Abstract
Ribosomes unfolded by the removal of Mg2+ at 25 degrees C were studied by Raman spectroscopy and electron microscopy. Raman spectra showed a reduction in the 813 cm-1 phosphodiester signal of 30S and 50S ribosomes compared to intact ribosomes, suggesting that a fraction of the ribose moieties had shifted from the 3' endo (ordered) to the 3' exo (disordered) conformation. The maximum diameters of unfolded 30S and 50S ribosomes, judged by electron microscopy, were 1.8 and 2.5-fold greater, respectively, than those of intact ribosomes. Most unfolded 30S ribosomes had three distinct structural domains and appeared "Y-shaped"; whereas most unfolded 50S ribosomes had four distinct domains and appeared "X-shaped". When ribosomes were partially unfolded (by brief exposure to 0.04 mM Mg2+ or EDTA), several possible intermediates in the unfolding process were observed. Both the shapes of particles and their Raman spectra reached the same final state in 0.04 mM Mg2+, where more than 50% of the rRNA phosphates are discharged by Mg2+, as in 10 mM EDTA, where less than 1% are discharged.
Full text
PDF

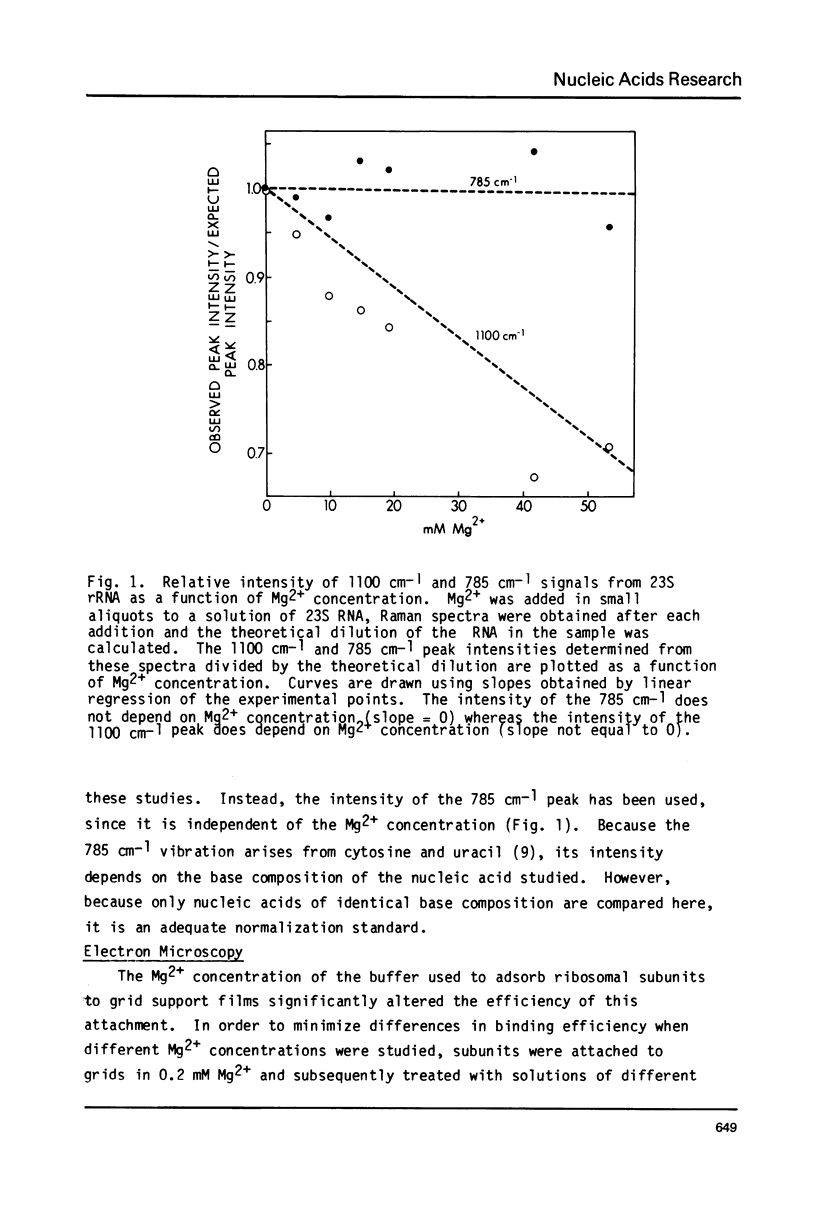

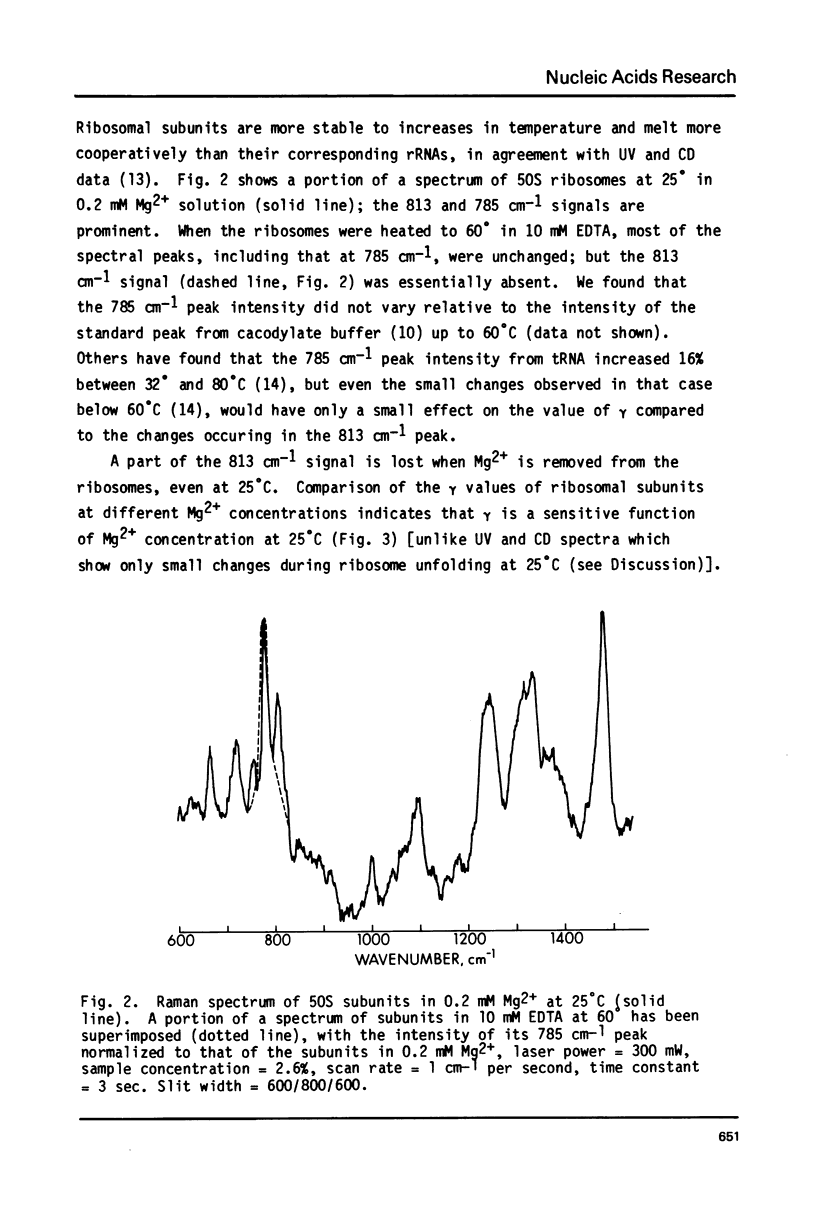
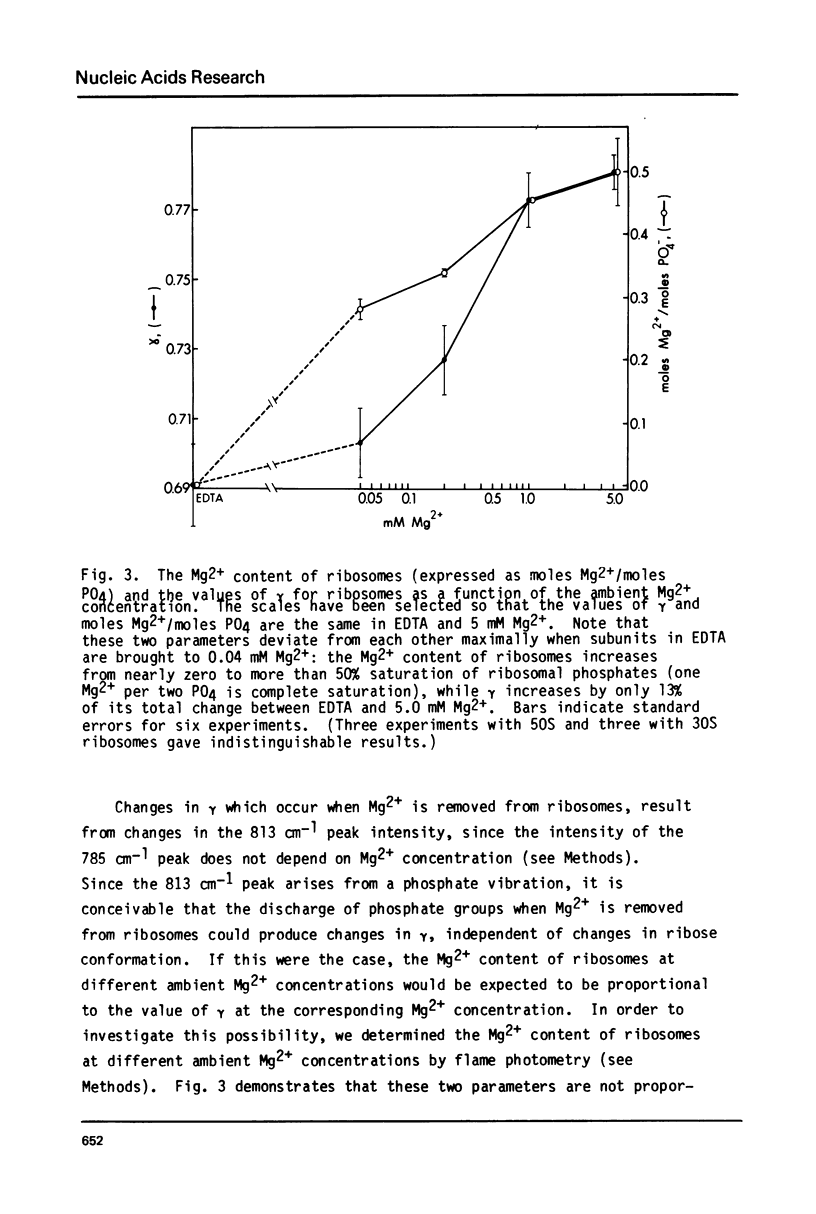

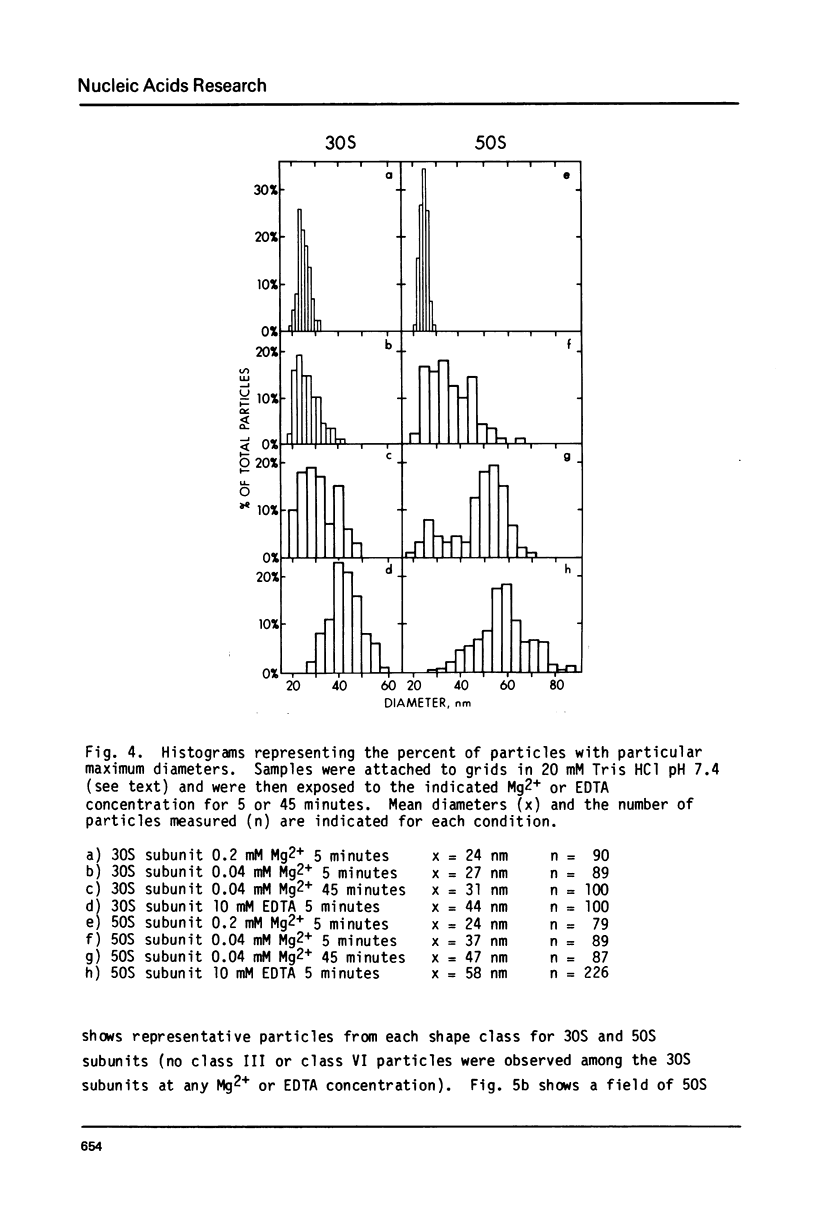
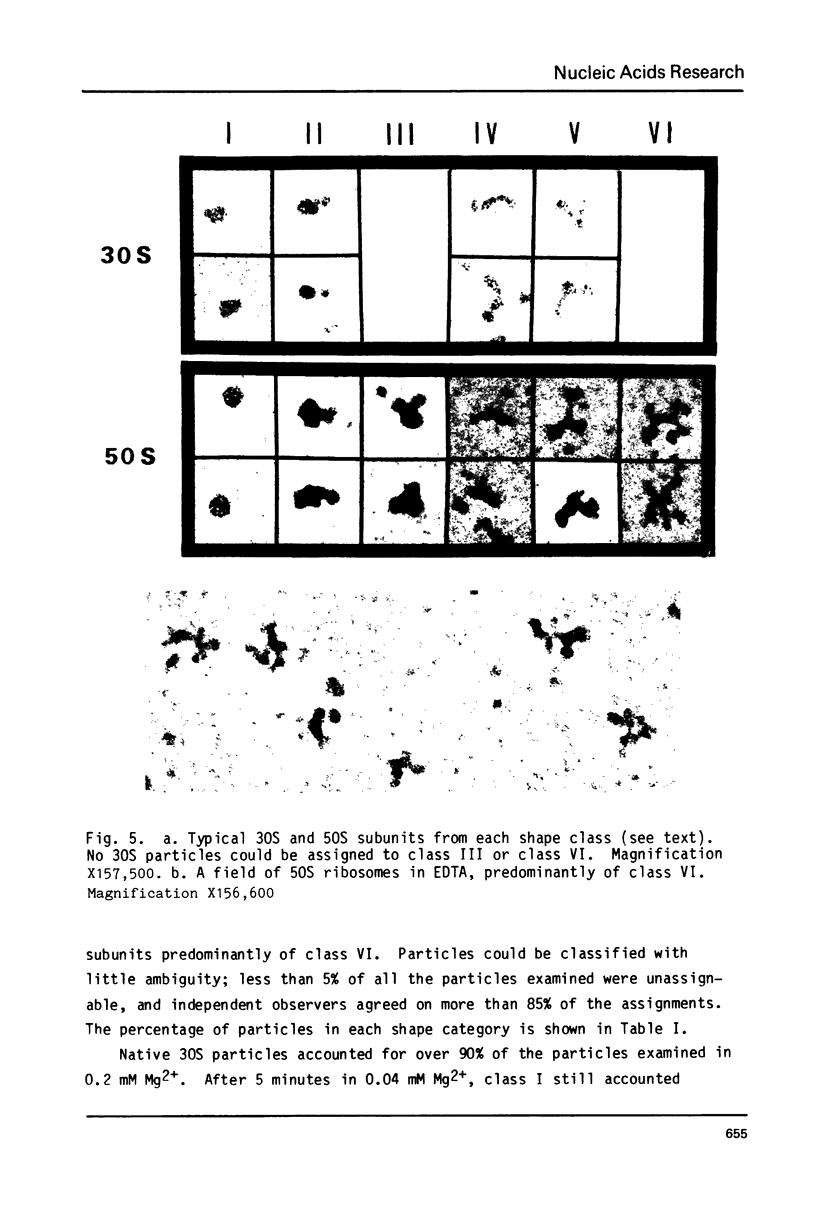
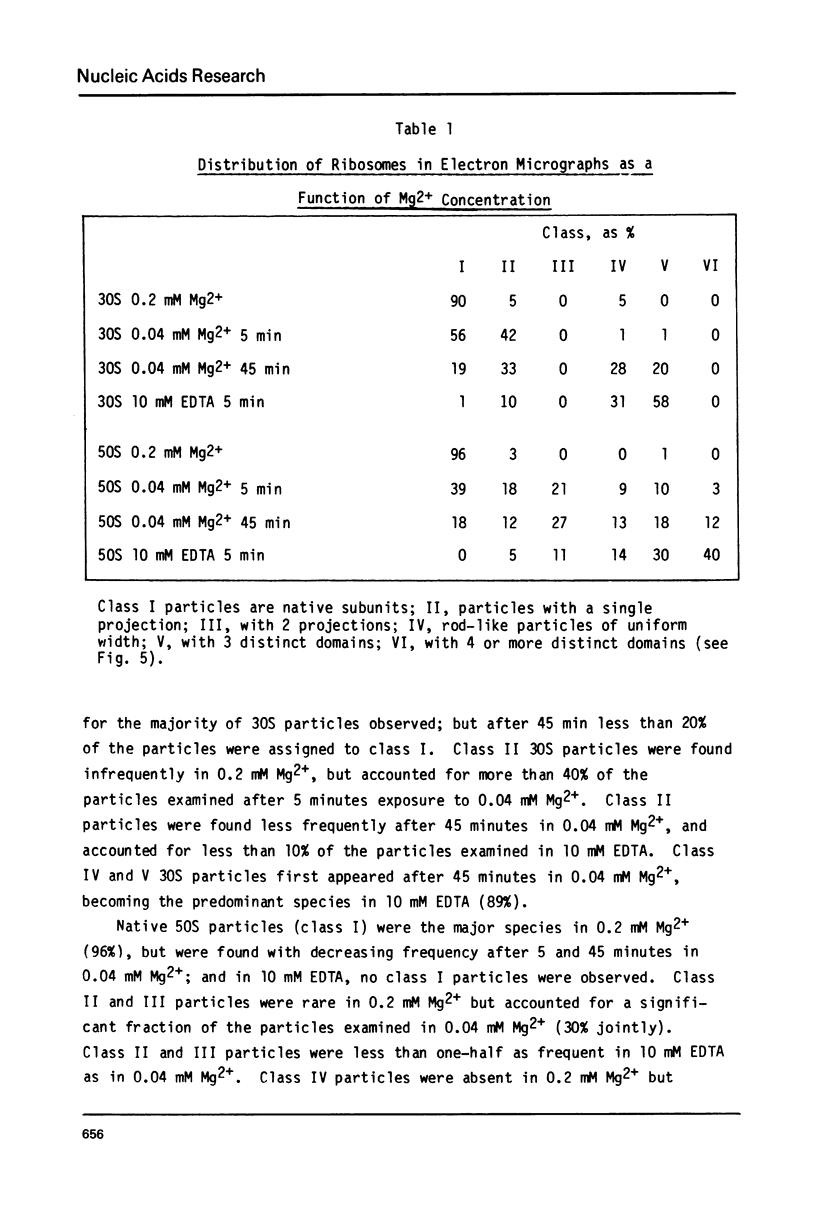





Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Adler A. J., Fasman G. D., Tal M. Circular dichroism of Escherichia coli ribosomes: effect of heating and metal ions. Biochim Biophys Acta. 1970 Aug 8;213(2):424–436. doi: 10.1016/0005-2787(70)90050-x. [DOI] [PubMed] [Google Scholar]
- Belli M., Onori G., Araco A., Giorgi C. Letter: Changes in the ultraviolet absorption of E. coli rRNA on ribosome unfolding. Biopolymers. 1976 Jun;15(6):1229–1232. doi: 10.1002/bip.1976.360150615. [DOI] [PubMed] [Google Scholar]
- Bielka H., Wahn K., Noll F., Lutsch G. Studies on the structure of animal ribosomes. II. Loosening and unlocking of rat liver ribosomes at low Mg ++ concentration: an electron microscopic study. Acta Biol Med Ger. 1972;29(4):607–619. [PubMed] [Google Scholar]
- Brown E. B., Peticolas W. L. Conformational geometry and vibrational frequencies of nucleic acid chains. Biopolymers. 1975 Jun;14(6):1259–1271. doi: 10.1002/bip.1975.360140614. [DOI] [PubMed] [Google Scholar]
- Cammack K. A., Wade H. E. The sedimentation behaviour of ribonuclease-active and -inactive ribosomes from bacteria. Biochem J. 1965 Sep;96(3):671–680. doi: 10.1042/bj0960671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen M. C., Thomas G. J., Jr Raman spectral studies of nucleic acids. XI. Conformations of yeast tRNAPhe and E. coli ribosomal RNA in aqueous solution and in the solid state. Biopolymers. 1974;13(3):615–626. doi: 10.1002/bip.1974.360130313. [DOI] [PubMed] [Google Scholar]
- Erfurth S. C., Kiser E. J., Peticolas W. L. Determination of the backbone structure of nucleic acids and nucleic acid oligomers by laser Raman scattering. Proc Natl Acad Sci U S A. 1972 Apr;69(4):938–941. doi: 10.1073/pnas.69.4.938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gabler R., Westhead E. W., Ford N. C. Studies of ribosomal diffusion coefficients using laser light-scattering spectroscopy. Biophys J. 1974 Jul;14(7):528–545. doi: 10.1016/S0006-3495(74)85933-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gavrilova L. P., Ivanov D. A., Spirin A. S. Studies on the structure of ribosomes. 3. Stepwise unfolding of the 50 s particles without loss of ribosomal protein. J Mol Biol. 1966 Apr;16(2):473–489. doi: 10.1016/s0022-2836(66)80186-9. [DOI] [PubMed] [Google Scholar]
- Haga J. Y., Hamilton M. G., Petermann M. L. Electron microscopic observations on the large subunit of the rat liver ribosome. J Cell Biol. 1970 Oct;47(1):211–221. doi: 10.1083/jcb.47.1.211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hill W. E., Thompson J. D., Anderegg J. W. X-ray scattering study of ribosomes from Escherichia coli. J Mol Biol. 1969 Aug 28;44(1):89–102. doi: 10.1016/0022-2836(69)90406-9. [DOI] [PubMed] [Google Scholar]
- King T. C., Schlessinger D., Milanovich F. Laser Raman studies of RNA backbone ordering in Escherichia coli ribosomes. Biophys J. 1980 Oct;32(1):456–458. doi: 10.1016/S0006-3495(80)84981-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lake J. A. Ribosome structure determined by electron microscopy of Escherichia coli small subunits, large subunits and monomeric ribosomes. J Mol Biol. 1976 Jul 25;105(1):131–139. doi: 10.1016/0022-2836(76)90200-x. [DOI] [PubMed] [Google Scholar]
- Luoma G. A., Marshall A. G. Laser Raman evidence for new cloverleaf secondary structures for eukaryotic 5.8S RNA and prokaryotic 5S RNA. Proc Natl Acad Sci U S A. 1978 Oct;75(10):4901–4905. doi: 10.1073/pnas.75.10.4901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miall S. H., Walker I. O. Structural studies on ribosomes. II. Denaturation and sedimentation of ribosomal subunits unfolded in EDTA. Biochim Biophys Acta. 1969 Feb 18;174(2):551–560. doi: 10.1016/0005-2787(69)90284-6. [DOI] [PubMed] [Google Scholar]
- Nanninga N. Electron microscopy of loosened 50 s ribosomal subunits of Bacillus subtilis. J Mol Biol. 1970 Mar 14;48(2):367–371. doi: 10.1016/0022-2836(70)90170-1. [DOI] [PubMed] [Google Scholar]
- Noll M., Hapke B., Schreier M. H., Noll H. Structural dynamics of bacterial ribosomes. I. Characterization of vacant couples and their relation to complexed ribosomes. J Mol Biol. 1973 Apr 5;75(2):281–294. doi: 10.1016/0022-2836(73)90021-1. [DOI] [PubMed] [Google Scholar]
- SPIRIN A. S., KISELEV N. A., SHAKULOV R. S., BOGDANOV A. A. IZUCHENIE STRUKTURY RIBOSOM; OBRATIMOE RAZVORACHIVANIE RIBOSOMNYKH CHASTITS V RIBONUKLEOPROTEIDNYE TIAZHI I MODEL' UKLADKI. Biokhimiia. 1963 Sep-Oct;28:920–930. [PubMed] [Google Scholar]
- Shen V., King T. C., Kumar V., Daugherty B. Monoclonal antibodies to Escherichia coli 50S ribosomes. Nucleic Acids Res. 1980 Oct 24;8(20):4639–4649. doi: 10.1093/nar/8.20.4639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Small E. W., Peticolas W. L. Conformational dependence of the Raman scattering intensities from polynucleotides. Biopolymers. 1971;10(1):69–88. doi: 10.1002/bip.360100107. [DOI] [PubMed] [Google Scholar]
- Spitnik-Elson P., Elson D. The fragmentation of ribosomes. Methods Enzymol. 1979;59:461–481. doi: 10.1016/0076-6879(79)59108-3. [DOI] [PubMed] [Google Scholar]
- Tal M. Metal ions and ribosomal conformation. Biochim Biophys Acta. 1969 Nov 19;195(1):76–86. doi: 10.1016/0005-2787(69)90604-2. [DOI] [PubMed] [Google Scholar]
- Thomas G. J., Jr, Prescott B., McDonald-Ordzie P. E., Hartman K. A. Studies of virus structure by laser-Raman spectroscopy. II. MS2 phage, MS2 capsids and MS2 RNA in aqueous solutions. J Mol Biol. 1976 Mar 25;102(1):103–124. doi: 10.1016/0022-2836(76)90076-0. [DOI] [PubMed] [Google Scholar]
- Wong K. P., Dunn J. M. Conformational studies of the unfolding of E. coli ribosome. FEBS Lett. 1974 Aug 15;44(1):50–54. doi: 10.1016/0014-5793(74)80303-0. [DOI] [PubMed] [Google Scholar]
- Yang J. T., Samejima T. Optical rotatory dispersion and circular dichroism of nucleic acids. Prog Nucleic Acid Res Mol Biol. 1969;9:223–300. doi: 10.1016/s0079-6603(08)60770-9. [DOI] [PubMed] [Google Scholar]



