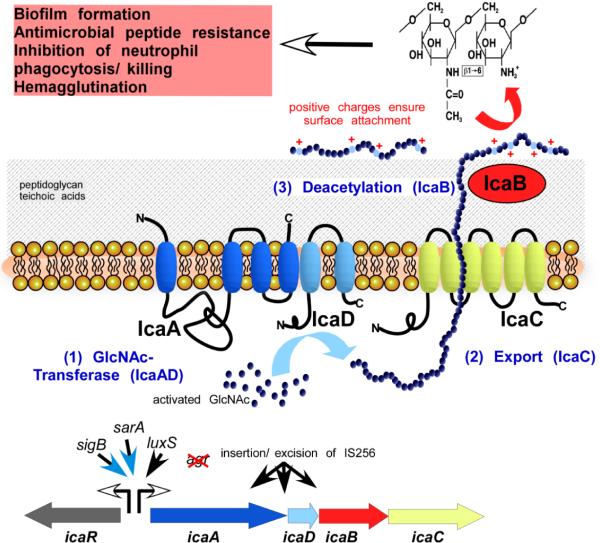
Fig. 1. The exopolysacharide PIA/PNAG.
PIA/PNAG is a partially de-acetylated homopolymer of N-acetylglucosamine residues in β1–6 linkage (top right). It is synthesized by the gene products of the ica operon (bottom). Transcription of ica is under control of multiple global regulators and other regulatory influences (but not Agr). It also may be inactivated by insertion of IS256. IcaA and IcaD form a glucosaminyltransferase. IcaC likely functions as exporter of the growing PIA chain. IcaB de-acetylates pre-PIA at the cell surface after PIA export. Positive charges introduced by deacetylation are crucial for surface location and the multiple functions of PIA shown at the top left.
PIA is synthesized by the gene products of the ica operon, icaA, icaD, icaB, and icaC [48]. Adjacent to the ica operon is a regulatory gene, icaR, that is involved in the multifaceted regulation of ica gene expression [49–54]. IcaA is a glucosaminyl transferase that was shown to transfer glucosamine residues from UDP-glucosamine to a growing chain of pre-PIA [55]. It requires IcaD for full activity, but the detailed role of IcaD is not known. Both proteins are located in the cytoplasmic membrane. Presumably, IcaC, another membrane protein, exports the growing chain of pre-PIA out of the cell. This was speculated based on membrane location and the fact that in an icaC mutant, chain elongation stops at about 15 residues, also suggesting that chain elongation and export are coupled and the IcaA, IcaD, and IcaC proteins may form a complex [55]. Pre-PIA is N-deacetylated after export by the IcaB enzyme, which is a surface-attached PIA N-deacetylase [56]. Deacetylation is crucial for the multiple roles of PIA discussed below [56]. It also significantly increases immunogenicity of the PIA polymer [57].
As expected, a correlation between PIA production and biofilm formation among a collection of clinical S. epidermidis strains was found early after discovery of the polymer [58], and PIA has long been regarded as indispensable for S. epidermidis biofilm formation. However, strains were found more recently, in which in-vitro and in-vivo biofilm formation is independent of PIA and in which proteins functionally substitute for PIA [59,60]. Interestingly, there is evidence indicating that PIA-dependent biofilms form more robust biofilms than those exclusively formed by proteinaceous factors [60]. Furthermore, PIA was found to have multiple, additional roles in immune evasion that exceed a mere contribution to intercellular aggregation in biofilms [56]. For example, PIA was described as the major S. epidermidis hemagglutinin [61]. Moreover, PIA reduces the activity of AMPs and killing by human neutrophils [56]. As the latter effects were observed after disruption of cell clusters by sonication, they are likely at least in part independent of intercellular aggregation, due only to the layer of PIA that surrounds a single S. epidermidis cell. PIA protects from the activity of both positively and negatively charged AMPs, indicating that different mechanisms may contribute to PIA-mediated resistance to AMPs, such as sequestration in addition to repulsion [56,32]. PIA was also reported to induce IL-8 expression in human astrocytes in a TLR2-dependent fashion [62]. However, this was not substantiated using isogenic ica deletion mutants, which is crucial when analyzing pro-inflammatory activities of bacterial compounds, because they almost never can be purified without leaving traces of known pro-inflammatory agents such as lipopeptides. This is especially true in case of the complicated purification of PIA [47,63].
The contribution of PIA to virulence has been monitored in a multitude of animal studies of device-related infection. These studies used mice, rats, rabbits, or Caenorhabditis elegans and mutants in the ica operon of S. aureus or S. epidermidis. A majority of studies found that PIA contributed to virulence in those models [64–67]. This includes a study in which specifically the role of IcaB-dependent deacetylation was investigated and found to have a significant impact on virulence [56]. However, some studies failed to detect a role of PIA in the pathogenesis of biofilm-associated infection [68,69]. While those latter studies are often exclusively cited when aiming to emphasize the importance of protein-dependent biofilm formation, it must be stressed that (i) differences in the experimental setup may have led to the negative outcome and (ii) those studies contradict a larger series of studies showing a significant role of PIA in biofilm-associated infection.
Teichoic acids
Teichoic acids (TA) are characteristic and virtually omnipresent surface components of Gram-positive bacteria. They consist of alternating phosphate and polyol (ribitol or glycerol) moieties. Teichoic acids occur in two major forms, lipoteichoic acids (LTA), which are linked to the cell membrane via a membrane-spanning lipid anchor, and wall teichoic acids (WTA), which are covalently linked to peptidoglycan [70]. Extracellular TA are loosely attached to the cell surface and may originate from WTA that have lost covalent anchoring. The structure of cell wall and extracellular TA of S. epidermidis have been analyzed in detail [71]. They consist of poly(glycerol phosphate) units, which are substituted at the 2-position of glycerol with alpha-glucose, alpha-glucosamine, D-alanine, or alpha-6-D-alanyl-glucose. Reports on “lipid S”, an alleged pro-inflammatory short-chain form of LTA in S. epidermidis [72], are most likely erroneous and caused by co-purification and misinterpretation of mass spectra originating from a phenol-soluble modulin peptide (PSM-mec) present in MRSE [73].
TA have multiple roles in staphylococcal physiology and pathogenesis, contributing to adhesion, colonization, and inflammation [74–76]. In S. epidermidis, TA were shown to enhance adhesion to fibronectin [77]. Furthermore, recent research substantiated the long suspected role of WTA in S. epidermidis biofilm formation by analyzing a tagO mutant of S. epidermidis that exhibited decreased attachment and aggregation phenotypes [78]. A direct role of TA in immune evasion of S. epidermidis has not yet been evaluated, but as a component that has a significant impact on biofilm formation, TA can be assumed to have such a function. Furthermore, S. epidermidis also contains the genes for D-alanylation of TA, a modification that is known from a series of other bacteria, including S. aureus, to protect from the activity of AMPs [79]. These genes, together with others that encode AMP protection systems, namely the VraFG transporter and the phospholipid lysylation enzyme MprF [80,81], are regulated by a system that senses AMP presence and was discovered in S. epidermidis [82].
Accumulation-associated protein (Aap)
In 1997, Hussain et al. described a 140-kilodalton extracellular protein that is essential for the accumulation of S. epidermidis on surfaces [83]. This protein was called accumulation-associated protein (Aap). Aap was later shown to form polymeric fibrils on the surface of S. epidermidis cells, also mediating intercellular adhesion [84]. One subpopulation of cells expresses fibrils, while another does not. The mechanism underlying this specification is not known. The formation of fibrils is zinc-dependent and proceeds via dimerization of so-called G5 domains and modular association of tandem G5 domains [85]. This mechanism is analogous to that involved in the formation of mammalian cadherin domains. Of note, Aap needs proteolytic processing to obtain its biofilm-active form [86]. In addition to mediating intercellular adhesion and adhesion to surfaces, Aap has also recently been shown to attach S. epidermidis to human corneocytes via its terminal A domain [87]. Aap is considered the most important factor contributing to protein-dependent (non-PIA-dependent) biofilm formation in S. epidermidis.
Embp
The extracellular matrix-binding protein (Embp) is a giant covalently attached surface protein of 1 MDa [41]. A truncated form of 460 kDa is necessary and sufficient to promote protein-dependent biofilm formation of S. epidermidis [88]. In addition, Embp mediates adherence to fibronectin and is thus involved in both adhesion and accumulation stages of biofilm formation [41,88].
Bap/Bhp
The biofilm-associated protein (Bap) is a 239 kDa surface protein involved in biofilm adhesion and accumulation [89]. While in S. aureus, Bap is present only in a minority of bovine mastitis isolates, located on a pathogenicity island, it is found frequently in S. epidermidis strains [90]. A protein with high similarity to Bap, the Bap homologue protein Bhp, occurs in some S. epidermidis strains, but does not appear to be essential for biofilm formation [91].
Structuring and detachment: PSMs. While molecular mechanisms contributing to aggregation during biofilm development have been intensely studied, we lack knowledge about cell-cell disruptive mechanisms. These are required for the formation of the characteristic biofilm structure with fluid-filled channels and mushroom-shaped cellular aggregations, and for the detachment of cells from a biofilm, which controls biofilm thickness and expansion. In several bacteria, including S. aureus and S. epidermidis, quorum-sensing systems have been implicated in the control of biofilm thickness and biofilm structuring [92–96]. However, the quorum-sensing controlled factors involved in those processes on a mechanistic level have remained largely undefined. In P. aeruginosa and B. subtilis, surfactant molecules with different chemical natures (rhamnolipid and surfactin, respectively) are involved in biofilm structuring [36,35]. In a recent study, the PSMβ peptides of S. epidermidis were shown to structure S. epidermidis biofilms and thus fulfill a task similar to that accomplished by rhamnolipid in P. aeruginosa [34]. Of note, in that study it was demonstrated for the first time, using the PSMβ peptides of S. epidermidis and a murine device-related infection model, that biofilm structuring molecules trigger the dissemination of biofilm-associated infection. Isogenic S. epidermidis psmβ deletion mutants showed significantly less systemic dissemination from the biofilm and anti-PSMβ antibodies blocked dissemination.
4.1.3 S. epidermidis biofilms have a specific, protective gene expression profile
Biofilms represent a strongly divergent mode of growth compared to that of free-floating (often called “planktonic”) bacteria, a situation that for many bacteria is encountered almost exclusively under laboratory conditions. Not surprisingly, gene expression profiles of biofilms show considerable differences when compared to planktonic bacteria in whole genome expression experiments using microarrays. In S. epidermidis, similar to other bacteria, gene expression in biofilms is characterized by a suppression of many active cell processes and an adaptation of metabolic processes to fermentative growth [27]. However, very specific adaptations were also observed. These comprised down-regulation of the Agr (accessory gene regulator) system and the Agr-controlled PSM peptides, which as discussed above were recently attributed a key role in biofilm development [34]. In addition, there was increased expression of genes encoding the biosynthetic machinery for production of the protective surface polymer poly-γ-glutamic acid PGA (see below), exemplifying that over-expression of protective factors not directly involved in aggregation may also contribute to the increased protective characteristics of biofilms.
4.2. Poly-γ-glutamic acid (PGA)
Poly-γ-glutamic acid (PGA) is a linear homopolymer of glutamic acid residues that are linked via the γ-carboxy group of glutamic acid [97]. Thus, it is strictly speaking a peptoid. PGA is found in many microorganisms, in particular in halophilic bacteria, where it is assumed to play a role in osmoprotection [98]. Until its discovery in S. epidermidis in 2005 [99], Bacillus anthracis was the only pathogenic organism in which PGA was attributed a role in immune evasion, namely in protecting from phagocytosis [100].
In S. epidermidis, PGA is composed of a roughly equal amount of D- and L-glutamic acid, while in B. anthracis, PGA is composed entirely of D-glutamic acid residues [99]. However, all forms of PGA (100% D-glutamic acid, 100% L-glutamic acid, and mixed) are found in different organisms in nature. Using isogenic deletion mutants in the cap locus of S. epidermidis, which encodes the PGA biosynthesis genes and is similar to the cap locus of B. anthracis, PGA of S. epidermidis was shown to protect from AMPs, neutrophil killing, and virulence in a mouse model of device-related infection [99]. Of note, while PIA and TA exhibit at least part of their role in protecting from host defenses via their contribution to biofilm formation, PGA did not have a detectable role in biofilm formation [99].
4.3 Toxins in S. epidermidis
S. epidermidis is a bacterial species that is commonly described as relatively innocuous, which is in part due to the notion that it lacks secreted toxins – in stark contrast to its aggressive cousin S. aureus, whose virulence is based on a large repertoire of secreted molecules that are toxic to humans, such as α-toxin, enterotoxins, and a series of leukocidins [101]. There are reports that describe the sporadic occurrence of toxic shock syndrome toxin (TSST) and enterotoxins in CNS including S. epidermidis [102,103]. Recently, a pathogenicity island, termed SePI, has been found and sequenced in one clinical S. epidermidis strain [104]. This island contains staphylococcal enterotoxin C3 (SEC3) and staphylococcal enterotoxin-like toxin L (SElL). However, the production of such toxins by S. epidermidis has to be regarded as exceptional and the acquisition of toxin gene-harboring MGEs, likely from S. aureus, as extremely rare. Possibly, this is due to the presence of so-called CRISPR sequences that appear to be common to S. epidermidis while they are absent from S. aureus [105]. These genetic elements provide a system that allows distinguishing between self and non-self DNA and the destruction of intruding foreign DNA sequences, such as phages. The mechanism of CRISPR interference has been studied intensively using S. epidermidis as a model organism [105].
4.3.1 Phenol-soluble modulins (PSMs)
Recently, the notion that S. epidermidis and other CNS are virtually toxin-free had to be somewhat revised with the identification and characterization of the PSMs, a family of genome-encoded peptides with frequently cytolytic character. The term phenol-soluble modulins was coined by the group of S. Klebanoff in 1999, when they described a peptide “complex” withb several pro-inflammatory activities that was purified from S. epidermidis culture filtrate using hot phenol extraction [106]. The PSM “complex”, which was then shown to comprise three components, δ-toxin and the newly described PSMα and PSMβ, activated the HIV1-long terminal repeat (LTR), induced cytokine release, activated nuclear factor B in cells of macrophage lineage [106], primed neutrophils and activated the neutrophil oxidative burst [107]. The same group also reported the pro-inflammatory activities of the PSM “complex” to be mediated via the Toll-like receptor 2 (TLR2) [108]. However, this was never substantiated using synthetic peptides or isogenic deletion mutants of psm genes, leaving the possibility that TLR2 induction may have been due, at least in part, to co-purified pro-inflammatory impurities. This is even more likely given the recent identification of the formyl peptide receptor 2 (FPR2) as a receptor that binds PSMs and triggers inflammatory responses to PSMs [109]. It still needs to be investigated whether TLR2 is involved in PSM pro-inflammatory activities using psm deletion mutants and pure PSM peptides.
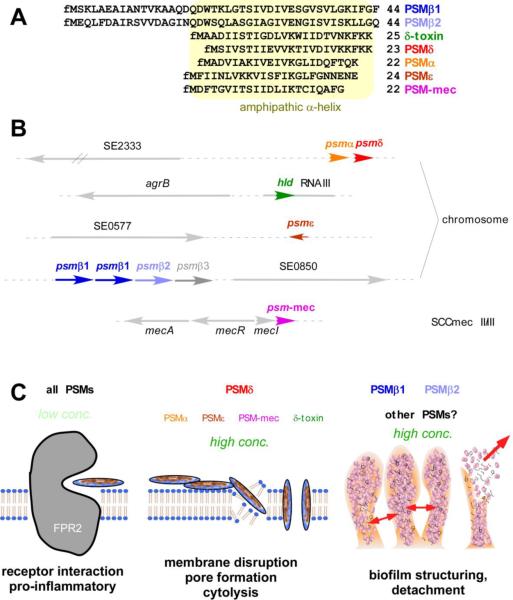
Members of the PSM peptide family show distinct physico-chemical characteristics [110–112]. All PSMs contain an amphipatic α-helix that confers surfactant-like properties. The shorter α-type PSMs are about 20–25 amino acids in length with the amphipathic α-helix stretching over the entire peptide. β-type PSMs are about double in size with the C-terminal part containing the α-helix. These characteristics also cause an unusual behavior of PSMs during reversed-phase chromatography compared to all other proteins found in culture filtrate, namely elution at extremely high percentages of organic solvent. This, together with the fact that concentrations of PSMs in culture filtrates are commonly very high, allowed the relatively easy detection of PSMs in several CNS [111]. In S. epidermidis, all genetic loci coding for PSMs detected by reversed-phase HPLC/mass spectrometry have been identified either using the detected masses or further purification and N-terminal amino acid sequencing [27,106,113] (Fig. 2). In addition to the 3 PSMs described by Mehlin et al., S. epidermidis was found to contain 3 additional PSMs: 2 α-type PSMs (PSMδ, PSMε) and an additional β-type PSM, which is now called PSMβ1, while the original PSMβ is now called PSMβ1. The S. epidermidis PSMs are encoded in 4 different genetic loci on the S. epidermidis chromosome: The genes coding for PSMα and PSMδ form an operon as do those coding for PSMβ1 and PSMβ2. Interestingly, the psmβ locus also contains a gene for an additional β-type PSM, which however was never found to be produced [110]. Furthermore, the gene coding for PSMβ1 is found in 2 copies in some S. epidermidis strains [34]. The δ-toxin (gene name hld) is encoded within RNAIII, the regulatory RNA of the Agr quorum-sensing system, similar to S. aureus and other staphylococci. The gene coding for PSMε is not linked to another psm gene. Of note, all PSMs are under strict regulation by Agr [113]. In addition to the aforementioned PSMs, a PSM, PSM-mec, was recently discovered on the staphylococcal cassette chromosomes (SCC) mec of types II, III, and VIII, which contain the genes responsible for resistance to methicillin [73]. As many clinical S. epidermidis isolates are MRSE and belong to one of those SCCmec types, PSM-mec production is frequent among S. epidermidis strains isolated from infection. PSM-mec significantly contributes to virulence in S. aureus strains that show relatively high production of that peptide [73]. The role of PSM-mec in virulence of S. epidermidis has not yet been investigated directly.
Fig. 2. Phenol-soluble modulins of S. epidermidis.
A, Amino acid sequences of all S. epidermidis PSMs. PSMs are exported as the primary translation products with N-terminal N-formyl-methionine (fM). B, Genetic location of psm genes in the chromosome or in the case of psm-mec, on SCCmec elements. Gene numbers are according to the genome of ATCC12228. C, Roles of PSMs in inflammation, cytolysis, and biofilm development. All PSMs trigger inflammatory responses by interaction with the FPR2 receptor. PSMδ and to some extent, some other PSMs, function as cytolysins. PSMβ peptides and possibly other PSMs contribute to biofilm structuring and detachment.
The PSM pattern is characteristic for a given staphylococcal species, with different species producing different PSM peptides and only occasionally occurring significant amino acid sequence similarity between PSMs from different species. For example, the δ-toxin of S. epidermidis is only different from that of S. aureus in 2 amino acids [114], suggesting a common ancestor. Similarity is also found among the PSMβ peptides of different species. PSMβ peptides were frequently annotated in staphylococcal genomes, including those of S. epidermidis that have been sequenced [115,116], whereas the shorter α-type PSMs usually missed the threshold for minimal length of open reading frames. Of note, PSMs lack a signal peptide and are therefore secreted as the primary translation product, harboring an N-terminal N-formyl methionine. The mechanism of PSM secretion is unknown.
The earlier reports on S. epidermidis PSMs passed without much appreciation by the staphylococcal research community, possibly because the pro-inflammatory capacities of PSMs were not further substantiated using deletion mutants and there was no evaluation of the impact that PSMs have on staphylococcal infection. This changed when PSMα peptides were discovered in S. aureus and recognized as key contributors to virulence in bacteremia and skin infection caused by community-associated methicillin-resistant S. aureus (CA-MRSA) [112]. In particular, S. aureus PSMα peptides were demonstrated to exhibit a strong capacity to lyse human neutrophils and other cell types. Mechanistically, there is evidence indicating that the cytolytic capacities of PSMs are not receptor-mediated, but likely caused by their surfactant characteristics [109].
The capacities of S. epidermidis PSM peptides to lyse neutrophils and erythrocytes have only recently been analyzed [110]. Similar to S. aureus PSMβ peptides, the PSMβ peptides of S. epidermidis are not cytolytic, indicating that the lack of cytolytic capacity may be a common feature of the β-type class of PSMs. Most α-type PSMs of S. epidermidis have low to moderate cytolytic activities with the notable exception of PSMδ, which reaches a cytolytic capacity comparable to that of the strongly cytolytic S. aureus PSMα3 [110].
The discovery that PSMδ is strongly cytolytic to human neutrophils and erythrocytes is of special importance, as it represents the first strongly potent toxin to be discovered in S. epidermidis that is conserved in all known members of the species. However, so far, PSMδ was found to be produced only at relatively low levels [110], in accordance with the commonly low overall aggressiveness of S. epidermidis as a colonizer and opportunistic pathogen. Possibly, the low production of cytolytic PSMs in S. epidermidis is linked to the much lower resistance of S. epidermidis compared to S. aureus against killing by human neutrophils. A detailed investigation of the contribution of S. epidermidis PSMs to infection has not yet been performed, mostly due to the fact that isogenic deletion mutants are difficult to produce in clinical S. epidermidis strains and have not yet been constructed except for the psmβ genes.
4.4. Exoenzymes
Several enzymes that S. epidermidis secretes have been implicated in virulence. S. epidermidis produces a series of secreted proteases, all of which may contribute to virulence via the destruction of host tissue and host proteins. Specific mechanisms could be attributed to SepA, which degrades human AMPs [117], and Esp, which degrades fibrinogen and complement factor C5 [118]. Esp may also contribute to interspecies interference during colonization (see below) [119]. The roles that secreted lipase plays are poorly understood [120,121]. Finally, S. epidermidis secretes a fatty acid-modifying enzyme activity (FAME), which detoxifies fatty acids that are harmful to bacteria [122,120,121]. The proteins or genes responsible for FAME activity are not known.
4.5 Competition with S. aureus
It has been speculated for a long time that colonization with S. epidermidis prevents overgrowth of the more aggressive S. aureus and thus, S. aureus infection, which is linked to S. aureus colonization [123]. However, evidence for such interference was not available. When cross-interfering quorum-sensing pheromones were discovered in staphylococci [124], notably including S. epidermidis pheromones that inhibited quorum-sensing activity of S. aureus [125], it was believed that quorum-sensing cross-inhibition could be the source for a potential S. epidermidis/S. aureus interference. However, this could not be verified in vivo [126].
Recently, Iwase et al. reported that S. epidermidis strains that express a certain secreted protease prevent nasal colonization with S. aureus [119]. In contrast, S. epidermidis lacking expression of that previously well characterized protease [118], termed Esp, do not interfere with S. aureus in vivo. The underlying mechanism was described as protease-dependent destruction of biofilms. While Esp showed such activity in vitro, it is debatable whether the observed in vivo phenomenon of interspecies competition is due to that mechanism [127]. Nevertheless, this study clearly demonstrated that interspecies competition between S. aureus and S. epidermidis exists in the nares and is, at least in part, responsible for the observed individual differences in nasal colonization with S. aureus.
4.6 Are there virulence factors in S. epidermidis that distinguish invasive from commensal strains?
It has been noted that the aggressiveness of S. aureus and the relatively benign relationship that S. epidermidis has with its host allows the two organisms to survive and spread, each by using a different approach [128]. The S. epidermidis approach would consist of a series of “passive” mechanisms to evade host defenses, such as the production of protective matrix polymers, without the production of aggressive toxins. This notion has been largely confirmed with the recent more thorough investigation of S. epidermidis virulence mechanisms. Interestingly, despite the discovery of PSMs as a series of novel potentially aggressive toxins in S. epidermidis, a detailed look at PSM expression in S. epidermidis confirmed rather than negated that notion: the expression pattern of PSM peptides is clearly shifted, compared to S. aureus, to those that lack cytolytic activity, such as the PSMβ peptides [110].
It has become clear that no virulence mechanism exists in S. epidermidis that clearly distinguishes invasive from colonizing strains. In a study using comparative genomic hybridization, a genome-wide comparison of strains isolated from infections of prostheses versus strains from healthy individuals, no factor could be identified that was solely present in infectious strains, but absent from control strains [129]. Nevertheless, several studies confirmed the in average more frequent presence of the ica genes encoding PIA synthesis and the insertion element IS256 in invasive strains [130–133]. These studies underscored the importance of PIA in infection and suggested that IS256 may contribute to genetic adaptations allowing optimization of gene content and expression to an infectious lifestyle. Insertion of IS256 was shown for example to occur in the ica operon, abolishing PIA synthesis [134]. Furthermore, a study with human volunteers indicated that ica-positive strains are less adapted to colonization [135]. Moreover, strains belonging to ST2, the by far most frequently found clonal type of hospital-associated invasive S. epidermidis, are all ica- and IS256-positive [132,136]. Then again, it has also been pointed out that when using higher stringency controls, ica and other virulence genes are not correlated with invasiveness [137]. Taken together, these studies indicate that there are no genetic markers that can be used to clearly predict the aggressiveness of S. epidermidis strains. In further support of that notion, the detailed investigation of virulence mechanisms of S. epidermidis in recent years has indicated that most if not all virulence factors in that bacterium have original roles in its commensal lifestyle. Likely, PSMs, PIA and other biofilm factors all have such original roles in establishing growth and allowing survival in microbial agglomerations on the skin [138].
As a consequence, any therapeutic directed against S. epidermidis virulence factors would also target commensal strains. Any such therapeutic would have to be evaluated for its impact on S. epidermidis colonization and its role in controlling the composition of the human microflora, especially in light of the recent findings regarding S. epidermidis-S. aureus interference.
Acknowledgments
This work was supported by the Intramural Research Program of the National Institute of Allergy and Infectious Diseases, National Institutes of Health.
Footnotes
This article is published as part of the Special Issue on Immunopathology of staphylococcal infections [34:3]
References
- 1.Kloos WE, Musselwhite MS. Distribution and persistence of Staphylococcus and Micrococcus species and other aerobic bacteria on human skin. Appl Microbiol. 1975;30(3):381–385. doi: 10.1128/am.30.3.381-395.1975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Grice EA, Segre JA. The skin microbiome. Nat Rev Microbiol. 2011;9(4):244–253. doi: 10.1038/nrmicro2537. doi:10.1038/nrmicro2537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kloos W, Schleifer KH. Staphylococcus. In: PHA S, S M, ME S, JG H, editors. Bergey's Manual of Systematic Bacteriology. Williams & Wilkins; Baltimore: 1986. [Google Scholar]
- 4.Rogers KL, Fey PD, Rupp ME. Coagulase-negative staphylococcal infections. Infect Dis Clin North Am. 2009;23(1):73–98. doi: 10.1016/j.idc.2008.10.001. [DOI] [PubMed] [Google Scholar]
- 5.Etienne J, Pangon B, Leport C, Wolff M, Clair B, Perronne C, Brun Y, Bure A. Staphylococcus lugdunensis endocarditis. Lancet. 1989;1(8634):390. doi: 10.1016/s0140-6736(89)91770-4. [DOI] [PubMed] [Google Scholar]
- 6.Zinkernagel AS, Zinkernagel MS, Elzi MV, Genoni M, Gubler J, Zbinden R, Mueller NJ. Significance of Staphylococcus lugdunensis bacteremia: report of 28 cases and review of the literature. Infection. 2008;36(4):314–321. doi: 10.1007/s15010-008-7287-9. [DOI] [PubMed] [Google Scholar]
- 7.Lina G, Etienne J, Vandenesch F. Biology and pathogenicity of staphylococci other than Staphylococcus aureus and Staphylococcus epidermidis. In: Fischetti VA, Novick RP, J.J. F, Portnoy DA, Rood JI, editors. Gram-positive pathogens. ASM Press; Washington, DC: 2000. [Google Scholar]
- 8.O'Grady NP, Alexander M, Dellinger EP, Gerberding JL, Heard SO, Maki DG, Masur H, McCormick RD, Mermel LA, Pearson ML, Raad II, Randolph A, Weinstein RA. Guidelines for the prevention of intravascular catheter-related infections. Centers for Disease Control and Prevention. MMWR Recomm Rep. 2002;51(RR-10):1–29. [PubMed] [Google Scholar]
- 9.Raad I, Hanna H, Maki D. Intravascular catheter-related infections: advances in diagnosis, prevention, and management. Lancet Infect Dis. 2007;7(10):645–657. doi: 10.1016/S1473-3099(07)70235-9. [DOI] [PubMed] [Google Scholar]
- 10.Wang A, Athan E, Pappas PA, Fowler VG, Jr., Olaison L, Pare C, Almirante B, Munoz P, Rizzi M, Naber C, Logar M, Tattevin P, Iarussi DL, Selton-Suty C, Jones SB, Casabe J, Morris A, Corey GR, Cabell CH. Contemporary clinical profile and outcome of prosthetic valve endocarditis. Jama. 2007;297(12):1354–1361. doi: 10.1001/jama.297.12.1354. [DOI] [PubMed] [Google Scholar]
- 11.Cheung GY, Otto M. Understanding the significance of Staphylococcus epidermidis bacteremia in babies and children. Curr Opin Infect Dis. 2010 doi: 10.1097/QCO.0b013e328337fecb. doi:10.1097/QCO.0b013e328337fecb. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Anday EK, Talbot GH. Coagulase-negative Staphylococcus bacteremia--a rising threat in the newborn infant. Ann Clin Lab Sci. 1985;15(3):246–251. [PubMed] [Google Scholar]
- 13.Diekema DJ, Pfaller MA, Schmitz FJ, Smayevsky J, Bell J, Jones RN, Beach M. Survey of infections due to Staphylococcus species: frequency of occurrence and antimicrobial susceptibility of isolates collected in the United States, Canada, Latin America, Europe, and the Western Pacific region for the SENTRY Antimicrobial Surveillance Program, 1997–1999. Clin Infect Dis. 2001;32(Suppl 2):S114–132. doi: 10.1086/320184. [DOI] [PubMed] [Google Scholar]
- 14.Chambers HF, Hartman BJ, Tomasz A. Increased amounts of a novel penicillin-binding protein in a strain of methicillin-resistant Staphylococcus aureus exposed to nafcillin. J Clin Invest. 1985;76(1):325–331. doi: 10.1172/JCI111965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Tomasz A, Nachman S, Leaf H. Stable classes of phenotypic expression in methicillin-resistant clinical isolates of staphylococci. Antimicrob Agents Chemother. 1991;35(1):124–129. doi: 10.1128/aac.35.1.124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Jones RN. Microbiological features of vancomycin in the 21st century: minimum inhibitory concentration creep, bactericidal/static activity, and applied breakpoints to predict clinical outcomes or detect resistant strains. Clin Infect Dis. 2006;42(Suppl 1):S13–24. doi: 10.1086/491710. [DOI] [PubMed] [Google Scholar]
- 17.Costerton JW, Lewandowski Z, Caldwell DE, Korber DR, Lappin-Scott HM. Microbial biofilms. Annu Rev Microbiol. 1995;49:711–745. doi: 10.1146/annurev.mi.49.100195.003431. [DOI] [PubMed] [Google Scholar]
- 18.Costerton JW, Stewart PS, Greenberg EP. Bacterial biofilms: a common cause of persistent infections. Science. 1999;284(5418):1318–1322. doi: 10.1126/science.284.5418.1318. [DOI] [PubMed] [Google Scholar]
- 19.Leid JG, Shirtliff ME, Costerton JW, Stoodley AP. Human leukocytes adhere to, penetrate, and respond to Staphylococcus aureus biofilms. Infect Immun. 2002;70(11):6339–6345. doi: 10.1128/IAI.70.11.6339-6345.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Thurlow LR, Hanke ML, Fritz T, Angle A, Aldrich A, Williams SH, Engebretsen IL, Bayles KW, Horswill AR, Kielian T. Staphylococcus aureus biofilms prevent macrophage phagocytosis and attenuate inflammation in vivo. J Immunol. 2011;186(11):6585–6596. doi: 10.4049/jimmunol.1002794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Mah TF, O'Toole GA. Mechanisms of biofilm resistance to antimicrobial agents. Trends Microbiol. 2001;9(1):34–39. doi: 10.1016/s0966-842x(00)01913-2. [DOI] [PubMed] [Google Scholar]
- 22.Walters MC, 3rd, Roe F, Bugnicourt A, Franklin MJ, Stewart PS. Contributions of antibiotic penetration, oxygen limitation, and low metabolic activity to tolerance of Pseudomonas aeruginosa biofilms to ciprofloxacin and tobramycin. Antimicrob Agents Chemother. 2003;47(1):317–323. doi: 10.1128/AAC.47.1.317-323.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Leite B, Gomes F, Teixeira P, Souza C, Pizzolitto E, Oliveira R. In vitro activity of daptomycin, linezolid and rifampicin on Staphylococcus epidermidis biofilms. Curr Microbiol. 2011;63(3):313–317. doi: 10.1007/s00284-011-9980-7. [DOI] [PubMed] [Google Scholar]
- 24.Dunne WM, Jr., Mason EO, Jr., Kaplan SL. Diffusion of rifampin and vancomycin through a Staphylococcus epidermidis biofilm. Antimicrob Agents Chemother. 1993;37(12):2522–2526. doi: 10.1128/aac.37.12.2522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Keren I, Kaldalu N, Spoering A, Wang Y, Lewis K. Persister cells and tolerance to antimicrobials. FEMS Microbiol Lett. 2004;230(1):13–18. doi: 10.1016/S0378-1097(03)00856-5. [DOI] [PubMed] [Google Scholar]
- 26.Shapiro JA, Nguyen VL, Chamberlain NR. Evidence for persisters in Staphylococcus epidermidis RP62a planktonic cultures and biofilms. J Med Microbiol. 2011;60(Pt 7):950–960. doi: 10.1099/jmm.0.026013-0. [DOI] [PubMed] [Google Scholar]
- 27.Yao Y, Sturdevant DE, Otto M. Genomewide analysis of gene expression in Staphylococcus epidermidis biofilms: insights into the pathophysiology of S. epidermidis biofilms and the role of phenol-soluble modulins in formation of biofilms. J Infect Dis. 2005;191(2):289–298. doi: 10.1086/426945. [DOI] [PubMed] [Google Scholar]
- 28.Rodgers J, Phillips F, Olliff C. The effects of extracellular slime from Staphylococcus epidermidis on phagocytic ingestion and killing. FEMS Immunol Med Microbiol. 1994;9(2):109–115. doi: 10.1111/j.1574-695X.1994.tb00481.x. [DOI] [PubMed] [Google Scholar]
- 29.Johnson GM, Lee DA, Regelmann WE, Gray ED, Peters G, Quie PG. Interference with granulocyte function by Staphylococcus epidermidis slime. Infect Immun. 1986;54(1):13–20. doi: 10.1128/iai.54.1.13-20.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Noble MA, Reid PE, Park CM, Chan VY. Inhibition of human neutrophil bacteriocidal activity by extracellular substance from slime-producing Staphylococcus epidermidis. Diagn Microbiol Infect Dis. 1986;4(4):335–339. doi: 10.1016/0732-8893(86)90074-x. [DOI] [PubMed] [Google Scholar]
- 31.Kristian SA, Birkenstock TA, Sauder U, Mack D, Gotz F, Landmann R. Biofilm formation induces C3a release and protects Staphylococcus epidermidis from IgG and complement deposition and from neutrophil-dependent killing. J Infect Dis. 2008;197(7):1028–1035. doi: 10.1086/528992. [DOI] [PubMed] [Google Scholar]
- 32.Otto M. Bacterial evasion of antimicrobial peptides by biofilm formation. Curr Top Microbiol Immunol. 2006;306:251–258. doi: 10.1007/3-540-29916-5_10. [DOI] [PubMed] [Google Scholar]
- 33.O'Toole G, Kaplan HB, Kolter R. Biofilm formation as microbial development. Annu Rev Microbiol. 2000;54:49–79. doi: 10.1146/annurev.micro.54.1.49. [DOI] [PubMed] [Google Scholar]
- 34.Wang R, Khan BA, Cheung GY, Bach TH, Jameson-Lee M, Kong KF, Queck SY, Otto M. Staphylococcus epidermidis surfactant peptides promote biofilm maturation and dissemination of biofilm-associated infection in mice. J Clin Invest. 2011;121(1):238–248. doi: 10.1172/JCI42520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Boles BR, Thoendel M, Singh PK. Rhamnolipids mediate detachment of Pseudomonas aeruginosa from biofilms. Mol Microbiol. 2005;57(5):1210–1223. doi: 10.1111/j.1365-2958.2005.04743.x. [DOI] [PubMed] [Google Scholar]
- 36.Branda SS, Gonzalez-Pastor JE, Ben-Yehuda S, Losick R, Kolter R. Fruiting body formation by Bacillus subtilis. Proc Natl Acad Sci U S A. 2001;98(20):11621–11626. doi: 10.1073/pnas.191384198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Foster TJ, Hook M. Surface protein adhesins of Staphylococcus aureus. Trends Microbiol. 1998;6(12):484–488. doi: 10.1016/s0966-842x(98)01400-0. [DOI] [PubMed] [Google Scholar]
- 38.Bowden MG, Chen W, Singvall J, Xu Y, Peacock SJ, Valtulina V, Speziale P, Hook M. Identification and preliminary characterization of cell-wall-anchored proteins of Staphylococcus epidermidis. Microbiology. 2005;151(Pt 5):1453–1464. doi: 10.1099/mic.0.27534-0. [DOI] [PubMed] [Google Scholar]
- 39.Davis SL, Gurusiddappa S, McCrea KW, Perkins S, Hook M. SdrG, a fibrinogen-binding bacterial adhesin of the microbial surface components recognizing adhesive matrix molecules subfamily from Staphylococcus epidermidis, targets the thrombin cleavage site in the Bbeta chain. J Biol Chem. 2001;276(30):27799–27805. doi: 10.1074/jbc.M103873200. [DOI] [PubMed] [Google Scholar]
- 40.Hartford O, O'Brien L, Schofield K, Wells J, Foster TJ. The Fbe (SdrG) protein of Staphylococcus epidermidis HB promotes bacterial adherence to fibrinogen. Microbiology. 2001;147(Pt 9):2545–2552. doi: 10.1099/00221287-147-9-2545. [DOI] [PubMed] [Google Scholar]
- 41.Williams RJ, Henderson B, Sharp LJ, Nair SP. Identification of a fibronectin-binding protein from Staphylococcus epidermidis. Infect Immun. 2002;70(12):6805–6810. doi: 10.1128/IAI.70.12.6805-6810.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Heilmann C, Hussain M, Peters G, Gotz F. Evidence for autolysin-mediated primary attachment of Staphylococcus epidermidis to a polystyrene surface. Mol Microbiol. 1997;24(5):1013–1024. doi: 10.1046/j.1365-2958.1997.4101774.x. [DOI] [PubMed] [Google Scholar]
- 43.Heilmann C, Thumm G, Chhatwal GS, Hartleib J, Uekotter A, Peters G. Identification and characterization of a novel autolysin (Aae) with adhesive properties from Staphylococcus epidermidis. Microbiology. 2003;149(Pt 10):2769–2778. doi: 10.1099/mic.0.26527-0. [DOI] [PubMed] [Google Scholar]
- 44.Bowden MG, Visai L, Longshaw CM, Holland KT, Speziale P, Hook M. Is the GehD lipase from Staphylococcus epidermidis a collagen binding adhesin? J Biol Chem. 2002;277(45):43017–43023. doi: 10.1074/jbc.M207921200. [DOI] [PubMed] [Google Scholar]
- 45.Mazmanian SK, Liu G, Ton-That H, Schneewind O. Staphylococcus aureus sortase, an enzyme that anchors surface proteins to the cell wall. Science. 1999;285(5428):760–763. doi: 10.1126/science.285.5428.760. [DOI] [PubMed] [Google Scholar]
- 46.Heilmann C. Adhesion mechanisms of staphylococci. Adv Exp Med Biol. 2011;715:105–123. doi: 10.1007/978-94-007-0940-9_7. [DOI] [PubMed] [Google Scholar]
- 47.Mack D, Fischer W, Krokotsch A, Leopold K, Hartmann R, Egge H, Laufs R. The intercellular adhesin involved in biofilm accumulation of Staphylococcus epidermidis is a linear beta-1,6-linked glucosaminoglycan: purification and structural analysis. J Bacteriol. 1996;178(1):175–183. doi: 10.1128/jb.178.1.175-183.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Heilmann C, Schweitzer O, Gerke C, Vanittanakom N, Mack D, Gotz F. Molecular basis of intercellular adhesion in the biofilm-forming Staphylococcus epidermidis. Mol Microbiol. 1996;20(5):1083–1091. doi: 10.1111/j.1365-2958.1996.tb02548.x. [DOI] [PubMed] [Google Scholar]
- 49.Conlon KM, Humphreys H, O'Gara JP. Regulation of icaR gene expression in Staphylococcus epidermidis. FEMS Microbiol Lett. 2002;216(2):171–177. doi: 10.1111/j.1574-6968.2002.tb11432.x. [DOI] [PubMed] [Google Scholar]
- 50.Conlon KM, Humphreys H, O'Gara JP. icaR encodes a transcriptional repressor involved in environmental regulation of ica operon expression and biofilm formation in Staphylococcus epidermidis. J Bacteriol. 2002;184(16):4400–4408. doi: 10.1128/JB.184.16.4400-4408.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Jefferson KK, Pier DB, Goldmann DA, Pier GB. The teicoplanin-associated locus regulator (TcaR) and the intercellular adhesin locus regulator (IcaR) are transcriptional inhibitors of the ica locus in Staphylococcus aureus. J Bacteriol. 2004;186(8):2449–2456. doi: 10.1128/JB.186.8.2449-2456.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Handke LD, Slater SR, Conlon KM, O'Donnell ST, Olson ME, Bryant KA, Rupp ME, O'Gara JP, Fey PD. SigmaB and SarA independently regulate polysaccharide intercellular adhesin production in Staphylococcus epidermidis. Can J Microbiol. 2007;53(1):82–91. doi: 10.1139/w06-108. [DOI] [PubMed] [Google Scholar]
- 53.Li M, Villaruz AE, Vadyvaloo V, Sturdevant DE, Otto M. AI-2-dependent gene regulation in Staphylococcus epidermidis. BMC Microbiol. 2008;8:4. doi: 10.1186/1471-2180-8-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Vuong C, Gerke C, Somerville GA, Fischer ER, Otto M. Quorum-sensing control of biofilm factors in Staphylococcus epidermidis. J Infect Dis. 2003;188(5):706–718. doi: 10.1086/377239. [DOI] [PubMed] [Google Scholar]
- 55.Gerke C, Kraft A, Sussmuth R, Schweitzer O, Gotz F. Characterization of the N-acetylglucosaminyltransferase activity involved in the biosynthesis of the Staphylococcus epidermidis polysaccharide intercellular adhesin. J Biol Chem. 1998;273(29):18586–18593. doi: 10.1074/jbc.273.29.18586. [DOI] [PubMed] [Google Scholar]
- 56.Vuong C, Kocianova S, Voyich JM, Yao Y, Fischer ER, DeLeo FR, Otto M. A crucial role for exopolysaccharide modification in bacterial biofilm formation, immune evasion, and virulence. J Biol Chem. 2004;279(52):54881–54886. doi: 10.1074/jbc.M411374200. [DOI] [PubMed] [Google Scholar]
- 57.Maira-Litran T, Kropec A, Goldmann DA, Pier GB. Comparative opsonic and protective activities of Staphylococcus aureus conjugate vaccines containing native or deacetylated Staphylococcal Poly-N-acetyl-beta-(1–6)-glucosamine. Infect Immun. 2005;73(10):6752–6762. doi: 10.1128/IAI.73.10.6752-6762.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Mack D, Haeder M, Siemssen N, Laufs R. Association of biofilm production of coagulase-negative staphylococci with expression of a specific polysaccharide intercellular adhesin. J Infect Dis. 1996;174(4):881–884. doi: 10.1093/infdis/174.4.881. [DOI] [PubMed] [Google Scholar]
- 59.Kogan G, Sadovskaya I, Chaignon P, Chokr A, Jabbouri S. Biofilms of clinical strains of Staphylococcus that do not contain polysaccharide intercellular adhesin. FEMS Microbiol Lett. 2006;255(1):11–16. doi: 10.1111/j.1574-6968.2005.00043.x. [DOI] [PubMed] [Google Scholar]
- 60.Rohde H, Burandt EC, Siemssen N, Frommelt L, Burdelski C, Wurster S, Scherpe S, Davies AP, Harris LG, Horstkotte MA, Knobloch JK, Ragunath C, Kaplan JB, Mack D. Polysaccharide intercellular adhesin or protein factors in biofilm accumulation of Staphylococcus epidermidis and Staphylococcus aureus isolated from prosthetic hip and knee joint infections. Biomaterials. 2007;28(9):1711–1720. doi: 10.1016/j.biomaterials.2006.11.046. [DOI] [PubMed] [Google Scholar]
- 61.Mack D, Riedewald J, Rohde H, Magnus T, Feucht HH, Elsner HA, Laufs R, Rupp ME. Essential functional role of the polysaccharide intercellular adhesin of Staphylococcus epidermidis in hemagglutination. Infect Immun. 1999;67(2):1004–1008. doi: 10.1128/iai.67.2.1004-1008.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Stevens NT, Sadovskaya I, Jabbouri S, Sattar T, O'Gara JP, Humphreys H, Greene CM. Staphylococcus epidermidis polysaccharide intercellular adhesin induces IL-8 expression in human astrocytes via a mechanism involving TLR2. Cell Microbiol. 2009;11(3):421–432. doi: 10.1111/j.1462-5822.2008.01264.x. [DOI] [PubMed] [Google Scholar]
- 63.Vuong C, Otto M. The biofilm exopolysaccharide polysaccharide intercellular adhesin--a molecular and biochemical approach. Methods Mol Biol. 2008;431:97–106. doi: 10.1007/978-1-60327-032-8_8. [DOI] [PubMed] [Google Scholar]
- 64.Rupp ME, Fey PD, Heilmann C, Gotz F. Characterization of the importance of Staphylococcus epidermidis autolysin and polysaccharide intercellular adhesin in the pathogenesis of intravascular catheter-associated infection in a rat model. J Infect Dis. 2001;183(7):1038–1042. doi: 10.1086/319279. [DOI] [PubMed] [Google Scholar]
- 65.Rupp ME, Ulphani JS, Fey PD, Bartscht K, Mack D. Characterization of the importance of polysaccharide intercellular adhesin/hemagglutinin of Staphylococcus epidermidis in the pathogenesis of biomaterial-based infection in a mouse foreign body infection model. Infect Immun. 1999;67(5):2627–2632. doi: 10.1128/iai.67.5.2627-2632.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Rupp ME, Ulphani JS, Fey PD, Mack D. Characterization of Staphylococcus epidermidis polysaccharide intercellular adhesin/hemagglutinin in the pathogenesis of intravascular catheter-associated infection in a rat model. Infect Immun. 1999;67(5):2656–2659. doi: 10.1128/iai.67.5.2656-2659.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Begun J, Gaiani JM, Rohde H, Mack D, Calderwood SB, Ausubel FM, Sifri CD. Staphylococcal biofilm exopolysaccharide protects against Caenorhabditis elegans immune defenses. PLoS Pathog. 2007;3(4):e57. doi: 10.1371/journal.ppat.0030057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Kristian SA, Golda T, Ferracin F, Cramton SE, Neumeister B, Peschel A, Gotz F, Landmann R. The ability of biofilm formation does not influence virulence of Staphylococcus aureus and host response in a mouse tissue cage infection model. Microb Pathog. 2004;36(5):237–245. doi: 10.1016/j.micpath.2003.12.004. [DOI] [PubMed] [Google Scholar]
- 69.Chokr A, Leterme D, Watier D, Jabbouri S. Neither the presence of ica locus, nor in vitro-biofilm formation ability is a crucial parameter for some Staphylococcus epidermidis strains to maintain an infection in a guinea pig tissue cage model. Microb Pathog. 2007;42(2–3):94–97. doi: 10.1016/j.micpath.2006.09.001. [DOI] [PubMed] [Google Scholar]
- 70.Glaser L. Bacterial cell surface polysaccharides. Annu Rev Biochem. 1973;42:91–112. doi: 10.1146/annurev.bi.42.070173.000515. [DOI] [PubMed] [Google Scholar]
- 71.Sadovskaya I, Vinogradov E, Li J, Jabbouri S. Structural elucidation of the extracellular and cell-wall teichoic acids of Staphylococcus epidermidis RP62A, a reference biofilm-positive strain. Carbohydr Res. 2004;339(8):1467–1473. doi: 10.1016/j.carres.2004.03.017. [DOI] [PubMed] [Google Scholar]
- 72.Lambert PA, Worthington T, Tebbs SE, Elliott TS. Lipid S, a novel Staphylococcus epidermidis exocellular antigen with potential for the serodiagnosis of infections. FEMS Immunol Med Microbiol. 2000;29(3):195–202. doi: 10.1111/j.1574-695X.2000.tb01523.x. [DOI] [PubMed] [Google Scholar]
- 73.Queck SY, Khan BA, Wang R, Bach TH, Kretschmer D, Chen L, Kreiswirth BN, Peschel A, DeLeo FR, Otto M. Mobile genetic element-encoded cytolysin connects virulence to methicillin resistance in MRSA. PLoS Pathog. 2009;5(7):e1000533. doi: 10.1371/journal.ppat.1000533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Gross M, Cramton SE, Gotz F, Peschel A. Key role of teichoic acid net charge in Staphylococcus aureus colonization of artificial surfaces. Infect Immun. 2001;69(5):3423–3426. doi: 10.1128/IAI.69.5.3423-3426.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Weidenmaier C, Kokai-Kun JF, Kristian SA, Chanturiya T, Kalbacher H, Gross M, Nicholson G, Neumeister B, Mond JJ, Peschel A. Role of teichoic acids in Staphylococcus aureus nasal colonization, a major risk factor in nosocomial infections. Nat Med. 2004;10(3):243–245. doi: 10.1038/nm991. [DOI] [PubMed] [Google Scholar]
- 76.Weidenmaier C, Peschel A. Teichoic acids and related cell-wall glycopolymers in Gram-positive physiology and host interactions. Nat Rev Microbiol. 2008;6(4):276–287. doi: 10.1038/nrmicro1861. [DOI] [PubMed] [Google Scholar]
- 77.Hussain M, Heilmann C, Peters G, Herrmann M. Teichoic acid enhances adhesion of Staphylococcus epidermidis to immobilized fibronectin. Microb Pathog. 2001;31(6):261–270. doi: 10.1006/mpat.2001.0469. [DOI] [PubMed] [Google Scholar]
- 78.Holland LM, Conlon B, O'Gara JP. Mutation of tagO reveals an essential role for wall teichoic acids in Staphylococcus epidermidis biofilm development. Microbiology. 2011;157(Pt 2):408–418. doi: 10.1099/mic.0.042234-0. [DOI] [PubMed] [Google Scholar]
- 79.Peschel A, Otto M, Jack RW, Kalbacher H, Jung G, Gotz F. Inactivation of the dlt operon in Staphylococcus aureus confers sensitivity to defensins, protegrins, and other antimicrobial peptides. J Biol Chem. 1999;274(13):8405–8410. doi: 10.1074/jbc.274.13.8405. [DOI] [PubMed] [Google Scholar]
- 80.Li M, Cha DJ, Lai Y, Villaruz AE, Sturdevant DE, Otto M. The antimicrobial peptide-sensing system aps of Staphylococcus aureus. Mol Microbiol. 2007;66(5):1136–1147. doi: 10.1111/j.1365-2958.2007.05986.x. [DOI] [PubMed] [Google Scholar]
- 81.Peschel A, Jack RW, Otto M, Collins LV, Staubitz P, Nicholson G, Kalbacher H, Nieuwenhuizen WF, Jung G, Tarkowski A, van Kessel KP, van Strijp JA. Staphylococcus aureus resistance to human defensins and evasion of neutrophil killing via the novel virulence factor MprF is based on modification of membrane lipids with llysine. J Exp Med. 2001;193(9):1067–1076. doi: 10.1084/jem.193.9.1067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Li M, Lai Y, Villaruz AE, Cha DJ, Sturdevant DE, Otto M. Gram-positive three-component antimicrobial peptide-sensing system. Proc Natl Acad Sci U S A. 2007;104(22):9469–9474. doi: 10.1073/pnas.0702159104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Hussain M, Herrmann M, von Eiff C, Perdreau-Remington F, Peters G. A 140-kilodalton extracellular protein is essential for the accumulation of Staphylococcus epidermidis strains on surfaces. Infect Immun. 1997;65(2):519–524. doi: 10.1128/iai.65.2.519-524.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Banner MA, Cunniffe JG, Macintosh RL, Foster TJ, Rohde H, Mack D, Hoyes E, Derrick J, Upton M, Handley PS. Localized tufts of fibrils on Staphylococcus epidermidis NCTC 11047 are comprised of the accumulation-associated protein. J Bacteriol. 2007;189(7):2793–2804. doi: 10.1128/JB.00952-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Conrady DG, Brescia CC, Horii K, Weiss AA, Hassett DJ, Herr AB. A zinc-dependent adhesion module is responsible for intercellular adhesion in staphylococcal biofilms. Proc Natl Acad Sci U S A. 2008;105(49):19456–19461. doi: 10.1073/pnas.0807717105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Rohde H, Burdelski C, Bartscht K, Hussain M, Buck F, Horstkotte MA, Knobloch JK, Heilmann C, Herrmann M, Mack D. Induction of Staphylococcus epidermidis biofilm formation via proteolytic processing of the accumulation-associated protein by staphylococcal and host proteases. Mol Microbiol. 2005;55(6):1883–1895. doi: 10.1111/j.1365-2958.2005.04515.x. [DOI] [PubMed] [Google Scholar]
- 87.Macintosh RL, Brittan JL, Bhattacharya R, Jenkinson HF, Derrick J, Upton M, Handley PS. The terminal A domain of the fibrillar accumulation-associated protein (Aap) of Staphylococcus epidermidis mediates adhesion to human corneocytes. J Bacteriol. 2009;191(22):7007–7016. doi: 10.1128/JB.00764-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Christner M, Franke GC, Schommer NN, Wendt U, Wegert K, Pehle P, Kroll G, Schulze C, Buck F, Mack D, Aepfelbacher M, Rohde H. The giant extracellular matrix-binding protein of Staphylococcus epidermidis mediates biofilm accumulation and attachment to fibronectin. Mol Microbiol. 2010;75(1):187–207. doi: 10.1111/j.1365-2958.2009.06981.x. [DOI] [PubMed] [Google Scholar]
- 89.Cucarella C, Solano C, Valle J, Amorena B, Lasa I, Penades JR. Bap, a Staphylococcus aureus surface protein involved in biofilm formation. J Bacteriol. 2001;183(9):2888–2896. doi: 10.1128/JB.183.9.2888-2896.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Tormo MA, Knecht E, Gotz F, Lasa I, Penades JR. Bap-dependent biofilm formation by pathogenic species of Staphylococcus: evidence of horizontal gene transfer? Microbiology. 2005;151(Pt 7):2465–2475. doi: 10.1099/mic.0.27865-0. [DOI] [PubMed] [Google Scholar]
- 91.Lasa I, Penades JR. Bap: a family of surface proteins involved in biofilm formation. Res Microbiol. 2006;157(2):99–107. doi: 10.1016/j.resmic.2005.11.003. [DOI] [PubMed] [Google Scholar]
- 92.Davies DG, Parsek MR, Pearson JP, Iglewski BH, Costerton JW, Greenberg EP. The involvement of cell-to-cell signals in the development of a bacterial biofilm. Science. 1998;280(5361):295–298. doi: 10.1126/science.280.5361.295. [DOI] [PubMed] [Google Scholar]
- 93.Yarwood JM, Bartels DJ, Volper EM, Greenberg EP. Quorum sensing in Staphylococcus aureus biofilms. J Bacteriol. 2004;186(6):1838–1850. doi: 10.1128/JB.186.6.1838-1850.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Vuong C, Saenz HL, Gotz F, Otto M. Impact of the agr quorum-sensing system on adherence to polystyrene in Staphylococcus aureus. J Infect Dis. 2000;182(6):1688–1693. doi: 10.1086/317606. [DOI] [PubMed] [Google Scholar]
- 95.Vuong C, Kocianova S, Yao Y, Carmody AB, Otto M. Increased colonization of indwelling medical devices by quorum-sensing mutants of Staphylococcus epidermidis in vivo. J Infect Dis. 2004;190(8):1498–1505. doi: 10.1086/424487. [DOI] [PubMed] [Google Scholar]
- 96.Boles BR, Horswill AR. Agr-mediated dispersal of Staphylococcus aureus biofilms. PLoS Pathog. 2008;4(4):e1000052. doi: 10.1371/journal.ppat.1000052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Ashiuchi M, Misono H. Biochemistry and molecular genetics of poly-gamma-glutamate synthesis. Appl Microbiol Biotechnol. 2002;59(1):9–14. doi: 10.1007/s00253-002-0984-x. [DOI] [PubMed] [Google Scholar]
- 98.Oppermann-Sanio FB, Steinbuchel A. Occurrence, functions and biosynthesis of polyamides in microorganisms and biotechnological production. Naturwissenschaften. 2002;89(1):11–22. doi: 10.1007/s00114-001-0280-0. [DOI] [PubMed] [Google Scholar]
- 99.Kocianova S, Vuong C, Yao Y, Voyich JM, Fischer ER, DeLeo FR, Otto M. Key role of poly-gamma-DL-glutamic acid in immune evasion and virulence of Staphylococcus epidermidis. J Clin Invest. 2005;115(3):688–694. doi: 10.1172/JCI23523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Makino S, Uchida I, Terakado N, Sasakawa C, Yoshikawa M. Molecular characterization and protein analysis of the cap region, which is essential for encapsulation in Bacillus anthracis. J Bacteriol. 1989;171(2):722–730. doi: 10.1128/jb.171.2.722-730.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Foster TJ. Immune evasion by staphylococci. Nat Rev Microbiol. 2005;3(12):948–958. doi: 10.1038/nrmicro1289. [DOI] [PubMed] [Google Scholar]
- 102.Bautista L, Gaya P, Medina M, Nunez M. A quantitative study of enterotoxin production by sheep milk staphylococci. Appl Environ Microbiol. 1988;54(2):566–569. doi: 10.1128/aem.54.2.566-569.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Marin ME, de la Rosa MC, Cornejo I. Enterotoxigenicity of Staphylococcus strains isolated from Spanish dry-cured hams. Appl Environ Microbiol. 1992;58(3):1067–1069. doi: 10.1128/aem.58.3.1067-1069.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Madhusoodanan J, Seo KS, Remortel B, Park JY, Hwang SY, Fox LK, Park YH, Deobald CF, Wang D, Liu S, Daugherty SC, Gill AL, Bohach GA, Gill SR. An Enterotoxin-Bearing Pathogenicity Island in Staphylococcus epidermidis. J Bacteriol. 2011;193(8):1854–1862. doi: 10.1128/JB.00162-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Marraffini LA, Sontheimer EJ. CRISPR interference limits horizontal gene transfer in staphylococci by targeting DNA. Science. 2008;322(5909):1843–1845. doi: 10.1126/science.1165771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Mehlin C, Headley CM, Klebanoff SJ. An inflammatory polypeptide complex from Staphylococcus epidermidis: isolation and characterization. J Exp Med. 1999;189(6):907–918. doi: 10.1084/jem.189.6.907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Liles WC, Thomsen AR, O'Mahony DS, Klebanoff SJ. Stimulation of human neutrophils and monocytes by staphylococcal phenol-soluble modulin. J Leukoc Biol. 2001;70(1):96–102. [PubMed] [Google Scholar]
- 108.Hajjar AM, O'Mahony DS, Ozinsky A, Underhill DM, Aderem A, Klebanoff SJ, Wilson CB. Cutting edge: functional interactions between toll-like receptor (TLR) 2 and TLR1 or TLR6 in response to phenol-soluble modulin. J Immunol. 2001;166(1):15–19. doi: 10.4049/jimmunol.166.1.15. [DOI] [PubMed] [Google Scholar]
- 109.Kretschmer D, Gleske AK, Rautenberg M, Wang R, Koberle M, Bohn E, Schoneberg T, Rabiet MJ, Boulay F, Klebanoff SJ, van Kessel KA, van Strijp JA, Otto M, Peschel A. Human formyl peptide receptor 2 senses highly pathogenic Staphylococcus aureus. Cell Host Microbe. 2010;7(6):463–473. doi: 10.1016/j.chom.2010.05.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Cheung GY, Rigby K, Wang R, Queck SY, Braughton KR, Whitney AR, Teintze M, DeLeo FR, Otto M. Staphylococcus epidermidis strategies to avoid killing by human neutrophils. PLoS Pathog. 2010;6(10) doi: 10.1371/journal.ppat.1001133. doi:10.1371/journal.ppat.1001133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Rautenberg M, Joo HS, Otto M, Peschel A. Neutrophil responses to staphylococcal pathogens and commensals via the formyl peptide receptor 2 relates to phenol-soluble modulin release and virulence. Faseb J. 2011;25(4):1254–1263. doi: 10.1096/fj.10-175208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Wang R, Braughton KR, Kretschmer D, Bach TH, Queck SY, Li M, Kennedy AD, Dorward DW, Klebanoff SJ, Peschel A, DeLeo FR, Otto M. Identification of novel cytolytic peptides as key virulence determinants for community-associated MRSA. Nat Med. 2007;13(12):1510–1514. doi: 10.1038/nm1656. [DOI] [PubMed] [Google Scholar]
- 113.Vuong C, Durr M, Carmody AB, Peschel A, Klebanoff SJ, Otto M. Regulated expression of pathogen-associated molecular pattern molecules in Staphylococcus epidermidis: quorum-sensing determines pro-inflammatory capacity and production of phenol-soluble modulins. Cell Microbiol. 2004;6(8):753–759. doi: 10.1111/j.1462-5822.2004.00401.x. [DOI] [PubMed] [Google Scholar]
- 114.McKevitt AI, Bjornson GL, Mauracher CA, Scheifele DW. Amino acid sequence of a deltalike toxin from Staphylococcus epidermidis. Infect Immun. 1990;58(5):1473–1475. doi: 10.1128/iai.58.5.1473-1475.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Gill SR, Fouts DE, Archer GL, Mongodin EF, Deboy RT, Ravel J, Paulsen IT, Kolonay JF, Brinkac L, Beanan M, Dodson RJ, Daugherty SC, Madupu R, Angiuoli SV, Durkin AS, Haft DH, Vamathevan J, Khouri H, Utterback T, Lee C, Dimitrov G, Jiang L, Qin H, Weidman J, Tran K, Kang K, Hance IR, Nelson KE, Fraser CM. Insights on evolution of virulence and resistance from the complete genome analysis of an early methicillin-resistant Staphylococcus aureus strain and a biofilm-producing methicillin-resistant Staphylococcus epidermidis strain. J Bacteriol. 2005;187(7):2426–2438. doi: 10.1128/JB.187.7.2426-2438.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Zhang YQ, Ren SX, Li HL, Wang YX, Fu G, Yang J, Qin ZQ, Miao YG, Wang WY, Chen RS, Shen Y, Chen Z, Yuan ZH, Zhao GP, Qu D, Danchin A, Wen YM. Genome-based analysis of virulence genes in a non-biofilm-forming Staphylococcus epidermidis strain (ATCC 12228) Mol Microbiol. 2003;49(6):1577–1593. doi: 10.1046/j.1365-2958.2003.03671.x. [DOI] [PubMed] [Google Scholar]
- 117.Lai Y, Villaruz AE, Li M, Cha DJ, Sturdevant DE, Otto M. The human anionic antimicrobial peptide dermcidin induces proteolytic defence mechanisms in staphylococci. Mol Microbiol. 2007;63(2):497–506. doi: 10.1111/j.1365-2958.2006.05540.x. [DOI] [PubMed] [Google Scholar]
- 118.Dubin G, Chmiel D, Mak P, Rakwalska M, Rzychon M, Dubin A. Molecular cloning and biochemical characterisation of proteases from Staphylococcus epidermidis. Biol Chem. 2001;382(11):1575–1582. doi: 10.1515/BC.2001.192. [DOI] [PubMed] [Google Scholar]
- 119.Iwase T, Uehara Y, Shinji H, Tajima A, Seo H, Takada K, Agata T, Mizunoe Y. Staphylococcus epidermidis Esp inhibits Staphylococcus aureus biofilm formation and nasal colonization. Nature. 2010;465(7296):346–349. doi: 10.1038/nature09074. [DOI] [PubMed] [Google Scholar]
- 120.Farrell AM, Foster TJ, Holland KT. Molecular analysis and expression of the lipase of Staphylococcus epidermidis. J Gen Microbiol. 1993;139(2):267–277. doi: 10.1099/00221287-139-2-267. [DOI] [PubMed] [Google Scholar]
- 121.Simons JW, van Kampen MD, Riel S, Gotz F, Egmond MR, Verheij HM. Cloning, purification and characterisation of the lipase from Staphylococcus epidermidis-comparison of the substrate selectivity with those of other microbial lipases. Eur J Biochem. 1998;253(3):675–683. doi: 10.1046/j.1432-1327.1998.2530675.x. [DOI] [PubMed] [Google Scholar]
- 122.Chamberlain NR, Brueggemann SA. Characterisation and expression of fatty acid modifying enzyme produced by Staphylococcus epidermidis. J Med Microbiol. 1997;46(8):693–697. doi: 10.1099/00222615-46-8-693. [DOI] [PubMed] [Google Scholar]
- 123.von Eiff C, Becker K, Machka K, Stammer H, Peters G. Nasal carriage as a source of Staphylococcus aureus bacteremia. Study Group. N Engl J Med. 2001;344(1):11–16. doi: 10.1056/NEJM200101043440102. [DOI] [PubMed] [Google Scholar]
- 124.Ji G, Beavis R, Novick RP. Bacterial interference caused by autoinducing peptide variants. Science. 1997;276(5321):2027–2030. doi: 10.1126/science.276.5321.2027. [DOI] [PubMed] [Google Scholar]
- 125.Otto M, Echner H, Voelter W, Gotz F. Pheromone cross-inhibition between Staphylococcus aureus and Staphylococcus epidermidis. Infect Immun. 2001;69(3):1957–1960. doi: 10.1128/IAI.69.3.1957-1960.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Lina G, Boutite F, Tristan A, Bes M, Etienne J, Vandenesch F. Bacterial competition for human nasal cavity colonization: role of Staphylococcal agr alleles. Appl Environ Microbiol. 2003;69(1):18–23. doi: 10.1128/AEM.69.1.18-23.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Krismer B, Peschel A. Does Staphylococcus aureus nasal colonization involve biofilm formation? Future Microbiol. 2011;6(5):489–493. doi: 10.2217/fmb.11.37. doi:10.2217/fmb.11.37. [DOI] [PubMed] [Google Scholar]
- 128.Massey RC, Horsburgh MJ, Lina G, Hook M, Recker M. The evolution and maintenance of virulence in Staphylococcus aureus: a role for host-to-host transmission? Nat Rev Microbiol. 2006;4(12):953–958. doi: 10.1038/nrmicro1551. [DOI] [PubMed] [Google Scholar]
- 129.Yao Y, Sturdevant DE, Villaruz A, Xu L, Gao Q, Otto M. Factors characterizing Staphylococcus epidermidis invasiveness determined by comparative genomics. Infect Immun. 2005;73(3):1856–1860. doi: 10.1128/IAI.73.3.1856-1860.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Kozitskaya S, Cho SH, Dietrich K, Marre R, Naber K, Ziebuhr W. The bacterial insertion sequence element IS256 occurs preferentially in nosocomial Staphylococcus epidermidis isolates: association with biofilm formation and resistance to aminoglycosides. Infect Immun. 2004;72(2):1210–1215. doi: 10.1128/IAI.72.2.1210-1215.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Gu J, Li H, Li M, Vuong C, Otto M, Wen Y, Gao Q. Bacterial insertion sequence IS256 as a potential molecular marker to discriminate invasive strains from commensal strains of Staphylococcus epidermidis. J Hosp Infect. 2005;61(4):342–348. doi: 10.1016/j.jhin.2005.04.017. [DOI] [PubMed] [Google Scholar]
- 132.Li M, Wang X, Gao Q, Lu Y. Molecular characterization of Staphylococcus epidermidis strains isolated from a teaching hospital in Shanghai, China. J Med Microbiol. 2009;58(Pt 4):456–461. doi: 10.1099/jmm.0.007567-0. [DOI] [PubMed] [Google Scholar]
- 133.Galdbart JO, Allignet J, Tung HS, Ryden C, El Solh N. Screening for Staphylococcus epidermidis markers discriminating between skin-flora strains and those responsible for infections of joint prostheses. J Infect Dis. 2000;182(1):351–355. doi: 10.1086/315660. [DOI] [PubMed] [Google Scholar]
- 134.Ziebuhr W, Krimmer V, Rachid S, Lossner I, Gotz F, Hacker J. A novel mechanism of phase variation of virulence in Staphylococcus epidermidis: evidence for control of the polysaccharide intercellular adhesin synthesis by alternating insertion and excision of the insertion sequence element IS256. Mol Microbiol. 1999;32(2):345–356. doi: 10.1046/j.1365-2958.1999.01353.x. [DOI] [PubMed] [Google Scholar]
- 135.Rogers KL, Rupp ME, Fey PD. The presence of icaADBC is detrimental to the colonization of human skin by Staphylococcus epidermidis. Appl Environ Microbiol. 2008;74(19):6155–6157. doi: 10.1128/AEM.01017-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Miragaia M, Thomas JC, Couto I, Enright MC, de Lencastre H. Inferring a population structure for Staphylococcus epidermidis from multilocus sequence typing data. J Bacteriol. 2007;189(6):2540–2552. doi: 10.1128/JB.01484-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Rohde H, Kalitzky M, Kroger N, Scherpe S, Horstkotte MA, Knobloch JK, Zander AR, Mack D. Detection of virulence-associated genes not useful for discriminating between invasive and commensal Staphylococcus epidermidis strains from a bone marrow transplant unit. J Clin Microbiol. 2004;42(12):5614–5619. doi: 10.1128/JCM.42.12.5614-5619.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Otto M. Staphylococcus epidermidis - the `accidental' pathogen. Nat Rev Microbiol. 2009;7:555–567. doi: 10.1038/nrmicro2182. [DOI] [PMC free article] [PubMed] [Google Scholar]