Abstract
Leukocyte telomere length is widely considered a biomarker of human age and in many studies indicative of health or disease. We have obtained quantitative estimates of telomere length from blood leukocytes in a population sample, confirming results of previous studies that telomere length significantly decreases with age. Telomere length was also positively associated with several measures of healthy aging, but this relationship was dependent on age. We screened two genes known to be involved in telomere maintenance for association with the age-related decline in telomere length observed in our population to identify candidate longevity-associated genes. A single-nucleotide polymorphism located in the SIRT1 gene and another in the 3′ flanking region of XRCC6 had significant effects on telomere length. At each bi-allelic locus, the minor variant was associated with longer telomeres, though the mode of inheritance fitting best differed between the two genes. No statistical interaction was detected for telomere length between the SIRT1 and XRCC6 variants or between these polymorphisms and age. The SIRT1 locus was significantly associated with longevity (P < 0.003). The frequency of the minor allele was higher in long-lived cases than in young controls, which coincides with the protective role of the minor variant for telomere length. In contrast, the XRCC6 variant was not associated with longevity. Furthermore, it did not affect the association of SIRT1 with exceptional survival. The association of the same variant of SIRT1 with longevity was near significant (P < 0.07) in a second population. These results suggest a potential role of SIRT1 in linking telomere length and longevity. Given the differences between this gene and XRCC6, they point to the distinct impact that alternate pathways of telomere maintenance may have on aging and exceptional survival.
Keywords: SIRT1, XRCC6, Ku, Telomeres, Aging, Longevity
Introduction
Telomere length shortens with each cell division in most somatic cells, due to the “end-replication problem” (Harley 1991). Telomere shortening also occurs with cellular senescence and aging in vivo, but its status as a biomarker of human aging is not settled (Shawi and Autexier 2008; Mather et al. 2011). Shortened telomeres are also found in cancer cells, and they are associated with high cancer incidence and mortality, although this association may be related to the presence of the disease and may not be a good predictor of cancer risk (Pooley et al. 2010; Willeit et al. 2010; von Zglinicki 2002). Indeed, longer telomeres appear to be a risk factor for development of cancer according to some studies (Arbeev et al. 2011). Telomere length has been associated with many other factors or conditions, such as oxidative damage (von Zglinicki 2002), UV irradiation (Rochette and Brash 2010), unhealthy lifestyle (Mirabello et al. 2009), and physical and mental functions (Grodstein et al. 2008; Cherkas et al. 2008; Ludlow et al. 2008). Among several age-related decrements, cognitive decline has been associated with decreased telomere length (Devore et al. 2011).
Circulating blood leukocytes have been a resource in telomere biology because of their abundance and the relative ease with which they are obtained. Leukocyte telomere length can be used as a reasonable estimator for the relative telomere length in other tissues, but this may not be applicable to all tissues (Friedrich et al. 2000; Thomas et al. 2008; Young 2010). It reflects the length of telomeres in cells throughout the hematopoietic hierarchy (Kimura et al. 2010). Leukocyte telomere length shows a high degree of heterogeneity among individuals at any given age (Iwama et al. 1998), and it has been associated with various physiological and pathological conditions (Aubert and Lansdorp 2008). Telomere length is heritable, with heritability estimates ranging from 44 to 80% (Slagboom et al. 1994; Andrew et al. 2006). Long-lived individuals and their offspring maintain longer telomeres compared with controls (Atzmon et al. 2010). Thus, leukocyte telomere length may serve as an endophenotype useful in developing candidate genes for association with longevity and healthy aging.
Many proteins are involved in telomere maintenance. They include the telomerase complex in proliferating cells and other capping proteins involved in protection of chromosome termini (Cohen et al. 2007; Palm and de Lange 2008). Telomere structure and function can be compromised by excessive telomere shortening in the absence of normal telomerase activity or by loss of one or more of the capping proteins. It can also be corrupted by various forms of DNA damage. Dysfunctional telomeres can also trigger damage responses, including pathways of double-strand break (DSB) repair that can lead to genomic instability (Lamarche et al. 2010). Unlike germ cells and stem cells, terminally differentiated somatic cells lack telomerase activity (Hastie et al. 1990; Qi et al. 2010).
Telomere length can also be maintained through alternative lengthening mechanisms in the absence of telomerase activity, as described in cancer cells (Bryan et al. 1997; Reddel et al. 2001; Reddel and Bryan 2003). These alternative mechanisms include homologous recombination (HR) or non-homologous end joining (NHEJ). In NHEJ, which involves the Ku heterodimer and DNA ligase complex, DNA ends are directly fused without the need for substantial homology (Moore and Haber 1996). Ku consists of Ku70 and Ku80 encoded by XRCC6 and XRCC5, respectively (Critchlow and Jackson 1998; Riha et al. 2006). In normal cells with intact telomeres, Ku promotes genomic stability by repressing intra- or inter-chromatid exchange via the telomeric repeats (Celli et al. 2006). In human somatic cells, induced loss of either Ku subunit results in cell death with substantial telomere loss, indicating that Ku is essential in maintaining normal telomere length (d’Adda di Fagagna et al. 2001; Wang et al. 2009).
The Ku complex interacts with a number of proteins. For example, Ku interacts with hTERT, the catalytic subunit of telomerase reverse transcriptase (Chai et al. 2002). Ku also interacts with SIRT1, as shown by co-immunoprecipitation (Jeong et al. 2007). SIRT1 is recruited to DSBs and is required for efficient DSB repair via HR (Oberdoerffer et al. 2008). Increased expression of SIRT1 in mice results in longer telomeres, an effect dependent on telomerase, and increased HR throughout the genome, including telomeres (Palacios et al. 2010). A catalytically inactive form of SIRT1 undermines deacetylation of Ku70 and its role in DNA repair (Jeong et al. 2007). These findings indicate that SIRT1 and Ku interact protecting from DNA damage and maintaining telomere integrity.
In this study, we have examined the association of two key genes in telomere maintenance pathways with age-related decline in telomere length. This telomere endophenotype was used to identify candidate genes for association with exceptional survival. Unlike XRCC6, SIRT1 was significantly associated with longevity.
Materials and methods
Study subjects
For the telomere analyses, we examined 357 Caucasian (European-origin) subjects who were ≥60 years old. The phenotypic summary characteristics of this sample are shown in Online Resource 1. For the longevity association study, we examined 517 Caucasians consisting of 293 young controls, which were 21–59 years old, and 224 nonagenarian cases, which were 90–103 years old. All of these relatively-healthy, community-dwelling individuals are part of the Louisiana Healthy Aging Study (LHAS) (Jazwinski et al. 2010). Only the Caucasian subjects in LHAS were included to avoid confounding by population admixture and because of sample size considerations. To replicate the results of the longevity association study, 390 Caucasian (European-origin) subjects from the Georgia Centenarian Study (GCS) were examined, including 170 subjects ≥98 years of age and 220 young controls 20–59 years old (Jazwinski et al. 2010). The samples for the longevity association and telomere studies are summarized in Table 1. Subjects provided informed consent according to protocols approved by the institutional review boards.
Table 1.
Demographic characteristics of population samples in longevity and telomere analysesa
| Sample | Caseb |
Controlc |
Additionald |
Total | |||
|---|---|---|---|---|---|---|---|
| Male | Female | Male | Female | Male | Female | ||
| LHASe | 84 | 140 | 100 | 193 | 0 | 0 | 517 |
| (80) | (130) | (0) | (0) | (68) | (79) | 357 | |
| GCSf | 25 | 145 | 84 | 136 | 0 | 0 | 390 |
Number of subjects in telomere analyses are in parentheses
Nonagenarian in LHAS, centenarian in GCS
20–59 years old
60–89 years old
Louisiana Healthy Aging Study
Georgia Centenarian Study
Population stratification
Ethnic affiliation for the LHAS (N = 869) and GCS (N = 650) subjects was inferred genetically. One hundred informative, identical-by-descent Alu insertion polymorphisms were used with 715 reference subjects of known geographic ancestry (Europe, Asia, Africa, India) (Ray et al. 2005) employing the Structure program (Pritchard et al. 2000), as described in detail previously (Jazwinski et al. 2010). Stratification of subjects was according to origin-population assignment probability greater than 0.80. Additionally, the mean frequencies of each of the individual Alu’s were compared in the Caucasian (European-origin) subjects thus stratified after grouping them according to case–control status and tertiles of telomere length in the longevity and telomere association analyses, respectively. No statistically significant differences were detected.
Telomere length determination
Average telomere length in white blood cells was estimated using a quantitative polymerase chain reaction method (q-PCR), as described (Cawthon 2002; Terry et al. 2008; Cawthon 2009) with modifications. Briefly, genomic DNA was extracted from whole blood and quantified using the Picogreen assay (Ahn et al. 1996). To estimate telomere length, two q-PCRs were applied to each sample, a single-copy-gene-specific q-PCR for β-globin and a telomere-specific q-PCR. A telomere/single-copy gene (T/S) ratio was calculated for each sample using the q-PCR results and normalized to the T/S ratio of a reference sample. The normalized T/S ratio was used as an estimate of the relative telomere length.
The telomere reaction mixture consisted of 15 ng genomic DNA, 1× Applied Biosystems, Inc. (ABI) SYBR Green PCR Master Mix, 2.5 mM DTT, 100 nM Tel-1b primer (5′-CGG TTT GTT TGG GTT TGG GTT TGG GTT TGG GTT TGG GTT-3′) and 900 nM Tel-2b primer (5′-GGC TTG CCT TAC CCT TAC CCT TAC CCT TAC CCT TAC CCT-3′). The reaction was processed in an ABI 7300 real-time PCR machine for 1 cycle at 95°C for 10 min, 30 cycles of 95°C, 15 s, and 54°C, 1 min. The β-globin reaction consisted of 15 ng genomic DNA, 1× ABI SYBR Green PCR Master Mix, 300 nM Hbb3 primer (5′-TGT GCT GGC CCA TCA CTT TG-3′) and 700 nM Hbb4 primer (5′-ACC AGC CAC CAC TTT CTG ATA GG-3′). The reaction was processed for 1 cycle at 95°C for 10 min, followed by 40 cycles at 95°C for 15 s, 58°C for 20 s, and 72°C for 28 s. All the reactions were done in triplicate. The relative T/S ratio was calculated by the comparative Ct (2−ΔΔCt) method. To determine the PCR efficiency, a series of two-fold dilutions of the reference DNA was added to every plate, and a standard curve was generated. According to the slopes of the standard curves, PCR amplification efficiencies for the target and reference samples were very similar, ranging from 92 to 108%.
In a given study, the relative T/S ratios correlate well with the mean terminal-restriction-fragment (TRF) lengths determined by Southern blot analysis (Cawthon 2002, 2009; Pavesi et al. 2009). However, there seems to be a large between-study variation in the correlation of the two measures: One unit of T/S corresponds to approximately 6–10 kb in TRF length, depending on the reference sample used and other experimental conditions.
Other phenotypic variables
Collection of data for health deficit variables and construction of the deficit index are essentially as described (Jazwinski et al. 2010). Briefly, the deficit index (DI) was derived as the un-weighted count of the number of deficits divided by the total number of possible deficits (N = 48) in an individual. The self-reported medical history questionnaire was administered to all study participants, and its components were used to construct the deficit index. This was supplemented with items from an activities-of-daily living (ADL) questionnaire. The continuous-scale physical function performance test involves the performance of ten standardized tasks (CS-PFP10) representing activities of daily living including carrying groceries, donning clothes, sweeping the floor, loading a dryer, climbing stairs along with a 6-min timed walk (Cress et al. 1996, 2005). The Yale Physical Activity Survey is to assess physical function and activity levels (Dipietro et al. 1993). We used one of its summary indices, Yale Energy Expenditure (YEE), which sums the time spent for each activity multiplied by an intensity code, expressed as kilocalories per week. Mini-Mental State Examination (MMSE) was used to screen for cognitive impairment (Folstein et al. 1975; Rovner and Folstein 1987). Age of subjects was verified using both documentary evidence and demographic questionnaires. Mortality data were collected using Social Security Death Index search.
Genotyping
The Illumina GoldenGate assay was performed according to the manufacturer’s instructions. SNPs were selected according to the Illumina Assay Design Tool. Following completion of the assay, all the samples were analyzed using Illumina GenomeStudio. Initial clustering was performed with the 10th percentile GenCall score set at 0.4 for both samples and SNPs. Samples with call rates below 90% were removed, and the remaining samples were re-clustered. SNPs not in Hardy–Weinberg proportions or with minor allele frequency lower than 1% were removed.
SNP rs7896005 (A/G; A is ancestral) is located in an intron of SIRT1. It is part of a large linkage disequilibrium (LD) block encompassing the entire gene (Online Resource 2). The A allele of rs7896005 shows a great ethnic variation in frequency: in the HapMap-CEU sample, its frequency is 0.288; whereas in Asian and African samples, it exceeds 0.8. The SNP rs132793 (G/A) is located in the intergenic region between the convergent XRCC6 and NHP2L1 genes on chromosome 22 (Online Resource 3). According to the dbSNP database, the G allele of rs132793 is ancestral with its frequency ranging from about 0.8 in the HapMap-CEU sample to almost unity in the HapMap-YRI. It is located about 3.6 kb downstream from the end of the last exon of XRCC6 and about 6.3 kb downstream from the end of the last exon of NHP2L1. XRCC6 is about 4,300 nucleotides long from the first exon to the last (including introns), and the NHP2L1 is about 1,500 nucleotides long. NHP2L1 is a human ortholog of the S. cerevisiae Nhp2 protein and binds to a stem-loop structure of an RNA component in the spliceosome assembly (Saito et al. 1996; Nottrott et al. 1999).
Pairwise LD comparison
LD blocks for SIRT1 and XRCC6 were examined using Haploview (http://www.broad.mit.edu/mpg/haploview). D′/LOD values are designated by: bright red (D′ = 1, LOD ≥ 2); blue (D′ = 1, LOD < 2); shades of pink/red (D′ < 1, LOD ≥ 2); white (D′ < 1, LOD < 2). The numbers shown inside the boxes are D′ values × 100 (empty boxes represent the D′ value of 1). Haplotype blocks are defined by the confidence bounds on D′. If the one-sided upper 95% confidence bound on D′ lies between 0.7 and 0.98, the SNP pairs involved are considered to be in strong LD (Gabriel et al. 2002).
Statistics
All the statistical analyses were performed in R (version R 2.11.1; R Development Team, 2008). Because of non-normality of the T/S data, either non-parametric tests or parametric tests on natural-log transformed T/S were performed. Agreement with Hardy–Weinberg proportions was tested using the χ2 goodness-of-fit method or an exact method for small sample sizes. A P-value < 0.05 was considered statistically significant. All statistical tests of significance reported in this study are two-sided. The genetic modes of inheritance for each SNP were based on the assumed effects of the minor allele. For example, additivity (add) of rs132793 assumes an increasing effect with the increasing dose of the minor allele A, whereas dominance (dom) and recessiveness (rec) were assumed when GA = AA and GG = GA, respectively. Survival analyses were performed using the Cox proportional hazards regression, with age, sex, and T/S as covariates. Multiple comparison corrections for the initial tests of association of alleles or genotypes with telomere length and longevity are inherent in the respective statistical tests or have been incorporated post hoc as indicated.
Results
Telomere length, age, and health status
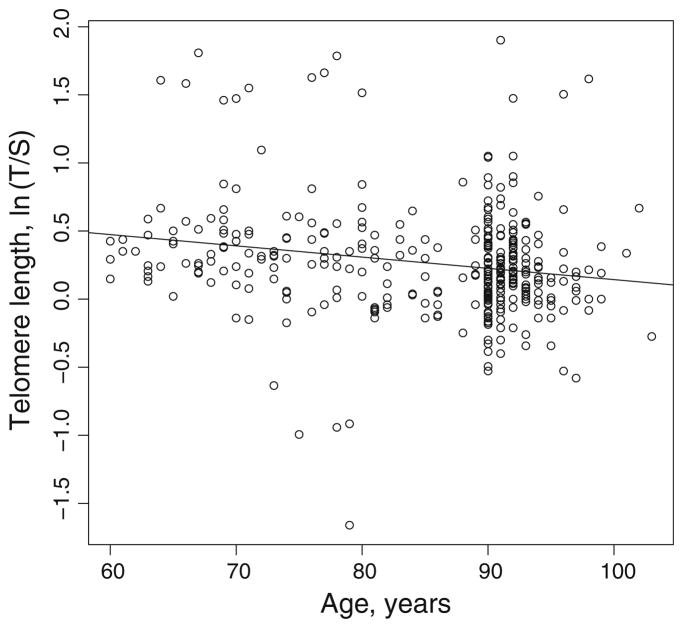
Telomere length was determined in DNA isolated from peripheral blood leukocytes as the T/S ratio, using a q-PCR assay (Cawthon 2002). The analysis was confined to the Caucasian (European-origin) subjects in the LHAS as determined by Structure analysis (0.80 assignment probability), because the sample size was sufficient. Consistent with other published studies, there was a significant negative correlation of T/S with age (ρ = −0.207; P = 8.09 × 10−5; Table 2; Fig. 1). On the other hand, T/S was positively correlated with better health status, represented by the CS-PFP10, the YEE, the MMSE, or collectively by the DI. Of these, CS-PFP10 was most prominent in its significance (ρ = 0.261; P = 7.78 × 10−5). This association of CS-PFP10 with telomere length was near significant (P < 0.07) after adjustment for age; however, the correlations of the remaining measures of healthy aging with longer telomeres did not survive this adjustment (Table 2), indicating that they were related to the decline in health status as a function of age. No significant differences in the mean T/S values were observed between male and female subjects (Online Resource 1). However, males performed significantly better than females in the CS-PFP10 (P = 1.59 × 10−4), which may well reflect the overall gender difference in health status, because males scored significantly better than females on the DI (P = 1.62 × 10−5). According to Cox regression, there was no association of T/S with survival (P ≫ 0.1), but survival was highly dependent on age (P = 9.14 × 10−12).
Table 2.
Spearman correlation of telomere length (T/S) with healthy aging
| Variable | Coefficient (ρ) | P-value | ρa | P-valuea |
|---|---|---|---|---|
| Age | −0.207 | 8.09 × 10−5 | n.a.b | n.a.b |
| CS-PFP10c | 0.261 | 7.78 × 10−5 | 0.125 | 6.12 × 10−2 |
| YEEd | 0.105 | 4.94 × 10−2 | 0.026 | 6.33 × 10−1 |
| MMSEe | 0.159 | 2.93 × 10−3 | 0.044 | 4.15 × 10−1 |
| DIf | −0.092 | 8.23 × 10−2 | −0.012 | 8.20 × 10−1 |
Partial correlation (adjusted for age)
Not applicable
Continuous-scale physical function performance test
Yale energy expenditure
Mini-mental state examination
Deficit index
Fig. 1.
Plot of normal log-transformed telomere length (T/S) versus age with a linear regression slope. Each circle represents an individual subject
SIRT1 and telomere length
We first examined SIRT1 as a potential modulator of the age-telomere length correlation. The SIRT1 SNP rs7896005, located in an intron and part of an extensive LD block in the gene (Online Resource 2) was chosen for analysis of association with telomere length. No significant differences in genotype distribution of the SNP were observed between males and females, and the population sample was in Hardy–Weinberg proportions for variation at this locus (P ≫ 0.1). The mean T/S values varied significantly among the three genotypes at rs7896005: AA (N = 45; 1.69 ± 1.098 [mean ± SD]), AG (N = 167; 1.36 ± 0.724), and GG (N = 145; 1.47 ± 0.876) (P = 0.0134, Kruskal–Wallis test). This difference was especially evident between the minor-allele homozygotes (AA) and the other two genotypes. This pattern of phenotypic variation is best fit by assuming the recessivity of the minor allele. Adjustment for age in a multiple linear regression model showed that the association of SIRT1 with T/S is independent of age (Table 3, model a).
Table 3.
Dependence of telomere length (T/S) on SIRT1 and XRCC6 variantsa
| Explanatory variablesb | Coefficient | SE | P-value | R2 |
|---|---|---|---|---|
| (a) age + SIRT1 rs7986005 (rec) | −8.867 × 10−3 (age) | 2.187 × 10−3 | 6.17 × 10−5 | 0.0571 |
| 1.73 × 10−1 (SIRT1) | 6.553 × 10−2 | 8.52 × 10−3 | ||
| (b) XRCC6 rs132793 (add) | 1.04 × 10−1 | 3.877 × 10−2 | 7.55 × 10−3 | 0.020 |
| (c) age + XRCC6 rs132793 (add) | −8.70 × 10−3 (age) | 2.073 × 10−3 | 3.42 × 10−5 | 0.066 |
| 1.05 × 10−1 (XRCC6) | 3.790 × 10−2 | 5.98 × 10−3 | ||
| (d) age + SIRT1 rs7896005 (rec) | −8.95 × 10−3 (age) | 2.162 × 10−3 | 4.35 × 10−5 | 0.0816 |
| + XRCC6 rs132793 (add) | 1.81 × 10−1 (SIRT1) | 6.482 × 10−2 | 5.50 × 10−3 | |
| 1.17 × 10−1 (XRCC6) | 3.804 × 10−2 | 2.31 × 10−3 |
Linear regression was performed on natural log-transformed T/S
SNPs are coded 0, 1, or 2 according to the inheritance mode of the minor alleles, recessive (rec) for rs7896005 and additive (add) for rs132793 (see Materials and methods)
Association of XRCC6 with telomere length
We next assessed the potential impact of XRCC6 on the age-telomere length correlation. Ku70, encoded by XRCC6 is part of the Ku70/80 complex which acts in concert with SIRT1 in telomere maintenance. The XRCC6 SNP rs132793 in the 3′ flanking region of this gene is in high LD (D′ = 1) with two SNPs in the promoter region and one in the first intron of XRCC6 (Online Resource 3). This SNP has been used as a marker for XRCC6 in several studies (Sobczuk et al. 2010; Li et al. 2011; Liu et al. 2007). No significant differences in genotype distribution of the SNP were observed between males and females, and the population sample was in Hardy–Weinberg proportions for variation at this locus (P ≫ 0.1).
The mean T/S ratios significantly differed between the subjects grouped according to the genotypes of rs132793 (P = 1.91 × 10−2, Table 4). The mean T/S values increased as the number of copies of the minor allele A increased, resulting in significant differences between GG and AA groups (P = 4.36 × 10−2) and GG and AG + AA combined groups (P = 4.42 × 10−2). The association of T/S with rs132793 was examined in detail using multiple linear regression (Table 3). The additive mode of inheritance for the genetic effect of the minor allele was statistically significance (model b), and the significance persisted after adjustment for age (model c). These results indicate that the shorter telomeres associated with rs132793 are independent of age. Indeed, we didn’t see any statistically significant interaction between age and rs132793 (not shown).
Table 4.
Comparison of telomere length (T/S) in different XRCC6 genotypesa
| Genotype | Number | Mean ± SD | P-value |
|---|---|---|---|
| GG | 240 | 1.393 ± 0.829 | |
| AG | 100 | 1.533 ± 0.877 | |
| AA | 15 | 1.745 ± 0.894 | |
| AA + AG | 115 | 1.561 ± 0.878 | |
| GG vs. AG vs. AA | 1.91 × 10−2b | ||
| GG vs. AA | 4.36 × 10−2c | ||
| GG vs. (AA + AG) | 4.42 × 10−2c |
XRCC6 SNP rs132793
Kruskal–Wallis test
Wilcoxon rank sum test (with Bonferroni correction)
To see whether the SIRT1 variant interacts with age or the XRCC6 SNP, we performed multiple linear regression (Table 3, model d). The SIRT1 SNP showed a significant association with T/S, and addition of this SNP to the model including age and the XRCC6 SNP rs132793 improved the model, as shown by the higher R2 values. However, we were not able to detect any statistical interactions between the predictors (not shown). These results suggest that SIRT1 and XRCC6 impinge on telomere length through separate pathways independently of age.
SIRT1 and longevity
Because SIRT1 passed our criterion of association with the aging endophenotype of telomere length, we next examined the association of this gene with longevity. To test its association with longevity, we compared the allele and genotype frequencies of rs7896005 in Caucasian (European-origin) subjects of the LHAS study population (0.80 assignment probability). We found that the frequency of the minor allele of the SIRT1 SNP rs7896005 was higher in the long-lived cases (≥90 years old) than in the young controls (20–59 years old) of the LHAS (Table 5). Logistic models testing effects of this SNP on case–control binary outcomes predicted that the odds of homozygotes of the minor allele being associated with the long-living case group is more than two-fold higher than the odds of other genotype carriers (Table 6). This recessive model for association of the SIRT1 variant with longevity mirrors the recessive genetic model for its association with telomere length (see text and Table 3).
Table 5.
Contingency of longevity upon SIRT1 and XRCC6 variantsa
| Gene (SNP) | Subject group | Genotype | P-valueb | Allele | P-valueb | |||
|---|---|---|---|---|---|---|---|---|
| SIRT1 (rs7896005) | AA | AG | GG | 8.0 × 10−3 | A | G | 5.6 × 10−3 | |
| Casec | 35 | 105 | 84 | 175 | 273 | |||
| Controld | 21 | 134 | 138 | 176 | 410 | |||
| XRCC6 (rs132793) | AA | AG | GG | 0.74 | A | G | 0.94 | |
| Casec | 12 | 60 | 152 | 84 | 364 | |||
| Controld | 12 | 84 | 197 | 108 | 478 | |||
Fisher’s exact test
With Bonferroni correction for SIRT1
Nonagenarians
20–59 years old
Table 6.
Effect of XRCC6 on association of SIRT1 with longevitya
| Sample | Gene (SNP) | OR | 95% CI | P-value |
|---|---|---|---|---|
| LHASb | SIRT1 (rs7896005)d | 2.382 | 1.355–4.282 | 2.96 × 10−3 |
| SIRT1 (rs7896005)d | 2.385 | 1.356–4.287 | 2.94 × 10−3 | |
| + XRCC6 (rs132793)e | 1.029 | 0.755–1.398 | 8.53 × 10−1 | |
| GCSc | SIRT1 (rs7896005)d | 1.334 | 0.978–1.824 | 6.96 × 10−2 |
Binary logistic regression for case, nonagenarian in LHAS and centenarian in GCS, versus control (20–59 years old) outcome adjusted for gender with SNPs as explanatory variables
Louisiana Healthy Aging Study
Georgia Centenarian Study
Recessive mode of inheritance
Additive mode of inheritance
Because of association of SIRT1 with longevity in LHAS, we replicated this analysis in a different population. Samples from the GCS were used. Caucasian (European-origin) subjects of the GCS study population (0.80 assignment probability) were chosen to correspond to the LHAS sample. In this sample, the cases were centenarians (≥98 years old) and the young controls were 20–59 years old. The LD block in SIRT1 of which rs7896005 is a part is similar in the GCS Caucasian population to the one in the LHAS population, though not identical (Online Resource 2). The logistic regression yielded odds one-third higher for cases based on homozygosity for the minor allele as compared to the other genotypes, which was near significant (Table 6), providing support for the association of SIRT1 with longevity.
XRCC6 and longevity
Finally, we tested whether rs132793 is associated with longevity, because XRCC6 fit the criterion of association with the aging endophenotype of telomere length. To test association of XRCC6 with longevity, we compared the allele and genotype frequencies of rs132793 between nonagenarian cases and young controls of the LHAS population, but found no significant differences (Table 5). Incorporation of the XRCC6 SNP rs132793 in the same logistic model with the SIRT1 SNP rs7896005 showed that the association with longevity of SIRT1 persists independently of XRCC6 (Table 6).
Discussion
Our study agrees with the consensus that telomere shortening occurs with age. We also found that function ability decreased with telomere shortening, which is likely due to the dependence of the function variables on age. There is a good deal of variability in the association of function and physiologic variables with telomere length in human studies, and additional examination of these associations is needed (Mather et al. 2011). Some of these discrepancies are likely due to the lack of adjustment for age. However, other differences include study design, population, function and disease status, and methods of assessment of telomere length and of function. We also made novel findings: (1) the SIRT1 and XRCC6 SNPs had significant effects on telomere length; (2) rs7896005 in SIRT1 had significant effects on longevity, whereas the XRCC6 SNP rs132793 did not; (3) the effects of the SNPs on telomere length were independent of age; (4) the effects of the SIRT1 and XRCC6 were independent of each other.
SNP functions
The SIRT1 SNP is in an intron. It is 35 nucleotides away from a nearby exon/intron junction and may affect splicing, though we couldn’t find any such putative function using the SNPinfo Web Server (Xu and Taylor 2009). However, using another web search tool, Pupasuite 3 (http://pupasuite.bioinfo.cipf.es) incorporated in GeneCards Version 3 (www.genecards.org), we found that the SNP is in the close proximity of other potentially functional SNPs. For example, rs7896005 is only 334 nucleotides downstream of rs11327756, which is embedded in a DNA triplex forming sequence. Triplex DNA structures have been suggested as regulatory regions for controlling gene expression (Goni et al. 2004), and SNPs located in these sequences can affect triplex formation and hence gene expression (Duca et al. 2008).
As for rs132793, this SNP lies between XRCC6 and NHP2L1 (Online Resource 3); see Materials and Methods for details. Upstream of NHP2L1 (farther downstream of XRCC6) lies a gene called MEI1, and we found that a few SNPs in MEI1 are also in high LD with rs132793 (not shown). This gene is expressed specifically in testis and is required for normal meiotic chromosome synapsis (Libby et al. 2003; Sato et al. 2006). Because we didn’t see any sex-specific phenotypes of rs132793, we rule out MEI1 as a gene responsible for the effects of rs132793 described in this article. SNP rs132793 is in high LD with four SNPs in or near XRCC6. Of these, according to our database search, rs132770 in the 5′ promoter region of XRCC6 has regulatory potential. It is located 30 nucleotides upstream of the start of the first exon, and the locus is part of a number of transcription factor binding sites, with the profile of predicted binding factors and their affinities substantially varying depending on the alleles present.
Online Resource 3 also shows that rs132793 is in LD with NHP2L1. NHP2L1 encodes a nuclear protein that is known to bind to the 5′-stem-loop of U4 snRNA, a component of the U4/U6 snRNP in the pre-mRNA splicing machinery (Nottrott et al. 1999; Watkins et al. 2002). Based on its exclusive affinity for RNA molecules, we exclude the possibility of NHP2L1 being directly involved in telomere length regulation. However, it could be indirectly involved in telomere metabolism by its post-transcriptional function. In a two-hybrid assay, a physical interaction between Ku70 and NHP2L1 was found (Stelzl et al. 2005), raising the possibility of such an indirect effect. Considering all these data and information together, we regard XRCC6 as the gene directly responsible for the phenotypes of rs132793 described in this article.
Telomere length maintenance
NHEJ is potentially harmful because it doesn’t involve extensive sequence homology. Fusion of an exposed telomere terminus with a DSB end in the middle of a chromosome would be catastrophic. Telomeric fusion of two chromatids would also be catastrophic if the resulting dicentric chromatids segregate inappropriately during mitosis. This questions the utility of this error-prone pathway as an alternative telomere lengthening mechanism. In any case, inactivation of either of the Ku subunits results in telomere shortening in various mouse primary cell types (d’Adda di Fagagna et al. 2001). Our findings indicate that telomere maintenance is more effective in homozygotes of the minor allele of rs132793. Therefore, we conclude that the A allele of rs132793 is coupled with upregulated expression of XRCC6 that results in more efficient telomere length maintenance. Regarding the SIRT1 SNP, increased expression of SIRT1 correlates with efficient DSB repair through HR and longer telomeres, but not with NHEJ (Oberdoerffer et al. 2008; Palacios et al. 2010). Therefore, we suggest that the intron SNP rs7896005 is coupled with upregulation of SIRT1 that results in more efficient HR-mediated telomere maintenance. Thus, we consider different pathways of telomere maintenance, involving either SIRT1 or XRCC6, even though these genes seem to overlap with each other in some of their functions.
The possibility that SIRT1 and XRCC6 are in different pathways for telomere maintenance could explain the lack of any statistical interaction between the two genes observed in this study. However, we cannot rule out the possibility that our study was not powered enough to detect such interactions due to either insufficient sample sizes or suboptimal statistical modeling. In this regard, we note the high variability of T/S values throughout the age range (Fig. 1), which resulted in the low R2 values in all the models of linear regression. Thus, the rate of telomere shortening is likely to vary among different individuals of the same age group, and this suggests additional factors, such as individual-specific environments, are in operation. It is also important to acknowledge the imperfect measure of global telomere shortening that leukocyte telomere length provides. Among the factors contributing to this limitation is the superposition of the state of the leukocyte population itself, which can undergo periodic fluctuations.
Our study suggests that telomere shortening occurs, at least in part, in an age-independent manner, and that SIRT1 and XRCC6 define two pathways impinging on this telomere decline. Obviously operation of multiple genetic pathways for telomere maintenance adds flexibility and complexity to the age-dependent default rate of telomere attrition. One can imagine that these pathways may act as modulators of environmental factors or conditions that bear the potential to afflict telomere integrity leading to genomic instability.
Telomere length and longevity
The yeast gene SIR2 and its orthologs are well characterized as lifespan determinants in model organisms (Guarente and Kenyon 2000), but the role of SIRT1 in human longevity hasn’t been established yet. Here, we presented statistical evidence for the involvement of SIRT1 in human longevity. Interestingly the same allele was found to be associated with longer telomeres, which tempts us to connect longer telomeres with longevity. However, we didn’t see any correlation of the XRCC6 SNP with longevity, even though it was associated with telomere length. One explanation is that the NHEJ in which Ku70 participates has deleterious side effects. It is also possible that there is no obligatory link between telomere attrition and lifespan and the association of SIRT1 with longevity depends on other factors. These other factors might act alone or in concert with telomere length. The elucidation of these factors, genetic and environmental, will be aided by studies in additional populations. We are aware of one such study (Flachsbart et al. 2006) in which no association of SIRT1 with longevity was detected. In this regard, it is noteworthy that the size of the SIRT1 effect in the Georgia population is substantially smaller than in the Louisiana population. While the LHAS was sufficiently powered to detect a statistically significant effect of the size seen, the GCS was not, and this is reflected in its near significance (P = 0.0696). The genetic architecture of the two populations differs, as evidenced even by the LD profile in the SIRT1 gene (Online Resource 2). Thus, the complex genetic and environmental nature of exceptional longevity must be taken into account. This complexity includes potential gene–gene and gene-environment interactions, as seen in other studies (Jazwinski et al. 2010; Bergman et al. 2007). This remains an area open to further investigation.
Acknowledgments
Supported by grants from the National Institute on Aging of the National Institutes of Health (P01AG022064 and P01AG017553) and by the Louisiana Board of Regents through the Millennium Trust Health Excellence Fund [HEF(2001-06)-02]. We thank the people of Louisiana and Georgia for participation in our study. We acknowledge the valuable recruitment, data acquisition, data management, and fiscal management efforts of M. Cervantes, D. Rouzan, T. Beard, M. Burgess, K. Grier, E. Jackson, E. McCarthy, K. Shaw, L. Strong, S. Reynolds, S. Anderson, E. Cassidy, M. Janke, T. Savla, and M. Poon. Additional authors include for The Louisiana Healthy Aging Study: M. Allen, I.E. Antikainen, A.M. Arce, J. Arceneaux, Mark A. Batzer, E.O. Boudreaux, L. Byerley, P.A. Callinan, C.M. Champagne, H. Cheng, Y.-W. Chiu, L. Cosenza, M.E. Cress, J.P. DeLany, J. Denver, A. Deutsch, M.J. deVeer, D.A. Dobrosielski, R. Ellis, A. Ermolao, M. Erwin, M. Erwin, J. Fabre, E. Fontham, M. Frisard, P. Geiselman, L. Goodwin, V. Greco, S. Hadie, T. Hall, M. Hamilton, S.W. Herke, K. Hawley, J. Hayden, K. Hebert, F. Holton, H.-C. Hsu, D. Johannsen, L. Kawasaki, B. Kimball, C. King-Rowley, M. Konkel, R. Kuhn, K. Landry, C. Lavie, D. LaVie, M. Leblanc, C. M. Lefante, L. Li, H.-Y. Lin, K. Lopez, B. McEvoy-Hein, J.D. Mountz, E. Olinde, J. Owens, K. Pedersen, A. Pellett, E. Ravussin, P. Remedios, Y. Robertson, J. Rood, H. Rothschild, R.A. Russell, E. Sandifer, B. Schmidt, R. Schwartz, D.K. Scott, J. Silva, F.N. Standberry, L.J. Su, J. Thomson, A. Tiwari, V. Toups, C. Traylor, C. Velasco-Gonzalez, J. Volaufova, C. Waguespack, J.A. Walker. X.-Y. Wang, R.H. Wood, Q. Yu, S. Zehr, and P. Zhang; for The Georgia Centenarian Study: R.C. Green, M. Gearing, W.R. Markesbery, J.L. Woodard, M.A. Johnson, J.S. Tenover, I.C. Siegler, P. Martin, M. MacDonald, C. Rott, W.L. Rodgers, D.B. Hausman, and A. Davey.
Footnotes
The study was conducted for The Louisiana Healthy Aging Study and The Georgia Centenarian Study.
Electronic supplementary material The online version of this article (doi:10.1007/s10522-011-9360-5) contains supplementary material, which is available to authorized users.
Contributor Information
Sangkyu Kim, Tulane Center for Aging and Department of Medicine, Tulane University Health Sciences Center, 1430 Tulane Ave., SL-12, New Orleans, LA 70112, USA.
Xiuhua Bi, Tulane Center for Aging and Department of Medicine, Tulane University Health Sciences Center, 1430 Tulane Ave., SL-12, New Orleans, LA 70112, USA.
Malwina Czarny-Ratajczak, Tulane Center for Aging and Department of Medicine, Tulane University Health Sciences Center, 1430 Tulane Ave., SL-12, New Orleans, LA 70112, USA.
Jianliang Dai, Tulane Center for Aging and Department of Medicine, Tulane University Health Sciences Center, 1430 Tulane Ave., SL-12, New Orleans, LA 70112, USA.
David A. Welsh, Department of Medicine, Louisiana State University Health Sciences Center, New Orleans, LA 70112, USA
Leann Myers, Department of Biostatistics, School of Public Health and Tropical Medicine, Tulane University Health Sciences Center, New Orleans, LA 70112, USA.
Michael A. Welsch, Department of Kinesiology, Louisiana State University, Baton Rouge, LA 70803, USA
Katie E. Cherry, Department of Psychology, Louisiana State University, Baton Rouge, LA 70803, USA
Jonathan Arnold, Genetics Department, University of Georgia, Athens, GA 30602, USA.
Leonard W. Poon, Institute of Gerontology, University of Georgia, Athens, GA 30602, USA
S. Michal Jazwinski, Email: sjazwins@tulane.edu, Tulane Center for Aging and Department of Medicine, Tulane University Health Sciences Center, 1430 Tulane Ave., SL-12, New Orleans, LA 70112, USA.
References
- Ahn SJ, Costa J, Emanuel JR. PicoGreen quantitation of DNA: effective evaluation of samples pre- or post-PCR. Nucleic Acids Res. 1996;24(13):2623–2625. doi: 10.1093/nar/24.13.2623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andrew T, Aviv A, Falchi M, Surdulescu GL, Gardner JP, Lu X, Kimura M, Kato BS, Valdes AM, Spector TD. Mapping genetic loci that determine leukocyte telomere length in a large sample of unselected female sibling pairs. Am J Hum Genet. 2006;78(3):480–486. doi: 10.1086/500052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arbeev KG, Hunt SC, Kimura M, Aviv A, Yashin AI. Leukocyte telomere length, breast cancer risk in the offspring: the relations with father’s age at birth. Mech Ageing Dev. 2011;132(4):149–153. doi: 10.1016/j.mad.2011.02.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Atzmon G, Cho M, Cawthon RM, Budagov T, Katz M, Yang X, Siegel G, Bergman A, Huffman DM, Schechter CB, Wright WE, Shay JW, Barzilai N, Govindaraju DR, Suh Y. Evolution in health and medicine Sackler colloquium: genetic variation in human telomerase is associated with telomere length in Ashkenazi centenarians. Proc Natl Acad Sci USA. 2010;107(Suppl 1):1710–1717. doi: 10.1073/pnas.0906191106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aubert G, Lansdorp PM. Telomeres and aging. Physiol Rev. 2008;88(2):557–579. doi: 10.1152/physrev.00026.2007. [DOI] [PubMed] [Google Scholar]
- Bergman A, Atzmon G, Ye K, MacCarthy T, Barzilai N. Buffering mechanisms in aging: a systems approach toward uncovering the genetic component of aging. PLoS Comput Biol. 2007;3(8):e170. doi: 10.1371/journal.pcbi.0030170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bryan TM, Englezou A, Dalla-Pozza L, Dunham MA, Reddel RR. Evidence for an alternative mechanism for maintaining telomere length in human tumors and tumor-derived cell lines. Nat Med. 1997;3(11):1271–1274. doi: 10.1038/nm1197-1271. [DOI] [PubMed] [Google Scholar]
- Cawthon RM. Telomere measurement by quantitative PCR. Nucleic Acids Res. 2002;30(10):e47. doi: 10.1093/nar/30.10.e47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cawthon RM. Telomere length measurement by a novel monochrome multiplex quantitative PCR method. Nucleic Acids Res. 2009;37(3):e21. doi: 10.1093/nar/gkn1027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Celli GB, Denchi EL, de Lange T. Ku70 stimulates fusion of dysfunctional telomeres yet protects chromosome ends from homologous recombination. Nat Cell Biol. 2006;8(8):885–890. doi: 10.1038/ncb1444. [DOI] [PubMed] [Google Scholar]
- Chai W, Ford LP, Lenertz L, Wright WE, Shay JW. Human Ku70/80 associates physically with telomerase through interaction with hTERT. J Biol Chem. 2002;277(49):47242–47247. doi: 10.1074/jbc.M208542200. [DOI] [PubMed] [Google Scholar]
- Cherkas LF, Hunkin JL, Kato BS, Richards JB, Gardner JP, Surdulescu GL, Kimura M, Lu X, Spector TD, Aviv A. The association between physical activity in leisure time and leukocyte telomere length. Arch Intern Med. 2008;168(2):154–158. doi: 10.1001/archinternmed.2007.39. [DOI] [PubMed] [Google Scholar]
- Cohen SB, Graham ME, Lovrecz GO, Bache N, Robinson PJ, Reddel RR. Protein composition of catalytically active human telomerase from immortal cells. Science. 2007;315(5820):1850–1853. doi: 10.1126/science.1138596. [DOI] [PubMed] [Google Scholar]
- Cress ME, Buchner DM, Questad KA, Esselman PC, deLateur BJ, Schwartz RS. Continuous-scale physical functional performance in healthy older adults: a validation study. Arch Phys Med Rehabil. 1996;77(12):1243–1250. doi: 10.1016/s0003-9993(96)90187-2. [DOI] [PubMed] [Google Scholar]
- Cress ME, Buchner DM, Prohaska T, Rimmer J, Brown M, Macera C, Dipietro L, Chodzko-Zajko W. Best practices for physical activity programs and behavior counseling in older adult populations. J Aging Phys Act. 2005;13(1):61–74. doi: 10.1123/japa.13.1.61. [DOI] [PubMed] [Google Scholar]
- Critchlow SE, Jackson SP. DNA end-joining: from yeast to man. Trends Biochem Sci. 1998;23(10):394–398. doi: 10.1016/s0968-0004(98)01284-5. [DOI] [PubMed] [Google Scholar]
- d’Adda di Fagagna F, Hande MP, Tong WM, Roth D, Lansdorp PM, Wang ZQ, Jackson SP. Effects of DNA non-homologous end-joining factors on telomere length and chromosomal stability in mammalian cells. Curr Biol. 2001;11(15):1192–1196. doi: 10.1016/s0960-9822(01)00328-1. [DOI] [PubMed] [Google Scholar]
- Devore EE, Prescott J, De Vivo I, Grodstein F. Relative telomere length and cognitive decline in the Nurses’ Health Study. Neurosci Lett. 2011;492(1):15–18. doi: 10.1016/j.neulet.2011.01.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dipietro L, Caspersen CJ, Ostfeld AM, Nadel ER. A survey for assessing physical activity among older adults. Med Sci Sports Exerc. 1993;25(5):628–642. [PubMed] [Google Scholar]
- Duca M, Vekhoff P, Oussedik K, Halby L, Arimondo PB. The triple helix: 50 years later, the outcome. Nucleic Acids Res. 2008;36(16):5123–5138. doi: 10.1093/nar/gkn493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flachsbart F, Croucher PJ, Nikolaus S, Hampe J, Cordes C, Schreiber S, Nebel A. Sirtuin 1 (SIRT1) sequence variation is not associated with exceptional human longevity. Exp Gerontol. 2006;41(1):98–102. doi: 10.1016/j.exger.2005.09.008. [DOI] [PubMed] [Google Scholar]
- Folstein MF, Folstein SE, McHugh PR. Mini-mental state”. A practical method for grading the cognitive state of patients for the clinician. J Psychiatr Res. 1975;12(3):189–198. doi: 10.1016/0022-3956(75)90026-6. [DOI] [PubMed] [Google Scholar]
- Friedrich U, Griese E, Schwab M, Fritz P, Thon K, Klotz U. Telomere length in different tissues of elderly patients. Mech Ageing Dev. 2000;119(3):89–99. doi: 10.1016/s0047-6374(00)00173-1. [DOI] [PubMed] [Google Scholar]
- Gabriel SB, Schaffner SF, Nguyen H, Moore JM, Roy J, Blumenstiel B, Higgins J, DeFelice M, Lochner A, Faggart M, Liu-Cordero SN, Rotimi C, Adeyemo A, Cooper R, Ward R, Lander ES, Daly MJ, Altshuler D. The structure of haplotype blocks in the human genome. Science. 2002;296(5576):2225–2229. doi: 10.1126/science.1069424. [DOI] [PubMed] [Google Scholar]
- Goni JR, de la Cruz X, Orozco M. Triplex-forming oligonucleotide target sequences in the human genome. Nucleic Acids Res. 2004;32(1):354–360. doi: 10.1093/nar/gkh188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grodstein F, van Oijen M, Irizarry MC, Rosas HD, Hyman BT, Growdon JH, De Vivo I. Shorter telomeres may mark early risk of dementia: preliminary analysis of 62 participants from the nurses’ health study. PLoS One. 2008;3(2):e1590. doi: 10.1371/journal.pone.0001590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guarente L, Kenyon C. Genetic pathways that regulate ageing in model organisms. Nature. 2000;408(6809):255–262. doi: 10.1038/35041700. [DOI] [PubMed] [Google Scholar]
- Harley CB. Telomere loss: mitotic clock or genetic time bomb? Mutat Res. 1991;256(2–6):271–282. doi: 10.1016/0921-8734(91)90018-7. [DOI] [PubMed] [Google Scholar]
- Hastie ND, Dempster M, Dunlop MG, Thompson AM, Green DK, Allshire RC. Telomere reduction in human colorectal carcinoma and with ageing. Nature. 1990;346(6287):866–868. doi: 10.1038/346866a0. [DOI] [PubMed] [Google Scholar]
- Iwama H, Ohyashiki K, Ohyashiki JH, Hayashi S, Yahata N, Ando K, Toyama K, Hoshika A, Takasaki M, Mori M, Shay JW. Telomeric length and telomerase activity vary with age in peripheral blood cells obtained from normal individuals. Hum Genet. 1998;102(4):397–402. doi: 10.1007/s004390050711. [DOI] [PubMed] [Google Scholar]
- Jazwinski SM, Kim S, Dai J, Li L, Bi X, Jiang JC, Arnold J, Batzer MA, Walker JA, Welsh DA, Lefante CM, Volaufova J, Myers L, Su LJ, Hausman DB, Miceli MV, Ravussin E, Poon LW, Cherry KE, Welsch MA. HRAS1 and LASS1 with APOE are associated with human longevity and healthy aging. Aging Cell. 2010;9(5):698–708. doi: 10.1111/j.1474-9726.2010.00600.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeong J, Juhn K, Lee H, Kim SH, Min BH, Lee KM, Cho MH, Park GH, Lee KH. SIRT1 promotes DNA repair activity and deacetylation of Ku70. Exp Mol Med. 2007;39(1):8–13. doi: 10.1038/emm.2007.2. [DOI] [PubMed] [Google Scholar]
- Kimura M, Gazitt Y, Cao X, Zhao X, Lansdorp PM, Aviv A. Synchrony of telomere length among hematopoietic cells. Exp Hematol. 2010;38(10):854–859. doi: 10.1016/j.exphem.2010.06.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lamarche BJ, Orazio NI, Weitzman MD. The MRN complex in double-strand break repair and telomere maintenance. FEBS Lett. 2010;584(17):3682–3695. doi: 10.1016/j.febslet.2010.07.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li R, Yang Y, An Y, Zhou Y, Liu Y, Yu Q, Lu D, Wang H, Jin L, Zhou W, Qian J, Shugart YY. Genetic polymorphisms in DNA double-strand break repair genes XRCC5, XRCC6 and susceptibility to hepatocellular carcinoma. Carcinogenesis. 2011;32(4):530–536. doi: 10.1093/carcin/bgr018. [DOI] [PubMed] [Google Scholar]
- Libby BJ, Reinholdt LG, Schimenti JC. Positional cloning and characterization of Mei1, a vertebrate-specific gene required for normal meiotic chromosome synapsis in mice. Proc Natl Acad Sci USA. 2003;100(26):15706–15711. doi: 10.1073/pnas.2432067100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y, Zhang H, Zhou K, Chen L, Xu Z, Zhong Y, Liu H, Li R, Shugart YY, Wei Q, Jin L, Huang F, Lu D, Zhou L. Tagging SNPs in non-homologous end-joining pathway genes and risk of glioma. Carcinogenesis. 2007;28(9):1906–1913. doi: 10.1093/carcin/bgm073. [DOI] [PubMed] [Google Scholar]
- Ludlow AT, Zimmerman JB, Witkowski S, Hearn JW, Hatfield BD, Roth SM. Relationship between physical activity level, telomere length, and telomerase activity. Med Sci Sports Exerc. 2008;40(10):1764–1771. doi: 10.1249/MSS.0b013e31817c92aa. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mather KA, Jorm AF, Parslow RA, Christensen H. Is telomere length a biomarker of aging? A review. The Journals of Gerontology Series A, Biological Sciences and Medical Sciences. 2011;66(2):202–213. doi: 10.1093/gerona/glq180. [DOI] [PubMed] [Google Scholar]
- Mirabello L, Huang WY, Wong JY, Chatterjee N, Reding D, Crawford ED, De Vivo I, Hayes RB, Savage SA. The association between leukocyte telomere length and cigarette smoking, dietary and physical variables, and risk of prostate cancer. Aging Cell. 2009;8(4):405–413. doi: 10.1111/j.1474-9726.2009.00485.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moore JK, Haber JE. Cell cycle and genetic requirements of two pathways of nonhomologous end-joining repair of double-strand breaks in Saccharomyces cerevisiae. Mol Cell Biol. 1996;16(5):2164–2173. doi: 10.1128/mcb.16.5.2164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nottrott S, Hartmuth K, Fabrizio P, Urlaub H, Vidovic I, Ficner R, Luhrmann R. Functional interaction of a novel 15.5kD [U4/U6.U5] tri-snRNP protein with the 5′ stem-loop of U4 snRNA. EMBO J. 1999;18(21):6119–6133. doi: 10.1093/emboj/18.21.6119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oberdoerffer P, Michan S, McVay M, Mostoslavsky R, Vann J, Park SK, Hartlerode A, Stegmuller J, Hafner A, Loerch P, Wright SM, Mills KD, Bonni A, Yankner BA, Scully R, Prolla TA, Alt FW, Sinclair DA. SIRT1 redistribution on chromatin promotes genomic stability but alters gene expression during aging. Cell. 2008;135(5):907–918. doi: 10.1016/j.cell.2008.10.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palacios JA, Herranz D, De Bonis ML, Velasco S, Serrano M, Blasco MA. SIRT1 contributes to telomere maintenance and augments global homologous recombination. J Cell Biol. 2010;191(7):1299–1313. doi: 10.1083/jcb.201005160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palm W, de Lange T. How shelterin protects mammalian telomeres. Annu Rev Genet. 2008;42:301–334. doi: 10.1146/annurev.genet.41.110306.130350. [DOI] [PubMed] [Google Scholar]
- Pavesi E, Avondo F, Aspesi A, Quarello P, Rocci A, Vimercati C, Pigullo S, Dufour C, Ramenghi U, Dianzani I. Analysis of telomeres in peripheral blood cells from patients with bone marrow failure. Pediatr Blood Cancer. 2009;53(3):411–416. doi: 10.1002/pbc.22107. [DOI] [PubMed] [Google Scholar]
- Pooley KA, Sandhu MS, Tyrer J, Shah M, Driver KE, Luben RN, Bingham SA, Ponder BA, Pharoah PD, Khaw KT, Easton DF, Dunning AM. Telomere length in prospective and retrospective cancer case-control studies. Cancer Res. 2010;70(8):3170–3176. doi: 10.1158/0008-5472.CAN-09-4595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pritchard JK, Stephens M, Donnelly P. Inference of population structure using multilocus genotype data. Genetics. 2000;155(2):945–959. doi: 10.1093/genetics/155.2.945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qi DL, Ohhira T, Oshimura M, Kugoh H. Human chromosome 5 carries a transcriptional regulator of human telomerase reverse transcriptase (hTERT) Biochem Biophys Res Commun. 2010;398(4):695–701. doi: 10.1016/j.bbrc.2010.07.003. [DOI] [PubMed] [Google Scholar]
- R Development Core Team. R: a language and environment for statistical computing. R Foundation for Statistical Computing; Vienna, Austria: 2008. http://www.R-project.org. [Google Scholar]
- Ray DA, Walker JA, Hall A, Llewellyn B, Ballantyne J, Christian AT, Turteltaub K, Batzer MA. Inference of human geographic origins using Alu insertion polymorphisms. Forensic Sci Int. 2005;153(2–3):117–124. doi: 10.1016/j.forsciint.2004.10.017. [DOI] [PubMed] [Google Scholar]
- Reddel RR, Bryan TM. Alternative lengthening of telomeres: dangerous road less travelled. Lancet. 2003;361(9372):1840–1841. doi: 10.1016/S0140-6736(03)13538-6. [DOI] [PubMed] [Google Scholar]
- Reddel RR, Bryan TM, Colgin LM, Perrem KT, Yeager TR. Alternative lengthening of telomeres in human cells. Radiat Res. 2001;155(1 Pt 2):194–200. doi: 10.1667/0033-7587(2001)155[0194:alotih]2.0.co;2. [DOI] [PubMed] [Google Scholar]
- Riha K, Heacock ML, Shippen DE. The role of the nonhomologous end-joining DNA double-strand break repair pathway in telomere biology. Annu Rev Genet. 2006;40:237–277. doi: 10.1146/annurev.genet.39.110304.095755. [DOI] [PubMed] [Google Scholar]
- Rochette PJ, Brash DE. Human telomeres are hypersensitive to UV-induced DNA damage and refractory to repair. PLoS Genet. 2010;6(4):e1000926. doi: 10.1371/journal.pgen.1000926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rovner BW, Folstein MF. Mini-mental state exam in clinical practice. Hosp Pract (Off Ed) 1987;22(1A):99, 103, 106, 110. [PubMed] [Google Scholar]
- Saito H, Fujiwara T, Shin S, Okui K, Nakamura Y. Cloning and mapping of a human novel cDNA (NHP2L1) that encodes a protein highly homologous to yeast nuclear protein NHP2. Cytogenet Cell Genet. 1996;72(2–3):191–193. doi: 10.1159/000134186. [DOI] [PubMed] [Google Scholar]
- Sato H, Miyamoto T, Yogev L, Namiki M, Koh E, Hayashi H, Sasaki Y, Ishikawa M, Lamb DJ, Matsumoto N, Birk OS, Niikawa N, Sengoku K. Polymorphic alleles of the human MEI1 gene are associated with human azoospermia by meiotic arrest. J Hum Genet. 2006;51(6):533–540. doi: 10.1007/s10038-006-0394-5. [DOI] [PubMed] [Google Scholar]
- Shawi M, Autexier C. Telomerase, senescence and ageing. Mech Ageing Dev. 2008;129(1–2):3–10. doi: 10.1016/j.mad.2007.11.007. [DOI] [PubMed] [Google Scholar]
- Slagboom PE, Droog S, Boomsma DI. Genetic determination of telomere size in humans: a twin study of three age groups. Am J Hum Genet. 1994;55(5):876–882. [PMC free article] [PubMed] [Google Scholar]
- Sobczuk A, Smolarz B, Romanowicz H, Zadrozny M, Baszczynski J, Westfal B, Pertynski T. Analysis of the polymorphisms in non-homologous DNA end joining (NHEJ) gene Ku70 and Ligase IV in sporadic breast cancer in women. Pol J Pathol. 2010;61(1):27–31. [PubMed] [Google Scholar]
- Stelzl U, Worm U, Lalowski M, Haenig C, Brembeck FH, Goehler H, Stroedicke M, Zenkner M, Schoenherr A, Koeppen S, Timm J, Mintzlaff S, Abraham C, Bock N, Kietzmann S, Goedde A, Toksoz E, Droege A, Krobitsch S, Korn B, Birchmeier W, Lehrach H, Wanker EE. A human protein–protein interaction network: a resource for annotating the proteome. Cell. 2005;122(6):957–968. doi: 10.1016/j.cell.2005.08.029. [DOI] [PubMed] [Google Scholar]
- Terry DF, Nolan VG, Andersen SL, Perls TT, Cawthon R. Association of longer telomeres with better health in centenarians. J Gerontol A Biol Sci Med Sci. 2008;63(8):809–812. doi: 10.1093/gerona/63.8.809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas P, O’Callaghan NJ, Fenech M. Telomere length in white blood cells, buccal cells and brain tissue and its variation with ageing and Alzheimer’s disease. Mech Ageing Dev. 2008;129(4):183–190. doi: 10.1016/j.mad.2007.12.004. [DOI] [PubMed] [Google Scholar]
- von Zglinicki T. Oxidative stress shortens telomeres. Trends Biochem Sci. 2002;27(7):339–344. doi: 10.1016/s0968-0004(02)02110-2. [DOI] [PubMed] [Google Scholar]
- Wang Y, Ghosh G, Hendrickson EA. Ku86 represses lethal telomere deletion events in human somatic cells. Proc Natl Acad Sci USA. 2009;106(30):12430–12435. doi: 10.1073/pnas.0903362106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watkins NJ, Dickmanns A, Luhrmann R. Conserved stem II of the box C/D motif is essential for nucleolar localization and is required, along with the 15.5 K protein, for the hierarchical assembly of the box C/D snoRNP. Mol Cell Biol. 2002;22(23):8342–8352. doi: 10.1128/MCB.22.23.8342-8352.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Willeit P, Willeit J, Mayr A, Weger S, Oberhollenzer F, Brandstatter A, Kronenberg F, Kiechl S. Telomere length and risk of incident cancer and cancer mortality. JAMA. 2010;304(1):69–75. doi: 10.1001/jama.2010.897. [DOI] [PubMed] [Google Scholar]
- Xu Z, Taylor JA. SNPinfo: integrating GWAS and candidate gene information into functional SNP selection for genetic association studies. Nucleic Acids Res. 2009;37 (Web Server issue):W600–W605. doi: 10.1093/nar/gkp290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Young NS. Telomere biology and telomere diseases: implications for practice and research. Hematol Am Soc Hematol Educ Program. 2010;2010:30–35. doi: 10.1182/asheducation-2010.1.30. [DOI] [PMC free article] [PubMed] [Google Scholar]