Abstract
Palindromati, the massive host-edited synthetic palindromic contamination found in GenBank, is illustrated and exemplified. Millions of contaminated sequences with portions or tandems of such portions derived from the ZAP adaptor or related linkers are shown (1) by the 12-bp sequence reported elsewhere, exon Xb, 5′ CCCGAATTCGGG 3′, (2) by a 22-bp related sequence 5′ CTCGTGCCGAATTCGGCACGAG 3′, and (3) by a longer 44-bp related sequence: 5′ CTCGTGCCGAATTCGGCACGAGCTCGTGCCGAATTCGGCACGAG 3′. Possible reasons for why those long contaminating sequences continue in the databases are presented here: (1) the recognition site for the plus strand (+) is single-strand self-annealed; (2) the recognition site for the minus strand (−) is not only single-strand self-annealed but also located far away from the single-strand self-annealed plus strand, rendering impossible the formation of the active EcoRI enzyme dimer to cut on 5′ G/AATTC 3′, its target sequence. As a possible solution, it is suggested to rely on at least two or three independent results, such as sequences obtained by independent laboratories with the use, preferably, of independent sequencing methodologies. This information may help to develop tools for bioinformatics capable to detect/remove these contaminants and to infer why some damaged sequences which cause genetic diseases escape detection by the molecular quality control mechanism of cells and organisms, being undesirably transferred unchecked through the generations.
Introduction
Apalindromic sequence is a sequence that says the same if read from the plus or the minus strand; that is, the numerous restriction sites of the double-helix DNA that are subject to enzymatic digestion. In grammar, this is equivalent to a sentence that reads the same from left to right than from right to left, that is, a 14-letter phrase: → MAPS DNA AND SPAM ← except that more complexity is to be found in the DNA in the form of a bi-dimensional complement of the double-helix (G-C and A-T), something even more complex than the closest examples obtained by using only words, examples lacking of such a differentially paired complement:
Similarly, palindromati is an artificial phenomenon of hetero-transcription contaminating millions of sequences in the GenBank, a nucleic acids database, through the sequence 5′ CTCGTGCCGAATTCGGCACGAG 3′ or its derivatives (Castro-Chavez, 2004). Palindromati is a laboratory artifact produced by a biological host (yeast or bacteria) interacting with a vector equipped with adaptors. Through host–vector interactions, those adaptors are generating either full or partial palindromes or palindromic sequences capable of single-strand self-annealing that are preventing them from being degraded by restriction enzymes such as the active dimer formed by EcoRI (Castro-Chavez, 2010, 2011). The normal digestion of EcoRI is represented in Figure 2A. The contamination by linkers may include the complete adaptor sequence or longer tandem repeats of it or of its fragments, contaminating DNA and/or RNA (cDNA) sequences, and/or any other technology depending on sequences of nucleic acids such as microarrays.
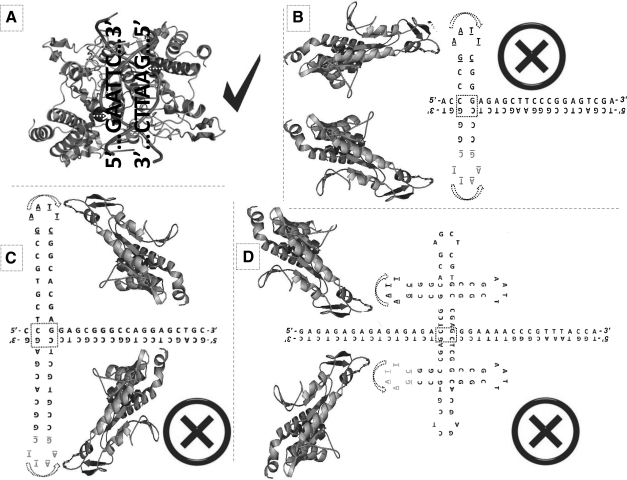
FIG. 2.
Representation of the lack of digestion by EcoRI: (A) The DNA restriction site where each monomer of the EcoRI dimer properly cuts: 5′- G/AATTC 3′. Lack of digestion of the single-strand self-annealed contaminating palindromes derived from the EcoRI linker ZAP adaptor: (B) the contaminated human ACAT1 L21934 (Li et al., 1999), a sequence that includes exon Xb (5′ CCGAATTCGG 3′). (C) Single-strand self-hybridizations of a 22-bp palindromati found in human phosphatase 1, catalytic subunit α cDNA, clone BC004482.2 (Strausberg et al., 2002). (D) Single-strand self-hybridizations of a 44-bp palindromati found in the alcohol dehydrogenase 7 (ADH7) cDNA AF195867.1 of Vitis vinifera (Or et al., 2000), its 5′ GA tandem also seems to be a contaminant.
Exon Xb is the name given by Li et al. (1999) to a palindromic linker of unknown origin (Zhao et al., 2009) represented by these authors as the 10-bp palindrome 5′ CCGAATTCGG 3′; however, due to its self-annealing properties, it actually takes the form 5′ CCCGAATTCGGG 3′, artificially adding the C at its 5′ side (apparently from human Chromosome 7), and the G at its 3′ side (apparently from the ACAT1 gene present in human Chromosome 1). It was named exon Xb by its authors to distinguish it from the exon Xa, a 1277-bp sequence at its 5′ side.
The most striking evidence pointing toward exon Xb being an artifact is when Chang et al. (1993) reported receiving the DNA library from Kodama, who donated his construct of a lambda-ZAP II library containing the cDNAs of the human macrophage line THP-1 (Kodama et al., 1990; Matsumoto et al., 1990); it was in 1990 when the ZAP adaptors reached the molecular research market (Yoshikawa et al., 1997). Additionally, “restriction fragments were subcloned into linker sites of pBluescript vectors (Stratagene) for sequencing” (Li et al., 1999).
Some of the current computational reviews of trans-splicing quoting the initial article of the exon Xb declare that to rule out the possibility of this hybrid mRNA as being a ligation artifact produced during cDNA synthesis in vitro, its authors performed RT–PCR experiments (of human brain, intestine, and liver), obtaining results apparently consistent with the trans-splicing hypothesis (Herai and Yamagishi, 2010); however, those RT-PCR experiments were done by the same group that initially reported exon Xb. Another article including Li's 1999 reference also used it as example of trans-splicing of “human acyl-CoA:cholesterol acyltransferase-1 (ACAT-1) and Xa exon, respectively on chromosomes 1 and 7” (Mayer and Floeter-Winter, 2005); however, in this case as in many others (Schoenfelder et al., 2010; Roy et al., 2011, etc.), the authors completely ignore the central palindromic exon Xb, which is apparently the responsible for the artificial phenomenon of hetero-transcription. We need to remember that every computer expert and data analyst is only going to receive what is deposited in the molecular databases; information that ought to be analyzed with strict stringency by both the submitter before uploading it online, and by the curator's careful cleansing and processing of such information publicly accessible to everybody.
The first sequence reported by Yoshikawa et al. (1997) in a Letter to the Editor as the most prominent contaminant, was one of five earlier instances of methodological nucleic acid contaminants; for example, it was the sequence under our consideration here that was presented in their Table 1 as: 5′ (G)AATTCGGCACGAG 3′ (Stratagene, ZAP library adaptor, commercially available in 1990). There, the G in parentheses of the 5′ end was not originally included in the manufacturers' sequence, but was added by Yoshikawa because the cDNA sequences studied by them contained an extra G at the 5′ end, added maybe by the host–vector interaction (see below).
Table 1.
Sequences with >22 Bases of Contaminating Palindromic Nucleotide Fragments in Tandem of the ZAP Adaptor
| # | GenBank ID | Position of the contaminating sequence of the EcoRI recognition site highlighted inbold |
|---|---|---|
| 1 | AF230097 | 3 AATTCGGCACGAGCTCGTGCCGAATTCGGCACGAG 37 |
| 2 | AY109498.1 | 1536 GAATTCGGCACGAGCTCGTGCCGAATTCGGCACGAG 1571 |
| 3 | AB205148.1 | 89 GAATTCGGCACGAGCTCGTGCCGAATTCGGCACGAG 124 |
| 4 | AJ250043.1 | 10 CGGCACGAGCTCGTGCCGAATTCGGCACGAG 40 |
| 5 | AM939570.1 | 9 CGGCACGAGCTCGTGCCGAATTCGGCACGAG 39 |
| 6 | AY074413.1 | 6 CTCGTGCCGAATTCGGCACGAGCTCGTGCCG 36 |
| 7 | AF069324 | 11 CTCGTGCCGAATTCGGCACGAGCTCGTGCCG 41 |
| 8 | AJ131096.1 | 156 GAGCTCGTGCCGAATTCGGCACGAG 180 |
| 9 | AY166797.1 | 48 GAGCTCGTGCCGAATTCGGCACGAG 72 |
| 10 | AF052221.1 | 185 GAGCTCGTGCCGAATTCGGCACGAG 209 |
| 11 | NM_001082374.1 | 39 GAGCTCGTGCCGAATTCGGCACGAG 63 |
| 12 | DQ118594.1 | 683 CTCGTGCCGAATTCGGCACGAGGCCTCGTGCCGAATTCGGCACGAG 728 |
| 13 | DQ394294.1 | 72 CTCGTGCCGAATTCGGCACGAGGCCTCGTGCCGAATTCGGCACGAG 117 |
| 14 | AF332963.1 | 100 CTCGTGCCGAATTCGGCACGAGGCCTCGTGCCGAATTCGGCACGAG 145 |
| 15 | AB298390.1 | 143 CTCGTGCCGAATTCGGCACGAGGCCTCGTGCCGAATTCGGCACGAG 188 |
| 16 | AF499715.1 | 1 AATTCGGCACGAGGCCTCGTGCCGAATTCGGCACGAG 37 |
| 17 | X69524.1 | 1 GAATTCGGCACGAGCTCGTGCCGAATTCGGCACGAG 36 |
| 18 | AF216582.1 | 1 GGCACGAGCTCGTGCCGAATTCGGCACGAG 30 |
| 19 | AF496666.1 | 1 GGCACGAGCTCGTGCCGAATTCGGCACGAG 30 |
| 20 | AY220737.1 | 1 GCACGAGCTCGTGCCGAATTCGGCACGAG 29 |
| 21 | M99575.1 | 1 GCACGAGCTCGTGCCGAATTCGGCACGAG 29 |
| 22 | AY488031.1 | 1 GGCACGAGCTCGTGCCGAATTCGGCACGAG 30 |
| 23 | AF308594.1 | 1 GGCACGAGCTCGTGCCGAATTCGGCACGAG 30 |
| 24 | AB104619.2 | 1 GGCACGAGCTCGTGCCGAATTCGGCACGAG 30 |
| 25 | AY029473.2 | 1 GGCACGAGCTCGTGCCGAATTCGGCACGAG 30 |
| 26 | U44430.1 | 1 GGCACGAGCTCGTGCCGAATTCGGCACGAG 30 |
| 27 | AF171857.1 | 1 GGCACGAGCTCGTGCCGAATTCGGCACGAG 30 |
| 28 | AF001136.1 | 1 GGCACGAGCTCGTGCCGAATTCGGCACGAG 30 |
| 29 | U60149.1 | 1 GGCACGAGCTCGTGCCGAATTCGGCACGAG 30 |
| 30 | AF487461.2 | 1 GCACGAGCTCGTGCCGAATTCGGCACGAG 29 |
| 31 | AY214171.1 | 1 GCACGAGCTCGTGCCGAATTCGGCACGAG 29 |
| 32 | AY531553.1 | 1 GCACGAGCTCGTGCCGAATTCGGCACGAG 29 |
| 33 | AF295637.1 | 1 GCACGAGCTCGTGCCGAATTCGGCACGAG 29 |
| 34 | AB294378.1 | 276 GAGCTCGTGCCGAATTCGGCACGAG 300 |
| 35 | DQ058828.1 | 1 CTCGTGCCGAATTCGGCACGAGCTC 25 |
| 36 | SJU50847 | 55 CTCGTGCCGAATTCGGCACGAGCTC 79 |
Examples taken from 1803 results of the Nucleotide collection (nr/nt) database [search done in Nov 2010]; only one example is presented per organism: 1. Homo sapiens, Hdac8, 2. Zea mays, CL28 mRNA, 3. Cynops pyrrhogaster, Wnt2b, 4. Anisakis simplex, mRNA for the IAA99-ASL3-15A gene, 5. Antheraea mylitta, fpi-1, 6. Trichosurus vulpecula, TrvuVK65 IgK-LCVR, 7. Mesembryanthemum crystallinum, 26S proteasome S5A, 8. Picea abies, microsatellite RNA, 9. Citrus unshiu, Ggps, 10. Lolium perenne, 4CL1, 11. Oryctolagus cuniculus, basigin BSG, 12. Karlodinium micrum, chloroplast psaE, 13. Mesostigma viride, ketol-acid reductoisomerase, 14. Polytomella sp., ferrochelatase, 15. Tamarix androssowii, MT1, 16. Thellungiella halophila, lipid transfer protein 4-like, 17. Mantoniella squamata, cabc1, 18. Avena sativa, Aldo, 19. Trypanosoma cruzi, unknown mRNA, 20. Hordeum vulgare, LoxA, 21. Babesia bovis, 85 kDa merozoite protein. 22. Capsicum annuum, RpL19 23. Crotalus durissus terrificus, bradykinin potentiating peptide, 24. Polyplastron multivesiculatum, celA, 25. Schizophyllum commune, ftt1, 26. Trametes versicolor, LccI, 27. D. melanogaster, clone 259, 28. Pinus radiata, PrCO, 29. Beta vulgaris, plasma membrane major intrinsic protein 3, 30. Gossypium hirsutum, Lecrk, 31. Paralichthys olivaceus (Japanese flounder fish) CPH, 32. Ginglymostoma cirratum, IgW, 33. Elaeis guineensis, calmodulin, 34. Dianthus caryophyllus, DcA93 glucosyltransferase, 35. Necator americanus, CLE, 36. Schistosoma japonicum, triosephosphate isomerase [all sequences, in the direction 5′–3′]. 20 examples more at Castro-Chavez (2011), 1268 additional examples taken from the GenBank can be found at www.reocities.com/plin9k/1200.zip, 166 examples including its contaminating translation into proteins can be found at www.reocities.com/plin9k/t2.htm; also see Table 2. Numbers at both sides of the sequences correspond to the position of the palindromic linker within the given sequence.
Yoshikawa et al. (1997) documented finding ∼88 sequences contaminated by the ZAP library adaptor in 1997 (here, in Appendices A–C you can find links to >1200 examples). In some of them “the match began with part or all of the EcoRI site (GAATTC) deleted”; the remarkable disappearance of the linker's core seems to be an additional host–vector interaction which is removing the palindrome by an apparent transposon-like mechanism. This mechanism seems to be producing heterogeneous transcripts as a result of its linking effects; for example, sequence AV728255 contains fragments of the vector only, lacking the linker. This additional and peculiar phenomenon of the palindromic linker's disappearance, or of portions of it, was noticed for the second time in the footnotes of their Table 1 as the “number of sequences that are identical to the adaptor sequence minus a complete EcoRI site (GAATTC)” (Yoshikawa et al., 1997); however, they did not provide the possible molecular mechanism for the intriguing palindromic formation and/or for the disappearance of the palindromic linker itself.
Seven years after Yoshikawa's initial findings and warnings, it was again reported that a dimer of the commercially available Stratagene ZAP adaptor was still contaminating hundreds of sequences of nucleic acids, presented in both Table 1 and in Figure 1 of a report published by Coker and Davies (2004) as 5′ CTCGTGCCG/AATTCGGCACGAG 3′ (EcoRI's cutting site underlined here as G/A). These authors also included in their drawing, and above this sequence, the same shorter double-stranded ZAP adaptor reported by Yoshikawa et al. (1997). Further, the Figure 1 done by Coker and Davies (2004) presented the EcoRI cutting space or gap between G and A in both cDNA strands. However, in their Table 1, Coker and Davies did not include the initial G added to the ZAP adaptor by Yoshikawa in parenthesis (G), nor did they provide any explanation for not including it, or for the possible origin of the palindrome-directed phenomenon of hetero-transcription. Here it will be shown that the most probable reason is that the cDNA strands of the palindromic adaptor quickly self-anneal independently from each other, preventing them from being digested by the EcoRI dimer.
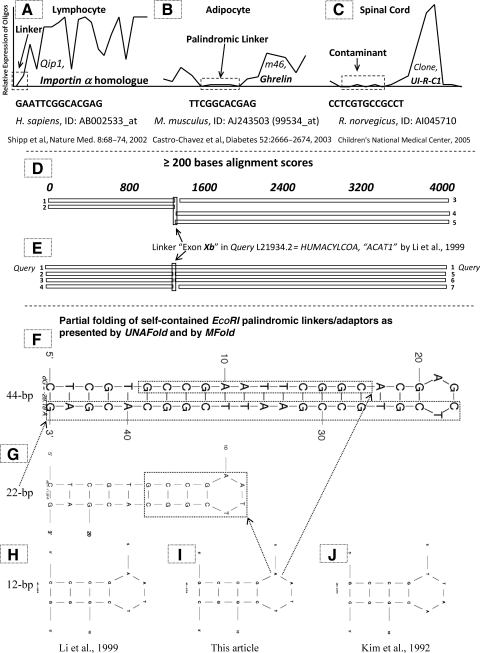
FIG. 1.
Microarray results of sequences including contaminating adaptor fragments, in a human gene (A), and in a mouse gene (B); for comparison, a related contaminant found in rat (C) is presented; similar to “the EcoRI site (GAATTC) deleted” described by Yoshikawa et al., 1997. >70 additional examples with the palindromic contaminant taken from the Affymetrix microarrays can be found at www.reocities.com/plin9k/affy70.zip. The drop of microarray expression to zero in (A), (B), and (C) demonstrates the absence of palindromic linkers or adaptors in the analyzed tissues (images stored by the author at www.iscid.org/papers/Chavez_Palindromati_101505.pdf). In (D) and (E) we see the BLAST analysis of sequence L21934.2 or HUMACYLCOA corresponding to the heterogeneous ACAT1 reported by Li et al. (1999). The gap on the sequences below and above it indicates the absence of exon Xb (Li et al., 1999) in ACAT1 human sequences reported by other groups. (D) Sequences present in the Human genomic plus transcript database: (D-1) Chr. 7 genomic contig, GRCh37 (NT_007933.15), (D-2) Chr. 7 genomic contig, alternate assembly by HuRef (NW_001839071.2), (D-3) Sterol O-acyltransferase 1 (SOAT1), transcript variant 688113 (NM_003101.4), (D-4) Chr. 1 genomic contig, GRCh37 (NT_004487.19), (D-5) Chr. 1 genomic contig, alternate assembly by HuRef (NW_001838533.2). (E) Sequences present in the Nucleotide collection (nr/nt) database: (E-1) HUMACYLCOA H. sapiens acyl-coenzyme A: cholesterol acyltransferase (L21934.2), (E-2) H. sapiens PAC clone RP4-797C5 from Chr. 7 (AC004888.1), (E-3) Pan troglodytes BAC clone CH251-572C18 from Chr. 7 (AC187744.3), (E-4) Pan troglodytes BAC clone RP43-28H17 from Chr. 7 (AC146259.4), (E-5) Pan troglodytes sterol O-acyltransferase 1, variant 2 (SOAT1) predicted (XM_514030.2), (E-6) H. sapiens cDNA: FLJ22958 fis, clone KAT09975, similar to HUMACYLCOA (AK026611.1), (E-7) H. sapiens sterol O-acyltransferase 1 (SOAT1), transcript variant 688113 (NM_003101.4, BC028940.1). (F) Partial self-annealing proposed by UNAFold for the 44-bp EcoRI contaminating palindrome, (G) Self-annealing of the 22-bp contaminant and in (H–J), of the 12-bp contaminants and/or synthetic palindromes. ACAT1, acyl-CoA:cholesterol acyltransferase-1.
The letter by Yoshikawa et al. (1997) cited by Coker and Davies (2004), was also cited in a book describing that sequence databases include contaminating sequences, pieces of foreign sequence that intentionally or accidentally were introduced at various steps of the cloning procedure or by recombination events in yeast or bacteria. These contaminations may cause problems for, for example, sequence analysis and database searching (Kampen and Horrevoets, 2006).
A recent work making reference to Coker and Davies (2004) was found in a software proposal (SeqTrim), which, according to its authors, “is under continuous development,” including its added purpose of removing artifacts caused by adaptors such as the ZAP DNA dimers (Falgueras et al., 2010). With this effort, plus others similar to it (Xu et al., 2007), we expect to help in restoring and cleansing the databases of nucleic acids while reducing as much as possible their contamination by adaptors.
However and apparently unbeknownst of the important warnings by Yoshikawa, a work based upon an artifact that included a palindromic sequence was published by Li et al. (1999). This methodological artifact was characterized in that article by its authors as if it were a biologically significant and naturally occurring phenomenon in Homo sapiens, being followed by more articles written by the same group that were based on the same artificial sequence (Yang et al., 2004; Yao et al., 2005; Chen et al., 2008; Chang et al., 2009; Zhao et al., 2009). Here, such sequence will be evaluated and deemed a methodological artifact while exemplifying and illustrating the possible reasons why the dimer of the restriction enzyme EcoRI is still unable of digesting these linking palindromes when they are present within these hetero-transcripts formed artificially.
Materials and Methods
General BLAST comparisons
For the online comparison of the 12, 22, and 44-bp palindromic contaminants, the BLAST collection of nucleotides was used (Wolfsberg and Madden, 2001). When searching for homologues of 5′ CTCGTGCCGAATTCGGCACGAGC TCGTGCCGAATTCGGCACGAG 3′, the 44-bp palindromic contaminant, 1803 sequences from the Nucleotide collection (nr/nt) database were obtained by using the default settings [search done in November, 2010]. However, if the Algorithm Parameters were adjusted to 20,000 for the Max target sequences and to 106 for the Expect threshold, the number of sequences increased at least approximately fivefold. However, in the new version of BLAST (http://blast.ncbi.nlm.nih.gov), when testing or probing the 44-bp palindromic sequence and in disregard of the adjustments made to the Algorithm Parameters, only 36 sequences from the current Human genomic+transcript collection, 26 sequences from the current Mouse genomic+transcript collection, and 393 sequences from the so-called Others (nr etc.) collection were obtained, but if we go back in time to 2005, and by using Blastn (nucleotide to nucleotide alignments) while selecting the nonredundant (nr) nucleic acid database sequences, a number of 6010 BLAST hits was obtained under the next query conditions: (a) 106 as the minimum expected number, and (b) 1000 as the number of descriptions and of alignments; it will be seen below that the high number of contaminated sequences continues today in the GenBank, even if their presence has been mostly masked by the current version of BLAST. The mRNA sequences present in the GenBank are single-strand products of double-helix cDNA experiments performed to increase stability, being this the methodological reason for the use of EcoRI in an attempt to individually cut these concatenated mRNA messengers and/or ORFs. The examples presented in this report include the basic core of the contaminated sequence, which is 5′ CCGTTAACGG 3′.
A specific full-sequence BLAST comparison
To exemplify and illustrate palindromati, the closest related matches for the GenBank contaminated ACAT1 sequence L21934 reported by Li et al. (1999) will be presented. The synthetic contaminant only appears in the sequence initially reported by Chang et al. since 1993 (L21934) and analyzed by Li et al. until 1999; this artificial sequence currently has two different names: L21934.2 and HUMACYLCOA. Three databases were compared to L21934: (1) the Human genomic plus transcript database, described by BLAST as Human genomic+transcript, or Human G+T, (2) the Nucleotide collection (nr/nt) database, literally described by BLAST as Others (nr etc.), and (3) the Mouse genomic plus transcript database, described by BLAST as Mouse genomic+transcript, or Mouse G+T.
Folding analysis of palindromic nucleotides
The folding of palindromic nucleic acids, their minimum energy requirements, and their melting temperatures were analyzed by using UNAFold and its sub-program Quickfold (Markham and Zuker, 2008).
Microarray analysis
Examples taken from microarray experiments done by the author with methodologies described elsewhere (Castro-Chavez et al., 2003; Castro-Chavez, 2004), and by others (Shipp et al., 2002, etc.), will be included as additional evidence demonstrating that palindromati is absent in living systems. The software used for the microarray analysis was dChip (Li and Wong, 2001).
GenBank data-mining
Given the masking of palindromic contaminants in the current BLAST, a third strategy was devised to find palindromic contaminants, consisting in applying directly to the NCBI nucleotide databases the words “EcoRI linker” [search done on July 07, 2011], finding 6289 sequences in the Expressed Sequence Tags (EST) sub-database, 51 in the Nucleotide database with at least 20 of them being cloning tools and/or patents. When the phrase “EcoRI adaptor” was used, only four sequences were obtained, three of them being patented sequences, and when the expression “EcoRI adapter” was used, four sequences were obtained with all of them being patents.
Results
One example will demonstrate the importance of a careful processing of these sequences of nucleic acids by an expert curator to prevent the danger of following an artificial palindromic (e.g., palindromati or palindromatic) sequence as if it were a natural phenomenon. Such a situation could be capable once more to lead toward erroneous biological or pharmaceutical conclusions, as seen in the biological aspect of the earliest article of a methodological palindromic artifact that was presented as natural heterogeneous trans-splicing (Li et al., 1999), and the several articles published by the same group based on it and/or derived from it (Yang et al., 2004; Yao et al., 2005; Chen et al., 2008; Chang et al., 2009; Zhao et al., 2009).
Two sets of experimental results will be presented (Fig. 1A–E), demonstrating that the palindromic linker is not present in cells and/or in tissues of living systems.
Figure 1 shows that the contaminated sequences present in the Affymetrix oligoarrays that were used as probes included contaminating palindromes or fragments of them, remaining at a zero (0) level of expression in humans (Fig. 1A), and in both rodents, the mouse and the rat (Fig. 1B, C, respectively). This is similar to the empty gap obtained by BLAST as a result of comparing the queried L21934 sequence to the rest of sequences in the database; L21934 contains the palindromic contaminant exon Xb (Fig. 1D, E).
Thus far, BLAST indicates that there has not been an independent sequence validation for the heterogeneous ACAT1 L21934 (Fig. 1E), or for its linking exon Xb palindrome 5′ CCGAATTCGG 3′ (Fig. 1D, E), which means that exon Xb was absent in all related sequences.
The result of the BLAST search in the Human genomic plus transcript shows a gap, or empty space, instead of exon Xb in all sequences compared (Fig. 1D, E).
As shown in Figure 1D, the names of the longest sequences resulting from this initial H. sapiens BLAST comparison are, either sequences only at the left side (5′) of the void left by the L21934's exon Xb, with a size of ∼1300-bp: D-1) Chr. 7 genomic contig GRCh37 (NT_007933.15), and D-2) Chr. 7 genomic contig alternate assembly by HuRef (NW_001839071.2); or sequences only at the right side (3′) of the void left by the L21934's exon Xb, with a size of ∼2700-bp: D-3) Sterol O-acyltransferase 1 (SOAT1) transcript variant 688113 (NM_003101.4), D-4) Chr. 1 genomic contig GRCh37 (NT_004487.19), and D-5) Chr. 1 genomic contig alternate assembly by HuRef (NW_001838533.2). Here, sequences D-2 and D-5 corresponded to whole genome shotgun sequencing, while sequences D-4 and D-5 had at least five interspersed regions of less than a 200-bp homology (≥80 but ≤200).
Again, in the Nucleotide collection (nr/nt) comparison, an empty space appeared instead of exon Xb. At the 5′ or left side of the empty space, there was a sequence of the human Chromosome 7 (Fig. E-2), and at its 3′ or right side, both a human mRNA sequence highly similar to HUMACYLCOA (Fig. E-6), and the human mRNA variant transcript for sterol O-acyltransferase 1 (SOAT1), Fig. E-7; both of them seem to be the genuine ACAT1 synonymous. Similarly, two chimpanzee sequences were clustered at the left side of the gap (Fig. E-3, E-4), while another chimpanzee sequence appeared at the right side of the empty space left by the L21934's contaminant exon Xb (Fig. E-5). Figure 1E shows the results, where the first sequence that appeared upon doing this second BLAST comparison was the query sequence itself (L21934.2, version 2 of the sequence under consideration which still includes the contaminant exon Xb and its long artificially bound heterogeneous sequences at its left (5′) and its right side or 3′). Only the matching results for longest sequences (≥200) were taken into consideration for this report. Fig. 1E shows the query (E-1) HUMACYLCOA H. sapiens acyl-coenzyme A: cholesterol acyltransferase with a size of ∼4000-bp, being this the only one including exon Xb (being it the query sequence itself, L21934.2). Sequences at the left side (5′) only of the void left by the L21934's exon Xb with a size of ∼1300-bp were: E-2) H. sapiens PAC clone RP4-797C5 from Chr. 7 (AC004888.1), E-3) Pan troglodytes BAC clone CH251-572C18 from Chr. 7 (AC187744.3), E-4) Pan troglodytes BAC clone RP43-28H17 from Chr. 7 (AC146259.4). Sequences only at the right side (3′) of the void left by the L21934's exon Xb with a size of ∼2700-bp were: E-5) Pan troglodytes sterol O-acyltransferase 1 variant 2 (SOAT1) predicted (XM_514030.2), E-6) H. sapiens cDNA: FLJ22958 fis clone KAT09975 similar to HUMACYLCOA (AK026611.1), and E-7) H. sapiens sterol O-acyltransferase 1 (SOAT1) transcript variant 688113 (NM_003101.4, BC028940.1).
Here again, any resemblance or homology to exon Xb (5′ CCGAATTCGG 3′) or its resulting hetero-transcription is missing. The same query (L21934) was used for both analyses.
The human sequence SOAT1 (NM_003101.4) appeared in both databases used [as number 3 in the Human genomic plus transcript (Fig. 1D), and as number 7 in the Nucleotide collection (nr/nt) (Fig. 1E)], but only at the right-side of the artificial exon Xb, whereas the contaminated sequence L21934 appeared only in the second database, the Nucleotide collection (nr/nt).
The artificial heterotranscript L21934 was also evaluated against the mouse database collection called Mouse genomic plus transcript, where its only match was a shorter sequence at the 3′ side (∼1700-bp, NM_009230.3, Mus musculus sterol O-acyltransferase 1, Soat1), with similarity only to the right-side nucleotides of L21934 (from ∼1400 to 3100); and again, matching only the right side of the heterogeneously contaminated sequence L21934.
Like in a Russian matrioshka toy, one 12-bp sequence (Fig. 1I) is self-contained within the 22-bp sequence (Fig. 1G), whereas both the 12-bp and the 22-bp sequence containing it, are respectively self-contained within the 44-bp sequence (Fig. 1F). Two additional 12-bp palindromic sequences are included for comparison: (1) The L21934's exon Xb with its two flanking nucleotides: C in the 5′ side and G in the 3′ side respectively (Li et al., 1999), and (2) The artificial sequence number 138166-98-0 registered by Kim et al. (1992) for their NMR studies.
Exon Xb, the short 10-bp EcoRI linker 5′ CCGAATTCGG 3′, seems to correspond to the inner part of the dimerized ZAP adaptor used to obtain the clone for the gene reported in 1993 as L21934.
The main question that arises from the millions of EcoRI linker-related heterogeneous chimeras still present in the GenBank is, if exon Xb is the 10-bp sequence 5′ CCGAA TTCGG 3′, why it was not cut when subjected to treatment with the restriction enzyme EcoRI to separate the individual cDNAs and/or ORFs? The logical explanation seems to be that the original 12-bp double-stranded sequence (the 10-bp palindrome plus its two, one for each side, flanking nucleotides C and G), ended up separating its plus (+) and minus (−) strands followed by a single-strand self-annealing performed within each individual strand separately. So each strand was independently self-annealed (see Fig. 2B), preventing these self-annealed single-strands from being cut by the EcoRI normal digestion between the nucleotides G and A (Fig. 2A) of the original double-stranded DNA adaptor palindrome 5′ G/AATTC 3′.
The 22-bp palindromic sequence 5′ CTCGTGCCGAAT TCGGCACGAG 3′ was reported earlier by the author of this article as a sequence contaminant (Castro-Chavez, 2004, 2010), and is illustrated in Figure 2C.
In the case of the 5′ CTCGTGCCGAATTCGGCACGA GCTCGTGCCGAATTCGGCACGAG 3′ (Castro-Chavez, 2011), a longer 44-bp palindromic sequence, the author of this article first thought that it was self-annealed in a full appendage or zipper-like conformation. This conformation was, and still is, the only result given online by the software UNAFold through its Quickfold sub-program. However, on revisiting palindromati more closely now, and considering how it was possible for the self contained 22-bp palindromic EcoRI linkers that are present at both sides of the 44-bp sequence, not to be digested, a double single-strand self-annealing was found to be the most plausible cause. A double single-strand self-annealing preventing the EcoRI dimer digestion was an event much more complex than what was initially expected. Figure 2D presents this 44-bp palindromic double single-strand self-annealing, improving our understanding even beyond the UNAFold results (Fig. 1F) while exhibiting this symmetrically independent single-strand self-annealing as the most plausible reason for not being digested by EcoRI (Fig. 2D).
UNAFold and its Quickfold sub-program were insensitive to the less-energy requirement of the independent double self-annealing described here for the 44-bp palindromic sequence illustrated in Figure 2D. Quickfold only folded the 44-bp as a long and single double-helix appendage (Fig. 1F), ignoring the less-energy requirement for the independent single-strand self-annealing of each 22-bp side of this long and palindromic cDNA double-helix, a requirement to prevent them from being cut by digestion enzymes; for such reason, the 44-bp sequence was subdivided into its two 22-bp identical halves (5′ CTCGTGCCGAATTCGGCACGAG 3′). The next results were obtained by Quickfold for the 22-bp loop free-energy decomposition: ΔG=−11.39 kcal/mol ΔH=−77.90 kcal/mol ΔS=−214.44 e.u. Tm=90.1°C (Fig. 1G).
It needs to be noticed that marked with a dotted rectangle in Figure 2B–D is the center of the palindrome, having three H-bonds, bonds that seem to be providing a stronger theoretical stability at the center of these CGs; the horizontal CG pair could be providing an extra resonating strength, but more research needs to be done on the biochemistry of these interactions.
For comparison, the UNAFold's unsolved 44-bp palindromic sequence (5′ CTCGTGCCGAATTCGGCACGAGCT CGTGCCGAATTCGGCACGAG 3′) gave a loop free-energy decomposition of: ΔG=−28.18 kcal/mol ΔH=−173.20 kcal/mol ΔS=−467.58 e.u. Tm=97.3°C (Fig. 1F).
While as expected for a shorter fragment, the 12-bp contaminating palindromic sequence 5′ CCCGAATTCGGG 3′ required less energy, having a loop free-energy decomposition of ΔG=−3.35 kcal/mol, ΔH=−32.60 kcal/mol, ΔS=−94.31 e.u. and Tm=72.5°C (Fig. 1H, being this the Exon Xb).
So, according to UNAFold we can conclude that the theoretical Tm for the shortest 12-bp sequence is 72.5°C, while the theoretical Tm for the 22-bp sequence is 90.1°C.
By using the NCBI GenBank's phrase search as my third and last experimental strategy, it was found that the EcoRI linker has been patented several times; that is, (1) as sequence A04694 by E. Degryse [Patent number EP0258118-A1/1, 02-Mar-1988. Transgene S.A., France], (2) as sequence E04017 by M. Mita et al. [Patent: JP 1992330279-A/1, 18-Nov-1992; Takara Shuzo Co. LTD], (3) as the 12-bp A29547 sequence 5′ GAGGAATTCCTC 3′ by W.C. Mackellar and C.S. Robey [Patent: EP 0523296-A1/16, 20-Jan-1993; Eli Lilly & Comp.]; while (4) the so-called EcoRI linker or E03862 patented by T. Kishimoto, S. Niwa, and A. Uno [Patent: JP 1992228075-A/1, 18-Aug-1992; Sumitomo Electric Ind. LTD], corresponded to the 19-bp sequence 5′ GCAACCATGCCTAAGTTTG 3′, a sequence lacking the EcoRI restriction site!, (5) a 18-bp synthetic nucleotide EcoRI linker, A10450, with the sequence 5′ TGCCATGAATTCATGGCA 3′, and (6) a 282-bp Patent, A27290, starting with 5′ GGAATTCCC 3′, etc.
Table 1 shows that the palindromati exon Xb joins together fragments that are not naturally connected (Fig. 1A–E). Table 1 also shows 36 sequences contaminated by the ZAP-Adaptor's EcoRI palindromic linker longer than 22-bp; 20 different sequences related to this analysis are also published (Castro-Chavez, 2011).
Table 1 starts again with the human sequence AF230097 to emphasize the importance of cleaning the sequences of genes; in this case, the contamination is affecting a key nuclear protein (Hdac8) that controls the gene expression for the rest of the genes (Hu et al., 2000).
When the word “adaptor” was used in the word search strategy applied to the GenBank, millions of results were obtained, a total of 4,663,104 sequences that were divided in three groups: Nucleotide (131,544), EST (3,896,223), and GSS (635,337). Subdividing the first or Nucleotide group, we obtained: Bacteria (5266), INSDC (GenBank) (90,174), mRNA (104,071), and RefSeq (41,275). Millions of results were obtained when the word “adapter” was used in the GenBank's word search strategy, obtaining a total of 4,768,553 nucleotide sequences! These included: Nucleotide (9444), EST (4,733,490), and GSS (25,619). Subdividing the Nucleotide group, we had: Bacteria (1227), INSDC (GenBank) (5950), mRNA (3866), and RefSeq (3467).
At the end of the analysis, the word “linker” was used for the last word search, and again the results were above one million, 1,532,240 nucleotide sequences were obtained and divided into: Nucleotide (51,945), EST (1,398,405), and GSS (81,890). When the Nucleotide group for this category was subdivided, the following were obtained: Bacteria (1,707), INSDC (GenBank) (45,721), mRNA (8467), and RefSeq (6012). These are only the millions of sequences reported by their submitters as containing contaminants, by using the words adaptor, adapter, or linker, respectively.
However, very few groups report the presence of the EcoRI linker equal to or longer than 12-bp; for example, from the published articles based on the submitted GenBank sequences, only Hirama et al. (1991) mentioned the longer extension of the EcoRI linker, saying: misc_feature 1.14/note=adaptor, reporting it as having a length of 14-bp while exposing it as contaminant in their reported sequence×56703, a sequence for the rearranged T-cell receptor alpha chain of Mus musculus; however, a longer segment of 16-bp seemed to be the contaminant originated in the linker (5′ GAATTCGGCACGAGCT 3′). The presence of a 14-bp EcoRI linker was also reported in the unpublished sequences Y14272 for Pisum sativum's KdsA, AJ243499 for Medicago sativa cyc2.2, and in the patented sequence BD413899 containing a different linker, the SalI linker. A possible host–vector interaction re-arrangement of 27-bp started with 5′ GAATTC 3′ and was followed by 21 C's at its−3′ side, being labeled EcoRI linker by Eisenberg et al. (1990) for their sequence×54230.
Inoue et al. (1996) mentioned a 8-bp length of EcoRI linker contamination for their sequence D83948 (Rattus norvegicus S1-1); however, the palindromic linker seemed to have a length of more than 16-bp: 5′ GGCACGAGCTCGTGCCG 3′ here, missing its palindromic core as reported for other sequences by Yoshikawa et al. (1997); similar to the unpublished sequence A27280 for Homo sapiens TGR-CL3 (Patent: WO 9219733-A1 23 12-Nov-1992). Savas et al. (1994) mentioned in his GenBank submission that the first 6 bases are part of the EcoRI linker; however, again, the host–vector rearrangement interaction seemed here to extend 21 bases inside the sequence×78445 for the mouse Cytochrome P450 Cyp1-b-1 (oligo 5′ GAATT CGGCACGAACTCGTGC 3′), and the same was seen twice in the unpublished sequence A01270, which is a patented sequence from Taenia ovis for the 45W antigen, while E10125, an unpublished sequence for a chicken's leucocytozoon immunogenicity protein (Patent: JP 1995284392-A 1 31-Oct-1995), mentioned twice, at its beginning and at its end, the presence of 25 nucleotides corresponding to a different EcoRI adaptor, at the start of this sequence, 5′ GAATTCGAGGATCCGGG TACCATGG 3′, and at the end of it, 5′ CCATGGTACCCGG ATCCTCGAATTC 3′, producing a plasmid-like structure with a 25-bp zipper binding its single-strand extremes.
Finally, by comparing the translation from codons to proteins with the rotating genetic code (Castro-Chavez, 2010, 2011), the translated palindromes were detected as contaminants. Exemplified in Table 2 is the seriousness of the contamination by EcoRI adaptors and linkers if they are translated as being part of a human exon or protein.
Table 2.
Examples of Human Sequences Contaminated with the EcoRI Linker Translated Into Proteins
| # | ID | Gene/protein (gene symbol) | Linker and its translation in amino acids (in bold) as presented in the GenBank | Corresponding references according to the GenBank and/or closest related match |
|---|---|---|---|---|
| 1 | U58090 | Cullin gene family member, Hs-cul-4A | 2_AATTCGGCACGAGCTCGTGCCGCT_25 NSARARAA | Kipreos et al. (1996) |
| 2 | U28831 | Protein immuno-reactive with anti-PTH polyclonal antibodies | 2_GCACGAGCTCGTGCCGAT_19 ARARAD | Kumar et al. (1995) |
| 3 | BC041619 [chimeric clone, 2008] | KIAA0404, for IMAGE:5923662 [R: hypothetical protein MGC16044] | 1397_CCCTCGTGCCGAATTCGGCACGAG_1420 PSCRIRHE | Strausberg et al. (2002) |
| 4 | AF176705 | F-box protein FBX10 (PINX1) [R: vector] | 1624_CCTCGTGCCGAATTC_1638 PRAEF | Winston et al. (1999) |
| 5 | X85792 | Vpr binding protein 1 | 1_TCGTGCCGAATTCGGCACGAG_21 SCRIRHE | Le Rouzic et al. (2002); Benichou et al. (Unpublished) |
| 6 | AF151109 | Putative BRCA1-interacting protein (BRIP1) | 1_GGCACGAGCTCGTGCCGC_18 GTSSCR | Wang et al. (2000); Wang et al. (Unpublished) |
| 7 | AF146697 | FOXP1 | 25_AAGAATTCGGCACGAGCT_42 KNSARA | Banham et al. (2001) |
| 8 | AY245868 | CDS for aldehyde oxidase-like protein (AOX2) pseudogene | 1_AAGAATTCGGCACGAGCA_18 LNSARA | Wright (Unpublished) |
| 9 | NM_173552* & BC037293 [437 to 457] | Predicted Hypothetical protein MGC33365 | *710_TTCGGCACGAGCTGGTGCCGC_730 FGTSWCR | Ota et al. (2004); Strausberg et al. (2002) |
Dozens of examples for other organisms can be found at www.reocities.com/plin9k/t2.htm. Numbers at both sides of the sequences correspond to the position of the palindromic linker within the sequences.
www.iscid.org/papers/Chavez_Palindromati_101505.pdf, where we can find 70 examples more.
Discussion
In the same way that a growing interest is currently underway to evaluate, to correct, and to prevent further errors in the submitted third dimensional (3D) structures of proteins (Chetty et al., 2009; Ginzinger et al., 2010), interest is developing for the quality control and cleansing of sequences of the nucleic acids deposited in the GenBank, which are used in research technologies fully depending on their quality, such as the Affymetrix microarrays and multiple others (Castro-Chavez, 2004).
As mentioned earlier, the proposal of the palindromati effect as being a palindromic single-strand self-annealing (Fig. 2B–D) was reinforced by the extra two nucleotides (underlined in italics below) that are present in the palindromic neighborhood of the 10-bp EcoRI restriction site of the adaptor nucleotide depicted in the Figure 1 published by Li et al. (1999): 5′ CCCGAATTCGGG 3′. Possible editing mechanisms of the host–vector interaction made it a 12-bp palindrome. The two extra flanks were noticed here by the dotted rectangle at the center of the symmetrical self-annealed structure derived from it (Fig. 2B), and a similar CG center was found in the 22-bp sequence (Fig. 2C), and in the 44-bp sequence (Fig. 2D).
Similar palindromes to the 12-bp palindromati containing exon Xb were found twice in the 2264-bp sequence×93499.1 reported by Vitelli et al. (1996) of the human RAB7 protein at positions 434 and 1584.
As seen in Results, thus far there has not been an independent corroboration for exon Xb and its artificially linked hetero-transcript L21934 in any of the BLAST databases evaluated, which were: the Human genomic plus transcript, the Nucleotide collection (nr/nt), and the Mouse genomic plus transcript.
Palindromati seems to be a sub-product of an edited and dimerized bi-tandem of the ZAP adaptor done by the host, present in its totality, or in portions or their tandems, in millions of GenBank sequences (Castro-Chavez, 2004, 2010, 2011), including basically every sequenced organism and every sequencing laboratory in the world.
The quality control of deposited DNA and RNA sequences is of vital importance to reach meaningful molecular and biological conclusions in our depiction of living systems, but also for their comparison to detect statistical outliers deemed as disease based on the classic circular genetic code (Castro-Chavez, 2010), for compatible genomics (Castro-Chavez, 2011), for our gene expression studies (Castro-Chavez et al., 2003; Vickers et al., 2010), for the study of its phenotypes in health and disease (Castro-Chavez, 2004), for the medical treatment of diseases, and/or for the discovery of new proteins and isoforms based solely on the sequences of nucleic acids present in the GenBank. The discovery of genes being expressed in a particular tissue can now be contrasted from artifacts produced by the methodologies used.
As shown in Figure 1A, B, microarray experiments were done on rodents and men in the past by the author and by others (with the corresponding RT-PCRs to cross-validate the results), without noticing the actual expression of any heterogeneous transcript or the presence of exon Xb, even when probing multiple heterogeneous sequences present in the GenBank that were different to the 5′ CCGAATTCGG 3′ palindromati reported here (i.e., Fig. 1C).
A previously reported sequence having some resemblance to the palindromati discussed here, except that it had the paired central nucleotides inverted (from AATT to TTAA), is 5′ GCCGTTAACGGC 3′, a synthetic DNA duplex dodecamer containing the HpaI restriction site GTT/AAC originally used as a model for the determination of nucleic acid backbone conformations done by NMR (Kim et al., 1992). Its authors registered their synthetic peptide (Registry No. d(GCCGTTAACGGC), 138166-98-0). In our analysis, this 12-bp sequence 5′ GCCGTTAACGGC 3′ (Fig. 1J) provided a loop of free-energy decomposition according to Quickfold of: ΔG=−3.75 kcal/mol ΔH=−34.40 kcal/mol ΔS=−98.82 e.u. Tm=75.0°C (Fig. 1J), identical to the loop of free-energy decomposition for the sister sequence 5′ GCCGAATTCGGC 3′ (Fig. 1I). Chao et al. (1990) presented a pair of unrelated “homologous palindromatic” sequences including the 12 core nucleotides of Fig. 1J interspersed “in the active promoter region” of BiP/GRP78 5′ GGCC--GCTTCGAATC-GGCA 3′ and of GAPDH 5′ TGCCCAG-TT-GAA-CCAGGCG 3′.
The hosts are living systems and as such, they do their own molecular modifying (dimerizing, editing, adding or removing nucleotides, tandemizing or performing transposon-like displacements, shortenings, and relocations, etc.). It is assumed that deposited sequences are without natural modification; however, a critical analysis and comparison of the nucleic acids will always be very helpful to succeed in obtaining biologically and medically meaningful results.
Related to the exon Xb present in the sequence L21934, a doubt remained since the beginning, because “DNA rearrangements have not been conclusively ruled out” (Finta and Zaphiropoulos, 2002). Since then, no group doing independent sequencing, preferably with independent methodologies, has corroborated the natural existence of the L21934 sequence. As noticed at the beginning, even its authors indicated recently that, “up to now, the formation of exon Xb is still unclear” (Zhao et al., 2009), being again exon Xb the EcoRI linker sequence 5′ CCGAATTCGG 3′ originally presented by Li et al. (1999), who since the beginning declared: “Within this region, an EcoRI site (nt 1282–1287) is present,” and also: “These two primers are located in regions flanking the EcoRI site described above,” failing to ask or explain why that EcoRI site remained uncut after being treated with the restriction enzyme.
Recognition of contaminating sequences different to EcoRI have been presented by Yoshikawa et al. (1997), and by Coker and Davies (2002); and because nucleic acid sequences are uploaded daily, contaminated heterogeneous transcripts are unfortunately expected to increase.
Conclusions
As a possible solution to prevent the presence of artificially contaminated heterogeneous sequences in databases of nucleic acids, it is highly recommended to have at least two or three independent research groups properly equipped for nucleic acid research, using preferably independent sequencing methodologies (Illumina [Solexa] sequencing, 454 pyrosequencing, SOLiD sequencing, etc.), as a way to validate the sequences deposited in the GenBank to prevent reaching the wrong biological and/or medical conclusions.
An example of a same method used by three independent laboratories was a GWAS finding showing obesity predisposing SNPs in the first intron of the little-known human FTO gene (chromosome 16); FTO encodes a non-heme dioxygenase ubiquitously expressed in fetal and adult tissues, especially in the brain (Dina et al., 2007; Frayling et al., 2007; Scuteri et al., 2007). On the other hand, the analysis of the atomic composition of the 9/11 extremely toxic dust (Clark et al., 2001), an example discussed by Castro-Chavez (2011) as a possible overload for the molecular quality control mechanisms of the human body, dealt with the unusually high presence of strontium that was completely underreported. In this case, one laboratory from the U.S.G.S. used two different methodologies, dust leachate and X-ray fluorescence, finding an average Sr=1.1 mg/L and 726.61 ppm, respectively. The thousands of unprotected first responders are still unaware of the extreme toxicity of the 9/11 WTC dust breathed by them, and of its high presence of Sr and other highly unusual, possibly unstable, atoms and of carbon nanotubes (Castro-Chavez, 2011).* However, it is encouraging to see a growing number of studies that use multiple groups and/or methodological platforms plus different data analysis tools and algorithms (Kidd et al., 2010; Halper-Stromberg et al., 2011; Pinto et al., 2011). The same validating strategy of two or three independent groups, materials, and/or methodologies could be used in any kind of research, or at least for the careful researcher to apply these principles by himself; for example, the critical analysis of two discrepant sequences for the same gene obtained by two independent groups was noticed by a third group (Coker and Davies, 2002), and it was reported and corrected, an action that avoided it from being steered in the wrong direction; unfortunately, this was not the case for Li et al. (1999), with ∼90 articles that are citing it to date.
Palindromati was initially reported as a 22-bp contaminating sequence 5′ CTCGTGCCGAATTCGGCACGAG 3′ (Castro-Chavez, 2004), being here revisited and illustrated, finding further evidence that the 44-bp longer contaminating sequence, 5′ CTCGTGCCGAATTCGGCACGAGCTCGTGC CGAATTCGGCACGAG 3′ (Castro-Chavez, 2011) included tandems derived from the dimerized ZAP adaptor that is currently contaminating millions of GenBank sequences. Here, the palindromic self-annealing was also illustrated by using the human ACAT1 L21934 as one example of an heterogeneous contaminated sequence containing fragments of human Chromosome 7 and of Chromosome 1 that was presented as a genuine event, bound together by the contaminating 10-bp EcoRI linker sequence 5′ CCGAATTCGG 3′ (Li et al., 1999), making impossible for EcoRI to cut or digest due to its actual single-strand self-annealed 12-bp size: 5′ CCCGAATTCGGG 3′.
Thus far, it seems that the human eye has been better than computers in tracking those contaminants present within the sequences of the nucleic acids (Castro-Chavez, 2011).
When the palindromes or their cores are absent, as mentioned earlier, the possible cause may be a transposon-like mechanism inside the host (yeast or bacteria), due to the removal from the heterogeneous artificial sequence of the palindromic linker that was responsible for the heterogeneous chimeric sequences; this may be something similar to the jumping genes found by Barbara McClintock in corn (Ringertz, 1983; Fedoroff, 2001), and such are the ongoing findings of retrotransposons in yeast (Guarraia et al., 2007; Stanley et al., 2010; Bleykasten-Grosshans et al., 2011, etc.). As the palindromes vanish, this vanishing makes harder to track the palindromic sequences that were responsible for those contaminated sequences, leaving only the artificially heterogeneous transcripts as evidence that a palindromati event did happen.
The host many times edits, transposes, or even removes the original adaptor's sequence, or fragments of it, leaving the mechanism for some contaminant sequences only to our indirect inference. Sequences lacking the linker cannot be detected through the GenBank's word search strategies used here, leaving them to be found through BLAST and/or microarrays or similar technologies, being noticed as an empty hole or gap in the BLAST multi-sequence alignment, or in microarrays as the sudden drop to zero in the signal of part of a sequence (Castro-Chavez, 2004). For example, as noticed earlier by Yoshikawa et al. (1997), some of the contaminated sequences lack a complete EcoRI site (GAATTC), or have a gap of one nucleotide, while other sequences have an addition of two nucleotides (GC) (Castro-Chavez, 2011), something also presented here in sequences 12 to 16 from Table 1, with some of them being 46-bp long; this in itself may indicate that novel and additional editing processes are taking place within the host.
Here, the palindrome contamination present in databases of nucleotides was illustrated while proposing a path to control the problem; in brief, the reason why EcoRI was not capable to digest the sequence products of the ZAP adaptor is graphically shown in Figure 2B–D, concluding it to be due to: (a) the EcoRI recognition site for the upper or plus strand (+) being single-strand self-annealed or self-folded, while (b) the recognition site for the lower or minus strand (−) is not only single-strand self-annealed, but it is also located far away from the folded upper strand, making it unreachable to the EcoRI or even making impossible its dimer formation.
It seems that a minimum palindromic size of 12-bp is sufficient to trigger the single-strand self-annealing separately for each strand (+and−), preventing them from being cut by restriction enzymes; that is, 5′ CCCGTTAACGGG 3′, or 5′ GCCGTTAACGGC 3′, being the last one the matrioshka contained within the longer palindromes of 22 and 44-bp reported here in detail. The 12-bp minimum length phenomenon presented here is suggested as extensive for the prevention of cDNAs to be cut by any other restriction enzyme's palindromic target site, or consensus site, hence its importance.
In conclusion, the sequence contamination illustrated in this report and its removal from the databases are imperative, especially now that it was also recognized that nearly 20% of the nonhuman genomes that are present in the databases are also contaminated with human DNA belonging to the researchers (Longo et al., 2011). These have been 21 years of an undetected and unchecked contamination by the ZAP adaptor and related EcoRI linkers, independent of the massive contamination by the other adaptors and linkers.
In a parallel way, these studies may also help us understand why damaged or contaminant sequences causing genetic diseases escape from being detected and removed by the molecular quality control mechanisms present in cells and organisms, being equally and undesirably transferred unchecked while causing hereditary genetic diseases generation after generation. Recently, it was reported an ∼20% of DNA insertions in 185 humans (Mills et al., 2011), with most of them (19.18%) being caused by mobile elements, and that, plus the findings reported here, will certainly add a third dimensional (3D) perspective to the study of genetic diseases, either being hereditary, or even somatic, such as in cancer or cancer-like events (Kannan et al., 2011; Li et al., 2008), also removing there the artificial contaminants done by polymerase and reverse-transcriptase (Houseley and Tollervey, 2010). This novel 3D DNA perspective will be an important aspect for the research of the molecular rules governing biological change, in both health (Castro-Chavez, 2010) and disease (Castro-Chavez, 2011), something certainly deserving a minutely thorough analysis.
Appendix A
1268 examples of contamination taken from the GenBank: www.reocities.com/plin9k/1200.zip
Appendix B
166 examples, including contaminants translated into proteins: www.reocities.com/plin9k/t2.htm
Appendix C
74 examples including the palindromic contaminants present in the Affymetrix microarrays: www.reocities.com/plin9k/affy70.zip
Footnotes
See for example: “911stealth 9/11 WTC 10th Anniversary: Doug Copp - Homage to volunteer First Responders” http://www.youtube.com/watch?v=vW6FDcQPozl.
Acknowledgments
To Rainer B. Lanz and Lawrence L. Rudel for their encouraging support and personal communications; to Ellen M. Carroll, Ellen Anderson, and Tracy L. Duncan for help in preparing the article; to the Indian Institute of Technology Roorkee, Uttarakhand, India, especially to its students Neha Bisht and Krati Verma, for their encouraging willingness to collaborate with this postdoctoral fellow, to ReoCities and others for saving my files; but most specially, to the kind Editor of DNA and Cell Biology and to its peer-reviewers, thank you! This work was sponsored in part by NIH grants ROI-HL-63090 and T32 HL-07812.
Disclosure Statement
No competing financial interests exist.
References
- Banham A.H. Beasley N. Campo E., et al. The FOXP1 winged helix transcription factor is a novel candidate tumor suppressor gene on chromosome 3p. Cancer Res. 2001;61:8820–8829. [PubMed] [Google Scholar]
- Bleykasten-Grosshans C. Jung P.P. Fritsch E.S. Potier S. de Montigny J. Souciet J.L. The Ty1 LTR-retrotransposon population in Saccharomyces cerevisiae genome: dynamics and sequence variations during mobility. FEMS Yeast Res. 2011;11:334–344. doi: 10.1111/j.1567-1364.2011.00721.x. [DOI] [PubMed] [Google Scholar]
- Castro-Chavez F. Microarrays, antiobesity and the liver. www.medigraphic.com/pdfs/hepato/ah-2004/ah044c.pdf. Ann Hepatol. 2004;3:137–145. [PMC free article] [PubMed] [Google Scholar]
- Castro-Chavez F. The rules of variation: amino acid exchange according to the rotating circular genetic code. http://www.ncbi.nlm.nih.gov/pubmed/20371250. J Theor Biol. 2010;264:711–721. doi: 10.1016/j.jtbi.2010.03.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castro-Chavez F. The rules of variation expanded, implications for the research on compatible genomics. http://www.ncbi.nlm.nih.gov/pubmed/21743816. Biosemiotics. 2011;2011:1–25. doi: 10.1007/s12304-011-9118-0. [In press] [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castro-Chavez F. Yechoor V.K. Saha P. Martinez-Botas J. Wooten E. Sharma S. O'Connell P. Taegtmeyer H. Chan L. Coordinated upregulation of oxidative pathways and downregulation of lipid biosynthesis underlie obesity resistance in Perilipin knockout mice. A microarray gene expression profile. Diabetes. 2003;52:2666–2674. doi: 10.2337/diabetes.52.11.2666. [DOI] [PubMed] [Google Scholar]
- Chang C.C. Huh H.Y. Cadigan K.M. Chang T.Y. Molecular cloning and functional expression of human acyl-coenzyme A:cholesterol acyltransferase cDNA in mutant Chinese hamster ovary cells. J Biol Chem. 1993;268:20747–20755. [PubMed] [Google Scholar]
- Chang T.Y. Li B.L. Chang C.C. Urano Y. Acyl-coenzyme A:cholesterol acyltransferases. Am J Physiol Endocrinol Metab. 2009;297:E1–E9. doi: 10.1152/ajpendo.90926.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chao C.C. Yam W.C. Lin-Chao S. Coordinated induction of two unrelated glucose-regulated protein genes by a calcium ionophore: human BiP/GRP78 and GAPDH. Biochem Biophys Res Commun. 1990;171:431–438. doi: 10.1016/0006-291x(90)91411-k. [DOI] [PubMed] [Google Scholar]
- Chen J. Zhao X.N. Yang L. Hu G.J. Lu M. Xiong Y., et al. RNA secondary structures located in the interchromosomal region of human ACAT1 chimeric mRNA are required to produce the 56-kDa isoform. Cell Res. 2008;18:921–936. doi: 10.1038/cr.2008.66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chetty P.S. Mayne L. Lund-Katz S. Stranz D., et al. Helical structure and stability in human apolipoprotein A-I by hydrogen exchange and mass spectrometry. Proc Natl Acad Sci USA. 2009;106:19005–19010. doi: 10.1073/pnas.0909708106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clark R.N. Green R.O. Swayze G.A., et al. Environmental Studies of the World Trade Center area after the September 11, 2001 attack. U.S. Geological Survey. 2001. http://www.webcitation.org/5wtEMG1Hp http://www.webcitation.org/5wtEMG1Hp
- Coker J.S. Davies E. Identifying adaptor contamination when mining DNA sequence data. Biotechniques. 2004;37:194–198. doi: 10.2144/04372BM03. [DOI] [PubMed] [Google Scholar]
- Coker J.S. Davies E. Ree A.H., et al. Expression of a novel factor in human breast cancer cells with metastatic potential. Cancer Res. 2002;62:4164–4165. Correspondence re: [PubMed] [Google Scholar]
- Dina C. Meyre D. Gallina S., et al. Variation in FTO contributes to childhood obesity and severe adult obesity. Nat Genet. 2007;39:724–726. doi: 10.1038/ng2048. [DOI] [PubMed] [Google Scholar]
- Eisenberg M. Gathy K. Vincent T. Rawls J. Molecular cloning of the UMP synthase gene rudimentary-like from Drosophila melanogaster. www.springerlink.com/index/t1t385q745231w2j.pdf. Mol Gen Genet. 1990;222:1–8. doi: 10.1007/BF00283015. [DOI] [PubMed] [Google Scholar]
- Falgueras J. Lara A.J. Fernández-Pozo N. Cantón F.R. Pérez-Trabado G., et al. SeqTrim: a high-throughput pipeline for preprocessing any type of sequence reads. BMC Bioinformatics. 2010;11:38. doi: 10.1186/1471-2105-11-38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fedoroff N. How jumping genes were discovered. Nat Struct Biol. 2001;8:300–301. doi: 10.1038/86161. [DOI] [PubMed] [Google Scholar]
- Finta C. Zaphiropoulos P.G. Intergenic mRNA molecules resulting from trans-splicing. J Biol Chem. 2002;277:5882–5890. doi: 10.1074/jbc.M109175200. [DOI] [PubMed] [Google Scholar]
- Frayling T.M. Timpson N.J. Weedon M.N., et al. A common variant in the FTO gene is associated with body mass index and predisposes to childhood and adult obesity. Science. 2007;316:889–894. doi: 10.1126/science.1141634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ginzinger S.W. Weichenberger C.X. Sippl M.J. Detection of unrealistic molecular environments in protein structures based on expected electron densities. J Biomol NMR. 2010;47:33–40. doi: 10.1007/s10858-010-9408-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guarraia C. Norris L. Raman A. Farabaugh P.J. Saturation mutagenesis of a +1 programmed frameshift-inducing mRNA sequence derived from a yeast retrotransposon. RNA. 2007;13:1940–1947. doi: 10.1261/rna.735107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Halper-Stromberg E. Frelin L. Ruczinski I., et al. Performance assessment of copy number microarray platforms using a spike-in experiment. Bioinformatics. 2011;27:1052–1060. doi: 10.1093/bioinformatics/btr106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herai R.H. Yamagishi M.E. Detection of human interchromosomal trans-splicing in sequence databanks. Brief Bioinform. 2010;11:198–209. doi: 10.1093/bib/bbp041. [DOI] [PubMed] [Google Scholar]
- Hirama T. Takeshita S. Matsubayashi Y. Iwashiro M. Masuda T. Kuribayashi K. Yoshida Y. Yamagishi H. Conserved V(D)J junctional sequence of cross-reactive cytotoxic T cell receptor idiotype and the effect of a single amino acid substitution. Eur J Immunol. 1991;21:483–488. doi: 10.1002/eji.1830210234. [DOI] [PubMed] [Google Scholar]
- Houseley J. Tollervey D. Apparent non-canonical trans-splicing is generated by reverse transcriptase in vitro. PLoS One. 2010;5:e12271. doi: 10.1371/journal.pone.0012271. (7 p.) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu E. Chen Z. Fredrickson T. Zhu Y. Kirkpatrick R. Zhang G.F. Johanson K. Sung C.M. Liu R. Winkler J. Cloning and characterization of a novel human class I histone deacetylase that functions as a transcription repressor. J Biol Chem. 2000;275:15254–15264. doi: 10.1074/jbc.M908988199. [DOI] [PubMed] [Google Scholar]
- Inoue A. Takahashi K.P. Kimura M. Watanabe T. Morisawa S. Molecular cloning of a RNA binding protein, S1-1. Nucleic Acids Res. 1996;24:2990–2997. doi: 10.1093/nar/24.15.2990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kampen A.H.C. Horrevoets A.J.G. The role of bioinformatics in genomic medicine. In: Pasterkamp G., editor; de Kleijn D.P.V., editor. Cardiovascular Research: New Technologies, Methods, and Applications. Springer; New York: 2006. pp. 103–119. Chapter 6. [Google Scholar]
- Kannan K. Wang L. Wang J. Ittman M.M. Li W. Yen L. Recurrent chimeric RNAs enriched in human prostate cancer identified by deep sequencing. ProcNatlAcad Sci USA. 2011;108:9172–9177. doi: 10.1073/pnas.1100489108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kidd J.M. Graves T. Newman T.L., et al. A human genome structural variation sequencing resource reveals insights into mutational mechanisms. Cell. 2010;143:837–847. doi: 10.1016/j.cell.2010.10.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim S.G. Lin L.J. Reid B.R. Determination of nucleic acid backbone conformation by proton (1H) NMR. Biochemistry. 1992;31:3564–3574. doi: 10.1021/bi00129a003. [DOI] [PubMed] [Google Scholar]
- Kipreos E.T. Lander L.E. Wing J.P. He W.W. Hedgecock E.M. Cul-1 is required for cell cycle exit in C. elegans and identifies a novel gene family. Cell. 1996;85:829–839. doi: 10.1016/s0092-8674(00)81267-2. [DOI] [PubMed] [Google Scholar]
- Kodama T. Freeman M. Rohrer L. Zabrecky J. Matsudaira P., et al. Type I macrophage scavenger receptor contains alpha-helical and collagen-like coiled coils. Nature. 1990;343:531–535. doi: 10.1038/343531a0. [DOI] [PubMed] [Google Scholar]
- Kumar R. Haugen J.D. Wieben E.D. Londowski J.M. Cai Q. Inhibitors of renal epithelial phosphate transport in tumor-induced osteomalacia and uremia. Proc Assoc Am Physicians. 1995;107:296–305. [PubMed] [Google Scholar]
- Le Rouzic E. Mousnier A. Rustum C. Stutz F. Hallberg E. Dargemont C. Benichou S. Docking of HIV-1 Vpr to the nuclear envelope is mediated by the interaction with the nucleoporin hCG1. J Biol Chem. 2002;277:45091–45098. doi: 10.1074/jbc.M207439200. [DOI] [PubMed] [Google Scholar]
- Li B.L. Li X.L. Duan Z.J. Lee O. Lin S. Ma Z.M., et al. Human acyl-CoA:cholesterol acyltransferase-1 (ACAT-1) gene organization and evidence that the 4.3-kilobase ACAT-1 mRNA is produced from two different chromosomes. J Biol Chem. 1999;274:11060–11071. doi: 10.1074/jbc.274.16.11060. [DOI] [PubMed] [Google Scholar]
- Li C. Wong W.H. Model-based analysis of oligonucleotide arrays: expression index computation and outlier detection. Proc Natl Acad Sci U S A. 2001;98:31–36. doi: 10.1073/pnas.011404098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H. Wang J. Mor G. Sklar J. A neoplastic gene fusion mimics trans-splicing of RNAs in normal human cells. Science. 2008;321:1357–1361. doi: 10.1126/science.1156725. [DOI] [PubMed] [Google Scholar]
- Longo M.S. O'Neill M.J. O'Neill R.J. Abundant human DNA contamination identified in non-primate genome databases. PLoS One. 2011;6:e16410. doi: 10.1371/journal.pone.0016410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Markham N.R. Zuker M. UNAFold: software for nucleic acid folding and hybridization. http://dinamelt.bioinfo.rpi.edu/twostate-fold.php. Methods Mol Biol. 2008;453:3–31. doi: 10.1007/978-1-60327-429-6_1. [DOI] [PubMed] [Google Scholar]
- Matsumoto A. Naito M. Itakura H. Ikemoto S. Asaoka H., et al. Human macrophage scavenger receptors: primary structure, expression, and localization in atherosclerotic lesions. Proc Natl Acad Sci USA. 1990;87:9133–9137. doi: 10.1073/pnas.87.23.9133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mayer M.G. Floeter-Winter L.M. Pre-mRNA trans-splicing: from kinetoplastids to mammals, an easy language for life diversity. Mem Inst Oswaldo Cruz. 2005;100:501–513. doi: 10.1590/s0074-02762005000500010. [DOI] [PubMed] [Google Scholar]
- Mills R.E. Walter K. Stewart C., et al. Mapping copy number variation by population-scale genome sequencing. Nature. 2011;470:59–65. doi: 10.1038/nature09708. 1000 Genomes Project. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Or E. Baybik J. Lavee S. Sadka A. Ogredovitch A. The electronic plant gene register: isolation and characterization of two cDNA clones (Accession nos. AF195866 and AF195867) encoding alcohol dehydrogenase from grape (Vitis vinifera cv Perlette) developing fruits (PGR 00–022) Plant Physiol. 2000;122:619–620. [Google Scholar]
- Ota T. Suzuki Y. Nishikawa T., et al. Complete sequencing and characterization of 21,243 full-length human cDNAs. Nat Genet. 2004;36:40–45. doi: 10.1038/ng1285. [DOI] [PubMed] [Google Scholar]
- Pinto D. Darvishi K. Shi X., et al. Comprehensive assessment of array-based platforms and calling algorithms for detection of copy number variants. Nat Biotechnol. 2011;29:512–520. doi: 10.1038/nbt.1852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ringertz N.R. The discovery of “jumping genes” in corn gave the entire Nobel prize to a 81-year woman (Barbara McClintock) Lakartidningen. 1983;80:3908–3910. [PubMed] [Google Scholar]
- Roy S.W. Hudson A.J. Joseph J. Yee J. Russell A.G. Numerous fragmented spliceosomal introns, AT-AC splicing, and an unusual dynein gene expression pathway in Giardia lamblia. Mol Biol Evol. 2011 doi: 10.1093/molbev/msr063. [Epub ahead of print] [DOI] [PubMed] [Google Scholar]
- Savas U. Bhattacharyya K.K. Christou M. Alexander D.L. Jefcoate C.R. Mouse cytochrome P-450EF, representative of a new 1B subfamily of cytochrome P-450s. Cloning, sequence determination, and tissue expression. J Biol Chem. 1994;269:14905–14911. [PubMed] [Google Scholar]
- Schoenfelder S. Clay I. Fraser P. The transcriptional interactome: gene expression in 3D. Curr Opin Genet Dev. 2010;20:127–133. doi: 10.1016/j.gde.2010.02.002. [DOI] [PubMed] [Google Scholar]
- Scuteri A. Sanna S. Chen W.M., et al. Genome-wide association scan shows genetic variants in the FTO gene are associated with obesity-related traits. PLoS Genet. 2007;3:1200–1210. doi: 10.1371/journal.pgen.0030115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shipp M.A. Ross K.N. Tamayo P., et al. Diffuse large B-cell lymphoma outcome prediction by gene-expression profiling and supervised machine learning. Nat Med. 2002;8:68–74. doi: 10.1038/nm0102-68. [DOI] [PubMed] [Google Scholar]
- Stanley D. Fraser S. Stanley G.A. Chambers P.J. Retrotransposon expression in ethanol-stressed Saccharomyces cerevisiae. Appl Microbiol Biotechnol. 2010;87:1447–1454. doi: 10.1007/s00253-010-2562-y. [DOI] [PubMed] [Google Scholar]
- Strausberg R.L. Feingold E.A. Grouse L.H. Derge J.G. Klausner R.D. Collins F.S., et al. Generation and initial analysis of more than 15,000 full-length human and mouse cDNA sequences. Proc Natl Acad Sci USA. 2002;99:16899–16903. doi: 10.1073/pnas.242603899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vickers K.C. Castro-Chavez F. Morrisett J.D. Lyso-phosphatidylcholine induces osteogenic gene expression and phenotype in vascular smooth muscle cells. Atherosclerosis. 2010;211:122–129. doi: 10.1016/j.atherosclerosis.2010.04.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vitelli R. Chiariello M. Lattero D. Bruni C.B. Bucci C. Molecular cloning and expression analysis of the human Rab7 GTP-ase complementary deoxyribonucleic acid. Biochem Biophys Res Commun. 1996;229:887–890. doi: 10.1006/bbrc.1996.1897. [DOI] [PubMed] [Google Scholar]
- Wang Q. Zhang H. Fishel R. Greene M.I. BRCA1 and cell signaling. Oncogene. 2000;19:6152–6158. doi: 10.1038/sj.onc.1203974. [DOI] [PubMed] [Google Scholar]
- Winston J.T. Koepp D.M. Zhu C. Elledge S.J. Harper J.W. A family of mammalian F-box proteins. Curr Biol. 1999;9:1180–1182. doi: 10.1016/S0960-9822(00)80021-4. [DOI] [PubMed] [Google Scholar]
- Wolfsberg T.G. Madden T.L. Sequence similarity searching using the BLAST family of programs. Curr Protoc Protein Sci. 2001 doi: 10.1002/0471140864.ps0205s15. Chapter 2:Unit2.5. [DOI] [PubMed] [Google Scholar]
- Xu H. Yang L. Xu P. Tao Y. Ma Z. cTrans: generating polypeptide databases from cDNA sequences. Proteomics. 2007;7:177–179. doi: 10.1002/pmic.200600508. [DOI] [PubMed] [Google Scholar]
- Yang L. Lee O. Chen J. Chen J. Chang C.C. Zhou P., et al. Human acyl-coenzyme A:cholesterol acyltransferase 1 (acat1) sequences located in two different chromosomes (7 and 1) are required to produce a novel ACAT1 isoenzyme with additional sequence at the N terminus. J Biol Chem. 2004;279:46253–46262. doi: 10.1074/jbc.M408155200. [DOI] [PubMed] [Google Scholar]
- Yao X.M. Wang C.H. Song B.L. Yang X.Y. Wang Z.Z. Qi W., et al. Two human ACAT2 mRNA variants produced by alternative splicing and coding for novel isoenzymes. Acta Biochim Biophys Sin. 2005;37:797–806. doi: 10.1111/j.1745-7270.2005.00118.x. [DOI] [PubMed] [Google Scholar]
- Yoshikawa T. Sanders A.R. Detera-Wadleigh S.D. Contamination of sequence databases with adaptor sequences. Am J Hum Genet. 1997;60:463–466. [PMC free article] [PubMed] [Google Scholar]
- Zhao X. Chen J. Lei L. Hu G. Xiong Y. Xu J., et al. The optional long 5'-untranslated region of human ACAT1 mRNAs impairs the production of ACAT1 protein by promoting its mRNA decay. Acta Biochim Biophys Sin. 2009;41:30–41. doi: 10.1093/abbs/gmn004. [DOI] [PubMed] [Google Scholar]