Abstract
We have recently shown that inhibition of HRR (homologous recombination repair) by Chk1 (checkpoint kinase 1) inhibition radiosensitizes pancreatic cancer cells, and others have demonstrated that Chk1 inhibition selectively sensitizes p53 mutant tumor cells. Furthermore, PARP1 [poly (ADP-ribose) polymerase-1] inhibitors dramatically radiosensitize cells with DNA double-strand break repair defects. Thus, we hypothesized that inhibition of HRR (mediated by Chk1 via AZD7762) and PARP1 [via olaparib (AZD2281)] would selectively sensitize p53 mutant pancreatic cancer cells to radiation. We also used two isogenic p53 cell models to assess the role of p53 status in cancer cells and intestinal epithelial cells to assess overall cancer specificity. DNA damage response and repair were assessed by flow cytometry, γH2AX and an HRR reporter assay. We found that the combination of AZD7762 and olaparib produced significant radiosensitization in p53 mutant pancreatic cancer cells and in all of the isogenic cancer cell lines. The magnitude of radiosensitization by AZD7762 and olaparib was greater in p53 mutant cells compared with p53 wild-type cells. Importantly, normal intestinal epithelial cells were not radiosensitized. The combination of AZD7762 and olaparib caused G2 checkpoint abrogation, inhibition of HRR and persistent DNA damage responses. These findings demonstrate that the combination of Chk1 and PARP1 inhibition selectively radiosensitizes p53 mutant pancreatic cancer cells. Furthermore, these studies suggest that inhibition of HRR by Chk1 inhibitors may be a useful strategy for selectively inducing a BRCA1/2 “deficient-like” phenotype in p53 mutant tumor cells, while sparing normal tissue.
Key words: pancreatic cancer, Chk1, PARP1, radiosensitization, p53
Introduction
Pancreatic cancer has the highest mortality rate of all major cancers, with 94% of patients succumbing to the disease within the first 5 years of diagnosis.1 While the lack of effective systemic disease control is a barrier to improved patient outcomes, local control is also an important aspect of pancreatic cancer treatment. This is supported by the following: local failure is responsible for up to 1/3 of the observed cancer related mortality,2 the addition of radiation to standard chemotherapy (gemcitabine) is superior to gemcitabine alone,3,4 and, finally, increasing the dose of radiation appears to improve outcome.5 Thus, strategies to improve local disease control while maintaining or improving systemic disease control are warranted.6–8
We have demonstrated that inhibition of Chk1 sensitizes pancreatic cancer cells and xenografts to gemcitabine and radiation.7,9 We recently found that inhibition of HRR and G2 checkpoint abrogation are mechanisms of radiosensitization in response to Chk1 inhibition. Abrogation of the G2 checkpoint (by Chk1 inhibition and other strategies) has been shown to preferentially sensitize p53 mutant tumor cells to chemotherapy and radiation.10–17 The prevailing model for tumor cell selectivity of Chk1 inhibition is that tumor cells harbor aberrations in other DNA damage response machinery (i.e., p53, p16, Rb) and, thus, do not G1 arrest in response to DNA damage leading to selective sensitization of tumor cells by Chk1 inhibition, while normal cells are protected from Chk1 inhibition by their other intact checkpoints (i.e., p53-mediated G1 arrest).
PARP inhibitors have generated great enthusiasm in the oncology community with regard to their use in BRCA1/2 mutant tumors, a concept known as synthetic lethality. Since BRCA1 and BRCA2 are required for HRR, and PARP is also required for repair, inhibition of both pathways results in synthetic lethality. PARP inhibitors have also been shown to sensitize to DNA damage in variety of cancer models, including those in which BRCA1 and BRCA2 are proficient.18,19 Radiosensitization occurs in a replication-dependent manner20 and more efficiently in cells with other double-strand break repair defects.21 One model to explain PARP inhibitor-mediated radiosensitization is that PARP inhibition delays repair of single-strand DNA breaks, which, when met by DNA replication forks, result in a collapsed fork and a double-strand break. While certain mutations are common in pancreatic cancers [k-Ras (100%), p16 (82%), p53 (76%)], BRCA1/2 mutations are rare.2,22 Therefore, “true” synthetic lethality from BRCA1/2 mutations and PARP inhibition would only be expected in the minority of pancreatic cancer cases. There has, however, been interest in extending synthetic lethality to tumors with defective HRR capabilities yet wild-type BRCA1/2, referred to as “BRCAness.”23,24 Based on the results of these studies, it seems plausible that combining a small molecule inhibitor of HRR (i.e., a Chk1 inhibitor) with a PARP inhibitor might extend synthetic lethality to tumor cells that do not have BRCA1/2 mutations/HRR defects, a concept referred to as induced synthetic lethality.25
Given the ability of Chk1 inhibition to block HRR and the efficacy of PARP1 inhibitors as radiation sensitizers in double-strand break repair defective tumor types, we hypothesized that the combination of Chk1 and PARP1 inhibitors would sensitize tumor cells to radiation. Furthermore, we hypothesized that p53 mutation would confer tumor cell selectivity for radiosensitization by Chk1 and PARP1 inhibition. To begin to test this hypothesis, we assessed radiosensitization in p53 mutant pancreatic cancers in response to the small molecule inhibitors of Chk1 and PARP1, AZD7762 and olaparib, respectively.15 When we found that Chk1 and PARP1 inhibition did produce significant radiosensitization in p53 mutant pancreatic cancer cells, we then went on to determine the roles of cell cycle checkpoints, DNA damage response and HRR in the mechanisms of sensitization. In order to begin to establish the potential mechanisms of tumor cell selectivity, we assessed radiosensitization by Chk1 and PARP1 inhibition in isogenic p53 models as well as in normal epithelial cells.
Results
Combined Chk1 and PARP1 inhibition radiosensitizes pancreatic cancer cells.
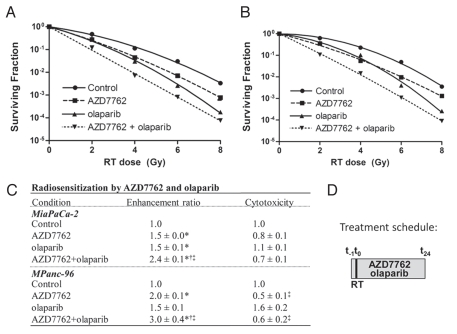
In order to begin to determine the radiosensitizing efficacy of combined Chk1 and PARP1 inhibition, MiaPaCa-2 and MPanc-96 pancreatic cancer cells were treated with AZD7762 and olaparib according to previously determined concentrations and schedules as illustrated (Fig. 1D).7,26 Consistent with previous reports,7,26 AZD7762 or olaparib alone produced comparable, significant radiosensitization (Fig. 1A–C). Radiosensitization by olaparib at clinically relevant doses was concentration-dependent and associated with inhibition of PARP activity as assessed by PAR27 (Fig. S1). More importantly however, the combination of AZD7762 with olaparib produced additive radiosensitization in MiaPaCa-2 (RER 2.4 ± 0.1) and MPanc-96 cells (RER 3.0 ± 0.4), which was greater than that produced by either agent alone (p > 0.2 under the null hypothesis that there is an additive effect of AZD7762 and olaparib; Fig. 1C). This substantial increase in radiosensitization was not accompanied by additional cytotoxicity over AZD7762 alone.
Figure 1.
Radiosensitization of pancreatic cancer cells in response to Chk1 and PARP1 inhibition. Representative radiation survival curves are shown for (A) MiaPaCa-2 and (B) MPanc-96 cell lines treated with AZD7762 (100 nM), olaparib (1 uM) and ionizing radiation (RT, 0–8 Gy) according to the illustrated schedule (D). Clonogenic survival plating efficiencies were 0.5 ± 0.1 (MiaPaCa-2) and 0.3 ± 0.1 (MPanc-96). Data are shown from a single representative experiment (A and B) or are the mean radiation enhancement ratio or cytotoxicity from n = 3 independent experiments ± SE. For (A and B), error bars are contained within the points. For (C), statistically significant differences (p < 0.05) are indicated vs. control (*), AZD7762 (†) and olaparib (‡).
Radiosensitization is associated with persistent DNA damage.
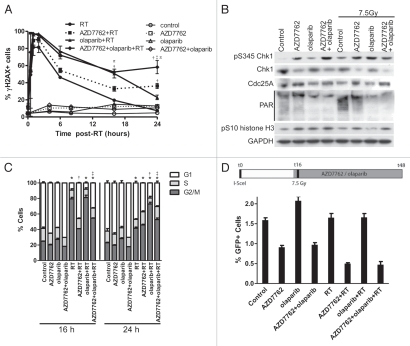
To explore the mechanisms of radiosensitization by the combination of AZD7762 and olaparib, we investigated their effects on γH2AX, cell cycle checkpoints and HRR. We hypothesized that the interaction between AZD7762 and olaparib leading to radiosensitization could be attributed to AZD7762-mediated checkpoint abrogation or inhibition of HRR, ultimately leading to persistent, unrepaired DNA damage. In order to determine the effects of AZD7762 and olaparib on DNA damage response/repair, we assessed γH2AX in MiaPaCa-2 cells at various time points following radiation and in response to AZD7762/olaparib. As anticipated, radiation produced a γH2AX signal as early as 30 min post-radiation, which resolved to near baseline by 24 h (Fig. 2A). Olaparib in combination with radiation caused a significant increase in γH2AX (16 h) that returned to baseline by 24 h. Consistent with our previous work, AZD7762 combined with radiation resulted in persistent γH2AX (24 h) compared with radiation alone. Most importantly, the combination of AZD7762 and olaparib resulted in a significant prolongation of γH2AX in response to radiation compared with radiation alone or in comparison with AZD7762 or olaparib (16 and 24 h). In the absence of radiation, olaparib and AZD7762 (alone or in combination), produced only a minor increase (p > 0.05) in the percentage of cells positive for γH2AX (Fig. 2A). Taken together, these results suggest that the radiosensitization produced through the interaction between AZD7762 and olaparib involves the presence of persistent, unrepaired DNA damage.
Figure 2.
DNA damage responses following AZD7762, olaparib and radiation treatments. (A) MiaPaCa-2 cells were treated with AZD7762, olaparib and RT (7.5 Gy) as illustrated (Fig. 1D). At the indicated times, post-RT cells were analyzed for γH2AX. Data are the percentage of cells staining positive for γH2AX. (B) At 24 h post-RT, cells were immunoblotted for the indicated proteins. (C) At 16 and 24 h post-RT, cell cycle distribution by DNA content was analyzed. The percentages of cells in each phase of the cell cycle were quantitated. (D) MiaPaCa-2-DR-GFP cells were treated as illustrated; at t = 48 h, the percentage of GFP-positive cells was measured by flow cytometry. As expected, radiation did not lead to an increase in HRR activity, as this assay only measures repair of I-SceI endonuclease-induced DNA double-strand breaks. Data are the mean ± SE of n = 3 experiments (A, C and D; except for the drug-only portion of Fig. 1A, which is n = 2) or are a single experiment representative of three independent experiments (B). (A, C and D) statistical significance (p < 0.05) is indicated vs. control (*), RT (†), AZD7762-RT (‡) and olaparib-RT (π). For cell cycle (C), statistical analysis was based on the G2/M population.
To further investigate the mechanisms underlying radiosensitization by AZD7762 and olaparib, we analyzed Chk1 and PARP1-mediated signaling. Consistent with inhibition of Chk1 by AZD7762,7 Cdc25A protein was accumulated in response to AZD7762 alone and in the presence of olaparib/radiation (Fig. 2B). Furthermore, pS345 Chk1, a recently identified pharmacodynamic biomarker of DNA damage in response to Chk1 inhibition,9 was elevated in response to AZD7762 alone and in combination with olaparib/radiation. Consistent with the finding that S345 Chk1 phosphorylation triggers ubiquitin-mediated proteosomal degradation of Chk1,28 we observed a decrease in Chk1 protein in response to AZD7762 alone and in combination with olaparib/radiation. In addition, the mitotic marker pS10 histone H3 was increased in response to radiation plus AZD7762 alone or in combination with olaparib, consistent with G2 checkpoint abrogation and mitotic entry. Finally, evidence that olaparib efficiently inhibits PARP1 is demonstrated by the decrease in PAR [poly(ADP-ribose)] in response to olaparib alone or in combination with AZD7762/radiation. Taken together, these results demonstrate that AZD7762 and olaparib, both alone and in combination, effectively block their respective targets.
In order to determine the mechanisms of interaction between Chk1 and PARP1 inhibition, which lead to persistent DNA damage and radiosensitization, we assessed radiation-induced cell cycle checkpoints and HRR in response to AZD7762 and olaparib. As anticipated, radiation caused a G2 arrest, which was abrogated by AZD7762 (16 h) (Fig. 2C). Olaparib prolonged the radiation-induced G2 checkpoint (24 h), which is likely a consequence of persistent DNA damage. The addition of AZD7762 to olaparib abrogated the radiation-induced G2 checkpoint, suggesting that G2 checkpoint abrogation is one possible mechanism of interaction between Chk1 and PARP1 inhibition that may lead to radiosensitization. Based on our previous finding that AZD7762 inhibits HRR, we wished to determine the effects of AZD7762 in combination with olaparib on HRR using a DR-GFP reporter that measures homology-directed repair of an I-SceI endonuclease-induced DNA double-strand break.7,29 We found that AZD7762 retained its ability to inhibit HRR in the presence of olaparib and/or radiation. Furthermore, olaparib had no effect on HRR activity (Fig. 2D). These results suggest that both inhibition of HRR as well as G2 checkpoint abrogation by AZD7762 may increase the sensitivity to olaparib and radiation.
Combined Chk1 and PARP1 inhibition preferentially sensitizes p53-defective cells.
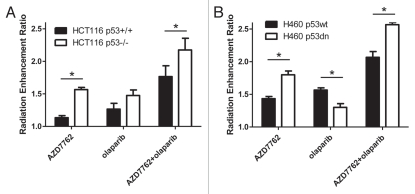
It has been shown that Chk1 inhibition preferentially radiosensitizes p53 mutant cancer cells.14 To begin to determine if this mechanism of selectivity might also be extended to radiosensitization by the combination of a Chk1 and PARP1 inhibitor, we utilized isogenic p53 models developed from the HCT116 and H460 cell lines.15 In HCT116 cells, as expected, AZD7762 produced significantly greater radiosensitization in HCT116 p53−/− cells than in HCT116 p53+/+ cells (RER 1.6 vs. 1.1, p < 0.05) (Figs. 3A, S2 and 3). p53 status conferred no selectivity in terms of olaparib-mediated radiosensitization. However, in response to the combination of AZD7762 and olaparib, while both HCT116 cell lines were radiosensitized, radiosensitization was significantly greater in the HCT116 p53−/− cells as compared with the p53+/+ cells (RER 2.2 vs. 1.8, p < 0.05). Furthermore, similar results were obtained in an independent p53 isogenic model wherein H460 p53dn cells were more radiosensitized by AZD7762 alone or in combination with olaparib than H460 p53wt cells (Fig. 3B and Fig. S4). Together, these data demonstrate preferential radiosensitization of p53 defective cells by AZD7762 and olaparib and suggest that Chk1 inhibition can confer selectivity when used in combination with a PARP1 inhibitor.
Figure 3.
Preferential radiosensitization by AZD7762 and olaparib in p53-defective cancer cells. HCT116 (A) and H460 (B) p53 isogenic cell lines were treated with AZD7762 and olaparib. Clonogenic survival plating efficiencies were 0.4 ± 0.1 (HCT116 p53+/+), 0.5 ± 0.03 (HCT116 p53−/−), 0.7 ± 0.03 (H460 p53wt) and 0.5 ± 0.2 (H460 p53 dn). The mean radiation enhancement ratios ± SE are shown for n = 3–4 independent experiments (See Figs. S2 and 4, respectively for representative clonogenic survival curves). Statistically significant differences between p53+/+ and p53−/− (HCT116) or p53 wt and p53 dn (H460) are indicated (*p < 0.05). Other significant differences not illustrated for both the HCT116 and H460 cell lines include control vs. AZD7762 (except HCT116 p53+/+), olaparib (except HCT116 p53+/+) or AZD7762 + olaparib as well as AZD7762 or olaparib alone vs. AZD7762 + olaparib.
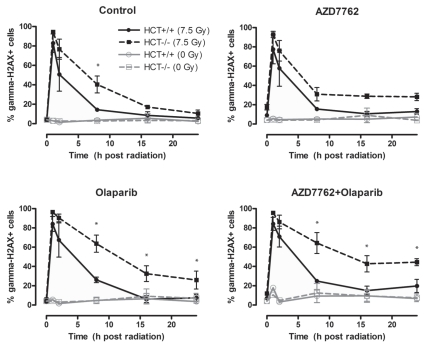
We then wished to determine whether the increased radiosensitization by Chk1 and PARP1 inhibition in p53-defective cells was associated with an increase in γH2AX in response to radiation. HCT116 p53−/− and p53+/+ cells were treated with AZD7762 and olaparib one hour before radiation and then assessed at various times following radiation for γH2AX. Consistent with the observation that p53−/− cells are more radiosensitized by AZD7762 than p53+/+ cells, AZD7762 combined with radiation led to prolonged γH2AX induction (24 h) in the p53−/− cells, whereas the p53+/+ cells had returned to baseline by 8 h (Fig. 4). Treatment with olaparib resulted in a similar profile with γH2AX in the p53+/+ cells, returning nearly to baseline after 8 h, and the p53−/− cells remaining elevated even after 24 h. The combination of AZD7762 and olaparib caused a persistent induction of γH2AX in response to radiation in the p53−/− cells that was resolved in the p53+/+ cells. The effects of AZD7762 and olaparib on γH2AX appeared to be dependent on radiation, as the drugs alone had minimal effects on γH2AX. These data suggest that p53-defective tumor cells encounter prolonged DNA damage in response to Chk1 and PARP1 inhibition, which is repaired in p53-proficient cells and is consistent with their observed differences in radiosensitization in response to Chk1 and PARP1 inhibition.
Figure 4.
Induction of γH2AX in HCT116 p53−/− or p53+/+ cells in response to AZD7762, olaparib and radiation. HCT116 cells were treated as indicated, for 1 h prior to radiation with AZD7762 and/or olaparib and analyzed for γH2AX at the indicated time points post-RT. Data are the mean ± SE of n = 3 experiments. Statistically significant differences are indicated (*p < 0.05) for HCT116 p53−/− (7.5 Gy) vs. HCT116 p53+/+ (7.5 Gy).
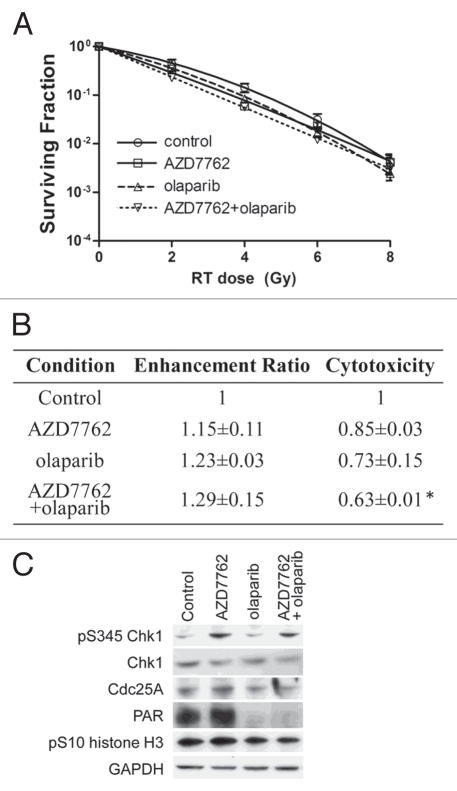
Given that the dose-limiting toxicity for radiation treatment of the pancreas is duodenum,3 we wished to determine the effects of Chk1 and PARP1 inhibition on the radiosensitization of normal small intestinal epithelial cells. Treatment of CCL-241 normal small intestinal epithelial cells with AZD7762 and/or olaparib did not produce significant radiosensitization under any of the treatment conditions despite producing cytotoxicity (Figs. 5A, B and S5), which was consistent with that observed in cancer cell lines. We confirmed that AZD7762 and olaparib did inhibit Chk1 and PARP1, respectively, as evidenced by Cdc25A stabilization and reduced PAR (Fig. 5C). Overall, these results show that AZD7762 combined with olaparib selectively sensitizes tumor cells, preferentially p53-defective, while sparing normal cells.
Figure 5.
Normal small intestinal epithelial cells are not radiosensitized by AZD7762 and/or olaparib. CCL-241 cells were treated as illustrated (Fig. 1D). Data are from a single representative experiment (A) or are the mean radiation enhancement ratio ± SE from n = 3 experiments (B). Statistically significant differences are indicated vs. control (*p < 0.05; there were no significant differences in the radiation enhancement ratios). (C) At the end of treatment, cells were analyzed by immunoblotting for the indicated proteins. Data are from a single representative experiment.
Discussion
In this study, we have found that combined inhibition of Chk1 and PARP1 produces profound radiosensitization of p53 mutant pancreatic cancer cells through mechanisms involving G2 checkpoint abrogation and HRR inhibition, resulting in accumulation of unrepaired DNA damage. Using isogenic p53 models, we found that p53-defective tumor cells are preferentially radiosensitized by Chk1 and PARP1 inhibition, while normal epithelial cells are not. These data suggest that inhibition of HRR by a Chk1 inhibitor can synergize with PARP1 inhibition to induce radiosensitization selectively in cancer cells and motivate the use of this combination in the treatment of unresectable pancreatic cancer.
While this is the first study to formally assess the combination of small molecule inhibitors of Chk1 and PARP1 as radiosensitizers, the concept of combining PARP inhibitors with small molecules that impair HRR is under active investigation. The Hsp90 inhibitor, 17-AAG, produces additive radiosensitization with PARP inhibition, most likely by mechanisms involving Rad51 and BRCA2 depletion, leading to inhibition of HRR.26 However, as Hsp90 inhibitors can affect hundreds of client proteins, other mechanisms could also underlie the resulting sensitization. In addition, Chk2 inhibition potentiates the cytotoxicity of PARP inhibition through mechanisms that may involve HRR inhibition.30 Similarly, PARP inhibitors have shown efficacy in tumors that contain wild-type BRCA1/2 but harbor other defects leading to HRR deficiency, a phenotype referred to as “BRCAness”.23,24 Finally, the use of mild hyperthermia to disrupt BRCA2 and inhibit HRR was shown to potentiate the cytotoxicity of PARP1 inhibitors.25
Although a hypothesis of this study is that the mechanism of interaction between Chk1 and PARP1 inhibition producing radiosensitization is via Chk1 inhibitor-mediated HRR inhibition, it is also possible that checkpoint abrogation plays a role. We and others have shown that PARP1 inhibition results in a greater accumulation of cells in the G2 phase of the cell cycle in response to radiation, likely due to persistent DNA damage.31 In this scenario, abrogation of the G2 checkpoint by Chk1 inhibition would be predicted to result in a greater degree of radiosensitization. Studies to decipher the contributions of HRR inhibition vs. checkpoint abrogation in the sensitizing mechanisms of Chk1 inhibitors are underway (Parsels, unpublished data) and will be important in determining the mechanisms of interaction between Chk1 and PARP1 as well as in identifying key pathways which could be exploited with novel therapeutic agents.
In the present study, while the combination of Chk1 and PARP1 inhibitors produced radiosensitization that was significantly greater than either agent alone and additive, we did not detect a significant synergistic effect between Chk1 and PARP1 inhibitors on radiosensitization. In our past and present studies, we have analyzed the interactions between two drugs and radiation by testing the null hypothesis that an additive effect between two drugs on radiosensitization holds for all radiation doses.7,32 However, it is conceivable that radiosensitization may be radiation dose-dependent, with optimal radiation doses varying for different agents and the interaction between two drugs presenting at higher doses or lower doses of radiation. In an effort to extend our understanding of the potential interactions between Chk1 and PARP1 inhibitors in the context of individual radiation doses, we investigated an improved model adapted from the Lindstrom method32 (unpublished data). In this model, we found that Chk1 and PARP1 inhibitors, although not synergistic, are complementary in terms of radiosensitization; sensitization by Chk1 inhibition predominates at lower radiation doses (2–4 Gy) and sensitization by PARP1 inhibition at higher radiation doses (6–8 Gy). Further development of this model will permit more informative estimations of interactions between two drugs with radiation (as well as without radiation) and will enhance our understanding of the biological mechanisms of radiosensitization and drug interactions.
Demonstrating tumor cell selectivity is a critical milestone in the preclinical development of novel therapeutic regimens. In the case of Chk1 inhibitors, the role of p53 in tumor cell selectivity has been extensively explored. The prevailing model11,14,15,33–35 suggests that p53 mutant tumor cells, unable to arrest in G1 in response to DNA damage, will rely entirely on the G2 checkpoint. Thus, abrogation of the G2 checkpoint by Chk1 inhibition will have a significantly greater impact on p53 mutant cancer cells than it would in normal cells (with an intact p53-mediated G1 checkpoint). Similarly, activation of wild-type p53 in normal cells has been shown to protect normal cells from DNA damage by initiation of the G1 checkpoint.36–38 In this study, we observed preferential radiosensitization of p53 mutant/defective tumor cells by Chk1 and PARP1 inhibition, which is consistent with the prevailing model regarding the tumor cell selectivity by Chk1 inhibitors. Our data also suggest that p53 mutation does not confer selectivity toward radiosensitization by PARP1 inhibition (Fig. 3) and are consistent with reports of PARP inhibitors radiosensitizing replicating cells.26 A central finding of this study is that both p53 mutant and wild-type tumor cells were radiosensitized by Chk1 and PARP1 inhibition, while normal cells were not. Our data demonstrate that p53 plays a role in this selectivity, but suggest that p53 mutation is not the only mechanism of tumor cell selectivity. Other likely mechanisms of tumor cell selectivity include the presence of mutant k-Ras39 and p16,40 which may drive the cell inappropriately into either S or M phase in the presence of unrepaired DNA damage.
Pharmacodynamic biomarkers of Chk1 and PARP1 inhibitors are being developed in order to monitor drug response and guide clinical trials. PAR, a product of PARP and γH2AX, a surrogate for DNA double-strand breaks are being widely used as pharmacodynamic biomarkers of PARP inhibition in the clinical setting.27,41,42 Although no clinical data have been published to date, the utility of pHistone H3, a marker of mitosis, and γH2AX as biomarkers of Chk1 inhibition have been supported by substantial preclinical data.7,43–45 In addition, we recently identified S345 Chk1 phosphorylation as a biomarker of Chk1 inhibition in vivo in tumor xenografts as well as in hair follicles and rectal biopsies.9 Our present findings demonstrating elevated γH2AX and S345 Chk1 phosphorylation as well as PAR inhibition in association with radiosensitization by Chk1 and PARP1 inhibition encourage the continued development of these pharmacodynamic endpoints as biomarkers of response to combined Chk1 and PARP1 inhibition.
Given the continued clinical development of several Chk1 (LY2606368, LY2603618, SCH900776) and PARP1 inhibitors (olaparib, ABT-888, MK-4827) and the demonstrated tumor cell selective radiosensitization demonstrated in our work, the combination of Chk1- and PARP1-targeted therapies with radiation represents a promising treatment strategy. Although this study focused primarily on pancreatic cancer, our finding that colon and lung cancer cells lines were also radiosensitized to this drug combination suggests that this strategy could be applicable to many types of cancers. Furthermore, although p53 status plays a role in radiosensitization, it will be important to determine the other mutations that mediate selective radiosensitization of tumor cells compared with normal intestinal cells. As both p53 mutant and wild-type tumor cells are radiosensitized by combined Chk1 and PARP1 inhibition, this novel therapeutic regimen would be predicted to have efficacy across a spectrum of cancer genotypes.
Materials and Methods
Cell culture.
MiaPaCa-2 and MPanc-96 pancreatic cancer cells were obtained from American Type Culture Collection (ATCC). HCT116 p53−/− or p53+/+ human colorectal carcinoma cells were a kind gift from Dr. Bert Vogelstein (John Hopkins University).46 H460 p53wt and p53dn human large cell lung carcinoma were obtained from AstraZeneca.15 CCL-241 (alternatively, FHs 74 Int) normal human small intestine epithelial cells were purchased from ATCC. Cells were grown in DMEM (MiaPaCa-2), RPMI (MPanc-96, H460), McCoy's (HCT116) or HybriCare (ATCC) with Hepes Buffer and 30 ng/ml epidermal growth factor (CCL-241) media supplemented with 10% fetal bovine serum (Invitrogen), 2 mmol/L L-glutamine (Sigma) and penicillin/streptomycin (Sigma). Cells were tested for Mycoplasma once every 3 mo and experiments were conducted on exponentially growing cells. Radiosensitization and cytotoxicity were assessed by clonogenic survival assays as previously described in references 8, 47 and 48.
Drug preparation.
AZD7762 was obtained from AstraZeneca and dissolved in DMSO. Olaparib (AZD2281) was obtained from Axon Medchem and dissolved in DMSO. For all experiments, 100 nM AZD7762 and 1 uM olaparib were used.
Flow cytometry.
Cell cycle was evaluated by propidium iodide-based flow cytometry, as previously described in reference 49. For γH2AX analysis, samples were processed as previously described in reference 50, and analyzed on a FACScan flow cytometer (Becton Dickinson) with FlowJo software (Tree Star).
Homologous recombination repair.
MiaPaCa-2 cells were transfected with the pDR-GFP plasmid29 using SuperFect transfection reagent (Qiagen) according to the manufacturer's protocol. Clones containing the DR-GFP reporter integrated chromosomally were isolated following puromycin selection. To measure repair of a DNA double-strand break, cells were infected with the adenovirus AdNGUS24i, expressing the I-SceI enzyme. I-SceI-induced homologous recombination was measured as the percentage of green fluorescent protein (GFP)-positive cells 48 h later by flow cytometry.29
Immunoblotting.
Cell pellets were lysed and immunoblotted as previously described in reference 8. Proteins were detected with Chk1 (S345), Chk1 (S296), Chk2 (T68), GAPDH, PARP1 (Cell Signaling), Chk2, PAR (Millipore), Chk1, Cdc25A, Rad51 (Santa Cruz) or β-actin (Calbiochem) antibodies.
Irradiation.
Irradiations were performed using a Philips RT250 (Kimtron Medical) at a dose rate of ∼2 Gy/min in the University of Michigan Comprehensive Cancer Center Experimental Irradiation Core. Dosimetry was performed using an ionization chamber connected to an electrometer system that is directly traceable to a National Institute of Standards and Technology calibration.
Statistical analysis.
For radiation enhancement, drug cytotoxicity, γH2AX, cell cycle and HRR assays, statistically significant differences were determined by one-way ANOVA with the Tukey post-comparison test in GraphPad PRISM version 5 (GraphPad software). Additivity was defined by the change in the radiation survival curve (across radiation doses) due to AZD7762-olaparib being the product of the change due to AZD7762 and olaparib alone, referred to as the multiplicative effect.32 Differences between p53 mutant of p53 wild type cells were determined by two-way ANOVA with a Bonferroni post-comparison test in GraphPad PRISM version 5.
Acknowledgments
This work was funded by NIH Grants R01CA78554, R01CA78554 10S1, R01CA138723, P50CA130810 and Cancer Center Core Grant P30 CA046592.
Abbreviations
- Chk1
checkpoint kinase 1
- RER
radiation enhancement ratio
- HRR
homologous recombination repair
- PARP1
poly (ADP-ribose) polymerase-1
- pS345 Chk1
phosphorylated S345 Chk1
- RT
radiation
Disclosure of Potential Conflicts of Interest
J.L.B. is an employee of AstraZeneca.
Supplementary Material
References
- 1.American Cancer Society, author. Cancer Facts & Figures 2010. Atlanta: American Cancer Society; 2010. [Google Scholar]
- 2.Iacobuzio-Donahue CA, Fu B, Yachida S, Luo M, Abe H, Henderson CM, et al. DPC4 gene status of the primary carcinoma correlates with patterns of failure in patients with pancreatic cancer. J Clin Oncol. 2009;27:1806–1813. doi: 10.1200/JCO.2008.17.7188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.McGinn CJ, Zalupski MM, Shureiqi I, Robertson JM, Eckhauser FE, Smith DC, et al. Phase I trial of radiation dose escalation with concurrent weekly full-dose gemcitabine in patients with advanced pancreatic cancer. J Clin Oncol. 2001;19:4202–4208. doi: 10.1200/JCO.2001.19.22.4202. [DOI] [PubMed] [Google Scholar]
- 4.Loehrer PJ, Powell ME, Cardenes HR, Wagner L, Brell JM, Ramanathan RK, et al. A randomized phase III study of gemcitabine in combination with radiation therapy versus gemcitabine alone in patients with localized, unresectable pancreatic cancer: E4201. Journal of Clinical Oncology; ASCO Meeting Abstracts; May 20; 2008. 4506. [Google Scholar]
- 5.Ben-Josef E, Griffith K, Francis G, Khan G, Lawrence T, Abrams R, et al. Phase I radiation dose-escalation trial of intensity-modulated radiotherapy (IMRT) with concurrent fixed dose-rate gemcitabine (FDR-G) for unresectable pancreatic cancer. Journal of Clinical Oncology. 2009;27:15. doi: 10.1016/j.ijrobp.2012.02.051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Morgan MA, Meirovitz A, Davis MA, Kollar LE, Hassan MC, Lawrence TS. Radiotherapy combined with gemcitabine and oxaliplatin in pancreatic cancer cells. Transl Oncol. 2008;1:36–43. doi: 10.1593/tlo.07106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Morgan MA, Parsels LA, Zhao LL, Parsels JD, Davis MA, Hassan MC, et al. Mechanism of Radiosensitization by the Chk1/2 Inhibitor AZD7762 Involves Abrogation of the G(2) Checkpoint and Inhibition of Homologous Recombinational DNA Repair. Cancer Res. 2010;70:4972–4981. doi: 10.1158/0008-5472.CAN-09-3573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Morgan MA, Parsels LA, Kollar LE, Normolle DP, Maybaum J, Lawrence TS. The combination of epidermal growth factor receptor inhibitors with gemcitabine and radiation in pancreatic cancer. Clin Cancer Res. 2008;14:5142–5149. doi: 10.1158/1078-0432.CCR-07-4072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Parsels LA, Qian Y, Tanska DM, Gross M, Zhao L, Hassan MC, et al. Assessment of chk1 phosphorylation as a pharmacodynamic biomarker of chk1 inhibition. Clin Cancer Res. 2011;17:3706–3715. doi: 10.1158/1078-0432.CCR-10-3082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Massey AJ, Borgognoni J, Bentley C, Foloppe N, Fiumana A, Walmsley L. Context-dependent cell cycle checkpoint abrogation by a novel kinase inhibitor. PLoS ONE. 2010;5:13123. doi: 10.1371/journal.pone.0013123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ma CX, Janetka JW, Piwnica-Worms H. Death by releasing the breaks: CHK1 inhibitors as cancer therapeutics. Trends Mol Med. 2011;17:88–96. doi: 10.1016/j.molmed.2010.10.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Levesque AA, Fanous AA, Poh A, Eastman A. Defective p53 signaling in p53 wild-type tumors attenuates p21waf1 induction and cyclin B repression rendering them sensitive to Chk1 inhibitors that abrogate DNA damage-induced S and G2 arrest. Mol Cancer Ther. 2008;7:252–262. doi: 10.1158/1535-7163.MCT-07-2066. [DOI] [PubMed] [Google Scholar]
- 13.Walton MI, Eve PD, Hayes A, Valenti M, De Haven Brandon A, Box G, et al. The preclinical pharmacology and therapeutic activity of the novel CHK1 inhibitor SAR-020106. Mol Cancer Ther. 2010;9:89–100. doi: 10.1158/1535-7163.MCT-09-0938. [DOI] [PubMed] [Google Scholar]
- 14.Mitchell JB, Choudhuri R, Fabre K, Sowers AL, Citrin D, Zabludoff SD, et al. In vitro and in vivo radiation sensitization of human tumor cells by a novel checkpoint kinase inhibitor, AZD7762. Clin Cancer Res. 2010;16:2076–2084. doi: 10.1158/1078-0432.CCR-09-3277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zabludoff SD, Deng C, Grondine MR, Sheehy AM, Ashwell S, Caleb BL, et al. AZD7762, a novel checkpoint kinase inhibitor, drives checkpoint abrogation and potentiates DNA-targeted therapies. Mol Cancer Ther. 2008;7:2955–2966. doi: 10.1158/1535-7163.MCT-08-0492. [DOI] [PubMed] [Google Scholar]
- 16.Powell SN, DeFrank JS, Connell P, Eogan M, Preffer F, Dombkowski D, et al. Differential sensitivity of p53(−) and p53(+) cells to caffeine-induced radiosensitization and override of G2 delay. Cancer Res. 1995;55:1643–1648. [PubMed] [Google Scholar]
- 17.Li J, Wang Y, Sun Y, Lawrence TS. Wild-type Tp53 inhibits G(2)-phase checkpoint abrogation and radiosensitization induced by PD0166285, a WEE1 kinase inhibitor. Radiat Res. 2002;157:322–330. doi: 10.1667/0033-7587(2002)157[0322:WTTIGP]2.0.CO;2. [DOI] [PubMed] [Google Scholar]
- 18.Calabrese CR, Almassy R, Barton S, Batey MA, Calvert AH, Canan-Koch S, et al. Anticancer chemosensitization and radiosensitization by the novel poly(ADP-ribose) polymerase-1 inhibitor AG14361. J Natl Cancer Inst. 2004;96:56–67. doi: 10.1093/jnci/djh005. [DOI] [PubMed] [Google Scholar]
- 19.Russo AL, Kwon HC, Burgan WE, Carter D, Beam K, Weizheng X, et al. In vitro and in vivo radiosensitization of glioblastoma cells by the poly (ADP-ribose) polymerase inhibitor E7016. Clin Cancer Res. 2009;15:607–612. doi: 10.1158/1078-0432.CCR-08-2079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Dungey FA, Loser DA, Chalmers AJ. Replication-dependent radiosensitization of human glioma cells by inhibition of poly(ADP-Ribose) polymerase: mechanisms and therapeutic potential. Int J Radiat Oncol Biol Phys. 2008;72:1188–1197. doi: 10.1016/j.ijrobp.2008.07.031. [DOI] [PubMed] [Google Scholar]
- 21.Chalmers AJ, Lakshman M, Chan N, Bristow RG. Poly(ADP-ribose) polymerase inhibition as a model for synthetic lethality in developing radiation oncology targets. Semin Radiat Oncol. 2010;20:274–281. doi: 10.1016/j.semradonc.2010.06.001. [DOI] [PubMed] [Google Scholar]
- 22.Rozenblum E, Schutte M, Goggins M, Hahn SA, Panzer S, Zahurak M, et al. Tumor-suppressive pathways in pancreatic carcinoma. Cancer Res. 1997;57:1731–1734. [PubMed] [Google Scholar]
- 23.McCabe N, Turner NC, Lord CJ, Kluzek K, Bialkowska A, Swift S, et al. Deficiency in the repair of DNA damage by homologous recombination and sensitivity to poly(ADP-ribose) polymerase inhibition. Cancer Res. 2006;66:8109–8115. doi: 10.1158/0008-5472.CAN-06-0140. [DOI] [PubMed] [Google Scholar]
- 24.Turner N, Tutt A, Ashworth A. Hallmarks of ‘BRCAness’ in sporadic cancers. Nat Rev Cancer. 2004;4:814–819. doi: 10.1038/nrc1457. [DOI] [PubMed] [Google Scholar]
- 25.Krawczyk PM, Eppink B, Essers J, Stap J, Rodermond H, Odijk H, et al. Mild hyperthermia inhibits homologous recombination, induces BRCA2 degradation and sensitizes cancer cells to poly (ADP-ribose) polymerase-1 inhibition. Proceedings of the National Academy of Sciences of the United States of America. 2011 doi: 10.1073/pnas.1101053108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Dungey FA, Caldecott KW, Chalmers AJ. Enhanced radiosensitization of human glioma cells by combining inhibition of poly(ADP-ribose) polymerase with inhibition of heat shock protein 90. Mol Cancer Ther. 2009;8:2243–2254. doi: 10.1158/1535-7163.MCT-09-0201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Fong PC, Boss DS, Yap TA, Tutt A, Wu P, Mergui-Roelvink M, et al. Inhibition of poly(ADP-ribose) polymerase in tumors from BRCA mutation carriers. N Engl J Med. 2009;361:123–134. doi: 10.1056/NEJMoa0900212. [DOI] [PubMed] [Google Scholar]
- 28.Zhang YW, Otterness DM, Chiang GG, Xie W, Liu YC, Mercurio F, et al. Genotoxic stress targets human Chk1 for degradation by the ubiquitin-proteasome pathway. Mol Cell. 2005;19:607–618. doi: 10.1016/j.molcel.2005.07.019. [DOI] [PubMed] [Google Scholar]
- 29.Pierce AJ, Johnson RD, Thompson LH, Jasin M. XRCC3 promotes homology-directed repair of DNA damage in mammalian cells. Genes Dev. 1999;13:2633–2638. doi: 10.1101/gad.13.20.2633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Anderson VE, Walton MI, Eve PD, Boxall KJ, Antoni L, Caldwell JJ, et al. CCT241533 is a potent and selective inhibitor of CHK2 that potentiates the cytotoxicity of PARP inhibitors. Cancer Res. 2011;71:463–472. doi: 10.1158/00085472.CAN-10-1252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lu HR, Wang X, Wang Y. A stronger DNA damage-induced G2 checkpoint due to over-activated CHK1 in the absence of PARP-1. Cell Cycle. 2006;5:2364–2370. doi: 10.4161/cc.5.20.3355. [DOI] [PubMed] [Google Scholar]
- 32.Lindstrom MJ, Kunugi KA, Kinsella TJ. Global comparison of radiation and chemotherapy dose-response curves with a test for interaction. Radiat Res. 1993;135:269–277. doi: 10.2307/3578305. [DOI] [PubMed] [Google Scholar]
- 33.Carrassa L, Broggini M, Erba E, Damia G. Chk1, but not Chk2, is involved in the cellular response to DNA damaging agents: differential activity in cells expressing or not p53. Cell Cycle. 2004;3:1177–1181. doi: 10.4161/cc.3.9.1080. [DOI] [PubMed] [Google Scholar]
- 34.Ganzinelli M, Carrassa L, Crippa F, Tavecchio M, Broggini M, Damia G. Checkpoint kinase 1 downregulation by an inducible small interfering RNA expression system sensitized in vivo tumors to treatment with 5-fluorouracil. Clin Cancer Res. 2008;14:5131–5141. doi: 10.1158/1078-0432.CCR-08-0304. [DOI] [PubMed] [Google Scholar]
- 35.Tao Y, Leteur C, Yang C, Zhang P, Castedo M, Pierre A, et al. Radiosensitization by Chir-124, a selective CHK1 inhibitor: Effects of p53 and cell cycle checkpoints. Cell Cycle. 2009;8:1196–1205. doi: 10.4161/cc.8.8.8203. [DOI] [PubMed] [Google Scholar]
- 36.van Leeuwen IM, Lain S. Pharmacological manipulation of the cell cycle and metabolism to protect normal tissues against conventional anticancer drugs. Oncotarget. 2011;2:274–276. doi: 10.18632/oncotarget.265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Apontes P, Leontieva OV, Demidenko ZN, Li F, Blagosklonny MV. Exploring long-term protection of normal human fibroblasts and epithelial cells from chemotherapy in cell culture. Oncotarget. 2011;2:222–233. doi: 10.18632/oncotarget.248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Rao B, van Leeuwen IM, Higgins M, Campbel J, Thompson AM, Lane DP, et al. Evaluation of an Actinomycin D/VX-680 aurora kinase inhibitor combination in p53-based cyclotherapy. Oncotarget. 2010;1:639–650. doi: 10.18632/oncotarget.198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Gilad O, Nabet BY, Ragland RL, Schoppy DW, Smith KD, Durham AC, et al. Combining ATR suppression with oncogenic Ras synergistically increases genomic instability, causing synthetic lethality or tumorigenesis in a dosage-dependent manner. Cancer Res. 2010;70:9693–9702. doi: 10.1158/0008-5472.CAN-10-2286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Shapiro GI, Edwards CD, Rollins BJ. The physiology of p16(INK4A)-mediated G1 proliferative arrest. Cell Biochem Biophys. 2000;33:189–197. doi: 10.1385/CBB:33:2:189. [DOI] [PubMed] [Google Scholar]
- 41.Redon CE, Nakamura AJ, Zhang YW, Ji JJ, Bonner WM, Kinders RJ, et al. Histone gammaH2AX and poly(ADP-ribose) as clinical pharmacodynamic biomarkers. Clin Cancer Res. 2010;16:4532–4542. doi: 10.1158/1078-0432.CCR-10-0523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kummar S, Kinders R, Gutierrez ME, Rubinstein L, Parchment RE, Phillips LR, et al. Phase 0 clinical trial of the poly (ADP-ribose) polymerase inhibitor ABT-888 in patients with advanced malignancies. J Clin Oncol. 2009;27:2705–2711. doi: 10.1200/JCO.2008.19.7681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Blasina A, Hallin J, Chen E, Arango ME, Kraynov E, Register J, et al. Breaching the DNA damage checkpoint via PF-00477736, a novel small-molecule inhibitor of checkpoint kinase 1. Mol Cancer Ther. 2008;7:2394–2404. doi: 10.1158/1535-7163.MCT-07-2391. [DOI] [PubMed] [Google Scholar]
- 44.McNeely S, Conti C, Sheikh T, Patel H, Zabludoff S, Pommier Y, et al. Chk1 inhibition after replicative stress activates a double strand break response mediated by ATM and DNA-dependent protein kinase. Cell Cycle. 2010;9:995–1004. doi: 10.4161/cc.9.5.10935. [DOI] [PubMed] [Google Scholar]
- 45.Morgan MA, Parsels LA, Parsels JD, Lawrence TS, Maybaum J. The relationship of premature mitosis to cytotoxicity in response to checkpoint abrogation and antimetabolite treatment. Cell Cycle. 2006;5:1983–1988. doi: 10.4161/cc.5.17.3184. [DOI] [PubMed] [Google Scholar]
- 46.Bunz F, Dutriaux A, Lengauer C, Waldman T, Zhou S, Brown JP, et al. Requirement for p53 and p21 to sustain G2 arrest after DNA damage. Science. 1998;282:1497–1501. doi: 10.1126/science.282.5393.1497. [DOI] [PubMed] [Google Scholar]
- 47.Lawrence TS. Ouabain sensitizes tumor cells but not normal cells to radiation. Int J Radiat Oncol Biol Phys. 1988;15:953–958. doi: 10.1016/0360-3016(88)90132-0. [DOI] [PubMed] [Google Scholar]
- 48.Fertil B, Dertinger H, Courdi A, Malaise EP. Mean inactivation dose: a useful concept for intercomparison of human cell survival curves. Radiat Res. 1984;99:73–84. doi: 10.2307/3576448. [DOI] [PubMed] [Google Scholar]
- 49.Morgan MA, Parsels LA, Parsels JD, Mesiwala AK, Maybaum J, Lawrence TS. Role of checkpoint kinase 1 in preventing premature mitosis in response to gemcitabine. Cancer Res. 2005;65:6835–6842. doi: 10.1158/0008-5472.CAN-04-2246. [DOI] [PubMed] [Google Scholar]
- 50.Huang X, Halicka HD, Darzynkiewicz Z. Detection of histone H2AX phosphorylation on Ser-139 as an indicator of DNA damage (DNA double-strand breaks) Curr Protoc Cytom. 2004;7:7–27. doi: 10.1002/0471142956.cy0727s30. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.