Abstract
Histone chaperones are proteins that shield histones from nonspecific interactions until they are assembled into chromatin. After their synthesis in the cytoplasm, histones are bound by different histone chaperones, subjected to a series of posttranslational modifications and imported into the nucleus. These evolutionary conserved modifications, including acetylation and methylation, can occur in the cytoplasm, but their role in regulating import is not well understood. As part of histone import complexes, histone chaperones may serve to protect the histones during transport, or they may be using histones to promote their own nuclear localization. In addition, there is evidence that histone chaperones can play an active role in the import of histones. Histone chaperones have also been shown to regulate the localization of important chromatin modifying enzymes. This review is focused on the role histone chaperones play in the early biogenesis of histones, the distinct cytoplasmic subcomplexes in which histone chaperones have been found in both yeast and mammalian cells and the importins/karyopherins and nuclear localization signals that mediate the nuclear import of histones. We also address the role that histone chaperone localization plays in human disease. This article is part of a Special Issue entitled: Histone chaperones and Chromatin assembly.
Keywords: Histone chaperones, importin, histone acetylation, Asf1, Nap1, NASP
1. Introduction
Following replication a mixture of new and old histones are deposited onto DNA to form nucleosomes. The new histones are made in the cytoplasm during S phase and are transported into the nucleus. The old histones are disassembled from DNA, presumably shielded and chaperoned until they are reassembled into nucleosomes. Nucleosome assembly and disassembly take place during the other phases of the cell cycle; nucleosomes are dismantled to make way for the transcription and DNA repair machinery and can be reassembled with both existing histones and new histones or variants. Histone chaperones function both inside and outside the nucleus in these processes. This review will focus on the histone chaperones that play important roles in the early biogenesis of histones, interact with histones in the cytoplasm and function in the nuclear import of histones. We will also discuss the import machinery, such as karyopherin proteins important for histone and histone chaperone import. While in the cytoplasm, histones are substrates for specific reversible posttranslational modifications, some of which correlate with their import and assembly into chromatin, and we will consider the function of these modifications. We will highlight histone chaperones that regulate the localization of important chromatin modifying enzymes. In addition, the correlation between mislocalization of histone chaperones and disease will be discussed.
1.1 Histone chaperones
Histones, the major protein components of chromatin, are among the most highly conserved proteins in eukaryotes. In addition, the structures of yeast and human nucleosomes are similar, suggesting the mechanisms that regulate chromatin structure may be highly conserved as well [1, 2]. To form a nucleosome, DNA is wound around a histone octamer, which comprises an H3-H4 heterotetramer flanked on both sides by H2A-H2B heterodimers. The nucleosome is an energetically favorable conformation due to the negative charge of the phosphate groups on DNA and the positively charged basic amino acids of the histone proteins, yet it does not self assemble. Combined in vitro under physiological ionic strength, DNA and histones form disordered aggregates rather than nucleosomes. By definition, histone chaperones are proteins that bind free histones to shield their positive charge to prevent non-specific interactions with the DNA and to guide the processes of chromatin assembly and disassembly. Assembly of a nucleosome is thought to occur in a stepwise manner. An H3-H4 tetramer (or two dimers) is likely to be deposited first because H3-H4 has a greater affinity for DNA than do the H2A-H2B dimers, followed by deposition of the H2A-H2B dimers, which have high affinity for H3-H4 bound to DNA [3-6]. Non-enzymatic histone chaperones, such as nucleosome assembly protein 1 (Nap1) can assemble nucleosome arrays in vitro in the absence of ATP [7]. Then, using the energy of ATP hydrolysis, ATP-dependent remodelers temporarily disrupt DNA-histone binding and reposition the nucleosomes, by sliding or transferring, until they are evenly spaced [8]. In contrast to their activities as histone ‘donors’ during assembly, in chromatin disassembly histone chaperones act as ‘acceptors’ for histones and shield them until they can be reassembled (for reviews, see [9-12]).
Most histone chaperones have a preference for binding to either H2A-H2B, such as Nap1 and nucleoplasmin, or H3-H4, namely Asf1, N1/N2, CAF-1, HIRA, Vps75, SET, RbAp46 and RbAp48 (Table 1, reviewed in [13, 14]). The nuclear histone chaperone FACT has been shown to act with both H2A-H2B and H3-H4, and there has been some evidence for Nap1 functioning as an H3-H4 chaperone as well [15-22]. In addition to preventing nonspecific interactions of histones and acting as donors and acceptors during assembly and disassembly of nucleosomes, histone chaperones are known to have a variety of other functions. Nucleoplasmin and N1/N2 are histone chaperones that have important functions in the storage of H2A-H2B and H3-H4, respectively, in Xenopus oocytes [23-25]. Histone chaperones may be specialized to function in chromatin assembly or disassembly during particular processes including transcription elongation or DNA repair [18, 26]. Some chaperones function in histone transport or in transfer of histones to other chromatin factors; Nap1 assists in the nuclear transport of H2A-H2B, and both Nap1 and Chz1 donate the H2A variant-containing dimer Htz1-H2B to the SWR1 remodeling complex for incorporation into chromatin [27-29]. Finally, Vps75, Asf1, RbAp46 and other histone chaperones are used to regulate the posttranslational modifications of histones (for reviews, see [30-32]). As histone chaperones modulate histone availability and localization, they can play key roles in chromatin regulation. We will focus on histone chaperones that have a role in nuclear transport, shuttle between the nucleus and cytoplasm or otherwise play a role in histone biogenesis.
Table 1.
Key players in histone and histone chaperone import. Listed are the histone chaperones, HATs, chromatin remodelers, importins and heat shock proteins discussed in detail in this review. Some histone chaperones, histones and importins are excluded for brevity.
| Family | Yeast protein | Metazoan counterpart human (h) or xenopus (x) | Function | Histone preference | Notes |
|---|---|---|---|---|---|
| CAF1 complex | Cac1 | p150 | Histone chaperone | H3-H4 | |
| Cac2 | p60 | Histone chaperone | H3-H4 | ||
| Cac3 | p48/RbAp48 | Histone chaperone | H3-H4 | homologous to RbAp46 | |
| HAT-B complex | Hat1 | HAT1 | HAT enzyme | H3-H4 | |
| Hat2 | RbAp46 | Chaperone-like accessory | H3-H4 | homologous to RbAp48 | |
| Hif1 | Histone chaperone | H3-H4 | some homology with NASP | ||
| N1/N2 family | NASP (tNASP, sNASP) (h) | Histone chaperone | H3-H4, H1 | some homology with Hif1 | |
| N1/N2 (x) | Histone chaperone | H3-H4 | |||
| Nap1 family | Nap1 | NAP1, NAP1L proteins | Histone chaperone | H2A/Z-H2B, H1 | |
| Vps75 | SET | Histone chaperone | H3-H4 | ||
| TSPY, TSPYL proteins | Histone chaperone | ||||
| NPM family | NPM1 (nucleophosmin | Histone) chaperone | H3-H4, H1 | ||
| Nucleoplasmin (NPM2) | Histone chaperone | H2A-H2B, H1 | |||
| Nucleolin | Histone chaperone | H2A-H2B | |||
| Asf1 | ASF1a, ASF1b | Histone chaperone | H3-H4 | ||
| Rtt109 | p300/CBP | HAT enzyme | H3-H4 | structural homologs only | |
| Chz1 | Histone chaperone | Htz1-H2B | HIRIP3 has Chz domain | ||
| Rtt106 | Histone chaperone | H3-H4 | some homology with Daxx | ||
| DAXX | Histone chaperone | H3.3-H4 | some homology with Rtt106 | ||
| ATRX | Chromatin remondeler | ||||
| HSP90 | Heat shock protein, ATPase | interacts with H3-H4 in cytoplasm | |||
| HSC70 | Heat shock protein | interacts with H3-H4 in cytoplasm | |||
| Karyopherin family | Kap60 | Impα | Importin | imports Chz1, Vps75, NPM2 | |
| Kap95 | Impβ | Importin | imports Chz1, Vps75, NPM2 | ||
| Kap114 Kap123 Kap121 | Imp9 Imp4 Imp5 | Importin Importin Importin | H2A/Z-H2B H3-H4 H2A-H2B, H3-H4 | Imports Nap1 |
1.2 Early Posttranslational modifications of histones
Chromatin structure presents a formidable barrier to DNA-templated processes such as transcription, replication and repair, and thus assembly and disassembly are continuously occurring within the cell. Other chromatin remodeling occurs to allow the exchange of core histones and histone variants, such as H2A.Z and H3.3, or to allow transcription factors or other proteins to bind DNA. Yet another dynamic aspect of chromatin structure is the pattern of posttranslational modifications on the histones, particularly their histone amino (N) terminal tails (reviewed in [33, 34]). Reversible covalent modifications such as acetylation, phosphorylation, methylation (mono, di and trimethylation), sumoylation and ubiquitylation can be rapidly added and removed during a single round of transcription, in different phases of the cell cycle or as environmental conditions change. Alternatively, they can be longer lasting, heritable epigenetic changes. Some of these marks have a specific biological outcome. For example, in metazoa, trimethylation of H3 lysine 9 (K9) leads to the recruitment of heterochromatin protein 1 (HP1) and other proteins, ultimately promoting gene silencing [35]. There are many possible modifications that can occur on each histone in a variety of combinations, and the functional significance of many of these modifications is not known. In fact, even H3 K9 methylation can be found outside of heterochromatin [35]. For these reasons, it has been suggested that often it is not just an individual modification that has an effect, but that a group of modifications can act combinatorially in a histone code [36, 37].
Histones H3 and H4 can be acetylated soon after translation, primarily on the N terminal tails, which also contain the nuclear localization signal (NLS). These marks are observed on newly synthesized histones prior to their assembly into chromatin [12, 38, 39]. Of these early marks, acetylation of H4 K5 and K12 and H3 K56 are evolutionarily conserved, raising the possibility that they play a role in the import or assembly processes [40-42]. In yeast, acetylation of K56 was demonstrated to promote H3 association with histone chaperones/chromatin assembly factors [43]. The majority of cytoplasmic H4 in human cells is thought to be diacetylated at K5 and K12 by the HAT-B enzyme complex [42, 44]. Early modifications observed on H3 are heterogeneous and the primary sites of acetylation on newly synthesized H3 vary between species [42]. In budding yeast, the primary sites of acetylation on H3 are K9, K14, K27 and K56 [45, 46]. In human cells, H3 can be acetylated on K14, K18 and K56 [44, 47]. H3 K56 acetylation (Ac) appears to be a rare event in human cells but is almost ubiquitous in newly synthesized H3 in yeast [44, 46]. This modification is catalyzed by the HAT Rtt109 in yeast and by its structural homolog, p300, in human cells [48-51]. In human cells one of the earliest marks seen on a proportion (∼30%) of soluble H3 is K9 monomethylation, suggesting that these histones may be marked for subsequent H3 K9 trimethylation and assembly into heterochromatin [52]. The methyltransferase implicated in this methylation is SETDB1, which has a significant cytoplasmic pool [52]. Interestingly, a recent paper has also identified poly ADP ribosylation of human H3 and H4 in the cytoplasm, but the significance of this modification is not known [53]. As we discuss below, it is unclear whether these modifications play a role in nuclear transport or signal different maturation steps, allowing interaction with subsequent proteins along the assembly pathway. There is also some debate as to whether these modifications occur on cytoplasmic histones before import, rather than a soluble nuclear pre-deposition fraction, because most of the modifying enzymes are found in both the nucleus and the cytoplasm [54].
1.3 Nuclear transport in eukaryotes
Histones and other proteins destined for the nucleus must pass through the nuclear pore complex (NPC). Unlike small molecules and ions, most macromolecules, including proteins and RNA, cannot diffuse through the NPC but must be actively transported by karyopherins (also called Kaps) (Table 1). These are members of the karyopherin β family of evolutionary conserved soluble transport factors that number ∼20 in humans and 14 in budding yeast (reviewed in [55, 56]). Most karyopherins are specialized for import (importins) or export (exportins), although karyopherins that act in both import and export have been identified [57, 58]. Karyopherin β family members are large (95-145 kDa) proteins composed of HEAT repeats [59]. Each karyopherin has many cargoes; Karyopherins have been shown to bind cargo over extended surfaces of the protein and to contain multiple binding sites for different cargoes [55]. Whether karyopherins transport multiple proteins at once and the conformation of these multimeric karyopherin-cargo complexes are not well understood; however, as large ribosomal subunits can transit the NPC, it is certainly conceivable that a single karyopherin could transport large multimeric complexes. In the cytoplasm, importins recognize and bind their cargoes via a nuclear localization signal (NLS). Classical NLSs can be monopartite or bipartite and are defined by loose consensus sequences: K-(K/R)-X-(K/R) for monopartite classical NLSs, where X is any amino acid, or two regions of basic residues separated by a 10 or more amino acid linker for bipartite [60-62]. Importin β (Kap95 in yeast) uses Importin α (Kap60), an adaptor protein, to bind cargo with a classical NLS. Non-classical nuclear localization signals tend to share less sequence conservation, can be much larger and are bound directly by other importin β family members [55]. Importins often have redundant functions and a single cargo can be transported by several importins, but whether these recognize discreet or shared NLSs is not known. Importins in complex with their cargoes translocate through the nuclear pore complex via transient interactions with nucleoporins. Once inside the nucleus, interaction with RanGTP causes dissociation of the import complex, resulting in release of the cargo. In some cases additional factors are required to stimulate this release [63, 64]. The importin is recycled to the cytoplasm for further rounds of import. Ran is a Ras-like guanosine triphosphatase (GTPase) that is primarily in the GTP bound form in the nucleus as a result of the chromatin bound RanGEF (guanine nucleotide exchange factor) [59]. Export of cargo containing a nuclear export signal (NES) occurs in manner complimentary to import. The most well defined NES is the leucine-rich NES recognized by the exportin Crm1 [65]. In the nucleus, the exportin binds NES-containing cargo and RanGTP to form an export complex [66]. Nuclear transport can be potentially regulated by different mechanisms, the best understood of which are reversible posttranslational modifications of the cargo NLS and mechanisms by which NLSs are reversibly masked or cargoes are tethered in a particular compartment. Other potential mechanisms may include availability of importins due to cargo competition, additional regulation of cargo dissociation and importin recycling.
2. H3 and H4 cytosolic histone chaperones
Histones undergo a series of modifications and interactions with different factors before their import into the nucleus and subsequent assembly into nucleosomes. Several studies carried out in both yeast and human tissue culture cells have identified cytoplasmic histone H3-H4 containing complexes [44, 52, 53, 67-71] (FIG 1). We initially identified the abundant H4 cytosolic interacting factors as H3, Hat1, Hat2, Kap123 and Kap121 [71]. In a later study we determined that this fraction also contained the histone chaperones Hif1 and Asf1 [69]. Hat1, Hat2, and Hif1 form the HAT-B holoenzyme (see section 2.1 below and Table 1), which is found in both the cytoplasm and nucleus and is responsible for the early acetylation of H4 K5 and K12 [54, 72-74]. Using recombinant proteins, we showed that Hat1 and Kap123, as well as Asf1 and Kap123, form complexes that are dependent on the presence of histones [69]. A subsequent paper indicated that Asf1 and the HAT-B proteins could be immunoprecipitated in the same complex [75]. These data suggest that Hat1 and Asf1 do not directly interact with Kap123 but interact via the histones. Interaction of HAT-B and Asf1 with H3-H4 is necessary for H4 and H3 acetylation, respectively, but the formation of stable complexes with histones and importins is likely not required for acetylation. It may be that these histone chaperones remain bound to histones in the import complexes to protect the histones from spurious interactions. In addition, they may play a more active role in histone import, similar to that of Nap1 [29]. Consistent with this model, a recent study in the slime mold Physarum polycephalum showed that H3-H4-Hat1 binding was necessary for the accumulation of H3 and H4 in the nucleus [76]. Alternatively the HAT-B complex and Asf1 are predominantly nuclear, and a stable complex with histones and importins would also promote their entry into the nucleus, although the HAT-B complex and Asf1 likely have other independent routes of nuclear import [77, 78].
Figure 1.
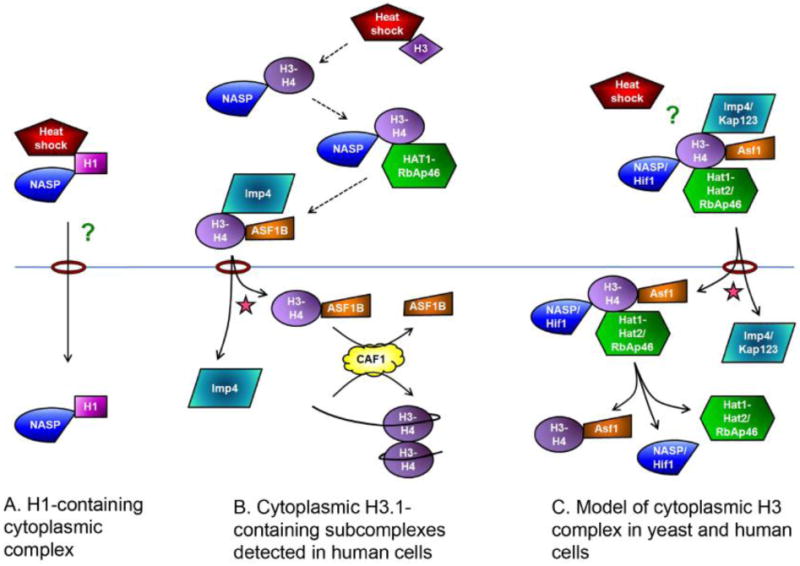
Cytoplasmic complexes of H1 and H3-H4 and their import pathways. Histones, importins and interacting proteins are labeled. The star represents Ran-GTP which promotes dissociation of the import complexes. The rings represent nuclear pore complexes in the nuclear membrane, separating the cytoplasm (upper) from the nucleus (lower). NASP refers to both sNASP and tNASP. A. H1 was found in a complex with NASP and a heat shock protein in the cytoplasm of murine cells. NASP promotes import of H1. Question mark denotes unknown import pathway. B. H3.1 containing subcomplexes detected in human cytosol. The proposed order of complex formation is indicated with dashed arrows based on references (45) and (73). Once inside the nucleus, ASF1B can donate H3.1-H4 for chromatin assembly by the CAF1 complex or other assembly proteins. C. Data from yeast and human cells indicates that H3 interacts with the proteins shown, suggesting the possibility of one large import complex. (Question mark indicates that heat shock proteins were found in human cells but not in yeast).
Several groups have reported the identification of mammalian H3 complexes. Human cells contain several isoforms of H3, the major ones being H3.1 and H3.3. H3.1 is expressed in S phase and is assembled into nucleosomes during replication, while H3.3 is expressed throughout the cell cycle and is incorporated into chromatin independently of replication [79]. H3.3 is believed to play an important role in transcriptional regulation. Although H3.1 and H3.3 are very similar (>90% identity), they localize to different domains of the genome [80]. Several studies have identified the proteins bound to the soluble forms of these histones that may be responsible for their specific modification and targeting. The earlier studies compared the interacting proteins of the H3.1 and H3.3 isoforms isolated from nuclei and found that both isoforms were bound to H4, Asf1a and Asf1b, tNASP and sNASP, Imp4, Hat1 and the CAF1 subunit p48 (also called RbAp48) (Table 1) [81]. H3.1, but not H3.3, interacted with the other CAF1 subunits, p150 and p60, while only H3.3 interacted with the nuclear chaperone Histone regulation A (HIRA). When H3.1 and H3.3 were affinity purified from cytosol, many of the abundant interacting proteins were the same as those found in the nucleus (see section 2.4 below) whereas the nuclear chaperones HIRA and CAF1 were absent [81].
2.1 HAT-B complex
The HAT-B complex protein, Hat1, can acetylate soluble histones and was originally thought to be cytoplasmic (reviewed in this issue [74]). In fact, further analysis indicated that Hat1 is predominantly nuclear but also present in the cytoplasm. Hat1 is conserved from yeast to humans and can acetylate H4 K5 and K12. Hat1 is part of a holoenzyme that includes the histone chaperone-like accessory factor Hat2 (RpAp46 in human cells) and the histone chaperone Hif1. Hat2 is necessary for the integrity of the complex as it bridges the Hat1-Hif1 interaction, and it may also be important for the localization of Hat1 to the nucleus [54, 82]. Human RbAp46 and its yeast homolog, Hat2, bind to and enhance the acetylation activity of Hat1 [78]. Hif1 can interact with H3 and H4 and has chromatin assembly activity in vitro, suggesting it is a histone chaperone [54, 82]. Some studies have detected Hif1 only in the nucleoplasmic fraction while others have observed it in both compartments; in addition Hif1 may have Hat1 independent functions in the nucleus [54, 69, 72, 83]. Hif1 has recently been shown to have modest homology with the abundant human histone chaperone sNASP [13, 72, 84].
The human protein RbAp48, which is 90% identical to RpAp46, is a component of the nuclear chromatin assembly/histone chaperone complex CAF1. Whereas both RpAp46 and RbAp48 participate in other chromatin modifying complexes, this exclusive partitioning between the HAT-B complex and the CAF1 complex is conserved from yeast to humans (reviewed in this issue [12]).
2.2 NASP
The mammalian gene NASP (nuclear autoantigenic sperm protein) encodes two variants, the full length tNASP that is highly expressed in testes and a shorter splice variant, somatic or sNASP [85-87]. Experiments have suggested that NASP can dimerize and form a complex with two H3-H4 dimers [68]. NASP shares homology with the N1/N2 family of chaperones [85]. Although NASP can be knocked down in tissue culture cells, the knockout in mice causes embryonic lethality [88]. NASP was originally identified as a chaperone for the linker histone H1; other H1 chaperones include nucleoplasmin and Nap1 [85, 86, 89-91]. In mouse spermatogenic cells, a cytosolic H1-NASP-Hsp90 complex was identified and proposed to be a pre-import complex. In a HeLa permeabilized cell assay, H1 import was promoted by NASP, but surprisingly no other cytosolic factors were required (FIG 1) [92]. NASP proteins have also been shown to interact with H3-H4 [87]. Studies by Campos et al. suggest that while tNASP may bind to H3-H4 immediately after biogenesis, the more abundant sNASP is likely one of the major histone chaperones for cytoplasmic H3 and H4, although this is likely cell type specific as tNASP is more abundant in some cell types [68]. Consistent with its homology to yeast Hif1, sNASP can be identified in a histone complex that includes Hat1 and RpAp46 [68]. sNASP does not seem to interact directly with Hat1 and RpAp46 as recruitment to the complex was dependent on the presence of histones. Knockdown of NASP led to a reduction in cytosolic histones, consistent with its role as a histone chaperone [68]. This finding is similar to that reported for Nap1 and Htz1-H2B (see section 5 below) [28].
2.3 Asf1
Asf1 is thought to assemble nucleosomes in a replication-independent manner with HIRA and in a replication-coupled manner through interactions with CAF1 [12, 81]. Asf1 was demonstrated to have an important role in buffering excess histones through studies of cells treated with hydroxyurea (HU), which halts the assembly of new nucleosomes during replication [93]. These cells accumulated Asf1 and newly synthesized histones (H3-H4) in the cytoplasm. Other histone chaperones such as RbAp46 and NASP were also found associated with these cytoplasmic Asf1-H3-H4 complexes [44]. It was not clear from these experiments whether the cytosolic Asf1 represented a dedicated cytoplasmic pool or whether Asf1, like Nap1, is a nucleocytoplasmic shuttling protein [93]. In a recent report by Jasenkova et al., Asf1 was isolated from cytosol and shown to be associated with Importin 4, NASP, RbAp46, RbAp48 and Hat1 [44]. In their analysis of posttranslational modifications, the authors observed that most new H4 bound to Asf1 was acetylated on K5 and K12, but only 20-30% of H3 was acetylated on K14 and K18. In addition, H3 K9 monomethylation was detected and the abundance of this mark was also increased after perturbation of replication, further suggesting that this increased cytoplasmic pool is composed of new histones. Lastly, they determined that only about 1% of the new/cytoplasmic H3 was acetylated on K56, in contrast to the ubiquitous K56 acetylation found on new H3 in yeast [14, 44].
Yeast Asf1 has two human homologs, ASF1a and ASF1b. ASF1a and ASF1b are highly conserved in their N terminal and core domains but more divergent in their carboxyl (C) termini [94]. While both proteins can interact with the p60 subunit of CAF1, only Asf1a appears to interact with HIRA. CAF1 p60 (Cac2) shares a conserved domain with HIRA or the homologous yeast protein, Hir1 [94, 95]. The crystal structure indicates that this domain mediates binding of both proteins to Asf1 and most likely explains why binding of CAF1 and HIRA to Asf1 is mutually exclusive [94] (for a review on HIRA, see [96] in this issue).
Asf1 preferentially associates with histones that have H4 K5 and K12 acetylation (in HeLa cytosol 70% of total H4 is diacetylated, whereas 95% of the H4 attached to Asf1 is diacetylated) [97]. However as the crystal structure of Asf1 revealed that Asf1 did not interact with the N terminal tails, it seemed unlikely that acetylation regulated the Asf1-histone interaction [98]. It has been shown that chromatin can be assembled in vitro with histones lacking N terminal tails, which raises the question of what function these early acetylation marks serve. Although CAF1 and Asf1 interact with diacetylated H4, these marks may not be important for their interaction. Instead, they may specify that the histone dimer has achieved a particular maturation step or regulate interaction with another partner [44, 99]. The studies described above did not elucidate whether the HAT-B complex and Asf1 bound H3 and H4 simultaneously. Structural studies indicate that the HAT-B component RbAp46 binds to helix 1 of H4 in the histone fold domain [100]. This interaction changes the conformation of the histone, such that helix 1 is released from the histone fold, likely making the N terminal tail more accessible to enzymes such as accompanying HATs or chromatin-remodeling complexes. Due to its high similarity to RbAp46, RbAp48 probably binds H4 in a similar manner [100]. In contrast, Asf1 interacts primarily with the C terminal helix of H3 but also makes some contacts with H4 [98, 101]. These findings suggest that both chaperones, Asf1 and the RpAp46 subunit of the HAT-B complex, could potentially interact with the same H3-H4 dimer.
2.4 H3 and H4 associate with several cytoplasmic chaperone complexes
Two recent studies have attempted to delineate the step-by-step regulation of newly synthesized histones through identification of cytoplasmic histone complexes (FIG 1) [53, 68]. Both groups purified H3.1 from HeLa cytosol and used immunoprecipitation and chromatography to define the different complexes that histones can form in the cytoplasm [53, 68]. These subcomplexes are likely to be relevant to our understanding of the mechanism by which histones are modified, imported and assembled into chromatin. Campos et al. isolated H3.1, the replication dependent H3 variant, from cytosol, then fractionated coeluting proteins by ion exchange chromatography and identified them [68]. The subcomplexes were subsequently analyzed by gel filtration. The resulting data suggested the existence of four cytoplasmic subcomplexes containing distinct histone modifications. Based on their components and histone posttranslational modifications, the authors have speculated on the order of formation of these complexes: (I) Hsc70 and H3, which is monomethylated on K9, (II) Hsp90, tNASP and H3-H4, in which H3 is monomethylated on K9 and H4 K12 acetylation is low, (III) sNASP, Hat1, RbAp46 and H3-H4, in which H4 K12 acetylation is high and H3 K9 monomethylation is low, and (IV) Imp4, Asf1b and H4. It is interesting that the authors identify particular heat shock proteins. While the specific role of heat shock proteins in these complexes is unknown, heat shock proteins have a general role in assisting protein folding and preventing aggregation, which could be important before stable histone dimers are formed and before more specialized histone chaperones are bound. Historically both Hsp70 and Hsp90 have been implicated in transport. Hsp70 overexpression has been shown to increase the rate of transport in yeast, and Hsp90 has been shown to be important for the recycling of exportins [102, 103]. Both Hsc70 and Hsp90 play important roles in the nucleocytoplasmic trafficking of steroid receptors [104].
These elegant fractionation experiments allow a glimpse of the pathway from H3-H4 synthesis to nuclear import. However, in vivo experiments will be necessary to confirm these results and determine whether the cofractionating proteins are truly in a complex. Surprisingly, the authors conclude that Imp4 is present only in the last complex (IV) with H3-H4 and Asf1, which would appear to contradict earlier studies where it is an abundant component of Hat1 containing complexes [44, 81]. Some Imp4 is evident in their other complexes, even after two purification steps, which leaves open the possibility that H3-H4, Asf1, HAT-B and Imp4/Kap123 could form a large cytoplasmic complex both in yeast and human cells; further experiments are needed to test this directly (FIG 1). Their temporal order is also dependent on histone modifications as they place the Hat1 complex, which is responsible for H4 K5 and K12 Ac, upstream of the Asf1 and importin complex; however, as indicated above these modifications are unlikely to be necessary for binding suggesting that Asf1 and importins could bind before HAT-B. In addition, as HAT-B is also present in the nucleus, some studies have suggested that their acetylation is a later event in the predeposition pathway [54]. Interestingly, they do not identify any cytoplasmic methyltransferase or H3 HATs in their complexes [44, 53, 68]. This may suggest that these interactions are quite transient and again raises the question as to why the cytoplasmic H3-H4-Hat1 complex interaction is so stable. Previous studies have identified a complex of SETDB1, CAF1 and HP1alpha and have shown that this complex monomethylates H3 K9 [52]. The presence of CAF1 suggests subsequent assembly of the monomethylated H3 into heterochromatin, however the presence of CAF1 also suggests this complex is nuclear, which is inconsistent with H3 being monomethylated in the cytoplasm. In a second paper, Alvarez et al. analyze H3.1 and H3.3 cytosolic complexes and observe similar results to those described above [53]. They do observe slight differences in the pattern of H3 cytoplasmic modifications, but the significance of this is not understood. As shown previously, modifications on cytoplasmic histones are heterogeneous, and only a proportion of the histones in cytosol are modified at each site. Whether there is a histone code of different modifications that specify where specific pools of histones will be targeted, e.g. to heterochromatin, or whether modifications are more random and are determined by time spent in the cytoplasm and relative access to modifying enzymes is not known.
3. Nuclear import of histones
The majority of histone import is presumed to take place during S phase, when the cell needs to double its histone content to package DNA after replication. Histones are well below the diffusion limit of the NPC (estimated to be ∼40 kDa) but are actively imported into the nucleus from the cytoplasm [56, 105]. As described above, histones are components of large cytoplasmic complexes, and active transport is likely required to ensure their rapid and efficient concentration in the nucleus during S phase.
The discovery of early modifications (H4 K5 and K12 Ac and H3 K9, K14 and K56 Ac) on histone tails led to much speculation as to their function. It was originally hypothesized that cytoplasmic acetylation may act to promote nuclear import [71]. To address this question, we and others analyzed the nuclear import pathways of histones [29, 69-71, 106-109]. Histones H2A and H2B are imported as dimers, and recent work suggests that H3 and H4 are also imported as dimers, although they can form tetramers. Evidence for the existence of H3-H4 dimers includes the demonstration that endogenous histone H3 does not copurify with epitope-tagged histone H3 from soluble extracts [81]. Furthermore, structural studies of the chaperone Asf1 bound to newly synthesized histones H3-H4 indicate that Asf1 occludes the surface of H3 that is necessary for tetramer formation and therefore can bind only H3-H4 dimers [98, 101].
Each histone amino terminal tail contains an NLS, and it has been demonstrated that the tails in each heterodimer or heterotetramer are redundant [110, 111]. Therefore, deletion of the NLS-containing tail of any single core histone does not prevent its import [69]. In S. cerevisiae we mapped the NLSs to the following residues: H2A 1-46, H2B 21-33, H3 1-28 and H4 1-34 [70, 71]. Other published data suggest that histones may also contain additional targeting sequences in their core domains, and studies in Physarum polycephalum indicate that the H4 tail NLS contains the critical domain responsible for targeting the H3-H4 dimer [76, 112]. The fact that the NLSs are redundant makes analysis of the contribution of individual histone NLSs complicated, necessitating the use of NLS-GFP fusions and mutant strains lacking the N terminus of one of the histones in the pair. Immunoprecipitation experiments with yeast histones purified from cytosolic extracts and in vitro binding assays with recombinant histones indicated that these histones could access several importin-mediated pathways [28, 69-71, 107]. This is borne out by the fact that individual importin deletions often do not result in strong histone localization defects. However biochemical experiments determined that much of the cytosolic H2A-H2B dimer was complexed with Kap114, suggesting it is the major import factor [70]. Kap121, Kap123, Kap95 and Kap104 were also shown to interact with H2A-H2B [70, 107]. Mislocalization of H2A and H2B NLS-GFP reporters was observed in strains with mutations in KAP114, KAP121, KAP123 and KAP95 [70, 107]. The in vitro binding of Kap114, Kap121 and Kap95 to H2A and H2B N terminal tails was abolished in the presence of RanGTP, further suggesting they are the cognate importins [70]. Similar experiments determined that the major import pathways for H3-H4 dimers are mediated by Kap123 and Kap121 [71, 106, 107]. There is genetic evidence that Kap123 is more important for the import of H3 than of H4, and this suggests that although the NLSs in the dimer are redundant they are not necessarily bound to the same repertoire of importins [69].
Studies in human cells have also identified multiple importins that likely mediate histone import. In vitro binding experiments and in vitro assays using permeabilized HeLa cells have identified Impβ, transportin, Imp5, Imp7, and Imp9 as importins for all four core histones [108, 109]. It is worth noting that there is some conservation of the major pathways between yeast and humans; Imp9, the Kap114 homolog, has been shown to play an important role in H2A-H2B import and Imp4, the Kap123 homolog, and Imp5, the Kap121 homolog, have been consistently identified in immunoprecipitation experiments using H3 and H4 [44, 53, 68, 108]. It is interesting that although Kap123 is the most abundant karyopherin and plays a major role in the import of histones and ribosomal proteins, deletion of this protein does not result in a significant growth defect, highlighting the redundancy of import pathways [113].
3.1 Role of posttranslational modifications in histone-importin interactions
Elucidation of histone transport pathways allowed investigation of the importin-histone interaction with regard to the role of posttranslational modifications. We showed that Kap123 directly interacts with H3 and H4 in yeast and that a proportion of the interacting histones were acetylated on H3 K9 and/or K14 and H4 K5 and K12 [71]. This suggested that acetylated histones can be imported into the nucleus. Surprisingly, further studies using acetylation site mutants, lysine to glutamine (Q) to mimic acetylation or to arginine to prevent acetylation, implied that acetylation had a negative effect on import [69]. Acetylation or substitution of the acetylation mimic leads to loss of the positive charge in the NLS. Histone-NLS GFP reporters with K to Q mutations at the relevant amino acids led to cytoplasmic localization of the GFP reporter and decreased interaction with importins as assessed by in vitro binding assays [69]. These data are in agreement with another published report that shows that strains with H4 K5, K8, K12 and K16 mutated to alanine had a lower concentration of H3 and H4 in the nucleus [106]. In a separate study, binding of recombinant importin α–importin β to acetylated and non-acetylated peptides was compared, revealing that importins bound with higher affinity to non-acetylated histone tail peptides than acetylated peptides [114]. Overall, these findings would suggest that acetylation is inhibitory to importin binding, which is inconsistent with the fact that acetylated histones are found in cytosolic importin-histone complexes. However these findings are consistent with the fact that many NLSs contain basic amino acids and are positively charged.
To investigate these findings in vivo, we generated strains in which the relevant acetylation sites were mutated in a background where the other redundant histone tail/NLS was deleted, e.g. H3-NLS mutant with H4ΔNLS [69]. These studies indicated that, as expected, each tail could act as an NLS. In the absence of the H4 tail, mutation of individual H3 acetylation sites (K9, K14 and K18) led to inviability. In the absence of the H3 tail, mutation of H4 K5, K8 and K12 together led to inviability [69, 115]. The requirement for these lysines could reflect a role in transport or a more general function in chromatin. Surprisingly, yeast cells in which H3 K9, K14 and K18 and H4 K5, K8 and K12 were simultaneously mutated to prevent acetylation were viable but slow growing [69]. This suggests that acetylation at these sites is not required for nuclear import, although it is possible that acetylation at additional sites may play a role. Overall there is not much evidence for acetylation promoting nuclear import in yeast, and these findings do not explain why histones are acetylated in the cytoplasm. In contrast to these findings in yeast, studies in Physarum using cellular fractionation showed that H3-H4 with acetylation mimic mutations (H4 K5Q, K12Q) accumulated in the nucleus to a greater degree than wild type H3-H4, whereas proteins with mutations that blocked acetylation at those sites were not observed in the nucleus or cytoplasm, suggesting they were degraded. These findings are consistent with acetylation promoting import, and further studies using approaches such as live cell imaging will clarify this [76]. A recent study using human proteins also indicated that acetylated histone H4 peptides (H42-20 K5, K8, K12, K16 Ac) competed for Imp4 binding better than unacetylated peptide [53]. This report also showed that recombinant H4-NLS-YFP fusion was imported less efficiently in a permeabilized cell assay than a similar fusion with acetylation mimic (K to Q) mutations. In vivo experiments are needed to confirm these findings and determine whether this evolutionary conserved pattern of acetylation plays different roles in yeast and higher eukaryotes. While it makes sense that acetylation may promote import because it is an early histone mark, acetylation would also change the overall charge of the NLS, which could be inhibitory for importin binding. The modifications may be important for promoting complex formation with chaperones and/or assembly factors in the deposition pathway or could confer specificity for different importins. In the nucleus, they may even stimulate dissociation of the histone from the transport receptor. Another possibility is that if some histones are modified and some are not, the modifications may prevent binding of multiple importins to each histone dimer or tetramer. Lastly, although H2A and H2B are modified, further studies are required to determine whether any of the modifications are added to soluble H2A-H2B and whether they affect the transport process.
4. The histone chaperone Nap1 promotes import of H2A-H2B by Kap114
In addition to Kap114, abundant Nap1 copurified with H2A or H2B isolated from yeast cytosol [70]. Further study of these interactions revealed simultaneous binding of both Kap114 and Nap1 to the NLS containing N terminal tails of H2A and H2B [29]. Nap1 and Kap114 also specifically interact, indicating the three components, Kap114, Nap1 and the H2A-H2B dimer, could form a complex in which each component can bind the other two. Nap1 increases the affinity of Kap114 for histones H2A and H2B and actually promotes histone import into the nucleus (FIG 2). Neither Kap114 nor Nap1 is essential for H2A-H2B nuclear localization, but there was a decrease in the ratio of nuclear to cytoplasmic H2A and H2B reporters in nap1Δ yeast [29]. While the presence of a histone chaperone in a histone import complex could simply result from the need for histones to be chaperoned at all times, in this case, Nap1 is also acting as an import cofactor. Some histone chaperones may exist in the cytoplasm only when newly synthesized and go through the import cycle once. Any effect these chaperones might have on histone import would likely be minimal due to the high ratio of histone synthesis to histone chaperone synthesis, particularly during S phase. A yeast cell has an estimated 1 million molecules of H4, whereas histone chaperones, which are produced throughout the cell cycle, number in the thousands [116]. In contrast, Nap1 and many of its homologs in metazoa are nucleocytoplasmic shuttling proteins, and as a result each molecule of Nap1 could assist in the import of many histone dimers [29, 117-119]. Therefore, it is understandable why the loss of the import cofactor Nap1 would significantly decrease import of H2A-H2B.
Figure 2.
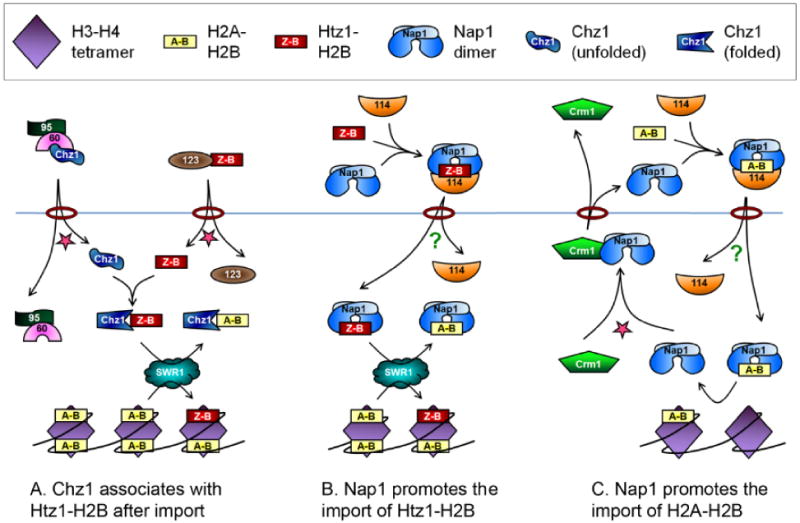
Import of H2A-H2B, Htz1-H2B, Chz1 and Nap1. The exportin Crm1 and the remodeling complex SWR1 are labeled, and the import Kaps are labeled by their numbers (Kap114, Kap123, Kap95 and Kap60) in the model. The histones and chaperones are labeled in the legend (top). The star represents Ran-GTP which either promotes association of the export complex, Crm1-Nap1, or the dissociation of the import complexes, Kap95-Kap60-Chz1 and Kap123-Htz1-H2B. The question marks represent dissociation of Kap114-Nap1-H2A-H2B or Kap114-Nap1-Htz1-H2B complexes, which is facilitated by additional factors. The rings represent nuclear pore complexes in the nuclear membrane, separating the cytoplasm (upper) from the nucleus (lower). A, B. Once inside the nucleus, the chaperones Chz1 or Nap1 can donate Htz1-H2B to the SWR1 complex which incorporates it into chromatin through exchange with H2A-H2B. C. After import, Nap1 is thought to be capable of depositing H2A-H2B dimers in chromatin. Nap1 export is mediated by Crm1, promoting cycles of histone import by Nap1.
The co-import of H2A-H2B with their chaperone, Nap1, has other effects. In the presence of Nap1, H2A-H2B preferentially associates with Kap114 rather than the other importins capable of binding them [29]. Intriguingly, whereas the interaction of the H2A-H2B NLSs with Kap114 is Ran-GTP sensitive, the interaction of Nap1 with Kap114 is resistant to Ran-GTP [29]. Because Nap1 can link Kap114 to H2A-H2B, the import complex with Nap1 is also Ran-GTP insensitive, meaning the histones and Kap114 do not dissociate in the presence of Ran-GTP. The exact conformation of the Nap1-histone-Kap114 complex is not known, however mutants of Kap114 that cannot bind Nap1 can still join a histone-Nap1 complex via histone interaction and in this conformation the complex is sensitive to RanGTP. In this way, the addition of the histone chaperone Nap1 to the Kap114-histone complex completely changes its character. It is currently unknown how this Kap114-H2A-H2B-Nap1 complex dissociates once inside the nucleus but most likely other factors are required. In addition, H2A-H2B can be imported by several importins including Kap123. Surprisingly when Kap123 associates with H2A and H2B, the complex lacks Nap1 and is RanGTP sensitive [29]. Nap1 could compete with Kap123 for H2A and H2B, which points to separate histone-import complexes.
Dynamic localization of Nap1 and Nap1-like proteins throughout the cell cycle has been demonstrated in a variety of metazoa. For example, in Drosophila embryos Nap1 is nuclear during S phase but otherwise cytoplasmic, consistent with its role as a histone import cofactor [7]. Similarly, human Nap1L4 is apparent in the nucleus only during S phase [117, 120]. In yeast, however, Nap1 appears to be predominantly cytoplasmic throughout all phases of the cell cycle [121]. Through mutational studies, we determined that yeast Nap1 shuttles via a leucine rich NES and its Kap114 binding domain [29]. The Kap114 binding domain includes a beta hairpin that points out of the protein and has been referred to as the Nap1 NLS, although NLS activity of the isolated domain has never been experimentally demonstrated [29, 122, 123]. The dynamic localization of Nap1 and some of the Nap1-like proteins in higher eukaryotes may be regulated by posttranslational modifications. In some metazoa, Casein kinase 2 (CK2) phosphorylation has been implicated in the dramatic relocalization of Nap1 or Nap1-like proteins into the nucleus in S phase [120, 124]. TSPY, which is a mammalian Nap1 family member less closely related to yeast Nap1, also shuttles into the nucleus in a manner dependent on phosphorylation of a CK2 target site [118]. In yeast, Nap1 phosphorylation by CK2 likely promotes its nuclear import, which was demonstrated through mutation of CK2 target sites in the context of an export deficient mutant of Nap1 [121]. It is possible phosphorylation changes the conformation of Nap1 and affects karyopherin binding to the NLS in some way.
5. Nap1, but not the Htz1-specific chaperone Chz1, promotes the nuclear import of the H2A variant Htz1
The H2A variant H2A.Z (Htz1 in yeast) is highly conserved throughout eukaryotes and functions in transcription regulation and silencing of heterochromatin [125]. In S. cerevisiae, Htz1 is commonly incorporated in one or two promoter nucleosomes just upstream of a gene's transcription start site. The SWR1 remodeling complex utilizes ATP to exchange H2A-H2B for Htz1-H2B heterodimers within chromatin [126, 127]. In vivo, unincorporated Htz1-H2B dimers are bound to Nap1 or Chz1, and either of these histone chaperones can deliver Htz1-H2B dimers to the SWR1 complex in vitro [27, 127] (FIG 2). Whereas Nap1 binds both H2A-H2B and Htz1-H2B dimers, Chz1 prefers binding Htz1-H2B and recognizes a domain unique to Htz1 [27, 29, 128]. Chz1 is unlike other histone chaperones because it appears to be unfolded until bound to a histone dimer, at which point it forms an extended, irregular fold that is unlike that of the other histone chaperones [10, 27, 129]. At only 17.5 kDa, Chz1 is slightly larger than histones and much smaller than most chaperones. There is no clear Chz1 homolog in higher eukaryotes, although the protein HIRIP3, which interacts with histones and the chaperone HIRA, and other metazoan proteins contain a region homologous to the histone binding domain of Chz1 [27, 81, 130].
When purified from yeast cytosol to identify possible Htz1 nuclear import factors, Htz1 was bound to Nap1 and to four importins, Kap114, Kap123, Kap95 and Kap108, but not to Chz1, suggesting that Nap1 is the predominant Htz1 chaperone in cytosol [28]. The NLS of Htz1 is located in its N terminus, similar to that of canonical H2A, and can bind directly to the importins Kap114, Kap123 and Kap95 in vitro [28]. The major import pathways of Htz1 are analogous to those of the canonical H2A-H2B: Htz1-H2B, Kap114 and Nap1 form a RanGTP insensitive import complex, whereas the Kap123-Htz1-H2B complex is independent of Nap1 and sensitive to RanGTP [28, 29]. Htz1 is also acetylated in the N terminus, but as with H3 and H4, acetylation at these sites is not required for import [28]. Surprisingly, in vitro binding assays indicated that Kap123, but not Kap114, bound preferentially to recombinant Htz1 NLS protein containing acetylation mimic (K to Q) mutations. Thus, Htz1 is imported by at least two pathways: One mediated by Kap114 and assisted by the histone chaperone Nap1 and the other mediated by Kap123 and likely regulated by acetylation. Experiments have shown that Nap1 is also necessary for maintaining a soluble pool of H2A.Z-H2B, and while this may seem counterintuitive, most studies of H2A.Z function suggest that in the nucleus it is constantly being exchanged with chromatin [28, 125]. It is likely that Nap1 is important for maintaining a soluble pool of H2A.Z available for exchange into chromatin, and as Nap1 is constantly shuttling it raises the possibility that it shuttles in and out of the nucleus with H2A.Z-H2B. It is interesting to note that H2A.Z is synthesized throughout the cell cycle and H2B is made predominantly in S phase [125]. As the cytoplasmic pool of H2B is likely small outside S phase, how newly synthesized H2A.Z finds free H2B with which to dimerize is not known.
In support of Chz1 import being independent of Htz1 import, Chz1 was found to contain a classical NLS, residues 36KPKR39, which was verified by mutational analysis [28, 62]. Chz1 is likely imported both by Kap60-Kap95 via this NLS, and also by other redundant importins. This data suggests that the histone chaperone Chz1 is not involved in Htz1 nuclear import and is independently transported into the nucleus.
6. Vps75 import by the classical import pathway promotes nuclear localization of the H3 HAT Rtt109
As Nap1 plays an important role in histone import, it seemed conceivable that the only paralog of Nap1 in budding yeast, Vps75, would also play a role in import. Vps75 is a histone H3-H4 chaperone whose domain and overall structures share more similarity with the human H3-H4 chaperone SET, a member of the human family of Nap1 like proteins, than with yeast Nap1 [131]. Vps75 associates with active genes, mediates histone exchange during transcription and may inhibit H3 replacement at nucleosomes that tend to be rapidly turned over [132, 133]. In vivo, Vps75 binds Rtt109, the sole histone acetyltransferase for H3 lysine 56, which is on the globular domain at the base of the H3 tail [49-51, 134, 135]. Rtt109 and H3 K56 acetylation are critical for robust growth in the presence of DNA damaging agents [134]. Acetylation of lysines 9 and 27 on the H3 tail is mediated by Gcn5 or Rtt109-Vps75 [75, 136-138]. Their acetylation is associated with actively transcribed genes, particularly the 5′ ends [139-141]. Vps75 binds to and stabilizes Rtt109, maintaining a higher concentration of Rtt109 in cells than is found in vps75Δ yeast [75]. Binding to Vps75 is required for Rtt109-mediated acetylation of H3 K9 and K27 but not of H3 K56 [142]. This requirement for Vps75 in acetylation of the H3 tail is likely due to the interaction of Vps75 with histone H3.
Vps75 nuclear import is mediated by Kap60-Kap95, as evidenced by significant mislocalization of Vps75-GFP reporters in a Kap60 mutant strain but not in other importin mutant yeast [142]. The classical NLS of Vps75 (256PSSKKRKV264) was demonstrated to be necessary and sufficient for nuclear localization of Vps75 and an unrelated protein, GFP [142]. Whereas there was no evidence for a significant role of Vps75 in H3 import, it was revealed that Vps75 nuclear localization promotes nuclear localization of the H3 HAT, Rtt109, presumably through nuclear retention facilitated by Vps75 binding or through Vps75 directly assisting in Rtt109 import [142]. If the latter were true, Vps75 and Nap1 would have similar roles as nonessential but nonetheless significant import cofactors for Rtt109 and H2A/H2A.Z-H2B, respectively. It will be interesting to determine if other histone chaperones promote nuclear import of their non-histone associated proteins. Consistent with this idea, Hat1 is in a complex with the accessory protein Hat2 and the chaperone Hif1, both of which are essential for its full function [54, 72].
7. Vertebrate histone chaperones and their association with disease
Considering the critical function of chromatin structure in regulation of gene expression and genome stability, it is not surprising that mounting evidence highlights the connection between cancer or other diseases and proteins that modify, remodel or otherwise regulate chromatin. In fact, the number of chromatin regulators that have been identified as diagnostic or therapeutic targets is continuously growing. Histone deacetylase (HDAC) inhibitors have become increasingly popular targets for cancer treatment (reviewed in [143]). Upregulation of key chromatin proteins has been associated with disease. Striking examples include: the link between a high level of H2A.Z expression, breast cancer progression and low survival (for review, see [144]); upregulation of the chromatin remodeler CAF1 and the histone chaperone Asf1b in breast cancer [145, 146]; and the upregulation of the Nap1 homolog TSPY in various germ cell tumors, which may be useful in diagnosis [147]. We are particularly interested in the correlation between cancer, notably leukemias, and histone chaperones that are mislocalized or overexpressed in one cellular compartment, altering the overall distribution of the protein. We present examples of such histone chaperones below. It has been proposed that it is the role of these histone chaperones in regulating the localization of other proteins that is relevant to disease.
7.1 Nucleophosmin and nucleoplasmin
The first histone chaperone to be discovered was nucleoplasmin [23, 148]. The nucleoplasmin family contains the histone chaperones NPM1 (also referred to as nucleophosmin, B23, numatrin or NO38) and NPM2 (nucleoplasmin) as well as other non-histone chaperone proteins (reviewed in [149]). The nucleocytoplasmic shuttling protein NPM1 binds rRNA in the nucleolus and exports ribosomal proteins to the cytoplasm to facilitate ribosome biogenesis [150]. As a histone chaperone, NPM1 can bind all four canonical histones but prefers to bind H3-H4. In addition to depositing the core histones for chromatin assembly in vitro, it can also deposit the linker histone H1, which was found to associate with NPM1 in vivo. Other histone chaperone functions of NPM1 include regulation of transcription by RNA Polymerases I and II and of chromatin structure within the nucleolus (for review, see [151])
NPM2/nucleoplasmin is a highly specialized histone chaperone present in the nuclei of eggs and oocytes of amphibians and mammals. In Xenopus laevis oocytes, H2A-H2B is bound by nucleoplasmin whereas H3-H4 is bound by another histone chaperone, N1/N2. The histone chaperones nucleoplasmin and N1/N2 are two of the most abundant proteins in Xenopus oocytes. They act as ‘histone sinks’ to maintain a high concentration of histones and cooperate to assemble chromatin during the rapid rounds of division that occur after fertilization. Hyperphosphorylated nucleoplasmin decondenses the sperm chromatin by removing protamines and subsequently inserting maternal H2A-H2B dimers to create the paternal pronucleus (for reviews, see [13, 152]). During cloning, somatic cell nuclei are injected into eggs and undergo a process called reprogramming. This process requires nucleoplasmin, which removes linker histone H1 and can regulate transcription [149].
Nucleoplasmin and NPM1 core subunits are eight-stranded beta barrels that form homopentamers and homodecamers that are capable of binding five separate histone complexes [153, 154]. Nucleoplasmin has a classical bipartite nuclear localization signal (KRX10KKK, where X represents any amino acid) that was among the first such NLSs experimentally verified to promote import via Importins α and β [155-157]. A pentamer of nucleoplasmin has five redundant NLSs and, based on structural studies, it can simultaneously bind five importin α-importin β complexes [158]. In vitro studies suggested that while one NLS is sufficient for nucleoplasmin nuclear accumulation, multiple NLSs increase the rate of nuclear import [159]. NPM1 has two leucine-rich, Crm1-dependent nuclear export sequences (NES), and its C terminal tail contains a nucleolar localization signal (NoLS) and a nucleic acid binding region [160, 161]. Strikingly, upregulation, mutation or translocation of NPM1 is observed in a wide variety of cancers. This may be due to the role of NPM1 in regulation of the tumor suppressors ARF and p53 (reviewed in [162, 163]). NPM1 is primarily localized in the nucleolus because the NLS is stronger than the NESs, but a mutation in NPM1 that creates a new, stronger NES is found in roughly one third of acute myeloid leukemia (AML) patients [164, 165]. This mutation causes aberrant cytoplasmic accumulation of NPM1 in the tumor cells and likely also mislocalization of NPM1 binding partners, which has been proposed to contribute to disease [165, 166]. Both nucleoplasmin and NPM1 are subject to posttranslational modifications that may regulate their localization and function. In the case of nucleoplasmin, which may have as many as 20 phosphate groups on each subunit, phosphorylation by Casein Kinase II likely regulates its nucleocytoplasmic distribution [167]. Phosphorylation of NPM1 targets it to nuclear speckles, affects its movement between the nucleolus and nucleoplasm and may also regulate its nuclear export [151]. In addition, NPM1 acetylation by p300 promotes its localization in the nucleoplasm, its binding of histones and its activation of transcription by RNA Polymerase II [168].
7.2 Nucleolin
Nucleolin, one of the most abundant proteins in the nucleolus, was first described in 1973 and recently characterized as a histone chaperone [169, 170]. Like NPM1, in addition to its nucleolar localization, it can shuttle between the nucleus and cytoplasm. Its functions include ribosome biogenesis, H2A-H2B turnover, and regulation of RNA Polymerase I transcription. Phosphorylation of nucleolin by casein kinase 2 promotes its transcriptional regulation function, whereas phosphorylation and dephosphorylation of p34cdc2 target sites promotes cytoplasmic and nuclear localization, respectively. High expression of nucleolin is associated with high rates of RNA Polymerase I transcription and cellular proliferation. Because it is expressed on the surface of cells and then internalized, nucleolin may promote infection through its interaction with pathogens or cellular transformation by binding to growth factors. Tumor cells or other rapidly dividing cells express the most surface nucleolin, and thus it can be utilized to deliver cargo into tumor cells. Studies have shown that nucleolin is overexpressed in the cytoplasm and on the cell surface of AML blasts. A nucleolin-specific aptamer was found to decrease expression of the anti-apoptotic Bcl2 and is being tested in phase II clinical trials for acute myeloid leukemia treatment.
7.3 DAXX
The interaction of the histone chaperone HIRA with H3.3 and of CAF1 with H3.1, is a relatively late pre-deposition event, as these proteins are not evident in cytosolic extracts suggesting they only associate with H3 in the nucleus. In a new study two additional proteins, DAXX and ATRX, were shown to bind to H3.3 independently of HIRA [80, 171, 172]. DAXX was previously identified as playing a role in apoptosis, while ATRX is a member of the Swi2/Snf2 family of ATP-dependent chromatin remodelers; mutations in ATRX are associated with alpha-thalassemia mental retardation X-linked syndrome [173]. DAXX is found only in higher eukaryotes but shares some homology with the yeast histone chaperone Rtt106 [171]. It appears that while HIRA plays an important role in the deposition of H3.3 at protein encoding genes, DAXX is likely important for deposition at promoters and intergenic regions [80, 171]. ATRX plays a role in targeting DAXX and H3.3. In embryonic stem cells ATRX appears to aid DAXX in the deposition of H3.3 at telomeres, ribosomal DNA and pericentric heterochromatin [80, 171]. A recent screen for mutations that are associated with pancreatic neuroendocrine tumors identified loss of function mutations in both ATRX and DAXX, suggesting that these proteins are tumor suppressors [174].
Although the interaction between DAXX and H3.3 is presumed to be nuclear and quite late in the assembly pathway, it is possible that the histone chaperone DAXX could play a more general role in histone localization. Surprisingly, DAXX has been shown to be a nucleocytoplasmic shuttling protein containing a Crm1-dependent NES sequence [175]. Future studies will determine whether this is related to its function in apoptosis or a role in shuttling histones.
7.4 DEK and SET
Another protein that was recently discovered in Drosophila to be an H3 histone chaperone is the DEK protein [176]. This is the homolog of the human DEK oncogene that is upregulated in several human cancers (for review, see [177]). In Drosophila, Dek associates with chromatin and acts as a transcriptional co-activator in Drosophila cells and also can assemble chromatin in vitro. Dek exists in a complex with histones, and with casein kinase 2, the kinase that also phosphorylates several other histone chaperones, including SET and Nap1. Dek bound to H3.3 with higher affinity than to H3.1, and its genome-wide localization overlapped with H3.3, consistent with a role for Dek in transcription rather than replication. Human DEK is also a histone chaperone and a DEK-CAN (Nup214) fusion is associated with onset of acute myeloid leukemia in patients [178]. This fusion leads to loss of casein kinase 2 binding and histone chaperone activity by the fused DEK. It is interesting that a similar translocation, SET-CAN, which results in the fusion of a histone chaperone (SET, the Vps75/Nap1 homolog) and CAN, the nucleoporin, results in acute undifferentiated leukemia [179]. The SET-CAN fusion protein is expressed under the SET promoter and contains all of the SET coding region, except the last 6 amino acids, fused to the C terminal two thirds of the Can/Nup214 protein [180]. The protein has a function distinct from that of either protein and is mislocalized to the NPC, suggesting that the SET function in particular is compromised [180]. It is interesting that two histone chaperone-nucleoporin translocations would give rise to cancer and argues that histone chaperones play important roles in the cell for which they must be correctly localized.
8. Discussion and perspectives
In conclusion, cytoplasmic pools of histone chaperones, including Nap1, Asf1 and NASP, interact with newly synthesized histones and play important roles in predeposition complexes. Most of these histone chaperones can be identified in large cytoplasmic complexes with histones, HATs and importins, however new experiments point to the existence of distinct subcomplexes. Histone chaperones may have one or more functions within these histone import complexes. As histones must be protected from nonspecific interactions at all times, it is plausible that some of these histone chaperones associate with newly synthesized histones in the cytoplasm, are coimported as part of their chaperone function and remain bound until the histone is transferred to another assembly factor in the nucleus. Most histone chaperones are predominantly nuclear and, like Asf1, have nuclear import pathways independent of histones. However, these chaperones may also “hitch a ride” into the nucleus by association with histones and importins. Furthermore, histone chaperones can play an active role in histone import by promoting the formation of the histone-importin complex, conferring importin binding specificity and/or altering dissociation requirements in the nucleus, as has been shown for Nap1. Nucleocytoplasmic shuttling has only been demonstrated for NPM1, NASP and Nap1, but it will be interesting to determine whether this is a common feature of all histone chaperones that can function in the nucleus and in the cytoplasm. Although it is an assembly factor, Nap1 is important in the maintenance of a soluble pool of histones and plays a role in preventing the misincorporation of H2A–H2B into chromatin, and whether this is a common function of other chaperones remains to be seen [181]. The roles of Vps75 and the chaperone-like protein Hat2 in nuclear localization of Rtt109 and Hat1, respectively, suggests that histone chaperones can also function in the import of HATs. The possibility that histone chaperones could simultaneously promote import of histones and histone modifying enzymes is particularly intriguing due to the role of early posttranslational modifications of histones.
The precise role of predeposition posttranslational modifications is still unclear. There is evidence to suggest that these early modifications occur in the cytoplasm, but a clear function in the regulation of import has not been shown. Modification of H4 K5 and K12 Ac and H3 K9 monomethylation may be an ongoing process that occurs on soluble histones in both the cytoplasm and nucleus, which would point to a role in dictating the formation of subsequent assembly complexes. In addition, it is possible that these modifications modulate histone binding with specific importins or trigger dissociation of histones from importins once inside the nucleus. The evolutionarily conserved acetylation of H4 at K5 and K12 is currently attributed to Hat1, but it is interesting to note that yeast Hat1 is not essential for viability or for H4 K5 and K12 acetylation, highlighting the redundancy of histone modifying enzymes.
It has been suggested that importins themselves can act as chaperones [182]. Although importins are generally redundant and several have been implicated in importing histones, some have a more significant, evolutionarily conserved role in histone import. The clearest example is Kap123/Imp4, which is important for the import of all histones, particularly H3-H4. In yeast, Kap123 is not essential but is the most abundant of the importins, and therefore it may be uniquely suited to accommodate the wave of histone synthesis in S phase. The recently described complexes of importins, histone dimers, histone chaperones and HATs have the potential to be very large, and each component may have its own NLS and binding sites for multiple proteins. Future studies will reveal the conformation of these complexes andb whether transport of large, multimeric cargoes through the NPC is more or less efficient than that of smaller ones. In vitro assays have shown that the transport of a nucleoplasmin pentamer was more efficient when its multiple NLSs were intact [159]. It may be that despite the disadvantage of larger size, multiple importins contribute to efficient import by outcompeting single importin complexes for NPC binding. Strikingly, at least one histone chaperone confers Ran-GTP insensitivity on a histone-importin complex, suggesting additional factors are required for cargo dissociation. In this way it appears that both acetylation of the NLS and histone chaperone binding could have a direct effect on the ability of the import machinery to function efficiently and so introduce another layer of regulation to histone import. For example, acetylation of the NLS could prevent too many importins binding to a single complex, and the presence of Nap1 could negatively regulate dissociation until specific factors that can stimulate this process were encountered. Future studies will dissect the spatial and temporal regulation of soluble cytoplasmic and nuclear subcomplexes of histone chaperones and histones and determine the function of posttranslational modifications in import. We expect that these studies will further indicate the intimate association between histone chaperones and the transport machinery. These studies are made challenging by the imperfect nature of biochemical fractionation and by the steady state localization of many of these factors, which often does not reveal the dynamic and shuttling pools. In addition, the functional redundancy of these factors hinders genetic analysis. Determining the function and localization of histone chaperones will greatly contribute to our understanding of chromatin biology and also the human diseases associated with mislocalized histone chaperones.
Highlights.
Evolutionarily conserved karyopherins/importins function in the nuclear import of histones and histone chaperones
Cytoplasmic histones are acetylated and monomethylated on their amino terminal tails, which may be relevant for nuclear import or chromatin assembly
Newly synthesized histones form multimeric complexes with different histone chaperones, chromatin modifying enzymes and karyopherins in the cytoplasm
Histone chaperones can function in the nuclear import of histones and chromatin modifying enzymes
Misregulation or mislocalization of human histone chaperones is associated with cancer
Acknowledgments
LFP is supported by research grant R01 GM65385 from the National Institutes of Health.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Luger K, Mader AW, Richmond RK, Sargent DF, Richmond TJ. Crystal structure of the nucleosome core particle at 2.8 A resolution. Nature. 1997;389:251–260. doi: 10.1038/38444. [DOI] [PubMed] [Google Scholar]
- 2.White CL, Suto RK, Luger K. Structure of the yeast nucleosome core particle reveals fundamental changes in internucleosome interactions. EMBO J. 2001;20:5207–5218. doi: 10.1093/emboj/20.18.5207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Nakagawa T, Bulger M, Muramatsu M, Ito T. Multistep chromatin assembly on supercoiled plasmid DNA by nucleosome assembly protein-1 and ATP-utilizing chromatin assembly and remodeling factor. J Biol Chem. 2001;276:27384–27391. doi: 10.1074/jbc.M101331200. [DOI] [PubMed] [Google Scholar]
- 4.Smith S, Stillman B. Stepwise assembly of chromatin during DNA replication in vitro. EMBO J. 1991;10:971–980. doi: 10.1002/j.1460-2075.1991.tb08031.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Annunziato AT. Assembling chromatin: The long and winding road. Biochim Biophys Acta. 2011 doi: 10.1016/j.bbagrm.2011.07.005. This issue. [DOI] [PubMed] [Google Scholar]
- 6.Elsasser SJ, D'Arcy S. Towards a mechanism for histone chaperones. Biochim Biophys Acta. 2011 doi: 10.1016/j.bbagrm.2011.07.007. This issue. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ito T, Bulger M, Kobayashi R, Kadonaga JT. Drosophila NAP-1 is a core histone chaperone that functions in ATP-facilitated assembly of regularly spaced nucleosomal arrays. Mol Cell Biol. 1996;16:3112–3124. doi: 10.1128/mcb.16.6.3112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bowman GD. Mechanisms of ATP-dependent nucleosome sliding. Curr Opin Struct Biol. 2010;20:73–81. doi: 10.1016/j.sbi.2009.12.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Corpet A, Almouzni G. Making copies of chromatin: the challenge of nucleosomal organization and epigenetic information. Trends Cell Biol. 2009;19:29–41. doi: 10.1016/j.tcb.2008.10.002. [DOI] [PubMed] [Google Scholar]
- 10.Das C, Tyler JK, Churchill ME. The histone shuffle: histone chaperones in an energetic dance. Trends Biochem Sci. 2010;35:476–489. doi: 10.1016/j.tibs.2010.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hargreaves DC, Crabtree GR. ATP-dependent chromatin remodeling: genetics, genomics and mechanisms. Cell Res. 2011;21:396–420. doi: 10.1038/cr.2011.32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Li Q, Burgess R, Zhang Z. All roads lead to chromatin: Multiple pathways for histone deposition. Biochim Biophys Acta. 2011 doi: 10.1016/j.bbagrm.2011.06.013. This issue. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Eitoku M, Sato L, Senda T, Horikoshi M. Histone chaperones: 30 years from isolation to elucidation of the mechanisms of nucleosome assembly and disassembly. Cell Mol Life Sci. 2008;65:414–444. doi: 10.1007/s00018-007-7305-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Rocha W, Verreault A. Clothing up DNA for all seasons: Histone chaperones and nucleosome assembly pathways. FEBS Lett. 2008;582:1938–1949. doi: 10.1016/j.febslet.2008.03.006. [DOI] [PubMed] [Google Scholar]
- 15.McBryant SJ, Park YJ, Abernathy SM, Laybourn PJ, Nyborg JK, Luger K. Preferential binding of the histone (H3-H4)2 tetramer by NAP1 is mediated by the amino-terminal histone tails. J Biol Chem. 2003;278:44574–44583. doi: 10.1074/jbc.M305636200. [DOI] [PubMed] [Google Scholar]
- 16.Ishimi Y, Kojima M, Yamada M, Hanaoka F. Binding mode of nucleosome-assembly protein (AP-I) and histones. Eur J Biochem. 1987;162:19–24. doi: 10.1111/j.1432-1033.1987.tb10535.x. [DOI] [PubMed] [Google Scholar]
- 17.Bowman A, Ward R, Wiechens N, Singh V, El-Mkami H, Norman DG, Owen-Hughes T. The histone chaperones Nap1 and Vps75 bind histones H3 and H4 in a tetrameric conformation. Mol Cell. 2011;41:398–408. doi: 10.1016/j.molcel.2011.01.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ransom M, Dennehey BK, Tyler JK. Chaperoning histones during DNA replication and repair. Cell. 2010;140:183–195. doi: 10.1016/j.cell.2010.01.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Stuwe T, Hothorn M, Lejeune E, Rybin V, Bortfeld M, Scheffzek K, Ladurner AG. The FACT Spt16 “peptidase” domain is a histone H3-H4 binding module. Proc Natl Acad Sci U S A. 2008;105:8884–8889. doi: 10.1073/pnas.0712293105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Belotserkovskaya R, Oh S, Bondarenko VA, Orphanides G, Studitsky VM, Reinberg D. FACT facilitates transcription-dependent nucleosome alteration. Science. 2003;301:1090–1093. doi: 10.1126/science.1085703. [DOI] [PubMed] [Google Scholar]
- 21.VanDemark AP, Xin H, McCullough L, Rawlins R, Bentley S, Heroux A, Stillman DJ, Hill CP, Formosa T. Structural and functional analysis of the Spt16p N-terminal domain reveals overlapping roles of yFACT subunits. J Biol Chem. 2008;283:5058–5068. doi: 10.1074/jbc.M708682200. [DOI] [PubMed] [Google Scholar]
- 22.Formosa T. The role of FACT in making and breaking nucleosomes. Biochim Biophys Acta. 2011 This issue. [PubMed] [Google Scholar]
- 23.Laskey RA, Honda BM, Mills AD, Finch JT. Nucleosomes are assembled by an acidic protein which binds histones and transfers them to DNA. Nature. 1978;275:416–420. doi: 10.1038/275416a0. [DOI] [PubMed] [Google Scholar]
- 24.Kleinschmidt JA, Fortkamp E, Krohne G, Zentgraf H, Franke WW. Co-existence of two different types of soluble histone complexes in nuclei of Xenopus laevis oocytes. J Biol Chem. 1985;260:1166–1176. [PubMed] [Google Scholar]
- 25.Kleinschmidt JA, Franke WW. Soluble acidic complexes containing histones H3 and H4 in nuclei of Xenopus laevis oocytes. Cell. 1982;29:799–809. doi: 10.1016/0092-8674(82)90442-1. [DOI] [PubMed] [Google Scholar]
- 26.Winkler DD, Luger K. The Histone Chaperone FACT: Structural Insights and Mechanisms for Nucleosome Reorganization. J Biol Chem. 2011;286:18369–18374. doi: 10.1074/jbc.R110.180778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Luk E, Vu ND, Patteson K, Mizuguchi G, Wu WH, Ranjan A, Backus J, Sen S, Lewis M, Bai Y, Wu C. Chz1, a nuclear chaperone for histone H2AZ. Mol Cell. 2007;25:357–368. doi: 10.1016/j.molcel.2006.12.015. [DOI] [PubMed] [Google Scholar]
- 28.Straube K, Blackwell JS, Jr, Pemberton LF. Nap1 and Chz1 have separate Htz1 nuclear import and assembly functions. Traffic. 2010;11:185–197. doi: 10.1111/j.1600-0854.2009.01010.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Mosammaparast N, Ewart CS, Pemberton LF. A role for nucleosome assembly protein 1 in the nuclear transport of histones H2A and H2B. EMBO J. 2002;21:6527–6538. doi: 10.1093/emboj/cdf647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Burgess RJ, Zhang Z. Histones, histone chaperones and nucleosome assembly. Protein Cell. 2010;1:607–612. doi: 10.1007/s13238-010-0086-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Avvakumov N, Nourani A, Cote J. Histone chaperones: modulators of chromatin marks. Mol Cell. 2011;41:502–514. doi: 10.1016/j.molcel.2011.02.013. [DOI] [PubMed] [Google Scholar]
- 32.Hansen JC, Nyborg JK, Luger K, Stargell LA. Histone chaperones, histone acetylation, and the fluidity of the chromogenome. J Cell Physiol. 2010;224:289–299. doi: 10.1002/jcp.22150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lee JS, Smith E, Shilatifard A. The language of histone crosstalk. Cell. 2010;142:682–685. doi: 10.1016/j.cell.2010.08.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Krebs JE. Moving marks: dynamic histone modifications in yeast. Mol Biosyst. 2007;3:590–597. doi: 10.1039/b703923a. [DOI] [PubMed] [Google Scholar]
- 35.Horn PJ, Peterson CL. Heterochromatin assembly: a new twist on an old model. Chromosome Res. 2006;14:83–94. doi: 10.1007/s10577-005-1018-1. [DOI] [PubMed] [Google Scholar]
- 36.Suganuma T, Workman JL. Signals and combinatorial functions of histone modifications. Annu Rev Biochem. 2011;80:473–499. doi: 10.1146/annurev-biochem-061809-175347. [DOI] [PubMed] [Google Scholar]
- 37.Jenuwein T, Allis CD. Translating the histone code. Science. 2001;293:1074–1080. doi: 10.1126/science.1063127. [DOI] [PubMed] [Google Scholar]
- 38.Ruiz-Carrillo A, Wangh LJ, Allfrey VG. Processing of newly synthesized histone molecules. Science. 1975;190:117–128. doi: 10.1126/science.1166303. [DOI] [PubMed] [Google Scholar]
- 39.Jackson V, Shires A, Tanphaichitr N, Chalkley R. Modifications to histones immediately after synthesis. J Mol Biol. 1976;104:471–483. doi: 10.1016/0022-2836(76)90282-5. [DOI] [PubMed] [Google Scholar]
- 40.Yuan J, Pu M, Zhang Z, Lou Z. Histone H3-K56 acetylation is important for genomic stability in mammals. Cell Cycle. 2009;8:1747–1753. doi: 10.4161/cc.8.11.8620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Xu F, Zhang K, Grunstein M. Acetylation in histone H3 globular domain regulates gene expression in yeast. Cell. 2005;121:375–385. doi: 10.1016/j.cell.2005.03.011. [DOI] [PubMed] [Google Scholar]
- 42.Sobel RE, Cook RG, Perry CA, Annunziato AT, Allis CD. Conservation of deposition-related acetylation sites in newly synthesized histones H3 and H4. Proc Natl Acad Sci U S A. 1995;92:1237–1241. doi: 10.1073/pnas.92.4.1237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Li Q, Zhou H, Wurtele H, Davies B, Horazdovsky B, Verreault A, Zhang Z. Acetylation of histone H3 lysine 56 regulates replication-coupled nucleosome assembly. Cell. 2008;134:244–255. doi: 10.1016/j.cell.2008.06.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Jasencakova Z, Scharf AN, Ask K, Corpet A, Imhof A, Almouzni G, Groth A. Replication stress interferes with histone recycling and predeposition marking of new histones. Mol Cell. 2010;37:736–743. doi: 10.1016/j.molcel.2010.01.033. [DOI] [PubMed] [Google Scholar]
- 45.Kuo MH, Brownell JE, Sobel RE, Ranalli TA, Cook RG, Edmondson DG, Roth SY, Allis CD. Transcription-linked acetylation by Gcn5p of histones H3 and H4 at specific lysines. Nature. 1996;383:269–272. doi: 10.1038/383269a0. [DOI] [PubMed] [Google Scholar]
- 46.Masumoto H, Hawke D, Kobayashi R, Verreault A. A role for cell-cycle-regulated histone H3 lysine 56 acetylation in the DNA damage response. Nature. 2005;436:294–298. doi: 10.1038/nature03714. [DOI] [PubMed] [Google Scholar]
- 47.Benson LJ, Gu Y, Yakovleva T, Tong K, Barrows C, Strack CL, Cook RG, Mizzen CA, Annunziato AT. Modifications of H3 and H4 during chromatin replication, nucleosome assembly, and histone exchange. J Biol Chem. 2006;281:9287–9296. doi: 10.1074/jbc.M512956200. [DOI] [PubMed] [Google Scholar]
- 48.Das C, Lucia MS, Hansen KC, Tyler JK. CBP/p300-mediated acetylation of histone H3 on lysine 56. Nature. 2009;459:113–117. doi: 10.1038/nature07861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Tsubota T, Berndsen CE, Erkmann JA, Smith CL, Yang L, Freitas MA, Denu JM, Kaufman PD. Histone H3-K56 acetylation is catalyzed by histone chaperone-dependent complexes. Mol Cell. 2007;25:703–712. doi: 10.1016/j.molcel.2007.02.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Han J, Zhou H, Horazdovsky B, Zhang K, Xu RM, Zhang Z. Rtt109 acetylates histone H3 lysine 56 and functions in DNA replication. Science. 2007;315:653–655. doi: 10.1126/science.1133234. [DOI] [PubMed] [Google Scholar]
- 51.Driscoll R, Hudson A, Jackson SP. Yeast Rtt109 promotes genome stability by acetylating histone H3 on lysine 56. Science. 2007;315:649–652. doi: 10.1126/science.1135862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Loyola A, Tagami H, Bonaldi T, Roche D, Quivy JP, Imhof A, Nakatani Y, Dent SY, Almouzni G. The HP1alpha-CAF1-SetDB1-containing complex provides H3K9me1 for Suv39-mediated K9me3 in pericentric heterochromatin. EMBO Rep. 2009;10:769–775. doi: 10.1038/embor.2009.90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Alvarez F, Munoz F, Schilcher P, Imhof A, Almouzni G, Loyola A. Sequential establishment of marks on soluble histones H3 and H4. J Biol Chem. 2011;286:17714–17721. doi: 10.1074/jbc.M111.223453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Poveda A, Pamblanco M, Tafrov S, Tordera V, Sternglanz R, Sendra R. Hif1 is a component of yeast histone acetyltransferase B, a complex mainly localized in the nucleus. J Biol Chem. 2004;279:16033–16043. doi: 10.1074/jbc.M314228200. [DOI] [PubMed] [Google Scholar]
- 55.Chook YM, Suel KE. Nuclear import by karyopherin-betas: recognition and inhibition. Biochim Biophys Acta. 2011;1813:1593–1606. doi: 10.1016/j.bbamcr.2010.10.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Fiserova J, Goldberg MW. Nucleocytoplasmic transport in yeast: a few roles for many actors. Biochem Soc Trans. 2010;38:273–277. doi: 10.1042/BST0380273. [DOI] [PubMed] [Google Scholar]
- 57.Mingot JM, Kostka S, Kraft R, Hartmann E, Gorlich D. Importin 13: a novel mediator of nuclear import and export. EMBO J. 2001;20:3685–3694. doi: 10.1093/emboj/20.14.3685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Yoshida K, Blobel G. The karyopherin Kap142p/Msn5p mediates nuclear import and nuclear export of different cargo proteins. J Cell Biol. 2001;152:729–740. doi: 10.1083/jcb.152.4.729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Cook A, Bono F, Jinek M, Conti E. Structural biology of nucleocytoplasmic transport. Annu Rev Biochem. 2007;76:647–671. doi: 10.1146/annurev.biochem.76.052705.161529. [DOI] [PubMed] [Google Scholar]
- 60.Lange A, McLane LM, Mills RE, Devine SE, Corbett AH. Expanding the definition of the classical bipartite nuclear localization signal. Traffic. 2010;11:311–323. doi: 10.1111/j.1600-0854.2009.01028.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Hodel MR, Corbett AH, Hodel AE. Dissection of a nuclear localization signal. J Biol Chem. 2001;276:1317–1325. doi: 10.1074/jbc.M008522200. [DOI] [PubMed] [Google Scholar]
- 62.Lange A, Mills RE, Lange CJ, Stewart M, Devine SE, Corbett AH. Classical nuclear localization signals: definition, function, and interaction with importin alpha. J Biol Chem. 2007;282:5101–5105. doi: 10.1074/jbc.R600026200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Lee DC, Aitchison JD. Kap104p-mediated nuclear import. Nuclear localization signals in mRNA-binding proteins and the role of Ran and Rna. J Biol Chem. 1999;274:29031–29037. doi: 10.1074/jbc.274.41.29031. [DOI] [PubMed] [Google Scholar]
- 64.Pemberton LF, Rosenblum JS, Blobel G. Nuclear import of the TATA-binding protein: mediation by the karyopherin Kap114p and a possible mechanism for intranuclear targeting. J Cell Biol. 1999;145:1407–1417. doi: 10.1083/jcb.145.7.1407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Xu D, Farmer A, Chook YM. Recognition of nuclear targeting signals by Karyopherin-beta proteins. Curr Opin Struct Biol. 2010;20:782–790. doi: 10.1016/j.sbi.2010.09.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Stewart M. Molecular mechanism of the nuclear protein import cycle. Nat Rev Mol Cell Biol. 2007;8:195–208. doi: 10.1038/nrm2114. [DOI] [PubMed] [Google Scholar]
- 67.Loyola A, Almouzni G. Marking histone H3 variants: how, when and why? Trends Biochem Sci. 2007;32:425–433. doi: 10.1016/j.tibs.2007.08.004. [DOI] [PubMed] [Google Scholar]
- 68.Campos EI, Fillingham J, Li G, Zheng H, Voigt P, Kuo WH, Seepany H, Gao Z, Day LA, Greenblatt JF, Reinberg D. The program for processing newly synthesized histones H3.1 and H4. Nat Struct Mol Biol. 2010;17:1343–1351. doi: 10.1038/nsmb.1911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Blackwell JS, Jr, Wilkinson ST, Mosammaparast N, Pemberton LF. Mutational analysis of H3 and H4 N termini reveals distinct roles in nuclear import. J Biol Chem. 2007;282:20142–20150. doi: 10.1074/jbc.M701989200. [DOI] [PubMed] [Google Scholar]
- 70.Mosammaparast N, Jackson KR, Guo Y, Brame CJ, Shabanowitz J, Hunt DF, Pemberton LF. Nuclear import of histone H2A and H2B is mediated by a network of karyopherins. J Cell Biol. 2001;153:251–262. doi: 10.1083/jcb.153.2.251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Mosammaparast N, Guo Y, Shabanowitz J, Hunt DF, Pemberton LF. Pathways mediating the nuclear import of histones H3 and H4 in yeast. J Biol Chem. 2002;277:862–868. doi: 10.1074/jbc.M106845200. [DOI] [PubMed] [Google Scholar]
- 72.Ai X, Parthun MR. The nuclear Hat1p/Hat2p complex: a molecular link between type B histone acetyltransferases and chromatin assembly. Mol Cell. 2004;14:195–205. doi: 10.1016/s1097-2765(04)00184-4. [DOI] [PubMed] [Google Scholar]
- 73.Parthun MR, Widom J, Gottschling DE. The major cytoplasmic histone acetyltransferase in yeast: links to chromatin replication and histone metabolism. Cell. 1996;87:85–94. doi: 10.1016/s0092-8674(00)81325-2. [DOI] [PubMed] [Google Scholar]
- 74.Parthun MR. Histone acetyltransferase 1: More than just an enzyme? Biochim Biophys Acta. 2011 doi: 10.1016/j.bbagrm.2011.07.006. This issue. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Fillingham J, Recht J, Silva AC, Suter B, Emili A, Stagljar I, Krogan NJ, Allis CD, Keogh MC, Greenblatt JF. Chaperone control of the activity and specificity of the histone H3 acetyltransferase Rtt109. Mol Cell Biol. 2008;28:4342–4353. doi: 10.1128/MCB.00182-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Ejlassi-Lassallette A, Mocquard E, Arnaud MC, Thiriet C. H4 replication-dependent diacetylation and Hat1 promote S-phase chromatin assembly in vivo. Molecular biology of the cell. 2011;22:245–255. doi: 10.1091/mbc.E10-07-0633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Ruiz-Garcia AB, Sendra R, Galiana M, Pamblanco M, Perez-Ortin JE, Tordera V. HAT1 and HAT2 proteins are components of a yeast nuclear histone acetyltransferase enzyme specific for free histone H4. J Biol Chem. 1998;273:12599–12605. doi: 10.1074/jbc.273.20.12599. [DOI] [PubMed] [Google Scholar]
- 78.Verreault A, Kaufman PD, Kobayashi R, Stillman B. Nucleosomal DNA regulates the core-histone-binding subunit of the human Hat1 acetyltransferase. Curr Biol. 1998;8:96–108. doi: 10.1016/s0960-9822(98)70040-5. [DOI] [PubMed] [Google Scholar]
- 79.Szenker E, Ray-Gallet D, Almouzni G. The double face of the histone variant H3.3. Cell Res. 2011;21:421–434. doi: 10.1038/cr.2011.14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Goldberg AD, Banaszynski LA, Noh KM, Lewis PW, Elsaesser SJ, Stadler S, Dewell S, Law M, Guo X, Li X, Wen D, Chapgier A, DeKelver RC, Miller JC, Lee YL, Boydston EA, Holmes MC, Gregory PD, Greally JM, Rafii S, Yang C, Scambler PJ, Garrick D, Gibbons RJ, Higgs DR, Cristea IM, Urnov FD, Zheng D, Allis CD. Distinct factors control histone variant H3.3 localization at specific genomic regions. Cell. 2010;140:678–691. doi: 10.1016/j.cell.2010.01.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Tagami H, Ray-Gallet D, Almouzni G, Nakatani Y. Histone H3.1 and H3.3 complexes mediate nucleosome assembly pathways dependent or independent of DNA synthesis. Cell. 2004;116:51–61. doi: 10.1016/s0092-8674(03)01064-x. [DOI] [PubMed] [Google Scholar]
- 82.Parthun MR. Hat1: the emerging cellular roles of a type B histone acetyltransferase. Oncogene. 2007;26:5319–5328. doi: 10.1038/sj.onc.1210602. [DOI] [PubMed] [Google Scholar]
- 83.Ge Z, Wang H, Parthun MR. Nuclear Hat1p complex (NuB4) components participate in DNA repair-linked chromatin reassembly. J Biol Chem. 2011;286:16790–16799. doi: 10.1074/jbc.M110.216846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Dunleavy EM, Pidoux AL, Monet M, Bonilla C, Richardson W, Hamilton GL, Ekwall K, McLaughlin PJ, Allshire RC. A NASP (N1/N2)-related protein, Sim3, binds CENP-A and is required for its deposition at fission yeast centromeres. Mol Cell. 2007;28:1029–1044. doi: 10.1016/j.molcel.2007.10.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Richardson RT, Batova IN, Widgren EE, Zheng LX, Whitfield M, Marzluff WF, O'Rand MG. Characterization of the histone H1-binding protein, NASP, as a cell cycle-regulated somatic protein. J Biol Chem. 2000;275:30378–30386. doi: 10.1074/jbc.M003781200. [DOI] [PubMed] [Google Scholar]
- 86.Finn RM, Browne K, Hodgson KC, Ausio J. sNASP, a histone H1-specific eukaryotic chaperone dimer that facilitates chromatin assembly. Biophys J. 2008;95:1314–1325. doi: 10.1529/biophysj.108.130021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Wang H, Walsh ST, Parthun MR. Expanded binding specificity of the human histone chaperone NASP. Nucleic Acids Res. 2008;36:5763–5772. doi: 10.1093/nar/gkn574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Richardson RT, Alekseev OM, Grossman G, Widgren EE, Thresher R, Wagner EJ, Sullivan KD, Marzluff WF, O'Rand MG. Nuclear autoantigenic sperm protein (NASP), a linker histone chaperone that is required for cell proliferation. J Biol Chem. 2006;281:21526–21534. doi: 10.1074/jbc.M603816200. [DOI] [PubMed] [Google Scholar]
- 89.Dimitrov S, Wolffe AP. Remodeling somatic nuclei in Xenopus laevis egg extracts: molecular mechanisms for the selective release of histones H1 and H1(0) from chromatin and the acquisition of transcriptional competence. EMBO J. 1996;15:5897–5906. [PMC free article] [PubMed] [Google Scholar]
- 90.Saeki H, Ohsumi K, Aihara H, Ito T, Hirose S, Ura K, Kaneda Y. Linker histone variants control chromatin dynamics during early embryogenesis. Proc Natl Acad Sci U S A. 2005;102:5697–5702. doi: 10.1073/pnas.0409824102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Shintomi K, Iwabuchi M, Saeki H, Ura K, Kishimoto T, Ohsumi K. Nucleosome assembly protein-1 is a linker histone chaperone in Xenopus eggs. Proc Natl Acad Sci U S A. 2005;102:8210–8215. doi: 10.1073/pnas.0500822102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Alekseev OM, Widgren EE, Richardson RT, O'Rand MG. Association of NASP with HSP90 in mouse spermatogenic cells: stimulation of ATPase activity and transport of linker histones into nuclei. J Biol Chem. 2005;280:2904–2911. doi: 10.1074/jbc.M410397200. [DOI] [PubMed] [Google Scholar]
- 93.Groth A, Ray-Gallet D, Quivy JP, Lukas J, Bartek J, Almouzni G. Human Asf1 regulates the flow of S phase histones during replicational stress. Mol Cell. 2005;17:301–311. doi: 10.1016/j.molcel.2004.12.018. [DOI] [PubMed] [Google Scholar]
- 94.Tang Y, Poustovoitov MV, Zhao K, Garfinkel M, Canutescu A, Dunbrack R, Adams PD, Marmorstein R. Structure of a human ASF1a-HIRA complex and insights into specificity of histone chaperone complex assembly. Nat Struct Mol Biol. 2006;13:921–929. doi: 10.1038/nsmb1147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Green EM, Antczak AJ, Bailey AO, Franco AA, Wu KJ, Yates JR, 3rd, Kaufman PD. Replication-independent histone deposition by the HIR complex and Asf1. Curr Biol. 2005;15:2044–2049. doi: 10.1016/j.cub.2005.10.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Amin AD, Vishnoi N, Prochasson P. A global requirement for the HIR complex in the assembly of chromatin. Biochim Biophys Acta. 2011 doi: 10.1016/j.bbagrm.2011.07.008. This issue. [DOI] [PubMed] [Google Scholar]
- 97.Loyola A, Bonaldi T, Roche D, Imhof A, Almouzni G. PTMs on H3 variants before chromatin assembly potentiate their final epigenetic state. Mol Cell. 2006;24:309–316. doi: 10.1016/j.molcel.2006.08.019. [DOI] [PubMed] [Google Scholar]
- 98.Natsume R, Eitoku M, Akai Y, Sano N, Horikoshi M, Senda T. Structure and function of the histone chaperone CIA/ASF1 complexed with histones H3 and H4. Nature. 2007;446:338–341. doi: 10.1038/nature05613. [DOI] [PubMed] [Google Scholar]
- 99.Tyler JK, Adams CR, Chen SR, Kobayashi R, Kamakaka RT, Kadonaga JT. The RCAF complex mediates chromatin assembly during DNA replication and repair. Nature. 1999;402:555–560. doi: 10.1038/990147. [DOI] [PubMed] [Google Scholar]
- 100.Murzina NV, Pei XY, Zhang W, Sparkes M, Vicente-Garcia J, Pratap JV, McLaughlin SH, Ben-Shahar TR, Verreault A, Luisi BF, Laue ED. Structural basis for the recognition of histone H4 by the histone-chaperone RbAp46. Structure. 2008;16:1077–1085. doi: 10.1016/j.str.2008.05.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.English CM, Adkins MW, Carson JJ, Churchill ME, Tyler JK. Structural basis for the histone chaperone activity of Asf1. Cell. 2006;127:495–508. doi: 10.1016/j.cell.2006.08.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Shulga N, Roberts P, Gu Z, Spitz L, Tabb MM, Nomura M, Goldfarb DS. In vivo nuclear transport kinetics in Saccharomyces cerevisiae: a role for heat shock protein 70 during targeting and translocation. J Cell Biol. 1996;135:329–339. doi: 10.1083/jcb.135.2.329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Kose S, Furuta M, Koike M, Yoneda Y, Imamoto N. The 70-kD heat shock cognate protein (hsc70) facilitates the nuclear export of the import receptors. J Cell Biol. 2005;171:19–25. doi: 10.1083/jcb.200506074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Pratt WB, Silverstein AM, Galigniana MD. A model for the cytoplasmic trafficking of signalling proteins involving the hsp90-binding immunophilins and p50cdc37. Cell Signal. 1999;11:839–851. doi: 10.1016/s0898-6568(99)00064-9. [DOI] [PubMed] [Google Scholar]
- 105.Wente SR, Rout MP. The nuclear pore complex and nuclear transport. Cold Spring Harb Perspect Biol. 2010;2:a000562. doi: 10.1101/cshperspect.a000562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Glowczewski L, Waterborg JH, Berman JG. Yeast chromatin assembly complex 1 protein excludes nonacetylatable forms of histone H4 from chromatin and the nucleus. Mol Cell Biol. 2004;24:10180–10192. doi: 10.1128/MCB.24.23.10180-10192.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Greiner M, Caesar S, Schlenstedt G. The histones H2A/H2B and H3/H4 are imported into the yeast nucleus by different mechanisms. Eur J Cell Biol. 2004;83:511–520. doi: 10.1078/0171-9335-00418. [DOI] [PubMed] [Google Scholar]
- 108.Muhlhausser P, Muller EC, Otto A, Kutay U. Multiple pathways contribute to nuclear import of core histones. EMBO Rep. 2001;2:690–696. doi: 10.1093/embo-reports/kve168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Baake M, Bauerle M, Doenecke D, Albig W. Core histones and linker histones are imported into the nucleus by different pathways. Eur J Cell Biol. 2001;80:669–677. doi: 10.1078/0171-9335-00208. [DOI] [PubMed] [Google Scholar]
- 110.Morgan BA, Mittman BA, Smith MM. The highly conserved N-terminal domains of histones H3 and H4 are required for normal cell cycle progression. Mol Cell Biol. 1991;11:4111–4120. doi: 10.1128/mcb.11.8.4111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Schuster T, Han M, Grunstein M. Yeast histone H2A and H2B amino termini have interchangeable functions. Cell. 1986;45:445–451. doi: 10.1016/0092-8674(86)90330-2. [DOI] [PubMed] [Google Scholar]
- 112.Thiriet C, Hayes JJ. A novel labeling technique reveals a function for histone H2A/H2B dimer tail domains in chromatin assembly in vivo. Genes Dev. 2001;15:2048–2053. doi: 10.1101/gad.910201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Rout MP, Blobel G, Aitchison JD. A distinct nuclear import pathway used by ribosomal proteins. Cell. 1997;89:715–725. doi: 10.1016/s0092-8674(00)80254-8. [DOI] [PubMed] [Google Scholar]
- 114.Johnson-Saliba M, Siddon NA, Clarkson MJ, Tremethick DJ, Jans DA. Distinct importin recognition properties of histones and chromatin assembly factors. FEBS Lett. 2000;467:169–174. doi: 10.1016/s0014-5793(00)01142-x. [DOI] [PubMed] [Google Scholar]
- 115.Ma XJ, Lu Q, Grunstein M. A search for proteins that interact genetically with histone H3 and H4 amino termini uncovers novel regulators of the Swe1 kinase in Saccharomyces cerevisiae. Genes Dev. 1996;10:1327–1340. doi: 10.1101/gad.10.11.1327. [DOI] [PubMed] [Google Scholar]
- 116.Ghaemmaghami S, Huh WK, Bower K, Howson RW, Belle A, Dephoure N, O'Shea EK, Weissman JS. Global analysis of protein expression in yeast. Nature. 2003;425:737–741. doi: 10.1038/nature02046. [DOI] [PubMed] [Google Scholar]
- 117.Rodriguez P, Munroe D, Prawitt D, Chu LL, Bric E, Kim J, Reid LH, Davies C, Nakagama H, Loebbert R, Winterpacht A, Petruzzi MJ, Higgins MJ, Nowak N, Evans G, Shows T, Weissman BE, Zabel B, Housman DE, Pelletier J. Functional characterization of human nucleosome assembly protein-2 (NAP1L4) suggests a role as a histone chaperone. Genomics. 1997;44:253–265. doi: 10.1006/geno.1997.4868. [DOI] [PubMed] [Google Scholar]
- 118.Krick R, Aschrafi A, Hasgun D, Arnemann J. CK2-dependent C-terminal phosphorylation at T300 directs the nuclear transport of TSPY protein. Biochem Biophys Res Commun. 2006;341:343–350. doi: 10.1016/j.bbrc.2005.12.190. [DOI] [PubMed] [Google Scholar]
- 119.Dong A, Liu Z, Zhu Y, Yu F, Li Z, Cao K, Shen WH. Interacting proteins and differences in nuclear transport reveal specific functions for the NAP1 family proteins in plants. Plant Physiol. 2005;138:1446–1456. doi: 10.1104/pp.105.060509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Rodriguez P, Pelletier J, Price GB, Zannis-Hadjopoulos M. NAP-2: histone chaperone function and phosphorylation state through the cell cycle. J Mol Biol. 2000;298:225–238. doi: 10.1006/jmbi.2000.3674. [DOI] [PubMed] [Google Scholar]
- 121.Calvert ME, Keck KM, Ptak C, Shabanowitz J, Hunt DF, Pemberton LF. Phosphorylation by casein kinase 2 regulates Nap1 localization and function. Mol Cell Biol. 2008;28:1313–1325. doi: 10.1128/MCB.01035-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Park YJ, McBryant SJ, Luger K. A beta-hairpin comprising the nuclear localization sequence sustains the self-associated states of nucleosome assembly protein 1. J Mol Biol. 2008;375:1076–1085. doi: 10.1016/j.jmb.2007.11.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Park YJ, Luger K. The structure of nucleosome assembly protein 1. Proc Natl Acad Sci U S A. 2006;103:1248–1253. doi: 10.1073/pnas.0508002103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Li M, Strand D, Krehan A, Pyerin W, Heid H, Neumann B, Mechler BM. Casein kinase 2 binds and phosphorylates the nucleosome assembly protein-1 (NAP1) in Drosophila melanogaster. J Mol Biol. 1999;293:1067–1084. doi: 10.1006/jmbi.1999.3207. [DOI] [PubMed] [Google Scholar]
- 125.Zlatanova J, Thakar A. H2A.Z: view from the top. Structure. 2008;16:166–179. doi: 10.1016/j.str.2007.12.008. [DOI] [PubMed] [Google Scholar]
- 126.Kobor MS, Venkatasubrahmanyam S, Meneghini MD, Gin JW, Jennings JL, Link AJ, Madhani HD, Rine J. A protein complex containing the conserved Swi2/Snf2-related ATPase Swr1p deposits histone variant H2A.Z into euchromatin. PLoS Biol. 2004;2:E131. doi: 10.1371/journal.pbio.0020131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Mizuguchi G, Shen X, Landry J, Wu WH, Sen S, Wu C. ATP-driven exchange of histone H2AZ variant catalyzed by SWR1 chromatin remodeling complex. Science. 2004;303:343–348. doi: 10.1126/science.1090701. [DOI] [PubMed] [Google Scholar]
- 128.Park YJ, Chodaparambil JV, Bao Y, McBryant SJ, Luger K. Nucleosome assembly protein 1 exchanges histone H2A-H2B dimers and assists nucleosome sliding. J Biol Chem. 2005;280:1817–1825. doi: 10.1074/jbc.M411347200. [DOI] [PubMed] [Google Scholar]
- 129.Zhou Z, Feng H, Hansen DF, Kato H, Luk E, Freedberg DI, Kay LE, Wu C, Bai Y. NMR structure of chaperone Chz1 complexed with histones H2A.Z-H2B. Nat Struct Mol Biol. 2008;15:868–869. doi: 10.1038/nsmb.1465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Lorain S, Quivy JP, Monier-Gavelle F, Scamps C, Lecluse Y, Almouzni G, Lipinski M. Core histones and HIRIP3, a novel histone-binding protein, directly interact with WD repeat protein HIRA. Mol Cell Biol. 1998;18:5546–5556. doi: 10.1128/mcb.18.9.5546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Park YJ, Luger K. Structure and function of nucleosome assembly proteins. Biochem Cell Biol. 2006;84:549–558. doi: 10.1139/o06-088. [DOI] [PubMed] [Google Scholar]
- 132.Kaplan T, Liu CL, Erkmann JA, Holik J, Grunstein M, Kaufman PD, Friedman N, Rando OJ. Cell cycle- and chaperone-mediated regulation of H3K56ac incorporation in yeast. PLoS Genet. 2008;4:e1000270. doi: 10.1371/journal.pgen.1000270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Selth LA, Lorch Y, Ocampo-Hafalla MT, Mitter R, Shales M, Krogan NJ, Kornberg RD, Svejstrup JQ. An rtt109-independent role for vps75 in transcription-associated nucleosome dynamics. Mol Cell Biol. 2009;29:4220–4234. doi: 10.1128/MCB.01882-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Schneider J, Bajwa P, Johnson FC, Bhaumik SR, Shilatifard A. Rtt109 is required for proper H3K56 acetylation: a chromatin mark associated with the elongating RNA polymerase II. J Biol Chem. 2006;281:37270–37274. doi: 10.1074/jbc.C600265200. [DOI] [PubMed] [Google Scholar]
- 135.Collins SR, Miller KM, Maas NL, Roguev A, Fillingham J, Chu CS, Schuldiner M, Gebbia M, Recht J, Shales M, Ding H, Xu H, Han J, Ingvarsdottir K, Cheng B, Andrews B, Boone C, Berger SL, Hieter P, Zhang Z, Brown GW, Ingles CJ, Emili A, Allis CD, Toczyski DP, Weissman JS, Greenblatt JF, Krogan NJ. Functional dissection of protein complexes involved in yeast chromosome biology using a genetic interaction map. Nature. 2007;446:806–810. doi: 10.1038/nature05649. [DOI] [PubMed] [Google Scholar]
- 136.Burgess RJ, Zhou H, Han J, Zhang Z. A role for Gcn5 in replication-coupled nucleosome assembly. Mol Cell. 2010;37:469–480. doi: 10.1016/j.molcel.2010.01.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Grant PA, Eberharter A, John S, Cook RG, Turner BM, Workman JL. Expanded lysine acetylation specificity of Gcn5 in native complexes. J Biol Chem. 1999;274:5895–5900. doi: 10.1074/jbc.274.9.5895. [DOI] [PubMed] [Google Scholar]
- 138.Berndsen CE, Tsubota T, Lindner SE, Lee S, Holton JM, Kaufman PD, Keck JL, Denu JM. Molecular functions of the histone acetyltransferase chaperone complex Rtt109-Vps75. Nat Struct Mol Biol. 2008;15:948–956. doi: 10.1038/nsmb.1459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Pokholok DK, Harbison CT, Levine S, Cole M, Hannett NM, Lee TI, Bell GW, Walker K, Rolfe PA, Herbolsheimer E, Zeitlinger J, Lewitter F, Gifford DK, Young RA. Genome-wide map of nucleosome acetylation and methylation in yeast. Cell. 2005;122:517–527. doi: 10.1016/j.cell.2005.06.026. [DOI] [PubMed] [Google Scholar]
- 140.Tie F, Banerjee R, Stratton CA, Prasad-Sinha J, Stepanik V, Zlobin A, Diaz MO, Scacheri PC, Harte PJ. CBP-mediated acetylation of histone H3 lysine 27 antagonizes Drosophila Polycomb silencing. Development. 2009;136:3131–3141. doi: 10.1242/dev.037127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Millar CB, Grunstein M. Genome-wide patterns of histone modifications in yeast. Nat Rev Mol Cell Biol. 2006;7:657–666. doi: 10.1038/nrm1986. [DOI] [PubMed] [Google Scholar]
- 142.Keck KM, Pemberton LF. Interaction with the histone chaperone Vps75 promotes nuclear localization and HAT activity of Rtt109 in vivo. Traffic. 2011;12:826–839. doi: 10.1111/j.1600-0854.2011.01202.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Lane AA, Chabner BA. Histone deacetylase inhibitors in cancer therapy. J Clin Oncol. 2009;27:5459–5468. doi: 10.1200/JCO.2009.22.1291. [DOI] [PubMed] [Google Scholar]
- 144.Rangasamy D. Histone variant H2A.Z can serve as a new target for breast cancer therapy. Curr Med Chem. 2010;17:3155–3161. doi: 10.2174/092986710792231941. [DOI] [PubMed] [Google Scholar]
- 145.Polo SE, Theocharis SE, Grandin L, Gambotti L, Antoni G, Savignoni A, Asselain B, Patsouris E, Almouzni G. Clinical significance and prognostic value of chromatin assembly factor-1 overexpression in human solid tumours. Histopathology. 2010;57:716–724. doi: 10.1111/j.1365-2559.2010.03681.x. [DOI] [PubMed] [Google Scholar]
- 146.Corpet A, De Koning L, Toedling J, Savignoni A, Berger F, Lemaitre C, O'Sullivan RJ, Karlseder J, Barillot E, Asselain B, Sastre-Garau X, Almouzni G. Asf1b, the necessary Asf1 isoform for proliferation, is predictive of outcome in breast cancer. EMBO J. 2011;30:480–493. doi: 10.1038/emboj.2010.335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Lau YF. Gonadoblastoma, testicular and prostate cancers, and the TSPY gene. Am J Hum Genet. 1999;64:921–927. doi: 10.1086/302353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Laskey RA, Earnshaw WC. Nucleosome assembly. Nature. 1980;286:763–767. doi: 10.1038/286763a0. [DOI] [PubMed] [Google Scholar]
- 149.Frehlick LJ, Eirin-Lopez JM, Ausio J. New insights into the nucleophosmin/nucleoplasmin family of nuclear chaperones. Bioessays. 2007;29:49–59. doi: 10.1002/bies.20512. [DOI] [PubMed] [Google Scholar]
- 150.Maggi LB, Jr, Kuchenruether M, Dadey DY, Schwope RM, Grisendi S, Townsend RR, Pandolfi PP, Weber JD. Nucleophosmin serves as a rate-limiting nuclear export chaperone for the Mammalian ribosome. Mol Cell Biol. 2008;28:7050–7065. doi: 10.1128/MCB.01548-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Okuwaki M. The structure and functions of NPM1/Nucleophsmin/B23, a multifunctional nucleolar acidic protein. J Biochem. 2008;143:441–448. doi: 10.1093/jb/mvm222. [DOI] [PubMed] [Google Scholar]
- 152.Prado A, Ramos I, Frehlick LJ, Muga A, Ausio J. Nucleoplasmin: a nuclear chaperone. Biochem Cell Biol. 2004;82:437–445. doi: 10.1139/o04-042. [DOI] [PubMed] [Google Scholar]
- 153.Namboodiri VM, Akey IV, Schmidt-Zachmann MS, Head JF, Akey CW. The structure and function of Xenopus NO38-core, a histone chaperone in the nucleolus. Structure. 2004;12:2149–2160. doi: 10.1016/j.str.2004.09.017. [DOI] [PubMed] [Google Scholar]
- 154.Dutta S, Akey IV, Dingwall C, Hartman KL, Laue T, Nolte RT, Head JF, Akey CW. The crystal structure of nucleoplasmin-core: implications for histone binding and nucleosome assembly. Mol Cell. 2001;8:841–853. doi: 10.1016/s1097-2765(01)00354-9. [DOI] [PubMed] [Google Scholar]
- 155.Robbins J, Dilworth SM, Laskey RA, Dingwall C. Two interdependent basic domains in nucleoplasmin nuclear targeting sequence: identification of a class of bipartite nuclear targeting sequence. Cell. 1991;64:615–623. doi: 10.1016/0092-8674(91)90245-t. [DOI] [PubMed] [Google Scholar]
- 156.Vancurova I, Vancura A, Lou W, Paine PL. A domain distinct from nucleoplasmin's nuclear localization sequence influences its transport. Biochem Biophys Res Commun. 1997;235:19–25. doi: 10.1006/bbrc.1997.6726. [DOI] [PubMed] [Google Scholar]
- 157.Dingwall C, Dilworth SM, Black SJ, Kearsey SE, Cox LS, Laskey RA. Nucleoplasmin cDNA sequence reveals polyglutamic acid tracts and a cluster of sequences homologous to putative nuclear localization signals. EMBO J. 1987;6:69–74. doi: 10.1002/j.1460-2075.1987.tb04720.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Falces J, Arregi I, Konarev PV, Urbaneja MA, Svergun DI, Taneva SG, Banuelos S. Recognition of nucleoplasmin by its nuclear transport receptor importin alpha/beta: insights into a complete import complex. Biochemistry. 2010;49:9756–9769. doi: 10.1021/bi101179g. [DOI] [PubMed] [Google Scholar]
- 159.Dingwall C, Sharnick SV, Laskey RA. A polypeptide domain that specifies migration of nucleoplasmin into the nucleus. Cell. 1982;30:449–458. doi: 10.1016/0092-8674(82)90242-2. [DOI] [PubMed] [Google Scholar]
- 160.Hingorani K, Szebeni A, Olson MO. Mapping the functional domains of nucleolar protein B23. J Biol Chem. 2000;275:24451–24457. doi: 10.1074/jbc.M003278200. [DOI] [PubMed] [Google Scholar]
- 161.Falini B, Bolli N, Shan J, Martelli MP, Liso A, Pucciarini A, Bigerna B, Pasqualucci L, Mannucci R, Rosati R, Gorello P, Diverio D, Roti G, Tiacci E, Cazzaniga G, Biondi A, Schnittger S, Haferlach T, Hiddemann W, Martelli MF, Gu W, Mecucci C, Nicoletti I. Both carboxy-terminus NES motif and mutated tryptophan(s) are crucial for aberrant nuclear export of nucleophosmin leukemic mutants in NPMc+ AML. Blood. 2006;107:4514–4523. doi: 10.1182/blood-2005-11-4745. [DOI] [PubMed] [Google Scholar]
- 162.Grisendi S, Mecucci C, Falini B, Pandolfi PP. Nucleophosmin and cancer. Nat Rev Cancer. 2006;6:493–505. doi: 10.1038/nrc1885. [DOI] [PubMed] [Google Scholar]
- 163.Lindstrom MS. NPM1/B23: A Multifunctional Chaperone in Ribosome Biogenesis and Chromatin Remodeling. Biochem Res Int. 2011;2011:195209. doi: 10.1155/2011/195209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.Bolli N, Nicoletti I, De Marco MF, Bigerna B, Pucciarini A, Mannucci R, Martelli MP, Liso A, Mecucci C, Fabbiano F, Martelli MF, Henderson BR, Falini B. Born to be exported: COOH-terminal nuclear export signals of different strength ensure cytoplasmic accumulation of nucleophosmin leukemic mutants. Cancer Res. 2007;67:6230–6237. doi: 10.1158/0008-5472.CAN-07-0273. [DOI] [PubMed] [Google Scholar]
- 165.Falini B, Mecucci C, Tiacci E, Alcalay M, Rosati R, Pasqualucci L, La Starza R, Diverio D, Colombo E, Santucci A, Bigerna B, Pacini R, Pucciarini A, Liso A, Vignetti M, Fazi P, Meani N, Pettirossi V, Saglio G, Mandelli F, Lo-Coco F, Pelicci PG, Martelli MF. Cytoplasmic nucleophosmin in acute myelogenous leukemia with a normal karyotype. N Engl J Med. 2005;352:254–266. doi: 10.1056/NEJMoa041974. [DOI] [PubMed] [Google Scholar]
- 166.Falini B, Martelli MP, Bolli N, Sportoletti P, Liso A, Tiacci E, Haferlach T. Acute myeloid leukemia with mutated nucleophosmin (NPM1): is it a distinct entity? Blood. 2011;117:1109–1120. doi: 10.1182/blood-2010-08-299990. [DOI] [PubMed] [Google Scholar]
- 167.Vancurova I, Paine TM, Lou W, Paine PL. Nucleoplasmin associates with and is phosphorylated by casein kinase II. J Cell Sci. 1995;108(Pt 2):779–787. doi: 10.1242/jcs.108.2.779. [DOI] [PubMed] [Google Scholar]
- 168.Shandilya J, Swaminathan V, Gadad SS, Choudhari R, Kodaganur GS, Kundu TK. Acetylated NPM1 localizes in the nucleoplasm and regulates transcriptional activation of genes implicated in oral cancer manifestation. Mol Cell Biol. 2009;29:5115–5127. doi: 10.1128/MCB.01969-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169.Angelov D, Bondarenko VA, Almagro S, Menoni H, Mongelard F, Hans F, Mietton F, Studitsky VM, Hamiche A, Dimitrov S, Bouvet P. Nucleolin is a histone chaperone with FACT-like activity and assists remodeling of nucleosomes. EMBO J. 2006;25:1669–1679. doi: 10.1038/sj.emboj.7601046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170.Orrick LR, Olson MO, Busch H. Comparison of nucleolar proteins of normal rat liver and Novikoff hepatoma ascites cells by two-dimensional polyacrylamide gel electrophoresis. Proc Natl Acad Sci U S A. 1973;70:1316–1320. doi: 10.1073/pnas.70.5.1316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171.Drane P, Ouararhni K, Depaux A, Shuaib M, Hamiche A. The death-associated protein DAXX is a novel histone chaperone involved in the replication-independent deposition of H3.3. Genes Dev. 2010;24:1253–1265. doi: 10.1101/gad.566910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172.Campos EI, Reinberg D. New chaps in the histone chaperone arena. Genes Dev. 2010;24:1334–1338. doi: 10.1101/gad.1946810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 173.Xue Y, Gibbons R, Yan Z, Yang D, McDowell TL, Sechi S, Qin J, Zhou S, Higgs D, Wang W. The ATRX syndrome protein forms a chromatin-remodeling complex with Daxx and localizes in promyelocytic leukemia nuclear bodies. Proc Natl Acad Sci U S A. 2003;100:10635–10640. doi: 10.1073/pnas.1937626100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 174.Jiao Y, Shi C, Edil BH, de Wilde RF, Klimstra DS, Maitra A, Schulick RD, Tang LH, Wolfgang CL, Choti MA, Velculescu VE, Diaz LA, Jr, Vogelstein B, Kinzler KW, Hruban RH, Papadopoulos N. DAXX/ATRX, MEN1, and mTOR pathway genes are frequently altered in pancreatic neuroendocrine tumors. Science. 2011;331:1199–1203. doi: 10.1126/science.1200609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 175.Song JJ, Lee YJ. Tryptophan 621 and serine 667 residues of Daxx regulate its nuclear export during glucose deprivation. J Biol Chem. 2004;279:30573–30578. doi: 10.1074/jbc.M404512200. [DOI] [PubMed] [Google Scholar]
- 176.Sawatsubashi S, Murata T, Lim J, Fujiki R, Ito S, Suzuki E, Tanabe M, Zhao Y, Kimura S, Fujiyama S, Ueda T, Umetsu D, Ito T, Takeyama K, Kato S. A histone chaperone, DEK, transcriptionally coactivates a nuclear receptor. Genes Dev. 2010;24:159–170. doi: 10.1101/gad.1857410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 177.Elsasser SJ, Allis CD, Lewis PW. Cancer. New epigenetic drivers of cancers. Science. 2011;331:1145–1146. doi: 10.1126/science.1203280. [DOI] [PubMed] [Google Scholar]
- 178.von Lindern M, Fornerod M, van Baal S, Jaegle M, de Wit T, Buijs A, Grosveld G. The translocation (6;9), associated with a specific subtype of acute myeloid leukemia, results in the fusion of two genes, dek and can, and the expression of a chimeric, leukemia-specific dek-can mRNA. Mol Cell Biol. 1992;12:1687–1697. doi: 10.1128/mcb.12.4.1687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 179.von Lindern M, van Baal S, Wiegant J, Raap A, Hagemeijer A, Grosveld G. Can, a putative oncogene associated with myeloid leukemogenesis, may be activated by fusion of its 3′ half to different genes: characterization of the set gene. Mol Cell Biol. 1992;12:3346–3355. doi: 10.1128/mcb.12.8.3346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 180.Fornerod M, Boer J, van Baal S, Morreau H, Grosveld G. Interaction of cellular proteins with the leukemia specific fusion proteins DEK-CAN and SET-CAN and their normal counterpart, the nucleoporin CAN. Oncogene. 1996;13:1801–1808. [PubMed] [Google Scholar]
- 181.Andrews AJ, Chen X, Zevin A, Stargell LA, Luger K. The histone chaperone Nap1 promotes nucleosome assembly by eliminating nonnucleosomal histone DNA interactions. Mol Cell. 2010;37:834–842. doi: 10.1016/j.molcel.2010.01.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 182.Jakel S, Mingot JM, Schwarzmaier P, Hartmann E, Gorlich D. Importins fulfil a dual function as nuclear import receptors and cytoplasmic chaperones for exposed basic domains. EMBO J. 2002;21:377–386. doi: 10.1093/emboj/21.3.377. [DOI] [PMC free article] [PubMed] [Google Scholar]


