Abstract
The enzyme Hpa II methylase extracted and partially purified from Haemophilus parainfluenza catalyzes the methylation of the tetranucleotide sequence CCGG at the internal cytosine. The enzyme will methylate this sequence if both DNA strands are unmethylated or if only one strand is unmethylated. Conditions have been developed for producing fully methylated DNA from various sources. In vitro methylation of this site protects the DNA against digestion by the restriction enzyme Hpa II as well as the enzyme Sma I which recognizes the hexanucleotide sequence CCCGGG. These properties make this enzyme a valuable tool for analyzing methylation in eukaryotic DNA where the sequence CCGG is highly methylated. The activity of this methylase on such DNA indicates the degree of undermethylation of the CCGG sequence. Several examples show that this technique can be used to detect small changes in the methylation state of eukaryotic DNA.
Full text
PDF


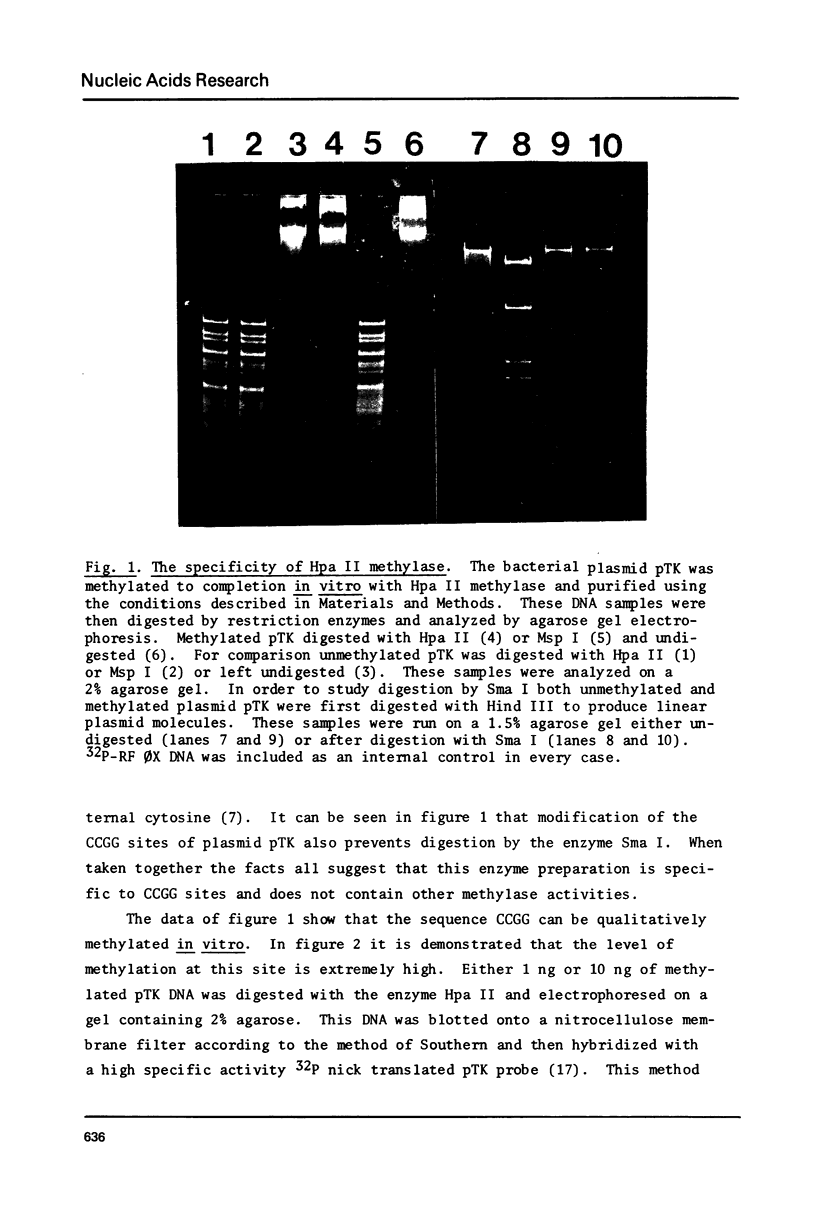

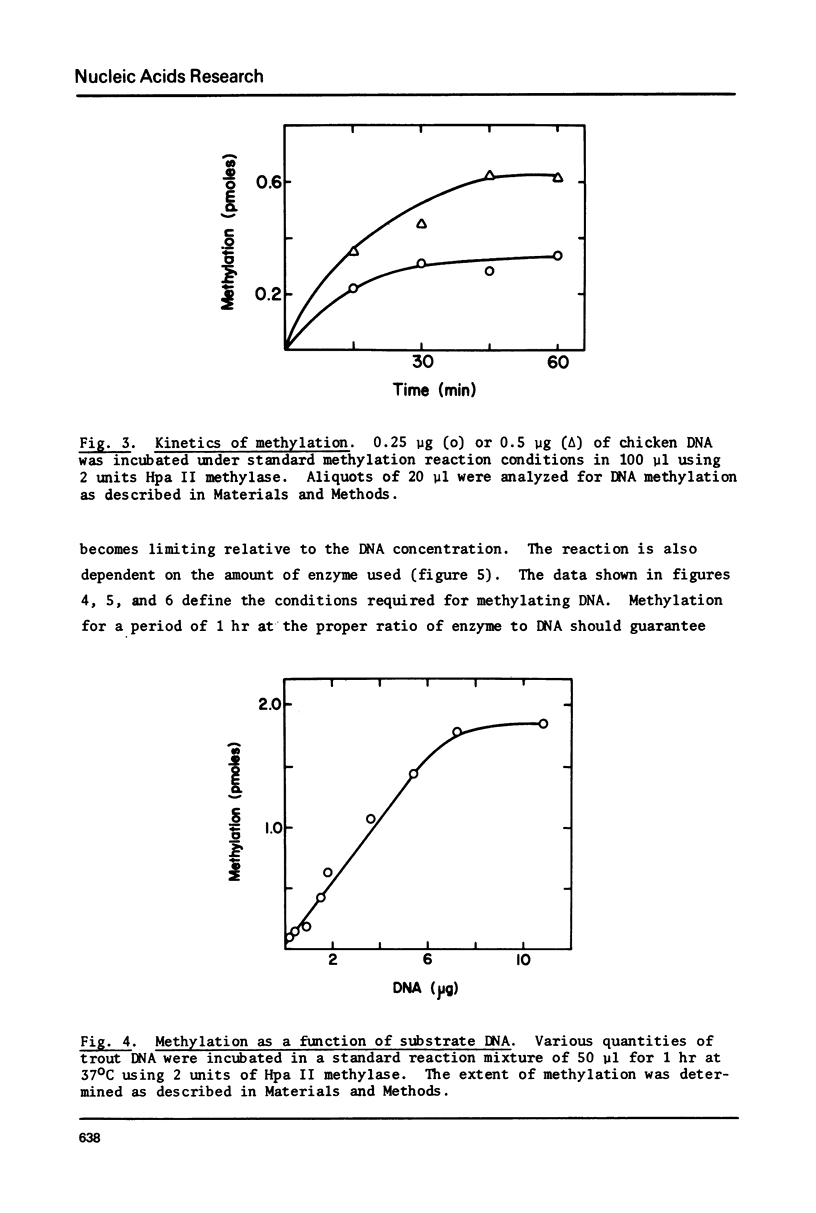





Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Cedar H., Solage A., Glaser G., Razin A. Direct detection of methylated cytosine in DNA by use of the restriction enzyme MspI. Nucleic Acids Res. 1979;6(6):2125–2132. doi: 10.1093/nar/6.6.2125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Desrosiers R. C., Mulder C., Fleckenstein B. Methylation of Herpesvirus saimiri DNA in lymphoid tumor cell lines. Proc Natl Acad Sci U S A. 1979 Aug;76(8):3839–3843. doi: 10.1073/pnas.76.8.3839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grippo P., Iaccarino M., Parisi E., Scarano E. Methylation of DNA in developing sea urchin embryos. J Mol Biol. 1968 Sep 14;36(2):195–208. doi: 10.1016/0022-2836(68)90375-6. [DOI] [PubMed] [Google Scholar]
- Harbers K., Harbers B., Spencer J. H. Nucleotide clusters in deoxyribonucleic acids. XII. The distribution of 5-methylcytosine in pyrimidine oligonucleotides of mouse L-cell satellite DNA and main band DNA. Biochem Biophys Res Commun. 1975 Sep 16;66(2):738–746. doi: 10.1016/0006-291x(75)90572-0. [DOI] [PubMed] [Google Scholar]
- Mandel J. L., Chambon P. DNA methylation: organ specific variations in the methylation pattern within and around ovalbumin and other chicken genes. Nucleic Acids Res. 1979 Dec 20;7(8):2081–2103. doi: 10.1093/nar/7.8.2081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mann M. B., Smith H. O. Specificity of Hpa II and Hae III DNA methylases. Nucleic Acids Res. 1977 Dec;4(12):4211–4221. doi: 10.1093/nar/4.12.4211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGhee J. D., Ginder G. D. Specific DNA methylation sites in the vicinity of the chicken beta-globin genes. Nature. 1979 Aug 2;280(5721):419–420. doi: 10.1038/280419a0. [DOI] [PubMed] [Google Scholar]
- Miller O. J., Schnedl W., Allen J., Erlanger B. F. 5-Methylcytosine localised in mammalian constitutive heterochromatin. Nature. 1974 Oct 18;251(5476):636–637. doi: 10.1038/251636a0. [DOI] [PubMed] [Google Scholar]
- Razin A., Cedar H. Distribution of 5-methylcytosine in chromatin. Proc Natl Acad Sci U S A. 1977 Jul;74(7):2725–2728. doi: 10.1073/pnas.74.7.2725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Solage A., Cedar H. Organization of 5-methylcytosine in chromosomal DNA. Biochemistry. 1978 Jul 11;17(14):2934–2938. doi: 10.1021/bi00607a036. [DOI] [PubMed] [Google Scholar]
- Sutcliffe J. G. pBR322 restriction map derived from the DNA sequence: accurate DNA size markers up to 4361 nucleotide pairs long. Nucleic Acids Res. 1978 Aug;5(8):2721–2728. doi: 10.1093/nar/5.8.2721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sutter D., Doerfler W. Methylation of integrated adenovirus type 12 DNA sequences in transformed cells is inversely correlated with viral gene expression. Proc Natl Acad Sci U S A. 1980 Jan;77(1):253–256. doi: 10.1073/pnas.77.1.253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waalwijk C., Flavell R. A. DNA methylation at a CCGG sequence in the large intron of the rabbit beta-globin gene: tissue-specific variations. Nucleic Acids Res. 1978 Dec;5(12):4631–4634. doi: 10.1093/nar/5.12.4631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weinstock R., Sweet R., Weiss M., Cedar H., Axel R. Intragenic DNA spacers interrupt the ovalbumin gene. Proc Natl Acad Sci U S A. 1978 Mar;75(3):1299–1303. doi: 10.1073/pnas.75.3.1299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van der Ploeg L. H., Flavell R. A. DNA methylation in the human gamma delta beta-globin locus in erythroid and nonerythroid tissues. Cell. 1980 Apr;19(4):947–958. doi: 10.1016/0092-8674(80)90086-0. [DOI] [PubMed] [Google Scholar]