Graphical abstract
Keywords: Nandrolone, Anabolic steroids, Androgens, Amyotrophic lateral sclerosis, Muscle, Motor neuron disease, TGFbeta1
Abstract
Anabolic/androgenic steroids (AAS) are drugs that enhance muscle mass, and are often illegally utilized in athletes to improve their performances. Recent data suggest that the increased risk for amyotrophic lateral sclerosis (ALS) in male soccer and football players could be linked to AAS abuse. ALS is a motor neuron disease mainly occurring in sporadic (sALS) forms, but some familial forms (fALS) exist and have been linked to mutations in different genes. Some of these, in their wild type (wt) form, have been proposed as risk factors for sALS, i.e. superoxide dismutase 1 (SOD1) gene, whose mutations are causative of about 20% of fALS. Notably, SOD1 toxicity might occur both in motor neurons and in muscle cells. Using gastrocnemius muscles of mice overexpressing human mutant SOD1 (mutSOD1) at different disease stages, we found that the expression of a selected set of genes associated to muscle atrophy, MyoD, myogenin, atrogin-1, and transforming growth factor (TGF)β1, is up-regulated already at the presymptomatic stage. Atrogin-1 gene expression was increased also in mice overexpressing human wtSOD1. Similar alterations were found in axotomized mouse muscles and in cultured ALS myoblast models. In these ALS models, we then evaluated the pharmacological effects of the synthetic AAS nandrolone on the expression of the genes modified in ALS muscle. Nandrolone administration had no effects on MyoD, myogenin, and atrogin-1 expression, but it significantly increased TGFβ1 expression at disease onset. Altogether, these data suggest that, in fALS, muscle gene expression is altered at early stages, and AAS may exacerbate some of the alterations induced by SOD1 possibly acting as a contributing factor also in sALS.
1. Introduction
ALS is an adult onset neurodegenerative disease characterized by motor neuron loss in the cortex, brain stem and spinal cord. Point mutations in the SOD1 gene have been found in about 20% of fALS cases, while alterations of wtSOD1 behavior have been reported in some cases of sALS [1,2]. sALS and fALS are clinically indistinguishable, and thus animal and cellular models expressing mutSOD1 are widely used to study the disease [3]. It is thought that SOD1 mutations destabilize protein conformation leading to misfolding, which may then result in protein accumulation, axonal transport alterations, mitochondrial and/or proteasome dysfunctions [3–5]. AAS have been proposed as a risk factor in ALS. Indeed, a variable male/female ratio has been reported: the ratio is 2.5 in younger groups (when males have elevated levels of androgens), declining to 1.4 in the older groups [6], and becoming 1:1 at ages above 60 (when androgen levels in males dramatically decrease) [7–9]. Furthermore, an older average age of ALS onset is reported in women [10] and in the mutSOD1 mouse model, disease progression is significantly more aggressive in males than in females [11,12]. Recently, AAS drug abuse has been suggested as one of the factors responsible for the increased ALS prevalence in Italian soccer and American football players [13–17]. A typical target of AAS is the skeletal muscle, particularly rich of the androgen receptor (AR), the mediator of the AAS action. For this reason, skeletal muscle mass and strength differ considerably in the two sexes. Notably, spinobulbar muscular atrophy, an ALS-related disease, is triggered by a mutation in the AR [18], while the selective overexpression of wt AR in mouse muscles induces an ALS-like phenotype with motor neuron dysfunctions and early death [19,20]. Thus, toxicity to motor neurons might also derive from their target muscle cells. In the mutSOD1 mouse model (expressing the G93A-hSOD1), the reduction of mutant protein in skeletal muscles has no effect on disease progression [21], but the selective expression of mutSOD1 in skeletal muscle results in progressive muscle atrophy [22–24]. Furthermore, muscle dysfunction and neuromuscular junction degeneration occur long before disease onset and motoneuronal death [25–27].
On this basis, we analysed the expression of a selected set of genes involved in skeletal muscle pathophysiology to evaluate early neuromuscular abnormalities that precede motor neuron death in ALS and the potential involvement of AAS drugs as a risk factor for ALS.
The results here obtained in mutSOD1 mice demonstrate that, already at the presymptomatic stage, the expression of MyoD, myogenin, atrogin-1, and transforming growth factor (TGF)β1 genes are up-regulated, and that AAS treatment resulted in a further increase of TGFβ1 expression levels.
2. Materials and methods
2.1. Animals and procedures
All the procedures involving animals and their care have been conducted following the institutional guidelines and in accordance with national (D.L. no. 116, G.U. suppl. 40, February 18, 1992) and international laws and policies (EEC Council Directives 86/609, OJ L 358, 1 DEC.12, 1987; NIH Guide for the Care and Use of Laboratory Animals, U.S. National Research Council, 1996). Mice were maintained at a temperature of 21 °C with 55 ± 10% relative humidity and 12 h of light. Food (standard pellets) and water were supplied ad libitum. Animals with substantial motor impairment had food on the bottom of the cage and water bottles with long drinking spouts. The time of death was defined when animals are unable to right themselves within 30 s after being placed on both sides. Animals were sacrificed by decapitation under anesthesia.
2.1.1. Experiment 1 – mutSOD1 effects on muscle gene expression at different stages of disease
We used transgenic mice carrying about 20 copies of mutant G93A-SOD1 or wt human SOD1 (hSOD1) originally obtained from Jackson Laboratories crossbred with a C57BL/6J mouse strain. The genotyping of the litters was conducted by PCR (Primer sequences: SOD1 Forward: CATCAGCCCTAATCCATCTGA; SOD1 Reverse: CGCGACTAACAATCAAAGTGA) on DNA extracted from tail biopsies. Non-transgenic (NTg) littermates were used as controls. To evaluate disease stages, starting from the 14th week of age and twice a week, mice were tested for deficit in grip strength and rotarod performance by the same operator. The symptoms onset was considered when the mice showed the first impairment in grip strength. Body weight loss was also monitored. Mice (n = 4 per group) were sacrificed at 80 (presymptomatic), 120 (on the average symptomatic), and 160 days (on the average end stage) [28].
2.1.2. Experiment 2 – effects of sciatic nerve resection on muscle gene expression
Three-month-old NTg male mice (C57BL/6 strain, n = 4) were anaesthetized with xylazine and ketamine. An incision was made through the skin and the upper region of the left gluteal muscle to expose the sciatic nerve, which was then cut 1–2 mm distal to the sciatic notch. The proximal portion of the nerve was sutured to prevent errant reinnervation of the gastrocnemius muscle. Right sciatic nerve was exposed and utilized as sham internal control in each animal. Mice were sacrificed 7 days later.
2.1.3. Experiment 3 – effects of nandrolone treatment on muscle gene expression
Mice carrying the mutSOD1 gene (strain designation: B6SLL-TgN[SOD1-G93A]1Gur) were purchased from Jackson Laboratories (Bar Harbor, ME, USA). Colonies with high-copy number of mutant SOD1 were crossbred with F1 B6SJL mice. F1 B6SJL mice were obtained by breeding C57BL6 mice with SJL mice in-house. mutSOD1 progeny was identified by specific polymerase chain reaction (see above). Male mutSOD1 and NTg control mice were assigned to different experimental groups (n = 4 per group) at 50–52 days of age. Two groups of mutSOD1 and two groups of age-matched NTg mice were injected subcutaneously with nandrolone dissolved in peanut oil (Sigma–Aldrich, Milan, Italy), at the dose of 10 mg/kg once a week. Nandrolone-treated mice were sacrificed at the onset of the disease (around 85–95 days in this colony), or at 120 days (on the average symptomatic); control groups of age-matched mutSOD1 and NTg mice were treated with vehicle and sacrificed at the same time points. Also in this experiment body weight was monitored; transgenic mice were assessed for the presence of tremors and lack of extension reflex, and tested for deficit in grip strength and rotarod performance twice weekly. Presence of tremors, lack of extension reflex, or failure in either of the two motor tests in 3 consecutive sessions indicated onset of disease.
For all the experiments, at sacrifice gastrocnemius muscle samples were rapidly dissected, washed in 0.01 phosphate-buffered saline, pH 7.4 (PBS), frozen on dry ice, and stored at −80 °C until RNA or protein extraction. Each experiment was carried out twice, with 4 independent samples.
2.2. Cell cultures and transfection
In vitro experiments were conducted on the C2C12 cell line, originally obtained from American Type Culture Collection (Rockville, MD) which represents a widely used myoblast cell line. C2C12 cells were routinely maintained in DMEM (Biochrom KG, Berlin, Germany) supplemented with 4 mM glutamine, 1 mM sodium pyruvate, 100 U/ml penicillin, 100 μg/ml streptomycin, and 10% fetal bovine serum (FBS, Invitrogen, San Giuliano Milanese, Italy) at 37 °C with 5% CO2. Differentiation was induced by replacing the growth medium (10% FBS) with the differentiation medium (2% horse serum, Invitrogen, in DMEM) after the cells reached 70% confluence. The plasmids pcDNA3-hSOD1, pcDNA3-mutSOD1 [29], and/or pCMV−AR.Q23 [30] were transiently transfected into C2C12 cells using Lipofectamine 2000TM (Invitrogen) according to the manufacturer's instructions. Briefly, 60,000 cells/ml were plated in 12-well dishes, and transfected with 1.6 μg of DNA, and 4 μl of lipofectamine/well. Controls were mock transfected. The medium was replaced with differentiation medium at 5 h after transfection. Cells were harvested for RNA isolation at 48 h after transfection.
2.3. Western blot analysis
Frozen samples of gastrocnemius muscles were homogenized in chilled PBS supplemented with a protease inhibitor cocktail (Sigma–Aldrich), with an ultra-turrax® homogenizer. Samples of C2C12 cells were harvested at 48 h after transfection, and centrifuged 5 min at 1200 rpm at 4 °C; cell pellets were resuspended in PBS plus protease inhibitor cocktail and homogenized using slight sonication. The supernatant protein concentration was assayed according to the Bradford method. Equal amount of each sample (containing 10 μg of proteins for gastrocnemius muscle samples, and 25 μg for C2C12 cells) was resolved on 12% SDS–polyacrylamide gel and electroblotted to nitrocellulose membrane (Trans-blot, Bio-Rad Laboratories, Segrate, Italy). Membranes were blocked with 5% non-fat dry milk in TBS-Tween for 1 h and then incubated overnight at 4 °C with rabbit polyclonal anti-SOD1 (SOD-100; Stressgen, Victoria, BC, Canada; dilution 1:1000) or rabbit polyclonal anti-GAPDH (Santa Cruz Biotechnology, Santa Cruz, CA, USA; dilution 1:1000). Immunoreactivity was revealed with goat anti-rabbit peroxidase-conjugated antibodies (Santa Cruz Biotechnology; dilution 1:5000), and visualized using ECL Plus reagents (GE Healthcare, Milan, Italy). Each experiment was carried out twice, with 4 independent samples.
2.4. RNA isolation and RT-qPCR
Frozen samples of gastrocnemius muscles were homogenized (with an ultra-turrax® homogenizer) in 4 M guanidium isothiocyanate (containing 25 mM sodium citrate pH 7.5, 0.5% sarcosyl and 0.1% 2-mercaptoethanol); C2C12 cells were harvested in the same buffer and total RNA isolated by phenol–chloroform extraction according to Chomczynski and Sacchi. Quantification was carried out by absorption at 260 nm. For Reverse Transcription (RT), an aliquot of total RNA (1 μg) was treated for 15 min at room temperature with 1U of DNaseI (Sigma–Aldrich). DNaseI was heat-inactivated and the samples were reverse-transcribed using the High-Capacity cDNA Reverse Transcription Kit (Applied Biosystems, Monza, Italy), according to the manufacturer's instructions, in a 25 μl volume. Primers for selected genes were designed via the primer Express software (PE Applied Biosystems, Foster City, CA, USA) and purchased from Eurofins MWG Operon (Ebersberg, Germany). The sequences of primers were as follows: MyoD: 5′-GGC TAC GAC ACC GCC TAC TA-3′ (forward), 5′-GTG GAG ATG CGC TCC ACT AT-3′ (reverse); myogenin: 5′-GGG CAA TGC ACT GGA GTT-3′ (forward), 5′-CAC GAT GGA CGT AAG GGA GT-3′ (reverse); atrogin-1: 5′-GAA GAG AGC AGT ATG GGG TCA-3′ (forward), 5′-CTT GAG GGG AAA GTG AGA CG-3′ (reverse); calpain-1: 5′-GCC GTG GAC TTT GAC AAC TT-3′ (forward), 5′-CAA CAC CAT CCA GGT CTG TG-3′ (reverse); TGFβ1: 5′-GAA GGA CCT GGG TTG GAA GT-3′ (forward), 5′-CGG GTT GTG TTG GTT GTA GA-3′ (reverse); GAPDH: 5′-CCA GAA CAT CAT CCC TGC AT-3′ (forward), 5′-CAG TGA GCT TCC CGT TCA-3′ (reverse). Efficiency of each set of primers was close to 100% for both target and reference genes. RT-qPCR was performed using the ABI Prism 7000 sequence detection system (PE Applied Biosystems) in a 25 μl total volume, using the iTaq SYBR Green Supermix (BioRad Laboratories), and with 500 nmol primers. PCR cycling conditions were as follows: 94 °C for 10 min, 35 cycles at 94 °C for 15 s, and 60 °C for 1 min. Melting curve analysis was performed at the end of each PCR assay to control specificity. Data was expressed as Ct values and used for the relative quantification of targets with the ΔΔCt calculation. To exclude potential bias due to averaging data transformed through the equation 2−ΔΔCt to give N-fold changes in gene expression, all statistics were performed with ΔCt values. Each experiment was carried out twice, with 4 independent samples. Each sample was run in duplicate wells.
2.5. Statistical analysis
Statistical analysis was performed through Student's t test (for the analyses between non-injured and axotomized muscles, and between undifferentiated and differentiated C2C12 cells), one way analysis of variance (ANOVA, when analyzing the effect of mutSOD1 in muscles) or two-way ANOVA (when studying the effect of androgen treatment on muscles of mutSOD1 mice), using the PRISM software (GraphPad, San Diego, CA, USA). Specific group pair(s) statistical difference were determined by the Tukey post hoc test for one-way ANOVA, and by Bonferroni post hoc test for two-way ANOVA.
3. Results
In murine fALS models, muscle denervation precedes motor neuron loss and muscles biochemistry is already altered at the presymptomatic stage [25,26,31–33]. Thus, in this study we initially characterized alterations in the expression of genes controlling muscle physiology and involved in muscle pathology. The analysis was conducted on muscle samples derived from fALS mouse models both at disease onset and at various stages of disease progression. Then, we analyzed the effects of pharmacological doses of a typical AAS, the nandrolone, on the expression of the same genes. In parallel, we set-up an ALS muscle cell model to validate and extend the observation obtained in vivo. This cellular model also allowed us to discriminate between cell-autonomous modifications of muscle pathophysiology and the effects due to muscle denervation associated to motor neuron death in the spinal cord.
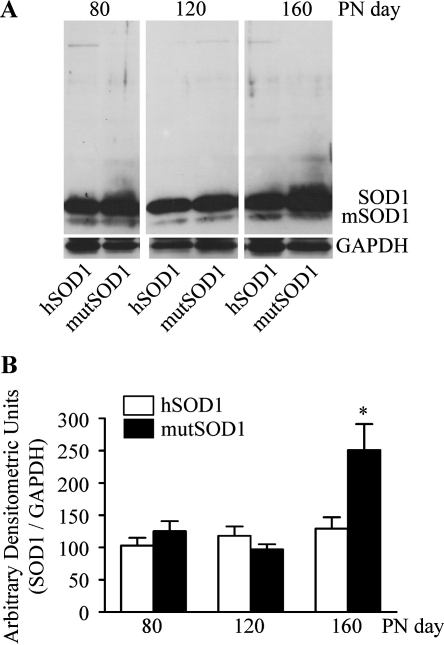
To evaluate muscle tissue modifications induced by mutSOD1, we firstly searched for SOD1 inclusions in muscle of mutSOD1 mice. Gastrocnemius muscles of male mutSOD1 tg mice was analysed at presymptomatic, symptomatic, and end stage (80, 120, and 160 day-old-mice, respectively), and comparison was made with both age-matched hSOD1 mice and NTg mice (data not shown). The levels of mutSOD1 were similar in muscles from animals at the presymptomatic and symptomatic stages and comparable to those of age-matched hSOD1 mice. In muscles from end stage mice the mutSOD1 levels increased two-fold over muscles from age matched hSOD1 mice (p < 0.05 vs. hSOD1-80 days; Fig. 1B). No high molecular weight insoluble species of SOD1 (oligomers or aggregates) were present (Fig. 1A); thus, in contrast to the observation performed in motor neurons [34], the potential mutSOD1-induced modification in muscle cells cannot be due to aggregate formation.
Fig. 1.
Biochemical properties of hSOD1 and mutSOD1 in the gastrocnemius muscle of transgenic mice. (A) Western blot assays were performed using the polyclonal anti-SOD1 antibody on gastrocnemius muscle lysates of mice expressing human wild type or G93A mutant SOD1 (hSOD1 and mutSOD1, respectively) at different ages. SOD1 indicates the human SOD1 monomeric form; mSOD1 indicates the endogenous mouse SOD1. PN day, postnatal day. (B) Quantitative data (after normalization with GAPDH levels) are expressed as percent versus the levels found in 80 day-old hSOD1 mice. Each bar represents the mean ± SEM of four independent replicates. *p < 0.05 vs. 80 days hSOD1.
3.1. Effects of mutSOD1 on gene expression in gastrocnemius muscle
To evaluate aggregate-independent mutSOD1 toxicity in muscle, we analyzed gene expression modifications of mRNAs coding for proteins controlling muscle pathways activated by nerve injury, stress, or muscular atrophy. These are MyoD and myogenin (two genes encoding myogenic regulatory factors), atrogin-1, calpain-1, and TGFβ1 considered markers for muscle fiber damage or atrophy in the gastrocnemius muscles. Their expression level was compared in mutSOD1 mice at different stages of disease, in age-matched hSOD1 and NTg mice.
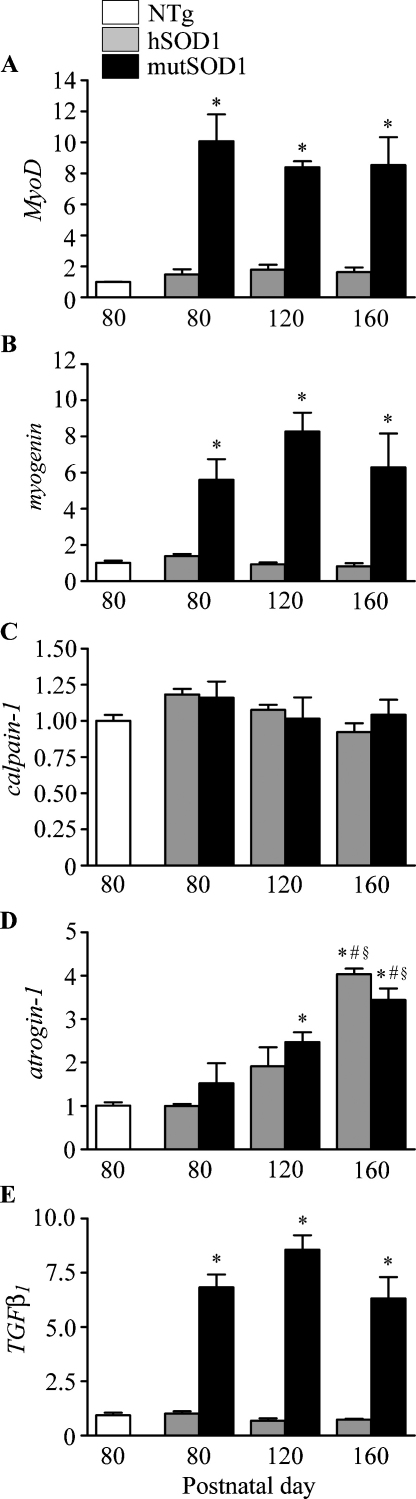
hSOD1 expression did not affect mRNA levels of MyoD and myogenin at all ages considered (Fig. 2A and B), while mutSOD1 expression induced a robust increase of MyoD and myogenin mRNA levels. Indeed, in the gastrocnemius muscles of mutSOD1 mice at presymptomatic, symptomatic and terminal stages, a 10-fold increase of MyoD and 6- to 8-fold increase of myogenin mRNA levels were observed in comparison to either hSOD1 or NTg mice at corresponding ages (p < 0.01 vs. NTg).
Fig. 2.
Effect of hSOD1 and mutSOD1 on gene expression in gastrocnemius muscle. RT-qPCRs were performed on total RNA extracted from gastrocnemius muscles of non-transgenic (NTg) mice, of mice expressing the wild type human SOD1 transgene (hSOD1), and of mice expressing the G93A mutant form of human SOD1 (mutSOD1) at different ages. All animals were age-matched. Data have been normalized to the amount of GAPDH mRNA, expressed relative to the levels determined in NTg mice (age-matched with 80-day-old mutSOD1 mice) taken as internal reference, and expressed as fold changes. Data are means ± SEM of four independent replicates. *p < 0.01 vs. NTg mice and vs. 80 days mutSOD1; #p < 0.05 vs. 120 days hSOD1; §p < 0.05 vs. 120 days mutSOD1.
Calcium-activated calpains, a class of proteins involved in the myofibrillar complex dissociation [35], are typically activated in atrophic muscles. We found that in gastrocnemius muscles calpain-1 mRNA levels are modulated by mutSOD1, but unaffected in all the other conditions tested (Fig. 2C).
The expression of the muscle-specific E3 ubiquitin ligase atrogin-1 (also called muscle atrophy F-box, MAFbx) [36] was increased in muscle of mutSOD1 mice during disease progression. At the symptomatic and terminal stages atrogin-1 mRNA levels were significantly higher than at the presymptomatic stage (p < 0.01 vs. NTg and 80 days mutSOD1; p < 0.05 vs. 120 days mutSOD1; Fig. 2D). Notably, also hSOD1 induced atrogin-1 up-regulation in the gastrocnemius muscle, but only at 160 days (*p < 0.01 vs. NTg and 80 days mutSOD1; #p < 0.05 vs.120 days hSOD1; §p < 0.05 vs. 120 days mutSOD1). No significant differences of atrogin-1 mRNA level were detected between presymptomatic mutSOD1 mice and age-matched hSOD1 mice.
Several in vivo studies suggest that TGFβ signaling is important in skeletal muscle repair [37], and we therefore analyzed the expression of TGFβ1. While TGFβ1 expression was similar in muscle of hSOD1 and NTg mice at all ages tested (Fig. 2E), we found that TGFβ1 expression is highly upregulated (up to 8-fold) in the gastrocnemius muscles of mutSOD1 mice at all disease stages (p < 0.01 vs. NTg).
3.2. Direct versus indirect effects of mutSOD1 in muscle
The modulation of activity parameters in mutSOD1 muscles may be a consequence of: (i) muscle denervation due to mutSOD1 – induced spinal motor neuron loss, and/or (ii) direct mutSOD1 toxicity in muscle cells. To investigate these alternative mechanisms (not mutually exclusive), we used two approaches. We first analyzed the mRNA levels expressed by the genes studied above in the gastrocnemius muscle of axotomized NTg mice and in a fALS muscle cell model.
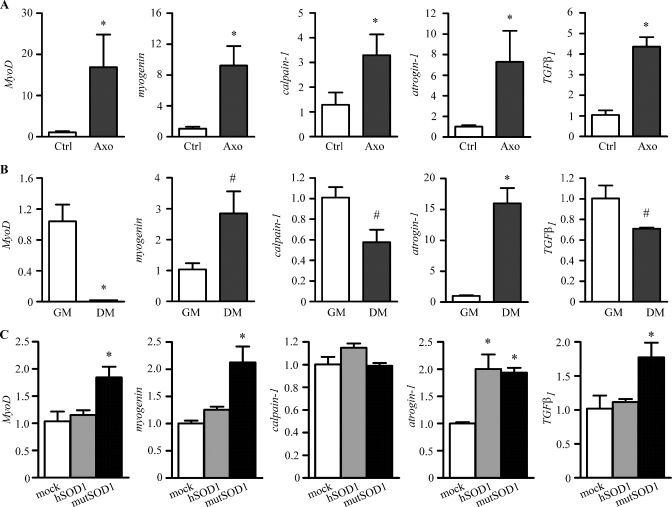
Muscles of axotomized NTg mice were collected 7 days after sciatic nerve transection, when muscle atrophy initiates as a consequence of denervation. All genes analyzed were found to be overexpressed in muscles denervated by axotomy when compared to control contralateral non-injured muscles (Fig. 3A). The variations paralleled those found in mutSOD1 mouse muscles, with the exception of calpain-1. Indeed, muscle expression of MyoD, myogenin, atrogin-1 and TGFβ1 were highly increased both in mutSOD1 and axotomized muscles, while calpain-1 showed a 3-fold increase in the denervated NTg muscle, but remained unchanged in mutSOD1 muscle.
Fig. 3.
Direct versus indirect effects of mutSOD1 toxicity in muscle. RT-qPCRs were performed on total RNA extracted from gastrocnemius muscles of left axotomized non transgenic mice (Axo). Right gastrocnemius muscles of the same animals were used as controls (Ctrl). Data have been normalized to the amount of GAPDH mRNA, expressed relative to the levels determined in control muscles, which are taken as internal reference, and expressed as fold changes. Each bar represents the mean ± SEM of four independent replicates. *p < 0.01 vs. Ctrl. RT-qPCRs were performed on total RNA extracted from C2C12 cells cultured in the growth medium (GM) or placed in the differentiation medium (DM) for 48 h. Data have been normalized to the amount of GAPDH mRNA, expressed relative to the levels determined in GM, which are taken as a reference, and expressed as fold changes. All the data shown are the means ± SEM of determinations performed (n = 4). *p < 0.001 vs. GM; #p < 0.05 vs. GM. RT-qPCRs were performed on total RNA extracted from C2C12 cells transfected with wild type human SOD1 (hSOD1) or the G93A mutant form of human SOD1 (mutSOD1) and cultured in the differentiation medium for 48 h. Mock transfected cells are used as controls. Data have been normalized to the amount of GAPDH-mRNA, expressed relative to the levels determined in control cells, which are taken as internal reference, and expressed as fold changes. Each bar represents the mean ± SEM of four independent replicates. *p < 0.01 vs. mock.
To test a potential direct toxicity of mutSOD1 in muscle cells we produced and characterized a cell model using the mouse myoblastic cell line C2C12, which provides a well-established in vitro model for the study of muscle differentiation [38]. To avoid modification linked to C2C12 differentiation after SOD1 expression, we initially quantified the expression levels of the five genes studied above in undifferentiated and differentiated C2C12 cells. Differentiation of C2C12 cells was induced by culturing cells in low serum concentrations for 48 h. C2C12 cell differentiation completely abolished MyoD mRNA expression, while myogenin levels was increased 3-fold (Fig. 3B); downregulation of calpain-1 and TGFβ1 expression was observed accompanied by a robust increase (about 15-fold) of atrogin-1 level (Fig. 3B). We then tested the potential detrimental effects of mutSOD1 in C2C12 fALS model. C2C12 were utilized in their differentiating phase since this better reflects the physiological condition of muscle in adult animals. In these cells mutSOD1 induced a significant increase of both MyoD and myogenin (Fig. 3C), while no variation were seen with hSOD1. Calpain-1 expression remained unchanged both in hSOD1- or mutSOD1-expressing C2C12 cells (Fig. 3C), as observed in mice (Fig. 2C). On the other hand, in C2C12 differentiating cells atrogin-1 mRNA (Fig. 3C) expression was increased both by hSOD1 and mutSOD1, while TGFβ1 mRNA levels were increased only by mutSOD1 (Fig. 3C).
Altogether these findings, which show high consistency, indicate that all the selected genes are upregulated by both denervation and mutSOD1 overexpression (either in cells and tissues), with the exception of calpain-1 which is modulated only by denervation.
3.3. Effect of androgen treatment on muscles of mutSOD1 mice
As mentioned previously, muscle atrophy is a major cause of disability in ALS, and increasing muscle strength might help to preserve functions in patients. Androgens are one of the factors that enhance muscle size and strength by activating many different mechanisms, and have been proposed as a risk factor for ALS [6]. To evaluate the impact of androgens on ALS muscles, we treated NTg or mutSOD1 mice with pharmacological doses of nandrolone. It is a synthetic testosterone derivative that exhibits much weaker androgenic properties and higher anabolic effects than testosterone [39]; it is used in some cases of osteoporosis in postmenopausal women as well as in cases of anemia associated to renal insufficiency. Unfortunately, this drug is often illegally used by body builders and other athletes to enhance their muscle mass.
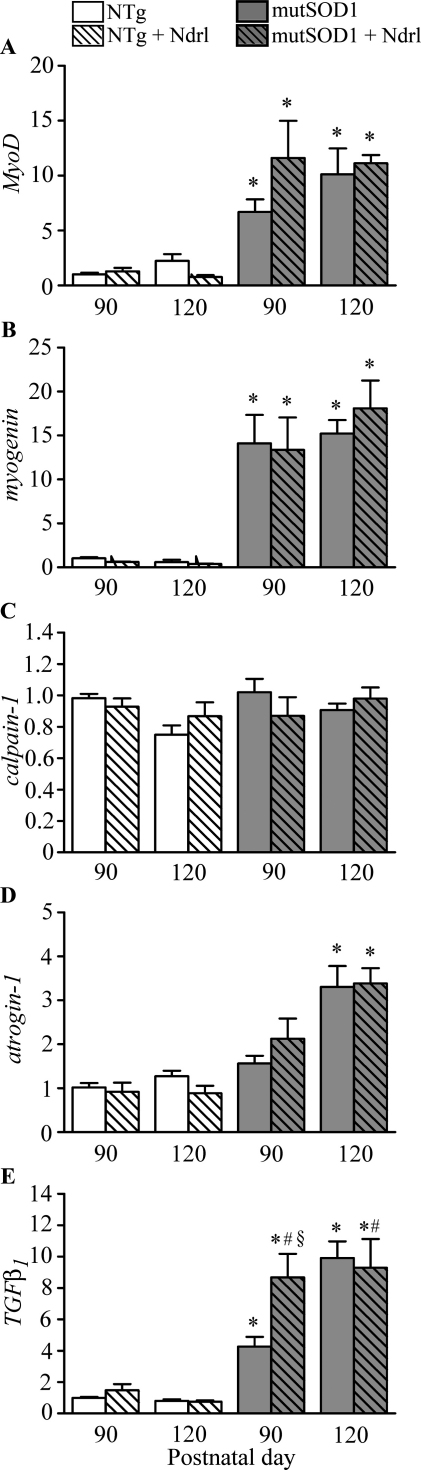
Mice were treated with nandrolone (10 mg/kg, subcutaneously once a week) from the 7th week of age until disease onset or the symptomatic stage (120 days). The clinical evaluation of disease onset showed that nandrolone did not affect the appearance of the symptoms in mutSOD1 mice (90 ± 7 days in vehicle-treated mutSOD1 mice and 89 ± 9 days in nandrolone-treated mutSOD1 mice). In addition, nandrolone treatment did not modify the massive effects of mutSOD1 on MyoD, myogenin and atrogin-1 expression (Fig. 4A–D), and did not alter the levels of calpain-1 expression. When TGFβ1 expression was compared in untreated and nandrolone-treated mice, we found that AAS administration greatly affects the overall TGFβ1 mRNA levels (Fig. 4E). Indeed, at disease onset, mutSOD1 mice treated with nandrolone showed TGFβ1 mRNA levels significantly higher than that detected in untreated mutSOD1 mice (p < 0.01 vs. NTg 90 days; p < 0.01 vs. untreated mutSOD1 mice at onset). Notably, TGFβ1 gene expression reached the same level in untreated and nandrolone-treated mutSOD1 mice at the symptomatic stage.
Fig. 4.
Effect of nandrolone treatment on gene expression in gastrocnemius muscles of mutSOD1 mice. RT-qPCRs were performed on total RNA extracted from gastrocnemius muscles of non transgenic (NTg) or mutSOD1 mice treated with vehicle or Nandrolone (Ndrl; see text for treatment parameters). Animals were age-matched. Data have been normalized to the amount of GAPDH mRNA, expressed relative to the levels determined in 90-day-old NTg mice, which are taken as internal reference, and expressed as fold changes. Each bar represents the mean ± SEM of 4 independent replicates. The two-way ANOVA found a significant effect of mutSOD1 (*p < 0.0001), no effect of nandrolone (p > 0.05), and no significant interaction between the two variables (p > 0.05) for MyoD, myogenin, calpain-1, and atrogin-1. For TGFβ1 the analysis indicated a significant effect of mutSOD1 (*p < 0.0001), and of nandrolone treatment (#p < 0.05), with a significant mutSOD1 by nandrolone treatment interaction at 90 days (§p < 0.05).
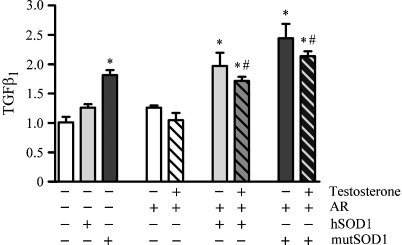
The effect of AAS on TGFβ1 expression was also tested in the C2C12 cell model of fALS. Since our line of C2C12 cells expresses very low levels of AR, we co-transfected the cells with AR (AR.Q23) and either hSOD1 or mutSOD1, and found that AR expression correlated with an increased TGFβ1 level in C2C12 co-expressing mutSOD1. Thus, AR exacerbated the detrimental effects of mutSOD1 in cultured muscle cells, whereas there was no apparent effect of testosterone treatment. Since C2C12 cells have been treated with testosterone 10−8 M for 48 h after transfection, this time period may not be sufficient to allow the muscle cells to directly respond to AAS (longer time could not be tested because our experiments have been performed in transient transfection) (Fig. 5). Very interestingly, C2C12 cells co-expressing hSOD1 and AR, even without androgens, showed TGFβ1 mRNA levels significantly higher than that of cells expressing only hSOD1 (Fig. 5), suggesting a synergistic detrimental effect of SOD1 and AR expressed at high levels on muscle physiology.
Fig. 5.
Effect of testosterone treatment on TGFβ1 gene expression in C2C12 cells transfected with hSOD1 or mutSOD1. RT-qPCRs were performed on total RNA extracted from C2C12 cells co-transfected with wild type human SOD1 (hSOD1) or G93A mutant form of human SOD1 (mutSOD1) and androgen receptor (AR). After transfection cells were cultured in the charcoal–dextran-treated horse serum differentiation medium for 48 h with or without testosterone 10−8 M. Mock transfected cells are used as controls. Data have been normalized to the amount of GAPDH-mRNA, expressed relative to the levels determined in control cells, which are taken as internal reference, and expressed as fold changes. Each bar represents the mean ± SEM of four independent replicates. The two-way ANOVA found a significant effect of hSOD1 and mutSOD1 (*p < 0.0001), and of testosterone (#p > 0.001), and no significant interaction (p > 0.05) between the two variables.
4. Discussion
In the present study we demonstrate in animal and cellular models that mutSOD1 modifies the expression of genes involved in muscle cell signaling pathways activated by nerve injury, stress, or atrophy. In addition, we show that nandrolone, a widely used synthetic ASS, is able to exacerbate the deleterious effects of mutSOD1 on TGFβ1 expression in the muscle of the murine model of fALS. The major goal of the present study was to identify the effect of mutSOD1 on the transcription of a set of genes relevant for muscle pathophysiology, and we therefore focused on the level of expression of these transcripts. Further verification at the protein level will help to understand the impact of such modifications, which, however, could be influenced by several factors (variations in the translational control, protein maturation, post-translational modifications, protein clearance).
The obtained data clearly indicate that mutSOD1 toxicity can be exerted independently of its tendency to aggregate. This is relevant to understand the cell specificity of the adverse effects of mutSOD1, since several observations demonstrated that proteinaceous inclusions rich in mutSOD1 are present in spinal cord tissues from mutSOD1 animals. These inclusions may alter SOD1 protein bioavailability and turnover [40], reducing the overall protection exerted by SOD1 against free radical reactive oxygen species [41]. In the present investigation, increased mutSOD1 levels were detectable in the gastrocnemius muscle of fALS mice, but only in the terminal stage of disease. However, we did not find SOD1 high molecular weight SDS-resistant species in muscle of mutSOD1 mice or mutSOD1-expressing C2C12 cells [38], in agreement with a recent report showing that the increased level of mutant protein in muscle is due to its soluble fraction [42]. On the contrary, in muscle C2C12 cells, at variance with immortalized motor neurons [34,41], mutSOD1 levels are significantly lower than those observed in hSOD1 C2C12 cells [38], suggesting that mutSOD1 clearance is accelerated in ALS muscle cells.
The upregulation of MyoD and myogenin transcripts indicates that muscles of fALS mice attempt to counteract the effect of mutSOD1. The expression of transcripts of myogenic regulatory factors in C2C12 cells paralleled that obtained in the gastrocnemius muscle, thus indicating that mutSOD1 provides a stimulus continuously upregulating MyoD, which is instead usually downregulated during the cell differentiation process. Indeed, MyoD up-regulation is necessary to activate quiescent satellite cells during the first phase of the satellite cell differentiation pathway, while MyoD down-regulation is then required to generate reserve cells [43]. This aberrant behavior induced by mutSOD1 might be at the basis of muscle atrophy in ALS, since prolonged increase of MyoD and myogenin levels could cause an irreversible commitment and terminal differentiation of satellite cells, depriving muscle of their cell reservoir, thus impairing its regenerative potential. In line with this hypothesis are the findings showing that cdk5 activity and cyclin D1 levels, which control cell cycle progression, are reduced in muscles of mutSOD1 mice [33], and that CDKN1 and RB1, which are MyoD downstream targets able to arrest cell cycle, are upregulated in preparalyzed SOD1G86R mouse muscles [44]. Of note, the present study reveals that nandrolone, despite its well-known potent anabolic properties on muscle tissue, was unable to revert the altered MyoD and myogenin transcription induced by mutSOD1 in ALS muscle. As a positive result, nandrolone did not exacerbate this deleterious action of mutSOD1 in muscle suggesting that AAS might not play a role on this particular aberrant pathway in muscle tissue.
The expression of calpain-1 was unchanged in cells transfected with either hSOD1 or mutSOD1, as well as in the gastrocnemius muscle of mutSOD1 mice. No changes of calpain-1 expression were induced by nandrolone, suggesting that this gene is not a target of the androgenic action in muscle. Surprisingly, calpain-1 expression significantly increased after muscle denervation in NTg mice. Thus, in fALS mice motor neuron damage or loss and consequent target muscle denervation are not the only cause of muscle dysfunctions and atrophy.
It is interesting to note that atrogin-1 level was robustly increased in muscles of both mutSOD1 at the end stage and age-matched hSOD1 mice, while at the symptomatic stage atrogin-1 was significantly induced in muscle of mutSOD1 mice but not in age-matched hSOD1 mice. Such data have been here confirmed also in the muscle cell model. This suggests that hSOD1 overexpression in muscle may in part mimic the intracellular response to mutSOD1. An increase of SOD1 levels could be interpreted by muscle cells as a stress signal, as supported by findings indicating that also aged hSOD1 animals may show symptoms reminiscent of alteration of muscle functions [45]. This hypothesis is also corroborated by the finding that muscle cells from ALS patients exhibit intrinsic increased sensitivity to oxidative stress [46]. Moreover, atrogin-1 expression is linked to normal protein turnover in muscles [36,47] and oxidative stress upregulates atrogin-1 expression in vivo [48–50], validating the hypothesis that high levels of hSOD1 represent a stress signal for skeletal muscle. As it will be discussed below in further details, our data have also shown that AR and wt SOD1 co-expression in muscle C2C12 cells results in a significant increase of TGFβ1 expression, in line with the hypothesis that wt SOD1 (and AR) may have a role also in cases of sALS. Moreover, previous data have indicated that both atrogin-1 mRNA and protein content are significantly increased in skeletal muscle of mutSOD1 mice and ALS patients compared with healthy control subjects [51]. As in the case of MyoD, myogenin and calpain-1, the AAS nandrolone had no effects on the level of atrogin-1 mRNA, suggesting that also this pathway is not influenced by the androgenic anabolic action in muscle tissue.
TGFβ1 is a diffusible factor that promotes motor neuron survival [52–54], and deeply modulates muscle functions. High levels of TGFβ1 are, however, detrimental for muscles. For example, mice deficient for the extracellular matrix protein fibrillin-1 are characterized by excessive TGFβ1 signaling which results in myopathy and inability to increase muscle mass despite exercise [37]. Dystrophin-deficient Mdx mice (a model of Duchenne muscular dystrophy) are characterized by degeneration of muscle fibers, increased skeletal muscle fibrosis and augmented TGFβ1 signaling [37,55]. In these murine models, neutralization of TGFβ1 signaling activates skeletal muscle regeneration, suggesting a direct role for this cytokine in skeletal muscle maintenance [37,55]. The present results showed that TGFβ1 expression is robustly increased by mutSOD1 in both gastrocnemius muscle and C2C12 cells. Interestingly, TGFβ1 inhibits MyoD transcription (and activity) through Smad3 [37,56–58], while attenuation of the TGFβ/pSmad3 signaling restores regeneration of old muscle satellite cells in vivo [59]. Therefore, the high levels of MyoD mRNA in muscle of mutSOD1 mice and in mutSOD1-transfected C2C12 cells strongly suggest the existence of an impairment of the TGFβ1 signaling pathway in ALS. In fact, several data indicates that a dysfunctional TGFβ/Smad signal transduction pathway could be involved in the pathogenesis of ALS, with impairment of TGFβ signal transduction presumably at the step of pSmad2/3 translocation into the nucleus [60]. Nucleocytoplasmic transport impairment has also been reported in fALS mice [61]. Furthermore, it has been reported that also in Spinal and Bulbar Muscular Atrophy (a polyglutamine disease dependent on the expansion of the CAG repeat within the AR) motor neuron damage is associated with disruption of TGFβ signaling [62]. The present data are also in agreement with the high TGFβ1 plasma levels detected in ALS patients, and with the significant positive correlation between TGFβ1 plasma concentration and disease duration [63]. Finally, it has been recently reported that the transcription factor ZNF512B, encoding an important positive regulator of TGFβ signaling, is a new susceptibility gene for ALS [64].
Axotomy was here found to up-regulate also muscle TGFβ1 mRNA levels. It is known that TGFβ1 doubles the size of acetylcholine receptor clusters at nerve–muscle contacts and increases the percentage of these contacts [65]. Thus, muscles may attempt to re-establish contacts with nerve endings even if the positive effects of a transiently increased TGFβ1 production at the neuromuscular junction may become detrimental.
Concerning androgenic steroids, we initially postulated that the anabolic actions of androgens (such as testosterone) on muscle could be due to its ability to repress the expression of atrogin-1 [66,67], or to promote myogenic differentiation of mesenchymal multipotent cells by inhibition of the TGFβ signaling pathway [68]. However, the present data clearly indicate that an excessive anabolic stimulation of AR, obtained with treatment with pharmacological doses of nandrolone, leads to increased TGFβ1 expression in the muscle of mutSOD1 mice. This evidence is also supported by our observation in C2C12 cultured muscle cells in which co-expression of AR (the mediator of AAS action) and mutSOD1 resulted in significant increase of TGFβ1 expression with respect to mutSOD1 expression only. Thus, the AR circuitry and particularly the anabolic action of nandrolone in ALS muscle seems to greatly enhance the detrimental effect of this growth factor. TGFβ1 exerts a key role in the trans-differentiation of myoblasts in myofibroblasts, thus hampering tissue repair [69]. Notably, targeted overexpression of AR in skeletal muscle fibers leads to muscle weakness and early death associated with motor neuron loss [19], strongly supporting the notion that aberrant androgenic stimulation of muscle may be detrimental for skeletal muscle and possibly for motor neurons.
Insulin-like Growth Factor-1 (IGF-1) expression in skeletal muscle increases the survival of mutSOD1 mice [70,71]; thus muscle trophic factors overproduction could help to rescue spinal motor neurons [12,72–74], suggesting that alterations of muscular origin could contribute to enhance axonal vulnerability in motor neurons. IGF-1 synthesis in muscle is regulated by AAS [75], and physical exercise may modify the production of endogenous AAS [76]. Interestingly, a cross-talk between androgen and IGF-1 has been described in several systems [77]. However, the role of androgens or the effect of androgen dysregulation on IGF-1 signaling in ALS remains to be clarified characterizing the molecular cross-talk between androgens and muscle-specific IGF-1 signaling in the maintenance of muscle phenotype.
Recent data indicate an increased risk for ALS in male soccer and football players, with a ALS prevalence 20 times higher than in general population [13,16]. This event suggests that there might be a link between sALS and sport. Although mechanical stress and repeated trauma of muscle have been implicated in sALS, no increased risk of developing ALS has been reported among professional road cyclists and basketball player, suggesting that extensive physical activity per se is not a risk for ALS [78]. It has been proposed that the increased risk of ALS in soccer players might be linked to AAS abuse [13,16]. The increase of TGFβ1 expression induced by androgenic treatment here reported seem to support this hypothesis, indicating that an extensive use of AAS may influence the effect of SOD1 protein becoming a risk factors for ALS.
5. Conclusions
Altogether the present findings support the notion that in mutSOD1 mice functionality of skeletal muscle, besides that of motor neurons, is altered and that these alterations precede severe motor neuron loss. Moreover, the molecular data we obtained in the fALS murine model suggest that mutSOD1 skeletal muscles might have impaired regenerative potential and increased susceptibility to atrophy. The significance of such data for sALS pathogenesis remains to be verified; however, the data obtained with the C2C12 cell model indicate that an increase of the AR (that in vivo was here obtained through stabilization of the receptor with chronic AAS treatment) may modify also the effect of hSOD1 leading to an increase of TGFβ1 expression. Future studies will be carried out to clarify the interaction of AR with SOD1.
No effective treatments are available for ALS patients, and strategies to ameliorate symptoms or positively affect the life of patients must be taken in consideration. Furthermore, the present findings suggest that TGFβ1 could be a new pharmacological target to delay muscles wasting in ALS.
Acknowledgements
The financial support of Telethon – Italy (GGP06063, and GGP07063), Fondazione CARIPLO (2008-2307), ARISLA, Italian Ministry of Labour, Health and Social Affairs (2007-36; 2008-15; and convenzione Fondazione Mondino/UNIMI), Fondation Thierry Latran (France), University of Milan are gratefully acknowledged.
References
- 1.Cova E., Bongioanni P., Cereda C., Metelli M.R., Salvaneschi L., Bernuzzi S. Time course of oxidant markers and antioxidant defenses in subgroups of amyotrophic lateral sclerosis patients. Neurochem Int. 2010;56:687–693. doi: 10.1016/j.neuint.2010.02.004. [DOI] [PubMed] [Google Scholar]
- 2.Gagliardi S., Cova E., Davin A., Guareschi S., Abel K., Alvisi E. Sod1 mrna expression in sporadic amyotrophic lateral sclerosis. Neurobiol Dis. 2010;39:198–203. doi: 10.1016/j.nbd.2010.04.008. [DOI] [PubMed] [Google Scholar]
- 3.Pasinelli P., Brown R.H. Molecular biology of amyotrophic lateral sclerosis: Insights from genetics. Nat Rev Neurosci. 2006;7:710–723. doi: 10.1038/nrn1971. [DOI] [PubMed] [Google Scholar]
- 4.Bendotti C., Carri M.T. Lessons from models of sod1-linked familial als. Trends Mol Med. 2004;10:393–400. doi: 10.1016/j.molmed.2004.06.009. [DOI] [PubMed] [Google Scholar]
- 5.Boillee S., Vande Velde C., Cleveland D.W. Als: A disease of motor neurons and their nonneuronal neighbors. Neuron. 2006;52:39–59. doi: 10.1016/j.neuron.2006.09.018. [DOI] [PubMed] [Google Scholar]
- 6.Manjaly Z.R., Scott K.M., Abhinav K., Wijesekera L., Ganesalingam J., Goldstein L.H. The sex ratio in amyotrophic lateral sclerosis: a population based study. Amyotroph Lateral Scler. 2010 doi: 10.3109/17482961003610853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Groeneveld G.J., Van Muiswinkel F.L., Sturkenboom J.M., Wokke J.H., Bar P.R., Van den Berg L.H. Ovariectomy and 17beta-estradiol modulate disease progression of a mouse model of als. Brain Res. 2004;1021:128–131. doi: 10.1016/j.brainres.2004.06.024. [DOI] [PubMed] [Google Scholar]
- 8.Haverkamp L.J., Appel V., Appel S.H. Natural history of amyotrophic lateral sclerosis in a database population. Validation of a scoring system and a model for survival prediction. Brain. 1995;118(Pt 3):707–719. doi: 10.1093/brain/118.3.707. [DOI] [PubMed] [Google Scholar]
- 9.Logroscino G., Traynor B.J., Hardiman O., Chio A., Mitchell D., Swingler R.J. Incidence of amyotrophic lateral sclerosis in europe. J Neurol Neurosurg Psychiatry. 2010;81:385–390. doi: 10.1136/jnnp.2009.183525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Rudnicki S.A. Estrogen replacement therapy in women with amyotrophic lateral sclerosis. J Neurol Sci. 1999;169:126–127. doi: 10.1016/s0022-510x(99)00234-8. [DOI] [PubMed] [Google Scholar]
- 11.Heiman-Patterson T.D., Deitch J.S., Blankenhorn E.P., Erwin K.L., Perreault M.J., Alexander B.K. Background and gender effects on survival in the tgn(sod1-g93a)1gur mouse model of als. J Neurol Sci. 2005;236:1–7. doi: 10.1016/j.jns.2005.02.006. [DOI] [PubMed] [Google Scholar]
- 12.Veldink J.H., Bar P.R., Joosten E.A., Otten M., Wokke J.H., van den Berg L.H. Sexual differences in onset of disease and response to exercise in a transgenic model of als. Neuromuscul Disord. 2003;13:737–743. doi: 10.1016/s0960-8966(03)00104-4. [DOI] [PubMed] [Google Scholar]
- 13.Abel E.L. Football increases the risk for lou gehrig's disease, amyotrophic lateral sclerosis. Percept Mot Skills. 2007;104:1251–1254. doi: 10.2466/pms.104.4.1251-1254. [DOI] [PubMed] [Google Scholar]
- 14.Beghi E., Logroscino G., Chio A., Hardiman O., Millul A., Mitchell D. Amyotrophic lateral sclerosis, physical exercise, trauma and sports: Results of a population-based pilot case-control study. Amyotroph Lateral Scler. 2010;11:289–292. doi: 10.3109/17482960903384283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Belli S., Vanacore N. Proportionate mortality of Italian soccer players: is amyotrophic lateral sclerosis an occupational disease. Eur J Epidemiol. 2005;20:237–242. doi: 10.1007/s10654-004-6879-7. [DOI] [PubMed] [Google Scholar]
- 16.Chio A., Benzi G., Dossena M., Mutani R., Mora G. Severely increased risk of amyotrophic lateral sclerosis among italian professional football players. Brain. 2005;128:472–476. doi: 10.1093/brain/awh373. [DOI] [PubMed] [Google Scholar]
- 17.Piazza O., Siren A.L., Ehrenreich H. Soccer, neurotrauma and amyotrophic lateral sclerosis: Is there a connection. Curr Med Res Opin. 2004;20:505–508. doi: 10.1185/030079904125003296. [DOI] [PubMed] [Google Scholar]
- 18.Johansen J.A., Yu Z., Mo K., Monks D.A., Lieberman A.P., Breedlove S.M. Recovery of function in a myogenic mouse model of spinal bulbar muscular atrophy. Neurobiol Dis. 2009;34:113–120. doi: 10.1016/j.nbd.2008.12.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Monks D.A., Johansen J.A., Mo K., Rao P., Eagleson B., Yu Z. Overexpression of wild-type androgen receptor in muscle recapitulates polyglutamine disease. Proc Natl Acad Sci USA. 2007;104:18259–18264. doi: 10.1073/pnas.0705501104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Johansen J.A., Troxell-Smith S.M., Yu Z., Mo K., Monks D.A., Lieberman A.P. Prenatal flutamide enhances survival in a myogenic mouse model of spinal bulbar muscular atrophy. Neurodegener Dis. 2011;8:25–34. doi: 10.1159/000313682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Miller T.M., Kim S.H., Yamanaka K., Hester M., Umapathi P., Arnson H. Gene transfer demonstrates that muscle is not a primary target for non-cell-autonomous toxicity in familial amyotrophic lateral sclerosis. Proc Natl Acad Sci USA. 2006;103:19546–19551. doi: 10.1073/pnas.0609411103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Corti S., Donadoni C., Ronchi D., Bordoni A., Fortunato F., Santoro D. Amyotrophic lateral sclerosis linked to a novel sod1 mutation with muscle mitochondrial dysfunction. J Neurol Sci. 2009;276:170–174. doi: 10.1016/j.jns.2008.09.030. [DOI] [PubMed] [Google Scholar]
- 23.Dobrowolny G., Aucello M., Rizzuto E., Beccafico S., Mammucari C., Boncompagni S. Skeletal muscle is a primary target of sod1g93a-mediated toxicity. Cell Metab. 2008;8:425–436. doi: 10.1016/j.cmet.2008.09.002. [DOI] [PubMed] [Google Scholar]
- 24.Wong M., Martin L.J. Skeletal muscle-restricted expression of human sod1 causes motor neuron degeneration in transgenic mice. Hum Mol Genet. 2010;19:2284–2302. doi: 10.1093/hmg/ddq106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Frey D., Schneider C., Xu L., Borg J., Spooren W., Caroni P. Early and selective loss of neuromuscular synapse subtypes with low sprouting competence in motoneuron diseases. J Neurosci. 2000;20:2534–2542. doi: 10.1523/JNEUROSCI.20-07-02534.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kennel P.F., Finiels F., Revah F., Mallet J. Neuromuscular function impairment is not caused by motor neurone loss in fals mice: an electromyographic study. Neuroreport. 1996;7:1427–1431. doi: 10.1097/00001756-199605310-00021. [DOI] [PubMed] [Google Scholar]
- 27.Fischer L.R., Culver D.G., Tennant P., Davis A.A., Wang M., Castellano-Sanchez A. Amyotrophic lateral sclerosis is a distal axonopathy: evidence in mice and man. Exp Neurol. 2004;185:232–240. doi: 10.1016/j.expneurol.2003.10.004. [DOI] [PubMed] [Google Scholar]
- 28.Pizzasegola C., Caron I., Daleno C., Ronchi A., Minoia C., Carri M.T. Treatment with lithium carbonate does not improve disease progression in two different strains of sod1 mutant mice. Amyotroph Lateral Scler. 2009;10:221–228. doi: 10.1080/17482960902803440. [DOI] [PubMed] [Google Scholar]
- 29.Tortarolo M., Crossthwaite A.J., Conforti L., Spencer J.P., Williams R.J., Bendotti C. Expression of sod1 g93a or wild-type sod1 in primary cultures of astrocytes down-regulates the glutamate transporter glt-1: lack of involvement of oxidative stress. J Neurochem. 2004;88:481–493. doi: 10.1046/j.1471-4159.2003.02208.x. [DOI] [PubMed] [Google Scholar]
- 30.Simeoni S., Mancini M.A., Stenoien D.L., Marcelli M., Weigel N.L., Zanisi M. Motoneuronal cell death is not correlated with aggregate formation of androgen receptors containing an elongated polyglutamine tract. Hum Mol Genet. 2000;9:133–144. doi: 10.1093/hmg/9.1.133. [DOI] [PubMed] [Google Scholar]
- 31.Pun S., Santos A.F., Saxena S., Xu L., Caroni P. Selective vulnerability and pruning of phasic motoneuron axons in motoneuron disease alleviated by cntf. Nat Neurosci. 2006;9:408–419. doi: 10.1038/nn1653. [DOI] [PubMed] [Google Scholar]
- 32.Willmann R., Pun S., Stallmach L., Sadasivam G., Santos A.F., Caroni P. Cholesterol and lipid microdomains stabilize the postsynapse at the neuromuscular junction. EMBO J. 2006;25:4050–4060. doi: 10.1038/sj.emboj.7601288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Park K.H., Vincent I. Presymptomatic biochemical changes in hindlimb muscle of g93a human cu/zn superoxide dismutase 1 transgenic mouse model of amyotrophic lateral sclerosis. Biochim Biophys Acta. 2008;1782:462–468. doi: 10.1016/j.bbadis.2008.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Crippa V., Sau D., Rusmini P., Boncoraglio A., Onesto E., Bolzoni E. The small heat shock protein b8 (hspb8) promotes autophagic removal of misfolded proteins involved in amyotrophic lateral sclerosis (als) Hum Mol Genet. 2010;19:3440–3456. doi: 10.1093/hmg/ddq257. [DOI] [PubMed] [Google Scholar]
- 35.Huang J., Forsberg N.E. Role of calpain in skeletal–muscle protein degradation. Proc Natl Acad Sci USA. 1998;95:12100–12105. doi: 10.1073/pnas.95.21.12100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Bodine S.C., Latres E., Baumhueter S., Lai V.K., Nunez L., Clarke B.A. Identification of ubiquitin ligases required for skeletal muscle atrophy. Science. 2001;294:1704–1708. doi: 10.1126/science.1065874. [DOI] [PubMed] [Google Scholar]
- 37.Schabort E.J., van der Merwe M., Loos B., Moore F.P., Niesler C.U. Tgf-beta's delay skeletal muscle progenitor cell differentiation in an isoform-independent manner. Exp Cell Res. 2009;315:373–384. doi: 10.1016/j.yexcr.2008.10.037. [DOI] [PubMed] [Google Scholar]
- 38.Onesto E., Rusmini P., Crippa V., Ferri N., Zito A., Galbiati M. Muscle cells and motoneurons differentially remove mutant sod1 causing familial amyotrophic lateral sclerosis. J Neurochem. 2011;118:266–280. doi: 10.1111/j.1471-4159.2011.07298.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Parr M.K., Flenker U., Schanzer W. Sports-related issues and biochemistry of natural and synthetic anabolic substances. Endocrinol Metab Clin North Am. 2010;39:45–57. doi: 10.1016/j.ecl.2009.11.004. [DOI] [PubMed] [Google Scholar]
- 40.Cozzolino M., Ferri A., Carri M.T. Amyotrophic lateral sclerosis: from current developments in the laboratory to clinical implications. Antioxid Redox Signal. 2008;10:405–443. doi: 10.1089/ars.2007.1760. [DOI] [PubMed] [Google Scholar]
- 41.Sau D., De Biasi S., Vitellaro-Zuccarello L., Riso P., Guarnieri S., Porrini M. Mutation of sod1 in als: a gain of a loss of function. Hum Mol Genet. 2007;16:1604–1618. doi: 10.1093/hmg/ddm110. [DOI] [PubMed] [Google Scholar]
- 42.Karch C.M., Borchelt D.R. An examination of alpha B-crystalline as a modifier of sod1 aggregate pathology and toxicity in models of familial amyotrophic lateral sclerosis. J Neurochem. 2010;113:1092–1100. doi: 10.1111/j.1471-4159.2010.06572.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Yoshida N., Yoshida S., Koishi K., Masuda K., Nabeshima Y. Cell heterogeneity upon myogenic differentiation: down-regulation of myod and myf-5 generates ‘reserve cells’. J Cell Sci. 1998;111(Pt 6):769–779. doi: 10.1242/jcs.111.6.769. [DOI] [PubMed] [Google Scholar]
- 44.Gonzalez de Aguilar J.L., Niederhauser-Wiederkehr C., Halter B., De Tapia M., Di Scala F., Demougin P. Gene profiling of skeletal muscle in an amyotrophic lateral sclerosis mouse model. Physiol Genomics. 2008;32:207–218. doi: 10.1152/physiolgenomics.00017.2007. [DOI] [PubMed] [Google Scholar]
- 45.Jaarsma D., Haasdijk E.D., Grashorn J.A., Hawkins R., van Duijn W., Verspaget H.W. Human cu/zn superoxide dismutase (sod1) overexpression in mice causes mitochondrial vacuolization, axonal degeneration, and premature motoneuron death and accelerates motoneuron disease in mice expressing a familial amyotrophic lateral sclerosis mutant sod1. Neurobiol Dis. 2000;7:623–643. doi: 10.1006/nbdi.2000.0299. [DOI] [PubMed] [Google Scholar]
- 46.Bradley L.J., Taanman J.W., Kallis C., Orrell R.W. Increased sensitivity of myoblasts to oxidative stress in amyotrophic lateral sclerosis peripheral tissues. Exp Neurol. 2009;218:92–97. doi: 10.1016/j.expneurol.2009.04.007. [DOI] [PubMed] [Google Scholar]
- 47.Gomes M.D., Lecker S.H., Jagoe R.T., Navon A., Goldberg A.L. Atrogin-1, a muscle-specific f-box protein highly expressed during muscle atrophy. Proc Natl Acad Sci USA. 2001;98:14440–14445. doi: 10.1073/pnas.251541198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Cai D., Frantz J.D., Tawa N.E., Jr., Melendez P.A., Oh B.C., Lidov H.G. Ikkbeta/nf-kappab activation causes severe muscle wasting in mice. Cell. 2004;119:285–298. doi: 10.1016/j.cell.2004.09.027. [DOI] [PubMed] [Google Scholar]
- 49.Li Y.P., Chen Y., Li A.S., Reid M.B. Hydrogen peroxide stimulates ubiquitin-conjugating activity and expression of genes for specific e2 and e3 proteins in skeletal muscle myotubes. Am J Physiol Cell Physiol. 2003;285:C806–C812. doi: 10.1152/ajpcell.00129.2003. [DOI] [PubMed] [Google Scholar]
- 50.Li Y.P., Chen Y., John J., Moylan J., Jin B., Mann D.L. Tnf-alpha acts via p38 mapk to stimulate expression of the ubiquitin ligase atrogin1/mafbx in skeletal muscle. FASEB J. 2005;19:362–370. doi: 10.1096/fj.04-2364com. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Leger B., Vergani L., Soraru G., Hespel P., Derave W., Gobelet C. Human skeletal muscle atrophy in amyotrophic lateral sclerosis reveals a reduction in akt and an increase in atrogin-1. FASEB J. 2006;20:583–585. doi: 10.1096/fj.05-5249fje. [DOI] [PubMed] [Google Scholar]
- 52.Krieglstein K., Strelau J., Schober A., Sullivan A., Unsicker K. Tgf-beta and the regulation of neuron survival and death. J Physiol Paris. 2002;96:25–30. doi: 10.1016/s0928-4257(01)00077-8. [DOI] [PubMed] [Google Scholar]
- 53.Martinou J.C., Le Van Thai A., Valette A., Weber M.J. Transforming growth factor beta 1 is a potent survival factor for rat embryo motoneurons in culture. Brain Res Dev Brain Res. 1990;52:175–181. doi: 10.1016/0165-3806(90)90233-o. [DOI] [PubMed] [Google Scholar]
- 54.Oppenheim R.W., Prevette D., Haverkamp L.J., Houenou L., Yin Q.W., McManaman J. Biological studies of a putative avian muscle-derived neurotrophic factor that prevents naturally occurring motoneuron death in vivo. J Neurobiol. 1993;24:1065–1079. doi: 10.1002/neu.480240806. [DOI] [PubMed] [Google Scholar]
- 55.Cohn R.D., van Erp C., Habashi J.P., Soleimani A.A., Klein E.C., Lisi M.T. Angiotensin ii type 1 receptor blockade attenuates tgf-beta-induced failure of muscle regeneration in multiple myopathic states. Nat Med. 2007;13:204–210. doi: 10.1038/nm1536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Florini J.R., Ewton D.Z., Magri K.A. Hormones, growth factors, and myogenic differentiation. Annu Rev Physiol. 1991;53:201–216. doi: 10.1146/annurev.ph.53.030191.001221. [DOI] [PubMed] [Google Scholar]
- 57.Vaidya T.B., Rhodes S.J., Taparowsky E.J., Konieczny S.F. Fibroblast growth factor and transforming growth factor beta repress transcription of the myogenic regulatory gene myod1. Mol Cell Biol. 1989;9:3576–3579. doi: 10.1128/mcb.9.8.3576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Liu D., Black B.L., Derynck R. Tgf-beta inhibits muscle differentiation through functional repression of myogenic transcription factors by smad3. Genes Dev. 2001;15:2950–2966. doi: 10.1101/gad.925901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Carlson M.E., Hsu M., Conboy I.M. Imbalance between psmad3 and notch induces cdk inhibitors in old muscle stem cells. Nature. 2008;454:528–532. doi: 10.1038/nature07034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Nakamura M., Ito H., Wate R., Nakano S., Hirano A., Kusaka H. Phosphorylated smad2/3 immunoreactivity in sporadic and familial amyotrophic lateral sclerosis and its mouse model. Acta Neuropathol. 2008;115:327–334. doi: 10.1007/s00401-007-0337-z. [DOI] [PubMed] [Google Scholar]
- 61.Zhang J., Ito H., Wate R., Ohnishi S., Nakano S., Kusaka H. Altered distributions of nucleocytoplasmic transport-related proteins in the spinal cord of a mouse model of amyotrophic lateral sclerosis. Acta Neuropathol. 2006;112:673–680. doi: 10.1007/s00401-006-0130-4. [DOI] [PubMed] [Google Scholar]
- 62.Katsuno M., Adachi H., Minamiyama M., Waza M., Doi H., Kondo N. Disrupted transforming growth factor-{beta} signaling in spinal and bulbar muscular atrophy. J Neurosci. 2010;30:5702–5712. doi: 10.1523/JNEUROSCI.0388-10.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Houi K., Kobayashi T., Kato S., Mochio S., Inoue K. Increased plasma tgf-beta1 in patients with amyotrophic lateral sclerosis. Acta Neurol Scand. 2002;106:299–301. doi: 10.1034/j.1600-0404.2002.01301.x. [DOI] [PubMed] [Google Scholar]
- 64.Iida A., Takahashi A., Kubo M., Saito S., Hosono N., Ohnishi Y. A functional variant in znf512b is associated with susceptibility to amyotrophic lateral sclerosis in Japanese. Hum Mol Genet. 2011 doi: 10.1093/hmg/ddr268. [DOI] [PubMed] [Google Scholar]
- 65.Feng Z., Ko C.P. Schwann cells promote synaptogenesis at the neuromuscular junction via transforming growth factor-beta1. J Neurosci. 2008;28:9599–9609. doi: 10.1523/JNEUROSCI.2589-08.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Zhao W., Pan J., Wang X., Wu Y., Bauman W.A., Cardozo C.P. Expression of the muscle atrophy factor muscle atrophy f-box is suppressed by testosterone. Endocrinology. 2008;149:5449–5460. doi: 10.1210/en.2008-0664. [DOI] [PubMed] [Google Scholar]
- 67.Zhao W., Pan J., Zhao Z., Wu Y., Bauman W.A., Cardozo C.P. Testosterone protects against dexamethasone-induced muscle atrophy, protein degradation and mafbx upregulation. J Steroid Biochem Mol Biol. 2008;110:125–129. doi: 10.1016/j.jsbmb.2008.03.024. [DOI] [PubMed] [Google Scholar]
- 68.Singh R., Bhasin S., Braga M., Artaza J.N., Pervin S., Taylor W.E. Regulation of myogenic differentiation by androgens: Cross talk between androgen receptor/beta-catenin and follistatin/transforming growth factor-beta signaling pathways. Endocrinology. 2009;150:1259–1268. doi: 10.1210/en.2008-0858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Cencetti F., Bernacchioni C., Nincheri P., Donati C., Bruni P. Transforming growth factor-beta1 induces transdifferentiation of myoblasts into myofibroblasts via up-regulation of sphingosine kinase-1/s1p3 axis. Mol Biol Cell. 2010;21:1111–1124. doi: 10.1091/mbc.E09-09-0812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Dobrowolny G., Giacinti C., Pelosi L., Nicoletti C., Winn N., Barberi L. Muscle expression of a local igf-1 isoform protects motor neurons in an als mouse model. J Cell Biol. 2005;168:193–199. doi: 10.1083/jcb.200407021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Kaspar B.K., Llado J., Sherkat N., Rothstein J.D., Gage F.H. Retrograde viral delivery of igf-1 prolongs survival in a mouse als model. Science. 2003;301:839–842. doi: 10.1126/science.1086137. [DOI] [PubMed] [Google Scholar]
- 72.Kaspar B.K., Frost L.M., Christian L., Umapathi P., Gage F.H. Synergy of insulin-like growth factor-1 and exercise in amyotrophic lateral sclerosis. Ann Neurol. 2005;57:649–655. doi: 10.1002/ana.20451. [DOI] [PubMed] [Google Scholar]
- 73.Kirkinezos I.G., Hernandez D., Bradley W.G., Moraes C.T. Regular exercise is beneficial to a mouse model of amyotrophic lateral sclerosis. Ann Neurol. 2003;53:804–807. doi: 10.1002/ana.10597. [DOI] [PubMed] [Google Scholar]
- 74.Deforges S., Branchu J., Biondi O., Grondard C., Pariset C., Lecolle S. Motoneuron survival is promoted by specific exercise in a mouse model of amyotrophic lateral sclerosis. J Physiol. 2009;587:3561–3572. doi: 10.1113/jphysiol.2009.169748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Serra C., Bhasin S., Tangherlini F., Barton E.R., Ganno M., Zhang A. The role of gh and igf-i in mediating anabolic effects of testosterone on androgen-responsive muscle. Endocrinology. 2011;152:193–206. doi: 10.1210/en.2010-0802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Vingren J.L., Kraemer W.J., Hatfield D.L., Anderson J.M., Volek J.S., Ratamess N.A. Effect of resistance exercise on muscle steroidogenesis. J Appl Physiol. 2008;105:1754–1760. doi: 10.1152/japplphysiol.91235.2008. [DOI] [PubMed] [Google Scholar]
- 77.Wang Y., Kreisberg J.I., Ghosh P.M. Cross-talk between the androgen receptor and the phosphatidylinositol 3-kinase/akt pathway in prostate cancer. Curr Cancer Drug Targets. 2007;7:591–604. doi: 10.2174/156800907781662248. [DOI] [PubMed] [Google Scholar]
- 78.Chio A., Calvo A., Dossena M., Ghiglione P., Mutani R., Mora G. Als in Italian professional soccer players: the risk is still present and could be soccer-specific. Amyotroph Lateral Scler. 2009;10:205–209. doi: 10.1080/17482960902721634. [DOI] [PubMed] [Google Scholar]