Abstract
Homologous recombination (HR) facilitates accurate repair of DNA double strand breaks (DSBs) during S and G2 phases of the cell cycle by using intact sister chromatids as sequence templates. HR capacity is maximized in S and G2 by Cyclin–Dependent Kinase (CDK) phosphorylation of CtIP, which subsequently interacts with BRCA1 and the Mre11–Rad50–NBS1 (MRN) complex. Here we show that Mre11 controls these events through a direct interaction with CDK2 that is required for CtIP phosphorylation and BRCA1 interaction in normally dividing cells. CDK2 binds the C–terminus of Mre11, which is absent in an inherited allele causing Ataxia–Telangiectasia Like Disorder. This newly uncovered role for Mre11 does not require ATM activation or nuclease activities. Therefore, functions of MRN are not restricted to DNA damage responses, but include regulating HR capacity during the normal mammalian cell cycle.
Introduction
Maintenance of genome stability is of crucial importance to cellular and organismal survival. Double strand breaks (DSBs) are highly toxic legions that can lead to loss or amplification of genetic information, chromosomal translocations, neoplastic transformation and cell death1. To counteract these lesions and mitigate their consequences, cells activate a complex network of repair and signaling pathways, collectively known as the DNA damage response (DDR)2. The DDR coordinates diverse processes such as cell cycle checkpoint signaling cascades, localized chromatin modifications, and functions of multiprotein DNA repair complexes. Individuals born with a defective DDR have syndromes with diverse sequelae including cancer predisposition, neurodegeneration, and immunodeficiency3.
The homologous recombination (HR) pathway facilitates highly accurate DSB repair by using homologous sequences on the sister chromatid as a replication template during repair4. Because of the need for a sister chromatid to be present, HR has a limited role in G1 and is the predominant pathway during S and G2 phases of the cell cycle. Recent evidence supports the notion that this is achieved through active control of the capacity to catalyze HR throughout the cell cycle. This active control takes place at the initiation step of HR, which entails nucleolytic resection of the DSB ends to generate single stranded DNA with 3' termini5. These termini subsequently serve to prime replication after strand invasion of the intact homologous duplex4.
In mammals, resection to initiate HR depends upon the Mre11–Rad50–NBS1 (MRN) complex6–9. MRN is a versatile protein complex that plays multiple roles in the DDR, including direct functions in repair as well as initiation of signaling cascades10. The core of MRN consists of a highly conserved Mre11–Rad50 heterotetramer which binds one or both sides of the DSB11,12. Once bound, coiled–coil arms of Rad50 stabilize the break over long distances13, followed by close range stabilization by an Mre11 dimer12. Within this dimer, Mre11 provides nuclease activities required to initiate resection6,12,14. The less conserved NBS1 subunit interacts with the ATM kinase, considered the primary signal transducer of the DDR15. Upon binding a DSB, structural alterations are transmitted through the MRN complex, leading to activation of ATM16–18.
Although required, the nuclease activities of Mre11 alone are not sufficient for resection. This requires the BRCA1 tumor suppressor and the CtIP protein, both bound to MRN9,19,20. CtIP shares a limited region of homology with the Sae2 nuclease in Saccharomyces cerevisiae9, but unlike Sae2 CtIP has not yet been shown to possess nuclease activities. Rather, CtIP interaction appears to be required to enhance Mre11 nuclease activity9,21. For efficient resection in vivo, CtIP must interact with BRCA119,20. It is the formation and dissolution of the MRN–CtIP–BRCA1 complex that provides the regulation of resection capacity (and thus HR capacity) throughout the cell cycle19,20. This complex is only found in S/G220.
In mammals, CtIP is the primary factor responsible for cell cycle regulation of resection19,20,22. This is accomplished by two types of cell phase specific posttranslational modifications of CtIP. First, CtIP protein levels are suppressed in G1 by proteasome mediated degradation, which is alleviated as cells enter S phase thereby permitting a rise in protein levels23. Second, CtIP is a substrate for S phase specific cyclin dependent kinase (CDK) activity19,22,24. This phosphorylation is required for assembly of the MRN–CtIP–BRCA1 resection complex20,22.
Phosphorylation of CtIP by S phase CDK provides a direct linkage between DNA resection capacity and the core cell cycle machinery25. The CDKs are a family of ser/thr protein kinases that are regulated by binding to their cyclin partners, in addition to posttranslational modifications and other interactions26. CtIP contains two well conserved CDK phosphorylation sites. Ser327 is phosphorylated as cells enter S phase and is necessary for interaction with BRCA119,20,24, and Thr847 is located in the Sae2 homology region and necessary for localization of CtIP to DSBs9,22,27. Phosphomimetic modification of the Thr847 site leads to hyperactivation of resection and increased genome instability22. In mammals, the primary active CDK in S phase is CDK2 bound to cyclin A26. Consistent with a role in controlling CtIP, deficiency of CDK2 in mammalian cells has been shown to confer a defect in HR28,29.
It is now apparent that eukaryotes have two overall strategies for managing the threat of DSBs. These are represented by the canonical DDR after damage occurs, and also by the regulation of repair capacity via interfacing the DNA repair and core cell cycle machineries. While the DDR itself has been studied extensively, comparatively little is known about CDK–dependent regulation of the DSB repair machinery. Hence, what factors govern preparation for damage and their relationship to DNA damage responses represents questions of fundamental importance. To gain understanding of the regulation of HR capacity and how CDK activity is involved, we investigated the relationship between CtIP and the MRN complex. We have discovered that the Mre11 component of MRN has a crucial role in regulating resection and HR capacity independent of previously characterized roles in the DDR.
Results
CtIP protein levels are reduced in Mre11 deficient cells
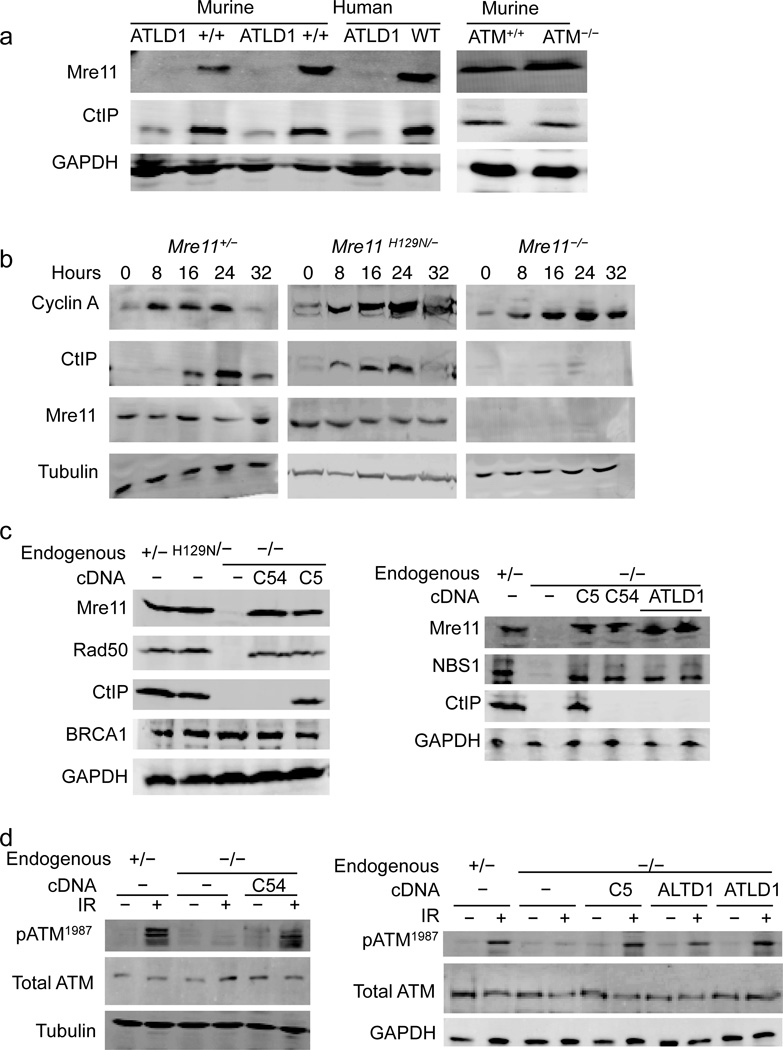
Ataxia telangiectasia like disorder (ATLD) features cerebellar degeneration leading to ataxia, as well as genomic instability and hypersensitivity to agents that cause DSBs. This syndrome results from inherited recessive hypomorphic alleles of Mre1130. Mre11ATLD1 is a nonsense mutation that truncates 78 amino acids from the C–terminus and causes very low levels of all three MRN components30,31. Given the relationship between MRN and CtIP, we determined if CtIP levels are impacted in this disorder. Indeed, CtIP levels are depleted in cells derived from a human patient or mouse model expressing only Mre11ATLD1 (Fig. 1a). Ataxia telangiectasia is a disorder similar to ATLD, and results from inherited mutation in the ATM gene32. Despite similar clinical outcomes and cellular phenotypes of the disorders, we find that ATM deficiency does not impact CtIP levels (Fig. 1a).
Figure 1. The MRN complex controls CtIP protein levels in mammals.
(a–d) Western blot analyses with primary antibodies indicated at left and genotype of cells at top. GAPDH or tubulin used as protein loading controls.
(a) Comparison of CtIP levels. (left) Murine cells are B lymphocyte lines from two Mre11ATLD1/ATLD1 and two Mre11+/+ littermate mice. Human cells are immortalized fibroblasts from an ATLD1 patient, and these cells complemented with human Mre11 cDNA (WT). (Right) ATM knockout (ATM−/−) and control (ATM+/+) murine embryonic fibroblasts (MEFs).
(b–d) Analyses of MEFs with endogenous Mre11 alleles as follows; wild–type (Mre11+), null (Mre11−), and nuclease deficient (Mre11H129N).
(b) CtIP deficiency in Mre11−/− cells is observed in the cyclin A positive population (S/G2 phase). MEFs were synchronized at G0/G1 and released from serum starvation for the indicated time (hours).
(c–d) Mre11 cDNA alleles expressed in Mre11−/− cells are as follows; empty expression vector (−), full length wild–type fused to a C–terminal tag of either 54 (C54) or 5 (C5) amino acids, or the ATLD1 78 amino acid C-terminal deletion (ATLD1).
(c) (left) A 54 amino acid C-terminal tag on Mre11 causes CtIP deficiency. (right) A 78 amino acid C-terminal deletion of Mre11 causes CtIP deficiency.
(d) The 54 amino acid C-terminal tag (left) or 78 amino acid C-terminal deletion (right) does not prevent ATM activation (ser1987 autophosphorylation) induced by 10 Gy ionizing radiation (+).
To further examine the MRN-CtIP relationship, we used murine cells harboring a Cre/LoxP conditional allele of Mre11 (Mre11cond/−) in which RAD50 and NBS1 protein levels are reduced when Mre11 is rendered deficient (Mre11−/−) (Supplemental Fig. 1)6. In MRN deficient cells (Mre11−/−) CtIP protein levels were markedly reduced (Supplemental Fig. 2). In contrast, BRCA1 levels were unaffected. CtIP levels were also assessed in murine cells expressing Mre11H129N (Mre11H129N/−) (Supplemental Fig. 1), a targeted germline missense mutation causing defective Mre11 nuclease activities while preserving integrity of the MRN complex6. No effect on CtIP levels was observed in these cells (Supplemental Fig 2).
The tight regulation of CtIP throughout the cell cycle raises the possibility that low CtIP levels could result from populations of cells arrested outside of S/G2 phase. However, prior work on cells harboring the Mre11−/− murine knockout or a variety of other spontaneous or engineered MRN mutations has demonstrated that MRN deficiency does not cause a significant arrest during the cell cycle6,7,31,33,34. Rather, these cells proliferate more slowly than control, but all phases of the cell cycle are equally represented. To further address this concern, we examined CtIP levels in cells synchronized by release from serum starvation. Cell cycle progression was tracked by the appearance of cyclin A, which is specific to S/G2. Upon release, cyclin A became readily detectable in control (Mre11+/−), MRN deficient (Mre11−/−), and Mre11 nuclease deficient (Mre11H129N/−) populations (Fig. 1b). CtIP was clearly deficient in Mre11−/− cells throughout the time course, including points with maximal cyclin A levels (S/G2 populations)(Fig. 1b). Thus, in mammals CtIP levels appear to be under control of the MRN complex. This hitherto unknown role for MRN is independent of Mre11 nuclease activities and the ATM kinase.
CtIP protein levels depend on the Mre11 C-terminus
We have engineered a system in which full length Mre11 cDNA is stably expressed from an integrated plasmid vector in cells that are Mre11cond/−. Endogenous Mre11 is subsequently removed via Cre-mediated conversion of Mre11cond/− to Mre11−/− and independent clones with Mre11 levels similar to endogenous are pursued for further study (Supplemental Fig. 3). Mre11 expressed from cDNA successfully reconstituted levels of Rad50 and NBS1 (Fig. 1c) and supported the ability of MRN to activate the ATM kinase as determined by relative levels of ATM autophosphorylation (Fig. 1d)15,35. Surprisingly, despite the apparent restoration of MRN, CtIP protein levels were not restored (Fig. 1c). This unexpected observation suggests the mechanisms by which Mre11 influences CtIP levels versus RAD50 or NBS1 levels are distinct.
The expression system fused 54 amino acids to the Mre11 C-terminus (hereafter termed Mre11C54), originating from three tags (his–V5–his). To test the possibility that this large addition prevents CtIP restoration, we reconstructed the plasmid to express Mre11 fused only to a short 5 amino acid histidine tag (Mre11C5). Indeed, Mre11C5 expression restored CtIP (Fig. 1c). Thus, we hypothesized that the C-terminus of Mre11 possesses an unknown function that controls CtIP levels. We therefore examined the impact of Mre11ATLD1 C-terminal deletion. Existing ATLD1 cell lines cannot be used to study specific roles of the Mre11 C-terminus since they also harbor low levels of the entire MRN complex30,31. To circumvent this limitation we stably expressed Mre11ATLD1 from cDNA in Mre11−/− cells (Fig. 1c). When expressed to approximately endogenous levels, Mre11ATLD1 showed a pattern identical to the impact of Mre11C54; restoration of NBS1 and Rad50 levels and ATM activation without restoration of CtIP (Fig. 1c and d). Thus, the C-terminus of Mre11 possesses a distinct function required to maintain normal CtIP levels.
Mre11 controls CtIP phosphorylation status
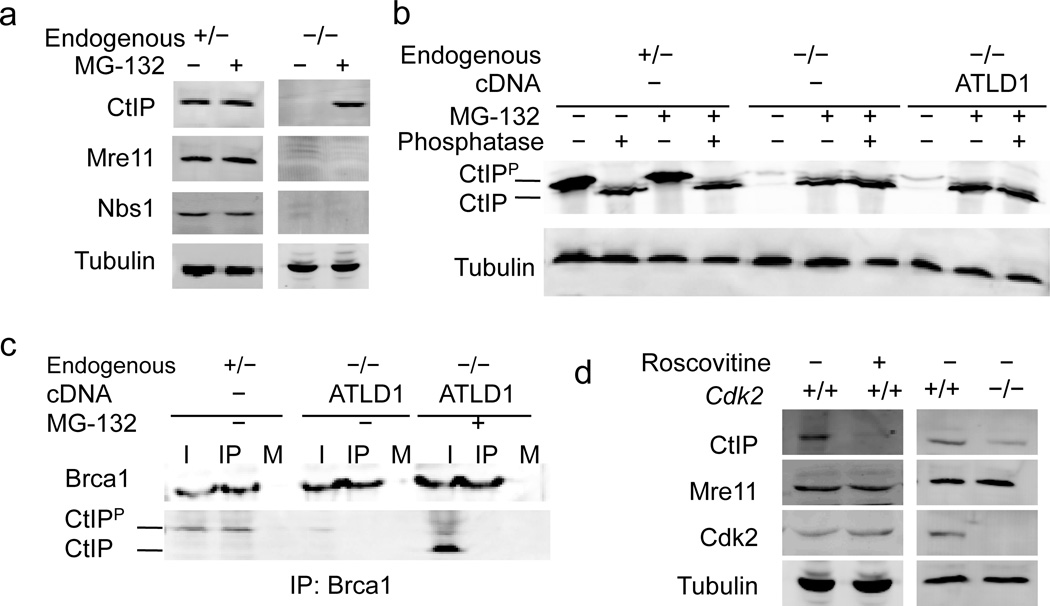
The dependence of CtIP on MRN might simply reflect the need for proper interactions to maintain protein levels, as is presumed to be the case for the mutual dependence among the three MRN components6,36. However, the cell cycle regulation of CtIP levels is distinct from MRN, and raises the specter of a more complex relationship. CtIP levels are suppressed in G1 through proteasome–mediated degradation23, hence we explored whether proteasome inhibition could reveal aspects of the CtIP–MRN relationship. We treated cells with the proteasome inhibitor MG–132, and observed that CtIP levels were restored in Mre11−/− cells (Fig 2a). Importantly, proteasome inhibition did not restore Nbs1 levels (Fig. 2a). Thus, physical association with MRN is not required to maintain CtIP levels.
Figure 2. Mre11 controls CtIP phosphorylation in normally dividing cells.
(a–d) Western blot analyses with primary antibodies indicated at left. Tubulin used as protein loading control. Endogenous and cDNA Mre11 alleles (top) are as described in the text and figure 1 legend.
(a) Proteasome inhibition restores CtIP levels in absence of MRN. MEFs were untreated (−) or treated (+) with MG–132 for 8 hrs.
(b) CtIP phosphorylation depends on the Mre11 C-terminus. MEFs of the indicated genotypes (top) were untreated (−) or treated (+) with MG–132 for 8 hours. Extracts were subsequently untreated (−) or treated (+) with lambda phosphatase. Phosphorylated (CtIPP) and hypophosphorylated (CtIP) forms of CtIP on the Western blot are indicated (left).
(c) BRCA1-CtIP complex formation depends on the Mre11 C-terminus. Western blot of co-immunoprecipitates using α–BRCA1 antibody (IP) or beads only (M). 3% of the lysate is shown for comparison (I). Phosphorylated (CtIPP) and hypophosphorylated (CtIP) forms of CtIP are indicated (left).
(d) CtIP levels are influenced by CDK2. Wild–type (+/+) or CDK2 knockout (−/−) MEFs were untreated (−) or treated with 10uM of the CDK inhibitor roscovitine (+) for 48 hours.
The return of CtIP to measurable levels by proteasome inhibition permitted further characterization of the protein in the absence of the Mre11 C-terminus. We utilized phage lambda phosphatase to analyze the phosphorylation status of CtIP in normally dividing cells. In control cells most detectable CtIP is phosphorylated, and proteasome inhibition had no detectable effect (Fig. 2b). In contrast, CtIP was clearly hypophosphorylated in the absence of MRN (Mre11−/−) or of the Mre11 C-terminus (Mre11ATLD1). Because CDK dependent phosphorylation of CtIP is required for interaction with BRCA119,20,24, we performed co–immunoprecipitation (coIP) experiments to assess CtIP–BRCA1 interaction in cells lacking the Mre11 C-terminus. Indeed, BRCA1–CtIP interaction is disrupted when MRN is deficient (Mre11−/−) or in the context of Mre11ATLD1 (Fig. 2c).
We further postulated that CDK activity might influence overall CtIP protein levels. To this end we determined the impact of the CDK inhibitor roscovitine on CtIP levels. This compound caused a dramatic reduction in CtIP levels, but no impact on Mre11 (Fig. 2d). We also examined murine fibroblasts homozygous for a CDK2 knockout allele (CDK2−/−)37. In this case CtIP levels were modestly lowered relative to controls (Fig. 2d). The minimal impact of CDK2 deficiency is not surprising given the ability of CDK family members to substitute for others when absent38. The greater impact of roscovitine is consistent with this notion, as it inhibits the CDK family broadly and would thus reduce substitution by other CDKs39.
We conclude that the phosphorylation status of CtIP in unperturbed dividing cells is under control of the Mre11 C-terminus. This newly uncovered function appears to govern both CtIP protein levels and interaction with BRCA1.
Mre11 associates with CDK2
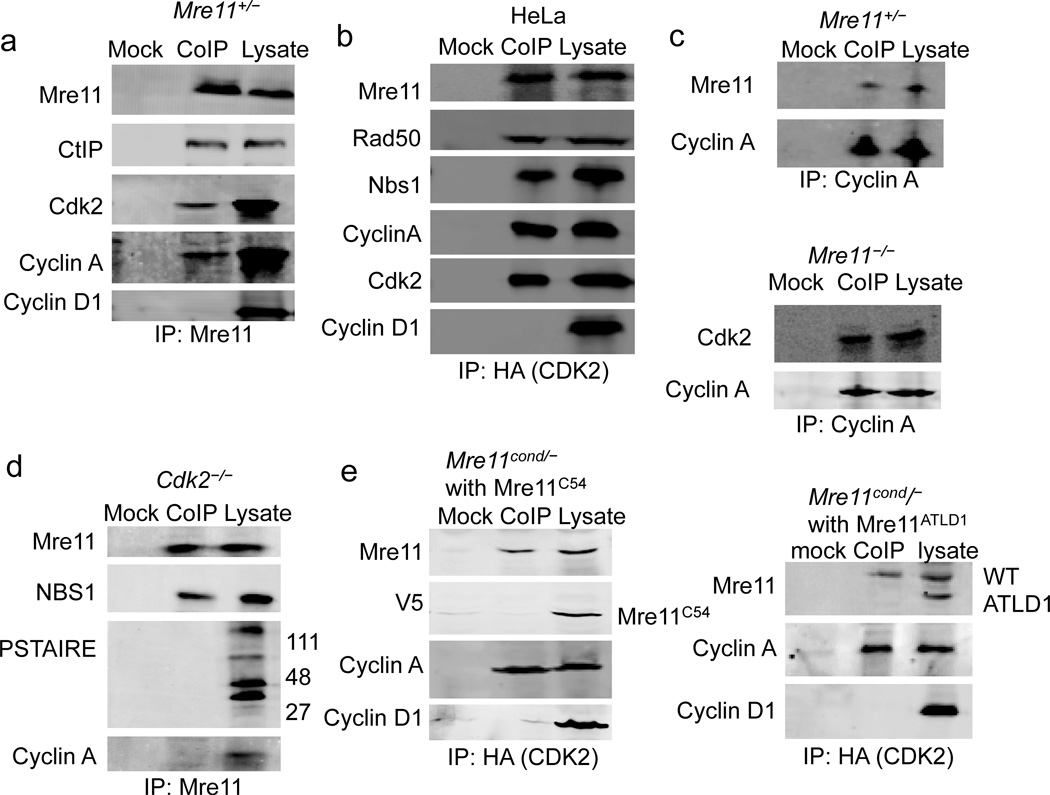
That CtIP levels are co–dependent on Mre11 and CDK2 suggests a functional relationship may exist among these factors. Hence, we performed a series of coIP experiments to explore the possibility that MRN and CDK2 function together in a multiprotein complex. First, endogenous Mre11 was immunoprecipitated from MEFs, and both CDK2 and cyclin A were observed to coIP (Fig 3a). To address the specificity of the coIPs and potential complexes, we blotted for cyclin D1, which is present in G1 and not known to associate with CDK2. Cyclin D1 was not detected in the coIP (Fig. 3a). Conversely, upon precipitation of HA-tagged CDK2 transiently expressed in human cells (HeLa), we detected Mre11, Rad50 and NBS1 (Fig. 3b). In addition, when cyclin A was precipitated, Mre11 was present in the coIP (Fig. 3c). Finally, in cells deficient for MRN (Mre11−/−), CDK2 maintained association with cyclin A (Fig 3c). Together these findings indicate that MRN associates in vivo with CDK2 and its S phase binding partner cyclin A.
Figure 3. Mre11 interacts with CDK2.
(a) Mre11 co-immunoprecipitates CDK2 and cyclin A. Western blot of coIPs from Mre11+/− MEFs, using α–Mre11 antibody (CoIP) or beads only (mock).
(b) CDK2 co–immunoprecipitates the MRN complex. Western blot analyses of coIPs from HeLa cells using α–HA antibody recognizing transiently expressed HA–tagged CDK2 (CoIP), or irrelevant α–GAPDH antibody control (mock).
(c) Cyclin A co–immunoprecipitates Mre11 from Mre11+/− MEFs (top). Cyclin A co–immunoprecipitates CDK2 from Mre11−/− MEFs (bottom).
(d) Mre11 does not co–immunoprecipitate CDKs other than CDK2. Western blot of co–IPs from CDK2−/− MEFs, using α–Mre11 antibody (CoIP) or beads only (mock).
(e) CoIP of Mre11 with CDK2/cyclin A requires an unperturbed Mre11 C–terminus. Western blot analyses of CoIPs using anti–HA antibody recognizing transiently expressed HA–tagged CDK2 (CoIP), or beads only control (mock). MEFs (Mre11cond/−) expressed endogenous wild–type Mre11 from the Mre11cond allele and cDNA encoding either Mre11C54 (left) or Mre11ATLD1 (right).
Next, endogenous Mre11 was immunoprecipitated from CDK2−/− MEFs to determine if other CDK family members associate with Mre11. While NBS1 was detected in the coIP, no CDKs were identified using a Western blot antibody to the highly conserved cyclin binding PSTAIR helix (Fig. 3d). In addition, cyclin A was not detected. Thus, our CoIPs appear to be quite specific, and support the notion that MRN does not associate globally with CDK–cyclins. We interpret the minimal impact on CtIP levels in CDK2−/− cells (Fig. 2d) to indicate that an alternative kinase acts without Mre11 interaction. However, we cannot rule out the possibility that weak interaction occurs below our detection limit, or that an uncharacterized kinase interacts that is not recognized by the PSTAIRE antibody.
Additional coIPs were performed to directly address the requirement for the Mre11 C–terminus. In this case, HA–tagged CDK2 was transiently expressed and precipitated from MEFs that expressed both wild–type endogenous Mre11 and either Mre11C54, or Mre11ATLD1 from cDNA (Fig. 3e). Whereas wild–type Mre11 was present in both coIPs, Mre11 with the large tag (Mre11C54) or C-terminal deletion (Mre11ATLD1) were absent. Thus, the in vivo association between CDK2–cyclin A and Mre11 requires an unperturbed Mre11 C-terminus.
Direct interaction between Mre11 and CDK2
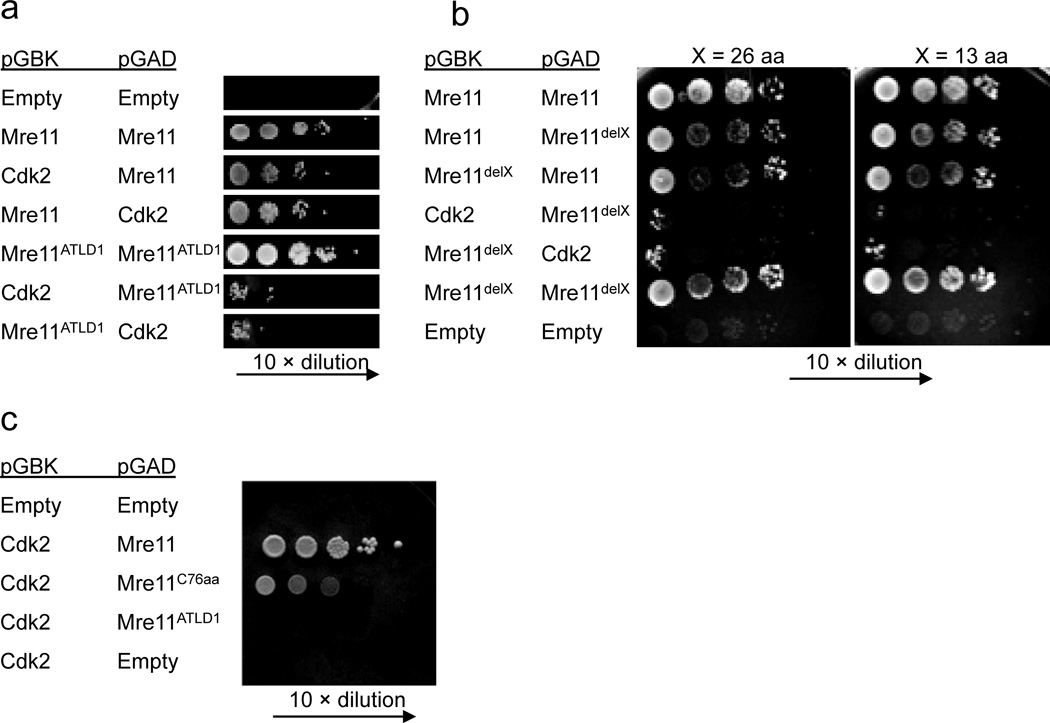
To determine if Mre11 and CDK2 interact directly we utilized the yeast two hybrid system40. When cloned into either configuration of bait and prey plasmid, Mre11 and CDK2 showed readily detectable interaction (Fig. 4a and Supplementary Fig. 4). Importantly, Mre11ATLD1 reduced the interaction with CDK2 to near background levels (Fig. 4a). In contrast, Mre11ATLD1 showed no reduction in the ability to homodimerize, indicating that the C-terminal deletion does not globally impact the Mre11 protein (Fig. 4a).
Figure 4. Direct interaction between Mre11 and CDK2.
(a–c) Yeast two hybrid analyses using pGBK (bait) and pGAD (prey) plasmids expressing the genes indicated (left). Mre11 dimerization is positive control. Empty vectors are negative control.
(a) Mre11 and CDK2 interact. The interaction requires the C-terminal 78 amino acids of Mre11 missing in Mre11ATLD1.
(b) Mre11–CDK2 interaction requires the very C-terminus of Mre11. delX (left) indicates deletion of 13 or 26 amino acids (top) from the Mre11 C–terminus.
(c) The C-terminus of Mre11 is sufficient for interaction with CDK2. C76aa indicates expression of the final 76 amino acids of Mre11.
We utilized this system to more precisely define Mre11 requirements, and found that deletion of the C-terminal 26 or 13 residues disrupted interaction (Fig. 4b). Furthermore, we addressed whether the Mre11 C-terminus alone is sufficient for interaction. In this case the C-terminal 76 amino acids of Mre11 (Mre11c76aa) was expressed in the yeast two hybrid system, along with CDK2. We found that the final 76 amino acids of Mre11 can interact with CDK2, although with lower apparent affinity than full length Mre11 (Fig. 4c).
These studies demonstrate that Mre11 and CDK2 interact directly. Interaction requires the final 13 amino acids of Mre11. Among mammals, this short sequence is well conserved (69% identity between human and mouse) and has a consensus sequence of PFM(N/S)o(S/N)ooRR(N/S)RR (where o denotes hydrophobic residues) (Supplementary Fig. 5).
CtIP DSB repair functions depend on the Mre11 C–terminus
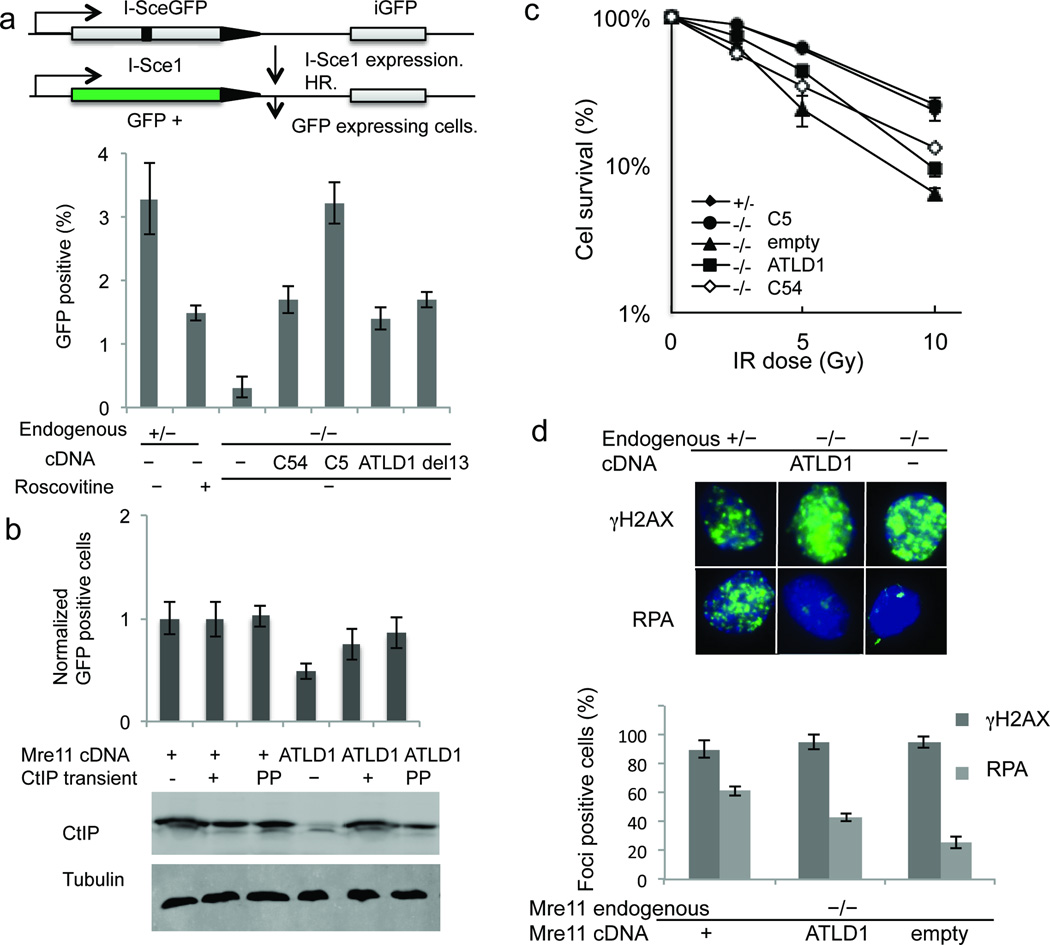
Our findings predict that through control of CtIP levels, the C-terminus of Mre11 regulates the ability of cells to repair DSBs by HR. We therefore determined the impact on DSB repair of Mre11 alleles that disrupt CDK2 interaction and cause low CTIP levels. First, we examined repair of a chromosomal DSB by the HR pathway using a single integrated copy of the DR-GFP reporter plasmid41. This construct contains a GFP gene inactivated by insertion of the I-SceI endonuclease recognition site (Fig. 5a). GFP is reconstructed only after the I-SceI induced DSB is repaired by HR, during which an adjacent GFP fragment is used as a template to restore missing sequences. We observed that Mre11C54 and Mre11ATLD1 each caused significant defects relative to control (p<.05 for both) (Fig. 5a). Even the short 13 amino acid deletion caused a similar defect (p<.05 for both) (Fig. 5a).
Figure 5. CtIP DSB repair functions are dependent on the Mre11 C-terminus.
(a) Schematic of DR-GFP assay for DSB repair by HR (top). Comparison of HR repair in MEFS of the indicated Mre11 genotypes (bottom). Error bars are average ± s.d. of at least 3 experiments.
(b) Complementation of HR deficiency by transient expression of CtIP (top). All MEFs were Mre11−/− at the endogenous locus. cDNAs expressed are indicated below the bar graph. Stable Mre11 expression was either wild–type (+) or ATLD1. Transient CtIP expression was either wild–type CtIP (+) or phosphomimetic mutant CtIP (pp). Western blot indicates levels of total CtIP protein (bottom). Tubulin is loading control.
(c) IR sensitivities of MEFS with Mre11 endogenous (left) and cDNA (right) genotypes indicated in the panel. Error bars are average ± s.d. of at least 3 experiments.
(d) Immunoflourescent foci induced by IR (10 Gy) (above). γH2AX (top) marks DSBs and RPA (bottom) indicates resection. Quantitation of foci is at bottom. Error bars are average ± s.d. of 3 experiments. Foci-positive is defined as cells with >10 foci.
To address the possibility that disruption of the Mre11 C-terminus causes an HR defect for reasons other than CtIP deficiency, we complemented CtIP via forced overexpression. We transfected Mre11WT or Mre11ATLD1 cells containing the DR–GFP reporter with exogenous murine CtIP using the pSport6–CMV expression plasmid. Indeed, CtIP expression caused a significant increase in HR in Mre11ATLD1 cells, but did not increase HR above normal in cells expressing wild–type Mre11 (p<.05) (Fig. 5b). While significant, the complementation by CtIP did not fully restore HR to wild–type levels. This might result from the forcibly expressed protein lacking appropriate post–translational modifications. We therefore expressed CtIP harboring established phosphomimetic mutations at two consensus CDK sites. Ser326 and Thr843 in mus musculus (corresponding to Ser327 and Thr847 in human) were changed to Glutamic acid (E) (CtIPpp)19,22. This construct complemented the recombination assay with greater apparent efficiency than did wild–type CtIP, although this difference was not statistically significant (p >.05). Side by side control transfections with a GFP expressing reporter indicate that ~40% of the cells can be transfected with plasmid (data not shown), thus the effectiveness of complementation is likely underestimated in these experiments. Therefore, the HR defect associated with disruption of the Mre11 C–terminus is a result of CtIP deficiency. However, a minor contribution by some other function of the Mre11 C-terminus cannot be entirely ruled out.
Next, we determined the impact of Mre11C54 and Mre11ATLD1 on ionizing radiation (IR) sensitivity to assess the overall ability to respond to DSBs. Both mutants showed significant defects relative to controls (p<.05) (Fig. 5c). Finally, to assess DSB resection capacity we examined DNA damage–induced immunoflourescent foci comprised of RPA, which coats single stranded DNA immediately following resection9. Consistent with a resection defect, there was a 30% reduction of IR induced RPA foci in Mre11ATLD1 cells (Fig. 5d).
Collectively, these studies support the notion that disruption of the Mre11 C-terminus causes a CtIP-dependent defect in DSB repair by HR, resulting from reduced capacity to resect DNA.
Discussion
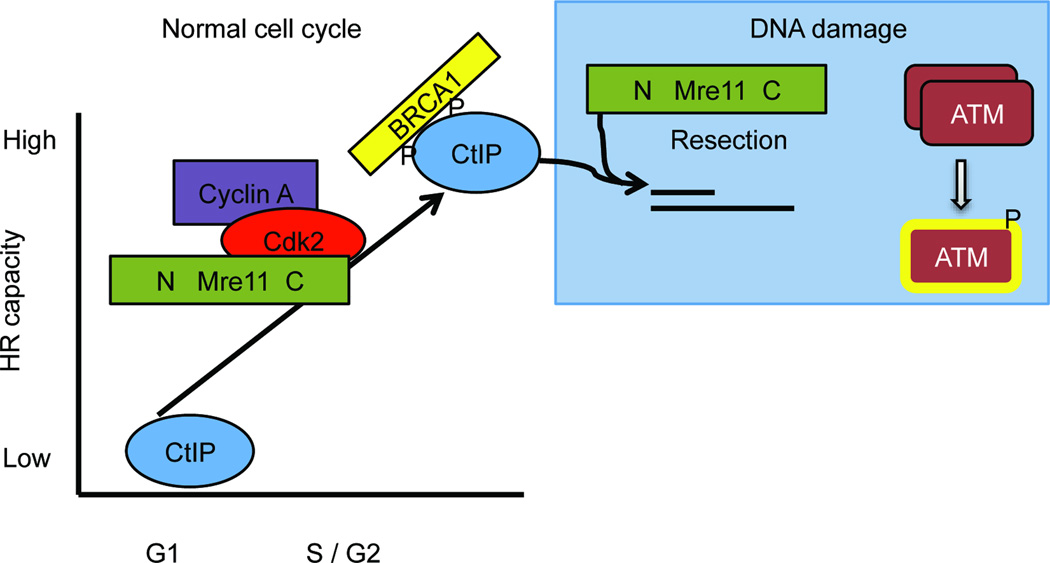
Here we have demonstrated that the C-terminus of Mre11 governs the phosphorylation status and overall levels of the CtIP protein, thereby controlling formation of the S/G2 phase MRN–CtIP–BRCA1 complex. We find that this same portion of Mre11 interacts directly with CDK2, forming a tripartite subcomplex with S/G2 specific cyclin A. Given that CtIP interactions in S/G2 require CDK activity19,20,22, our work supports the notion that interaction between Mre11 and CDK2–cyclin A facilitates phosphorylation of CtIP in normally dividing cells. This newly uncovered function of Mre11 serves to control the capacity of cells to initiate resection at DSBs, thereby restricting use of the HR pathway to phases of the cell cycle when sister chromatids are present (Fig. 6).
Figure 6. Model depicting control of HR capacity by Mre11.
Cells increase the capacity to repair DSBs by HR upon entry into S phase to take advantage of the presence of sister chromatids, which serve as replication templates during repair. The findings in this manuscript demonstrate that Mre11 controls this process by controlling signaling from the core cell cycle apparatus to the DSB repair machinery. This is accomplished by direct interaction between CDK2–cyclin A and the C-terminus of Mre11, which facilitates phosphorylation of CtIP leading to increased CtIP levels and interaction with BRCA1. This phosphorylation is regulated in the cell cycle by the requirement of CDK2 for S/G2-specific cyclin A. These events occur during the normal cell cycle and are distinct from the DNA damage response that is initiated upon recognition of DSBs by the MRN complex. (Rad50 and NBS1 are present with Mre11 during interaction with CDK2, but are not shown in the figure for simplicity).
Classically, MRN is considered a central player in the canonical DDR10. The complex binds rapidly to DNA ends and then engages in several downstream processes42,43. These include stabilizing the two sides of a DSB in close proximity and processing the ends via the nuclease activities of Mre116,12. These functions are encoded by the most highly conserved portions of Mre11 and Rad50, which are apparently present in all terrestrial life forms44. The third component is only found in eukaryotes and is less well conserved. This subunit has evolved as a flexible linker thought to transmit information from the Rad50–Mre11 core to binding partners such as the ATM kinase17,18. Remarkably, the newly uncovered function of the Mre11 C-terminus revealed in our studies appears to act independently of the previously characterized roles of MRN. Perturbations of the Mre11 C-terminus that disrupt interaction with CDK2 and cause CtIP deficiency did not prevent activation of the ATM kinase after exposure to ionizing radiation. This strongly suggests that DDR functions of MRN are intact, as ATM activation requires DSB recognition and conformational alterations by all three MRN components6,33,45. Like the newly uncovered function of the Mre11 C-terminus, the nuclease activities encoded by the Mre11 N-terminus (Supplementary Fig. 1) also act independently of ATM activation6. However, these two functions of Mre11 are separable, as evidenced by normal CtIP levels being present in cells expressing the nuclease-dead Mre11H129N.
The C-terminus of mammalian Mre11 represents a previously uncharacterized domain capable of binding CDK2. This region is conserved amongst mammals but shows no sequence homology to Mre11 of other evolutionary groups. Intriguingly, despite lack of sequence conservation, Mre11 from eukaryotes all contain a C-terminus extending several hundred amino acids beyond the highly conserved N terminal nuclease domains. Prokaryotes and archaea, which do not possess eukaryotic–like CDKs, lack an extended Mre11 C-terminus46,47. This parallels the evolutionary pattern of the third subunit (NBS1 in mammals), which also arose in eukaryotes and interacts directly with a kinase (ATM in mammals). At present there is no structural information available for the C-terminus of eukaryotic Mre11, nor does the sequence contain recognizable domains. We speculate that this portion of Mre11 is a relatively unstructured flexible tether with CDK2 binding capabilities. This would parallel the flexible tethers that have been proposed for NBS1 interactions, and are a common strategy for transient protein interactions that allow for varied and intricate conformational changes18. The precise structure of the mammalian Mre11–CDK2 interface and the extent to which Mre11 interacts with a CDK family member in other eukaryotes represent important questions for future studies.
Collectively, our findings demonstrate that through control of CtIP, Mre11 and CDK2 provide the molecular switch whereby mammalian cells induce the capacity to catalyze HR upon entry into S phase (Fig. 6). Given that CtIP interacts with NBS1, we postulate that the MRN complex serves as a bridge to facilitate the kinase-substrate relationship between CDK2 and CtIP. This occurs in unperturbed dividing cells and is distinct from the roles of MRN in response to DNA damage. Therefore this newly uncovered relationship between Mre11 and CDK2 represents a fundamental feature of the mammalian cell cycle.
Methods
MEFs harboring targeted Mre11 alleles (Mre11cond, Mre11− and Mre11H129N) were described previously6. Mouse B lymphocytes harboring Mre11 engineered alleles and CD19–Cre were as described48. CDK2−/− and control MEFS (Philipp Kaldis–IMCB Singapore)37. MEFs were derived from ATM+/− mouse intercrosses49. The following cells were provided as kind gifts: mouse ATLD1 and control Abelson transformed B lymphocyte lines50 (Barry Sleckman – Wash. U. St. Louis), human ATLD1 and Mre11–complemented fibroblast lines (Matthew Weitzman –Salk). Expression of mouse Mre11 cDNA in immortalized MEFs was achieved using pEF6b (Invitrogen) with blasticidin resistance selection for stable integrants. Mutagenesis was carried out with QuikChange II Mutagenesis kit (Stratagene). Cells exposed to 10µM MG–132 (Sigma Aldrich) were treated for 8 hours. Western blots and immunoprecipitations were performed via standard procedures using antibodies listed below. Yeast two hybrid analyses were performed with Matchmaker II System (Clonetech) using pGBKT7 and pGADT7 vectors and yeast strain Y190. P values were two tailed and calculated by unpaired student T test.
Western blots and antibodies
Cell extracts were prepared in Laemmli buffer (4% (w/v) SDS, 20% (w/v) glycerol, 120 mM Tris–HCl [pH 6.8]), resolved by SDS–PAGE (6, 8, 10, or 12% gel), and transferred via standard procedures. Only SDS–PAGE gels of <8% readily detect size difference of Mre11–54aa tag. SDS–PAGE gels for band shift of CtIP were run on 6% gel at 70 volts until a 75 kDa marker was run to bottom of a gel. Primary antibodies were the following: Mre11 (Cell Signaling)1:1000, NBS1 (Novus) 1:1000, Rad50 (Bethyl) 1:500, p–ATM Ser 1981(Rockland) 1:500, γ–tubulin (AbCam)1:2000, GAPDH (AbCam) 1:2000, CtIP (Santa cruz) 1:100, BRCA1 (Santa Cruz) 1:200, CDK2 (cell signaling) 1:1000, cyclin A (Santa Cruz) 1:100, cyclin D1 (Santa Cruz) 1:100, pSTAIRE (Santa Cruz) (1:100). Secondary antibodies for Western blots were IRDye800CW–conjugated goat anti–rabbit or anti–mouse (Li–Cor Biosciences) 1:5000.
Immunoprecipitations
Whole cell extracts were prepared from 10×106 cells lysed in buffer containing 20mM Tris (pH 7.5), 150mM NaCl, 1mM EDTA, 1mM EGTA, 1% (w/v) trition X–100, 2.5mM Sodium pyrophosphate, 1mM β–glycerphosphate, 1mM Na3VO4, 1mM Leutpeptin. Extracts were performed with 40µL protein G sepharose beads (GE healthcare) with 3 mg protein extract incubated with anti–Mre11 antibody (Cell Signaling) or anti–HA (Roche) for 16 hr at 4°C. Extracts were washed 5 times and Western blots performed as above. MEFs were transfected transiently with a cDNA construct using Liptofectamine 2000 (PKB722 Cdk2– HA in pBabe puro, BamHI) at 3:1.
IR sensitivity
300,000 cells were plated in 6 well dishes and treated with 0, 2.5, 5, 10 Gy of ionizing radiation, cells recovered for 72 hours and were counted via hemocytometer.
DR–GFP assays
Previously described MEFs6 harbored a single copy of the DR–GFP reporter41. I–SceI was transiently expressed via infection with adenovirus (AdNGUS24i), and recombination frequency was determined by percentage of GFP–positive cells. GFP fluorescence detection was carried out using an Accuri C6 Flow Cytometer with CFlow Software (Accuri Cytometers). Transient CtIP transfections were carried out using murine CtIP cDNA in the pSport6 CMV vector and Lipofectamine 2000 while simultaneously infecting with adenovirus.
Immunoflourescent foci
As previously described6, MEFs were grown on glass slides and treated with 10 Gy IR followed by 4 hours recovery. Cells were treated with permiabilizaton buffer and fixed in paraformaldehyde. Cells were incubated 1 hour in TBST containing 5% (w/v) bovine albumin serum followed overnight incubation with primary antibody; RPA (1 :50, Roche) and γ–H2AX (1:100, Upstate). Cells were washed and incubated with secondary antibody. (1:50, Sigma, FITC)
Preparation of Images
Images were cropped and, when appropriate, placed in composites with Adobe Photoshop Software (Adobe).
Supplementary Material
Acknowledgements
The authors thank Drs. Matthew Weitzman (Salk Institute), Philipp Kaldis (A*STAR, IMCB) and Barry Sleckman (Washington University) for providing cell lines, JoAnn Sekiguchi (University of Michigan) for providing ATM−/− mice, and Maria Jasin (Sloan–Kettering) for providing DR–GFP plasmid. We thank Drs. Tom Wilson, Xiaochun Yu, JoAnn Sekiguchi, Greg Dressler and Christine Canman for helpful discussions regarding the manuscript. Support for this work was provided by NIH R01–HL079118 (DOF), the Leukemia and Lymphoma Society (DOF), the University of Michigan Cancer Center Support Grant 5–P30–CA46592 (DOF), NIH F32–GM087073 (JB) and NIH T32–AI007413 (ES).
Footnotes
Author Contributions
JB planned and performed all experiments except two hybrid and B lymphocyte analyses, analyzed and interpreted data from all experiments, and participated in writing the manuscript. TS performed and interpreted two hybrid analyses. ES performed B lymphocyte western blot analysis from spleens of mice from complex breedings. DF participated in design of all experiments, analyses and interpretation of data, and writing of the manuscript.
Author Information
The authors declare no competing financial interests.
References
- 1.Jackson SP, Bartek J. The DNA-damage response in human biology and disease. Nature. 2009;461:1071–1078. doi: 10.1038/nature08467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Matsuoka S, et al. ATM and ATR substrate analysis reveals extensive protein networks responsive to DNA damage. Science. 2007;316:1160–1166. doi: 10.1126/science.1140321. [DOI] [PubMed] [Google Scholar]
- 3.McKinnon PJ, Caldecott KW. DNA strand break repair and human genetic disease. Annu Rev Genomics Hum Genet. 2007;8:37–55. doi: 10.1146/annurev.genom.7.080505.115648. [DOI] [PubMed] [Google Scholar]
- 4.San Filippo J, Sung P, Klein H. Mechanism of eukaryotic homologous recombination. Annu Rev Biochem. 2008;77:229–257. doi: 10.1146/annurev.biochem.77.061306.125255. [DOI] [PubMed] [Google Scholar]
- 5.Mimitou EP, Symington LS. DNA end resection-Unraveling the tail. DNA Repair (Amst) 2011;10:344–348. doi: 10.1016/j.dnarep.2010.12.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Buis J, et al. Mre11 nuclease activity has essential roles in DNA repair and genomic stability distinct from ATM activation. Cell. 2008;135:85–96. doi: 10.1016/j.cell.2008.08.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Yamaguchi-Iwai Y, et al. Mre11 is essential for the maintenance of chromosomal DNA in vertebrate cells. EMBO J. 1999;18:6619–6629. doi: 10.1093/emboj/18.23.6619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Takeda S, Nakamura K, Taniguchi Y, Paull TT. Ctp1/CtIP and the MRN complex collaborate in the initial steps of homologous recombination. Mol Cell. 2007;28:351–352. doi: 10.1016/j.molcel.2007.10.016. [DOI] [PubMed] [Google Scholar]
- 9.Sartori AA, et al. Human CtIP promotes DNA end resection. Nature. 2007;450:509–514. doi: 10.1038/nature06337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Williams GJ, Lees-Miller SP, Tainer JA. Mre11-Rad50-Nbs1 conformations and the control of sensing, signaling, and effector responses at DNA double-strand breaks. DNA Repair (Amst) 2010;9:1299–1306. doi: 10.1016/j.dnarep.2010.10.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hopfner KP, et al. Structural biochemistry and interaction architecture of the DNA double-strand break repair Mre11 nuclease and Rad50-ATPase. Cell. 2001;105:473–485. doi: 10.1016/s0092-8674(01)00335-x. [DOI] [PubMed] [Google Scholar]
- 12.Williams RS, et al. Mre11 dimers coordinate DNA end bridging and nuclease processing in double-strand-break repair. Cell. 2008;135:97–109. doi: 10.1016/j.cell.2008.08.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.de Jager M, et al. Human Rad50/Mre11 is a flexible complex that can tether DNA ends. Mol Cell. 2001;8:1129–1135. doi: 10.1016/s1097-2765(01)00381-1. [DOI] [PubMed] [Google Scholar]
- 14.Nimonkar AV, et al. BLM-DNA2-RPA-MRN and EXO1-BLM-RPA-MRN constitute two DNA end resection machineries for human DNA break repair. Genes Dev. 2011;25:350–362. doi: 10.1101/gad.2003811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lee JH, Paull TT. Direct activation of the ATM protein kinase by the Mre11/Rad50/Nbs1 complex. Science. 2004;304:93–96. doi: 10.1126/science.1091496. [DOI] [PubMed] [Google Scholar]
- 16.Lee JH, Paull TT. ATM activation by DNA double-strand breaks through the Mre11-Rad50-Nbs1 complex. Science. 2005;308:551–554. doi: 10.1126/science.1108297. [DOI] [PubMed] [Google Scholar]
- 17.Lloyd J, et al. A supramodular FHA/BRCT-repeat architecture mediates Nbs1 adaptor function in response to DNA damage. Cell. 2009;139:100–111. doi: 10.1016/j.cell.2009.07.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Williams RS, et al. Nbs1 flexibly tethers Ctp1 and Mre11-Rad50 to coordinate DNA double-strand break processing and repair. Cell. 2009;139:87–99. doi: 10.1016/j.cell.2009.07.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Yun MH, Hiom K. CtIP-BRCA1 modulates the choice of DNA double-strand-break repair pathway throughout the cell cycle. Nature. 2009;459:460–463. doi: 10.1038/nature07955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Chen L, Nievera CJ, Lee AY, Wu X. Cell cycle-dependent complex formation of BRCA1.CtIP.MRN is important for DNA double-strand break repair. J Biol Chem. 2008;283:7713–7720. doi: 10.1074/jbc.M710245200. [DOI] [PubMed] [Google Scholar]
- 21.You Z, et al. CtIP links DNA double-strand break sensing to resection. Mol Cell. 2009;36:954–969. doi: 10.1016/j.molcel.2009.12.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Huertas P, Jackson SP. Human CtIP mediates cell cycle control of DNA end resection and double strand break repair. J Biol Chem. 2009;284:9558–9565. doi: 10.1074/jbc.M808906200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Germani A, et al. SIAH-1 interacts with CtIP and promotes its degradation by the proteasome pathway. Oncogene. 2003;22:8845–8851. doi: 10.1038/sj.onc.1206994. [DOI] [PubMed] [Google Scholar]
- 24.Yu X, Chen J. DNA damage-induced cell cycle checkpoint control requires CtIP, a phosphorylation-dependent binding partner of BRCA1 C-terminal domains. Mol Cell Biol. 2004;24:9478–9486. doi: 10.1128/MCB.24.21.9478-9486.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Cerqueira A, et al. Overall Cdk activity modulates the DNA damage response in mammalian cells. J Cell Biol. 2009;187:773–780. doi: 10.1083/jcb.200903033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Satyanarayana A, Kaldis P. Mammalian cell-cycle regulation: several Cdks, numerous cyclins and diverse compensatory mechanisms. Oncogene. 2009;28:2925–2939. doi: 10.1038/onc.2009.170. [DOI] [PubMed] [Google Scholar]
- 27.Huertas P, Cortes-Ledesma F, Sartori AA, Aguilera A, Jackson SP. CDK targets Sae2 to control DNA-end resection and homologous recombination. Nature. 2008;455:689–692. doi: 10.1038/nature07215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Muller-Tidow C, et al. The cyclin A1-CDK2 complex regulates DNA double-strand break repair. Mol Cell Biol. 2004;24:8917–8928. doi: 10.1128/MCB.24.20.8917-8928.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Deans AJ, et al. Cyclin-dependent kinase 2 functions in normal DNA repair and is a therapeutic target in BRCA1-deficient cancers. Cancer Res. 2006;66:8219–8226. doi: 10.1158/0008-5472.CAN-05-3945. [DOI] [PubMed] [Google Scholar]
- 30.Stewart GS, et al. The DNA double-strand break repair gene hMRE11 is mutated in individuals with an ataxia-telangiectasia-like disorder. Cell. 1999;99:577–587. doi: 10.1016/s0092-8674(00)81547-0. [DOI] [PubMed] [Google Scholar]
- 31.Theunissen JW, et al. Checkpoint failure and chromosomal instability without lymphomagenesis in Mre11(ATLD1/ATLD1) mice. Mol Cell. 2003;12:1511–1523. doi: 10.1016/s1097-2765(03)00455-6. [DOI] [PubMed] [Google Scholar]
- 32.Frappart PO, McKinnon PJ. Ataxia-telangiectasia and related diseases. Neuromolecular Med. 2006;8:495–511. doi: 10.1385/NMM:8:4:495. [DOI] [PubMed] [Google Scholar]
- 33.Difilippantonio S, et al. Role of Nbs1 in the activation of the Atm kinase revealed in humanized mouse models. Nat Cell Biol. 2005;7:675–685. doi: 10.1038/ncb1270. [DOI] [PubMed] [Google Scholar]
- 34.Adelman CA, De S, Petrini JH. Rad50 is dispensable for the maintenance and viability of postmitotic tissues. Mol Cell Biol. 2009;29:483–492. doi: 10.1128/MCB.01525-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Bakkenist CJ, Kastan MB. DNA damage activates ATM through intermolecular autophosphorylation and dimer dissociation. Nature. 2003;421:499–506. doi: 10.1038/nature01368. [DOI] [PubMed] [Google Scholar]
- 36.Williams BR, et al. A murine model of Nijmegen breakage syndrome. Curr Biol. 2002;12:648–653. doi: 10.1016/s0960-9822(02)00763-7. [DOI] [PubMed] [Google Scholar]
- 37.Berthet C, Aleem E, Coppola V, Tessarollo L, Kaldis P. Cdk2 knockout mice are viable. Curr Biol. 2003;13:1775–1785. doi: 10.1016/j.cub.2003.09.024. [DOI] [PubMed] [Google Scholar]
- 38.Santamaria D, et al. Cdk1 is sufficient to drive the mammalian cell cycle. Nature. 2007;448:811–815. doi: 10.1038/nature06046. [DOI] [PubMed] [Google Scholar]
- 39.Meijer L, Raymond E. Roscovitine and other purines as kinase inhibitors. From starfish oocytes to clinical trials. Acc Chem Res. 2003;36:417–425. doi: 10.1021/ar0201198. [DOI] [PubMed] [Google Scholar]
- 40.Chien CT, Bartel PL, Sternglanz R, Fields S. The two-hybrid system: a method to identify and clone genes for proteins that interact with a protein of interest. Proc Natl Acad Sci U S A. 1991;88:9578–9582. doi: 10.1073/pnas.88.21.9578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Pierce AJ, Johnson RD, Thompson LH, Jasin M. XRCC3 promotes homology-directed repair of DNA damage in mammalian cells. Genes Dev. 1999;13:2633–2638. doi: 10.1101/gad.13.20.2633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Mirzoeva OK, Petrini JH. DNA damage-dependent nuclear dynamics of the Mre11 complex. Mol Cell Biol. 2001;21:281–288. doi: 10.1128/MCB.21.1.281-288.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Lisby M, Barlow JH, Burgess RC, Rothstein R. Choreography of the DNA damage response: spatiotemporal relationships among checkpoint and repair proteins. Cell. 2004;118:699–713. doi: 10.1016/j.cell.2004.08.015. [DOI] [PubMed] [Google Scholar]
- 44.Rupnik A, Lowndes NF, Grenon M. MRN and the race to the break. Chromosoma. 2010;119:115–135. doi: 10.1007/s00412-009-0242-4. [DOI] [PubMed] [Google Scholar]
- 45.Morales M, et al. The Rad50S allele promotes ATM-dependent DNA damage responses and suppresses ATM deficiency: implications for the Mre11 complex as a DNA damage sensor. Genes Dev. 2005;19:3043–3054. doi: 10.1101/gad.1373705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Hopfner KP, et al. Mre11 and Rad50 from Pyrococcus furiosus: cloning and biochemical characterization reveal an evolutionarily conserved multiprotein machine. J Bacteriol. 2000;182:6036–6041. doi: 10.1128/jb.182.21.6036-6041.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Cromie GA, Leach DR. Recombinational repair of chromosomal DNA double-strand breaks generated by a restriction endonuclease. Mol Microbiol. 2001;41:873–883. doi: 10.1046/j.1365-2958.2001.02553.x. [DOI] [PubMed] [Google Scholar]
Methods-only references
- 48.Dinkelmann M, et al. Multiple functions of MRN in end-joining pathways during isotype class switching. Nat Struct Mol Biol. 2009;16:808–813. doi: 10.1038/nsmb.1639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Zha S, Sekiguchi J, Brush JW, Bassing CH, Alt FW. Complementary functions of ATM and H2AX in development and suppression of genomic instability. Proc Natl Acad Sci U S A. 2008;105:9302–9306. doi: 10.1073/pnas.0803520105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Helmink BA, et al. MRN complex function in the repair of chromosomal Rag-mediated DNA double-strand breaks. J Exp Med. 2009;206:669–679. doi: 10.1084/jem.20081326. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.