Abstract
Homologous recombination (HR)-defective cells, such as those lacking BRCA1/2, are hypersensitive to poly (ADP-ribose) polymerase (PARP) inhibition. However, BRCA-deficient tumors represent only a small fraction of adult cancers, potentially restricting the therapeutic utility of PARP inhibitor monotherapy. We previously showed that cyclin-dependent kinase (cdk)1 phosphorylates BRCA1, an event essential for efficient BRCA1 focus formation. Here, we show that cdk1 depletion or inhibition compromises the cellular capacity to repair DNA by HR. Combined cdk1 and PARP inhibition in BRCA wild-type cancer cells results in reduced colony formation, delayed human tumor xenograft growth and tumor regression with prolonged survival in a mouse lung adenocarcinoma model. Cdk1 inhibition did not sensitize non-transformed cells or tissues to PARP inhibition. Because reduced cdk1 activity impairs BRCA1 function and HR repair, cdk1 inhibition represents a plausible strategy for expanding the utility of PARP inhibitors to the BRCA-proficient cancer population.
Cyclin-dependent kinase (cdk)1 is a core component of the cell cycle machinery, and forms complexes with cyclins A and B to promote S, G2 and M phase progression1–3. Recently, cdk1, as well as other family members, has been shown to participate upstream in DNA damage response pathways4–8. We previously established that the function of BRCA1 in S phase checkpoint control is compromised in cdk1-depleted cells; consequently, cancer cells are sensitized to a range of DNA damaging agents. Cdk1 phosphorylates BRCA1 at S1497 and at the double phosphorylation site S1189/S1191, events necessary for BRCA1 to efficiently form foci at sites of DNA damage and facilitate checkpoint activation8.
BRCA1 is also critical for HR-mediated DNA repair9. BRCA-negative and other HR-deficient cells are highly susceptible to PARP inhibition10–13, a finding now clinically validated14–16. Here, we demonstrate that cdk1 is necessary not only for BRCA1-mediated S phase checkpoint activation, but also for HR repair. Consequently, cdk1-depleted or -inhibited cancer cells are HR-defective and sensitized to PARP inhibition both in vitro and in vivo. Furthermore, we did not observe similar sensitization of non-transformed cells or tissues to PARP inhibitor treatment. Therefore, cdk1 depletion or inhibition creates a state of ‘BRCAness’17 in transformed cells and represents a rational approach for expanding the efficacy of PARP inhibitors to BRCA-proficient cancer populations.
RESULTS
Compromised cdk1 activity reduces HR DNA repair
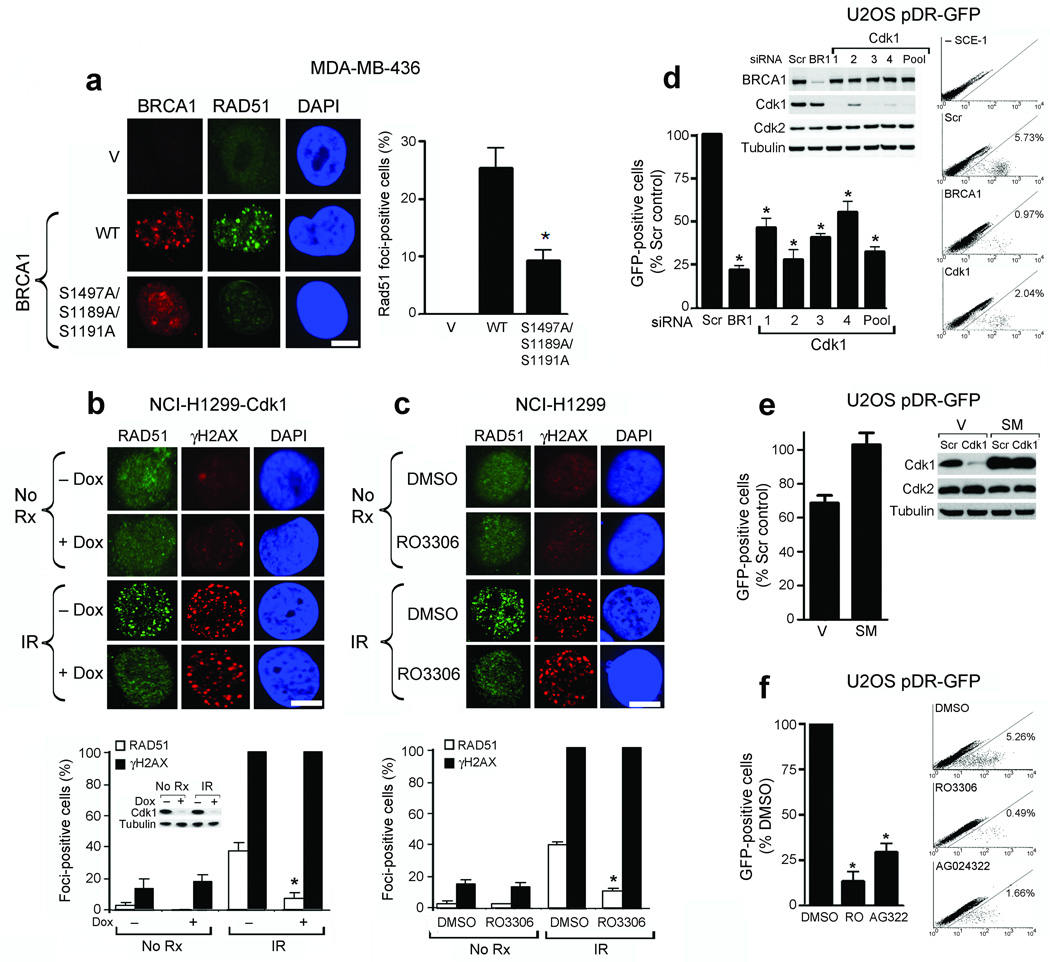
BRCA1-deficient cells do not efficiently form Rad51 foci11,18, a crucial component of the HR repair machinery. We therefore hypothesized that in addition to checkpoint activation8, cdk1-mediated phosphorylation of BRCA1 would also be required for HR DNA repair. We measured the ability of wild-type and S1189A/S1191A/S1497A triple-mutant forms of BRCA1, as well as an empty vector control, to restore Rad51 foci in response to γ-irradiation (IR) in the MDA-MB-436 breast cancer cell line harboring a deleterious BRCA1 mutation19. Rad51 foci could not be detected in parental or empty vector cells under any condition. Compared to cells expressing wild-type BRCA1, there was a 64% reduction (P = 0.015) in formation of Rad51 foci in response to IR in cells expressing the S1189A/S1191A/S1497A mutant (Fig. 1a). Therefore, cdk1-mediated phosphorylation of BRCA1 is required for efficient recruitment of both BRCA1 and Rad51 to sites of DNA damage.
Figure 1.
Cdk1 depletion or inhibition reduces Rad51 focus formation and HR. (a) Detection of BRCA1, Rad51 and DAPI by immunofluorescence after IR in empty vector (V), wild-type (WT) or S1189A/S1191A/S1497A mutant HA-tagged BRCA1-expressing MDA-MB-436 cells. (Left) Representative foci-containing cells; (Right) Mean number of BRCA1- expressing cells with ≥ five Rad51 foci ± standard error (SE) over three experiments. (b) Detection of Rad51, γ-H2AX and DAPI by immunofluorescence in NCI-H1299 cells inducibly expressing shRNA targeting cdk1, untreated or treated with IR ± doxycycline. Western blots demonstrate cdk1 knockdown. (c) NCI-H1299 cells, untreated or treated with IR with DMSO or RO-3306 and stained as in (b). For (b and c): (Upper Panels) Representative foci-containing cells. (Lower Panels) Mean number of cells containing ≥ five Rad51 and γ-H2AX foci ± SE over three experiments. (d) Detection and quantification of GFP-positive U2OS pDR-GFP cells after treatment with scrambled siRNA (Scr), or siRNAs targeting BRCA1 (BR1) or cdk1 (1–4 individual siRNAs and 1–4 pooled). Western blots demonstrate cdk1 knockdown. (e) Quantification of GFP-positive U2OS pDR-GFP cells expressing empty vector (V) or cdk1 containing a silent mutation (SM) after treatment with scrambled siRNA (Scr) or cdk1 siRNA. Western blots demonstrate protein knockdown. (f) Detection and quantification of GFP-positive U2OS pDR-GFP cells after treatment with DMSO, RO-3306 or AG024322. For (d–f), mean ± SE number of GFP-positive cells is expressed as a percentage of scrambled siRNA or DMSO-treated controls over three experiments. *’s indicate statistically significant P values. Scale bars, 10 µM.
To determine whether Rad51 focus formation is also reduced in cdk1 depleted cells, where BRCA1 does not efficiently form foci8, we utilized NCI-H1299 non-small cell lung cancer (NSCLC) cells engineered to inducibly express shRNA targeting cdk1 or cdk2 upon doxycycline exposure20. Cdk1 depletion resulted in an 80% reduction (P = 0.001) in Rad51 focus formation after IR compared to cells with normal cdk1 expression (Fig. 1b). In contrast, cdk2 depletion did not affect Rad51 focus formation (Supplementary Fig. 1). The small molecule cdk1 inhibitor RO-330621 also reduced the focus forming capacity of BRCA1 following DNA damage8. Compared to parental NCI-H1299 cells pre-treated with vehicle, 71% fewer (P = 0.0001) cells pre-treated with RO-3306 efficiently formed Rad51 foci in response to IR (Fig. 1c). Neither cdk1 depletion nor RO-3306 affected the formation of γ-H2AX foci (Fig. 1b,c).
To further assess the impact of cdk1 depletion or inhibition on HR directly, we used a gene conversion assay in which GFP expression indicates the occurrence of HR repair22. Depletion of cdk1 using individual or pooled siRNAs resulted in a 44% (P = 0.0035) to 72% (P = 0.0018) reduction in GFP expression compared to control siRNA-treated U2OS pDR-GFP cells (Fig. 1d). In contrast, siRNA-mediated depletion of cdk2 did not routinely reduce GFP expression (Supplementary Fig. 2). To account for possible ‘off-target’ effects of cdk1 siRNA, we reconstituted U2OS pDR-GFP cells with empty vector or a cdk1 expression construct containing a silent mutation conferring cdk1 siRNA resistance. Compared to control siRNA, cdk1 siRNA resulted in a 32% (P = 0.019) reduction in GFP expression in empty vector containing cells. However, cdk1 siRNA did not reduce exogenous silent mutation-containing cdk1 protein expression and subsequently there was no reduction in GFP expression (Fig. 1e). The small molecule cdk1 inhibitor RO-3306 also reduced GFP expression 87% (P = 0.013) compared to to DMSO-treated control cells. Similar data were obtained with the cdk inhibitor AG02432223 (Fig. 1f), which also preferentially inhibits cdk120 (Supplementary Fig. 3).
Depletion of cdk1 with PARP inhibition results in cell death
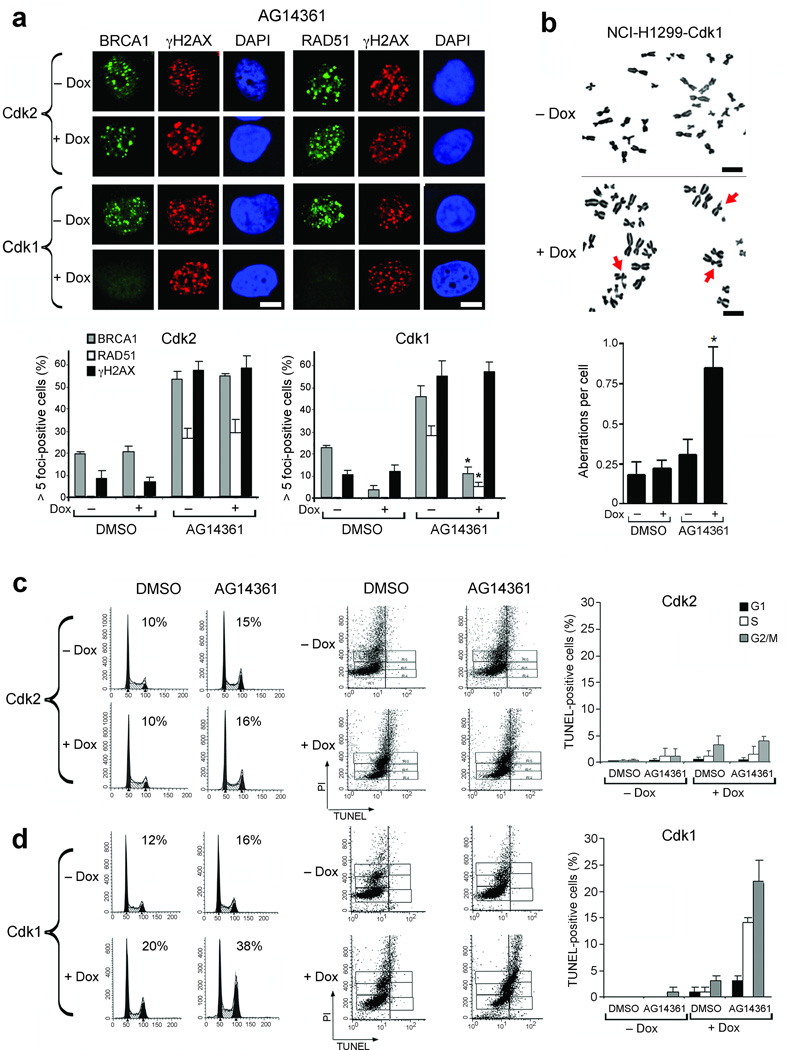
The failure to repair double-strand breaks (DSBs) by HR underlies the PARP inhibitor sensitivity of BRCA1-deficient cells11. We reasoned that cdk1-depleted cells would be similarly sensitive to PARP inhibition. First, we examined the ability of cdk1 depleted cells to recruit BRCA1 and Rad51 to sites of DNA damage after treatment with the PARP-1 and 2 inhibitor AG1436124. Treatment of NCI-H1299 cells expressing normal amounts of cdk1 with AG14361 for 24 hours resulted in DNA DSBs that were repaired by HR, demonstrated by the formation of γ-H2AX, BRCA1 and Rad51 foci. However, as for IR treatment, depletion of cdk1, but not cdk2, caused a 76% (P = 0.0013) and 82% (P = 0.0004) reduction in the number of cells with BRCA1 and Rad51 foci, respectively. Formation of γ-H2AX foci was intact (Fig. 2a). Furthermore, when cdk1-depleted NCI-H1299 cells were treated with AG14361, the number of chromosome aberrations per cell detected by metaphase spread analyses increased by 3.8-fold (P = 0.011) compared with vehicle, or 2.7-fold (P = 0.029) compared to AG14361 treatment in cells with normal cdk1 expression (Fig. 2b). Consequently, after 24 hours AG14361 treatment, cdk1-depleted cells accumulated at the G2/M boundary, in contrast to cells with normal cdk1 expression, or cells depleted of cdk2, which had little change in cell cycle profile (Fig. 2c,d). At later time points after AG14361 treatment, cells with normal cdk1 expression or depleted of cdk2 did not undergo cell death. In contrast, cdk1-depleted cells began to die out of the S and G2/M phases of the cell cycle, as indicated by increased TUNEL-positivity 72 hours after AG14361 treatment (Fig. 2c,d).
Figure 2.
Cdk1 depletion results in reduced Rad51 foci, multiple chromosome aberrations, G2/M accumulation and cell death following PARP inhibition. (a) Detection of BRCA1, Rad51, γ-H2AX and DAPI by immunofluorescence in NCI-H1299 cells inducibly expressing shRNA targeting cdk2 or cdk1 treated with AG14361 ± doxycycline. (Upper Panels) Representative foci-containing cells. (Lower Panels) Mean number of cells containing ≥ foci ± SE over three experiments. (b) Metaphase spread analyses of NCI-H1299-cdk1 cells analyzed for chromosomal breaks after 24 hours treatment with AG14361. (Upper panels) Representative metaphase spreads; chromosomal aberrations are indicated by arrows. (Lower panels) mean number of chromosome aberrations per cell, ± SE over three experiments. For (a and b), *’s indicate statistically significant P values. (c) Cell cycle profiles (left) and detection of TUNEL-positive (middle) NCI-H1299-cdk2 cells treated with DMSO or AG14361 ± doxycycline. Vertical lines indicate the TUNEL-positive threshold. (Right) Mean percent of TUNEL-positive cells in G1, S and G2/M ± SE over three experiments. (d) NCI-H1299-cdk1 cells treated and analyzed as in (c). Scale bars, 10 µM.
Reduced cdk1 activity sensitizes to PARP inhibition
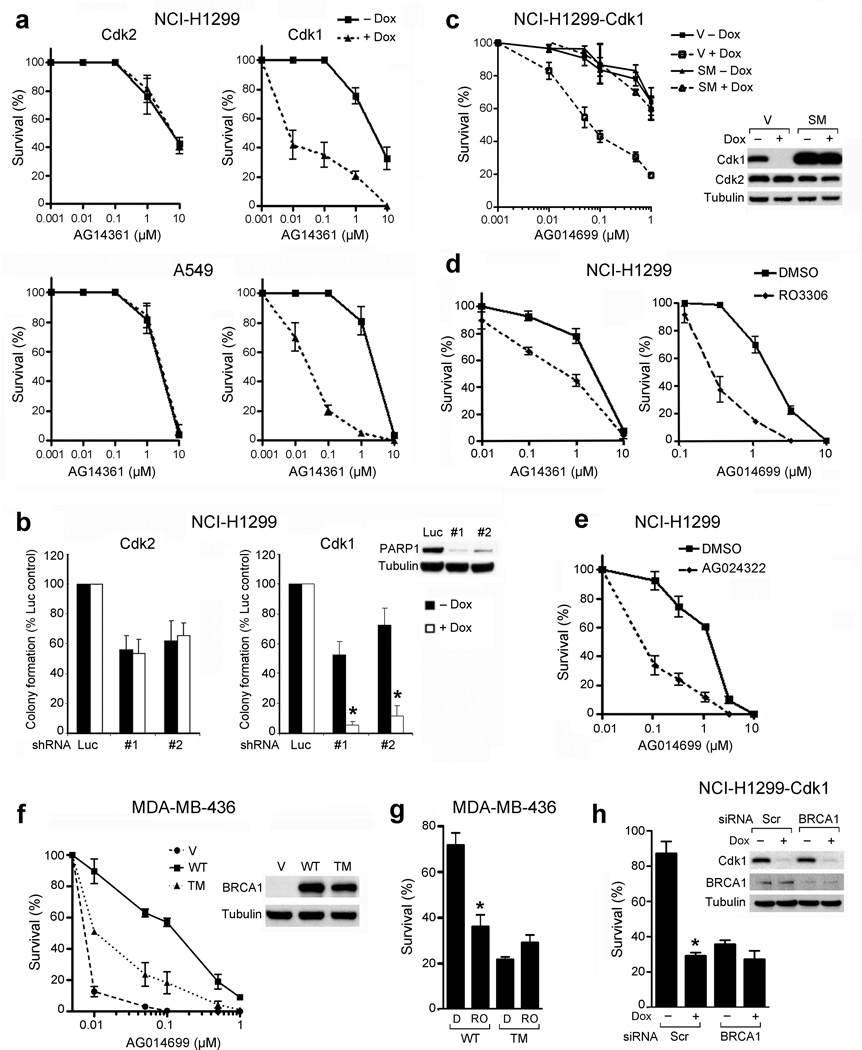
We next examined whether cdk1 depletion could sensitize NSCLC cells to PARP inhibition in long-term colony assays. NCI-H1299-cdk1 and A549-cdk1 cells were 220-fold (P = 0.006) and 110-fold (P = 0.0019) more sensitive to AG14361 (representing 99.5% and 99.1% reductions in AG14361 LC50, respectively), in the presence compared to the absence of doxycycline, whereas cdk2 depletion did not sensitize these cells (Fig. 3a). In addition, multiple cdk1, but not cdk2, siRNA constructs sensitized NCI-H1299 cells to treatment with AG014699, a newer generation PARP inhibitor currently in clinical trial25,26 (Supplementary Fig. 4), and shRNA-mediated depletion of PARP-1 from NCI-H1299 cells only demonstrated substantial reduction in colony formation when cdk1 was concomitantly depleted (Fig. 3b). Furthermore, to account for possible cdk1 shRNA ‘off-target’ effects, we engineered NCI-H1299-cdk1 shRNA inducible cells to express an empty vector or an exogenous cdk1 protein containing a silent mutation that confers resistance to cdk1 targeting shRNA. In empty vector-containing cells, the addition of doxycycline resulted in a 97.8% (P = 0.0066) reduction in the LC50 value of AG014699, compared to cells grown in the absence of doxcycline. In contrast, the presence of doxycyline did not sensitize cells expressing a cdk1 protein containing a silent mutation to AG014699 treatment (Fig. 3c). The effects of cdk1 knockdown were replicated with small molecule cdk1 inhibitors. RO-3306 reduced AG14361 and AG014699 LC50 by 82% (P = 0.001) and 84% (P = 0.0012), respectively (Fig. 3d). Additionally, the degree of RO-3306-mediated cdk1 inhibition correlated with the degree of sensitization to PARP-1 inhibition (Supplementary Fig. 5). AG024322 also reduced the AG014699 LC50 value by 95% (P = 0.0001) (Fig. 3e).
Figure 3.
Cdk1-depleted or -inhibited cells are highly sensitive to PARP inhibition. (a) Colony formation of NCI-H1299 and A549 cells inducibly expressing shRNAs targeting cdk2 (left) or cdk1 (right) treated with AG14361 ± doxycycline. (b) Colony formation of NCI-H1299-cdk2 or -cdk1 cells treated with shRNA targeting luciferase or PARP-1 ± doxycycline. Western blot demonstrates PARP-1 knockdown. *, P ≤ 0.0011. (c) Colony formation of NCI-H1299-cdk1 cells expressing empty vector (V) or cdk1 containing a silent mutation (SM) treated with AG014699 ± doxycycline. Western blot demonstrates cdk1 knockdown. (d) Colony formation of NCI-H1299 cells treated with DMSO or RO-3306 and AG14361 or AG014699. (e) Colony formation of NCI-H1299 cells treated with DMSO or AG024322 and AG014699. (f) Colony formation of MDA-MB-436 cells expressing empty vector (V), wild-type (WT) or S1189A/S1191A/S1497A (TM) BRCA1 treated with AG014699. Western blot demonstrates BRCA1 protein expression. (g) Colony formation of MDA-MB-436 cells expressing WT or S1189A/S1191A/S1497A mutant BRCA1 treated with DMSO + AG014699 (D) or RO-3306 + AG014699 (RO). Survival is expressed as a percentage of colonies formed ± SE compared to the corresponding DMSO or RO-3306-treated control. *, P = 0.0094, comparing RO-3306 + AG014699 to DMSO + AG014699 in WT cells. (h) Colony formation of NCI-H1299-cdk1 cells transfected with either scrambled (Scr) or BRCA1 siRNA, treated with vehicle or AG014699 ± doxycycline. Survival is expressed as a percentage of colonies formed ± SE compared to the corresponding vehicle-treated control. *, P = 0.0154 for scrambled siRNA, comparing the presence to absence of doxycycline.
We further investigated the mechanism by which cdk1 depletion sensitizes cells to PARP inhibition and assessed the ability of wild-type and S1189A/S1191A/S1497A triple-mutant forms of BRCA1 to render the MDA-MB-436 cell line19 PARP inhibitor-resistant. MDA-MB-436 cells containing an empty vector construct were highly sensitive to AG014699 treatment (LC50 0.006 µM). When cells expressed wild-type BRCA1, the LC50 value for AG014699 treatment increased 32-fold (P = 0.0018) compared to empty vector cells. In contrast, when cells expressed the triple mutant form of BRCA1, the LC50 value increased 5-fold (P = 0.001) compared to empty vector cells (Fig. 3f). Furthermore, when MDA-MB-436 cells were treated concomitantly with RO-3306 with AG014699, cells reconstituted with wild-type, but not triple mutant BRCA1 were sensitized to AG014699 treatment (Fig. 3g). Additionally, if reduced cdk1 activity sensitizes cells to PARP inhibition primarily through abrogation of BRCA1 function, then cdk1 depletion should not further sensitize BRCA1-deficient cells. In the absence of doxycycline, BRCA1 depletion sensitized NCI-H1299 cells to AG014699 treatment to a similar degree as doxycycline-induced cdk1 depletion. However, there was no further reduction in colony formation after AG014699 treatment in cells that were depleted of BRCA1 and cdk1 together (Fig. 3h).
Non-transformed cells are not sensitized to PARP inhibition
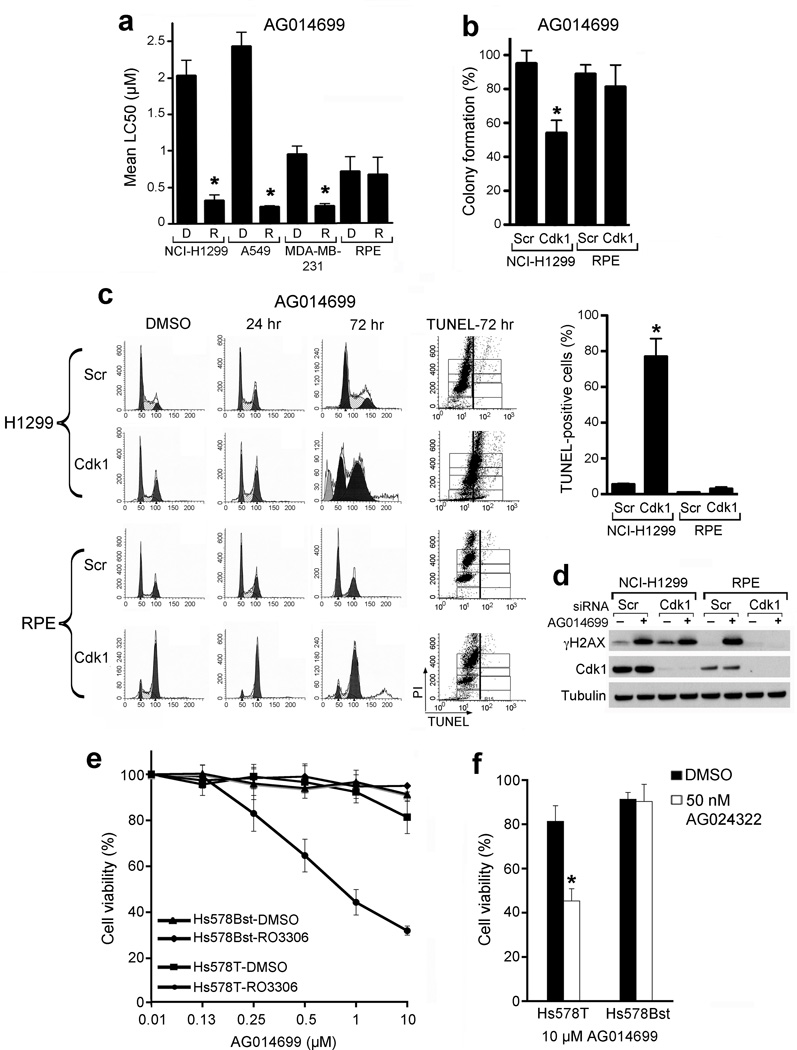
In addition to NCI-H1299, colony formation was significantly reduced in A549 and MDA-MB-231 cell lines concomitantly treated with AG014699 and RO-3306 compared to AG014699 treatment alone (Fig. 4a). In contrast to transformed cells, non-transformed Retinal Pigment Epithelial (RPE) cells were less sensitive to combined RO-3306 and AG014699 (Fig. 4a) or cdk1 siRNA and AG014699 treatment than cancer cell lines (Fig. 4b). Unlike NCI-H1299 cells, cdk1 depletion resulted in potent and prolonged G2/M cell cycle arrest in RPE cells. Subsequently, RPE cells were not exposed to PARP inhibitor mediated S-phase specific DNA damage and few TUNEL-positive cells could be detected (Fig. 4c). AG014699 treatment resulted in an increase in γ-H2AX in control siRNA-treated, but not in the G2/M-accumulated cdk1-depleted RPE cells (Fig. 4d). Furthermore, we treated Hs578T breast cancer cells and a non-transformed breast epithelial cell line derived from the same patient, Hs578Bst27, with RO-3306 and AG014699. Only Hs578T cells were sensitized to AG014699 by RO-3306 (Fig. 4e). Similar data were obtained with the cdk inhibitor AG024322 (Fig. 4f).
Figure 4.
Cdk1 depletion or inhibition protects non-transformed cells from PARP inhibitor treatment. (a) Colony formation of cell lines treated with DMSO or RO-3306 and AG014699. Mean ± SE LC50 values for AG014699 from DMSO (D)- or RO-3306 (R)-treated cells. *, P ≤ 0.003. (b) Colony formation of RPE and NCI-H1299 cells treated with scrambled siRNA (Scr) or cdk1 siRNA prior to treatment with AG014699 for 72 hours followed by replating for colony formation. *, P = 0.018. (c) Cell cycle profiles (left) and TUNEL-positive (middle) NCI-H1299 cells transfected with scrambled siRNA (Scr) or cdk1 siRNA for 48 hours, and subsequently treated with DMSO or AG014699 for the indicated times. (Right) Mean percent of TUNEL-positive cells ± SE at 120 hrs over three experiments. *, P = 0.0191. (d) Western blot analyses of cells treated as in (c) after 24 hours AG014699 treatment. (e) Viability of HS578T and HS578Bst cell lines treated with DMSO or RO-3306 and increasing concentrations of AG014699. (f) Viability of HS578T and HS578TBst cell lines treated with DMSO or AG024322 and AG014699. *, P = 0.0023.
Compromise of cdk1 and PARP activities delays tumor growth
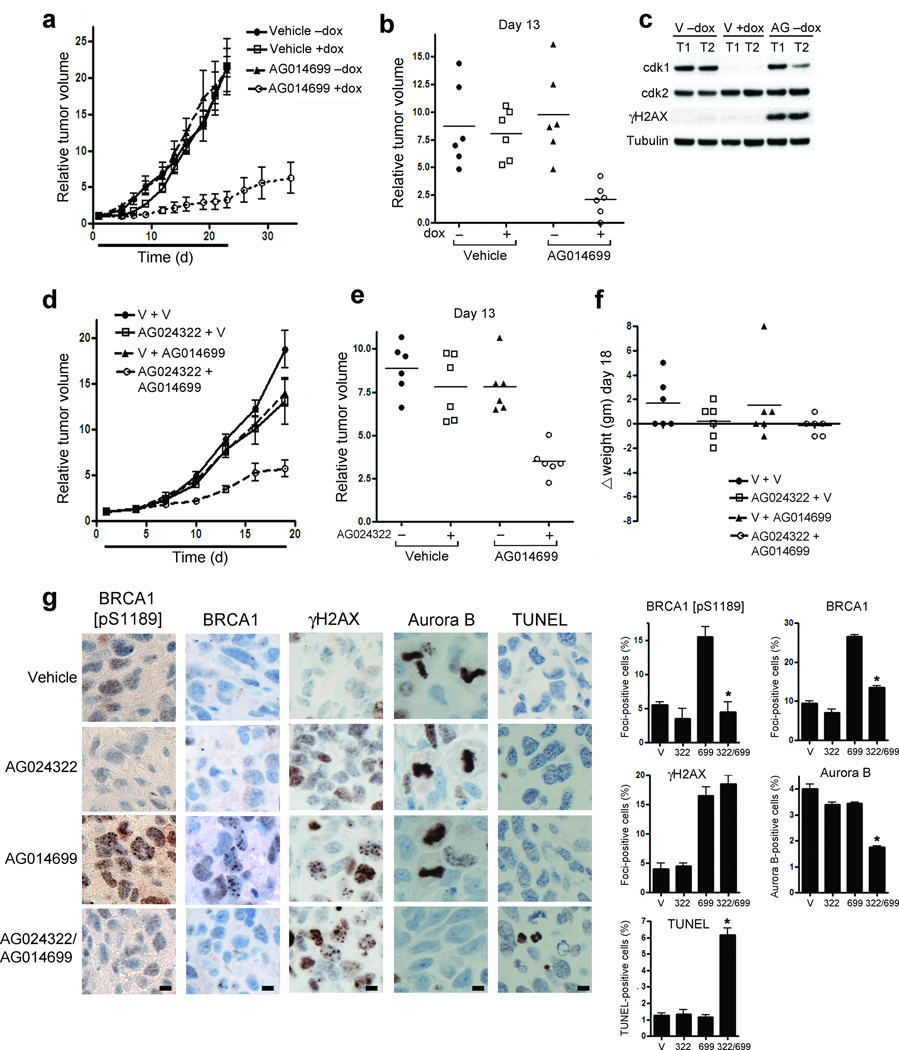
To measure the efficacy of cdk1 and PARP inhibitor combinations in vivo, xenografts of NCIH1299 cells inducibly expressing shRNA targeting cdk1 upon exposure to doxycycline were established in athymic nu/nu mice. Mice were subsequently fed either normal or doxycycline-containing diets and treated for 23 days with either vehicle or AG014699. Neither doxycycline nor AG014699 alone affected xenograft growth. However, when mice were fed doxycycline-containing food and treated with AG014699, tumor growth was substantially delayed (Fig. 5a). The mean relative tumor volume (RTV) of mice on doxycycline-containing diets and treated with AG014699 was 80% smaller (P = 0.0013) than of mice treated with AG014699 without doxycycline (Fig. 5b). Cdk1 knockdown in xenografts was confirmed by western blot. Additionally, AG014699 treatment resulted in increased γ-H2AX in tumors (Fig. 5c).
Figure 5.
Cdk1 inhibition sensitizes cancer cells to PARP inhibition in vivo. (a) Growth of NCI-H1299-cdk1 xenografts in mice administered regular or doxycycline-containing diets, treated with vehicle or AG014699 over days 1–23. Mean relative tumor volume (RTV) (n=6), is expressed compared to tumor volumes on day 1. (b) RTV for individual mice treated in (a) at day 13. (c) Cdk1, cdk2, γ-H2AX and tubulin protein expression from representative tumors in mice sacrificed on day 23 measured by western blot. (d) Growth of NCI-H1299 xenografts over 19 days in mice receiving the indicated treatments. V, vehicle. Tumor size is presented as the mean tumor volume (n = 6) relative to day 1. (e) RTV for individual mice treated in (d) at day 13. (f) Change in weight of individual mice treated as in (d) at day 18. (g) Immunohistochemical analysis for BRCA1 [pS1189], total BRCA1 and γ-H2AX focus formation, and Aurora B and TUNEL staining in NCI-H1299 xenografts harvested from mice treated for 5 continuous days with the indicated treatments. (Left) Representative sections, stained as indicated. (Right) mean ± SE foci- or staining-positive cells. *’s indicate statistically significant P values. Scale bars, 10 µM.
RO-3306 is rapidly cleared from plasma, so that drug concentrations necessary to inhibit cdk1 cannot be sustained (data not shown). To test the combination of systemic cdk and PARP inhibition, we utilized AG024322, which has suitable pharmacokinetic properties for in vivo studies23. Mice bearing NCI-H1299 xenografts were treated with vehicle or AG024322 followed by vehicle or AG014699 daily for 18 days. Individually, AG024322 and AG014699 had modest effects on tumor growth. However, in mice treated with both compounds, tumor growth was significantly delayed (Fig. 5d), with a 60% (P = 0.0005) 55% (P = 0.02) and 55% (P = 0.0007) reduction in RTV at day 13 in mice treated with both AG024322 and AG014699 compared to mice treated with vehicle, AG024322 alone or AG014699 alone, respectively (Fig. 5e). Additionally, the combination of AG024322 and AG014699 did not induce significant weight loss over the treatment time course (Fig. 5f).
We next assessed pharmacodynamic markers of AG024322 and AG014699 activity in tumor xenografts. AG014699-mediated PARP inhibition resulted in an increase in both BRCA1 [pS1189] and total BRCA1 foci-containing cells. However, after combined AG024322 and AG014699 treatment, the percentages of BRCA1 [pS1189] and total BRCA1 foci-containing cells were reduced 71% (P = 0.035) and 49% (P = 0.0029), respectively, compared to AG014699 treatment alone. In contrast, AG024322 did not affect the percentage of γ-H2AX foci-containing cells. Furthermore, only the combination of AG024322 and AG014699 significantly reduced the mitotic count (56% reduction compared to vehicle treated tumors; P = 0.0083) while increasing the apoptotic count (4.96-fold compared to vehicle treated tumors; P = 0.0092), measured by Aurora B and TUNEL staining, respectively (Fig. 5g).
Combined cdk1 and PARP inhibition prolongs survival
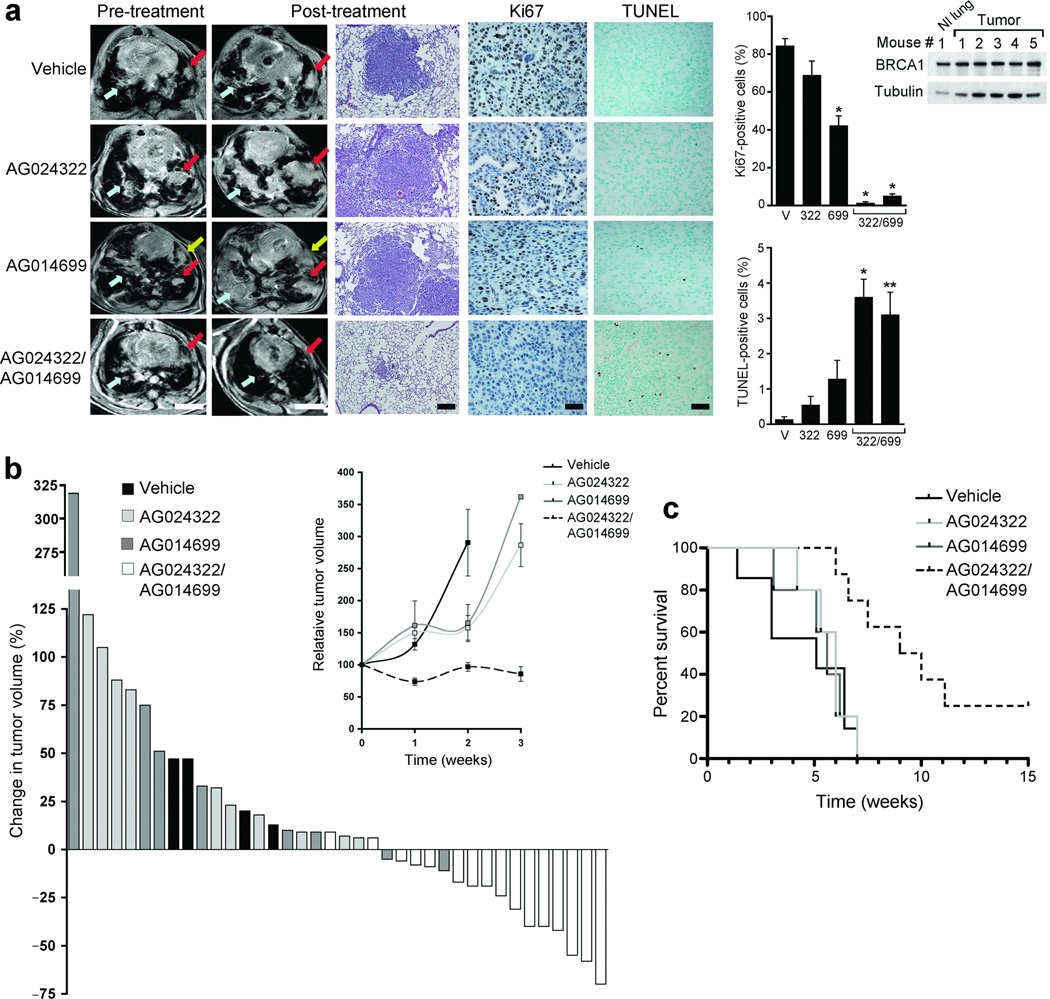
We further assessed the therapeutic efficacy of combined cdk1 and PARP inhibition in the KrasG12D p53L/L mouse lung adenocarcinoma model28. BRCA1 expression in these lung tumors was validated by western blot (Fig. 6a). Mice treated for 1 week with vehicle or AG024322 all demonstrated tumor growth by magnetic resonance imaging (MRI). Similarly, only 2 out of 8 (25%) mice treated with AG014699 had a small reduction in tumor volume after one week of treatment (up to 11% regression). In contrast, 14 out of 16 (87.5%) treated for 1 week with both AG024322 and AG014699 demonstrated tumor volume reduction (up to 70% regression) (Fig. 6a,b). At 3 weeks of treatment, all mice treated with vehicle, AG024322 or AG014699 alone had substantial tumor growth. However, 9 out of 13 (69%) mice treated with combined AG024322 and AG014699 maintained reduced tumor volume (up to 82% regression) (Fig. 6b and Supplementary Fig. 6). By 6 weeks, 2 mice treated with combined AG024322 and AG014699 continued to have low tumor volume (Supplementary Fig. 6). Combined AG024322 and AG014699 treatment resulted in a marked reduction in Ki67 staining and increased TUNEL staining in residual tumor compared to vehicle or individual treatments (Fig. 6a). Kaplan-Meier analyses indicated that the median survival of mice treated with both AG024322 and AG014699 increased by 86% (P = 0.0014), 58% (P = 0.0031) and 70% (P = 0.0027) compared to that of mice treated with vehicle, AG024322 alone or AG014699 alone, respectively (Fig 6c). Of note, the 2 mice treated with both AG024322 and AG014699 with reduced tumor volume at 6 weeks were alive at 15 weeks post-treatment (Fig. 6c) with response maintained. No toxicity or damage to normal mouse tissues and organs was found after 1, 2 or 4 weeks of combination treatment by pathologic assessment (Supplementary Fig. 7 and 8).
Figure 6.
Combined inhibition of cdk1 and PARP causes tumor regression and prolongs survival in the KrasG12D p53L/L mutant lung cancer mouse model. (a) (Left) Representative MRI images of lung tumor volumes before and after one week of the indicated treatments. Colored arrows show matched lesions in the pre- and post-treatment images; scale bars, 4.5 mm. (Middle) Representative H and E stains (scale bars, 500 µM), as well as Ki67 and TUNEL staining of tumors after the indicated treatments (scale bars, 100 µM). Graphs show the mean ± SE number of positive cells; results for two mice treated with both AG024322 and AG014699 are shown. *, P ≤ 0.0002; **, P < 0.002 of treatment compared to vehicle. (Right) Western blot demonstrates BRCA1 expression in mouse normal lung or tumor tissue. (b) Waterfall plot showing percent change in tumor volume after 1 week of treatment measured by MRI compared to start of treatment for each mouse. (Inset) Mean relative tumor volume ± SE over the first 3 weeks of treatment for mice treated as indicated. At 1 and 3 weeks, each data point represents the average of 4–16 mice; at 2 weeks, 2–4 mice in each group were analyzed. At 3 weeks, the SE for mice treated with AG014699 was ± 171.2, based on one tumor that was increased by > 900-fold. (c) Kaplan-Meier analyses demonstrating median survival times from the start of treatment of mice treated with vehicle, AG024322, AG014699 or both of 5.1, 6, 5.6 and 9.5 weeks, respectively.
DISCUSSION
We previously demonstrated that cdk1 depletion or inhibition in lung cancer cells reduces BRCA1 focus formation and the activation of DNA damage-induced checkpoint control8. We have now implicated cdk1 in HR repair in these cells. In response to PARP inhibition, reduced cdk1 activity results in chromosomal aberrations and cell death, in agreement with previous studies demonstrating that HR-deficient cells are hypersensitive to PARP inhibitor therapy10–12. Furthermore, cdk1 was previously identified in an siRNA library screen designed to identify proteins that when depleted cause sensitivity to PARP inhibitors29. In contrast to cdk1, cdk2 phosphorylates BRCA2, impairing its interaction with Rad51, thereby limiting HR until cell cycle arrest is accomplished and cdk2 activity extinguished30. Consistent with these data, depletion of cdk2 did not significantly reduce HR in the cell lines examined, and in several instances, increased the percentage of GFP-positive cells in the gene conversion assay.
In yeast, cdk1 is essential for multiple steps of HR4. Although cdk1 may directly influence the function of other HR proteins, it is likely that reduced cdk1 activity sensitizes cells to PARP inhibition through disruption of BRCA1 function in lung cancer cells. Cdk1 depletion afforded an increase in sensitivity to PARP inhibition by >100-fold, similar to what is seen in BRCA1-deficient cells11, and combined depletion of cdk1 and BRCA1 did not sensitize cells to a greater degree than depletion of either alone. Additionally, we previously showed that selective cdk1 inhibition does not affect DNA-end resection in these cells, likely because cdk2 can compensate in this process; such compensation does not occur at the level of BRCA1 focus formation8.
Our in vitro observations were translated in xenograft models, where cdk1 inhibition resulted in a reduction in the PARP inhibitor-mediated increase in BRCA1 but not γ-H2AX foci-containing cells. We also studied mice with lung-specific conditional activating Kras and inactivating p53 mutations that develop highly aggressive lung adenocarcinomas with short latency compared to those driven by KrasG12D alone28,31. KrasG12D tumors with concomitant p53 inactivation are also less responsive to cytotoxic therapy than those with wild-type p5332. The cdk and PARP inhibitor combination induced regression and disease stabilization over 1–3 weeks of treatment in established tumors. Although resistance was documented by 6 weeks, a subset of mice demonstrated sustained response, so that combination therapy significantly increased median survival. Our results suggest a possible approach to lung cancers harboring this genotype that frequently have poor outcomes33, as well as to other BRCA-proficient tumors.
Importantly, mice treated with both AG024322 and AG014699 had no organ or normal tissue toxicities. In accordance with these observations, cdk1 depletion or inhibition did not sensitize RPE cells or non-transformed HS578TBst breast epithelial cells to PARP inhibition in vitro. Of note, cdk2 cannot compensate for the loss of cdk1 in cellular proliferation in non-transformed cells to the same degree as in cancer cells34; consequently, RPE cells were arrested at G2/M when cdk1 was depleted. Following PARP inhibition, single-strand breaks (SSBs) degenerate to DSBs during S phase traversal; non-transformed cells, arrested in G2/M after cdk1 depletion, likely do not accumulate SSBs followed by DSBs, demonstrated by a failure to accumulate γ-H2AX, and are therefore not sensitive to combined cdk1 and PARP inhibition. The data therefore suggest that cdk and PARP inhibitor combinations will have a therapeutic window.
Our data support the clinical development of combined cdk1 and PARP inhibition. Analysis of cdk-mediated BRCA1 phosphorylation suggests that 70–90% reduction in cdk1 activity by small molecule inhibitors results in sensitization to PARP inhibition in vitro, translating to substantial anti-tumor activity in vivo, and serves as a guide for the target degree of inhibition desirable in clinical trials.
In summary, the present study is the first to use targeted kinase inhibition to inactivate BRCA1, handicap the HR DNA repair machinery and selectively sensitize transformd cells to PARP inhibition. This approach avoids the use of toxic DNA damaging chemotherapeutic drugs, thus providing the potential to extend well tolerated PARP inhibition to treatment for BRCA-proficient cancers.
Methods
Cell lines and reagents
We obtained cell lines from the ATCC. We pretreated cells expressing shRNA for cdk1 or cdk2 with 5 µg/mL doxycycline for 3 days to achieve cdk knockdown8,20, and used RO-3306 (Calbiochem) at selective cdk1 inhibitory concentrations, ranging from 0.4–2 µM, depending on exposure time. We purchased cdk1 and cdk2 constructs 1–4 (individual and pooled) from Dharmacon. We introduced silent G129T and T135C mutations (Agilent) into a cdk1 cDNA pENTR(tm)221 expression construct (Invitrogen) that provided resistance to siRNA targeting cdk1 (GGGGTTCCTAGTACTGCAA) (Qiagen) Pfizer provided AG14361, AG014699 (PF-01367338) and AG024322 (PF-00176275).
Colony formation, cell viability assays and western blotting
Cells maintained in doxycycline, 0.8 µM RO-3306, 50 nM AG024322 or treated with siRNA were replated at 1×104 in a 10-cm dish with AG14361 or AG014699 for 2 weeks before counting colonies. Mean survival from three experiments was expressed as percentage of colonies ± SE (standard error) relative to vehicle-treated cells in the absence or presence of either doxycycline, siRNA or cdk inhibitor. For shRNA experiments, we treated cells ± doxycycline with shRNA targeting PARP-1 or luciferase. Mean ± SE survival is expressed as the percentage of luciferase shRNA-treated control colonies. LC50 indicates the value at which colony formation was reduced by 50% of that of control-treated cells. We seeded HS578T and HS578TBst cells at 5,000 well−1 (96-well plate), cultured in the presence of drugs or vehicle for 6 days, and performed the CCK-8 colorimetric assay (Dojindo). Western blot analysis and antibodies utilized are previously described8,20.
Immunofluoresence and focal microscopy
Preparation of cells is previously described8,35. BRCA1, γ-H2AX [pS139] (Upstate Biotechnology) and Rad51 (Santa Cruz Biotechnology) antibodies were followed by secondary antibodies conjugated to FITC or Texas Red (Jackson ImmunoResearch Laboratories). We acquired confocal immunofluorescence images using Andor iQ software. For IR experiments, we fixed cells 4 hours after treatment with 10 Gy. For metaphase spreads, we exposed cells for 2 hours to colcemid, harvested, and stained with Wright’s stain. We scored fifty metaphase spreads for aberrations, captured using CytoVision software (Applied Imaging).
Fluorescence-activated cell sorting analysis and detection of apoptosis and GFP
Cell cycle and apoptosis analyses are previously described8. For measurement of HR, we transfected U2OS pDR-GFP cells with an SCE-1 cutting enzyme22 for 72 hours and analyzed for GFP expression by flow cytometry using CellQuest software (BD Biosciences).
Studies of xenograft-bearing and KrasG12D p53L/L mice
The Harvard Medical Area Standing Committee on Animals approved experiments utilizing xenografts and the genetically engineered mouse model. For xenograft studies, we subcutaneously implanted 0.5 × 106 cells (1:1 in matrigel; BD Biosciences) in female nu/nu nude mice on both flanks. Two weeks later, mice bearing NCI-H1299-cdk1 xenografts received either doxycycline-containing or normal diets. After tumors reached 100–200 mm3, animals were randomized to treatment with vehicle or AG014699 (10 mg kg−1), by intraperitoneal injection (IP) daily for 23 days. We treated mice bearing parental NCI-H1299 xenografts IP with vehicle, AG024322 (10 mg kg–1), AG014699 (10 mg kg–1) or both daily for 19 days. Tumor volume by caliper measurement was formulated as (length × width2)/2. Growth curves were plotted as the mean relative tumor volume (RTV) for each group; RTV indicates change in tumor volume at a given time point relative to that at initial dosing (= 1).
We treated KrasG12D p53L/L mice with 5 × 106 p.f.u. adeno-Cre (U. Iowa) intranasally28 and imaged by MRI 8–9 weeks later. Animals with similar tumor volumes received vehicle, AG024322 (18 mg kg–1), AG014699 (25 mg kg–1) or both drugs. MRI measurements were performed as described previously36. On each image, areas indicating tumor were manually segmented and measured to calculate tumor volumes using NIH ImageJ (version 1.33; http://rsb.info.nih.gov/ij/). Tumor volume at the beginning of treatment was considered 100%. Median survival was determined using Kaplan-Meier analyses (GraphPad Prism Software).
Histologic and immunohistochemical staining
We treated mice bearing NCI-H1299 xenografts with vehicle, AG024322, AG014699 or both for 5 days. We stained formalin-fixed paraffin-embedded sections of harvested xenografts with p[S1189]BRCA1 (Novus, Littleton, Colorado), BRCA1, γ-H2AX [pS139]8, TUNEL (ApopTag Peroxidase In Situ Apoptosis Detection Kit; Millipore) and Aurora B (Abcam) antibodies. At least 2 xenografts, each with at least five 40X fields, were scored for each treatment. For BRCA1 or γ-H2AX [pS139], cells containing ≥ 5 foci were considered positive. For KrasG12D p53L/L mice, tumor and surrounding lung tissue from one mouse from each treatment group was harvested 1 or 2 weeks post-treatment and stained with H & E and for Ki67 (Dako) and TUNEL. At least five 40X fields were scored. The mean (± SE) percentage positive cells from 5 images in each treatment group was calculated.
Statistical analysis
P < 0.05 was considered statistically significant in two-tailed, unpaired Student’s t tests. *’s indicate statistically significant P values.
Supplementary Material
Acknowledgments
This work was supported by US National Institutes of Health (NIH) Grants R01 CA090687 (G.I.S.), P50 CA089393 [Dana-Farber/Harvard Cancer Center (DF/HCC) Specialized Program of Research Excellence (SPORE) in Breast Cancer], including Developmental Project Funding (G.I.S.) and a Career Development Award (N.J.), as well as Susan G. Komen Post-Doctoral Fellowship Award KG080773 (N.J., G.I.S. and N.J.C.) and NIH Grant P50 CA090578 (DF/HCC SPORE in Lung Cancer) (G.I.S., K.-K.W.). H.D.T., N.J.C. and D.R.N. were supported by a Programme Grant from Cancer Research U.K. K.-K.W. was also supported by a Uniting Against Lung Cancer grant and NIH grants U01 CA141576, R01 AG2400401, R01 CA122794, R01 CA140594 and 1RC2 CA147940-01. We thank Mansi Arora and Jeffrey D. Parvin of the Department of Biomedical Informatics and the Comprehensive Cancer Center of Ohio State University for assistance with preliminary experiments. We thank Donna Skinner of the Dana-Farber/Harvard Cancer Center Research Pathology Core and members of the Confocal and Light Microscopy Core Facility at the Dana-Farber Cancer Institute for technical assistance with tissue block and slide preparation and acquisition of confocal microscopy images, respectively. We also thank Maria Jasin (Memorial Sloan-Kettering Cancer Center) for U2OS pDR-GFP cells and Kenneth Hook, Don Madren and Zdenek Hostomsky of Pfizer for supplying AG14361, AG014699 (PF-01367338) and AG024322 (PF-00176275).
Footnotes
Author Contributions
N.J. and G.I.S. designed this study. N.J., Y-C.L., L.A.M., D.L., Z.W., C.U., R.T.B., and H.D.T. performed the experiments. N.J. and G.I.S. analyzed the data. N.J. and G.I.S. communicated with S.J.R., K.K.W., D.R.N., A.D.A. and N.J.C. about the data interpretation and wrote the manuscript. G.I.S. supervised the project.
Competing Financial Interests
The authors declare no competing financial interests.
References
- 1.Shapiro GI. Cyclin-dependent kinase pathways as targets for cancer treatment. J Clin Oncol. 2006;24:1770–1783. doi: 10.1200/JCO.2005.03.7689. [DOI] [PubMed] [Google Scholar]
- 2.Malumbres M, Barbacid M. Cell cycle, CDKs and cancer: a changing paradigm. Nat Rev Cancer. 2009;9:153–166. doi: 10.1038/nrc2602. [DOI] [PubMed] [Google Scholar]
- 3.Hochegger H, et al. An essential role for Cdk1 in S phase control is revealed via chemical genetics in vertebrate cells. J Cell Biol. 2007;178:257–268. doi: 10.1083/jcb.200702034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ira G, et al. DNA end resection, homologous recombination and DNA damage checkpoint activation require CDK1. Nature. 2004;431:1011–1017. doi: 10.1038/nature02964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Jazayeri A, et al. ATM- and cell cycle-dependent regulation of ATR in response to DNA double-strand breaks. Nat Cell Biol. 2006;8:37–45. doi: 10.1038/ncb1337. [DOI] [PubMed] [Google Scholar]
- 6.Tian B, Yang Q, Mao Z. Phosphorylation of ATM by Cdk5 mediates DNA damage signalling and regulates neuronal death. Nat Cell Biol. 2009;11:211–218. doi: 10.1038/ncb1829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Myers JS, Zhao R, Xu X, Ham AJ, Cortez D. Cyclin-dependent kinase 2 dependent phosphorylation of ATRIP regulates the G2-M checkpoint response to DNA damage. Cancer Res. 2007;67:6685–6690. doi: 10.1158/0008-5472.CAN-07-0495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Johnson N, et al. Cdk1 participates in BRCA1-dependent S phase checkpoint control in response to DNA damage. Mol Cell. 2009;35:327–339. doi: 10.1016/j.molcel.2009.06.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Moynahan ME, Chiu JW, Koller BH, Jasin M. Brca1 controls homology-directed DNA repair. Mol Cell. 1999;4:511–518. doi: 10.1016/s1097-2765(00)80202-6. [DOI] [PubMed] [Google Scholar]
- 10.Bryant HE, et al. Specific killing of BRCA2-deficient tumours with inhibitors of poly(ADP-ribose) polymerase. Nature. 2005;434:913–917. doi: 10.1038/nature03443. [DOI] [PubMed] [Google Scholar]
- 11.Farmer H, et al. Targeting the DNA repair defect in BRCA mutant cells as a therapeutic strategy. Nature. 2005;434:917–921. doi: 10.1038/nature03445. [DOI] [PubMed] [Google Scholar]
- 12.McCabe N, et al. Deficiency in the repair of DNA damage by homologous recombination and sensitivity to poly(ADP-ribose) polymerase inhibition. Cancer Res. 2006;66:8109–8115. doi: 10.1158/0008-5472.CAN-06-0140. [DOI] [PubMed] [Google Scholar]
- 13.Ashworth A. A synthetic lethal therapeutic approach: poly(ADP) ribose polymerase inhibitors for the treatment of cancers deficient in DNA double-strand break repair. J Clin Oncol. 2008;26:3785–3790. doi: 10.1200/JCO.2008.16.0812. [DOI] [PubMed] [Google Scholar]
- 14.Fong PC, et al. Inhibition of poly(ADP-ribose) polymerase in tumors from BRCA mutation carriers. N Engl J Med. 2009;361:123–134. doi: 10.1056/NEJMoa0900212. [DOI] [PubMed] [Google Scholar]
- 15.Tutt A, et al. Oral poly(ADP-ribose) polymerase inhibitor olaparib in patients with BRCA1 or BRCA2 mutations and advanced breast cancer: a proof-of-concept trial. Lancet. 2010;376:235–244. doi: 10.1016/S0140-6736(10)60892-6. [DOI] [PubMed] [Google Scholar]
- 16.Audeh MW, et al. Oral poly(ADP-ribose) polymerase inhibitor olaparib in patients with BRCA1 or BRCA2 mutations and recurrent ovarian cancer: a proof-of-concept trial. Lancet. 2010;376:245–251. doi: 10.1016/S0140-6736(10)60893-8. [DOI] [PubMed] [Google Scholar]
- 17.Turner N, Tutt A, Ashworth A. Hallmarks of 'BRCAness' in sporadic cancers. Nat Rev Cancer. 2004;4:814–819. doi: 10.1038/nrc1457. [DOI] [PubMed] [Google Scholar]
- 18.Bhattacharyya A, Ear US, Koller BH, Weichselbaum RR, Bishop DK. The breast cancer susceptibility gene BRCA1 is required for subnuclear assembly of Rad51 and survival following treatment with the DNA cross-linking agent cisplatin. J Biol Chem. 2000;275:23899–23903. doi: 10.1074/jbc.C000276200. [DOI] [PubMed] [Google Scholar]
- 19.Elstrodt F, et al. BRCA1 mutation analysis of 41 human breast cancer cell lines reveals three new deleterious mutants. Cancer Res. 2006;66:41–45. doi: 10.1158/0008-5472.CAN-05-2853. [DOI] [PubMed] [Google Scholar]
- 20.Cai D, Latham VM, Jr, Zhang X, Shapiro GI. Combined depletion of cell cycle and transcriptional cyclin-dependent kinase activities induces apoptosis in cancer cells. Cancer Res. 2006;66:9270–9280. doi: 10.1158/0008-5472.CAN-06-1758. [DOI] [PubMed] [Google Scholar]
- 21.Vassilev LT, et al. Selective small-molecule inhibitor reveals critical mitotic functions of human CDK1. Proc Natl Acad Sci U S A. 2006;103:10660–10665. doi: 10.1073/pnas.0600447103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Pierce AJ, Johnson RD, Thompson LH, Jasin M. XRCC3 promotes homology-directed repair of DNA damage in mammalian cells. Genes Dev. 1999;13:2633–2638. doi: 10.1101/gad.13.20.2633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Brown AP, et al. Toxicity and toxicokinetics of the cyclin-dependent kinase inhibitor AG-024322 in cynomolgus monkeys following intravenous infusion. Cancer Chemother Pharmacol. 2008;62:1091–1101. doi: 10.1007/s00280-008-0771-1. [DOI] [PubMed] [Google Scholar]
- 24.Calabrese CR, et al. Anticancer chemosensitization and radiosensitization by the novel poly(ADP-ribose) polymerase-1 inhibitor AG14361. J Natl Cancer Inst. 2004;96:56–67. doi: 10.1093/jnci/djh005. [DOI] [PubMed] [Google Scholar]
- 25.Thomas HD, et al. Preclinical selection of a novel poly(ADP-ribose) polymerase inhibitor for clinical trial. Mol Cancer Ther. 2007;6:945–956. doi: 10.1158/1535-7163.MCT-06-0552. [DOI] [PubMed] [Google Scholar]
- 26.Plummer R, et al. Phase I study of the poly(ADP-ribose) polymerase inhibitor, AG014699, in combination with temozolomide in patients with advanced solid tumors. Clin Cancer Res. 2008;14:7917–7923. doi: 10.1158/1078-0432.CCR-08-1223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hackett AJ, et al. Two syngeneic cell lines from human breast tissue: the aneuploid mammary epithelial (Hs578T) and the diploid myoepithelial (Hs578Bst) cell lines. J Natl Cancer Inst. 1977;58:1795–1806. doi: 10.1093/jnci/58.6.1795. [DOI] [PubMed] [Google Scholar]
- 28.Ji H, et al. LKB1 modulates lung cancer differentiation and metastasis. Nature. 2007;448:807–810. doi: 10.1038/nature06030. [DOI] [PubMed] [Google Scholar]
- 29.Turner NC, et al. A synthetic lethal siRNA screen identifying genes mediating sensitivity to a PARP inhibitor. Embo J. 2008;27:1368–1377. doi: 10.1038/emboj.2008.61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Esashi F, et al. CDK-dependent phosphorylation of BRCA2 as a regulatory mechanism for recombinational repair. Nature. 2005;434:598–604. doi: 10.1038/nature03404. [DOI] [PubMed] [Google Scholar]
- 31.Jackson EL, et al. The differential effects of mutant p53 alleles on advanced murine lung cancer. Cancer Res. 2005;65:10280–10288. doi: 10.1158/0008-5472.CAN-05-2193. [DOI] [PubMed] [Google Scholar]
- 32.Oliver TG, et al. Chronic cisplatin treatment promotes enhanced damage repair and tumor progression in a mouse model of lung cancer. Genes Dev. 2010;24:837–852. doi: 10.1101/gad.1897010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Herbst RS, Heymach JV, Lippman SM. Lung cancer. N Engl J Med. 2008;359:1367–1380. doi: 10.1056/NEJMra0802714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Santamaria D, et al. Cdk1 is sufficient to drive the mammalian cell cycle. Nature. 2007;448:811–815. doi: 10.1038/nature06046. [DOI] [PubMed] [Google Scholar]
- 35.Garcia-Higuera I, et al. Interaction of the Fanconi anemia proteins and BRCA1 in a common pathway. Mol Cell. 2001;7:249–262. doi: 10.1016/s1097-2765(01)00173-3. [DOI] [PubMed] [Google Scholar]
- 36.Ji H, et al. The impact of human EGFR kinase domain mutations on lung tumorigenesis and in vivo sensitivity to EGFR-targeted therapies. Cancer Cell. 2006;9:485–495. doi: 10.1016/j.ccr.2006.04.022. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.