Abstract
Disruption of chromatin during replication poses a major challenge to the maintenance and integrity of genome organization. It creates the need to accurately reconstruct the chromatin landscape following DNA duplication but there is little mechanistic understanding of how chromatin based modifications are restored on newly synthesized DNA. ATP-dependent chromatin remodeling activities serve multiple roles during replication and recent work underscores their requirement in the maintenance of proper chromatin organization. A new component of chromatin replication, the SWI/SNF-like chromatin remodeler SMARCAD1, acts at replication sites to facilitate deacetylation of newly assembled histones. Deacetylation is a pre-requisite for the restoration of epigenetic signatures in heterochromatin regions following replication. In this way, SMARCAD1, in concert with histone modifying activities and transcriptional repressors, reinforces epigenetic instructions to ensure that silenced loci are correctly perpetuated in each replication cycle. The emerging concept is that remodeling of nucleosomes is an early event imperative to promote the re-establishment of histone modifications following DNA replication.
Key words: epigenetics, chromatin, ATP-dependent chromatin remodeling, replication, histone modifications, SMARCAD1
Introduction
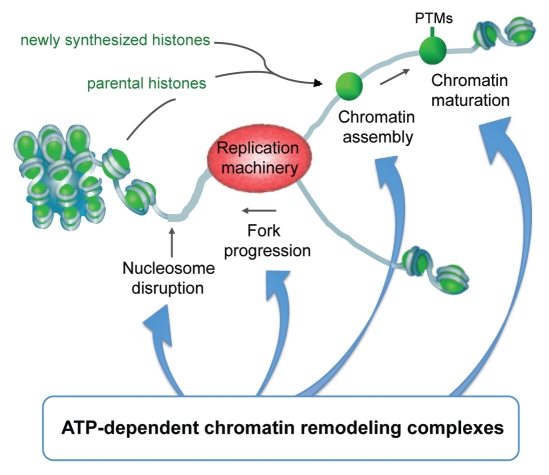
During normal growth and cell division eukaryotic chromosomes and all their structural and functional features must be duplicated to pass an intact genome from one generation to the next. This involves both the accurate replication of DNA sequences and the subsequent faithful reconstruction of chromatin including vital specialized domains such as centromeres and telomeres.1 Indeed, replication must be considered in the context of chromatin where nucleosomes block access to the underlying DNA. It is becoming increasingly apparent that remodeling of nucleosomes accompanies all steps of the replication process (Fig. 1).2,3 To plough through chromatin the replication machinery needs to mobilize and evict nucleosomes and there is evidence that nucleosome remodeling helps to clear the path for efficient progression of the replication fork.4–6 Remodeling may also be involved in the initiation of replication and origin definition, although less is known about this aspect.2,7 Behind the replication fork, nucleosome remodeling contributes to the reformation of higher order chromatin structures.8 This includes adjustment of appropriate nucleosome spacing and post-translational histone modification patterns to match the original patterns of a given domain. Restoring the local chromatin organization is a critical step that ensures genome stability and preserves the proliferation status and identity of a cell. Consequently, chromatin remodeling is an essential function for the faithful maintenance of both genetic and epigenetic information in dividing cells. Fully understanding the underlying remodeling pathways and their specific players is an important goal as these are candidates for regulating DNA/chromatin replication and either preserving or switching an epigenetic state. Here we illustrate emerging principles by considering specific remodeling factors involved in replication. The emphasis of this review is on new work that places nucleosome remodeling as one of the earliest steps in the reassembly of functional chromatin domains after replication and provides mechanistic insights into how this is achieved.9,10
Figure 1.
ATP-dependent chromatin remodeling complexes play important roles during all steps of replication: they facilitate the disassembly of nucleosomes ahead of the replication fork, efficient progression of replication, subsequent proper assembly of chromatin onto newly synthesized DNA, the copying of epigenetic information onto the replicated chromatin (PTM: post-translational modification) as well as the repair of DNA during replication.
ATP-Dependent Remodelers Play Integral Roles during Replication
ATP-dependent chromatin remodeling is typically performed by multi-protein complexes with a conserved catalytic core related to the yeast SWI/SNF ATPase.3 This constitutes a large group of enzymes which exert their distinct functions upon hydrolysis of ATP to drive transitions in chromatin structure. There are at least four different families of SWI/SNF-like factors, defined by sequence variations in both their ATPase and flanking domains.3 The role of ATP-dependent chromatin remodelers is most intensely studied in the context of transcriptional regulation and DNA repair, yet members of all remodeler families are also targeted to sites of ongoing replication in yeast and metazoans where they serve multiple roles.2,3 Among them, INO80 facilitates S-phase progression and also functions in the restart of stalled replication forks.5,11–13 The ACF complex, consisting of the ATPase SNF2h and Acf1, appears to facilitate the progression of replication through highly condensed mammalian chromatin; its depletion causes a delay in S-phase progression that is reversible upon chromatin decondensation.4 This suggests a function for ACF in establishing an accessible chromatin structure ahead of the replication machinery.4 Similarly, the SWI/SNF remodeler Brg1 apparently accelerates replication elongation since Brg1 mutants show reduced incorporation of nucleotide analogs into nascent DNA.6
The requirement for specific remodelers during replication is likely to depend on the nature of the chromatin structure to be duplicated and perhaps on the cell type. Condensed heterochromatin has been proposed to pose a greater challenge for replication both in terms of accessibility, and in terms of restoring the transcriptionally unfavorable environment after DNA duplication is completed.8,14 Indeed, several remodeling activities are associated predominantly with replicating constitutive heterochromatin, although their targeting to euchromatic sites could not always be ruled out.4,15,16 Mi-2/NuRD operates during replication of pericentric heterochromatin, specifically in rapidly proliferating lymphoid cells.16 This could indicate the need for specialized remodeling activities in cell types with an accelerated S phase to deal with the demands of speed and fidelity.
Remodeling complexes that serve roles related to chromatin assembly and maturation include Mi2/NURD, WICH and SMARCAD1.9,16–19 Depletion of the WICH complex, comprising SNF2h and the Williams Syndrome Transcription Factor WSTF, is accompanied by an increase in heterochromatin marks following S phase.17 WICH seems to be responsible for keeping chromatin accessible after its assembly. It was proposed that WICH could contribute to epigenetic inheritance by allowing the binding of factors involved in replication of chromatin states in the wake of the replication fork.18 Defining the function of SMARCAD1 has reinforced the idea that ATP-dependent chromatin remodeling factors play a significant part in the transmission of epigenetic information as discussed further below.
Maintaining Silence through Remodeling: A New Player
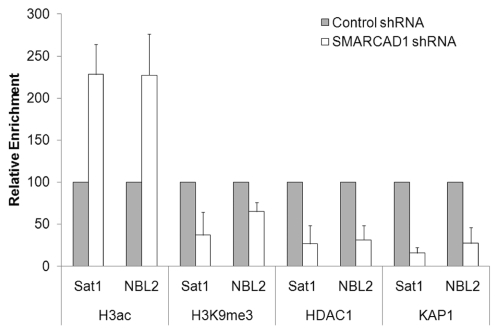
SMARCAD1 is a mammalian SWI/SNF-like protein and we have recently reported that it is required for the restoration of heterochromatin organization after replication.9,10 Heterochromatin promotes genome stability by repressing transcription and illegitimate recombination between repetitive DNA elements.20,21 In particular, heterochromatic repeats flanking centromeres are important for centromere function, mediating proper chromosome segregation. Maintaining silence through replication is thus of fundamental importance, as failure to do so can trigger aberrant gene expression programs and genome instability with implications for development and disease.22,23 Previous studies on heterochromatin replication have emphasized the role of histone chaperones and their complex interplay with histone modifiers and chromatin binding proteins.24–29 Depletion of SMARCAD1 in human cancer cell lines leads to the global accumulation of acetylated histones H3 and H4, a characteristic of an open chromatin structure.9 Concomitantly, heterochromatin features are lost. These changes depend on an intact ATPase domain, consistent with a function of this remodeling activity in silencing. Loci that display elevated levels of histone acetylation include pericentric heterochromatin repeats which are normally hypoacetylated (Sat1 and NBL2, Fig. 2).9 The significance of this is emphasized by chromosome segregation defects triggered upon SMARCAD1 depletion. Methylation of lysine 9 on H3 (H3K9me), a hallmark of heterochromatin, is markedly reduced upon SMARCAD1 knock down (Fig. 2). Accordingly, factors required for transcriptional silencing such as heterochromatin protein 1 (HP1), histone deacetylase 1 (HDAC1) and the co-repressor KAP1 (TIF1β) are delocalized from chromatin (Fig. 2).9 SMARCAD1 function is not restricted to pericentric heterochromatin but impacts on other transcriptionally silent regions such as telomeres, although the specific underlying pathways may be distinct. Of note, budding and fission yeast SMARCAD1 homologs have also been implicated in gene silencing and maintenance of heterochromatin structures.31–33 The fact that interfering with SMARCAD1 function perturbs heterochromatin organization so profoundly is in line with the recognized prerequisite for histone deacetylation in establishing silent chromatin structures and maintaining chromosome stability from yeast to man.22,34–36
Figure 2.
SMARCAD1 knockdown (KD) affects histone modifications and protein occupancy at pericentric repeats. Chromatin immunoprecipitation of H3ac, H3K9me3, HDAC1 and KAP1 at satellite repeats from SMARCAD1 KD and control HeLa cells. %IP from KD cells is shown relative to %IP from control cells which is normalized to 100. Error bars denote standard deviation from 3 independent experiments. Primers: Sat1 (this study) forward 5′-TTG AAG GTA TAT TCA TAC TGG CC-3′ reverse 5′-TTC AAA GGT ACT CTG CTT GGT ACA-3′ NBL230 forward 5′-TCC CAC AGC AGT TGG TGT TA-3′ reverse 5′-TTG GCA GAA ACC TCT TTG CT-3′.
Interestingly, the global changes in histone modification patterns observed in SMARCAD1 knockdown cells coincide with S-phase progression.9 SMARCAD1 is tightly associated with chromatin in S phase and associates with sites of replication. Collectively, these characteristics link SMARCAD1's role in preserving heterochromatin organization to replication. It is noteworthy that depletion of SMARCAD1 has no severe impact on S-phase progression, implying that it is not essential for DNA replication per se.9 Given that SMARCAD1 has been shown to frequently bind in the vicinity of transcriptional start sites, it could act by controlling the expression of factors involved in chromatin replication.37 However, since it co-localizes to newly synthesized DNA we favor the idea that SMARCAD1's main role is in the assembly or maturation of heterochromatin at replication sites.
Rebuilding Chromatin on Replicated DNA
On newly synthesized DNA, nucleosomes are formed using both recycled and newly synthesized histones (Fig. 1).38,39 While parental histones carry posttranslational modifications that characterize the local domain, new histones are mainly unmethylated but are acetylated at specific lysine residues on H4 and H3.40–45 Once assembled into nucleosomes, they become rapidly deacetylated by a mechanism that is poorly understood.39,43,46 Typical acetylation marks of new histones, for instance acetylation on H4, lysine 12 or on H3 at lysines 14 and/or 18, persist upon SMARCAD1 depletion.9,40,42,43,45 SMARCAD1 is therefore a prime candidate for mediating deacetylation of newly deposited histones. The association of SMARCAD1 with both late replicating heterochromatin and early replicating euchromatin supports a general role in chromatin replication.9 We propose therefore that SMARCAD1 mediates deacetylation of newly assembled histones at all replication sites. Intriguingly, even histone acetylation marks that previously have not been unambiguously connected with replication-coupled chromatin assembly accumulate in SMARCAD1 knock down cells. The reason for this is unclear, but it is conceivable that certain acetylated lysines may not have been detected in earlier studies because they become apparent only upon interfering with SMARCAD1 function. In principle, some hyperacetylation could be a consequence of increased transcription caused by impaired silencing. S-phase specific histone hyperacetylation could also reflect a requirement for an accessible chromatin structure just after replication, for instance to facilitate binding of factors involved in chromatin maturation or DNA repair. Indeed, quantitative mass spectrometry analysis on newly synthesized histones suggests that additional acetylation events take place following their incorporation into nucleosomes.43
Restoring Silence after Replication
What is the role of deacetylation during chromatin replication? Deacetylation is an acknowledged requirement for chromatin maturation and formation of higher order structures.39 It is thought to promote the stable deposition of histone H1 and is a critical step to allow the re-establishment of heterochromatin marks such as H3K9 di- and tri-methylation, which are absent from newly synthesized histones.39 In fact, most histone methylation occurs slowly and stepwise after nucleosome assembly is completed.42,43,47 Acetylation on H3 at lysines 9 and 14, which is increased upon SMARCAD1 deletion, blocks the activity of H3K9 specific histone methyltransferases. Removal of acetylation must occur before methylation can take place, not least because of the antagonism between these modifications at the same lysine residue. SMARCAD1 directed deacetylation might therefore prime new nucleosomes for H3K9 methylation, contributing to propagation of H3K9 methylation and maintenance of heterochromatin.
Di-and tri-methylation of H3K9 are known to stabilize HP1 binding.48,49 In turn, HP1 favors heterochromatin formation by bridging neighboring nucleosomes and recruiting other repressive components such as H3K9 histone methyltransferases and KAP1.28,50–53 An attractive idea is that remodeling by SMARCAD1 initiates replication coupled histone deacetylation and restoration of silencing by facilitating subsequent repressive modifications to lysines. Such a model can account for the observation that SMARCAD1 deficiency leads to increased histone acetylation and a decrease in H3K9 methylation and HP1 binding.
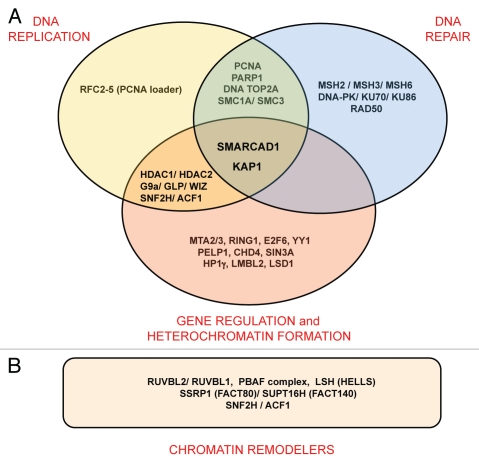
This model fits well with biochemical observations that place SMARCAD1 into a network of proteins that coordinate chromatin duplication by coupling chromatin modifying activities to replication.9 SMARCAD1 predominantly associates with factors linked to transcriptional repression, replication and repair (Fig. 3A). Among them are enzymes that catalyze modifications that are altered in SMARCAD1 depleted cells, namely histone deacetylases HDAC1 and HDAC2 and histone H3K9 methyltransferase G9a/GLP (EHMT1–2). Precisely how G9a aids heterochromatin formation has yet to be established as it has both enzymatic and non-enzymatic silencing functions.54 Association of SMARCAD1 with histone modifying enzymes, proteins that bind modified histones (i.e., HP1γ), transcriptional repressors and factors involved in replication (i.e., PCNA) suggests a potential mechanism by which nucleosome remodeling and re-establishment of appropriate histone modification patterns could be coordinated in vivo during replication (Fig. 4).
Figure 3.
Summary of SMARCAD1 interacting proteins (listed in full in ref. 9). (A) Several SMARCAD1 interaction partners have overlapping functions in gene silencing and heterochromatin formation, replication and repair. (B) A number of chromatin remodelers co-purify with SMARCAD1.
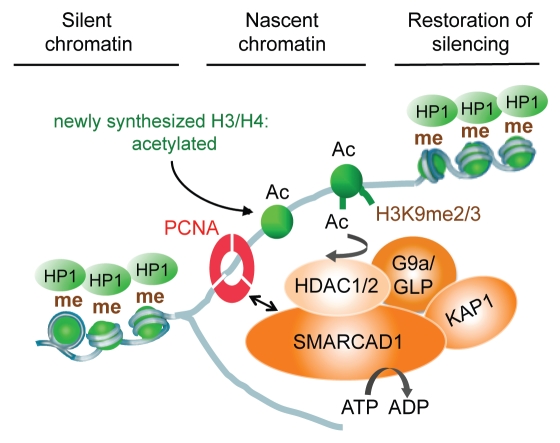
Figure 4.
Model of SMARCAD1 function in chromatin replication: SMARCAD1 is recruited to replication sites by PCNA where it functions in a complex with KAP1, HDAC1, HDAC2 and the histone methyltransferase G9a/GLP. Deacetylation of newly assembled histones is facilitated by SMARCAD1 nucleosome remodeling and primes new nucleosomes for further modifications, promoting the inheritance of H3K9 methylation and the formation of heterochromatin.
While biochemical analysis has identified factors which SMARCAD1 can interact with, open questions concern what subset of proteins function together with SMARCAD1 in a particular context. KAP1 emerged as a stoichiometric component of SMARCAD1 complexes.9,37 This protein is an important regulator of chromatin organization during differentiation and development.55 It has been previously linked to heterochromatin replication and is thought to function as a scaffold that integrates multiple activities required for transcriptional repression.28,55 Moreover, ATM-mediated phosphorylation of KAP1 promotes chromatin relaxation required for repair of heterochromatin.56,57 Interactions of SMARCAD1 with DNA repair proteins (see Fig. 3) could reflect the intimate link between replication and repair, especially at stalled replication forks, and are consistent with the outcome of a screen for DNA damage response proteins.58 Notably, SMARCAD1 co-purifies with several other remodeling factors (Fig. 3B). These remodelers could act consecutively and/or cooperatively to break histone DNA contacts and introduce changes in the position and conformation of nucleosomes as required during the stepwise maturation of newly assembled chromatin. The developing picture is one of replication involving the concerted action of a large number of ATP-dependent remodelers. This raises the important question of how remodelers are targeted specifically to sites of ongoing replication.
Directing Remodeling to Sites of Replication
It is appealing to evoke a targeting strategy that directly couples remodeling enzymes to the replication machinery. This would ensure rapid restoration of chromatin domains and allow the remodeler access to newly replicated chromatin regardless of the sequence context. PCNA is an essential player at the replication fork that ensures processivity of DNA polymerases and orchestrates processes related to replication.59 Many factors implicated in DNA and chromatin replication bind directly to PCNA or through proteins that bind PCNA such as the chromatin assembly factor CAF1.28,59,60 In earlier studies WSTF, a subunit of the WICH remodeling complex, had been shown to bind to replication sites by interacting with PCNA.17 We found that SMARCAD1 also physically interacts with PCNA both in vitro and in vivo, supporting a model in which SMARCAD1 acts at replication sites to restore heterochromatin organization through a recruitment mechanism involving PCNA.9
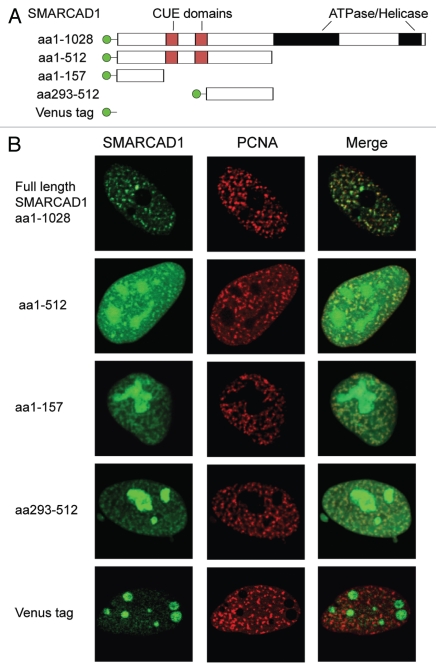
If this model holds, disrupting the SMARCAD1-PCNA interaction should prevent restoration of repressive chromatin. Most PCNA interacting factors, including WSTF, bind via a conserved motif referred to as the PCNA interacting protein (PIP) box.17,61 Several putative PIP boxes were identified in SMARCAD1, yet their mutagenesis had no apparent effect on the observed co-localization of SMARCAD1 with PCNA (data not shown). SMARCAD1 may thus belong to a group of proteins that bind PCNA via different, non-conserved sequences.62,63 In an effort to systematically map the interaction site(s) we discovered that there are at least two different regions within SMARCAD1 that can independently colocalize with PCNA (Fig. 5). Multiple interactions with PCNA may help to stabilize SMARCAD1's association with replication sites, similar to what has been observed for CAF1 and RFCp140.64–66
Figure 5.
Multiple independent regions of SMARCAD1 co-localize with PCNA. (A) Cartoon representation of tagged SMARCAD1 protein and truncations, amino acids (aa) are indicated. (B) Confocal microscopy of SMARCAD1 knockdown HeLa cells expressing Venus-SMARCAD1 proteins and CFP-PCNA. Panels on the right shows a merge of the PCNA and SMARCAD1 channels. Images depict the autofluorescence of the transfected proteins in fixed cells. Representative cells are shown, images were pseudo-colored and adjusted for brightness and contrast.
Besides PCNA there may be additional mechanisms acting in parallel to target SMARCAD1 to ongoing replication. A precedent comes from DNMT1, whose interaction with PCNA is not essential for recruitment to replication foci while interaction with another protein, UHRF1/NP95, is critical.67,68 Factors or events that increase the affinity for SMARCAD1 within replication sites are likely crucial determinants of its localization. It is noteworthy that a proteomic analysis of SMARCAD1 complexes identified several proteins that are also targeted to replication sites including mismatch repair proteins (MSH2/3/6), HDAC1, HDAC2 and G9a.69–72
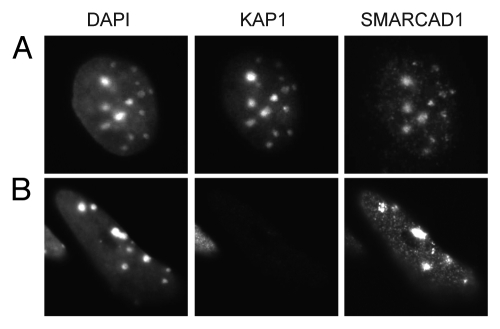
It is important to keep in mind that PCNA and other key replication factors are present at all replication foci. This brings into question whether SMARCAD1 has specificity for particular chromatin domains and if so, how could this be achieved? The major interaction partner of SMARCAD1, KAP1, is a prime candidate that could promote targeting of SMARCAD1 to heterochromatin domains. KAP1 is proposed to silence transcription by assembling HDACs and other chromatin modifiers and remodelers at specific loci.50,51,55 Moreover, KAP1 is a component of the CAF1-HP1α complex involved in heterochromatin replication and associates with pericentric heterochromatin during the retinoic acid induced differentiation of F9 embryonal carcinoma and embryonic stem cells.28,73 Yet loss of KAP1 does not abolish the S-phase dependent localization of SMARCAD1 to pericentric heterochromatin, suggesting that association of SMARCAD1 with heterochromatin is not critically dependent on KAP1 (Fig. 6). This does not exclude the existence of other mechanisms that could direct SMARCAD1 activity toward heterochromatin. Nevertheless, we favor a model in which this remodeler acts at all replication sites. This is consistent with SMARCAD1 localization to early, mid- and late replicating DNA and its interaction with proteins that operate in heterochromatin and euchromatin.9
Figure 6.
Localization of SMARCAD1 to pericentric heterochromatin is not dependent on KAP1 levels. (A) F9 embryonic carcinoma cells and (B) F9 cells that were engineered to express low levels of KAP1/TIF1β (TIF1β-/-/rTA-f.TIF1β) were differentiated for 7 d by exposure to 1 µM retinoic acid as described by Cammas et al. representative cells stained for KAP1 (ab22553) and SMARCAD1 9 are shown, images were adjusted for brightness and contrast. DAPI bright foci mark pericentric heterochromatin.
Frequently, chromatin remodelers recognize post-translational modifications on proteins through dedicated protein domains present in either the ATPase itself or their accessory proteins.74 SMARCAD1 and its homologs in other species contain potential mono-ubiquitin binding motifs, termed CUE domains, raising the possibility that SMARCAD1 could target ubiquitinylated proteins like PCNA or H2A at replication sites.32,75,76
Outlook
It is likely that SMARCAD1 directs histone deacetylation at replication sites through its physical association with HDAC1 and HDAC2. Coupling HDACs with ATP-dependent chromatin remodeling activities within a multi-enzyme complex is a strategy also employed by Mi-2/NURD, NCoR1, NoRC and the yeast SHREC complexes involved in transcriptional repression.77–82 The fact that a functional ATPase domain in SMARCAD1 is required for mediating global histone acetylation levels in cells suggests that chromatin remodeling occurs prior to or concomitantly with deacetylation and emphasizes a functional link between these activities. Remodeling may facilitate histone deacetylation and subsequent methylation by exposing substrates. This is in line with the observation that HDAC1 alone can deacetylate histones in nucleosomes but not in oligonucleosomes in vitro.83 Likewise, nucleosomal histone H3 is a poor substrate for all characterized H3K9 histone methyltransferases compared with free histone H3.84–86 Further support comes from the demonstration that ATP stimulates deacetylase activity of the NuRD remodeler in vitro.77,87 As HDAC enzymes display broad substrate specificity one important question is how SMARCAD1 activity is controlled to avoid deacetylation at sites where it may provoke inappropriate silencing. It would also be interesting to know whether SMARCAD1 performs the same basic activity (remodeling coupled histonedeacetylation) or other specialized roles at different loci and cell types and during different cell cycle or developmental stages.
A mutation in a skin-specific isoform of SMARCAD1 is causally linked to adermatoglyphia, also dubbed “immigration delay disease” as it causes the lack of epidermal ridges and consequently fingerprints.88 SMARCAD1 knock out mice have reduced viability and show growth retardation, skeletal dysplasia and impaired fertility.89 It will be important to address whether these phenotypes are related to SMARCAD1's role in chromatin restoration after replication. It is tempting to speculate that SMARCAD1 may also have a role outside of S phase in repair and transcription, since all these processes involve chromatin disruption and require rebuilding of chromatin structures.90
Insights into how chromatin organization may be altered by SMARCAD1 come from studies of the S. cerevisae ortholog Fun30, which revealed histone H2A-H2B dimer exchange and weak nucleosome sliding activity in vitro.91 The most pertinent challenge ahead is to uncover the specific substrate(s) for SMARCAD1 remodeling. In principle this could be a particularly modified histone. One possibility is mono-ubiquitinylation which is known to occur on H2A/H2B, though Fun30 has no apparent preference for binding to ubiquitinylated chromatin in vitro.75,91 Alternative targets include histone variants and the linker histone H1 which co-purifies with SMARCAD1 complexes.9
Our characterization of SMARCAD1 has revealed that the repertoire of processes during replication which require SWI/SNF-like proteins is greater that previously appreciated.
Acknowledgments
We are grateful to Florence Cammas, Inserm, France, for providing F9 cells that express low levels of KAP1/TIF1β (TIF1β-/-/rTA-f.TIF1β). We thank Wendy Dean, Colin Dingwall, Louise Matheson, Fatima Santos and Nicola Stead for comments on the manuscript. This work was supported by the UK Biotechnology and Biological Sciences Research Council (BBSRC), the UK Medical Research Council, the European Union FP6 Network of Excellence “The Epigenome” and a BBSRC studentship to S.R.
Abbreviation
- NuRD
nucleosome remodeling and histone deacetylation
- KAP1
Krüppel-associated box-associated protein 1
- PCNA
proliferating cell nuclear antigen
- SMARCAD1
SWI/SNF-related, matrix-associated actindependent regulator of chromatin, subfamily a, containing DEAD/H box 1
- WICH
WSTF-ISWI chromatin remodeling complex
References
- 1.Probst AV, Dunleavy E, Almouzni G. Epigenetic inheritance during the cell cycle. Nat Rev Mol Cell Biol. 2009;10:192–206. doi: 10.1038/nrm2640. [DOI] [PubMed] [Google Scholar]
- 2.Falbo KB, Shen X. Chromatin remodeling in DNA replication. J Cell Biochem. 2006;97:684–689. doi: 10.1002/jcb.20752. [DOI] [PubMed] [Google Scholar]
- 3.Clapier CR, Cairns BR. The biology of chromatin remodeling complexes. Annu Rev Biochem. 2009;78:273–304. doi: 10.1146/annurev.biochem.77.062706.153223. [DOI] [PubMed] [Google Scholar]
- 4.Collins N, Poot RA, Kukimoto I, Garcia-Jimenez C, Dellaire G, Varga-Weisz PD. An ACF1-ISWI chromatin-remodeling complex is required for DNA replication through heterochromatin. Nat Genet. 2002;32:627–632. doi: 10.1038/ng1046. [DOI] [PubMed] [Google Scholar]
- 5.Papamichos-Chronakis M, Peterson CL. The Ino80 chromatin-remodeling enzyme regulates replisome function and stability. Nat Struct Mol Biol. 2008;15:338–345. doi: 10.1038/nsmb.1413. [DOI] [PubMed] [Google Scholar]
- 6.Cohen SM, Chastain PD, 2nd, Rosson GB, Groh BS, Weissman BE, Kaufman DG, et al. BRG1 colocalizes with DNA replication factors and is required for efficient replication fork progression. Nucleic Acids Res. 2010;38:6906–6919. doi: 10.1093/nar/gkq559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Sugimoto N, Yugawa T, Iizuka M, Kiyono T, Fujita M. The chromatin remodeler sucrose nonfermenting 2 homolog (SNF2H) is recruited onto DNA replication origins through interaction with CDC10-dependent transcript 1 (CDT1) and promotes the pre-replication complex formation. J Biol Chem. 2011;286:39200–39210. doi: 10.1074/jbc.M111.256123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Corpet A, Almouzni G. Making copies of chromatin: the challenge of nucleosomal organization and epigenetic information. Trends Cell Biol. 2009;19:29–41. doi: 10.1016/j.tcb.2008.10.002. [DOI] [PubMed] [Google Scholar]
- 9.Rowbotham SP, Barki L, Neves-Costa A, Santos F, Dean W, Hawkes N, et al. Maintenance of silent chromatin through replication requires SWI/SNF-like chromatin remodeler SMARCAD1. Mol Cell. 2011;42:285–296. doi: 10.1016/j.molcel.2011.02.036. [DOI] [PubMed] [Google Scholar]
- 10.Jasencakova Z, Groth A. Broken silence restored—remodeling primes for deacetylation at replication Forks. Mol Cell. 2011;42:267–269. doi: 10.1016/j.molcel.2011.04.007. [DOI] [PubMed] [Google Scholar]
- 11.Vincent JA, Kwong TJ, Tsukiyama T. ATP-dependent chromatin remodeling shapes the DNA replication landscape. Nat Struct Mol Biol. 2008;15:477–484. doi: 10.1038/nsmb.1419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Shimada K, Oma Y, Schleker T, Kugou K, Ohta K, Harata M, et al. Ino80 Chromatin Remodeling Complex Promotes Recovery of Stalled Replication Forks. Current biology: CB. 2008;18:566–575. doi: 10.1016/j.cub.2008.03.049. [DOI] [PubMed] [Google Scholar]
- 13.Hur SK, Park EJ, Han JE, Kim YA, Kim JD, Kang D, et al. Roles of human INO80 chromatin remodeling enzyme in DNA replication and chromosome segregation suppress genome instability. Cell Mol Life Sci. 2010;67:2283–2296. doi: 10.1007/s00018-010-0337-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.de la Serna IL, Imbalzano AN. Unfolding heterochromatin for replication. Nat Genet. 2002;32:560–562. doi: 10.1038/ng1202-560. [DOI] [PubMed] [Google Scholar]
- 15.Yan Q, Cho E, Lockett S, Muegge K. Association of Lsh, a regulator of DNA methylation, with pericentromeric heterochromatin is dependent on intact heterochromatin. Mol Cell Biol. 2003;23:8416–8428. doi: 10.1128/MCB.23.23.8416-28.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Helbling Chadwick L, Chadwick B, Jaye D, Wade P. The Mi-2/NuRD complex associates with pericentromeric heterochromatin during S phase in rapidly proliferating lymphoid cells. Chromosoma. 2009;118:445–457. doi: 10.1007/s00412-009-0207-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Poot RA, Bozhenok L, van den Berg DLC, Steffensen S, Ferreira F, Grimaldi M, et al. The Williams syndrome transcription factor interacts with PCNA to target chromatin remodelling by ISWI to replication foci. Nat Cell Biol. 2004;6:1236–1244. doi: 10.1038/ncb1196. [DOI] [PubMed] [Google Scholar]
- 18.Poot RA, Bozhenok L, van den Berg DL, Hawkes N, Varga-Weisz PD. Chromatin remodeling by WSTF-ISWI at the replication site: opening a window of opportunity for epigenetic inheritance? Cell Cycle. 2005;4:543–546. doi: 10.4161/cc.4.4.1624. [DOI] [PubMed] [Google Scholar]
- 19.Sims JK, Wade PA. Mi-2/NuRD complex function is required for normal S phase progression and assembly of pericentric heterochromatin. Mol Biol Cell. 2011;22:3094–3102. doi: 10.1091/mbc.E11-03-0258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Craig JM. Heterochromatin—many flavours, common themes. Bioessays. 2005;27:17–28. doi: 10.1002/bies.20145. [DOI] [PubMed] [Google Scholar]
- 21.Grewal SI, Jia S. Heterochromatin revisited. Nat Rev Genet. 2007;8:35–46. doi: 10.1038/nrg2008. [DOI] [PubMed] [Google Scholar]
- 22.Taddei A, Maison C, Roche D, Almouzni G. Reversible disruption of pericentric heterochromatin and centromere function by inhibiting deacetylases. Nat Cell Biol. 2001;3:114–120. doi: 10.1038/35055010. [DOI] [PubMed] [Google Scholar]
- 23.Peters AHFM, O'Carroll D, Scherthan H, Mechtler K, Sauer S, Schöfer C, et al. Loss of the Suv39 h histone methyltransferases impairs mammalian heterochromatin and genome stability. Cell. 2001;107:323–337. doi: 10.1016/S0092-8674(01)00542-6. [DOI] [PubMed] [Google Scholar]
- 24.Kaufman PD, Kobayashi R, Stillman B. Ultraviolet radiation sensitivity and reduction of telomeric silencing in Saccharomyces cerevisiae cells lacking chromatin assembly factor-I. Genes Dev. 1997;11:345–357. doi: 10.1101/gad.11.3.345. [DOI] [PubMed] [Google Scholar]
- 25.Quivy JP, Roche D, Kirschner D, Tagami H, Nakatani Y, Almouzni GA. CAF-1 dependent pool of HP1 during heterochromatin duplication. EMBO J. 2004;23:3516–3526. doi: 10.1038/sj.emboj.7600362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Dohke K, Miyazaki S, Tanaka K, Urano T, Grewal SI, Murakami Y. Fission yeast chromatin assembly factor 1 assists in the replication-coupled maintenance of heterochromatin. Genes Cells. 2008;13:1027–1043. doi: 10.1111/j.1365-2443.2008.01225.x. [DOI] [PubMed] [Google Scholar]
- 27.Quivy JP, Gerard A, Cook AJL, Roche D, Almouzni G. The HP1-p150/CAF-1 interaction is required for pericentric heterochromatin replication and S-phase progression in mouse cells. Nat Struct Mol Biol. 2008;15:972–979. doi: 10.1038/nsmb.1470. [DOI] [PubMed] [Google Scholar]
- 28.Loyola A, Tagami H, Bonaldi T, Roche D, Quivy JP, Imhof A, et al. The HP1[alpha]-CAF1-SetDB1-containing complex provides H3K9me1 for Suv39-mediated K9me3 in pericentric heterochromatin. EMBO Rep. 2009;10:769–775. doi: 10.1038/embor.2009.90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Yamane K, Mizuguchi T, Cui B, Zofall M, Noma K, Grewal SI. Asf1/HIRA facilitate global histone deacetylation and associate with HP1 to promote nucleosome occupancy at heterochromatic loci. Mol Cell. 2011;41:56–66. doi: 10.1016/j.molcel.2010.12.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Fraga MF, Ballestar E, Villar-Garea A, Boix-Chornet M, Espada J, Schotta G, et al. Loss of acetylation at Lys16 and trimethylation at Lys20 of histone H4 is a common hallmark of human cancer. Nat Genet. 2005;37:391–400. doi: 10.1038/ng1531. [DOI] [PubMed] [Google Scholar]
- 31.Yu Q, Zhang X, Bi X. Roles of chromatin remodeling factors in the formation and maintenance of heterochromatin structure. J Biol Chem. 2011;286:14659–14669. doi: 10.1074/jbc. M110.183269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Neves-Costa A, Will WR, Vetter AT, Miller JR, Varga-Weisz P. The SNF2-Family Member Fun30 Promotes Gene Silencing in Heterochromatic Loci. PLoS ONE. 2009;4:8111. doi: 10.1371/journal.pone.0008111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Strålfors A, Walfridsson J, Bhuiyan H, Ekwall K. The FUN30 Chromatin remodeler, Fft3, protects centromeric and subtelomeric domains from euchromatin formation. PLoS Genet. 2011;7:1001334. doi: 10.1371/journal.pgen.1001334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ekwall K, Olsson T, Turner BM, Cranston G, Allshire RC. Transient inhibition of histone deacetylation alters the structural and functional imprint at fission yeast centromeres. Cell. 1997;91:1021–1032. doi: 10.1016/S0092-8674(00)80492-4. [DOI] [PubMed] [Google Scholar]
- 35.Grewal SI, Bonaduce MJ, Klar AJ. Histone deacetylase homologs regulate epigenetic inheritance of transcriptional silencing and chromosome segregation in fission yeast. Genetics. 1998;150:563–576. doi: 10.1093/genetics/150.2.563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Maison C, Bailly D, Peters AHFM, Quivy JP, Roche D, Taddei A, et al. Higher-order structure in pericentric heterochromatin involves a distinct pattern of histone modification and an RNA component. Nat Genet. 2002;30:329–334. doi: 10.1038/ng843. [DOI] [PubMed] [Google Scholar]
- 37.Okazaki N, Ikeda S, Ohara R, Shimada K, Yanagawa T, Nagase T, et al. The novel protein complex with SMARCAD1/KIAA1122 binds to the vicinity of TSS. J Mol Biol. 2008;382:257–265. doi: 10.1016/j.jmb.2008.07.031. [DOI] [PubMed] [Google Scholar]
- 38.Jasencakova Z, Groth A. Restoring chromatin after replication: how new and old histone marks come together. Semin Cell Dev Biol. 2010;21:231–237. doi: 10.1016/j.semcdb.2009.09.018. [DOI] [PubMed] [Google Scholar]
- 39.Annunziato AT. Assembling chromatin: The long and winding road. Biochim Biophys Acta. 2011 doi: 10.1016/j.bbagrm.2011.07.005. In press. [DOI] [PubMed] [Google Scholar]
- 40.Sobel RE, Cook RG, Perry CA, Annunziato AT, Allis CD. Conservation of deposition-related acetylation sites in newly synthesized histones H3 and H4. Proc Natl Acad Sci USA. 1995;92:1237–1241. doi: 10.1073/pnas.92.4.1237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Benson LJ, Gu Y, Yakovleva T, Tong K, Barrows C, Strack CL, et al. Modifications of H3 and H4 during chromatin replication, nucleosome assembly and histone exchange. J Biol Chem. 2006;281:9287–9296. doi: 10.1074/jbc.M512956200. [DOI] [PubMed] [Google Scholar]
- 42.Loyola A, Bonaldi T, Roche D, Imhof A, Almouzni G. PTMs on H3 variants before chromatin assembly potentiate their final epigenetic state. Mol Cell. 2006;24:309–316. doi: 10.1016/j.molcel.2006.08.019. [DOI] [PubMed] [Google Scholar]
- 43.Scharf AND, Barth TK, Imhof A. Establishment of Histone Modifications after Chromatin Assembly. Nucleic Acids Res. 2009;37:5032–5040. doi: 10.1093/nar/gkp518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Campos EI, Fillingham J, Li G, Zheng H, Voigt P, Kuo WH, et al. The program for processing newly synthesized histones H3.1 and H4. Nat Struct Mol Biol. 2010;17:1343–1351. doi: 10.1038/nsmb.1911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Jasencakova Z, Scharf AN, Ask K, Corpet A, Imhof A, Almouzni G, et al. Replication stress interferes with histone recycling and predeposition marking of new histones. Mol Cell. 2010;37:736–743. doi: 10.1016/j.molcel.2010.01.033. [DOI] [PubMed] [Google Scholar]
- 46.Taddei A, Roche D, Sibarita JB, Turner BM, Almouzni G. Duplication and maintenance of heterochromatin domains. J Cell Biol. 1999;147:1153–1166. doi: 10.1083/jcb.147.6.1153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Scharf AN, Imhof A. Every methyl counts—epigenetic calculus. FEBS Lett. 2011;585:2001–2007. doi: 10.1016/j.febslet.2010.11.029. [DOI] [PubMed] [Google Scholar]
- 48.Lachner M, O'Carroll D, Rea S, Mechtler K, Jenuwein T. Methylation of histone H3 lysine 9 creates a binding site for HP1 proteins. Nature. 2001;410:116–120. doi: 10.1038/35065132. [DOI] [PubMed] [Google Scholar]
- 49.Bannister AJ, Zegerman P, Partridge JF, Miska EA, Thomas JO, Allshire RC, et al. Selective recognition of methylated lysine 9 on histone H3 by the HP1 chromo domain. Nature. 2001;410:120–124. doi: 10.1038/35065138. [DOI] [PubMed] [Google Scholar]
- 50.Nielsen AL, Ortiz JA, You J, Oulad-Abdelghani M, Khechumian R, Gansmuller A, et al. Interaction with members of the heterochromatin protein 1 (HP1) family and histone deacetylation are differentially involved in transcriptional silencing by members of the TIF1 family. EMBO J. 1999;18:6385–6395. doi: 10.1093/emboj/18.22.6385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Ryan RF, Schultz DC, Ayyanathan K, Singh PB, Friedman JR, Fredericks WJ, et al. KAP-1 corepressor protein interacts and colocalizes with heterochromatic and euchromatic HP1 proteins: a potential role for Kruppel-associated box-zinc finger proteins in heterochromatin-mediated gene silencing. Mol Cell Biol. 1999;19:4366–4378. doi: 10.1128/mcb.19.6.4366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Aagaard L, Laible G, Selenko P, Schmid M, Dorn R, Schotta G, et al. Functional mammalian homologues of the Drosophila PEV-modifier Su(var)3-9 encode centromere-associated proteins which complex with the heterochromatin component M31. EMBO J. 1999;18:1923–1938. doi: 10.1093/emboj/18.7.1923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Canzio D, Chang EY, Shankar S, Kuchenbecker KM, Simon MD, Madhani HD, et al. Chromodomain-mediated oligomerization of HP1 suggests a nucleosome-bridging mechanism for heterochromatin assembly. Mol Cell. 2011;41:67–81. doi: 10.1016/j.molcel.2010.12.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Shinkai Y, Tachibana M. H3K9 methyltransferase G9a and the related molecule GLP. Genes Dev. 2011;25:781–788. doi: 10.1101/gad.2027411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Iyengar S, Farnham PJ. KAP1 protein: an enigmatic master regulator of the genome. J Biol Chem. 2011;286:26267–26276. doi: 10.1074/jbc. R111.252569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Ziv Y, Bielopolski D, Galanty Y, Lukas C, Taya Y, Schultz DC, et al. Chromatin relaxation in response to DNA double-strand breaks is modulated by a novel ATM- and KAP-1 dependent pathway. Nat Cell Biol. 2006;8:870–876. doi: 10.1038/ncb1446. [DOI] [PubMed] [Google Scholar]
- 57.Goodarzi AA, Noon AT, Deckbar D, Ziv Y, Shiloh Y, Löbrich M, et al. ATM signaling facilitates repair of DNA double-strand breaks associated with heterochromatin. Mol Cell. 2008;31:167–177. doi: 10.1016/j.molcel.2008.05.017. [DOI] [PubMed] [Google Scholar]
- 58.Matsuoka S, Ballif BA, Smogorzewska A, McDonald ER, 3rd, Hurov KE, Luo J, et al. ATM and ATR substrate analysis reveals extensive protein networks responsive to DNA damage. Science. 2007;316:1160–1166. doi: 10.1126/science.1140321. [DOI] [PubMed] [Google Scholar]
- 59.Moldovan GL, Pfander B, Jentsch S. PCNA, the maestro of the replication fork. Cell. 2007;129:665–679. doi: 10.1016/j.cell.2007.05.003. [DOI] [PubMed] [Google Scholar]
- 60.Kohn KW, Aladjem MI, Weinstein JN, Pommier Y. Chromatin challenges during DNA replication: a systems representation. Mol Biol Cell. 2008;19:1–7. doi: 10.1091/mbc.E07-06-0528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Warbrick E, Lane DP, Glover DM, Cox LS. A small peptide inhibitor of DNA replication defines the site of interaction between the cyclin-dependent kinase inhibitor p21WAF1 and proliferating cell nuclear antigen. Curr Biol. 1995;5:275–282. doi: 10.1016/S0960-9822(95)00058-3. [DOI] [PubMed] [Google Scholar]
- 62.Fan J, Otterlei M, Wong HK, Tomkinson AE, Wilson DM. XRCC1 co-localizes and physically interacts with PCNA. Nucleic Acids Res. 2004;32:2193–2201. doi: 10.1093/nar/gkh556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Guo C, Sonoda E, Tang TS, Parker JL, Bielen AB, Takeda S, et al. REV1 Protein interacts with PCNA: Significance of the REV1 BRCT domain in vitro and in vivo. Mol Cell. 2006;23:265–271. doi: 10.1016/j.molcel.2006.05.038. [DOI] [PubMed] [Google Scholar]
- 64.Fotedar R, Mossi R, Fitzgerald P, Rousselle T, Maga G, Brickner H, et al. A conserved domain of the large subunit of replication factor C binds PCNA and acts like a dominant negative inhibitor of DNA replication in mammalian cells. EMBO J. 1996;15:4423–4433. [PMC free article] [PubMed] [Google Scholar]
- 65.Uhlmann F, Cai J, Gibbs E, O'Donnell M, Hurwitz J. Deletion analysis of the large subunit p140 in human replication factor C reveals regions required for complex formation and replication activities. J Biol Chem. 1997;272:10058–10064. doi: 10.1074/jbc.272.15.10058. [DOI] [PubMed] [Google Scholar]
- 66.Rolef Ben-Shahar T, Castillo AG, Osborne MJ, Borden KLB, Kornblatt J, Verreault A. Two fundamentally distinct PCNA interaction peptides contribute to chromatin assembly factor 1 function. Mol Cell Biol. 2009;29:6353–6365. doi: 10.1128/MCB.01051-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Spada F, Haemmer A, Kuch D, Rothbauer U, Schermelleh L, Kremmer E, et al. DNMT1 but not its interaction with the replication machinery is required for maintenance of DNA methylation in human cells. J Cell Biol. 2007;176:565–571. doi: 10.1083/jcb.200610062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Sharif J, Muto M, Takebayashi S-i, Suetake I, Iwamatsu A, Endo TA, et al. The SRA protein Np95 mediates epigenetic inheritance by recruiting Dnmt1 to methylated DNA. Nature. 2007;450:908–912. doi: 10.1038/nature06397. [DOI] [PubMed] [Google Scholar]
- 69.Rountree MR, Bachman KE, Baylin SB. DNMT1 binds HDAC2 and a new co-repressor, DMAP1, to form a complex at replication foci. Nat Genet. 2000;25:269–277. doi: 10.1038/77023. [DOI] [PubMed] [Google Scholar]
- 70.Kleczkowska HE, Marra G, Lettieri T, Jiricny J. hMSH3 and hMSH6 interact with PCNA and colocalize with it to replication foci. Genes Dev. 2001;15:724–736. doi: 10.1101/gad.191201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Milutinovic S, Zhuang Q, Szyf M. Proliferating cell nuclear antigen associates with histone deacetylase activity, integrating DNA replication and chromatin modification. J Biol Chem. 2002;277:20974–20978. doi: 10.1074/jbc.M202504200. [DOI] [PubMed] [Google Scholar]
- 72.Estève PO, Chin HG, Smallwood A, Feehery GR, Gangisetty O, Karpf AR, et al. Direct interaction between DNMT1 and G9a coordinates DNA and histone methylation during replication. Genes Dev. 2006;20:3089–3103. doi: 10.1101/gad.1463706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Cammas F, Oulad-Abdelghani M, Vonesch JL, Huss-Garcia Y, Chambon P, Losson R. Cell differentiation induces TIF1beta association with centromeric heterochromatin via an HP1 interaction. J Cell Sci. 2002;115:3439–3448. doi: 10.1242/jcs.115.17.3439. [DOI] [PubMed] [Google Scholar]
- 74.Erdel F, Krug J, Langst G, Rippe K. Targeting chromatin remodelers: Signals and search mechanisms. Biochim Biophys Acta. 2011;1809:497–508. doi: 10.1016/j.bbagrm.2011.06.005. [DOI] [PubMed] [Google Scholar]
- 75.Vassilev AP, Rasmussen HH, Christensen EI, Nielsen S, Celis JE. The levels of ubiquitinated histone H2A are highly upregulated in transformed human cells: partial colocalization of uH2A clusters and PCNA/cyclin foci in a fraction of cells in S-phase. J Cell Sci. 1995;108:1205–1215. doi: 10.1242/jcs.108.3.1205. [DOI] [PubMed] [Google Scholar]
- 76.Kannouche PL, Wing J, Lehmann AR. Interaction of human DNA polymerase eta with monoubiquitinated PCNA: a possible mechanism for the polymerase switch in response to DNA damage. Mol Cell. 2004;14:491–500. doi: 10.1016/S1097-2765(04)00259-X. [DOI] [PubMed] [Google Scholar]
- 77.Xue Y, Wong J, Moreno GT, Young MK, Cote J, Wang W. NURD, a novel complex with both ATP-dependent chromatin-remodeling and histone deacetylase activities. Mol Cell. 1998;2:851–861. doi: 10.1016/S1097-2765(00)80299-3. [DOI] [PubMed] [Google Scholar]
- 78.Kehle J, Beuchle D, Treuheit S, Christen B, Kennison JA, Bienz M, et al. dMi-2, a hunchback-interacting protein that functions in polycomb repression. Science. 1998;282:1897–1900. doi: 10.1126/science.282.5395.1897. [DOI] [PubMed] [Google Scholar]
- 79.Sugiyama T, Cam HP, Sugiyama R, Noma K, Zofall M, Kobayashi R, et al. SHREC, an effector complex for heterochromatic transcriptional silencing. Cell. 2007;128:491–504. doi: 10.1016/j.cell.2006.12.035. [DOI] [PubMed] [Google Scholar]
- 80.Zhou Y, Santoro R, Grummt I. The chromatin remodeling complex NoRC targets HDAC1 to the ribosomal gene promoter and represses RNA polymerase I transcription. EMBO J. 2002;21:4632–4640. doi: 10.1093/emboj/cdf460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Guetg C, Lienemann P, Sirri V, Grummt I, Hernandez-Verdun D, Hottiger MO, et al. The NoRC complex mediates the heterochromatin formation and stability of silent rRNA genes and centromeric repeats. EMBO J. 2010;29:2135–2146. doi: 10.1038/emboj.2010.17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Tai HH, Geisterfer M, Bell JC, Moniwa M, Davie JR, Boucher L, et al. CHD1 associates with NCoR and histone deacetylase as well as with RNA splicing proteins. Biochem Biophys Res Commun. 2003;308:170–176. doi: 10.1016/S0006-291X(03)01354-8. [DOI] [PubMed] [Google Scholar]
- 83.Hassig CA, Tong JK, Fleischer TC, Owa T, Grable PG, Ayer DE, et al. A role for histone deacetylase activity in HDAC1-mediated transcriptional repression. Proc Natl Acad Sci USA. 1998;95:3519–3524. doi: 10.1073/pnas.95.7.3519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Ogawa H, Ishiguro K-i, Gaubatz S, Livingston DM, Nakatani Y. A complex with chromatin modifiers that occupies E2F- and Myc-responsive genes in G0 cells. Science. 2002;296:1132–1136. doi: 10.1126/science.1069861. [DOI] [PubMed] [Google Scholar]
- 85.Wang H, An W, Cao R, Xia L, Erdjument-Bromage H, Chatton B, et al. mAM facilitates conversion by ESET of dimethyl to trimethyl lysine 9 of histone H3 to cause transcriptional repression. Mol Cell. 2003;12:475–487. doi: 10.1016/j.molcel.2003.08.007. [DOI] [PubMed] [Google Scholar]
- 86.Schotta G, Lachner M, Sarma K, Ebert A, Sengupta R, Reuter G, et al. A silencing pathway to induce H3-K9 and H4-K20 trimethylation at constitutive heterochromatin. Genes Dev. 2004;18:1251–1262. doi: 10.1101/gad.300704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Tong JK, Hassig CA, Schnitzler GR, Kingston RE, Schreiber SL. Chromatin deacetylation by an ATP-dependent nucleosome remodelling complex. Nature. 1998;395:917–921. doi: 10.1038/27699. [DOI] [PubMed] [Google Scholar]
- 88.Nousbeck J, Burger B, Fuchs-Telem D, Pavlovsky M, Fenig S, Sarig O, et al. A mutation in a skin-specific isoform of SMARCAD1 causes autosomal-dominant adermatoglyphia. Am J Hum Genet. 2011;89:302–307. doi: 10.1016/j.ajhg.2011.07.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Schoor M, Schuster-Gossler K, Roopenian D, Gossler A. Skeletal dysplasias, growth retardation, reduced postnatal survival and impaired fertility in mice lacking the SNF2/SWI2 family member ETL1. Mech Dev. 1999;85:73–83. doi: 10.1016/S0925-4773(99)00090-8. [DOI] [PubMed] [Google Scholar]
- 90.Xu Y, Price BD. Chromatin dynamics and the repair of DNA double strand breaks. Cell Cycle. 2011;10:261–267. doi: 10.4161/cc.10.2.14543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Awad S, Ryan D, Prochasson P, Owen-Hughes T, Hassan AH. The Snf2 homolog Fun30 acts as a homodimeric ATP-dependent chromatin-remodeling enzyme. J Biol Chem. 2010;285:9477–9484. doi: 10.1074/jbc.M109.082149. [DOI] [PMC free article] [PubMed] [Google Scholar]