Abstract
Aflatoxin is among the most potent naturally occurring carcinogens known. Previous studies demonstrated that endosomes in the filamentous fungus Aspergillus parasiticus carry enzymes that catalyze the final two steps in aflatoxin synthesis and these structures also play a role in aflatoxin storage and export. We hypothesized that endosomes house a complete and functional aflatoxin biosynthetic pathway. To address this hypothesis, we purified a cellular fraction containing endosomes, transport vesicles, and vacuoles (V fraction) from A. parasiticus grown under aflatoxin inducing and non-inducing conditions. We also added (fed) aflatoxin pathway intermediates to V fraction to test the functional status of aflatoxin pathway enzymes. High throughput LC/MS/MS analysis of proteins in V fraction detected 8 aflatoxin enzymes with high reliability and 8 additional enzymes at lower reliability suggesting that most aflatoxin pathway enzymes are present. Purified V fraction synthesized aflatoxin and addition of the pathway intermediate versicolorin A increased aflatoxin synthesis confirming that middle and late aflatoxin enzymes in V fraction are functional. Of particular significance, proteomic and biochemical analysis strongly suggested that additional secondary metabolic pathways as well as proteins involved in response to heat, osmotic, and oxidative stress are housed in V fraction.
Keywords: transport vesicles, endosomes, secondary metabolism, aflatoxin, stress response
Introduction
Filamentous fungi produce a wide array of secondary metabolites with detrimental or beneficial properties and several of these are thought to serve specific biological functions for the producing organism1,2. Aspergillus parasiticus synthesizes the polyketide mycotoxin aflatoxin which is among the most potent naturally occurring carcinogens known3. Aspergillus flavus, a close relative of A. parasiticus, carries more than 50 gene clusters associated with the synthesis of polyketide and non-ribosomal peptide secondary metabolites4. The end product associated with only a small number of these clusters has been identified.
In previous studies, gene disruption of avaA (encodes a GTPase and functional component of endosome membranes), addition of sortin 3a (specific inhibitor of vps16, a tethering complex protein that mediates endosome fusion), and feeding of a purified cell fraction (V fraction contains transport vesicles, endosomes, and vacuoles) with the late aflatoxin pathway intermediate sterigmatocystin, demonstrated that the aflatoxin enzymes OmtA and OrdA catalyze the late steps in aflatoxin biosynthesis in endosomes5. Subsequent studies demonstrated that endosomes also store and export aflatoxin2,5,6. Western blot analysis of V fraction proteins using specific polyclonal antibodies demonstrated that Nor-1 (early aflatoxin enzyme), VBS, and Ver-1 (middle enzymes) are also present in V fraction in A. parasiticus grown under aflatoxin-inducing growth conditions (YES medium) although the functionality of these proteins was not demonstrated6. Based on these previous studies, we hypothesized that a complete and functional aflatoxin biosynthetic pathway localizes to V fraction. To test this hypothesis, in the current study we conducted biochemical and high throughput proteomic (LC/MS/MS) analyses of V fraction purified from A. parasiticus grown under aflatoxin inducing (YES) and non-inducing conditions (YEP medium).
Materials and Methods
Fungal strains and growth conditions
Aspergillus parasiticus SU-1 (ATCC 56775), a wild type aflatoxin producer, was used in this study. YES liquid medium (contains 2% yeast extract and 6% sucrose; pH 5.8) was used as an aflatoxin inducing growth medium. YEP liquid medium (contains 2% yeast extract and 6% peptone, pH 5.8) was used as an aflatoxin non-inducing growth medium. A. parasiticus conidiospores (105 spores/ml) were inoculated into flasks each containing 100 ml liquid medium and 5 glass beads; flasks were incubated for 24 or 36 h at 30°C with shaking at 150 rpm in the dark. Two biological replicate experiments were conducted on cultures incubated in YEP media for 24 or 36 h or in YES media for 24 h after conidiospore inoculation. Four biological replicates were conducted on cultures incubated in YES media for 36 h after conidiospore inoculation.
Purification of V fraction under aflatoxin inducing and non-inducing growth conditions
The method for V fraction purification was based on a published method6. Briefly, A. parasiticus conidiospores (105 spores/ml) were inoculated into 12 flasks each containing 100 ml liquid YES or YEP media and incubated for 24 or 36 h in the dark at 150 rpm. Mycelia were harvested through Miracloth (EMD Biosciences, Inc., San Diego, CA, USA) and excess growth medium was removed by spreading the mycelium on a bed of paper towels. The mycelia were suspended in protoplast buffer (1.2 M MgSO4, 10 mM KH2PO4, pH 5.8) supplemented with 8 mg/ml Drizelase and 8 mg/ml Lysing Enzymes (both from Sigma-Aldrich, St. Louis, MO, USA). These enzymes were prepared and filter sterilized through a 0.45 μm filter just before use. The mycelial suspension was incubated at 30°C with gentle shaking for 5 to 6 h. Protoplasts were separated from undigested mycelia by filtration through a multiple layer filter consisting of 1 layer of Nylon mesh (30 μm; SpectrumLabs, Rancho Dominguez, CA, USA), 2 layers of Miracloth (EMD Biosciences, Inc., San Diego, CA, USA), and 2 layers of cheesecloth. The protoplast suspension (25 ml) was overlaid with 15 ml of buffer A (0.6 M sorbitol, 100 mM Tris-HCl, pH 7.0) in a 50 ml polypropylene centrifuge tube (Corning Inc., Corning, NY, USA) and protoplasts (5 ml) were collected at the interface after centrifugation at 1500 × g for 15 min at RT. After dilution with an equal volume (5 ml) of buffer B (1.2 M sorbitol, 10 mM Tris-HCl, pH 7.5), the protoplast suspension was centrifuged at 1000 × g for 10 min at RT, and pellets containing protoplasts were combined in a total volume 1 ml. Release of vacuoles, endosomes, and transport vesicles was initiated by addition of 1 ml of protoplast lysis buffer (0.6 M sorbitol, 10 mM Tris-HCl, 0.025% Triton-X 100, pH 7.5). After 10 min, 1 ml of solution containing lysed protoplasts was applied onto a 0.9 ml sucrose cushion (3 M sucrose, 1.2 M sorbitol, 10 mM Tris-HCl, pH 7.5) in a 2 ml microcentrifuge tube, centrifuged at 3000 × g for 30 min at RT, and pure V fraction was harvested from just above the interface in a volume of 100 to 200 μl per tube (200 to 400 μl total volume). The remainder of the preparation above the sucrose cushion was also collected (non V fraction). To generate a re-band sample, V fraction was purified from 2 of the 4 replicate cultures grown for 36 h in YES and this was mixed with 0.6M sorbitol, 10 mM Tris-HCl, pH 7.5 (protoplast lysis solution minus triton X-100) to make a final volume of 1 ml. This solution was layered onto a 1 ml sucrose cushion in a 2 ml microcentrifuge tube and centrifuged at 3000 × g for 30 min at RT. Reband V fraction was collected as described above.
The contents of V fraction were analyzed in previous work using Cell Tracker Blue CMAC (fluorescent vacuolar peptidase stain) and MDY64 (fluorescent vesicle and vacuole membrane stain)6. Membrane bound, sub-cellular organelles including transport vesicles (defined as membrane bound sub-cellular organelles < 2.5 μM), endosomes, and vacuoles (sub-cellular organelles >2.5 μM) were observed and the purity was judged to be >95% based on the absence of cytoplasmic and mitochondrial marker enzymes and the absence of nuclei upon microscopic examination. V fraction was highly enriched for alpha mannosidase, a key marker enzyme for CVT transport vesicles and vacuoles6.
Feeding of V fraction with early and middle aflatoxin pathway intermediates
Ten μl containing 243 μg of versicolorin A (VerA) or 40 μg of norsolorinic acid (NA) were added to 100 μl of pure V fraction or 100 μl of non V fraction and the mixture was incubated for 16 h at 30°C. Untreated controls were harvested immediately or after 16 h incubation at 30C. Aflatoxin was extracted with 500 μl chloroform and then with 100 μl of chloroform. The chloroform extracts were combined, evaporated under N2, and re-dissolved in 100 μl of 70% methanol. Aflatoxin B1 was determined by ELISA7 and data were presented as ng of aflatoxin produced per μg of total protein. Protein in the water-soluble fraction remaining after aflatoxin extraction was determined by a commercial Bradford assay (BioRad Laboratories, Hercules, CA).
High throughput LC/MS/MS analysis of V fraction
An aliquot of pure V fraction containing 250 μg total protein was precipitated from a total of 10 samples (2 biological replicate samples of cultures incubated in YES [24 h] or YEP [24 h or 36 h]; 2 biological replicate samples of cultures incubated in 36 h YES; and 2 biological replicate samples of cultures incubated in YES for 36 h and V fraction rebanded) using chloroform: methanol (1:4). Pellets were re-suspended in 30 μl of SDS-PAGE sample buffer and heated to 60°C for 15 min. The samples were then electrophoresed into a 12.5% Tris-HCl Criterion pre-cast 1D SDS polyacrylamide gel from BioRad (www.bio-rad.com) at 50V constant for 15 min until the dye just entered the stacking gel. Here, electrophoresis simply was used as a preparative step to further purify proteins contained in lysed transport vesicles, endosomes, and vacuoles in V fraction. Electrophoresis was stopped and the gel fixed in 40% methanol/20% acetic acid overnight followed by staining with coomassie blue protein stain for 4 h. The gel was de-stained with repeated washes using 10% acetic acid until protein bands were seen on a clear background. Protein bands were excised with a razor blade and placed into individual microcentrifuge tubes. Gel bands were then subjected to in-gel tryptic digestion. The extracted peptides were re-suspended in a solution of 2% acetonitrile/0.1% trifluoroacetic acid to a final volume of 20 μl. From this, 10μl were automatically injected by a Waters nanoAcquity Sample Manager (www.waters.com) and loaded for 5 min onto a Waters Symmetry C18 peptide trap (5μm, 180μm × 20mm) at 4 μl/min in 2% acetotnitrile/0.1% formic acid. The bound peptides were then eluted using a Waters nanoAcquity UPLC (Buffer A = 99.9% water/0.1% formic acid, Buffer B = 99.9% acetonitrile/0.1% formic acid) onto a Michrom MAGIC C18AQ column (3 u, 200A, 100U × 150 mm, www.michrom.com) and eluted over 120 min using a gradient ranging from 5% buffer B to 35% buffer B in 108 min at a flow rate of 1 μl/min. Eluted peptides were sprayed into a ThermoFisher LTQ mass spectrometer (www.thermo.com) using a Michrom ADVANCE nanospray source. The top five ions in each survey scan were then subjected to data-dependant zoom scans followed by low energy collision induced dissociation (CID). The resulting MS/MS spectra were converted to peak lists using BioWorks Browser v3.3.1 (ThermoFisher) software with default parameters and searched against a database of Aspergillus flavus protein sequences downloaded 4-20-09 from NCBI (www.ncbi.nlm.nih.gov), using the Mascot searching algorithm, v 2.2 (www.matrixscience.com). The Mascot output (see Supplemental Table 1) was analyzed using Scaffold v 3.08 (www.proteomesoftware.com) to probabilistically validate protein identifications using the ProteinProphet computer algorithm (see Supplemental Tables 2 and 3). Assignments validated above the Scaffold 95% confidence filter were considered true. In all analyses, a 0.2% protein % false discovery rate (FDR) was observed. This high throughput method using state of the art technology generated a complete proteome for V fraction at 24 and 36 h under aflatoxin inducing (YES) and non inducing (YEP) growth conditions. MS/MS data supporting all protein identifications were formatted by Scaffold version 3.08 and submitted to Tranche (ProteomeCommons.org; Filename, Linz Data 2009 24–36hr Analyses 20110412.tranche.text; Tranche Hash, ++DERCA7dheGvYGFsGYN4/ mUFJ8lh 5wIvcdxu6JhsGwEDGRwpgnSCutg Q9 ngDX+VgYF/XvEQqd60thpoLevviURPRo AAAAAA zp8AA==). These data are summarized in Supplemental Table 2. The vast majority of protein identifications in this manuscript were based on a 95% confidence threshold (including 6 aflatoxin enzymes presented in Table 2) and these are based on multiple peptide matches (at least 3). Table 2 also presents 10 aflatoxin enzymes that were identified at confidence levels ranging from <1% to 91% - these represent the only exceptions to the 95% confidence threshold rule. Table 3 presents enzymes involved in stress response and other secondary metabolic pathways that were all identified at a confidence level of 95%.
Table 2.
Tryptic peptide matches (from LC/MS/MS analysis) with aflatoxin enzymes
| Aflatoxin enzyme | Peptide sequence (percent reliability) |
|---|---|
| CypA/P450 monooxygenase | TITTITFMLAGLK (1%) |
| PksA | AQMLQSSMNTVDH (4%) |
| Fas-1 | ADNDLDQSR (91%) |
| Fas-2 | TPVGACATGVESIDSGYESIMAG (4%) |
| QDTMNEDAMASSAIDPAVVEFS (<1%) | |
| SSMISVTSRPSSRSSTSSEVSDK (<1%) | |
| Nor-1 | LMGRPQAPTTVADSVAGICAR (<1%) |
| NorA | VLYLGVSDTP (95%) |
| AflR | DSCTSCASSK (0–3%) |
| Vbs | SPQLLMVSGIGPK (95%) |
| Ver-1 | HFGYLDIVSSNAGIVSFGHLK (95%) |
| GlcA | AVGFDRGELAMAFQFEIMDIDHG (82%) |
| NadA | FDQFANLAAAARANGSLILAQISH (13%) |
| Hypothetical protein | GDNPMHAGGITSGSAKLP (6%) |
| EstA | LIDQDMADK (95%) |
| OmtB | ESDTPITLAEIVK (95%) |
| OmtA | SILHDYPDAACVEILSNIVTAMDPS (95%) |
| VerB | LPLAPNGVMSILVSDTK (12%) |
| VerA | SCIGQTLAMLELRIALAMTIR (<1%) |
Table 3.
Tryptic peptide matches (from LC/MS/MS analysis) with secondary metabolism and stress response enzymes
| Secondary metabolism enzyme | Peptide sequence (percent reliability) |
|---|---|
| PksL 2 (6 methylsalicylic acid synthase) | KNMAEAVKNGDHILATLKG (95%) |
| Diphosphomevalonate decarboxylase | RMAAIETAIQNRD (95%) |
| Spermidine synthase | RASFVLPNFARK (95%) |
| Pentafunctional polypeptide AroM | KIAAISSEEEFTALEDNAEAILTAVRS (95%) |
| Stress response enzyme | |
|---|---|
| Catalase A | KIAGEDPDVLIRD (95%) |
| Catalase | KFGFDLFDPTKI (95%) |
| Superoxide dismutase Mn | KAAIEAQYGSVEKF (95%) |
| Superoxide dismutase Cu/Zn | RNYGDLGNFKT (95%) |
| Superoxide dismutase Fe | KAIDESFGSLGEFQSKM (95%) |
| Hsp70 (heat stress) | KDAGAISGLNVLR (95%) |
| Trehalose synthase | RLGPSDQLLNALLSKA (95%) |
| Xylulose reductase | MPIPVPSANSLTDLLSKG (95%) |
Analysis of proteome data
Potential function and sub-cellular location of proteins were assigned based on information in a Aspergillus flavus database (www.aspegillusflavus.org), a UNIPROT protein database, and by review of the literature. The A. flavus protein database is built on the complete genome sequence originally released in 2005 and re-annotated in 2009 (http://www.aspergillusflavus.org/genomics/). The genome carries 13,487 genes and the vast majority of proteins in the organism are thought to be represented in this database (5× sequence coverage). Classification of proteins involved in synthesis of aflatoxin and other polyketides was conducted using an A. parasiticus protein database available in NCBI. The comparative proteome analysis (Figure 1) presents data from one of two independent duplicate experiments that we performed which exhibited similar trends. The number of peptide matches to specific groups of proteins is listed at the top of each bar in the graphs shown in Figures 1b–d. Two criteria were utilized to assign significance to differences in the number of peptide matches dependent on growth medium, time, or sub-cellular location. Differences were considered significant if: 1) the difference in the number of peptide matches was greater than 2 fold; and 2) differences of similar magnitude were observed in two independent replicates of the experiment. Based on these criteria, we identified a subset of proteins that was enriched after re-banding of V fraction. Because of the conservative nature of this analysis, this subset of proteins was considered to be tightly associated with the V fraction.
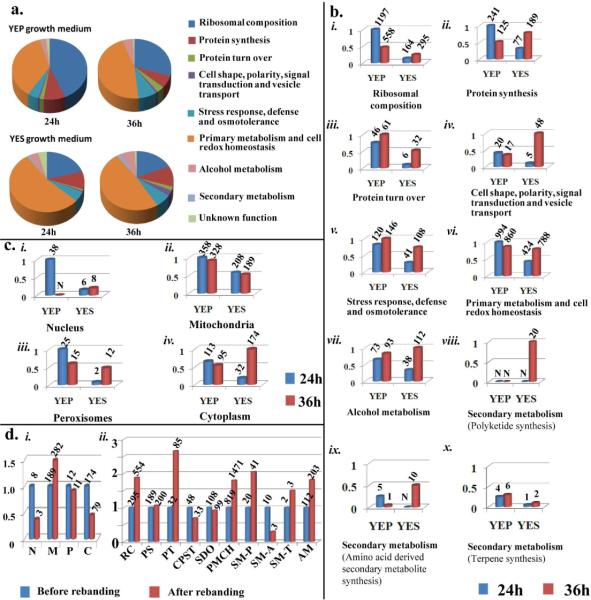
Figure 1. Analysis of the A. parasiticus vesicle-vacuole (V fraction) proteome.
A. parasiticus was cultured under standard growth conditions for 24 or 36 h under aflatoxin inducing (YES) or non-inducing (YEP) conditions (see Methods). V - fraction was purified from mycelial samples and high throughput LC/MS/MS conducted. a. Relative distribution of predicted protein functions in the V proteome. Peptides identified by LC/MS/MS analysis were matched to specific proteins in the A. flavus genome using Scaffold v3.08 software. Each unique protein was assigned to a cellular function category by Scaffold as illustrated by color-codes in the pie chart. b. The relative number of peptides that matched A. flavus proteins in each functional category as observed in (a). c. The relative number of peptides that match A. flavus proteins that function in specific sub-cellular organelles based on studies conducted in filamentous fungi or other biological models. d. Effect of “rebanding” on the V proteome of A. parasiticus grown in YES for 36h: i) The relative number of peptides functionally linked to specific sub-cellular organelles (see c). Abbreviations: N, nucleus; M, mitochondria; P, peroxisomes; C, cytoplasm. ii) The relative number of peptides assigned to functional groups as described in (b). Abbreviations: RC, ribosomal composition; PS, protein synthesis; PT, protein turn over; CPST, cell shape, polarity, signal transduction and vesicle transport; SDO, stress response, defense and osmoregulation; PMCH, primary metabolism and cell redox homeostasis; SM-P, polyketides; SM-A, amino acid-derived secondary metabolites; SM-T, terpenes; AM, alcohol metabolism. The entire analysis was performed on two biological replicates with similar results. Numbers on bars in figures (b), (c) and (d) represent the number of the peptides observed in a single experiment. Only peptide matches reported with a 95% confidence level were considered in this analysis. The false discovery rate (FDR) for these analyses was 0.2%.
Results
Transport vesicles, endosomes, and vacuoles in V fraction carry protein cargo associated with several biological functions
A. parasiticus was grown for 24 or 36 h under standard aflatoxin inducing (YES) and non-inducing conditions (YEP)5,8. Aflatoxin synthesis initiates at approximately 30 h in YES medium, reaches maximum rates between 36 and 48 h, and then declines; neither aflatoxin nor aflatoxin enzymes are synthesized at detectable levels at any time in YEP or at 24 h in YES5,8,9,10. In contrast, aflatoxin and aflatoxin enzymes are detected at high levels by 36 h in YES5,8,9,10. A purified fraction containing transport vesicles, endosomes, and vacuoles which we designated V fraction was prepared from cultures incubated for 24 or 36 h in YEP or YES and high throughput LC/MS/MS analysis was conducted (see Methods). Proteome analysis for 2 biological replicates for YES 24 h, YEP 24 h, YEP 36h samples and 4 biological replicates for YES 36 h sample (including 2 V fraction reband samples) showed similar trends; representative data from one biological replicate are presented to illustrate the analyses.
We included a protein in the V fraction proteome if 3 peptides in a sample matched a single protein with 100% identity in the A. flavus genome database. LC/MS/MS analysis combined with Scaffold Software v3.08 identified 252 proteins in the 24 and 36 h YEP samples combined, 207 proteins in the 24 and 36 h YES samples combined, and 276 proteins in the YES 36 h re-band samples. The vast majority of these proteins (> 90%) were detected in both the 24 and 36 h samples and they were identified at a reliability > 95%. The number of peptides in a sample that matched a particular protein in the A. flavus genome ranged from 75 (pyruvate decarboxylase in YES) to a single peptide (protein transport protein sec61 in YES 36 h).
We distributed proteins identified in V fraction among 8 groups (Fig. 1A) based on predicted biological function and a small number were placed in a ninth category when their function was unknown. Specific enzyme markers typical of a fraction containing transport vesicles, enodosomes, and vacuoles were consistently identified in V fraction samples including vpsA (Ras-like GTPase involved in transport vesicle fusion), vacuolar transporters, stomatin, coatamer, clathrin, autophagic serine protease, metallopeptidase, calnexin (an endoplasmic reticulum membrane marker), woronin body major protein, 2 catalases, SNARE domain protein, 3 aminopeptidases, secretory pathway gdp dissociation protein, sorbitol and xylulase reductase, trehalose synthase, and 3 superoxide dismutases, (Mn, Zn, Fe)11,12,13. Of particular interest, enzymes involved in secondary metabolism were detected primarily at 36 h in YES (including a group of aflatoxin enzymes discussed below), although progressively smaller numbers of peptides associated with secondary metabolic enzymes could be detected at 24 h in YES and at 24 and 36 h in YEP, respectively.
V fraction proteome composition is dependent on nutrients in the growth medium; a comparison of aflatoxin inducing (YES) and non-inducing (YEP) growth conditions
We distributed V fraction proteins to groups of different biological function (Fig. 1B). The numbers on each bar represent the total number of peptides from a sample that match a protein within that particular group. Because one A. flavus protein within a group may match several peptides in the sample, the total number of peptide matches exceeds the total number of proteins detected. The proteins were sorted into groups i through vii based on biological function (see Fig. 1A) and group viii proteins involved in secondary metabolism were further sub-divided into three subgroups (viii – x) based on the end-product of the biosynthetic pathway (polyketides, terpenes, and amino acid- derived metabolites). YEP supported synthesis of a larger number and wider array of proteins carried by V fraction than YES. YES supported increased numbers of proteins involved in synthesis of polyketide- and amino-acid-derived secondary metabolites (the two largest groups of biosynthetic enzymes among secondary metabolic pathways) whereas enzymes involved in synthesis of terpene secondary metabolites were present predominantly in YEP. Similar to enzymes involved in secondary metabolism, peptides derived from other proteins associated with the transition between active growth and stationary phase were present at relatively low levels at 24 h in YES but these proteins appeared to be strongly induced (at least a 2 fold increase in the number of peptides detected) by 36 h; these groups included protein turnover [iii], cell shape, polarity, signal transduction and vesicle transport [iv], stress response, defense, and osmotolerance [v], and alcohol metabolism [vii].
V fraction carries protein cargo associated with multiple sub-cellular organelles
One primary function of transport vesicles is to carry protein cargo to and from sub-cellular organelles within the cell2, 11. Consistent with this role, a large number of proteins detected in V fraction were assigned to nuclei, mitochondria, and peroxisomes. We previously demonstrated that intact nuclei and mitochondria are not present in V fraction and key cytoplasmic (lactate dehydrogenase) and mitochondrial (succinate dehydrogenase) marker enzymes are barely detected6. These data confirm that many of the membrane bound sub-cellular compartments in V fraction are transport vesicles which transport a complex mixture of enzymes involved in primary and secondary metabolism to their destination within the cell. At present, it is not clear if these enzymes co-exist in the same membrane bound compartment or if separate compartments carry out unique functions.
Re-banding of V fraction enriches enzymes involved in polyketide synthesis
To further assess function and purity, we purified V fraction isolated from 2 biological replicates samples (cultures incubated in YES for 36 h) on a sucrose cushion using the standard protocol, harvested this fraction, and then re-banded it on a fresh sucrose cushion (see Methods). The number of proteins in the re-banded V fraction proteome increased by approximately 25% as compared to the 2 biological replicate samples of cultures incubated for 36 h in YES obtained by the standard procedure. Most enzymes present in V fraction isolated at 36 h in YES were present in the 36 h re-band although new proteins were detected and a small number of proteins present in the standard sample were absent in the 36 h re-band. Consistent with this observation, re-banding enriched enzymes destined for the mitochondria (Fig. 1d [i]), enzymes associated with protein turnover, and enzymes involved in alcohol metabolism. In addition, the number of peptides associated with ribosome composition and stress response, defense, and osmotolerance did not decrease (Fig. 1d [ii]) supporting their inclusion in the V fraction proteome. Of particular significance, enzymes associated with synthesis of polyketide and terpene secondary metabolites (Fig. 1d [ii]) were enriched by re-banding strongly supporting their inclusion within V fraction.
V fraction carries early, middle, and late aflatoxin enzymes
Consistent with Western blot analysis conducted in previous studies, no enzymes associated with aflatoxin biosynthesis were detected in V fraction purified from cells grown for 24 or 36 h in YEP medium or in cells grown for 24 h in YES5,8,9,10. However, LC/MS/MS analysis detected 8 aflatoxin enzymes in duplicate 36 h YES samples with high reliability including NorA, VBS (early enzyme), Ver-1, EstA, (middle enzymes), OmtB, OmtA (all greater than >95%), Fas-1 (91%) (early enzyme), GlcA (82%) and 8 additional enzymes at lower reliability (ranging from 1 to 13%) including CypA, PksA, Fas-2, Nor-1, AflR, NadA, hypothetical protein, VerB, and VerA (Table 1 and 2). Most of these enzymes were enriched at least 2 fold in the reband sample supporting the hypothesis that many early, middle, and late aflatoxin enzymes are carried in V fraction.
Table 1.
Proteins enriched in V fraction (at least two-fold) after reband
| Aflatoxin enzymes | GI number | Mass (KDa) |
|---|---|---|
|
| ||
| 1. Fatty acid synthase, alpha subunit (Fas-2) | 45477379 | 182.2 |
| 2. Fatty acid synthase – beta subunit (Fas-1) | 45477380 | 181.4 |
| 3. Polyketide synthase (PksA) | 45477381 | 230.7 |
| 4. Norsolorinic acid reductase (Nor-1) | 45477382 | 29.6 |
| 5. Norsolorinic acid reductase/dehydrogensase (NorA) | 45477383 | 43.7 |
| 6. P450 monooxygenase (AvnA) | 45477385 | 56.3 |
| 7. Esterase (EstA) | 45477388 | 34.6 |
| 8. Versicolorin B synthase (Vbs) | 45477389 | 70.2 |
| 9. Dehydrogenase(Ver-1) | 45477391 | 27.9 |
| 10. O-methyltransferaseB (OmtB) | 45477393 | 43.1 |
| 11. O-methyltransferaseA (OmtA) | 45477394 | 46.3 |
| 12. Oxidoreductase (OrdA) | 45477408 | 60.2 |
| Non-aflatoxin enzymes | GI number | Mass (KDa) |
|---|---|---|
|
| ||
| 1. Aspartic endopeptidase Pep2 | 220699514 | 43.4 |
| 2. ADP-ribosylation factor, putative | 220693637 | 21.0 |
| 3. Phosphoribosylaminoimidazolecarboxamide formyltransferase/IMP | 220697497 | 65.2 |
| 4. Glucose-6-phosphate isomerase | 220696438 | 61.2 |
| 5. Sorbitol/xylulose reductase Sou1-like, putative | 220697966 | 28.8 |
| 6. Pyruvate dehydrogenase E1 beta subunit PdbA, putative | 220692495 | 40.7 |
| 7. Phosphoglycerate mutase, 2,3-bisphosphoglycerate-independent | 220701504 | 57.4 |
| 8. Enolase/allergen Asp F 22 | 220700135 | 47.4 |
| 9. ATP synthase F1, beta subunit, putative | 220689386 | 55.4 |
| 10. Ubiquinol-cytochrome C reductase complex core protein 2, putative | 220691708 | 32.9 |
| 11. Succinate dehydrogenase subunit Sdh1, putative | 220689985 | 71.7 |
| 12. ATP synthase gamma chain, mitochondrial precursor, putative | 220700969 | 32.4 |
| 13. Outer mitochondrial membrane protein porin | 220697586 | 37.2 |
| 14. Mitochondrial F1 ATPase subunit alpha, putative | 220700832 | 60.1 |
| 15. Mitochondrial processing peptidase beta subunit, putative | 220698109 | 53.2 |
| 16. Pyruvate decarboxylase PdcA, putative | 220699546 | 63.5 |
| 17. Alcohol dehydrogenase, putative | 220690467 | 37.4 |
| 18. Aldehyde reductase (AKR1), putative | 220696972 | 36.7 |
| 19. Potential toxin transporter (AflT) | 45477397 | 55.0 |
| 20. Polyketide synthase (PksL2) | 1762234 | 192.1 |
V fraction synthesizes aflatoxin and feeding a middle pathway intermediate enhances the level of synthesis
We previously demonstrated that OmtA and OrdA catalyze the final 2 steps in aflatoxin biosynthesis in endosomes carried in V fraction5. To determine whether the additional aflatoxin enzymes identified by LC/MS/MS analysis are enzymatically functional, we conducted a feeding experiment by adding early (norsolorinic acid) and middle (versicolorin A) aflatoxin pathway intermediates to V fraction and measured aflatoxin synthesized by the fraction using ELISA5. We reasoned that if aflatoxin enzymes are present in V fraction, adding pathway intermediates should enhance the synthesis of aflatoxin by this fraction. This concept was demonstrated for the late aflatoxin enzymes OmtA and OrdA in previous work5. We observed that versicolorin A increased aflatoxin synthesis in 3 independent experiments while norsolorinic acid did not. These data support the hypothesis that many aflatoxin pathway enzymes (at least from Ver-1 to OrdA) are present and functional in V fraction. LC/MS/MS analysis also detected the early aflatoxin enzymes Fas and Pks (convert acetyl CoA to hexanoyl CoA and then to norsolorinic acid14) supporting the hypothesis that a complete and functional aflatoxin pathway is carried in V fraction. Approximately 6 fold less norsolorinic acid was used in the feeding analysis perhaps explaining our inability to consistently observe an increase in aflatoxin synthesis with this pathway intermediate.
V fraction carries enzymes involved in stress response and additional secondary metabolic pathways
LC/MS/MS analysis detected enzymes involved in several additional secondary metabolic pathways in V fraction including those involved in the biosynthesis of patulin (PksL encodes 6 methyl salicylic acid synthase [6MSAS]) involved in synthesis of patulin), isoprenes (diphosphmevalonate decarboxylase), polyamines (spermidine synthase), and shikimate (pentafunctional polypeptide AroM). These data strongly suggest that many Aspergillus secondary metabolites are synthesized, stored, and/or exported by endosomes in V fraction. Several enzymes involved in cellular response to oxidative (catalase, three different superoxide dismutases), heat (HSP70) and osmotic stress (sorbitol and xylulose reductase, trehalose synthase) were also consistently detected at high reliability (>95%) in V fraction. Moreover, we observed that catalase specific activity increased over 20 fold during V fraction purification supporting the presence of a functional catalase in V fraction.
Discussion
V fraction carries out multiple biological functions associated with primary and secondary metabolism
In filamentous fungi, transport vesicles carry out many important biological functions. They transport protein cargo from ribosomes located in the cytoplasm to sub-cellular organelles via the carboxypeptidase Y (CPY) pathway and the cytoplasm to vacuole transport [CVT] pathway, from endoplasmic reticulum-associated ribosomes to the cell exterior via the classical secretory pathway and exocytosis, from one organelle to another, and from the cytoplasmic membrane to the vacuole (endocytosis)2,13,15,16,17,18. Upon nutrient limitation, transport vesicles also mediate the recycling of proteins and organelles by transporting them to vacuoles (autophagy)19. These processes occur simultaneously in fungal cells and there is significant overlap in the machinery that carries out these functions, especially for autophagy and the CVT pathway13,15,20. Vesicles functionally linked with specific transport pathways carry diagnostic marker proteins including aminopeptidase I (API) and alpha-mannosidase for the CVT pathway and carboxypeptidase Y and proteinase A for the CPY pathway13,15,20. Transport vesicles fuse with each other to generate endosomes and this process is driven by a specific recognition mechanism called tethering21.
We previously demonstrated that interfering with the activity of two HOPS (class C) tethering complex proteins encoded by avaA (homologue of yeast ypt7) and vps16 results in accumulation of large numbers of small, membrane bound organelles presumably because they fail to fuse with each other5. This increase in organelle number accompanies increases in synthesis, storage, and export of aflatoxin. Because ypt7 in yeast mediates fusion of late endosomes with vacuoles, these data confirmed that V fraction is composed primarily of transport vesicles and endosomes; a small number of vacuoles was also observed in V fraction. Treatments that block endosome fusion with vacuoles increased the number of small sub-cellular compartments (and not larger compartments) as well as the synthesis, storage, and export of aflatoxin which suggests that transport vesicles, endosomes, but not vacuoles play a major role in aflatoxin synthesis. V fraction proteome data support this idea.
V fraction proteome is affected by nutrient composition and shifts during transition from active growth to stationary phase
The number and composition of proteins in the V fraction proteome varied in response to a change in the nutrients in the growth medium (YEP versus YES) as well as in response to time of growth (24 versus 36 h). This observation is consistent with our previous study that demonstrated that the number of transport vesicles and endosomes in the fungal mycelium dramatically increases during the transition from active growth to stationary phase2,5 in YES growth medium. In contrast, this increase in transport vesicle and endosome number is not observed in YEP medium. This high endosome number shift is at least in part regulated by a global regulator of fungal secondary metabolism and development called VeA2,5; the shift was not observed in a strain carrying a veA deletion. We also demonstrated that transcript accumulation for two key proteins involved in fusion of endosomes with vacuoles (Vps16 and AvaA) appear to be regulated by VeA providing a potential mechanistic link between VeA and accumulation of transport vesicles and endosomes. In summary, previous data and the current study confirm that intracellular transport of protein cargo is highly influenced by growth conditions and this influence is likely mediated, at least in part, by VeA.
Aflatoxin enzymes localize to and are functional in V fraction
LC/MS/MS analysis combined with feeding studies confirmed that many functional aflatoxin enzymes are present in V fraction. This observation is supported by the detection of SAM synthase in V fraction. This enzyme synthesizes S-adenosyl-methionine, a cofactor in two methyl transferase reactions catalyzed by the late aflatoxin enzymes, OmtA and OmtB. From a functional standpoint, it is reasonable to hypothesize that aflatoxin enzymes co-localize to the same sub-cellular compartment allowing efficient synthesis of the mycotoxin. Future work will focus on testing this hypothesis.
V fraction is an important site for synthesis of secondary metabolites
One of the most important and novel findings of the current work is that enzymes involved in several secondary metabolic pathways localize to transport vesicles and endosomes within V fraction. This observation implies that there is a functional and regulatory link between several secondary metabolic pathways and that V fraction houses the physical machinery to accomplish multiple tasks involved in secondary metabolism. This also implies that understanding the complex regulatory circuits that mediate endosome biogenesis can promote our ability to control synthesis of many secondary metabolites simultaneously; this has important implications to control of negative and positive impacts of secondary metabolites on agriculture and human health.
A functional and regulatory link, stress response and secondary metabolism
We also detected several enzymes involved in the ability of fungal cells to respond to heat (heat shock proteins), osmotic (trehalose synthase), and oxidative stress (catalase, superoxide dismutase) in V fraction. Consistent with this observation, heat shock protein and superoxide disumutase were previously identified in the vacuole proteome in Saccharomyces cerevisiae12. The number of proteins (386) and the overall protein composition of V fraction in our work was consistent with the number of proteins (360) identified in a “vacuole” fraction in S. cerevisiae12. The total number of V fraction proteins was also consistent the number of proteins (265) identified in endosomes isolated from macrophage and dendritic cells22 supporting the idea that our LC/MS/MS analysis generated a complete proteome of Aspergillus V fraction.
We previously demonstrated that a cAMP response element binding protein (CRE1bp) binds to many aflatoxin promoters and is associated with their activation during aflatoxin synthesis2,7. We also demonstrated that CRE1bp exhibits 99% identity to AtfB in Aspergillus orzyae. The bZIP transcription factor AtfB regulates development of conidiospores as well as their ability to respond to osmotic and oxidative stress. These observations prompt us to hypothesize that CRE1bp is a functional homolog of AtfB and that this transcription factor regulates activation of aflatoxin synthesis and stress response via interaction at cAMP response elements (CRE) sites present in the respective genes. Addressing this hypothesis represents an important focus area of future work23. We also speculate that AtfB and VeA play a broader role in regulation of endosome biogenesis, secondary metabolism, and stress response and that functional interaction of these cellular processes drive redox signaling within endosomes. Future studies therefore will focus on the role of secondary metabolism oxidases in generating a redox signal in endosomes and on the downstream impacts of redox signaling on cell development (asexual spore development).
Supplementary Material
Figure 2. Aflatoxin intermediate feeding experiments.
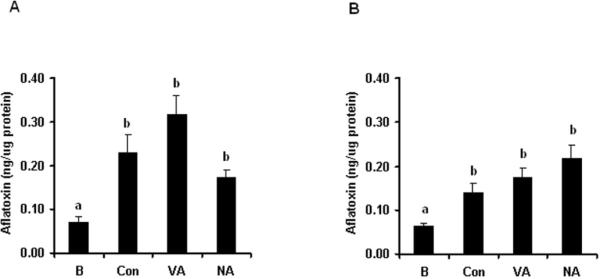
A. parasiticus SU-1 was cultured in 100 ml of YES liquid medium for 36 h under standard growth conditions and a vesicle/vacuole (V) fraction and a non-vesicle/vacuole (NV) fraction were purified. VA and NA were fed to the V or NV fraction for 16 h (see Methods). (A) Feeding VA or NA to V fraction. (B) Feeding VA or NA to NV fraction. Bar designations on graphs: B, sample analyzed immediately without feeding and without incubation; Con, sample without feeding and incubated for 16 h; VA, NA, samples fed with versicolorin A or norsolorinic acid, respectively and incubated for 16 h. Aflatoxin concentration was measured in triplicate in each experiment. Feeding experiments of V and NV fractions were repeated 3 times with similar trends. Statistical analysis was performed using Student's t-test. Bars labeled with different lower case letters specify a statistically significant difference between two measurements. Similar lower case letters indicate no statistically significant difference between two measurements. P values (compared to B sample) are as follows: Con (P=0.020), VA (P=0.006), NA (P=0.007) in panel A; Con (P=0.015), VA (P=0.009), NA (P=0.008) in panel B.
Acknowledgment
This work was supported by a grant from NIH (NCI) R01 CA52003-20, Michigan AgBio Research (formerly known as the Agricultural Experiment Station), and the Internal Research Grants Program (MSU).
Footnotes
Supporting information available. This material is available free of charge via the Internet at http://pubs.acs.org.
References
- 1.Miller MJ, Linz JE. Genetic mechanisms involved in regulation of mycotoxin biosynthesis. In: Shetty K, editor. Food Biotechnology. Second Edition. Taylor and Francis Group LLC; Boca Raton FL: 2006. Chapter 13. [Google Scholar]
- 2.Chanda A, Roze LV, Linz JE. A possible role for exocytosis in aflatoxin export in Aspergillus parasiticus. Eukaryot. Cell. 2010;11:1724–1727. doi: 10.1128/EC.00118-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Cullen JM, Newberne PM. Acute hepatotoxity of aflatoxins. In: Eaton DL, Groopman JD, editors. The toxicology of aflatoxins; human health, veterinary, and agricultural significance. Academic Press; San Diego CA: 1994. pp. 3–21. [Google Scholar]
- 4.Georgianna DR, Fedorova ND, Burroughs JL, Dolezal AL, Bok JW, Horowitz-Brown S, Woloshuk CP, Yu J, Keller NP, Payne G. Beyond aflatoxin; four distinct expression patterns and functional roles associated with Aspergillus flavus secondary metabolism gene clusters. Mol. Plant Pathol. 2010;11:213–226. doi: 10.1111/j.1364-3703.2009.00594.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chanda A, Roze LV, Kang S, Artymovich KA, Hicks GR, Raikel N. A key role for vesicles in fungal secondary metabolism. PNAS. 2009;106:19533–19538. doi: 10.1073/pnas.0907416106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chanda A, Roze LV, Pastor A, Frame MK, Linz JE. Purification of a vesiclevacuole fraction functionally linked to aflatoxin synthesis in Aspergillus parasiticus. J Microbiol. Methods. 2009;78:28–33. doi: 10.1016/j.mimet.2009.03.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Roze LV, Miller MJ, Rarick M, Mahanti N, Linz JE. A novel cAMP-response element, CRE1, modulates expression of nor-1 in Aspergillus parasiticus. J. Biol. Chem. 2004;279:27428–27439. doi: 10.1074/jbc.M400075200. [DOI] [PubMed] [Google Scholar]
- 8.Roze LV, Arthur AE, Hong SY, Chanda A, Linz JE. The initiation and pattern of spread of histone H4 acetylation parallel the order of transcriptional activation of genes in the aflatoxin cluster. Mol. Microbiol. 2007;66:713–726. doi: 10.1111/j.1365-2958.2007.05952.x. [DOI] [PubMed] [Google Scholar]
- 9.Hong SY, Linz JE. Functional expression and sub-cellular localization of the aflatoxin pathway enzyme Ver-1 fused to enhanced green fluorescent protein. Appl. Environ. Microbiol. 2008;74:6385–6396. doi: 10.1128/AEM.01185-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hong SY, Linz JE. Functional expression and sub-cellular localization of the early aflatoxin pathway enzyme Nor-1 in Aspergillus parasiticus. Mycol. Res. 2009;113:591–601. doi: 10.1016/j.mycres.2009.01.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wiederhold E, Gandhi T, Permentier HP, Breitling R, Poolman B, Slotboom DJ. The yeast vacuolar membrane proteome. Mol. Cellular Proteomics. 2009;8:380–392. doi: 10.1074/mcp.M800372-MCP200. [DOI] [PubMed] [Google Scholar]
- 12.Sarry J-E, Chen S, Collum RP, Liang S, Peng M, Lang A, Naumann B, Dzierszinski F, Yuan C-X, Hippler M, Rea P. Analysis of the vacuolar luminal proteome of Saccharomyces cerevisiae. FEBS J. 2007;274:4287–4305. doi: 10.1111/j.1742-4658.2007.05959.x. [DOI] [PubMed] [Google Scholar]
- 13.Klionsky DJ, Herman PK, Emr SD. The fungal vacuole - composition, function, and biogenesis. Microbiol. Rev. 1990;54:266–292. doi: 10.1128/mr.54.3.266-292.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Watanabe CM, Wilson D, Linz JE, Townsend CA. Demonstration of the catalytic roles and evidence for the physical association of type I fatty acid synthases and a polyketide synthase in the biosynthesis of aflatoxin B1. Chem. Biol. 1996;3:463–469. doi: 10.1016/s1074-5521(96)90094-0. [DOI] [PubMed] [Google Scholar]
- 15.Klionsky DJ. Nonclassical protein sorting to the yeast vacuole. J. Biol. Chem. 1998;273:10807–10810. doi: 10.1074/jbc.273.18.10807. [DOI] [PubMed] [Google Scholar]
- 16.Oda M, Scott S, Hefner-Gravnik A, Caffarelli A, Klionsky DJ. Identification of a cytoplasm to vacuole targeting determinant in aminopeptidase I. J. Cell Biol. 1996;132:999–1010. doi: 10.1083/jcb.132.6.999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Read ND, Kalkman ER. Does endocytosis occur in fungal hyphae? Fungal Genet. Biol. 2003;39:199–203. doi: 10.1016/s1087-1845(03)00045-8. [DOI] [PubMed] [Google Scholar]
- 18.Higuchi Y, Nakahama T, Shoji J-Y, Arioka M, Kitamoto K. Visualization of the endocytic pathway in the filamentous fungus Aspergillus oryzae using an EGFP-fused plasma membrane proein. Biochem. Biosphys. Res. Commun. 2006;340:784–791. doi: 10.1016/j.bbrc.2005.12.077. [DOI] [PubMed] [Google Scholar]
- 19.Cebollero E, Reggiori F. Regulation of autophagy in yeast Saccharomyces cerevisiae. Biochim. Biophys. Acta. 2009;1793:1413–1421. doi: 10.1016/j.bbamcr.2009.01.008. [DOI] [PubMed] [Google Scholar]
- 20.Yorimitsu T, Klionsky DJ. Autophagy: molecular machinery for self-eating. Cell Death and Differentiation. 2005;12:1542–1552. doi: 10.1038/sj.cdd.4401765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Cai H, Reinisch K, Ferro-Novik S. Coats, tethers, Rabs, and SNAREs work together to mediate the intracellular destination of a transport vesicle. Develop. Cell. 2007;12:671–682. doi: 10.1016/j.devcel.2007.04.005. [DOI] [PubMed] [Google Scholar]
- 22.Duclos S, Clavarino G, Rousserie G, Goyette G, Boulais J, Camosetto V, Gatti E, LaBoissiere S, Desjardins M. The endosomal proteome of macrophage and dendritic cells. Proteomics. 2011;11:854–864. doi: 10.1002/pmic.201000577. [DOI] [PubMed] [Google Scholar]
- 23.Roze LV, Chanda A, Linz JE. Compartmentalization and molecular traffic in secondary metabolisms: a new understanding of established cellular processes. Fungal Genet. Biol. 2011;48:35–48. doi: 10.1016/j.fgb.2010.05.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.