Abstract
In the developing vertebrate nervous system, bHLH proneural factors, such as Ascl1, are known to play important regulatory roles at different stages of the neurogenic differentiation process. In spite of the wealth of information gathered on the cellular functions of proneural factors, little was known of the molecular basis for their activities and, in particular, of the identity of their target genes. The development of genomic approaches is making possible the characterization of transcriptional programs at an unprecedented scale. Recently, we have used a combination of genomic location analysis by ChIP-on-chip and expression profiling in order to characterize the proneural transcription program regulated by Ascl1 in the ventral telencephalon of the mouse embryonic brain. Our results demonstrate that Ascl1 directly controls successive steps of neurogenesis and provide a molecular frame for previously described Ascl1 functions. In addition, we uncovered an important but previously unrecognized role for Ascl1 in promoting the proliferation of neural progenitors. Here we discuss our recent findings and review them in light of efforts from other laboratories to characterize the transcriptional programs downstream various proneural factors.
Key words: Ascl1/Mash1, proneural gene, transcriptional program, bHLH transcription factor, chromatin immunoprecipitation
Introduction
The generation of new neurons by progenitor cells in the developing vertebrate central nervous system requires a number of precisely orchestrated steps. As they undergo their last cell division, progenitors become committed to their neuronal fate and select a unique neuronal subtype identity.1 Soon after becoming post mitotic, neurons migrate out of the progenitor zone along specific routes2 and begin to differentiate. Proneural transcription factors of the bHLH family are expressed in proliferating progenitors and are thought to play a pivotal role in the coordinate regulation of the neurogenic differentiation program.3,4 Since their identification in vertebrates, extensive phenotypic analysis of various mouse mutants in proneural genes has implicated them in the regulation of multiple steps of neurogenesis, including the activation of the Notchmediated process of lateral inhibition, the neuronal commitment of progenitors and their acquisition of neuronal subtype-specific traits and neuronal migration.3,5,6 In spite of the wealth of information on their cellular functions, the molecular mechanisms underpinning the activities of proneural proteins, including the identity of the genes that they directly regulate, have remained largely unknown. In particular, it has been unclear whether they regulate successive phases of neurogenesis directly through the activation of multiple effector genes, or indirectly through the mobilization of a transcription factor cascade. In addition, it has been unclear whether their functions have been entirely elucidated by genetic analysis, or whether some new functions, masked by the complexity of the proneural mutant phenotypes, have remained unidentified. A series of recent studies combining genetics with novel genomic approaches has finally begun to provide a much needed molecular frame for the various roles of this important family of transcription factors in neural development and, in some cases, has also revealed novel functions.
Diverse Components of the Neurogenic Program are Directly Controlled by Ascl1
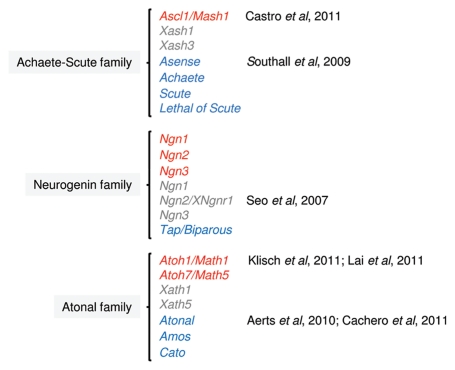
The search for vertebrate transcription factors with determination functions in the nervous system led to the identification two decades ago of Ascl1/Mash1,7 a murine homolog of the Drosophila genes of the achaete-scute complex which encodes the first bHLH proneural factor to be identified in vertebrates (Fig. 1). In order to gather novel insights into the molecular mechanisms underpinning the various cellular functions of Ascl1, we chose to perform a genome-wide characterization of its transcriptional program in the developing ventral telencephalon, where Ascl1 has been implicated in the generation and specification of GABAergic interneurons, the main neuronal population produced in that region.8–10 Aiming at performing a large-scale identification of direct targets of Ascl1, we used chromatin immunoprecipitation of Ascl1 from ventral telencephalon tissue dissected at E12.5 of development, followed by hybridization onto genomic microarrays (ChIP-on-chip), tiling approximately 17,000 of well-characterized proximal promoter regions.11 The genomic location analysis was combined with expression profiling data of ventral telencephalon tissue either mutant for, or overexpressing Ascl1, leading to the identification of 339 Ascl1 direct targets defined by their association with Ascl1 binding event and their deregulation when Ascl1 expression is manipulated. This strategy, which probably underestimated the total number of genes directly regulated by Ascl1 (due to the exclusion of genes regulated by Ascl1 binding to a distal enhancer and to genetic redundancy in null mutant embryos) allowed for a first glance at a proneural program directly governed by Ascl1. Functional annotation of Ascl1 targets by gene ontology (GO) showed great diversity of functions, with most phases of neurogenesis being directly regulated by this proneural factor (Table 1). Overrepresented biological process terms are associated with the early steps of lateral inhibition (e.g., “Notch signaling pathway”), cell fate decisions (e.g., “neuron fate commitment”) and control of cell proliferation (e.g., “regulation of cell cycle”), but also later steps of neuronal differentiation (e.g., “neurotransmitter biosynthetic process”) and neurite outgrowth (e.g., “cell projection organization”). A large fraction of Ascl1 target genes encode transcription factors or other proteins with transcription regulatory activity (48%), but many other encode signal transduction components (36%), or structural proteins, such as cytoskeleton components (11%).11 Thus, Ascl1's role does not rely solely on the activation of downstream transcriptional cascades, as many of its functions (including late steps in the neurogenic process) are directly controlled by activation of downstream effectors. Altogether, this study provides a useful molecular frame to better understand previously identified cellular functions of Ascl1.
Figure 1.
Neural bHLH proteins that display proneural function activity in mouse (red), frog (gray) and fly (blue) can be group into distinct families, based on the similarity in their bHLH domain. Neural bHLH factors of the NeuroD family that are usually involved in steps of differentiation in postmitotic neurons, and, therefore, that do not have a proneural function, are not shown. References of the published work discussed here are indicated in the figure.
Table 1.
Selection of enriched Gene Ontology (GO) terms associated with Ascl1 target genes in ventral telencephalon of developing mouse embryo
| GO Number | Biological Process | Genes | p value |
| GO:0045449 | Regulation of transcription | 69 | 1.8 × 10−8 |
| GO:0022008 | Neurogenesis | 27 | 5.2 × 10−7 |
| GO:0042136 | Neurotransmitter biosynthetic process | 3 | 9.2 × 10−5 |
| GO:0051726 | Regulation of cell cycle | 11 | 9.6 × 10−5 |
| GO:0010646 | Regulation of cell communication | 25 | 1.1 × 10−4 |
| GO:0016337 | Cell-cell adhesion | 11 | 1.1 × 10−4 |
| GO:0030030 | Cell projection organization | 17 | 1.2 × 10−4 |
| GO:0009966 | Regulation of signal transduction | 22 | 1.8 × 10−4 |
| GO:0006928 | Cell motion | 18 | 2.8 × 10−4 |
| GO:0048663 | Neuron fate commitment | 6 | 2.8 × 10−4 |
| GO:0007417 | Central nervous system development | 16 | 3.1 × 10−4 |
| GO:0007411 | Axon guidance | 8 | 3.4 × 10−4 |
| GO:0019226 | Transmission of nerve impulse | 13 | 4.0 × 10−4 |
| GO:0007409 | Axonogenesis | 11 | 5.0 × 10−4 |
| GO:0016568 | Chromatin modification | 12 | 5.2 × 10−4 |
| GO:0007219 | Notch signaling pathway | 6 | 8.4 × 10−4 |
| GO:0051960 | Regulation of nervous system development | 9 | 8.7 × 10−4 |
| GO:0048667 | Cell morphogenesis involved in neuron differentiation | 11 | 8.8 × 10−4 |
Transcriptional profiling of Ascl1 gain- and loss-of-function, combined with genomic location analysis using ChIP-on-chip, was used to identify 339 direct Ascl1 trasncriptional targets.11 Enriched terms describing biological processes were identified with GoToolbox,12 using a hypergeometric distribution.
A Novel Function for Ascl1 in Proliferation of Neural Progenitors
Experiments showing that Ascl1 overexpression in vivo or in cultured progenitors results in a rapid cell cycle exit (sometimes shown to be associated with the induction of cyclin-dependent kinase (Cdk) inhibitors), have provided evidence of an anti-proliferative function of Ascl1.13–15 However, the identification of a molecular link between Ascl1 and regulators of proliferation of neural progenitors has remained elusive and its function in cell proliferation unclear. In line with earlier predictions, we did find among Ascl1 target genes, candidate mediators of its anti-proliferative function (e.g., Prmt2, Ccng2 and Gadd45 g). Surprisingly, and in apparent contradiction with the aforementioned observations, we also found that Ascl1 positively regulates, in both embryonic and cultured progenitors, many genes that are known to promote cell proliferation through various mechanisms. Most notable among those is the transcription factor E2f1, widely known for its role in entry and transition through S phase of the cell cycle.16 Such a proliferation role of E2f1 has been well-defined in telencephalic progenitors, where it is regulated by a physical interaction with the tumor suppressor retinoblastoma.17 Also among Ascl1 target genes is FoxM1, another important regulator of cell cycle progression in a variety of cell types, which regulates G2/M transition in cerebellar ganule neuron precursors (CGNPs).18 The direct regulation by Ascl1 of downstream targets of both E2f1 (Cdca7) and FoxM1 (Skp2) and of some of their interacting partners (Ep400/Prmt2 and Cdca2, respectively) exemplifies a recurrent feature of the Ascl1 program, which often contains various members of the same pathway. Other important mediators of proliferation directly regulated by Ascl1 in embryonic telencephalon are the transcription factors Tead1/2 and their coactivator Taz, downstream mediators of the Hippo signaling pathway, which regulates cell proliferation in multiple contexts, including neural progenitors.19 The large-scale identification of Ascl1 target genes, therefore, suggested that Ascl1 plays an unanticipated role in promoting the proliferation of progenitors, a function further verified by manipulating the activity of Ascl1 in both embryonic brain and neural stem cell cultures. In Ascl1 null embryos, decreased expression of genes that promote proliferation is associated with a severe reduction in number of dividing sub-ventricular zone (SVZ) progenitors. A more causal link with cell proliferation was established by acute deletion of Ascl1 by in utero delivery of Cre recombinase in embryos carrying an Ascl1 conditional null allele, which results in premature withdrawal from cell cycle of a significant numbers of both ventricular zone (VZ) and SVZ progenitors. Finally, the acute knock down of Ascl1 in cultured neural progenitors resulted in a reduced rate of proliferation, again uncovering a pro-division function of Ascl1 in neural progenitors. Although the decreased proliferation associated with the various Ascl1 loss-of-function experiments is only partial and suggests redundancy with other pathways promoting cell proliferation, overall the functional data strongly support the initial prediction drawn from the identification of Ascl1 targets.
Thus, our unexpected findings suggest that Ascl1 directly links a cell type specific developmental program with pathways (such as E2f1 and its targets) more universally used in the control of cell proliferation. It is also tempting to speculate that the regulation by Ascl1 of proliferation genes during embryonic development may provide a molecular basis for its previously suggested role in tumors of neuroendocrine origin, such as small cell lung carcinomas.20 Ascl1 expression positively correlates with tumorigenic capacity, suggesting a role in cancer development, also supported by knock down experiments of Ascl1.21,22 Future work should thus investigate to which extent the genetic program used to maintain proliferation in the embryo is used by Ascl1 in a cancer context.
The provocative finding that Ascl1 activates both genes promoting cell cycle progression and genes promoting cell cycle-exit suggests that Ascl1 might regulate distinct genetic modules with opposing activities with regard to cell proliferation, at distinct stages of neurogenesis. Interestingly, this dual role may be evolutionary conserved, as suggested by the genome wide characterization of a proneural transcriptional network in the developing CNS of Drosophila.23 Southall and Brand used the in vivo expression of transcription factors tagged with an Escherichia coli DNA adenine methyltransferase (a technique named DamID), to map genome-wide binding sites of several transcriptional regulators in the CNS of the developing embryo. This approach identified a large number of promoter regions bound by Asense, a proneural factor expressed in proliferating neuroblasts and in a subset of their differentiating daughter cells (called Ganglion Mother Cells or GMC). The combination of this approach with expression profiling of microdissected neuroblasts and GMCs from Asense mutants, led to the identification of a large number of Asense target genes in the two cell types. This elegant work has revealed a dual regulatory role for Asense in the self-renewal of neuroblasts and the differentiation of GMCs, respectively.
Characterization of Additional Transcriptional Programs of Neurogenesis
In the developing vertebrate neural tube, proneural genes are expressed by distinct, and often non-overlapping, populations of neural progenitors that generate different types of neuronal cells. How divergent are the transcriptional programs regulated by different proneural proteins and producing different types of neurons has remained until recently a matter of speculation.24,25 A recent study identifying the transcriptional targets of the proneural factor Atoh1/Math1 in the cerebellum revealed striking similarities in the types of targets controlled by Ascl1 and Atoh1 in two distinct CNS populations.26 Klisch and colleagues combined chromatin immunoprecipitation followed by deep sequencing (ChIP-seq) with transcription profiling, to characterize the whole range of genes regulated by Atoh1 in post-natal CGNPs, a population that undergoes a clonal expansion followed by differentiation during the first week after birth. Similar to our findings with Ascl1, Klish and colleagues found that Atoh1 directly controls genes that regulate successive steps of neurogenesis, such as proliferation, migration and differentiation. An important role of Atoh1 in progenitor physiology was revealed by the high number of Atoh1 targets controlling cell metabolism and ribosome biogenesis, presumably catering for the high energy demands of cycling cells. Among the many Atoh1 targets involved in cell cycle regulation are both E2f1 and FoxM1, suggesting significant overlap with pathways used by Ascl1 in the control of cell proliferation.
Inducing cell cycle promoting genes may be a common activity of all vertebrate proneural genes, or it may reflect a mitogenic role shared only by a subset of proneural proteins, in particular factors such as Ascl1 and Atoh1 that are expressed in neuronal lineages undergoing extensive progenitor divisions (e.g., in the ventral telencephalon and cerebellar primordium, respectively). To address this question, it will be interesting to determine whether members of the Neurogenin family of proneural factors share cell cycle promoting targets with Ascl1 and Atoh1. Strikingly, some neuronal lineages undergoing limited progenitor expansion, e.g., in the dorsal telencephalon, ventral midbrain or dorsal spinal cord, express both Ascl1 and Neurogenin2 in rapid succession. These expression patterns together with mutation analyses9,27 suggest that Neurogenin2 does not share the proliferation promoting activity of Ascl1 and that its expression following that of Ascl1 may counteract Ascl1 mitotic activity and suppress progenitor divisions. Seo and colleagues have used a gain of function approach in Xenopus and mouse cells, together with an in silico analysis of consensus binding motifs, to identify downstream targets of Neurogenin.28 The genes identified in this screen are predicted to regulate neuronal differentiation and cell migration rather than cell proliferation. However, the strategy used in this study may have biased the type of Neurogenin targets identified and future genome-wide studies should clarify whether Neurogenin factors also regulate, positively or negatively, genes involved in progenitor cell divisions.
Coordination of Various Neurogenic Steps by Proneural Factors
The ongoing genome-wide characterization of proneural transcriptional targets is greatly advancing our understanding of how proneural factors regulate complex developmental events. The emerging picture of proneural proteins directly regulating sequential steps of neurogenesis provides a mechanistic explanation for their preponderant role in the coordination of the neurogenic process. This may also help explain the unique ability of ectopically expressed proneural factors to re-program various cell types into neurons (e.g., 14 and 29–31). On the other hand, such studies pose important questions concerning the molecular mechanisms by which proneural proteins regulate gene expression. For example, how can a single transcription factor expressed at an early stage in a developmental program, directly control differentiation events that take place much later in time? This issue has been addressed by a study dissecting the transcriptional program regulated by the proneural factor Atonal in the Drosophila peripheral nervous system, where pioneering studies had uncovered a role for proneural genes in the specification of sensory organ identity.4,32 Cachero and colleagues have shown that, similar to a vertebrate proneural factor, Atonal directly regulates both early events of cell specification and later differentiation events resulting in the acquisition of terminally differentiated neuronal features.33 Thus, Atonal regulates a program of cilliogenesis specific to the mechanosensory neurons of chordotonal organs. Some of this activity is mediated by intermediary transcriptional regulators, including Rfx, a well-established regulator of ciliogenesis.34 However, Atonal also directly regulates dilatory, a gene encoding a downstream component of the ciliogenesis process. Interestingly, a detailed analysis of the temporal profile of gene expression in the mechanosensory neuron lineage showed that the expression of many differentiation genes, including genes in the ciliary differentiation pathway, can be detected much earlier than previously anticipated, in still proliferating precursors. This result is reminiscent of the earlier observation, made in neural and hematopoietic cell lineages, that some genes associated with postmitotic functions are already expressed in dividing progenitors prior to their overt differentiation.35,36 Pursuing this line of research should shed light on the mechanisms by which the early expression of a transcription factor governs events that take place at great distance in time.
Coming back to our study of the target genes of Ascl1, some targets expressed at “late” steps in neurogenesis may have already begun to be transcribed at an earlier stage, but a more complex model involving more intricate regulatory mechanisms is likely to be required to explain the temporal expression profile of many of the genes contributing to the transcriptional program regulated by Ascl1. Results from a large screen of publicly available in situ hybridization data suggest that Ascl1 targets display very distinct patterns of expression and show in particular very different timings of onset of expression. While most targets are expressed in progenitors of the ventricular and/or subventricular zone (58%), similarly to Ascl1, others are only expressed at later stages, for example in differentiating neurons of the mantle zone (15%).11 These results have been replicated in a study examining the temporal profiles of gene expression in cultured neural stem cells induced to differentiate into neurons by overexpression of Ascl1 (Drechsel D, François G, Castro DS, unpublished results).
What kind of mechanisms could be involved in the temporal patterning of Ascl1 transcriptional program? Functional interactions with other transcription factors co-regulating Ascl1 target genes could be involved. In particular, the fact that many Ascl1 regulated genes encode themselves transcription factors suggests that feed-back and feed-forward loop motifs, which are common features of transcriptional networks, may regulate the timing of expression of direct Ascl1 targets. Identification of co-regulators of Ascl1 target genes and characterization of their own targets will shed light on how transcriptional networks control the progression of the gene expression program operating during neurogenesis. In addition, chromatin organization at Ascl1 target genes could control the efficiency with which Ascl1 binds to their regulatory elements and/or activate their transcription. It will thus be important to determine the extent to which distinct parts of the Ascl1 program depend on chromatin modifying/remodeling activities for their regulation. Last, Ascl1 itself might be translationally modified over time, which could result in different timings of expression for Ascl1 target genes, if modified versions of Ascl1 vary in their affinities for target promoters. Elucidating the molecular mechanisms underpinning Ascl1 regulatory functions is, therefore, an important goal, a task made easier by the large-scale identification of its targets.
Context Dependence of Proneural Proteins
Each proneural factor is expressed by multiple progenitor populations, distributed in diverse regions of the nervous system and generating different kind of neurons, suggesting that its activity might be influenced by regional cues, adding a new layer of complexity to proneural gene regulatory functions.4,37 The idea that the same factor can regulate distinct targets in different lineages is supported by the observation that proneural proteins regulate genes specific to particular neuronal subtypes, as best shown recently by Lai and colleagues, who identified a subset of Atoh1 targets that in the dorsal spinal cord are specific to the dI1 population of interneurons.38 Similarly, we have found among Ascl1 targets in the telencephalon, the genes encoding the enzymes Gad1 and Gad2, which are specific of GABAergic neurons. What fraction of the transcriptional targets of a particular proneural protein differs, for example, between regions of the brain or spinal cord, is however not yet known. The importance of the cellular context is of equal relevance in Drosophila, exemplified by the different roles of Atonal in distinct locations. Work by Aerts and colleagues have combined expression profiling with in silico scanning of promoter regions of candidate targets, leading to the identification of a large number of Atonal target genes and their regulatory regions in precursors of R8 photoreceptors.39 Strikingly, most identified Atonal targets were expressed in all atonal lineages while others were expressed in subsets of lineages, but none of the targets were specific to a single sensory organ, suggesting that subtype-specific differentiation programs are activated by the expression of different combinations of Atonal target genes. Whether a similar strategy is used in vertebrates to generate diverse proneural programs remains to be determined.
Concluding Remarks
The study of proneural factors has greatly benefited from the use of novel genomic approaches, which have allowed the genome-scale identification of their transcriptional targets. Identification of these targets has in turn helped better understand the cellular functions of these transcription factors. Old functions are being finally provided with a molecular underpinning, while previously unrecognized activities have been discovered. The complexity of the transcriptional programs activated by proneural factors, exemplified by the direct control by Ascl1 of multiple steps of neurogenesis, raise intriguing questions about the mechanisms involved in their regulation. Addressing these questions will keep the research community busy for many years to come.
Acknowledgments
Our work was supported by a project grant from the Wellcome Trust (08234/Z/07/Z) and institutional funds from the Medical Research Council to F.G. (U117570528). Work in D.S.C. laboratory is supported by a grant from Fundação para a Ciência e Tecnologia (PTDC/SAU-NEU/100208/2008) and the Oeiras Municipality (Começar em Oeiras).
References
- 1.Edlund T, Jessell TM. Progression from extrinsic to intrinsic signaling in cell fate specification: a view from the nervous system. Cell. 1999;96:211–224. doi: 10.1016/S0092-8674(00)80561-9. [DOI] [PubMed] [Google Scholar]
- 2.Marín O, Valiente M, Ge X, Tsai LH. Guiding neuronal cell migrations. Cold Spring Harb Perspect Biol. 2010;2:1834. doi: 10.1101/cshperspect.a001834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bertrand N, Castro DS, Guillemot F. Proneural genes and the specification of neural cell types. Nat Rev Neurosci. 2002;3:517–530. doi: 10.1038/nrn874. [DOI] [PubMed] [Google Scholar]
- 4.Powell LM, Jarman AP. Context dependence of proneural bHLH proteins. Curr Opin Genet Dev. 2008;18:411–417. doi: 10.1016/j.gde.2008.07.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ge W, He F, Kim KJ, Blanchi B, Coskun V, Nguyen L, et al. Coupling of cell migration with neurogenesis by proneural bHLH factors. Proc Natl Acad Sci USA. 2006;103:1319–1324. doi: 10.1073/pnas.0510419103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Heng JI, Nguyen L, Castro DS, Zimmer C, Wildner H, Armant O, et al. Neurogenin 2 controls cortical neuron migration through regulation of Rnd2. Nature. 2008;455:114–118. doi: 10.1038/nature07198. [DOI] [PubMed] [Google Scholar]
- 7.Johnson JE, Birren SJ, Anderson DJ. Two rat homologues of Drosophila achaete-scute specifically expressed in neuronal precursors. Nature. 1990;346:858–861. doi: 10.1038/346858a0. [DOI] [PubMed] [Google Scholar]
- 8.Casarosa S, Fode C, Guillemot F. Mash1 regulates neurogenesis in the ventral telencephalon. Development. 1999;126:525–534. doi: 10.1242/dev.126.3.525. [DOI] [PubMed] [Google Scholar]
- 9.Fode C, Ma Q, Casarosa S, Ang SL, Anderson DJ, Guillemot F. A role for neural determination genes in specifying the dorsoventral identity of telencephalic neurons. Genes Dev. 2000;14:67–80. [PMC free article] [PubMed] [Google Scholar]
- 10.Parras CM, Schuurmans C, Scardigli R, Kim J, Anderson DJ, Guillemot F. Divergent functions of the proneural genes Mash1 and Ngn2 in the specification of neuronal subtype identity. Genes Dev. 2002;16:324–338. doi: 10.1101/gad.940902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Castro DS, Martynoga B, Parras C, Ramesh V, Pacary E, Johnston C, et al. A novel function of the proneural factor Ascl1 in progenitor proliferation identified by genome-wide characterization of its targets. Genes Dev. 2011;25:930–945. doi: 10.1101/gad.627811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Martin D, Brun C, Remy E, Mouren P, Thieffry D, Jacq B. GOToolBox: functional analysis of gene datasets based on Gene Ontology. Genome Biol. 2004;5:101. doi: 10.1186/gb-2004-5?12-r101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Castro DS, Skowronska-Krawczyk D, Armant O, Donaldson IJ, Parras C, Hunt C, et al. Proneural bHLH and Brn proteins coregulate a neurogenic program through cooperative binding to a conserved DNA motif. Dev Cell. 2006;11:831–844. doi: 10.1016/j.devcel.2006.10.006. [DOI] [PubMed] [Google Scholar]
- 14.Farah MH, Olson JM, Sucic HB, Hume RI, Tapscott SJ, Turner DL. Generation of neurons by transient expression of neural bHLH proteins in mammalian cells. Development. 2000;127:693–702. doi: 10.1242/dev.127.4.693. [DOI] [PubMed] [Google Scholar]
- 15.Nakada Y, Hunsaker TL, Henke RM, Johnson JE. Distinct domains within Mash1 and Math1 are required for function in neuronal differentiation versus neuronal cell-type specification. Development. 2004;131:1319–1330. doi: 10.1242/dev.01008. [DOI] [PubMed] [Google Scholar]
- 16.DeGregori J, Johnson DG. Distinct and Overlapping Roles for E2F Family Members in Transcription, Proliferation and Apoptosis. Curr Mol Med. 2006;6:739–748. doi: 10.2174/1566524010606070739. [DOI] [PubMed] [Google Scholar]
- 17.McClellan KA, Ruzhynsky VA, Douda DN, Vanderluit JL, Ferguson KL, Chen D, et al. Unique requirement for Rb/E2F3 in neuronal migration: evidence for cell cycle-independent functions. Mol Cell Biol. 2007;27:4825–4843. doi: 10.1128/MCB.02100-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Schüller U, Zhao Q, Godinho SA, Heine VM, Medema RH, Pellman D, et al. Forkhead transcription factor FoxM1 regulates mitotic entry and prevents spindle defects in cerebellar granule neuron precursors. Mol Cell Biol. 2007;27:8259–8270. doi: 10.1128/MCB.00707-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Cao X, Pfaff SL, Gage FH. YAP regulates neural progenitor cell number via the TEA domain transcription factor. Genes Dev. 2008;22:3320–3334. doi: 10.1101/gad.1726608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ball DW. Achaete-scute homolog-1 and Notch in lung neuroendocrine development and cancer. Cancer Lett. 2004;204:159–169. doi: 10.1016/S0304-3835(03)00452-X. [DOI] [PubMed] [Google Scholar]
- 21.Jiang T, Collins BJ, Jin N, Watkins DN, Brock MV, Matsui W, et al. Achaete-scute complex homologue 1 regulates tumor-initiating capacity in human small cell lung cancer. Cancer Res. 2009;69:845–854. doi: 10.1158/0008-5472.CAN-08-2762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Linnoila RI, Zhao B, DeMayo JL, Nelkin BD, Baylin SB, DeMayo FJ, et al. Constitutive achaete-scute homologue-1 promotes airway dysplasia and lung neuroendocrine tumors in transgenic mice. Cancer Res. 2000;60:4005–4009. [PubMed] [Google Scholar]
- 23.Southall TD, Brand AH. Neural stem cell transcriptional networks highlight genes essential for nervous system development. EMBO J. 2009;28:3799–3807. doi: 10.1038/emboj.2009.309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Anderson D, JaJ YN. The determination of the neuronal phenotype. In: Cowan WM, editor. Molecular and cellular approaches to neural development. New York: Oxford University Press; 1997. pp. 26–63. [Google Scholar]
- 25.Brunet JF, Ghysen A. Deconstructing cell determination: proneural genes and neuronal identity. Bioessays. 1999;21:313–318. doi: 10.1002/(SICI)1521-1878 (199904)21: 4 <313 : :AIDBIES7>3.0.CO;2-C. [DOI] [PubMed] [Google Scholar]
- 26.Klisch TJ, Xi Y, Flora A, Wang L, Li W, Zoghbi HY. In vivo Atoh1 targetome reveals how a proneural transcription factor regulates cerebellar development. Proc Natl Acad Sci USA. 2011;108:3288–3293. doi: 10.1073/pnas.1100230108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Nieto M, Schuurmans C, Britz O, Guillemot F. Neural bHLH genes control the neuronal versus glial fate decision in cortical progenitors. Neuron. 2001;29:401–413. doi: 10.1016/S0896-6273(01)00214-8. [DOI] [PubMed] [Google Scholar]
- 28.Seo S, Lim JW, Yellajoshyula D, Chang LW, Kroll KL. Neurogenin and NeuroD direct transcriptional targets and their regulatory enhancers. EMBO J. 2007;26:5093–5108. doi: 10.1038/sj.emboj.7601923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Berninger B, Guillemot F, Gotz M. Directing neurotransmitter identity of neurones derived from expanded adult neural stem cells. Eur J Neurosci. 2007;25:2581–2590. doi: 10.1111/j.1460-9568.2007.05509.x. [DOI] [PubMed] [Google Scholar]
- 30.Geoffroy CG, Critchley JA, Castro DS, Ramelli S, Barraclough C, Descombes P, et al. Engineering of dominant active basic helix-loop-helix proteins that are resistant to negative regulation by postnatal central nervous system antineurogenic cues. Stem Cells. 2009;27:847–856. doi: 10.1002/stem.17. [DOI] [PubMed] [Google Scholar]
- 31.Vierbuchen T, Ostermeier A, Pang ZP, Kokubu Y, Sudhof TC, Wernig M. Direct conversion of fibroblasts to functional neurons by defined factors. Nature. 2010;463:1035–1041. doi: 10.1038/nature08797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Chien CT, Hsiao CD, Jan LY, Jan YN. Neuronal type information encoded in the basic-helix-loop-helix domain of proneural genes. Proc Natl Acad Sci USA. 1996;93:13239–13244. doi: 10.1073/pnas.93.23.13239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Cachero S, Simpson TI, Zur Lage PI, Ma L, Newton FG, Holohan EE, et al. The gene regulatory cascade linking proneural specification with differentiation in Drosophila sensory neurons. PLoS Biol. 2011;9:1000568. doi: 10.1371/journal.pbio.1000568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Thomas J, Morle L, Soulavie F, Laurencon A, Sagnol S, Durand B. Transcriptional control of genes involved in ciliogenesis: a first step in making cilia. Biol Cell. 2010;102:499–513. doi: 10.1042/BC20100035. [DOI] [PubMed] [Google Scholar]
- 35.Orkin SH. Diversification of haematopoietic stem cells to specific lineages. Nat Rev Genet. 2000;1:57–64. doi: 10.1038/35049577. [DOI] [PubMed] [Google Scholar]
- 36.Tapscott SJ, Bennett GS, Holtzer H. Neuronal precursor cells in the chick neural tube express neurofilament proteins. Nature. 1981;292:836–838. doi: 10.1038/292836a0. [DOI] [PubMed] [Google Scholar]
- 37.Guillemot F. Spatial and temporal specification of neural fates by transcription factor codes. Development. 2007;134:3771–3780. doi: 10.1242/dev.006379. [DOI] [PubMed] [Google Scholar]
- 38.Lai HC, Klisch TJ, Roberts R, Zoghbi HY, Johnson JE. In vivo neuronal subtype-specific targets of Atoh1 (Math1) in dorsal spinal cord. J Neurosci. 2011;31:10859–10871. doi: 10.1523/JNEUROSCI.0445-11.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Aerts S, Quan XJ, Claeys A, Naval Sanchez M, Tate P, Yan J, et al. Robust target gene discovery through transcriptome perturbations and genomewide enhancer predictions in Drosophila uncovers a regulatory basis for sensory specification. PLoS Biol. 2010;8:1000435. doi: 10.1371/journal.pbio.1000435. [DOI] [PMC free article] [PubMed] [Google Scholar]