Abstract
Here, we report genome-wide analysis of the tumor suppressor p53 binding sites in normal human cells. 743 high-confidence ChIP-seq peaks representing putative genomic binding sites were identified in normal IMR90 fibroblasts using a reference chromatin sample. More than 40% were located within 2 kb of a transcription start site (TSS), a distribution similar to that documented for individually studied, functional p53 binding sites and, to date, not observed by previous p53 genome-wide studies. Nearly half of the high-confidence binding sites in the IMR90 cells reside in CpG islands in marked contrast to sites reported in cancer-derived cells. The distinct genomic features of the IMR90 binding sites do not reflect a distinct preference for specific sequences, since the de novo developed p53 motif based on our study is similar to those reported by genome-wide studies of cancer cells. More likely, the different chromatin landscape in normal, compared with cancer-derived cells, influences p53 binding via modulating availability of the sites. We compared the IMR90 ChIP-seq peaks to the recently published IMR90 methylome1 and demonstrated that they are enriched at hypomethylated DNA. Our study represents the first genome-wide, de novo mapping of p53 binding sites in normal human cells and reveals that p53 binding sites reside in distinct genomic landscapes in normal and cancer-derived human cells.
Key words: p53, whole genome binding, tumor suppressor, transcription factor, CpG island, ChIP-seq
Introduction
The tumor suppressor p53 is a transcription factor that, after stabilization and activation through a variety of posttranslational modifications in response to stress,2 regulates the expression of an extensive network of genes.3 Upon sequence-specific binding, p53 can activate transcription at some targets4 and repress it at others.5 p53-dependent transcription drives the cellular responses to stress, such as DNA repair, cell cycle arrest, senescence and apoptosis, but also a number of recently recognized programs in cell metabolism and development, including fertility, neural development and cell differentiation.6,7 p53 binds as a tetramer to a consensus sequence motif containing two decamers RRRCWWGYYY separated by a spacer of 0–14 nt.8,9 More recently, p53 was shown to modulate transcription by binding to half or three-quarter sites.10 It also can bind to simple repeats and a variety of DNA structures and mismatches, although these have not been shown to affect transcription.11,12 Binding of p53 to DNA is highly cooperative,13 which has been proposed to be particularly important at imperfect binding sites.14 Despite the fact that 20% of the putative p53 binding sites contain one or more CpG dinucleotides, and p53 binding was proposed to be affected by their methylation state,15 the impact of DNA methylation on p53 genome-wide binding has not been investigated in vivo.
Given the importance of site-specific binding to DNA for p53-mediated transcriptional regulation (the majority of p53 cancer-associated mutations alter its DNA binding domain), extensive efforts are being directed toward the experimental identification of p53 genomic binding sites. The question of how p53 discriminates between the hundreds of thousands of potential p53 binding sites predicted in the human genome by bioinformatics analyses still remains unresolved.16 Over the past 20 years, studies, primarily on individual genes, have identified and experimentally validated about 200 functional p53 response elements (p53 REs) in the human genome.3,17 Recent advances in sequencing technologies combined with chromatin immunoprecipitation (ChIP) have lead to the identification of thousands of p53 binding sites, providing unmatched opportunities for analysis and comparison of the global genomic p53 binding pattern under different experimental conditions. Notably, all de novo p53 binding studies published to date18–25 have used cancer-derived cell lines. Binding of transcription factors (TF) to DNA is known to be context-dependent, governed in vivo by chromatin architecture and epigenetic modifications,26 and these are subjected to major changes during tumor development.27 Thus, p53 binding in cancer-derived cell lines may not accurately reflect binding in normal cells, and this may be a major limitation to understanding p53 functions in normal cells. Such a hypothesis is supported by recent findings that upon exposure to genotoxic stress, the majority of p53 binding sites examined by ChIP-chip were found occupied in cancer or immortalized cells but not in primary normal cells.28 Genome-wide binding studies combined with histone modification and DNA methylation maps in both normal and cancer cell types can help place p53 binding in the context of chromatin in vivo and address the impact of the epigenetic landscape on p53-dependant transcriptional regulation.
Here, we report the first genome-wide analysis of p53 binding in normal human cells by ChIP-seq. The identified 743 high-confidence peaks represent putative genomic p53 binding sites in IMR90 fibroblasts29 treated for 6 h with 5-fluorouracil (5-FU). After comparing these with binding sites previously reported in the cancer cell lines HCT116 23 and U2OS,24,25 we found that the p53 sites in the normal fibroblasts reside in a distinct genomic landscape significantly different from that previously reported in the cancer cells, pointing at chromatin as a critical factor in the selection of the occupied sites. We analyzed the p53 binding profile in IMR90 in the context of the genome-wide map of methylated cytosines from the same cells1 and demonstrate that in these normal cells, p53 binding sites are enriched in hypomethylated DNA.
Results and Discussion
Genome-wide mapping of p53 binding sites by ChIP-seq.
To identify stress-induced p53 genomic binding sites, we used highthroughput sequencing of chromatin immunoprecipitated DNA (ChIP-seq) from IMR90 normal human lung fibroblasts treated for 6 h with 375 µM 5-FU. Before preparing the sequencing libraries, we verified p53 induction (Fig. S1A) and binding at known target sites (Fig. S1B). Because of systematic biases in the data, accurate interpretation of the ChIP-seq results requires a reference control sample, and we used an Input control (chromatin taken before ChIP) to correct for genomic copy number variations, sonication-introduced fragmentation bias and chromatin accessibility.30 The p53-specific ChIP library obtained with the monoclonal antibody DO1 and the corresponding Input library were sequenced on Illumina GA 2x platform (Fig. S2A and Sup. Methods). About 69% of the 7.4 million p53 ChIP-seq reads and 80% of the 11.7 million Input-seq reads obtained were distinct sequences that mapped uniquely to the human genome (hg18), and these were further analyzed (Table S1).
To identify genomic locations enriched for sequencing reads, we used a peak-finding method (Fig. S2B, Methods) based on the approach of Rozowsky et al.31 A total of 6,789 p53 ChIP-seq peaks and 2,537 Input peaks were defined. To identify the peaks most likely to represent genomic p53 binding sites and to correct for the systematic biases noted above, we compared the ChIP-seq and the Input-seq data by applying two independent confidence tests based on binomial or Poisson distributions, requiring a minimum score of 99% in both, to generate a set of 1,732 ChIP-seq peaks significantly enriched above the Input background. After removing artifactual peaks (see Sup. Methods), a refined set of 1,687 ChIP-seq peaks was obtained. These were ranked by peak height, and a minimum height of 10 was required to derive the final set of 743 high-confidence p53 ChIP-seq peaks (Table S2 and Sup. Methods). An overall low correlation between ChIP-seq and Input-seq peaks was obtained in our study; for example, see the overview of chromosome 6 (Fig. 1A). Four major types of peak profiles were observed (Fig. S2C–G). About 73% of the 743 high-confidence peaks were classified as sharp (Fig. S2C), ∼21% as wide (Fig. S2D) and ∼6% as low enrichment regions (Fig. S2E). The fourth type, ChIP-seq peaks correlating with Input-seq (Fig. S2F), were removed from the high-confidence set. The region of chromosome 22 used as a negative control for p53 binding (Fig. S2G) showed a low Input-seq background and no p53 ChIP-seq enrichment.
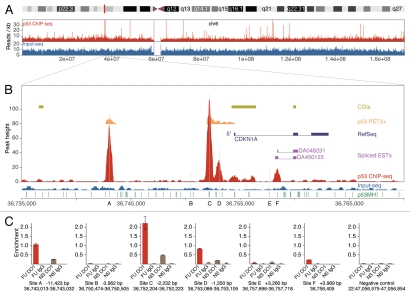
Figure 1.
p53 ChIP-seq map at the CDKN1A locus. (A) Overview of chromosome 6. Plotted are ChIP-seq (red) and Input-seq (blue) coverage maps. (B) ChIP-seq map across a 35 kb genomic region of the CDKN1A locus. Previously reported p53 binding sites A–E (marked with vertical gray lines) and the newly identified site F are shown. Other features plotted include CGIs (olive green), RefSeq genes (dark blue), spliced ESTs (magenta) and p53 PET3+ clusters23 (orange), all downloaded from the UCSC Genome Browser (hg18) and p53 binding sites predicted by p53MH algorithm16 (green). For details, see Supplemental Methods. (C) qPCR validation of p53 binding at CDKN1A. For each site, the hg18 coordinates and the distance to the TSS (CDKN1A transcript NM_000389) are shown. The position of the peak maximum is shown for site F. The indicated region from chr22 was used as a negative control for p53 binding. Enrichment is calculated as percentage of Input; average results are shown from duplicate qPCR samples. FU, 6 h treatment with 5-FU; NS, no stimulation; DO1, ChIP with p53 specific DO1 antibody; IgG, ChIP with non-specific IgG.
Validation of p53 ChIP-seq results.
We validated the ChIP-seq findings by analyzing the p53 binding sites at the canonical p53 target CDKN1A, by comparing the high-confidence peaks to 168 reference p53 REs (individually studied functional binding sites) and by a small-scale qPCR validation on independent ChIP samples.
p53 binding at the CDKN1A locus.
CDKN1A (p21) is a cyclin-dependent kinase inhibitor that controls cell cycle progression and has been extensively studied for transcriptional regulation by p53. Five p53 binding sites (A–E, Fig. 1B) have been reported in the CDKN1A locus.32 Two of them, at −2,241 bp and −1,354 bp from the major TSS (sites C and D, Fig. 1B), were first described by El-Deiry et al.8 and have since been confirmed by many others. A third site was reported at −111 bp from an alternative TSS33 corresponding to position 3,256 bp downstream of the major TSS (site E, Fig. 1B). A fourth site (B, Fig. 1B) was reported at position −3,969 bp relative to the major TSS,34 and a site at −11.7 kb from the major TSS (A, Fig. 1B) was identified and confirmed in the ChIP-PET study23 and was later shown to regulate p53-dependent lincRNA-p21 transcription.35
Among the 743 high-confidence ChIP-seq peaks, we identified four in the CDKN1A locus. Three were located at sites A, C and D (Fig. 1B), and one (site F), not characterized previously, was located 3,956 bp downstream of the major TSS, close to spliced human ESTs (Fig. 1B). Sites B and E were not identified by ChIP-seq peaks. p53 enrichment was analyzed at each of the reported sites (A–E) and at the newly found site F by qPCR on independent ChIP samples (Fig. 1C). Validating the ChIP-seq results, we confirmed that sites A, C, D and F were occupied by p53, and we observed a good correlation between the qPCR enrichment and the ChIP-seq peak height. Consistent with our ChIP-seq data, no qPCR enrichment was detected at sites B and E. These two sites also were not identified by other genome-wide studies,23,24 nor were they found to be occupied by p53 when the CDKN1A locus was analyzed by in cellulo genomic footprinting.36 However, these two sites were occupied when naked DNA was used in a microsphere assay but not when tested in the context of chromatin by a ChIP assay.37
ChIP-seq identifies previously reported, reference p53 REs.
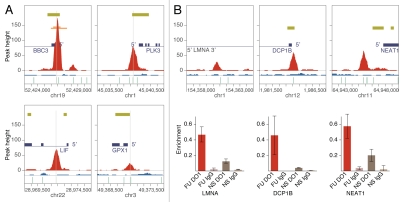
To verify that the genomic sites mapped by the 743 high-confidence ChIP-seq peaks are, indeed, enriched for p53 binding sites, we compared them to previously reported functional p53 response elements (p53 REs). We compiled a list of 168 individually analyzed p53 REs based on two reports17,38 and updated with more recently published sites (Table S3). Of these, 48 p53 REs were identified by high-confidence peaks (Table S4A), and a strong correlation was observed between the peak maximum and the center of the reference p53RE (see BBC3, PLK3, LIF and GPX1 in Fig. 2A). For 79% of the identified p53 REs, the distance between the peak maximum and the reference p53 RE center was less than 50 nt, and for 52%, it was less than 20 nt (Table S4A and Fig. S3A). In addition to these 48 reference p53 REs, another 14 were identified by peaks excluded from the final set of 743 high-confidence peaks (see Table S4B), suggesting that bona fide p53 binding sites of low enrichment may be underreported.
Figure 2.
p53 ChIP-seq validation. (A) Examples of ChIP-seq peaks that coincide with the reference p53 REs (see Table S4A) at the target genes BBC3, PLK3, LIF and GPX1. ChIP-seq (red) and Input-seq (blue) coverage maps in 5 kb regions are plotted and centered at the reference REs (marked with vertical gray lines). All annotated features shown (e.g., RefSeq genes, CGIs, p53 PET3+ and p53MH predicted binding sites) are as indicated on Figure 1. (B) Examples of newly identified binding sites validated by qPCR at the genes LMNA, DCP1B and NEAT1. Average enrichment is calculated as a percentage of Input; results shown are from duplicate qPCR samples. FU, 6 h treatment with 5-FU; NS, no stimulation; DO1, ChIP with p53 specific DO1 antibody; IgG, ChIP with non-specific IgG.
Next, we looked for peaks in the vicinity of reference p53 REs not identified by ChIP-seq that might represent “alternative” binding sites. We examined the corresponding target genes (including the 25 kb upstream and 5 kb downstream regions) and found nine ChIP-seq peaks at such “alternative” sites (see examples in Fig. S3B). All of them had at least one p53 binding site within 20 nt of the peak maximum predicted by the p53MH algorithm;16 four of them were also identified by the p53 ChIP-PET genome-wide study23 (e.g., PHLDA3 and ATF3, Fig. S3B), and two of them were targeted for validation and confirmed by qPCR (MAD1L1 and NOTCH1, Fig. S4). Therefore, these nine sites, located in the vicinity of previously reported reference p53 REs, are good candidates for alternative binding sites occupied in IMR90 cells.
The fact that we did not find p53 binding at or near 97 reference p53 REs may be due to using only one cell type under one stress condition, while the list of 168 reference REs was compiled based on studies in various cell types and treatments, or it may be due to technical limitations. By comparison, our analysis indicated that 25 of the 168 reference p53 REs were identified by high-confidence PET3+ clusters in the genome-wide study of Wei et al.23 and 32 by high-confidence sites in the ChIP-chip study of Smeenk et al.24
qPCR validation.
We chose 45 high-confidence ChIP-seq peaks for validation of p53 binding by qPCR; 22 of them were selected randomly among peaks with height of 25 or below (see Fig. S5), 7 overlapped known p53 REs, and 16 were not reported previously, some of which are discussed below. A good correlation was observed between ChIP-seq peak height and the enrichment level measured by qPCR on independently obtained ChIP samples, supporting the semi-quantitative nature of the ChIP-seq data (Fig. S7). Enrichment levels obtained from mock ChIP samples using non-specific IgG were low and showed no such correlation. Our qPCR validation, although done on a small scale, suggests that the majority of the 743 high-confidence peaks represent enriched p53 binding locations under the conditions of the experiment.
Examples of newly identified, confirmed p53 binding sites.
High-confidence peak ID 99 (Table S2) located in the first intron of LMNA (Fig. 2B) was validated by qPCR. This gene encodes for lamins A and C, the major components of the nuclear lamina, which provide a scaffold for the assembly and regulation of a variety of protein complexes involved in the control of nuclear integrity and gene expression.39 Mutations in LMNA cause laminopathies in humans, including aging-associated pathologies. Activation of p53 by mitomycin C in HCT116 cells was reported to induce transcription of lamin A and C, but p53 binding at that locus was not examined.40
At the promoter of DCP1B (Fig. 2B), we confirmed high-confidence peak ID 42 (Table S2). DCP1B is homologous to DCP1A, which encodes a core component of the decapping complex involved in nonsense-mediated mRNA decay,41 a quality-control pathway found in all eukaryotes that is responsible for degrading mRNAs harboring a premature termination codon, in which p53 has been previously implicated only indirectly.42
The validated high-confidence peak ID 84 (Fig. S6A and Table S2) belongs to the group of 31 peaks found at bidirectional promoters (see below and Table S5); it is located at the 5′ end of both C18orf5 and TYMS, the latter of which encodes the enzyme thymidylate synthase, a primary target for anticancer therapy with 5-FU.43 This finding is interesting not only because it is widely accepted that the TYMS expression level and the p53 status are the major determinants of a cancer cell's response to 5-FU therapy, but also because p53 was shown to repress the mouse Tyms gene transcription.44
At the promoter of NEAT1, which encodes for an abundant ncRNA that is an integral component of the nuclear paraspecles,45 high-confidence ChIP-seq peak ID 37 was confirmed (Fig. 2B and Table S2). Paraspeckles are discrete ribonucleoprotein bodies in the nucleoplasm of mammalian cells that are associated with hyperediting of mRNAs containing inverted repeats (such as Alu) performed by ADARs (adenosine deaminases acting on RNA).46 Transcription of NEAT1 recently was shown to be essential for both de novo assembly of paraspeckles and their maintenance.47 p53 is known to bind to some Alu repeats, which have been implicated in shaping the p53 transcriptional network.48,49 Now, our finding of p53 binding at the NEAT1 promoter opens up the possibility for potential p53-mediated regulation of the NEAT1 transcription, with a possible impact on the expression of hundreds of genes that contain Alu repeats in their 3′-UTRs50 through RNA editing. Interestingly, p53 was recently implicated in the only other known pathway for RNA editing, which is performed by APOBEC51 (restricted to the small intestine in humans, affecting few genes). In addition, a low level of NEAT1 mRNA was speculated to be a marker of pluripotency,46 while reducing the p53 activity was shown to improve the efficiency of inducing pluripotent stem cells.52
p53 ChIP-seq peaks are highly enriched for putative p53 binding sites.
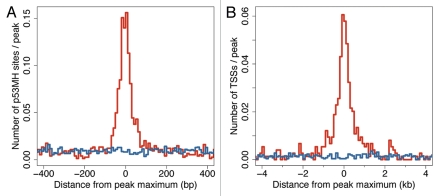
Several algorithms have been developed for predicting putative p53 binding sites.16,23,24,53,54 For analysis of the identified ChIP-seq peaks, we used the p53MH algorithm16 and demonstrated that the ChIP-seq peaks are enriched for putative p53MH sites, unlike the Input peaks. The enrichment was tightly confined within a 100 nt window centered at the peak maximum (Fig. 3A). Half of the 743 high-confidence ChIP-seq peaks (50.3%) had a predicted p53MH site within 50 nt of the peak maximum, more than 7-fold enrichment above the Input (6.7%). In the context of the peak height, ∼31% of the peaks with a height of 10–15 contained a putative p53MH site within 50 nt of the peak maximum; this fraction went up to 74% for the group with peak height 16–50 and reached 94% for the peaks with height above 50. Taking into account that the p53MH algorithm was trained on a relatively small set of p53 binding sites and the ability of p53 to bind half-sites and sites significantly deviating from the consensus binding motif,10,38 the p53MH algorithm may have missed actual binding sites.
Figure 3.
p53 ChIP-seq peaks from IMR90 are strongly enriched for predicted p53 binding sites and for TSSs. (A) Distribution of p53 binding sites, predicted by the p53MH algorithm16 as a function of distance (nt) to the peak maximum. p53 ChIP-seq peaks are highly enriched for p53MH sites within 50 nt of the peak maximum. (B) Distribution of TSSs as a function of distance (kb) to the peak maximum. p53 ChIP-seq peaks, unlike Input-seq peaks, are enriched for TSSs within 2 kb of the peak maximum.
p53 ChIP-seq peaks are highly enriched at TSSs.
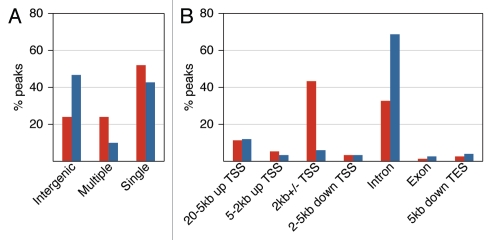
The functions of p53 as a transcription factor are associated with binding to gene promoters.3 Thus, we analyzed the peaks' proximities to TSSs (UCSC). Compared with the Input-seq peaks, the ChIP-seq peaks demonstrated a clear enrichment within 2 kb of a TSS (Fig. 3B). Most ChIP-seq peaks (76%) were gene-associated, and only 24% were intergenic, unlike the Input-seq peaks (Table S6). More than half of the intergenic ChIP peaks were close to human ESTs, and 20% were within 5 kb of the 5′ end of a spliced human EST (UCSC), thus some may be associated with unannotated, alternative TSSs. For analysis of the distribution of gene-associated peaks, we considered 12 non-overlapping regions relative to RefSeq genes (e.g., 10-5 kb to TSS, 5-2 kb to TSS, intron, exon, etc.; for details, see Table S6A). About half of the high-confidence ChIP-seq peaks were associated with a single region of the 12 analyzed, while 24% were associated with multiple regions (Fig. 4A). The genomic distribution of peaks associated with a single region is presented in Figure 4B. Strikingly, 43% of these ChIP-seq peaks reside within 2 kb of a TSS, but only 6% of the Input-seq peaks do. The major difference between the ChIP-seq and Input-seq peaks' distributions is in the immediate vicinity of a TSS, otherwise they were similar (see also Tables S6B and S6C). Such high enrichment close to TSSs is characteristic of functional p53 binding sites. By our analysis, 72% of the 168 reference p53 REs (Table S3) are located within 2 kb of their target gene TSS.
Figure 4.
Genomic distribution of p53 binding sites in IMR90 cells. (A) Gene-associated ChIP-seq (red) and Input-seq (blue) peaks are categorized as either “single,” located in only one of the genic regions defined below and typically associated with a single transcript, or “multiple,” located in more than one genic region due to proximity to more than one transcript. Intergenic peaks are located outside the boundaries of any gene extended 20 kb upstream and 5 kb downstream. (B) Breakdown of the relative positions for the peaks associated with single genic regions shows ChIP-seq peaks tightly clustered in the immediate vicinity of TSSs. The following non overlapping genic regions were considered for this analysis: 20-5 kb to TSS, 5-2 kb to TSS, 2 kb ± TSS, 2–5 kb downstream of TSS, intron or exon 0 (if > 5 kb downstream of TSS) and within 5 kb downstream of transcription end site (TES) (see Table S6 for details).
About 4% of the high-confidence peaks in IMR90 cells were located at bidirectional promoters (at genes arranged head-to-head on opposite strands with closely spaced TSSs, Table S5 and Fig. S6). Although a significant number of human genes share bidirectional promoters, the mechanisms for their regulation are largely unknown; while some gene pairs are co-, others are anti-regulated.55 Among the genes identified by high-confidence ChIP-seq peaks, two pairs of well known p53 target genes, APAF1 and IKBIP56 and PTEN and KILIN,57 were experimentally shown to share promoter elements. Another identified pair was FAS and ACTA2 (Fig. S6B). While the short ACTA2 transcript is known to be regulated by p53 via a functional RE,58 the longer ACTA2 transcript (shown on Fig. S6B) shares a bidirectional promoter with the FAS gene. Notably, among the ChIP-seq-identified genes with bidirectional promoters, those involved in apoptotic responses are well-represented, while apoptotic genes overall are underrepresented among the human genes with bidirectional promoters.59
p53 binding patterns in IMR90 and HCT116 cells are distinct.
Previous studies have pointed out the limitations of using cancer-derived cells for studying wild-type p53 functions in normal cells.60 Since ours is the first genome-wide study to map p53 binding sites in normal human cells, we compared them with sites previously reported in cancer-derived cell lines, choosing the ChIP-PET study in HCT116,23 because both sequencing studies were conducted under the same conditions (6 h treatment with 375 µM 5-FU), and the annotated locations from both were ranked (by peak height and number of cluster members), allowing for a more accurate comparison. According to Wei et al.,23 PET3+ clusters (composed of at least three overlapping ChIP fragments) are most likely to represent true p53 binding sites; thus, we compared them with the high-confidence ChIP-seq peaks identified in IMR90 cells (Table S2).
Among the 743 ChIP-seq peaks, 130 overlapped with PET3+ clusters (Table S8), identifying p53 binding locations common to IMR90 and HCT116; 613 ChIP-seq peaks were not identified by PET3+ clusters, and 181 PET3+ clusters were not identified by high-confidence ChIP-seq peaks. Twenty-one of the 130 binding sites common to both studies intersected reference p53 REs (from the list of 168, Table S3) and at four locations ChIP-seq and ChIP-PET agreed on “alternative” binding positions in proximity to the reference REs (for examples, see Figs. S3B and S4B). The overlapping sites tended to be the strongest binding sites in both studies (28% of PET3 clusters and 52% of PET5 clusters overlapped with high-confidence ChIP-seq peaks, Table S7A). Similarly, ChIP-seq peaks with height of 31–50 were more likely to overlap with PET3+ clusters than peaks with height of 16–20 (Table S7B). Greater overlap between the higher ranked sites is expected in genome-wide studies.61 Interestingly, 30% of the top ranked PET7+ and 36% of the top ranked ChIP-seq peaks with a height of 51–205, did not overlap (see Tables S7A and S7B), suggesting differences in the p53 binding site occupancies between IMR90 and HCT116 cells.
Next, we compared the three sets of sites (the 130 sites common to the two cell types, the 181 PET3+ identified sites found only in HCT116 and the 613 sites identified by ChIP-seq only in IMR90 cells) with respect to various genomic features and found significant differences (Table S9). The sites found only in IMR90 cells were less enriched in repeats (24%) and significantly more enriched at genes (74%); notably, 52% of them resided in CpG islands (CGIs). Those found only in HCT116 cells were far more enriched at repeats (65%), less enriched at genes (47%) and were virtually outside CGIs (1%). The sites common to the two sets had intermediate characteristics with respect to proximity to genes, CGIs and repeats (Table S9); however, they had higher enrichment for p53MH-predicted sites.16 To verify that the differences observed with respect to proximity to genes, CGIs and repeats reflect the general behavior of the data sets and not just a small fractions of those, we repeated the analysis using various subsets (including the top ranked sites) and obtained similar results.
We conclude, therefore, that while many strong p53 binding sites are likely to be occupied after stress in both IMR90 and HCT116 cells, not only are the majority of the identified sites different, but more importantly, they have distinct genomic characteristics (e.g., proximity to genes and repeats, distance to TSS and enrichment in CGIs).
p53 binding sites in normal IMR90 cells are enriched at CGIs unlike in cancer cells.
p53 binding sites' differences between IMR90 and HCT116 cells could reflect differences between normal and cancer cells or differences between cell types. If the former is correct, we should see similar results upon comparing the IMR90 sites with the binding sites reported in cancer cells derived from other tissues. To test this hypothesis, we analyzed the two sets of binding sites identified in U2OS osteosarcoma cells by ChIP-chip24 and by ChIP-seq.25 Both studies used cells treated for 24 h with actinomycin D.
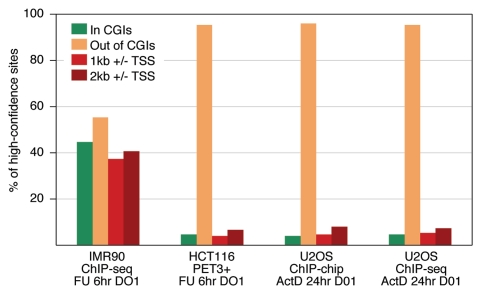
Nearly half (44.5%) of the 743 high-confidence binding sites in IMR90 cells resided in CGIs in marked contrast to less than 5% of the sites in any of the cancer cell studies (Fig. 5). A similar striking disparity is seen in the fraction of sites near TSSs. Thirty-seven percent of the high-confidence sites in IMR90 cells are located within 1 kb of a TSS, which is true for only 4% of the PET3+ clusters (HCT116), 4.7% of the ChIP-chip sites (U2OS) and 5.4% of the ChIP-seq sites (U2OS). Further analysis of the top and bottom fractions of these sets demonstrated that despite the variations seen between the highest and lowest rank sites in IMR90, their distributions remain clearly different from the cancer data sets (Fig. S8).
Figure 5.
Distribution of p53 binding sites with respect to CGIs and TSSs. Compared were high-confidence sites we identified in IMR90 (743 ChIP-seq peaks) with those reported in HCT116 (310 PET3+ loci23) and U2OS (1516 ChIP-chip sites;24 2137 ChIP-seq sites25). p53 binding sites are highly enriched at TSSs and CGIs only in the normal IMR90 cells. Cells treated with 5-FU for 6 h (FU, 6 h) or with actinomycin D for 24 h (ActD, 24 h). ChIPs in all four experiments were done with the p53-specific antibody DO1.
The similar features of the p53 binding sites in HCT116 and U2OS cells (depletion at CGIs, away from TSSs), although they were derived from different types of cancers and treated with different agents and times, are in striking contrast with the features of the IMR90 sites (close to TSSs, enriched at CGIs). These results are consistent with our hypothesis that the differences in the p53 binding between IMR90 and HCT116/U2OS cells likely reflect differences between normal and cancer cells rather than differences between fibroblasts and epithelial cells. Such an interpretation is supported by the findings of Shaked et al.,28 who used an array of 62 functional p53 REs and 540 PET loci to examine p53 binding by ChIP-chip in the cancer cell lines HCT116 and U2OS, in immortalized human fibroblasts and in primary peripheral blood mononuclear cells (PBMC). Importantly, the immortalized fibroblasts behaved as the cancer cells and did not show selective p53 binding at the majority of the tested sites, while the normal PBMC cells showed distinct p53 binding in a stress-dependent manner. The finding that in normal IMR90 cells, the p53 binding sites are close to TSSs and in CGIs is consistent with the fact that 70% of the annotated human promoters are associated with CGIs, representing the most common promoter type in the vertebrate genome.62
p53 binding sites in normal IMR90 cells are enriched at hypomethylated DNA.
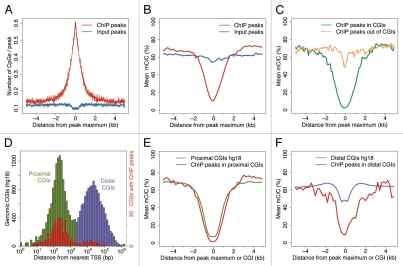
Given the strong enrichment of the ChIP-seq peaks for CpGs in IMR90 (Fig. 6A), we were interested in determining if there is a correlation between the p53 binding sites and the CpG methylation status. For this analysis we used the complete DNA methylation map at a single nucleotide resolution generated in IMR90 cells.1 The p53 ChIP-seq peaks correlated with DNA hypomethylation, unlike the Input-seq peaks (Fig. 6B). This is true for ChIP-seq peaks both in CGIs and outside CGIs (Fig. 6C). The hypomethylation effect at the peaks in CGIs is profound, centered at the peak maximum with a wide profile, suggesting that it is a feature of the islands. Peaks out of CGIs show reduced, although obvious hypomethylation in a narrow, more focused region, sharply centered at the peak maximum.
Figure 6.
High-confidence p53 ChIP-seq peaks are enriched at hypomethylated DNA in IMR90 cells. (A) p53 ChIP-seq peaks (red), unlike Input-seq peaks (blue), are enriched at CGIs. The number of genomic CpG dinucleotides (hg18) is plotted in 10 nt bins as a function of the distance to the peak maximum. (B) p53 ChIP-seq peaks (red), but not Input-seq peaks (blue), are enriched at hypomethylated DNA. Relative methylation density (mean mC/C ratio) calculated from the data reported for IMR90 by Lister et al.1 is plotted as a function of distance to the peak maximum (ChIP-seq peaks) or to CGI centers (B, C, E and F). (C) p53 ChIP-seq peaks in CGIs (green) and out of CGIs (orange) are enriched at hypomethylated DNA. (D) Distribution of all human CGIs (UCSC definition) with respect to TSS. The distance (bp) from CGI center to the nearest TSS is plotted on a log scale. Proximal CGIs (within 2 kb of a TSS) are shown in green and distal CGIs (away from TSSs) in purple. CGIs, at which high-confidence p53 ChIP-seq peaks, are found are plotted in red (note the change in the scale). (E) The hypomethylation level of ChIP-seq peaks in proximal CGIs (red) is similar to that of all human proximal CGIs (green). (F) ChIP-seq peaks in distal CGIs (red) are far more hypomethylated than the human distal CGIs (purple).
Human CGIs can be divided in two groups, proximal and distal, based on their distance to the nearest TSS (Fig. 6D). Proximal CGIs (close to TSSs) typically are hypomethylated, while distal CGIs (far from TSSs, also known as “orphans”) are more likely to be methylated in normal somatic cells.63 Most p53 ChIP-seq peaks are in proximal although some are in distal CGIs (Figs. 6D and S9). ChIP-seq peaks, both in proximal (Fig. 6E) and in distal CGIs (Fig. 6F), are hypomethylated. More importantly, there is a significant decrease in the relative methylation density around the p53 peaks in distal CGIs compared with the bulk of the genomic distal CGIs (Fig. 6F), suggesting that p53 binding at CGIs is generally enriched at hypomethylated tracts.
Mammalian TFs often contain CpGs in their recognition sequences, and their methylation state can directly affect protein binding. Preference for hypomethylated binding sites has been documented for proteins such as SP1, CREB and CTCF.64,65 Limited data are available for p53; however, estimates suggest that about 20% of the predicted genomic binding sites contain a CpG. Analysis of p53 binding in vitro using fluorescence anisotropy titration showed similar p53 affinity to methylated and unmethylated oligonucleotides, and in some cases, when multiple CpGs were present, stronger binding was reported to methylated DNA.15 We, on the other hand, looked at p53 genomic binding in cells, where the binding patterns might be significantly modulated by the chromatin structure. In the context of chromatin, p53 binding is not determined purely by DNA binding affinity.37
The hypomethylation observed at the p53 ChIP-seq peaks in IMR90 cells has a broad profile, spanning ∼3 kb for the peaks in CGIs (Fig. 6E and F) and ∼1 kb for the peaks outside CGIs (Fig. 6C), arguing that the hypomethylation is a property of the genomic regions surrounding these binding sites. Our data suggest that in the normal IMR90 cells p53 binding sites primarily reside in hypomethylated CGIs and other hypomethylated genomic regions. This in vivo binding preference is supported by a recent study which showed that in endometrial tumor samples, CGI promoter methylation at survivin/BIRC5 prevented p53 from binding.66 Cancer development is known to be associated with substantial epigenetic changes, DNA hypomethylation on a global genomic scale (particularly at repetitive sequences) and hypermethylation at CGIs.27 CpG methylation may induce a more compact and rigid nucleosome structure67 that would significantly affect the CGIs, which normally are relatively nucleosome-deficient and permissive for TF binding.62 Perhaps more significantly, hypomethylation of the bulk of the genome might open access to many potential binding sites that are not accessible in normal cells. In this context, our findings that in IMR90 cells, the p53 binding sites are enriched at CGIs and in hypomethylated regions, while in cancer cells they are more enriched at repeats and outside CGIs (Table S9) may reflect the substantial epigenetic differences in these genomes. We suggest, therefore, that changes in promoter methylation during cancer progression may lead to changes in the chromatin structure responsible for the differences in p53 binding sites occupancy in normal and cancer cells.
p53 binding motif analysis.
The historical p53 consensus sequence contains two decameric half-sites with a variable length spacer;8,9 several algorithms have been developed for predicting putative binding sites.16,23,24,53,54 Because the p53 binding sites in IMR90 cells are enriched at CGIs, we asked if they contained sequence motif(s) distinct from those reported by genome-wide studies in cancer cells. A de novo motif search was conducted on the 743 high-confidence peaks (350 nt regions centered at the peak maxima) using MEME 4.6.1 (see Sup. Methods). The identified 20 nt sequence motif (spacer 0), designated as p53CSI (ChIP-seq identified, Fig. 7A), strongly resembled the established p53 consensus sequence, was detected in 62% of the high-confidence ChIP-seq peaks and was highly enriched in a 100 bp window centered at the peak maxima (Fig. 7B). Comparison of the motifs determined by the three genome-wide studies, p53CSI (IMR90), p53PET (HCT116) and p53scan (U2OS), showed that they were very similar. The two core elements (CATG) were well defined in all three, although p53CSI had different weights at some positions (Fig. 7A).
Figure 7.
Motif analysis of the 743 high-confidence p53 ChIP-seq peaks identified in IMR90 cells using MEME. (A) Sequence logo depicting the p53CSI motif (E-value 1.2e−1059). (B) p53CSI motif distribution in the 743 high-confidence peaks. Number of p53CSI motifs found (in 20 bp bins) is plotted vs. the distance (bp) between the motif center and the peak maximum. Strong enrichment of the p53CSI motif is observed within 100 nt centered at the peak maximum.
Finding p53CSI in 461 out of 743 high-confidence peaks left 282 peaks without an obvious p53 binding motif. We then searched these with the p53MH algorithm and found putative sites in 53 peaks (all but two with spacer >0), leaving 229 peaks without a recognized motif. Searching these with p53scan revealed only five more putative sites. Although we tried additional de novo motif development approaches, no significant motif was identified in the remaining 224 peaks. Since p53 could be targeted to certain locations by some of its many binding partners,2 we searched the 224 peaks for known TF binding sites (see Sup. Methods). The most significant motif reported was for SP1; it was broadly enriched around the peaks, more likely reflecting its association with CGIs. We also considered the possibility that p53 binds directly in a sequence independent manner to repetitive or low complexity sequences that might serve as a structural binding motif.11 However, this is unlikely because only 6.9% of the peaks with no motif were in repeats (UCSC repeat masker) compared with 28% of the 743 peaks found in repeats.
In conclusion, the de novo developed p53CSI motif was very similar to those reported by genome-wide studies in cancer cells; therefore, the distinct p53 binding pattern in normal IMR90 cells does not reflect a distinct binding preference to specific sequences. More likely, the genomic landscape (DNA methylation and associated chromatin changes) modulates the availability of the binding sites. The lack of a p53CSI motif in 30% of the IMR90 high-confidence peaks could be due to general algorithm limitations for identifying sites that significantly deviate from the consensus, supported by our finding that a similar fraction (24%) of the 168 reference p53 REs was neither recognized by p53MH nor matched by p53CSI.
Functional annotation clustering of genes associated with p53 binding sites.
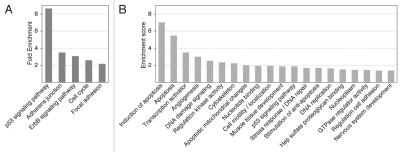
p53 binding sites in normal IMR90 cells are tightly associated with TSSs, a distribution similar to that of functional reference p53 REs. We looked for the most enriched signaling pathways among the 686 genes associated with the 743 ChIP-seq peaks using DAVID annotation chart analysis (see Sup. Methods). The p53 signaling pathway showed the highest enrichment (8.7 fold), demonstrating that the binding sites in IMR90 cells are indeed associated with p53-related genes. The five most enriched pathways are shown in Figure 8A.
Figure 8.
DAVID functional annotation analysis of the genes associated with high-confidence p53 ChIP-seq peaks in IMR90 cells. (A) Most highly enriched KEGG pathways (p-value < 0.01). Fold enrichment is shown as calculated by DAVID. (B) Most highly enriched clusters of genes with enrichment score above 1.3. See Table S10 for all enriched clusters.
Functional clustering of the ChIP-seq-associated genes was performed with DAVID, which ranks the overall enrichment of entire groups of enriched gene ontology terms based on functional similarity (Table S10, see Sup. Methods). The 20 most highly enriched clusters (score > 1.3) are shown in Figure 8B and include categories related to apoptosis, transcription activation, angiogenesis and DNA damage signaling, all well known to be modulated by p53.7 Clusters with a score 0.5–1.3 (Fig. S10), though less enriched, clearly contained p53-related terms, such as response to ionizing radiation, transcription repressor activity and cell cycle regulation.
In addition to the well-known functions in tumor suppression, p53 is implicated in regulating normal physiological processes, such as fertility68 and immunity,69 and similar categories are present among the less enriched clusters (Fig. S10). p53 is known to affect maternal fertility by regulating LIF expression through a characterized p53RE,68 which we detected (Fig. 2A). Interestingly, a spermatogenesis-associated gene (SPATA18) was identified in our study. SPATA18 recently was reported to be regulated by p53 through a binding site in intron 1 (identified by ChIP-seq peak ID 41, Table S2) and is expressed in a p53-dependent manner in IMR90 cells.70 The expression of the same gene (under the name Mieap), was characterized in several human cell lines and was shown to play a role in mitochondria quality control.71 Notably, SPATA18 is silenced by DNA methylation in several cancer cell lines, including HCT116. We believe that the functional clusters provided here represent a good source for examining potential p53-related functions in normal cells.
Methods
Normal human lung fibroblasts (IMR90) were obtained from ATCC (CCL-186) at PDL 24. Data were generated using cells at PDL 38–40. The p53-specific antibody DO1 (sc125X, Santa Cruz) and normal mouse IgG (sc2025, Santa Cruz) were used for the ChIP experiments. The ChIP-seq library was prepared from p53 ChIP DNA using 1.5 × 106 IMR90 cells treated for 6 h with 375 µM 5-fluorouracil (5-FU, SIGMA). The Input-seq library was prepared from 30 ng Input DNA (chromatin sample taken before ChIP). After polishing and Illumina adaptor ligation, ChIP and Input DNA were PCR amplified and gel purified. Small-scale sequencing was used to validate the libraries, which were then sequenced on the Illumina Genome Analyzer 2x.
Ilumina 36 nt-reads were mapped to the reference human genome (hg18) using in-house-developed software. Enriched locations were identified by adapting the method of Rozowsky et al.,31 whereby mapped ChIP-seq reads were used to construct a coverage map at each nucleotide at each genome position, yielding peaks at the enriched locations. These were then filtered by applying statistical confidence tests, requiring enrichment over the Input-seq reads (background) to correct for systematic biases present in the data. High-confidence peak locations were entered into an SQL database for positional correlation with various features analyzed (such as RefSeq genes, CGIs, repeats and reference p53 REs). Peak locations were used to retrieve the neighboring genomic DNA sequence for scanning with the p53MH algorithm or for correlating with the IMR90 methylome.1 MEME 4.6.1 was used for de novo motif analysis. DAVID 6.7 was used for the functional annotation clustering of the genes identified by the high-confidence p53 ChIP-seq peaks.
Conclusion
Despite intensive research, the mechanisms by which p53 selects its binding sites in vivo among all genomic sites available remain unclear. It is becoming more clear that conclusions from studying p53 functions in cancer or immortalized cells can be misleading if directly extrapolated to normal cells.60 In support of this, we found profound differences in the genomic distribution of the p53 binding sites in normal IMR90 cells (this ChIP-seq study) compared with those reported by three previous genome-wide studies using the cancer cell lines HCT116 and U2OS (ChIP-PET,23 ChIP-chip24 and ChIP-seq25). In normal IMR90, but not in the cancer cells, the p53 binding sites are highly enriched close to TSSs and in CGIs (Fig. 5). Such p53 enrichment at CGIs, although not reported previously, is consistent with their proposed role as TF “landing lights,” as reviewed by Illingworth and Bird.72 The distinct p53 binding pattern in IMR90 cells does not reflect affinity toward specific sequences, because the de novo developed motif p53CSI is similar to p53PET (HCT116) and p53scan (U2OS) reported by genome-wide studies in cancer cells.
In IMR90 cells, about half of the p53 binding sites reside in CGIs and mainly in proximal CGIs at promoters (Fig. 6D). Two types of promoters are recognized based on their core promoter elements, TATA-containing, focused (usually with single TSS) and TATA-less, dispersed (with multiple TSSs), typically found in CGIs.73 Although it has been known for a while that p53 targets diverse promoters, only recently was it demonstrated that core promoter elements directly affect p53 transcriptional regulation.74 Besides that study, little is known about the core promoter elements “signatures” of the promoters occupied by p53.75 Here we showed not only that in vivo p53 is enriched at promoters in normal cells, but specifically at CGI promoters. It remains to be seen how the core promoter elements globally impact the p53 transcriptional network.
Promoter CGIs have a specific chromatin architecture that acts as a platform for transcriptional regulation and epigenetic control. Although predicted to have high intrinsic nucleosome affinity,76 in vivo studies in somatic cells showed that CGIs have low nucleosome occupancy and a chromatin environment permissive for transcription, while promoters that lack CGIs have high nucleosome occupancy and require chromatin remodeling for transcription activation.62,77 A recent study in the breast cancer cell line MCF7 reported that p53 was bound to regions with predicted high nucleosome occupancy,78 although it was unclear how close and to what promoters these sites were located. Promoter CGIs typically are hypomethylated in an otherwise heavily methylated genome, unlike distal (orphan) CGIs, which are more often methylated in normal somatic cells.62 Our analysis of the IMR90 ChIP-seq peaks in the context of the IMR90 methylome1 showed that p53 binding sites are enriched in hypomethylated genomic regions (Fig. 6). Since the major difference in the p53 binding between the normal IMR90 and cancer HCT116/U2OS cells is at CGIs, and cancer development is associated with global genomic hypomethylation and local CGI hypermethylation; the distribution of p53 away from CGI promoters in cancer cells might be due to aberrant DNA methylation and associated chromatin changes.
Genome-wide studies are no longer just tools for identifying of new protein-DNA binding sites; they represent a powerful approach for chromatin profiling, annotation and regulatory predictions.79 The p53 transcriptional network is remarkably complex and difficult to decipher one gene at a time. Genome-wide studies on different types of cells (normal and cancer), combined with histone modification and DNA methylation maps, can help place p53 binding in the context of chromatin in vivo to address the epigenetic impact on p53 dependent transcriptional regulation and to provide a global view of the p53 network changes during cancer development.
Acknowledgments
We thank D. Turner (Wellcome Trust Sanger Institute, UK), J. Rozowsky (Yale University) and M. Regulski (CSHL) for the helpful discussions. We acknowledge the work of D. Kotov during his summer internship (SULI, BNL), M. Blewitt for helping with the small-scale sequencing at BNL and E. Ghiban (CSHL) for the help with the Illumina sequencing. We thank P. Freimuth and P. Wilson for their critical reading of the manuscript. This work was performed under the auspices of the US Department of Energy by Brookhaven National Laboratory under contract DE-AC02-98CH10886 and was supported by a grant from the US. Department of Energy Low Dose Radiation Research Program (C.W.A.). These sequence data have been submitted to the NCBI/GEO database under accession number GSE31558; human genome reference sequence hg18 (NCBI Build 36.1) was used for the analysis.
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
Supplementary Material
References
- 1.Lister R, Pelizzola M, Dowen RH, Hawkins RD, Hon G, Tonti-Filippini J, et al. Human DNA methylomes at base resolution show widespread epigenomic differences. Nature. 2009;462:315–322. doi: 10.1038/nature08514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Anderson CW, Appella E. Signaling to the p53 tumor suppressor through pathways activated by genotoxic and non-genotoxic stresses. In: Bradshaw RA, Dennis EA, editors. Handbook of Cell Signaling. Elsevier; 2009. [Google Scholar]
- 3.Riley T, Sontag E, Chen P, Levine A. Transcriptional control of human p53-regulated genes. Nat Rev Mol Cell Biol. 2008;9:402–412. doi: 10.1038/nrm2395. [DOI] [PubMed] [Google Scholar]
- 4.Kruse JP, Gu W. Modes of p53 regulation. Cell. 2009;137:609–622. doi: 10.1016/j.cell.2009.04.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Rinn JL, Huarte M. To repress or not to repress: This is the guardian's question. Trends Cell Biol. 2011;21:344–353. doi: 10.1016/j.tcb.2011.04.002. [DOI] [PubMed] [Google Scholar]
- 6.Levine AJ, Oren M. The first 30 years of p53: growing ever more complex. Nat Rev Cancer. 2009;9:749–758. doi: 10.1038/nrc2723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Vousden KH, Prives C. Blinded by the Light: The Growing Complexity of p53. Cell. 2009;137:413–431. doi: 10.1016/j.cell.2009.04.037. [DOI] [PubMed] [Google Scholar]
- 8.El-Deiry WS, Kern SE, Pietenpol JA, Kinzler KW, Vogelstein B. Definition of a consensus binding site for p53. Nat Genet. 1992;1:45–49. doi: 10.1038/ng0492-45. [DOI] [PubMed] [Google Scholar]
- 9.Funk WD, Pak DT, Karas RH, Wright WE, Shay JW. A transcriptionally active DNA-binding site for human p53 protein complexes. Mol Cell Biol. 1992;12:2866–2871. doi: 10.1128/mcb.12.6.2866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Jordan JJ, Menendez D, Inga A, Noureddine M, Bell DA, Resnick MA. Noncanonical DNA motifs as trans-activation targets by wild type and mutant p53. PLoS Genet. 2008;4:1000104. doi: 10.1371/journal.pgen.1000104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kim E, Deppert W. The versatile interactions of p53 with DNA: when flexibility serves specificity. Cell Death Differ. 2006;13:885–889. doi: 10.1038/sj.cdd.4401909. [DOI] [PubMed] [Google Scholar]
- 12.Bakhanashvili M, Hizi A, Rahav G. The interaction of p53 with 3′-terminal mismatched DNA. Cell Cycle. 2010;9:1380–1389. doi: 10.4161/cc.9.7.11201. [DOI] [PubMed] [Google Scholar]
- 13.Weinberg RL, Veprintsev DB, Fersht AR. Cooperative binding of tetrameric p53 to DNA. J Mol Biol. 2004;341:1145–1159. doi: 10.1016/j.jmb.2004.06.071. [DOI] [PubMed] [Google Scholar]
- 14.Schlereth K, Charles JP, Bretz AC, Stiewe T. Life or death: p53-induced apoptosis requires DNA binding cooperativity. Cell Cycle. 2010;9:4068–4076. doi: 10.4161/cc.9.20.13595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Petrovich M, Veprintsev DB. Effects of CpG methylation on recognition of DNA by the tumour suppressor p53. J Mol Biol. 2009;386:72–80. doi: 10.1016/j.jmb.2008.11.054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hoh J, Jin S, Parrado T, Edington J, Levine AJ, Ott J. The p53MH algorithm and its application in detecting p53-responsive genes. Proc Natl Acad Sci USA. 2002;99:8467–8472. doi: 10.1073/pnas.132268899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Horvath MM, Wang X, Resnick MA, Bell DA. Divergent evolution of human p53 binding sites: cell cycle versus apoptosis. PLoS Genet. 2007;3:127. doi: 10.1371/journal.pgen.0030127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Cawley S, Bekiranov S, Ng HH, Kapranov P, Sekinger EA, Kampa D, et al. Unbiased mapping of transcription factor binding sites along human chromosomes 21 and 22 points to widespread regulation of noncoding RNAs. Cell. 2004;116:499–509. doi: 10.1016/S0092-8674(04)00127-8. [DOI] [PubMed] [Google Scholar]
- 19.Chen J, Sadowski I. Identification of the mismatch repair genes PMS2 and MLH1 as p53 target genes by using serial analysis of binding elements. Proc Natl Acad Sci USA. 2005;102:4813–4818. doi: 10.1073/pnas.0407069102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hearnes JM, Mays DJ, Schavolt KL, Tang L, Jiang X, Pietenpol JA. Chromatin immunoprecipitation-based screen to identify functional genomic binding sites for sequence-specific transactivators. Mol Cell Biol. 2005;25:10148–10158. doi: 10.1128/MCB.25.22.10148-10158.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kaneshiro K, Tsutsumi S, Tsuji S, Shirahige K, Aburatani H. An integrated map of p53-binding sites and histone modification in the human ENCODE regions. Genomics. 2007;89:178–188. doi: 10.1016/j.ygeno.2006.09.001. [DOI] [PubMed] [Google Scholar]
- 22.Krieg AJ, Hammond EM, Giaccia AJ. Functional analysis of p53 binding under differential stresses. Mol Cell Biol. 2006;26:7030–7045. doi: 10.1128/MCB.00322-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wei CL, Wu Q, Vega VB, Chiu KP, Ng P, Zhang T, et al. A global map of p53 transcription-factor binding sites in the human genome. Cell. 2006;124:207–219. doi: 10.1016/j.cell.2005.10.043. [DOI] [PubMed] [Google Scholar]
- 24.Smeenk L, van Heeringen SJ, Koeppel M, van Driel MA, Bartels SJ, Akkers RC, et al. Characterization of genome-wide p53-binding sites upon stress response. Nucleic Acids Res. 2008;36:3639–3654. doi: 10.1093/nar/gkn232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Smeenk L, van Heeringen SJ, Koeppel M, Gilbert B, Janssen-Megens E, Stunnenberg HG, et al. Role of p53 serine 46 in p53 target gene regulation. PLoS ONE. 2011;6:17574. doi: 10.1371/journal.pone.0017574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Dowell RD. Transcription factor binding variation in the evolution of gene regulation. Trends Genet. 2010;26:468–475. doi: 10.1016/j.tig.2010.08.005. [DOI] [PubMed] [Google Scholar]
- 27.Jones PA, Baylin SB. The epigenomics of cancer. Cell. 2007;128:683–692. doi: 10.1016/j.cell.2007.01.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Shaked H, Shiff I, Kott-Gutkowski M, Siegfried Z, Haupt Y, Simon I. Chromatin immunoprecipitation-on-chip reveals stress-dependent p53 occupancy in primary normal cells but not in established cell lines. Cancer Res. 2008;68:9671–9677. doi: 10.1158/0008-5472.CAN-08-0865. [DOI] [PubMed] [Google Scholar]
- 29.Nichols WW, Murphy DG, Cristofalo VJ, Toji LH, Greene AE, Dwight SA. Characterization of a new human diploid cell strain, IMR-90. Science. 1977;196:60–63. doi: 10.1126/science.841339. [DOI] [PubMed] [Google Scholar]
- 30.Park PJ. ChIP-seq: advantages and challenges of a maturing technology. Nat Rev Genet. 2009;10:669–680. doi: 10.1038/nrg2641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Rozowsky J, Euskirchen G, Auerbach RK, Zhang ZD, Gibson T, Bjornson R, et al. PeakSeq enables systematic scoring of ChIP-seq experiments relative to controls. Nat Biotechnol. 2009;27:66–75. doi: 10.1038/nbt.1518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Jung YS, Qian Y, Chen X. Examination of the expanding pathways for the regulation of p21 expression and activity. Cell Signal. 2010;22:1003–1012. doi: 10.1016/j.cellsig.2010.01.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Nozell S, Chen X. p21B, a variant of p21Waf1/Cip1, is induced by the p53 family. Oncogene. 2002;21:1285–1294. doi: 10.1038/sj.onc.1205191. [DOI] [PubMed] [Google Scholar]
- 34.Saramäki A, Banwell CM, Campbell MJ, Carlberg C. Regulation of the human p21waf1/cip1 gene promoter via multiple binding sites for p53 and the vitamin D3 receptor. Nucleic Acids Res. 2006;34:543–554. doi: 10.1093/nar/gkj460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Huarte M, Guttman M, Feldser D, Garber M, Koziol MJ, Kenzelmann-Broz D, et al. A large intergenic noncoding RNA induced by p53 mediates global gene repression in the p53 response. Cell. 2010;142:409–419. doi: 10.1016/j.cell.2010.06.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Millau JF, Bastien N, Bouchard EF, Drouin R. p53 Pre- and post-binding event theories revisited: stresses reveal specific and dynamic p53-binding patterns on the p21 gene promoter. Cancer Res. 2009;69:8463–8471. doi: 10.1158/00085472.CAN-09-2036. [DOI] [PubMed] [Google Scholar]
- 37.Millau JF, Bandele OJ, Perron J, Bastien N, Bouchard EF, Gaudreau L, et al. Formation of stress-specific p53 binding patterns is influenced by chromatin but not by modulation of p53 binding affinity to response elements. Nucleic Acids Res. 2011;39:3053–3063. doi: 10.1093/nar/gkq1209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wang B, Xiao Z, Ren EC. Redefining the p53 response element. Proc Natl Acad Sci USA. 2009;106:14373–14378. doi: 10.1073/pnas.0903284106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Gruenbaum Y, Margalit A, Goldman RD, Shumaker DK, Wilson KL. The nuclear lamina comes of age. Nat Rev Mol Cell Biol. 2005;6:21–31. doi: 10.1038/nrm1550. [DOI] [PubMed] [Google Scholar]
- 40.Rahman-Roblick R, Johannes Roblick U, Hellman U, Conrotto P, Liu T, Becker S, et al. p53 targets identified by protein expression profiling. Proc Natl Acad Sci USA. 2007;104:5401–5406. doi: 10.1073/pnas.0700794104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Cougot N, Babajko S, Séraphin B. Cytoplasmic foci are sites of mRNA decay in human cells. J Cell Biol. 2004;165:31–40. doi: 10.1083/jcb.200309008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Barbier J, Dutertre M, Bittencourt D, Sanchez G, Gratadou L, de la Grange P, et al. Regulation of H-ras splice variant expression by cross talk between the p53 and nonsense-mediated mRNA decay pathways. Mol Cell Biol. 2007;27:7315–7333. doi: 10.1128/MCB.00272-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Boni V, Bitarte N, Cristobal I, Zarate R, Rodriguez J, Maiello E, et al. miR-192/miR-215 influence 5-fluorouracil resistance through cell cycle-mediated mechanisms complementary to its post-transcriptional thymidilate synthase regulation. Mol Cancer Ther. 2010;9:2265–2275. doi: 10.1158/1535-7163.MCT-10-0061. [DOI] [PubMed] [Google Scholar]
- 44.Lee Y, Chen Y, Chang LS, Johnson LF. Inhibition of mouse thymidylate synthase promoter activity by the wild-type p53 tumor suppressor protein. Exp Cell Res. 1997;234:270–276. doi: 10.1006/excr.1997.3605. [DOI] [PubMed] [Google Scholar]
- 45.Clemson CM, Hutchinson JN, Sara SA, Ensminger AW, Fox AH, Chess A, et al. An architectural role for a nuclear noncoding RNA: NEAT1 RNA is essential for the structure of paraspeckles. Mol Cell. 2009;33:717–726. doi: 10.1016/j.molcel.2009.01.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Chen LL, Carmichael GG. Altered nuclear retention of mRNAs containing inverted repeats in human embryonic stem cells: functional role of a nuclear noncoding RNA. Mol Cell. 2009;35:467–478. doi: 10.1016/j.molcel.2009.06.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Mao YS, Sunwoo H, Zhang B, Spector DL. Direct visualization of the co-transcriptional assembly of a nuclear body by noncoding RNAs. Nat Cell Biol. 2011;13:95–101. doi: 10.1038/ncb2140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Zemojtel T, Kielbasa SM, Arndt PF, Chung HR, Vingron M. Methylation and deamination of CpGs generate p53-binding sites on a genomic scale. Trends Genet. 2009;25:63–66. doi: 10.1016/j.tig.2008.11.005. [DOI] [PubMed] [Google Scholar]
- 49.Cui F, Sirotin MV, Zhurkin VB. Impact of Alu repeats on the evolution of human p53 binding sites. Biol Direct. 2011;6:2. doi: 10.1186/1745-6150-6-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Chen LL, DeCerbo JN, Carmichael GG. Alu element-mediated gene silencing. EMBO J. 2008;27:1694–1705. doi: 10.1038/emboj.2008.94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Ashur-Fabian O, Har-Zahav A, Shaish A, Wiener Amram H, Margalit O, Weizer-Stern O, et al. apoB and apobec1, two genes key to lipid metabolism, are transcriptionally regulated by p53. Cell Cycle. 2010;9:3761–3770. doi: 10.4161/cc.9.18.12993. [DOI] [PubMed] [Google Scholar]
- 52.Puzio-Kuter AM, Levine AJ. Stem cell biology meets p53. Nat Biotechnol. 2009;27:914–915. doi: 10.1038/nbt1009-914. [DOI] [PubMed] [Google Scholar]
- 53.Veprintsev DB, Fersht AR. Algorithm for prediction of tumour suppressor p53 affinity for binding sites in DNA. Nucleic Acids Res. 2008;36:1589–1598. doi: 10.1093/nar/gkm1040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Riley T, Yu X, Sontag E, Levine A. The p53HMM algorithm: using profile hidden markov models to detect p53-responsive genes. BMC Bioinformatics. 2009;10:111. doi: 10.1186/1471-2105-10-111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Trinklein ND, Aldred SF, Hartman SJ, Schroeder DI, Otillar RP, Myers RM. An abundance of bidirectional promoters in the human genome. Genome Res. 2004;14:62–66. doi: 10.1101/gr.1982804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Hofer-Warbinek R, Schmid JA, Mayer H, Winsauer G, Orel L, Mueller B, et al. A highly conserved proapoptotic gene, IKIP, located next to the APAF1 gene locus, is regulated by p53. Cell Death Differ. 2004;11:1317–1325. doi: 10.1038/sj.cdd.4401502. [DOI] [PubMed] [Google Scholar]
- 57.Cho YJ, Liang P. Killin is a p53-regulated nuclear inhibitor of DNA synthesis. Proc Natl Acad Sci USA. 2008;105:5396–5401. doi: 10.1073/pnas.0705410105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Comer KA, Dennis PA, Armstrong L, Catino JJ, Kastan MB, Kumar CC. Human smooth muscle α-actin gene is a transcriptional target of the p53 tumor suppressor protein. Oncogene. 1998;16:1299–1308. doi: 10.1038/sj.onc.1201645. [DOI] [PubMed] [Google Scholar]
- 59.Yang MQ, Elnitski LL. A computational study of bidirectional promoters in the human genome. Lect Notes Comput Sci. 2007;4463:361–371. doi: 10.1007/978-3-540-72031-7_33. [DOI] [Google Scholar]
- 60.Millau JF, Mai S, Bastien N, Drouin R. p53 functions and cell lines: have we learned the lessons from the past? Bioessays. 2010;32:392–400. doi: 10.1002/bies.200900160. [DOI] [PubMed] [Google Scholar]
- 61.Euskirchen GM, Rozowsky JS, Wei CL, Lee WH, Zhang ZD, Hartman S, et al. Mapping of transcription factor binding regions in mammalian cells by ChIP: comparison of array- and sequencing-based technologies. Genome Res. 2007;17:898–909. doi: 10.1101/gr.5583007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Deaton AM, Bird A. CpG islands and the regulation of transcription. Genes Dev. 2011;25:1010–1022. doi: 10.1101/gad.2037511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Illingworth RS, Gruenewald-Schneider U, Webb S, Kerr ARW, James KD, Turner DJ, et al. Orphan CpG islands identify numerous conserved promoters in the mammalian genome. PLoS Genet. 2010;6:1001134. doi: 10.1371/journal.pgen.1001134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Mancini DN, Singh SM, Archer TK, Rodenhiser DI. Site-specific DNA methylation in the neurofibromatosis (NF1) promoter interferes with binding of CREB and SP1 transcription factors. Oncogene. 1999;18:4108–4119. doi: 10.1038/sj.onc.1202764. [DOI] [PubMed] [Google Scholar]
- 65.Renda M, Baglivo I, Burgess-Beusse B, Esposito S, Fattorusso R, Felsenfeld G, et al. Critical DNA binding interactions of the insulator protein CTCF: a small number of zinc fingers mediate strong binding, and a single finger-DNA interaction controls binding at imprinted loci. J Biol Chem. 2007;282:33336–33345. doi: 10.1074/jbc.M706213200. [DOI] [PubMed] [Google Scholar]
- 66.Nabilsi NH, Broaddus RR, Loose DS. DNA methylation inhibits p53-mediated survivin repression. Oncogene. 2009;28:2046–2050. doi: 10.1038/onc.2009.62. [DOI] [PubMed] [Google Scholar]
- 67.Choy JS, Wei S, Lee JY, Tan S, Chu S, Lee TH. DNA methylation increases nucleosome compaction and rigidity. J Am Chem Soc. 2010;132:1782–1783. doi: 10.1021/ja910264z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Hu W, Feng Z, Teresky AK, Levine AJ. p53 regulates maternal reproduction through LIF. Nature. 2007;450:721–724. doi: 10.1038/nature05993. [DOI] [PubMed] [Google Scholar]
- 69.Menendez D, Shatz M, Azzam K, Garantziotis S, Fessler MB, Resnick MA. The Toll-like receptor gene family is integrated into human DNA damage and p53 networks. PLoS Genet. 2011;7:1001360. doi: 10.1371/journal.pgen.1001360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Bornstein C, Brosh R, Molchadsky A, Madar S, Kogan-Sakin I, Goldstein I, et al. SPATA18, a spermatogenesis-associated gene, is a novel transcriptional target of p53 and p63. Mol Cell Biol. 2011;31:1679–1689. doi: 10.1128/MCB.01072-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Miyamoto Y, Kitamura N, Nakamura Y, Futamura M, Miyamoto T, Yoshida M, et al. Possible existence of lysosome-like organella within mitochondria and its role in mitochondrial quality control. PLoS ONE. 2011;6:16054. doi: 10.1371/journal.pone.0016054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Illingworth RS, Bird AP. CpG islands—‘a rough guide’. FEBS Lett. 2009;583:1713–1720. doi: 10.1016/j.febslet.2009.04.012. [DOI] [PubMed] [Google Scholar]
- 73.Juven-Gershon T, Hsu JY, Theisen JWM, Kadonaga JT. The RNA polymerase II core promoter—the gateway to transcription. Curr Opin Cell Biol. 2008;20:253–259. doi: 10.1016/j.ceb.2008.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Morachis JM, Murawsky CM, Emerson BM. Regulation of the p53 transcriptional response by structurally diverse core promoters. Genes Dev. 2010;24:135–147. doi: 10.1101/gad.1856710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Gomes NP, Espinosa JM. Differential regulation of p53 target genes: it's (core promoter) elementary. Genes Dev. 2010;24:111–114. doi: 10.1101/gad.1893610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Tillo D, Kaplan N, Moore IK, Fondufe-Mittendorf Y, Gossett AJ, Field Y, et al. High nucleosome occupancy is encoded at human regulatory sequences. PLoS ONE. 2010;5:9129. doi: 10.1371/journal.pone.0009129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Ramirez-Carrozzi VR, Braas D, Bhatt DM, Cheng CS, Hong C, Doty KR, et al. A unifying model for the selective regulation of inducible transcription by CpG islands and nucleosome remodeling. Cell. 2009;138:114–128. doi: 10.1016/j.cell.2009.04.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Lidor Nili E, Field Y, Lubling Y, Widom J, Oren M, Segal E. p53 binds preferentially to genomic regions with high DNA-encoded nucleosome occupancy. Genome Res. 2010;20:1361–1368. doi: 10.1101/gr.103945.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Ernst J, Kheradpour P, Mikkelsen TS, Shoresh N, Ward LD, Epstein CB, et al. Mapping and analysis of chromatin state dynamics in nine human cell types. Nature. 2011;473:43–49. doi: 10.1038/nature09906. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.