Abstract
Processivity, the ability of single molecules to move continuously along a track, is a fundamental requirement of cargo-transporting molecular motors. Here, we investigate how cytoplasmic dynein, a homodimeric, microtubule-based motor, achieves processive motion. To do this we developed a versatile method for assembling Saccharomyces cerevisiae dynein heterodimers using complementary DNA oligonucleotides covalently linked to dynein monomers labeled with different organic fluorophores. Using two-color, single-molecule microscopy and high-precision, two-dimensional tracking, we find dynein has a highly variable stepping pattern that is distinct from all other processive cytoskeletal motors, which use “hand-over-hand” mechanisms. Uniquely, dynein stepping is stochastic when its two motor domains are close together. However, coordination emerges as the distance between motor domains increases, implying a tension-based mechanism governs these steps. This plasticity may allow tuning of dynein for the diversity of cellular functions it performs.
Introduction
The microtubule-based motor cytoplasmic dynein (referred to here as dynein) powers the transport of a diverse array of cargos, allowing cells to organize their contents, move, divide, and respond to stimuli. Neurons and other long cells are especially sensitive to defects in transport; mutations in dynein motor-associated subunits lead to neurodevelopmental and neurodegenerative diseases1,2. Like other motors that move cargo over long distances, single dynein molecules move processively along their microtubule track3-8. Dynein is the last class of cytoskeletal motor for which the mechanism of processive motility remains unknown.
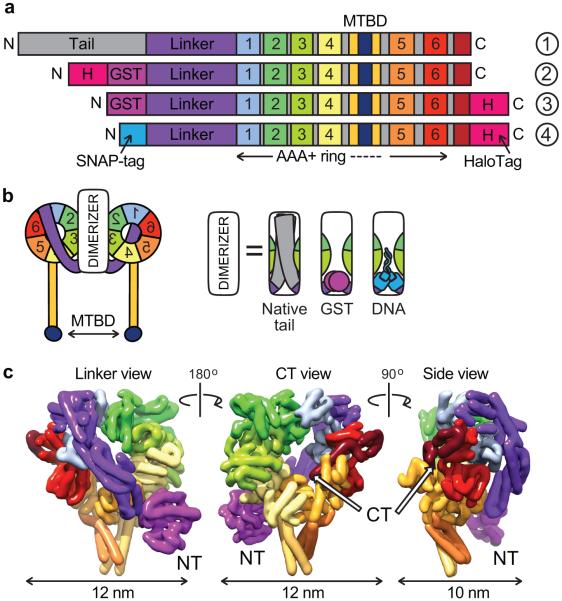
Dynein’s mechanism continues to be mysterious due to its enormous size and complexity9. The dynein holoenzyme is composed of two ~500 kDa motor (or “head”) containing heavy chain subunits and at least 6 other polypeptides. The domain structure of the dynein heavy chain is shown in Figure 1a. The N-terminal “tail” domain represents ~30% of the entire mass of the heavy chain and is required for dimerization and the interaction of most dynein subunits and associated proteins. Connected to the tail is the “linker” domain, which is thought to amplify structural changes during dynein’s ATPase cycle and is required for motility5,10-12. Following the linker domain are six concatenated AAA+ (ATPase Associated with diverse cellular Activities) domains, which fold into a ring. As a member of the AAA+ superfamily, dynein is evolutionarily distinct from kinesin and myosin, which are distantly related to G-proteins13. Dynein’s first AAA+ domain is the primary site of ATP hydrolysis14, but AAA+ domains 2–4 are also expected to bind ATP or ADP based on mutant phenotypes15-18. Projecting from the fourth AAA+ domain is a 15 nm, antiparallel, coiled-coil “stalk” capped by a globular microtubule-binding domain19-21.
Figure 1.
Dynein structure and constructs used in this study. (a) Linear diagrams of (1) native dynein’s domain structure, and constructs used in this study: (2) GST-dimerized dynein with an N-terminal HaloTag (H) for tail-labeled experiments, (3) GST-dimerized dynein with a C-terminal HaloTag for motor domain-labeled experiments, and (4) dynein monomer with an N-terminal SNAP-tag for DNA dimerization, and C-terminal HaloTag for motor domain labeling. MTBD is the microtubule-binding domain. (b) 2-D schematic of dimeric dynein. Dimerization (white box) can be achieved using the native protein dimerization domain, GST or complementary DNA oligomers attached via a SNAP-tag. (c) 3-D structure of yeast dynein (3QMZ) from Carter, Cho et al.19, filtered to 8 Å resolution. Views from left to right: the linker face, the opposite face of the ring containing the C-terminal (CT) alpha helix, and the side of the ring. Dimerization is achieved via GST (magenta) at the N terminus (NT).
Despite the complexity of the dynein motor, a dimer of two truncated Saccharomyces cerevisiae dynein heavy chains is sufficient for processive motility5,22. Previously we showed that dynein monomers lacking most of the tail domain were not processive on their own, but moved processively when they were linked together with glutathione S-transferase (GST, a stable homodimer)5 (Fig. 1b). GST–dynein homodimers behaved similarly to native yeast dynein with respect to velocity, processivity, stepping behavior, and force production in vitro5,23, demonstrating that the native dimerization interface is not required for motility and suggesting that the basic motile mechanism is insensitive to the method of dimerization.
However, how dynein achieves processive motility remains unknown. For the well-studied kinesin-1 and myosin-V motors, nucleotide-driven conformational changes of their mechanical elements power the sequential “hand-over-hand” stepping of their two identical motor domains24-28. Previously, to investigate the dynein stepping mechanism, we labeled GST–dynein homodimers with a single Quantum dot (Qdot) on a single motor domain or on the tail domain (approximate center of mass). High-precision one-dimensional stepping analyses revealed that the motor domain step size was nearly twice the size of the tail step size, consistent with a model in which dynein’s two motor domains alternate their position in time and pass each other in space5. However, unlike kinesin, dynein takes steps of variable size and direction4,5,8, making other stepping patterns theoretically possible5. Dynein’s variable stepping behavior is likely due its large size10,12,29, which allows the motor rings to separate and access multiple microtubule binding sites. While advances in understanding the architecture of the dynein motor domain have come from two near-atomic resolution crystal structures19,21 (Fig. 1c), how dynein’s two motors are arranged on the microtubule when moving processively also remains unknown. Here, we set out to determine how dynein achieves processive motility.
A major barrier to determining the dynein stepping pattern and its structural basis is the lack of an efficient system to make high-affinity, functional heterodimers, so that each protomer can be probed independently. We have created S. cerevisiae dynein heterodimers labeled with two distinct fluorophores through base-pairing of covalently-attached, complementary DNA oligonucleotides. DNA-dimerized dynein behaves indistinguishably from native dynein and protein-based dynein homodimers5,23. Using two-color, single-molecule microscopy coupled with high-precision, two-dimensional particle tracking, we find that dynein has a highly unusual stepping pattern compared to processive kinesins and myosins. We show that dynein’s two motor domains can step both alternately and non-alternately in time, and can either pass or not pass each other in space. Surprisingly, we have found that many dynein steps are uncoordinated, but become coordinated as the distance between the two motor domains increases. These results suggest that dynein can switch between stochastic and tension-based stepping, making it distinct from all two-headed processive motors.
Results
Two-dimensional analysis of dynein stepping
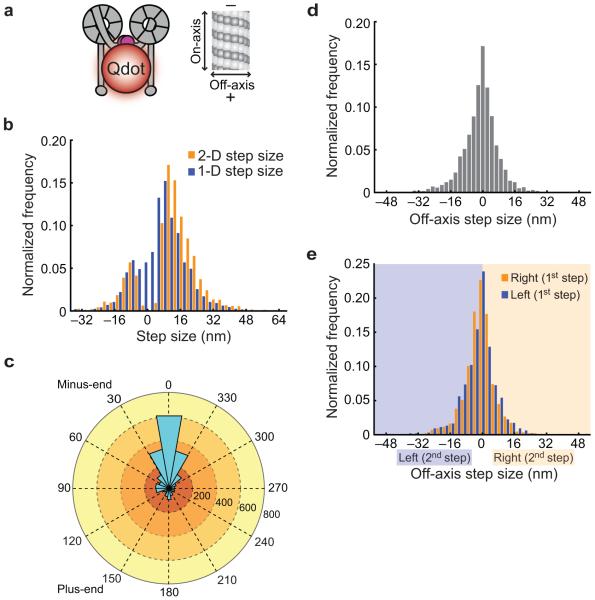
Because dynein’s steps are known to have an off-axis component5,8, analysis of stepping projected onto one dimension (1-D) along the microtubule axis (as is standard in the field) could yield an underestimate of dynein’s true step size. To determine the step size of dynein in two dimensions (2-D), we implemented a custom step-finding program (see Supplementary Methods). Before analyzing dynein’s stepping behavior in 2-D, we first determined the measurement precision of our total internal reflection fluorescence (TIRF) microscope to be ~1.5 nm for Qdot 655 and ~3.5 nm for the organic fluorophores Atto647N and Cy3B (Supplementary Fig. 1a–c; Supplementary Methods). As an additional control for the precision of our methods, we performed 1-D and 2-D stepping experiments with the yeast kinesin-8/Kip330. We found that kinesin-8/Kip3 labeled on a single motor domain takes ~16 nm steps (Supplementary Fig. 2a–d), similar to other kinesin family members28.
To determine the 2-D step size of dynein, we tracked the stepping of GST–dynein homodimers labeled with a single Qdot 655 placed on either the tail domain (via an N-terminal HaloTag; Figs. 1a and 2a–e) or on a single motor domain (via a C-terminal HaloTag; Fig. 1a and Supplementary Fig. 2e–j)5. Our new analysis method revealed that the 2-D step size of tail-labeled dynein was ~10 nm (Fig. 2b), which is larger than the originally reported 1-D step size. However, when the 2-D data was projected onto the direction of motion along the microtubule axis, we observed an ~8 nm 1-D step size (Fig. 2b), in agreement with previous 1-D on-axis step sizes reported for dynein5,7,23. Observation of a fluorophore on a single motor domain (head-labeled) resulted in a 2-D step size of ~14–16 nm, while the 1-D on-axis step size was slightly smaller (Supplementary Fig. 2i). As reported previously, we found that the majority of dynein steps were in the forward direction (Fig. 2c and Supplementary Fig. 2j)5. Analysis of the 2-D stepping data allowed us to determine the step size and angle of dynein’s off-axis steps for the first time. We found that many steps taken by the tail-labeled dynein and the single motor domain-labeled dynein contained an off-axis component > 6 nm (Fig. 2d, Supplementary Fig. 2g,). By contrast, most kinesin-8/Kip3 steps did not contain an off-axis component (Supplementary Fig. 2c), similar to other kinesins31,32. The percentage of off-axis steps observed here was higher than previously reported5, most likely due to our development and implementation of the 2-D stepping algorithm, which allows a more accurate and thorough classification of the off-axis component of dynein stepping. However, both the curvature of the microtubule and the geometry associated with the distance between the fluorophore on the dynein motor domain and dynein’s microtubule-binding domain could introduce additional sources of error for measurements in the off-axis direction. Thus, distances measured in the off-axis direction may be an underestimate and dynein may take more frequent and larger off-axis steps than we can detect.
Figure 2.
Two-dimensional stepping analysis of GST–dynein homodimers. (a) Schematic of a GST–dynein homodimer labeled with a Qdot via an N-terminal (tail domain) HaloTag and a diagram of a microtubule showing on- and off-axis directions of movement. (b) Histograms of dynein’s step sizes in 1-D and 2-D. N = 1391 steps for all panels. (c) An angle histogram (or rose plot) of the step angles. The stepping angle is defined as the angle between the stepping vector and the direction of on-axis movement. Steps to the left or right of the direction of motion are between 0° and 180° or 180° and 360°, respectively. Steps between 90° and 270° are backwards steps. 77% of steps are forward steps. (d) Histogram of off-axis step sizes. Percentages of step sizes larger than 4, 6, and 8 nm are 53%, 38%, and 28%, respectively. (e) Histograms of leftward or rightward steps after a previous left or right step. Leftward and rightward steps are shown as steps with negative and positive off-axis components, respectively.
The 2-D analysis also allowed us to investigate whether dynein has a preference for stepping to the left or right. We calculated the probability that an off-axis step is followed by another off-axis step in the same direction (for example, how likely it is that a leftward step is followed by another leftward step). For tail-labeled dynein, for which every step is observed, dynein is equally likely to step to the left or the right (Fig. 2e), irrespective of the direction of a prior off-axis step. In summary, by analyzing dynein stepping in two dimensions, we have found that dynein’s true step size is larger than the previously reported 1-D step size, many steps contain an off-axis component, and steps are equally likely to be to the left or to the right.
Development of DNA-dynein heterodimers
To determine how dynein’s two motor domains move processively, we next wanted to examine the stepping behavior of each of dynein’s two motor domains independently. Our prior work used a rapamycin–FKBP–FRB-mediated heterodimer5, however this complex has lower affinity33 than is desirable for creating robust and stable heterodimers at the low protein concentrations necessary for single molecule experiments. Instead, we chose a DNA-based dimerization approach that achieves high affinity (subfemtomolar for a 21 base-pair duplex at 22°C34), combinatorial flexibility, and allows individual modification of each protomer within the dimer35. We reasoned that the dynein molecule would be amenable to this method, since its dimerization interface exhibits great plasticity5. Furthermore, the DNA dimerization interface should be stable under load as yeast dynein’s maximum force generation has been measured to be ~7 pN23, whereas “unzipping” DNA requires a force of ~14 pN36.
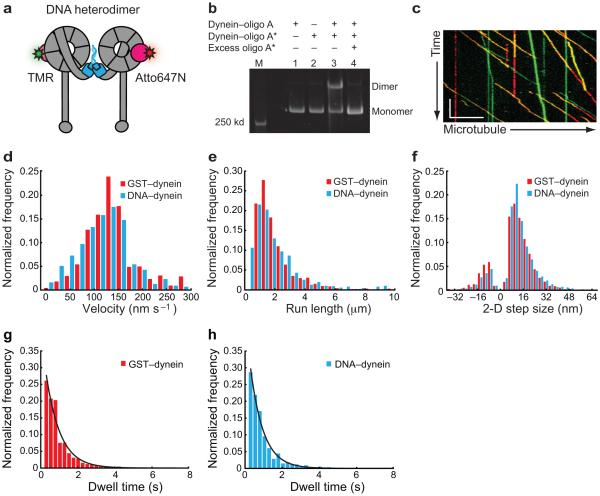
To make DNA-based dynein dimers, we engineered a dynein monomer fusion protein in which the SNAP-tag replaced the N-terminal dimerization domain of the endogenous dynein heavy chain (Fig. 1a,b). Like the Qdot-labeled dynein motors, this dynein monomer is also fused to a HaloTag at the C terminus of the motor domain to enable fluorophore labeling (Figs. 1a and 3a). Both the SNAP-tag and HaloTag are small enzymes that form covalent bonds with substrates that can be coupled to fluorophores, biotin, or reactive chemical groups. Taking advantage of the flexibility of the SNAP-tag, we next coupled the 5′ or 3′ end of complementary, 21 nucleotide DNA oligonucleotides to the SNAP substrate, benzylguanine (BG), and the DNA–BG molecules were then mixed with purified, SNAP-tagged dynein monomers. We found that this dimerization method was highly specific, with dynein monomers attached to complementary oligomers forming stable dimers but not in the presence of excess, competing oligomers (Fig. 3b, compare lanes 3 and 4).
Figure 3.
DNA-based dynein heterodimers are functional and step similarly to protein-based dynein homodimers. (a) Schematic of a DNA-based dynein heterodimer labeled with Atto647N (red star) and TMR (green star) via a C-terminal HaloTag (pink circles). The SNAP-tag and DNA oligomers (attached via the N-terminal SNAP-tag) are shown in blue. (b) LDS-PAGE gel showing dimerization of dynein monomers via DNA hybridization. (c) Kymograph of the motility of DNA-based dynein dimers labeled with TMR (green) and Atto647N (red), with overlapping, dual-labeled heterodimers in yellow. Scale bars: y (1 min), × (10 μm). (d) Histograms of the velocity of GST- and DNA-based dynein dimers. GST–dynein velocity is 134 ± 60.4 nm s−1 (mean ± s.d., N = 943), and DNA–dynein velocity is 125 ± 56.1 nm s−1 (mean ± s.d., N = 866). (e) Histograms of the run length of GST- and DNA-based dynein dimers. GST–dynein run length is 1.06 ± .044 μm (mean ± s.e.m., N = 943), and DNA–dynein run length is 1.45 ± .063 μm (mean ± s.e.m., N = 866). (f) Histograms of the 2-D step size of GST- and DNA-based dynein dimers labeled with a single Qdot 655 on the N-terminal tail domain. (g,h) Histograms of the dwell time distribution of GST–dynein homodimers (g) and DNA–dynein heterodimers (h) labeled with a single Qdot 655 on the N-terminal tail domain. The distributions are fit to single exponential functions with stepping rates of k = 1.78 ± 0.13 s−1 and 1.43 ± 0.10 s−1, respectively.
To determine if DNA-dimerized dynein was functional, we compared its motility and stepping pattern to GST–dynein homodimers. Each dynein monomer was labeled with a different small organic fluorophore (TMR or Atto647N) via the C-terminal HaloTag prior to dimerization (Fig. 3a). Using TIRF microscopy, we found that the majority of moving motors were dual-labeled (Fig. 3c), and their velocities and run lengths were similar to GST–dynein homodimers (Fig. 3d,e). As a more stringent test of functionality, we determined the 2-D step size and dwell time distribution of DNA–dynein dimers by labeling their tail domains with a Qdot 655. Again, we found these parameters to be comparable to that of GST-dynein homodimers (Fig. 3f–h). Importantly, the stepping behavior of DNA–dynein heterodimers was also similar to native yeast dynein analyzed at both rate limiting and cellular ATP concentrations5,23, a further indication that both the method of dimerization and low ATP concentrations (to slow the speed of the motor) used in our experiments do not alter the dynein stepping behavior. Together, these results indicate that DNA–dynein heterodimers are an excellent model system for dissecting the dynein stepping mechanism.
Dynein’s stepping is distinct from kinesin and myosin
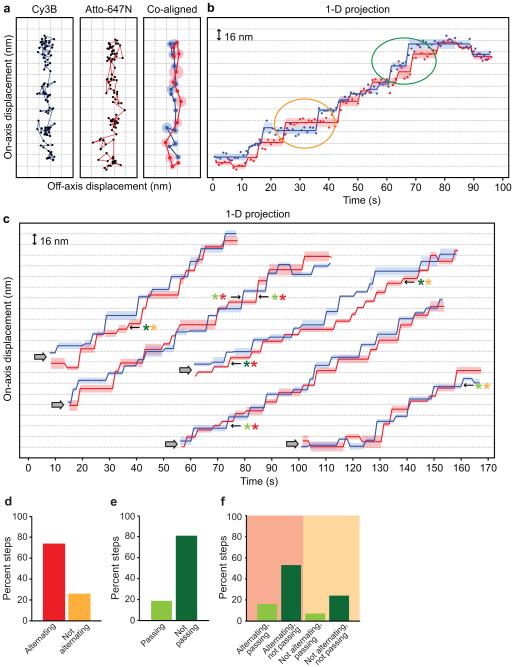
All dimeric processive myosin and kinesin motors studied to date achieve processive motility by alternating the position of their two motor domains in both space and time (hand-over-hand stepping). We sought to determine if dynein had a similar spatial and temporal pattern of stepping. To do so, we used our DNA dimerization method to construct dynein heterodimers labeled with the bright fluorophores Cy3B and Atto647N, which are much smaller (~ 2 nm in size) than Qdots (typically > 15 nm, larger than a dynein motor domain), and thus unlikely to interfere with dynein’s motion. Dual-labeled dynein motors were imaged using near-simultaneous, alternating-excitation, high-precision TIRF microscopy under rate-limiting ATP conditions. We located the centroid position of each fluorophore-labeled motor domain with high precision by applying a 2-D Gaussian fit to the data from each channel37, allowing for a position measurement precision of ~3.5 nm in both the x and y directions for both the Atto647N and Cy3B channels (Supplementary Fig. 1b,c; see Supplementary Methods). To precisely co-localize the Cy3B and Atto647N data, we applied the Single molecule High REsolution Co-localization (SHREC) method38, which yielded a mean mapping error of ~4 nm in both the x and y directions, and an overall uncertainty in our measurements of ~6 nm (Supplementary Fig. 3a,b; see Supplementary Methods).
Images from the Cy3B and Atto647N channels were screened for dual-labeled motile molecules with clearly defined 2-D stepping clusters (see Supplementary Methods). We analyzed 27 different two-color dynein stepping trace pairs, containing 708 steps (Fig. 4a,b; Supplementary Fig. 3c–l). The average 1-D, 2-D (compare Supplementary Fig. 2i with Supplementary Fig. 3g,h) and off-axis step sizes (compare Supplementary Fig. 2g with Supplementary Fig. 3i,j) were similar to the motor domain step sizes we observed for Qdot labeled GST-homodimers. Additionally, the 1-D on-axis step sizes (Supplementary Fig. 3g,h) we observed in each channel for the DNA–dynein were also very similar to those that we previously measured for full-length native yeast cytoplasmic dynein5, another indication that the DNA-dimerization method is an excellent model system for examining the dynein stepping mechanism.
Figure 4.
High-precision, two-color tracking of dynein stepping. (a) Representative two-color stepping trace of a DNA–dynein heterodimer. The raw 2-D positions (black dots in left and center panels) from a DNA–dynein heterodimer labeled with Cy3B (left panel, blue line) and Atto647N (center panel, red line). Co-alignment of the motor domain traces from each channel is shown in the right panel, with darker solid blue (Cy3B) and red (Atto647N) dots representing steps determined by a 2-D step finding algorithm, and larger, lighter-colored blue and red circles representing the s.d. of individual steps. (b) 1-D on-axis projection of the 2-D data from (a), with lighter blue and red bars representing the s.d. of individual steps along the projection axis. Ovals highlight examples of hand-over-hand (orange) or inchworm (green) steps. Y-axis grid lines are spaced 16 nm apart in all panels. See Supplementary Figures 3c–f for additional stepping traces. (c) Examples of 1-D on-axis projections of two-color stepping trace pairs from dual-labeled DNA-dynein heterodimers. The grey arrows indicate the start of each trace. Pairs of solid blue and red lines represent the 1-D on-axis projection of steps, determined by a 2-D step finding algorithm for the Cy3B and Atto647N traces, respectively. Lighter blue and red bars represent the s.d. of individual steps along the projection axis. Examples of the four different types of steps are as follows: light green and red asterisks represent alternating and passing steps (“hand-over-hand”); dark green and red asterisks represent alternating and not passing steps (“inchworm”); light green and light orange asterisks represent not alternating and passing steps; dark green and light orange asterisks represent not alternating and not passing steps. (d) Temporal analysis of the relative frequency of stepping events. Alternating events are defined as current and previous stepping events originating from different heads, while non-alternating events are current and previous stepping events originating from the same head. N = 268. (e) Spatial analysis of the relative frequency of passing or not passing stepping events. N = 233. (f) Combined temporal and spatial analysis of stepping events. N =135.
Labeling each of dynein’s two motor domains with different colored fluorophores allowed us to observe spatial and temporal relationships of the motor domains during processive motion (Fig. 4a,b; Supplementary Fig. 3c–f). Here we use the terms alternating or not alternating to describe the motor domains relative temporal behavior, and passing or not passing to describe their relative spatial behavior. We found that the majority (~74%) of dynein steps alternated in time (each head “taking turns” stepping), but non-alternating events (a single head taking multiple steps in a row) were also observed (Fig. 4b–d; Supplementary Fig. 3d,f). In our spatial analysis of dynein stepping, we found that the majority (~83%) of dynein steps did not pass each other (leading and lagging heads maintained their identity), although passing events (one head switching from the leading to the lagging position) were also observed (Fig. 4b,c,e; Supplementary Fig. 3d,f). These results (Fig. 4f) are in marked contrast to the well-studied two-headed processive kinesin and myosin motors, which alternate their steps in time and pass one another in space, the combination of which results in hand-over-hand stepping27,28.
Spatial relationship of dynein’s two motor domains
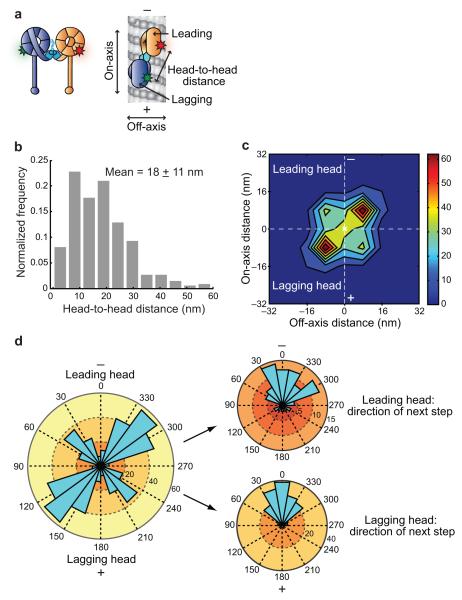
Despite recent reports of dynein’s motor domain structure at near atomic resolution19,21, the position and orientation of each motor domain within the dimer when bound to microtubules remain unknown. Therefore, we next determined the distance between dynein’s motor domains in the “two-head-bound state”, when both heads were simultaneously bound to the microtubule (Fig. 5a, b, and Supplementary Fig. 4a–c). Given that the dimensions of a dynein motor domain are ~12 nm (diameter of the AAA+ ring) × 10 nm (thickness of the AAA+ ring and linker) (Fig. 1c), our data suggest that the dynein motor domains are positioned close together. However, to accommodate the largest distances we observed between motor domains (4% of head-to-head distances were > 30 nm), linker domain undocking from the motor domain likely occurs, a phenomena that has been observed in electron microscopy studies of both cytoplasmic and axonemal dyneins10,12. In addition, unlike other cytoskeletal motors, the distribution of head-to-head distances for dynein was broad and varied widely within individual traces (Fig. 4a–c and Supplementary Fig. 3c–f), further highlighting the unusual nature of dynein’s stepping mechanism.
Figure 5.
Spatial arrangement of dynein motor domains during the two-head-bound state. (a) Schematic of dual-labeled DNA-dimerized dynein bound to a microtubule. Other arrangements of the motor domains are possible. Fluorophores are represented by red and green stars. (b) Histogram of dynein’s head-to-head distances during the two-head-bound state. N = 523. (c) Contour plot showing the left and right asymmetry between the leading and lagging heads. Orientation of the microtubule axis is vertical, as indicated by − and +, with the centroid position of each dynein molecule placed at the origin of the axes (white X). The number of occurrences of each position is indicated by the color bar on the right, with bin edges at 8 nm increments from −32 nm to +32 nm on both axes. N = 256 dimers or 512 heads. (d) On the left, an angle histogram of the position of the leading and lagging heads of individual dynein dimers relative to their respective centroid position (placed at the origin of the axes). Locations to the left or right of the direction of motion are between 0° and 180°, or 180° and 360°, respectively. 64% of the leading heads are to the right of the microtubule axis. N = 256 dimers or 512 heads. On the right, the angular distributions of the next step taken by the leading (top) or lagging (bottom) head. Steps between 90° and 270° are backwards steps.
Our 2-D tracking of dynein stepping also allowed us to determine the spatial relationship of each dynein motor domain in the two-head-bound state relative to the direction of motion along the microtubule (Fig. 5a). Analysis of the position of each of dynein’s motor domains revealed that the leading head was more likely to be to the right of the axis of motion, while the lagging head was more likely to be to the left of the axis of motion (Fig. 5c). This analysis included only motors with statistically resolvable leading or lagging and left or right positions (two-tailed Student’s t-test with alpha 0.05). We also performed this analysis on our entire data set and observed the same trend (Supplementary Fig. 4d). This observation suggests that the two motor domains of a dynein dimer do not typically reside on the same protofilament of a microtubule. Additionally, these finding support the idea that each dynein motor domain maintains a relatively stable identity of being a right leading head or a left lagging head. Although the dynein heads have a distinct left or right identity, the direction of the next step taken by either a leading or lagging head is predominantly forward, with no off-axis bias (Fig. 5d), suggesting that dynein’s two motor domains typically straddle at least one microtubule protofilament, but then move forward toward the microtubule’s minus-end.
Dynein’s steps can be either stochastic or coordinated
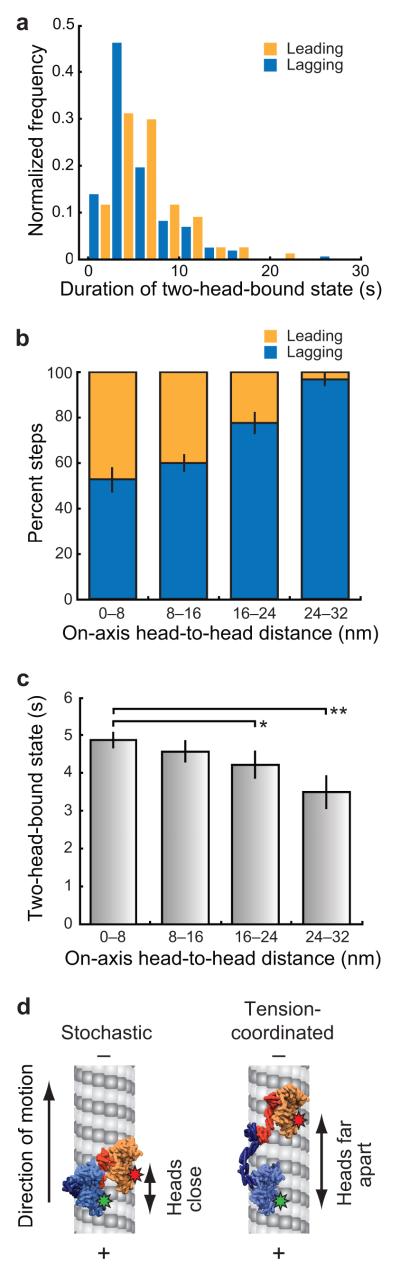
The spatial asymmetry between the leading and lagging motor domains suggested that dynein’s two heads have distinct identities when microtubule-bound. Additionally, optical trapping studies have shown that dynein responds asymmetrically to rearward and forward forces23. To determine whether tension played a role in the dynein stepping mechanism, we analyzed the duration of two-head-bound states associated with leading or lagging head stepping events. The mean duration for two-head-bound states terminated by a lagging head stepping is significantly shorter than that of two-head-bound states terminated by a leading head stepping (Fig. 6a). We hypothesized that the asymmetric response of leading and lagging heads is due to the difference in direction of the respective force vectors acting upon them along the microtubule axis (forward-directed force for the lagging heads and rearward-directed force for the leading heads).
Figure 6.
Dynein steps are stochastic at short head-to-head spacing and coordinated as head-to-head spacing increases. (a) Histograms of the duration of the two-head-bound states that are terminated by a leading head stepping event or a lagging head stepping event. The mean durations are 4.3 s or 5.4 s for two-head-bound states terminated by a lagging head stepping (N = 228) or leading head stepping (N = 119), respectively; the means are significantly different (P = 9.7e−5, alpha 0.05, one-tailed KS test). (b) Relative stepping frequency of the leading and lagging heads as a function of the on-axis distance between motor domains. Error bars represent the s.e.m. and were generated by bootstrapping each bin. N = 352. (c) The duration of the two-head-bound state plotted as a function of the on-axis head-to-head distance. Mean durations ± s.e.m. are shown (*P = 0.0139, **P = 0.0094; two-tailed KS test, alpha value 0.05, N = 485). (d) Model for the dynein stepping mechanism. The 3-D structure of dynein (3QMZ)19 filtered to 8 Å resolution was used to generate two microtubule-bound models of dimeric GST–dynein. The dynein rings are shown parallel to the long-axis of the microtubule and parallel to each other based on electron microscopy reconstructions52-55. Stepping is stochastic when dynein’s motor domains are close together (left panel). Large distances between the two motor domains result in a tension-based mechanism that coordinates stepping (right panel).
Since motor domains separated by larger distances may experience increased tension, we examined whether dynein’s stepping pattern changed as a function of the head-to-head distance. When the dynein motor domains were close together, we found that there was an equal probability of the leading or lagging head stepping (Fig. 6b). However, as the heads became separated by larger distances in the on-axis direction the lagging head was increasingly more likely to step (Fig. 6b). This trend was also observable when we examined the duration of the two-head-bound state as a function of distance between motor domains in the on-axis direction. As larger distances separated the motor domains, the duration of the two-head-bound state decreased (Fig. 6c). Additionally, we found that the direction of the force is important for this effect, as neither the percentage of leading vs. lagging heads stepping, nor the duration of the two-head-bound state varied as a function of the distance between motor domains in the off-axis direction (Supplementary Fig. 5a,b).
Our results suggest that stochastic, uncoordinated stepping dominates when dynein’s motor domains are close together, but when dynein’s two motor domains are separated by larger distances, stepping becomes increasingly coordinated (Fig. 6d). We hypothesize that when large distances separate dynein’s two motor domains, the dynein microtubule-binding domain may respond asymmetrically to force in the direction of movement along the microtubule (on-axis), but not across the microtubule (off-axis). This is consistent with reports that dynein responds asymmetrically to forward- and rearward-directed forces5,23, as well as studies that demonstrate that dynein’s step size is smaller under increased load4,23.
Discussion
A new model for the dynein stepping mechanism
By combining two-color, single-molecule microscopy with high-precision, two-dimensional tracking, we have shown that dynein’s stepping mechanism is distinct from all cytoskeletal molecular motors characterized to date. Although many of dynein’s steps alternate in time, most stepping events do not switch the leading or lagging spatial identities of dynein’s two heads, a distinction from the two-headed processive myosin and kinesin motors that use alternating and passing (hand-over-hand) mechanisms. Strikingly, our data suggest that dynein’s steps are uncoordinated when the distance between motor domains is small, presumably because the intramolecular strain is low. However, as the distance between motor domains increases, our data show that dynein becomes increasingly coordinated, likely through a tension-based mechanism. These findings demonstrate that dynein is the first two-headed processive cytoskeletal motor that can alternate between stochastic- and tension-based stepping to achieve processivity (Fig. 6d).
In addition to tension governing dynein’s stepping behavior, the large size of the dynein motor domains (Fig. 1c) may impart steric constraints on the stepping pattern. Our data suggest that structural limitations influence the range and location of dynein on the microtubule lattice. We have found that dynein has an inherent left or right asymmetry, with the right motor domain of the dimer more likely to be the leading head and the left motor domain more likely to be the lagging head. It is unlikely that this asymmetry is generated by DNA-based dimerization, as the linkers we have included between dynein and the DNA contain multiple freely rotatable bonds. Therefore, a possible source of asymmetry could be the position of the linker domain, which lies across the face of the AAA+ ring and moves in response to the nucleotide occupancy at AAA110,12. We propose that the linker domain of the motor on the right is “sandwiched” between the dynein rings, while the linker domain of the motor on the left is not bound by another motor domain, imparting a structural and functional asymmetry (Fig. 6d). Our observation of large separations between dynein’s two motor domains also suggests that the linker domain can undock from the motor domain (most likely the lagging motor as in Fig. 6d, panel 2), as has been observed in electron microscopy studies of both cytoplasmic and axonemal dyneins10,12
The stochastic nature of dynein’s stepping behavior raises the intriguing question of how a partially uncoordinated motor achieves processive motility. In the case of kinesin-1, myosin-V and myosin-VI, processivity is accomplished by maintaining the leading head in a strong, filament-bound state until the lagging head detaches from the filament to pass the bound head and becomes the new leading head. Intramolecular tension between the motor domains provides a nucleotide gating mechanism that favors the detachment of the lagging motor domain and promotes a biased, diffusion-based search of this head for the next filament binding site39-44. For dynein, it is possible that dynein’s high duty ratio22 substantially reduces the likelihood that both motor domains simultaneously dissociate from their microtubule track. This idea is corroborated by recent findings showing that mutant myosin-V and -VI motors are still processive even when their tension-based gating mechanisms are impaired or destroyed45,46. However, our data do not rule out the possibility of nucleotide gating for dynein. For example, our finding that 74% of the steps we observed were alternating would be consistent with gating of some steps, as has been suggested by studies of Dictyostelium cytoplasmic dynein47.
The similarity between the motility of the DNA-dynein heterodimer, the GST-dynein homodimer, and native yeast dynein5,23 suggests that the basic stepping mechanism will be the same for native dynein and the model systems that are currently being used to study the dynein mechanism. Thus, the DNA-based technique of heterodimer formation that we developed will be a powerful tool for orthogonal control over each dynein protomer for the study of additional heterodimer combinations in the future. However, our data do not rule out the possibility that dynein’s native dimerization interface, dynein’s associated subunits (intermediate, light intermediate, and light chains), cofactors (dynactin, Lis1, and Nudel), or cargo may impart an additional layer of regulation on the dynein stepping mechanism. For instance, given that tension appears to coordinate dynein stepping, an interesting area for future study will be to determine if dynein’s stepping mechanism becomes coordinated under the load of moving large cargo.
In vivo implications
Compared to other cytoskeletal motors, dynein’s stepping behavior shows great variability and flexibility. Many dynein steps have an off-axis component, some steps are backwards, and dynein’s two motor domains can step independently of each other. We propose that this flexibility allows dynein to navigate a crowded cytoplasm as well as obstacles on microtubules. Our results provide a molecular explanation for the observation that dynein is better able to navigate obstacles than kinesin motors48,49.
This apparent plasticity of the dynein stepping mechanism suggests that layers of regulation may be used to accomplish different cell biological functions. In eukaryotic cells, dynein transports dozens, if not hundreds, of different cargo, but there is only a single gene encoding cytoplasmic dynein 1 in all sequenced eukaryotic genomes (with the exception of flowering plants and some algae, which lack dynein genes50,51). Given the variability of the type, size, and loads imparted by different cargo (ranging from endosomes to the mitotic spindle), a number of mechanisms for regulating cytoplasmic dynein’s stepping behavior may have evolved. Future studies on the function of dynein-associated subunits and cofactors, the affects of cargo load on motility, as well as how multiple motors may coordinate to move cargo, will determine whether this partially uncoordinated motor is regulated to step coordinately for some functions.
Supplementary Material
Acknowledgements
We thank Xiaolei Su and David Pellman (Dana Farber Cancer Institute and Department of Cell Biology, Harvard Medical School, Boston Massachusetts) for providing purified kinesin-8; Sirui Zou for technical assistance; Andrew Carter, Stirling Churchman, Arne Gennerich, Yale Goldman, Adam Hendricks, Julie Huang, Andres Leschziner, and Anthony Roberts for critical comments on the manuscript; Francois Aguet, Achem Besser, Marco Vilela, and Gaudenz Danuser for discussions of data analysis; Maria Bagonis for early work on oligomer-SNAP linking; Andres Leschziner for help with figure design; and Andrew Carter for providing MATLAB code. W.Q. is supported by a postdoctoral fellowship from the American Heart Association. S.R.P. is funded by the Rita Allen Foundation, the Harvard Armenise Foundation and an NIH New Innovator award (1 DP2 OD004268-01).
APPENDIX
Methods
Yeast strains
Modification of the endogenous Saccharomyces cerevisiae dynein heavy chain gene was accomplished by insertion of the Kluyveromyces lactis URA3 selectable marker into the DYN1 gene at the location of the desired change. The K. lactis URA3 gene was subsequently replaced with the SNAP-tag, SNAPf-tag (New England Biolabs), HaloTag (Promega), or GFP. Yeast strains used in this study are listed in Supplementary Table 1.
Preparation of HaloTag ligand–fluorophores
HaloTag ligand–fluorophore conjugates not commercially available were prepared as follows. Atto647N was conjugated to the HaloTag ligand by mixing 10 mM Atto647N NHS ester (Atto-Tec), 20 mM HaloTag amine (O4) Ligand (Promega), and 30 mM N,N-diisopropylethylamine in dimethylformamide, and nutating at 30°C for 24 hrs. The HaloTag-Atto647N conjugate was separated from unreacted material by HPLC using a reverse-phase C18 column with a methanol: water gradient. Final product purity was > 85% as assessed by mass spectrometry. Cy3B–Halotag was prepared by Bio-synthesis, Inc (Lewisville, TX) from Cy3B-NHS (GE Healthcare Lifesciences) and HaloTag amine (O4) ligand (Promega).
Benzylguanine-conjugated DNA oligonucleotides
Benzylguanine-conjugated DNA oligonucleotides (BG–oligos) were prepared by mixing 10 mM BG–GLA–NHS (New England Biolabs) in anhydrous DMSO with 0.33 mM PAGE-purified amine-functionalized oligos (Bioneer) in 67 mM HEPES (pH 8.5) and 50% DMSO (v/v) for 30 min at room temperature. Unreacted BG–GLA–NHS was removed using Micro Bio-Spin 6 Columns (Bio-Rad), pre-equilibrated with 10 mM Tris (pH 8.0), 150 mM KCl, and 10% (v/v) glycerol. Linkage of BG to oligos was confirmed by gel shift assays on 20% TBE gels (Invitrogen). Oligo sequences used for dynein dimerization are listed in the Supplementary Methods.
Protein purification and labeling
Dynein motors were purified as described previously5 with modifications detailed in the Supplementary Methods. Motors were labeled with BG–oligos and HaloTag ligand–fluorophores during dynein purification (see Supplementary Methods). Oligos used for labeling dynein, and dynein monomers used in different experiments are listed in Supplementary Table 2.
TIRF microscopy
Motility assays were performed using an inverted objective type Olympus IX-81 TIRF microscope with a 100X 1.45 N.A. oil immersion TIRF objective (Olympus) equipped with four continuous-wave diode-pumped solid state lasers: 405 nm and 640 nm cubic lasers (Coherent Inc.) and 491 nm and 561 nm lasers (Cobolt). Signals were detected with a back-thinned electron multiplier CCD camera (Hamamatsu). For near-simultaneous, two-color imaging, the microscope was modified to include a dual-band laser polychroic mirror (z561/635rpc, Chroma) and a dual-band sputtered emission filter (etCy3/Cy5m, Chroma) in the main optical path. The excitation path of the 561 nm laser was controlled by an acousto-optical tunable filter (NEOS; response time of 10 ms), while that of the 640 nm laser was controlled by a fast mechanical shutter (SmartShutter, Sutter Inc.; response time of 25 ms).
Stepping analysis
A custom program was written in MATLAB (Mathworks, Inc.) to analyze dynein stepping behavior in both the temporal and spatial domains and determine correlated events. Briefly, all valid dwells included in the analysis were at least 3 frames long in their respective channels. A step was counted only if the dwells before and after contained at least 3 frames in that step’s channel. Two-head-bound states were defined as containing 3 points or more (two in one channel and one in the other), where all points were part of a valid dwell location in their respective channels. Alternating and non-alternating steps were assigned only when both the current and previous steps were resolvable in time (i.e. more than one frame away from each another). Leading or lagging identities were assigned only if both head’s positions were different as determined by a two-tailed Student’s t-test with an alpha level of 5%. Position data is normally distributed about the mean making a t test appropriate. Classification of passing and not-passing steps was assigned only if the leading vs. lagging identities before and after the step could be determined. See Supplementary Methods for details.
Footnotes
Author Contributions
W.Q. and N.D.D. contributed equally. W.Q., N.D.D, W.S., and S.R.P. designed the experiments. W.Q., N.D.D., and B.S.G. performed the experiments and analyzed the data. W.Q., N.D.D., B.S.G., and S.R.P. wrote the paper. E.V. and D.W. wrote the 2-D particle tracking code.
References
- 1.Eschbach J, Dupuis L. Cytoplasmic dynein in neurodegeneration. Pharmacol Ther. 2011;130:348–363. doi: 10.1016/j.pharmthera.2011.03.004. [DOI] [PubMed] [Google Scholar]
- 2.Wynshaw-Boris A. Lissencephaly and LIS1: insights into the molecular mechanisms of neuronal migration and development. Clin Genet. 2007;72:296–304. doi: 10.1111/j.1399-0004.2007.00888.x. [DOI] [PubMed] [Google Scholar]
- 3.King SJ, Schroer TA. Dynactin increases the processivity of the cytoplasmic dynein motor. Nat Cell Biol. 2000;2:20–24. doi: 10.1038/71338. [DOI] [PubMed] [Google Scholar]
- 4.Mallik R, Carter BC, Lex SA, King SJ, Gross SP. Cytoplasmic dynein functions as a gear in response to load. Nature. 2004;427:649–652. doi: 10.1038/nature02293. [DOI] [PubMed] [Google Scholar]
- 5.Reck-Peterson SL, et al. Single-molecule analysis of dynein processivity and stepping behavior. Cell. 2006;126:335–348. doi: 10.1016/j.cell.2006.05.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ross JL, Wallace K, Shuman H, Goldman YE, Holzbaur EL. Processive bidirectional motion of dynein-dynactin complexes in vitro. Nat Cell Biol. 2006;8:562–570. doi: 10.1038/ncb1421. [DOI] [PubMed] [Google Scholar]
- 7.Toba S, Watanabe TM, Yamaguchi-Okimoto L, Toyoshima YY, Higuchi H. Overlapping hand-over-hand mechanism of single molecular motility of cytoplasmic dynein. Proc Natl Acad Sci USA. 2006;103:5741–5745. doi: 10.1073/pnas.0508511103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wang Z, Khan S, Sheetz MP. Single cytoplasmic dynein molecule movements: characterization and comparison with kinesin. Biophys J. 1995;69:2011–2023. doi: 10.1016/S0006-3495(95)80071-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kardon JR, Vale RD. Regulators of the cytoplasmic dynein motor. Nat Rev Mol Cell Biol. 2009;10:854–865. doi: 10.1038/nrm2804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Burgess SA, Walker ML, Sakakibara H, Knight PJ, Oiwa K. Dynein structure and power stroke. Nature. 2003;421:715–718. doi: 10.1038/nature01377. [DOI] [PubMed] [Google Scholar]
- 11.Kon T, et al. Helix sliding in the stalk coiled coil of dynein couples ATPase and microtubule binding. Nat Struct Mol Biol. 2009;16:325–333. doi: 10.1038/nsmb.1555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Roberts AJ, et al. AAA+ Ring and linker swing mechanism in the dynein motor. Cell. 2009;136:485–495. doi: 10.1016/j.cell.2008.11.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Vale RD. Switches, latches, and amplifiers: common themes of G proteins and molecular motors. J Cell Biol. 1996;135:291–302. doi: 10.1083/jcb.135.2.291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gibbons IR, et al. Photosensitized cleavage of dynein heavy chains. Cleavage at the “V1 site” by irradiation at 365 nm in the presence of ATP and vanadate. J Biol Chem. 1987;262:2780–2786. [PubMed] [Google Scholar]
- 15.Cho C, Reck-Peterson SL, Vale RD. Regulatory ATPase sites of cytoplasmic dynein affect processivity and force generation. J Biol Chem. 2008;283:25839–25845. doi: 10.1074/jbc.M802951200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kon T, Nishiura M, Ohkura R, Toyoshima YY, Sutoh K. Distinct functions of nucleotide-binding/hydrolysis sites in the four AAA modules of cytoplasmic dynein. Biochemistry. 2004;43:11266–11274. doi: 10.1021/bi048985a. [DOI] [PubMed] [Google Scholar]
- 17.Reck-Peterson SL, Vale RD. Molecular dissection of the roles of nucleotide binding and hydrolysis in dynein’s AAA domains in Saccharomyces cerevisiae. Proc Natl Acad Sci U S A. 2004;101:1491–1495. doi: 10.1073/pnas.2637011100. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 18.Silvanovich A, Li MG, Serr M, Mische S, Hays TS. The third P-loop domain in cytoplasmic dynein heavy chain is essential for dynein motor function and ATP-sensitive microtubule binding. Mol Biol Cell. 2003;14:1355–1365. doi: 10.1091/mbc.E02-10-0675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Carter AP, Cho C, Jin L, Vale RD. Crystal structure of the dynein motor domain. Science. 2011;331:1159–1165. doi: 10.1126/science.1202393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Carter AP, et al. Structure and functional role of dynein’s microtubule-binding domain. Science. 2008;322:1691–1695. doi: 10.1126/science.1164424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kon T, Sutoh K, Kurisu G. X-ray structure of a functional full-length dynein motor domain. Nat Struct Mol Biol. 2011;18:638–642. doi: 10.1038/nsmb.2074. [DOI] [PubMed] [Google Scholar]
- 22.Shima T, Imamula K, Kon T, Ohkura R, Sutoh K. Head-head coordination is required for the processive motion of cytoplasmic dynein, an AAA+ molecular motor. J Struct Biol. 2006;156:182–189. doi: 10.1016/j.jsb.2006.03.014. [DOI] [PubMed] [Google Scholar]
- 23.Gennerich A, Carter AP, Reck-Peterson SL, Vale RD. Force-induced bidirectional stepping of cytoplasmic dynein. Cell. 2007;131:952–965. doi: 10.1016/j.cell.2007.10.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Gennerich A, Vale RD. Walking the walk: how kinesin and dynein coordinate their steps. Curr Opin Cell Biol. 2009;21:59–67. doi: 10.1016/j.ceb.2008.12.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sellers JR, Veigel C. Walking with myosin V. Curr Opin Cell Biol. 2006;18:68–73. doi: 10.1016/j.ceb.2005.12.014. [DOI] [PubMed] [Google Scholar]
- 26.Sweeney HL, Houdusse A. Myosin VI rewrites the rules for myosin motors. Cell. 2010;141:573–582. doi: 10.1016/j.cell.2010.04.028. [DOI] [PubMed] [Google Scholar]
- 27.Yildiz A, et al. Myosin V walks hand-over-hand: single fluorophore imaging with 1.5-nm localization. Science. 2003;300:2061–2065. doi: 10.1126/science.1084398. [DOI] [PubMed] [Google Scholar]
- 28.Yildiz A, Tomishige M, Vale RD, Selvin PR. Kinesin walks hand-over-hand. Science. 2004;303:676–678. doi: 10.1126/science.1093753. [DOI] [PubMed] [Google Scholar]
- 29.Samso M, Koonce MP. 25 Angstrom resolution structure of a cytoplasmic dynein motor reveals a seven-member planar ring. J Mol Biol. 2004;340:1059–1072. doi: 10.1016/j.jmb.2004.05.063. [DOI] [PubMed] [Google Scholar]
- 30.Su X, et al. Mechanisms underlying the dual-mode regulation of microtubule dynamics by Kip3/kinesin8. Mol Cell. 2011;43:751–763. doi: 10.1016/j.molcel.2011.06.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ray S, Meyhofer E, Milligan RA, Howard J. Kinesin follows the microtubule’s protofilament axis. J Cell Biol. 1993;121:1083–1093. doi: 10.1083/jcb.121.5.1083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ray S, Wolf SG, Howard J, Downing KH. Kinesin does not support the motility of zinc-macrotubes. Cell Motil Cytoskeleton. 1995;30:146–152. doi: 10.1002/cm.970300206. [DOI] [PubMed] [Google Scholar]
- 33.Banaszynski LA, Liu CW, Wandless TJ. Characterization of the FKBP.rapamycin.FRB ternary complex. J Am Chem Soc. 2005;127:4715–4721. doi: 10.1021/ja043277y. [DOI] [PubMed] [Google Scholar]
- 34.Markham NR, Zuker M. DINAMelt web server for nucleic acid melting prediction. Nucleic Acids Res. 2005;33:W577–581. doi: 10.1093/nar/gki591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Miyazono Y, Hayashi M, Karagiannis P, Harada Y, Tadakuma H. Strain through the neck linker ensures processive runs: a DNA-kinesin hybrid nanomachine study. EMBO J. 2010;29:93–106. doi: 10.1038/emboj.2009.319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Essevaz-Roulet B, Bockelmann U, Heslot F. Mechanical separation of the complementary strands of DNA. Proc Natl Acad Sci U S A. 1997;94:11935–11940. doi: 10.1073/pnas.94.22.11935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Thompson RE, Larson DR, Webb WW. Precise nanometer localization analysis for individual fluorescent probes. Biophys J. 2002;82:2775–2783. doi: 10.1016/S0006-3495(02)75618-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Churchman LS, Okten Z, Rock RS, Dawson JF, Spudich JA. Single molecule high-resolution colocalization of Cy3 and Cy5 attached to macromolecules measures intramolecular distances through time. Proc Natl Acad Sci U S A. 2005;102:1419–1423. doi: 10.1073/pnas.0409487102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Yildiz A, Tomishige M, Gennerich A, Vale RD. Intramolecular strain coordinates kinesin stepping behavior along microtubules. Cell. 2008;134:1030–1041. doi: 10.1016/j.cell.2008.07.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Rosenfeld SS, Sweeney HL. A model of myosin V processivity. J Biol Chem. 2004;279:40100–40111. doi: 10.1074/jbc.M402583200. [DOI] [PubMed] [Google Scholar]
- 41.Purcell TJ, Sweeney HL, Spudich JA. A force-dependent state controls the coordination of processive myosin V. Proc Natl Acad Sci U S A. 2005;102:13873–13878. doi: 10.1073/pnas.0506441102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Veigel C, Schmitz S, Wang F, Sellers JR. Load-dependent kinetics of myosin-V can explain its high processivity. Nat Cell Biol. 2005;7:861–869. doi: 10.1038/ncb1287. [DOI] [PubMed] [Google Scholar]
- 43.Sweeney HL, et al. How myosin VI coordinates its heads during processive movement. EMBO J. 2007;26:2682–2692. doi: 10.1038/sj.emboj.7601720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Dunn AR, Chuan P, Bryant Z, Spudich JA. Contribution of the myosin VI tail domain to processive stepping and intramolecular tension sensing. Proc Natl Acad Sci U S A. 2010;107:7746–7750. doi: 10.1073/pnas.1002430107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Baboolal TG, et al. The SAH domain extends the functional length of the myosin lever. Proc Natl Acad Sci U S A. 2009;106:22193–22198. doi: 10.1073/pnas.0909851106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Elting MW, Bryant Z, Liao JC, Spudich JA. Detailed tuning of structure and intramolecular communication are dispensable for processive motion of myosin VI. Biophys J. 2011;100:430–439. doi: 10.1016/j.bpj.2010.11.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Numata N, Shima T, Ohkura R, Kon T, Sutoh K. C-sequence of the Dictyostelium cytoplasmic dynein participates in processivity modulation. FEBS Lett. 2011;585:1185–1190. doi: 10.1016/j.febslet.2011.03.036. [DOI] [PubMed] [Google Scholar]
- 48.Dixit R, Ross JL, Goldman YE, Holzbaur EL. Differential regulation of dynein and kinesin motor proteins by tau. Science. 2008;319:1086–1089. doi: 10.1126/science.1152993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Ross JL, Shuman H, Holzbaur EL, Goldman YE. Kinesin and dynein-dynactin at intersecting microtubules: motor density affects dynein function. Biophys J. 2008;94:3115–3125. doi: 10.1529/biophysj.107.120014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Lawrence CJ, Morris NR, Meagher RB, Dawe RK. Dyneins have run their course in plant lineage. Traffic. 2001;2:362–363. doi: 10.1034/j.1600-0854.2001.25020508.x. [DOI] [PubMed] [Google Scholar]
- 51.Wickstead B, Gull K. Dyneins across eukaryotes: a comparative genomic analysis. Traffic. 2007;8:1708–1721. doi: 10.1111/j.1600-0854.2007.00646.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Ishikawa T, Sakakibara H, Oiwa K. The architecture of outer dynein arms in situ. J Mol Biol. 2007;368:1249–1258. doi: 10.1016/j.jmb.2007.02.072. [DOI] [PubMed] [Google Scholar]
- 53.Mizuno N, Narita A, Kon T, Sutoh K, Kikkawa M. Three-dimensional structure of cytoplasmic dynein bound to microtubules. Proc Natl Acad Sci USA. 2007;104:20832–20837. doi: 10.1073/pnas.0710406105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Nicastro D, et al. The molecular architecture of axonemes revealed by cryoelectron tomography. Science. 2006;313:944–948. doi: 10.1126/science.1128618. [DOI] [PubMed] [Google Scholar]
- 55.Ueno H, Yasunaga T, Shingyoji C, Hirose K. Dynein pulls microtubules without rotating its stalk. Proc Natl Acad Sci U S A. 2008;105:19702–19707. doi: 10.1073/pnas.0808194105. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.