Abstract
Yeast NAD+-specific isocitrate dehydrogenase (IDH) is an octameric enzyme composed of four each of regulatory IDH1 and catalytic IDH2 subunits that share 42% sequence identity. IDH2 contains catalytic isocitrate/Mg2+ and NAD+ binding sites whereas IDH1 contains homologous binding sites, respectively, for cooperative binding of isocitrate and for allosteric binding of AMP. Ligand binding is highly ordered in vitro, and IDH exhibits the unusual property of half-site binding for all ligands. The structures of IDH solved in the absence or presence of ligands have shown: (a) a heterodimer to be the basic structural/functional unit of the enzyme, (b) the organization of heterodimers to form tetramer and octamer structures, (c) structural differences that may underlie cooperative and allosteric regulatory mechanisms, and (d) the possibility for formation of a disulfide bond that could reduce catalytic activity. In vivo analyses of mutant enzymes have elucidated the physiological importance of catalytic activity and allosteric regulation of this tricarboxylic acid cycle enzyme. Other studies have established the importance of a disulfide bond in regulation of IDH activity in vivo, as well as contributions of this bond to the property of half-site ligand binding exhibited by the wild-type enzyme.
Keywords: Isocitrate dehydrogenase, Tricarboxylic acid cycle, Allosteric regulation, Half-site ligand binding, Disulfide bond
Introduction
Mitochondrial NAD+-specific isocitrate dehydrogenase (IDH) catalyzes the oxidative decarboxylation of isocitrate to produce α-ketoglutarate and NADH. This rate-liming reaction in the tricarboxylic acid (TCA) cycle is essentially irreversible under physiological conditions [1], and IDH is subject to extensive allosteric regulation. A hypothesis that rates of respiratory metabolism are finely controlled at the level of this reaction by cellular energy levels [2] is based on dramatic allosteric activation of the yeast enzyme by AMP or of the mammalian enzyme by ADP, and of inhibition of the latter enzyme by ATP [3, 4]. Both enzymes are inhibited by NADH [1, 5]. Thus, flux through the TCA cycle would be attenuated at the level of IDH when cellular ratios of [ATP]:[AMP or ADP] and of [NADH]:[NAD+] are high, allowing diversion of isocitrate and citrate into biosynthetic pathways. These allosteric controls are also believed to balance relative rates of energy production by oxidative pathways and glycolysis [6]. Despite the implications from these early kinetic analyses that IDH may be a major point for control of flux through respiratory metabolism, until relatively recently there were few analyses of function in vivo and little understanding of structural/functional relationships for this TCA cycle enzyme. Much of this review focuses on work in these areas, in particular analyses of IDH from Saccharomyces cerevisiae by this and other groups.
Functions of IDH subunits
Barnes et al. [7] suggested that yeast IDH was an octameric enzyme containing similar subunits of Mr = ~38,000. However, two-dimensional gel electrophoresis and amino-terminal sequence analysis [8] showed that the octameric enzyme contains equal amounts of two different types of subunits. The genes encoding the two subunits were cloned and sequence analysis showed that IDH1 (Mr = 38,001) and IDH2 (Mr = 37,755) subunits share substantial primary sequence identity (42%) [9, 10]. Disruption of either the IDH1 or IDH2 genes eliminated cellular IDH activity although, as described below, IDH2 primarily contributes to catalytic function while IDH1 primarily contributes to regulatory properties of the enzyme. The major growth phenotypes exhibited by yeast strains lacking IDH1 and/or IDH2 were the inability to grow with acetate as the carbon source [9, 10] and the absence of mitochondrial respiration with citrate or isocitrate as the respiratory substrate [11]. The absence of growth with acetate as the carbon source is a phenotype shared with several other yeast TCA cycle mutants [12], and both phenotypes provide convincing evidence that the physiological functions of IDH in respiration cannot be provided by the residual mitochondrial NADP+-specific isocitrate dehydrogenase. Yeast mitochondria lack a transhydrogenase capable of interconversion of NADPH and NADH [13]. Mammalian cells do have such a transhydrogenase, perhaps explaining the proposed compensation by human mitochondrial NADP+-specific isocitrate dehydrogenase in some families with retinitis pigmentosa that exhibit defects in IDH [14].
The IDH1 and IDH2 protein sequences were found to be ~32% identical with that of Escherichia coli NADP+-specific isocitrate dehydrogenase, for which a crystal structure [15, 16] was available during early work on the yeast enzyme. Alanine replacement of a serine residue in IDH1 (Ser-92) that corresponds to a serine residue (Ser-113) in the bacterial enzyme known to be essential for isocitrate binding and catalytic activity [17] produced a yeast enzyme exhibiting a slight decrease in Vmax, a decrease in cooperativity, and a loss of activation by AMP [18]. A similar replacement of the corresponding serine residue in IDH2 (Ser-98) substantially reduced Vmax but had no effect on activation by AMP. These results suggested that IDH2 contributes catalytic isocitrate binding sites while IDH1 contributes to the regulatory binding of isocitrate and AMP. Similarly, based on NAD(P)H+ binding sites in bacterial enzymes [16, 19, 20], alanine replacements for adjacent residues in IDH1 (Asp-279 and Ile-280) resulted in a loss of activation by AMP, whereas similar replacements for corresponding residues in IDH2 (Asp-286 and Ile-287) resulted in a dramatic reduction in Vmax primarily due to a 70-fold increase in the S value for NAD+ 0.5 [21]. Thus, it was proposed that the subunits of the yeast enzyme have co-evolved to preserve homologous catalytic and regulatory ligand binding sites: both subunits contain isocitrate binding sites (for catalysis in IDH2 and for cooperativity in IDH1) and nucleotide binding sites (for NAD+ in IDH2 and for AMP in IDH1).
Ligand binding sites and ordered ligand binding in IDH
An ultrafiltration system was developed to investigate ligand binding properties of affinity-purified wild-type and mutant forms of yeast IDH. Enzymes containing the isocitrate binding site alterations in IDH1 (IDH1S92A/IDH2) and in IDH2 (IDH1/IDH2S98A) were obvious candidates for these assays. In addition, a comparison of IDH sequences with those for the E. coli enzyme suggested that nine residues in the catalytic site of the bacterial enzyme were conserved in IDH2 but only five of the nine were conserved in IDH1. With the goal of preserving isocitrate binding but not function in IDH1 and IDH2 subunits, reciprocal replacements of the four non-identical residues in putative isocitrate binding sites were made [22]. This generated IDH1A108R,F136Y,T241D,N245D/IDH2 and IDH1/IDH2R114A,Y142F,D248T,D252N enzymes. Kinetic and ligand binding properties determined for these two sets of mutant enzymes [23] are summarized in Table 1 and suggested that the wild-type yeast enzyme has four isocitrate binding sites. Mutations interfering with isocitrate binding (IDH1S92A or IDH2S98A) reduced this number to two, whereas mutations that mimicked the isocitrate binding site of the other subunit (IDH1A108R,F136Y,T241D,N245D or IDH2R114A,Y142F,D248T,D252N) preserved four isocitrate binding sites. Both types of mutations were primarily detrimental to the functions of each subunit, catalysis by IDH2 (with reductions in Vmax values of ~150-fold) and cooperativity plus AMP activation by IDH1 (with Hill coefficients of 1 and no allosteric regulation by AMP).
Table 1.
Relative effects of residue substitutions on velocity and isocitrate binding properties.
| Enzyme | Relative Vmax appa | Isocitrate Binding sites |
Hill Coefficientb |
AMP effectc |
|---|---|---|---|---|
| IDH1/IDH2 | 1 | 4 | 4 | 3.5 |
| IDH1S92A/IDH2 | 0.090 | 2 | 1 | none |
| IDH1/IDH2S98A | 0.007 | 2 | 3 | 1.8 |
| IDH1A108R,F136Y,T241D,N245D/IDH2 | 0.060 | 4 | 1 | none |
| IDH1/IDH2R114A,Y142F,D248T,D252N | 0.007 | 4 | 3 | 1.8 |
Kinetic Vmax values were determined with respect to concentrations of D-isocitrate. The Vmax app value measured for the wild-type (IDH1/IDH2) enzyme was ~30 units/mg [23].
Hill coefficients were determined in isocitrate binding assays.
In ligand binding assays, the affinity of the wild-type enzyme for isocitrate was increased ~3.5 fold in the presence of 100 μM.
Similar mutagenesis experiments were utilized to investigate nucleotide binding sites [24]. Residues corresponding to those in the cofactor binding site of the E. coli enzyme or in the putative cofactor binding site of mammalian IDH [16, 25] were replaced by alanines in IDH1 and IDH2 subunits of the yeast enzyme. In IDH2 these substitutions, IDH2H281A and IDH2D286A,I287A, produced a dramatic decrease in the apparent affinity for NAD+ in kinetic assays (Table 2), but only the latter substitution resulted in a substantial reduction in Vmax as described above. The IDH1/IDH2H281A max enzyme exhibited no binding sites for NAD+ under the concentrations used for ligand binding analyses, and the IDH1/IDH2D286A,I287A enzyme was too unstable to obtain the quantity of purified enzyme needed for binding assays. Corresponding residue replacements in IDH1 (IDH1R274A and IDH1D279A,I280A) had relatively little effect on Vmax but eliminated allosteric activation by AMP. As shown in Table 2, loss of the kinetic AMP effect was due to elimination of AMP binding. These results suggested that the nucleotide cofactor binding site is primarily contributed by IDH2 whereas a homologous nucleotide binding site in IDH1 has evolved for allosteric binding of AMP.
Table 2.
Relative effects of residue substitutions on kinetic parameters and binding sites for NAD+ and AMP.
| Enzyme | Relative Vmax appa |
S0.5NAD+ (mM) |
NAD+ binding sites |
AMP binding sites |
|---|---|---|---|---|
| IDH1/IDH2 | 1 | 0.2 | 2 | 2 |
| IDH1/IDH2H281A | 0.900 | 6.1 | 0 | 2 |
| IDH1/IDH2D286A,I287A | 0.006 | 8.2 | NDb | NDb |
| IDH1R274A/IDH2 | 0.500 | 0.8 | 2 | 0 |
| IDH1D279A,I280A/IDH2 | 0.500 | 0.2 | 2 | 0 |
Kinetic Vmax app values were determined with respect to concentrations of NAD+. The Vmax app value measured for the wild-type enzyme (IDH1/IDH2) was ~38 units/mg [24].
ND = not determined. The IDH1/IDH2D286A,I287A enzyme was refractive to purification in amounts needed for ligand binding assays.
Mutant enzymes used to investigate isocitrate and nucleotide binding sites were also used to examine the order of ligand binding [23, 24]. It was determined that binding of isocitrate by the catalytic IDH2 site but not by the regulatory IDH1 site requires Mg2+, that binding of isocitrate/Mg2+ by the catalytic IDH2 site is a prerequisite for binding of NAD+, and that binding of isocitrate by the regulatory site in IDH1 is a prerequisite for binding of AMP. Thus, ligand binding is strictly ordered, with isocitrate binding by IDH1 being the only binding that can occur in the absence of another ligand of the enzyme. Based again on the bacterial enzyme structure [16], putative ligands for Mg2+ in the catalytic IDH2 site (Asp248 and a neighboring Asp252) are represented by other residues in the active site of IDH1 (Thr241 and Asn245, respectfully), suggesting a structural basis for the binding of Mg2+ by IDH2 but not by IDH1. The role of Mg2+ in catalysis appears to be stabilization of a negative charge formed on the C2 hydroxyl of isocitrate during dehydrogenation [16]. The structural basis for ordered ligand binding by IDH was partially revealed by crystallographic analyses [26] as described below.
Basic heterodimer unit of IDH
Several lines of evidence suggested that the basic structural/functional unit of octameric yeast IDH is a heterodimer of IDH1 and IDH2 subunits. For example, application of the yeast two hybrid system indicated substantial interaction between the different subunits but not between identical subunits [27]. Also, mutagenesis studies again based on the bacterial enzyme structure [15, 16] suggested that two residues (Lys-183 and Asp-217) from the regulatory IDH1 subunit participate in isocitrate binding in the catalytic IDH2 site, and that corresponding residues (Lys-189 and Asp-222) from the catalytic IDH2 subunit participate in isocitrate binding in the regulatory IDH1 site [27]. These interactions in each heterodimer could provide a direct mechanism for communication between regulatory and catalytic sites.
Effects of expression of mutant forms of yeast IDH in vivo
Several studies [28, 29] have shown that expression of IDH mutant enzymes with substantial defects in catalytic activity (e.g. the IDH1/IDH2S98A enzyme in Table 1 or the IDH1/IDH2D286A,I287A enzyme in Table 2) results in an inability to grow with acetate as the carbon source and in an increase in production of respiratory deficient progeny. These phenotypes are also observed for yeast strains lacking IDH1 and/or IDH2 [9, 10], suggesting that only dramatic reductions in IDH activity interfere with enzymatic function in the TCA cycle.
In contrast, expression of mutant enzymes with primary defects in regulatory properties (e.g. the IDH1S92A/IDH2 enzyme in Table 1 or the IDH1D279A,I280A/IDH2 enzyme in Table 2) had little effect on growth with acetate or on production of respiratory deficient progeny [29]. However, expression in yeast of the regulatory IDH mutant enzymes or of a NAD+-specific bacterial enzyme that is not allosterically regulated resulted in very slow transitions (relative to strains expressing the wild-type or catalytic mutant enzymes) in rates of oxygen consumption following a shift of cells from medium with glucose to medium with ethanol as the carbon source [30]. These results suggest that allosteric properties of IDH provide a physiological advantage during changes in environmental conditions but may be less important for steady state growth.
The growth properties (no growth with acetate as the carbon source and an increase in respiratory progeny) associated with loss of IDH (e.g. in an idhΔ mutant containing disruptions in genes encoding both subunits) are shared with a yeast mutant (aco1Δ) lacking aconitase [31, 32]. These growth properties were shown to be alleviated or moderated by co-disruption of the gene (CIT1) encoding mitochondrial citrate synthase [33, 34]. Using a gas chromatography/mass spectrometry approach, we found that cellular levels of citrate were dramatically elevated in idhΔ and aco1Δ mutants, and that these levels were substantially lower in idhΔcit1Δ and in aco1Δcit1Δ mutants [33]. Addition of citrate to cultures of the parental strain partially recapitulated effects observed in the idhΔ and aco1Δ mutants, suggesting that elevated levels of this TCA cycle metabolite is responsible for detrimental physiological effects in these mutants.
Allosteric motions revealed by structural analyses
Development of a bacterial expression system for yeast IDH facilitated purification of sufficient quantities of the enzyme for crystallization trials. Crystallographic structures of IDH were obtained in the absence of bound ligand, in the presence of bound citrate (a substrate analogue), and in the presence of citrate plus AMP [26].1 The latter two structures were similar, but the ligand-free and ligand-bound forms of IDH exhibited many differences permitting a description of allosteric changes in the enzyme.
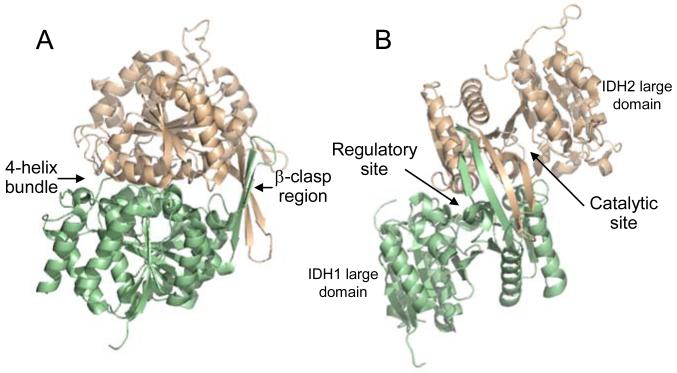
The structures confirmed that the basic structural unit within IDH is a heterodimer of IDH1 and IDH2 subunits (shown respectively in light green and light gold in Fig. 1A and 1B). The subunits are related by a pseudo two-fold axis. Major regions of interaction include a 4-helix bundle and a β-clasp region formed by two anti-parallel β-strands from each of the small domains of IDH1 and IDH2 (Fig. 1A). Residues in (iso)citrate ligand-binding sites are located between large and small domains of each subunit (Fig. 1B), and the identities of these residues verified results from previous mutagenesis studies [18, 21-24, 27]. In the ligand-free structure of IDH, the iso(citrate) and AMP binding sites in IDH1 are largely inaccessible, whereas, in the citrate-bound structure, a helix blocking the (iso)citrate binding site forms an extended loop, making both (iso)citrate and AMP sites accessible, partially explaining the ordered binding of isocitrate prior to AMP in the active site of IDH1. In the citrate plus AMP structure, AMP appears to prop open these sites, hindering reformation of the obstructive helix. The major difference in the IDH2 catalytic site between ligand-free and citrate-bound structures is the repositioning of Tyr-142, a critical residue for substrate binding [23, 35], Numerous structural changes in the 4-helix bundle suggest that changes in IDH1 upon citrate binding are likely communicated through the 4-helix bundle to the catalytic (iso)citrate binding site in IDH2, suggesting a mechanism for cooperative and allosteric interactions. In support of this idea, mutagenesis and kinetic studies [36] demonstrated the importance of this 4-helix bundle in allosteric regulation.
Fig. 1.
The yeast IDH heterodimer. The regulator y IDH1 subunit is shown in light green and the catalytic IDH2 subunit is shown in light gold. (A) Shown are major sites of interaction between the subunits, which are related by a pseudo two-fold axis. Two anti-parallel β-strands of each subunit align with the same strands of the other subunit forming a β-clasp region. A four-helix bundle is formed by two analogous alpha helices from each subunit. (B) A 90o rotation of the structure is used to indicate the positions of large domains (which contribute little to subunit interactions) and of active sites in each subunit. The heterodimer structure is from the ligand-free structure of IDH (PDB ID: 3BLX), and the orientation of IDH1 and IDH2 subunits in the heterodimer in (A) is maintained in upper left portions of structures shown in Fig. 2.
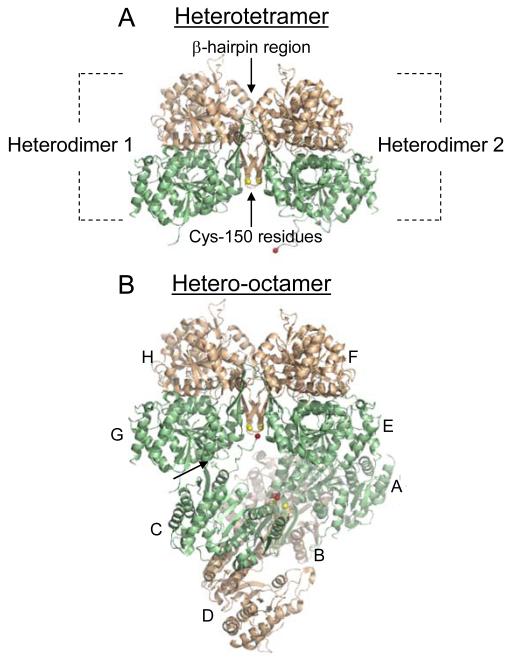
Two IDH1/IDH2 heterodimers associate to form a heterotetramer (Fig. 2A). The heterodimers are related by a two-fold axis, with the major interface being a β-hairpin region formed by the β-clasp regions of each heterodimer. Substantial differences in residue interactions in the β-hairpin region were noted in a comparison of the ligand-free and citrate-bound structures of IDH, suggesting this region is important in allosteric communication. An unexpected finding from the crystal structure was proximity of Cys-150 residues (shown in yellow in Fig. 2) from adjacent IDH2 subunits in the β-hairpin structure. The significance of interactions between the side chains of these residues to ligand binding and allosteric regulation are described below.
Fig. 2.
The IDH heterotetramer and hetero-octamer. Regulatory IDH1 subunits are shown in light green and catalytic IDH2 subunits are shown in light gold. (A) The major interaction between heterodimers in the heterotetramer is a β-hairpin structure formed by the β-clasp regions from each heterodimer. Cys-150 residues from IDH2 subunits located on one side of the β-hairpin structure are shown in yellow. (B) Heterodimers (A/B, C/D, E/F, and G/H) that form the octameric enzyme are indicated. In the octamer, one heterotetramer (A/B and C/D) is twisted relative to the other heterotetramer (E/F and G/H). The heterotetramers interact primarily by a protrusion of the amino terminus from an IDH1 subunit in one tetramer (e.g. C) into a pocket of an IDH1 subunit in the other tetramer (e.g. G), as indicated by an arrow, positioning the amino terminus of the C subunit (shown in red) in the vicinity of the IDH2 Cys-150 residues in the other tetramer. The heterotetramer and hetero-octamer structures are from the citrate-bound structure of IDH (PDB ID: 3BLV).
Two heterotetramers associate to form the octameric structure in a manner that deviates from pseudo-222 symmetry (Fig. 2B) [26]. The interactions between two heterotetramers are less substantial than those that form heterodimers or heterotetramers. The heterotetramer interactions are limited to a protrusion of ~16 residues from the amino terminus of an IDH1 subunit from one tetramer into the other tetramer. In Fig. 2B, this is illustrated by a tight turn of residues 12-16 from IDH subunit C into a pocket of IDH subunit G in the other tetramer (indicated by an arrow). In the citrate-bound structure, the remainder of the amino terminus of the protruding IDH1 subunit extends toward the other tetramer, placing residue 1 (shown in red) in close proximity of IDH2 Cys-150 residues which appear to be well separated with reduced side chains. In the ligand-free structure, the amino terminus of IDH subunit C is removed from the proximity of the IDH2 Cys-150 residues which are sufficiently close to form a disulfide bond.
Based on the structural interactions between heterotetramers in the IDH octamer, three stable tetrameric forms of IDH were constructed by slight truncation (-5 residues) of the amino terminus of IDH1 and/or by introduction of residue substitutions (in particular IDH1G15D) [37]. The tetrameric enzymes exhibited half of the values for Vmax and cooperativity observed for the octameric enzyme. However, the tetramers retained allosteric activation of activity by AMP, suggesting that this property is controlled at the level of the tetramer (or of component heterodimers) in the wild-type holoenzyme.
Disulfide bond formation in IDH
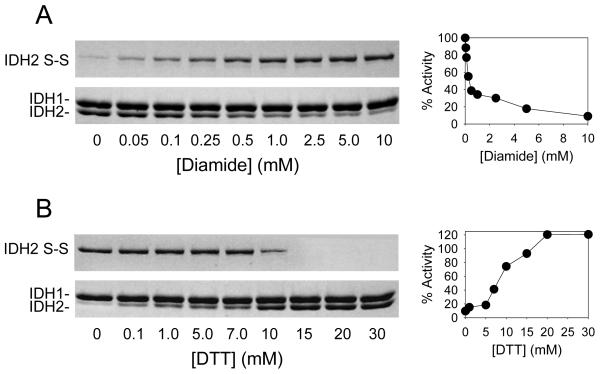
To examine the possibility and relevance of formation of a disulfide bond between IDH2 Cys-150 residues in IDH, two mutant enzymes were constructed, one with a C150S substitution in IDH2 and another with C56S/C242S substitutions in IDH2 that leaves IDH2 Cys-150 as the only cysteine residue in IDH [38]. Treatment of affinity purified wild-type and IDH1/IDH2C56S/C242S enzymes with diamide (a sulfhydral oxidant) resulted in formation of disulfide bonds between IDH2 subunits (with concomitant reduction in levels of the free IDH2 subunit) and in reductions of activities for the wild-type and IDH1/IDH2C56S/C242S enzymes (as shown in Fig. 3A for the IDH1/IDH2C56S/C242S enzyme to illustrate effects due solely to IDH2 Cys-150). In cont rast, treatment with diamide had no effect on the IDH1/IDH2C150S enzyme. These results suggested that the IDH2 Cys-150 residue is essential for formation of a disulfide bond that inhibits IDH activity. The formation of the disulfide bond and loss of IDH activity in wild-type and IDH1/IDH2C56S/C242S enzymes were completely reversible by subsequent addition of dithiothreitol, a disulfide bond reductant (as shown in Fig. 3B for the IDH1/IDH2C56S/C242S enzyme).
Fig. 3.
Effects of diamide and dithiothreitol on IDH. (A) The IDH1/IDH2C56S/C242S mutant enzyme which contains IDH2 Cys-150 as the only cysteine residue was incubated with increasing concentrations of diamide prior to non-denaturing gel electrophoresis (with staining using Coomassie blue) and activity assays. (B) The diamide-treated IDH1/IDH2C56S/C242S mutant enzyme was incubated with increasing concentrations of dithiothreitol (DTT) prior to non-denaturing gel electrophoresis and activity assays. The disulfide bond form of IDH2 migrates aberrantly slowly under these conditions [38, 40], so only portions of the gels are shown.
More importantly, formation of the IDH2 Cys-150 disulfide bond was found to occur naturally in vivo with a concomitant decrease in cellular IDH activity as yeast strains expressing the wild-type or IDH1/IDH2C56S/C242S enzymes (but not the IDH1/IDH2C150S enzyme) entered the stationary phase of growth [38]. The strain expressing the IDH1/IDH2C150S enzyme demonstrated reduced viability in stationary phase, suggesting that the normal formation of the IDH2 Cys-150 bond and associated decrease in IDH activity in the parental strain supports metabolic changes advantageous for survival under this condition. A remaining question under investigation is whether formation of the IDH2 Cys-150 disulfide bond during stationary phase occurs directly in response to changes in the redox environment of the mitochondrial matrix or is the result of enzymatic catalysis.
The tetrameric forms of IDH mentioned above were found to be particularly sensitive to diamide-induced formation of the Cys-150 disulfide bond and reduction of activity [37]. Pre-incubation of the wild-type enzyme with ligands, but not of the tetrameric enzymes, was found to reduce sensitivity to diamide. The effect of expression of the tetrameric enzymes on survival in stationary phase is under investigation. Collectively, however, these results support structural data suggesting that the octameric structure of wild-type IDH has in part evolved for regulation of disulfide bond formation and activity by ensuring the proximity of the amino terminus of an IDH subunit from one tetramer to the IDH2 Cys-150 residues in the other tetramer.
Basis for half-site ligand binding in IDH
The ligand binding studies described above confirmed previous results from Kuehn et al. [39] that yeast IDH exhibits half-site binding of all ligands. The composition of the holoenzyme with four IDH1 and four IDH2 subunits suggests there should be eight isocitrate, four NAD+, and four AMP binding sites. However, only half of these sites are measurable in standard ligand binding analyses [23, 24, 39].
Based on the potential interaction between side chains of Cys-150 residues in IDH2 subunits in both tetramers of IDH, ligand-binding properties of wild-type and IDH1/IDH2C150S enzymes were re-examined in the presence or absence of dithiothreitol, a thiol reductant [40]. As illustrated in Table 3, eight isocitrate and four AMP binding sites were observed for the wild-type enzyme when measured in the presence of DTT, and the same numbers were observed for the IDH1/IDH2C150S enzyme when measured in the absence or in the presence of DTT. However, only two NAD+ binding sites were measurable for either enzyme. A tetrameric form of IDH (the IDH1G15D/IDH2 enzyme) exhibited half-site binding (two sites) for isocitrate in the absence of DTT and full-site binding (four sites) in the presence of DTT (Table 3). Only one NAD+ site was observed for the tetramer under both conditions. In the context of the structure of IDH, these results suggest interactions between Cys-150 residues contribute to the property of half-site binding of isocitrate, but that some form of negative cooperativity may limit access to apparently equivalent NAD+ binding sites.
Table 3.
Numbers of ligand binding sites determined in the absence or the presence of 0.1 mM DTT.
| IDH1/IDH2 | IDH1/IDH2C150S | IDH1G15D/IDH2 | ||||
|---|---|---|---|---|---|---|
| −DTT | +DTT | −DTT | +DTT | −DTT | +DTT | |
| Isocitratea | 4.0 | 8.2 | 8.1 | 8.1 | 2.0 | 3.9 |
| AMP | 2.2 | 3.9 | 4.1 | 4.1 | 2.2 | 2.2 |
| NAD+ | 2.0 | 2.1 | 2.0 | 2.1 | 1.0 | 1.0 |
Isocitrate binding sites measured in the absence or presence of 0.1 mM AMP were similar [40], and average numbers are shown.
Despite the implication from these studies that the native IDH enzyme exhibits half-site binding of isocitrate (and reduced activity) due to formation of the IDH2 Cys-150 bond, there is no indication that substantial levels of the disulfide bond are present in the isolated native enzyme even during catalytic turnover [40]. Thus, formation of this bond may normally be rapid and transient in vitro and in vivo, with some mechanism for stabilization of the disulfide bond in vivo during the stationary phase of growth.
Evolutionary perspective
Data described above suggest that yeast IDH is regulated both by allostery and by covalent formation of a disulfide bond, and that these regulatory mechanisms contribute to modulation of respiratory metabolism in vivo. The corresponding TCA cycle enzyme in Escherichia coli is a homdimeric NADP+ specific enzyme that exhibits no allosteric regulation but that is controlled by covalent modification. In bacterial cells shifted to medium with acetate as the carbon source, ~80% of the isocitrate dehydrogenase molecules are rapidly inactivated by phosphorylation of a serine residue (i.e. Ser-113 as mentioned above) in the catalytic site [17, 41]. This reversible modification, catalyzed by a specific kinase/phosphatase [42], results in redirection of ~30% of the total carbon flux from the TCA cycle into the biosynthetic glyoxylate cycle [43-45]. The residual active isocitrate dehydrogenase molecules support oxidative energy production and provide NADPH for biosynthesis. The glyoxylate cycle, present in many bacteria and in non-mitochondrial compartments of yeast and plant cells, permits synthesis of four-carbon metabolites (succinate or malate) from two molecules of acetyl CoA. We suggest that the formation of an IDH2 Cys-150 disulfide bond during stationary phase, which reduces IDH activity by 50% [38], may similarly control metabolic flux.
The subunit composition of TCA cycle isocitrate dehydrogenases in plant and mammalian mitochondria appears to be more complex than in the yeast enzyme. There are apparently one gene encoding a catalytic IDHa subunit and two genes encoding regulatory IDHb and IDHc enzymes in tobacco (Nicotiana tabacum) [46]. It was shown that expression of a combination of a and b or of a and c subunits, but not the single subunits, could complement mitochondrial respiration in a yeast mutant lacking IDH1 and IDH2. This implied a multimeric subunit structure for the plant enzyme. In Arabidopsis thaliana, there are five genes encoding two potential catalytic and three potential regulatory subunits that are differentially expressed in various tissues [47]. Expression of any catalytic and regulatory subunit pair in a yeast idhΔ mutant was sufficient to restore growth with acetate as the carbon source. However, the relevance of subunit composition to distinct catalytic or regulatory functions has not been established. It would be of interest to kinetically characterize the different forms of IDH in plant cells to determine how these different functions might contribute to metabolic properties of various tissues.
The mammalian NAD+-specific isocitrate dehydrogenase is an octamer composed of four catalytic α, and two each of regulatory β and γ subunits [48-50; unpublished observations]. The sizes of the mammalian enzyme subunits are quite similar to those of the yeast enzyme, and respective catalytic and regulatory subunits share 40-50% sequence identity [51, 52]. The specific functions of the regulatory subunits are largely unknown, although there is evidence that the β subunit may contribute to allosteric activation by ADP [53]. Other potential functions to be determined for the different regulatory subunits include activation by Ca2+ and inhibition by ATP. It will also clearly be of interest to examine if the mammalian enzyme is additionally regulated by some reversible covalent mechanism in light of the regulation of bacterial and yeast enzymes by such mechanisms.
Highlights.
Yeast IDH is composed of four catalytic IDH2 and four regulatory IDH1 subunits.
IDH structures demonstrate changes associated with allosteric regulation.
A disulfide bond in IDH can decrease catalytic activity in vitro and in vivo.
The disulfide bond contributes to half-site ligand binding in IDH.
Acknowledgments
This work was supported by NIH Grant GM051265. Previous contributors to this work include Daniel A. Keys, Jill R. Cupp-Vickery, Robert J. Haselbeck, Wen-Ning Zhao, Ellen A. Panisko, Mark T. McCammon, Gang Hu, and Joshua A. Garcia. Current contributors include An-Ping Lin, Karyl I. Minard, and Sondra L. Anderson. Collaborators at the UTHSCSA include P. John Hart, Alex B. Taylor, Susan T. Weintraub, Kevin W. Hakala, Borries Demeler, and Virgil Schirf.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Protein Data Bank accession numbers are 3BLX for ligand-free IDH, 3BLV for citrate-bound IDH, and 3BLW for citrate plus AMP-bound IDH.
References
- [1].Barnes LD, McGuire JJ, Atkinson DE. Biochemistry. 1972;11:4322–4329. doi: 10.1021/bi00773a019. [DOI] [PubMed] [Google Scholar]
- [2].Hathaway JA, Atkinson DE. J. Biol. Chem. 1963;238:2875–2881. [PubMed] [Google Scholar]
- [3].Chen RF, Plaut GWE. Biochemistry. 1963;2:1023–1032. doi: 10.1021/bi00905a020. [DOI] [PubMed] [Google Scholar]
- [4].Plaut GWE. Curr. Top. Cell Regul. 1970;2:1–27. [Google Scholar]
- [5].Gabriel JL, Zervos PR, Plaut GW. Metabolism. 1986;35:661–667. doi: 10.1016/0026-0495(86)90175-7. [DOI] [PubMed] [Google Scholar]
- [6].Sols A, Gancedo C, DelaFuente G. Energy-yielding metabolism in yeast. In: Rose AH, Harrison JH, editors. The Yeasts. II. Academic Press; N.Y.: 1973. [Google Scholar]
- [7].Barnes LD, Kuehn GD, Atkinson DE. Biochemistry. 1971;10:3939–3944. doi: 10.1021/bi00797a022. [DOI] [PubMed] [Google Scholar]
- [8].Keys DA, McAlister-Henn L. J. Bacteriol. 1990;172:4280–4287. doi: 10.1128/jb.172.8.4280-4287.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Cupp JR, McAlister-Henn L. J. Biol. Chem. 1991;266:22199–22205. [PubMed] [Google Scholar]
- [10].Cupp JR, McAlister-Henn L. J. Biol. Chem. 1992;267:16417–16423. [PubMed] [Google Scholar]
- [11].Haselbeck RJ, McAlister-Henn L. J. Biol. Chem. 1993;268:12116–12122. [PubMed] [Google Scholar]
- [12].McCammon MT. Genetics. 1996;144:57–69. doi: 10.1093/genetics/144.1.57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Rydstrom J. Biochim. Biophys. Acta. 1977;463:155–184. doi: 10.1016/0304-4173(77)90007-6. [DOI] [PubMed] [Google Scholar]
- [14].Hartong DT, Dange M, McGee TL, Berson EL, Dryja TP, Colman RF. Nat. Genet. 2008;40:1230–1234. doi: 10.1038/ng.223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Hurley JH, Dean AM, Sohl JL, Koshland DE, Jr., Stroud RM. Science. 1990;249:1012–1016. doi: 10.1126/science.2204109. [DOI] [PubMed] [Google Scholar]
- [16].Hurley JH, Dean AM, Koshland DE, Jr., Stroud RM. Biochemistry. 1991;30:8671–8678. doi: 10.1021/bi00099a026. [DOI] [PubMed] [Google Scholar]
- [17].Thorsness PE, Koshland DE., Jr. J. Biol. Chem. 1987;262:10422–10425. [PubMed] [Google Scholar]
- [18].Cupp JR, McAlister-Henn L. Biochemistry. 1993;32:9323–9328. doi: 10.1021/bi00087a010. [DOI] [PubMed] [Google Scholar]
- [19].Imada K, Sato M, Tanaka N, Katsube Y, Matsuura Y, Oshima T. J. Mol. Biol. 1991;222:725–738. doi: 10.1016/0022-2836(91)90508-4. [DOI] [PubMed] [Google Scholar]
- [20].Chen R, Greer A, Dean AM. Proc. Natl. Acad. Sci. U.S.A. 1995;92:11666–11670. doi: 10.1073/pnas.92.25.11666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Zhao WN, McAlister-Henn L. J. Biol. Chem. 1997;272:21811–21817. doi: 10.1074/jbc.272.35.21811. [DOI] [PubMed] [Google Scholar]
- [22].Panisko EA, University of Texas Health Science Center . Ph.D. dissertation. San Antonio, TX: 2000. [Google Scholar]
- [23].Lin AP, McAlister-Henn L. J. Biol. Chem. 2002;277:22475–22483. doi: 10.1074/jbc.M202534200. [DOI] [PubMed] [Google Scholar]
- [24].Lin AP, McAlister-Henn L. J. Biol. Chem. 2003;278:12864–12872. doi: 10.1074/jbc.M300154200. [DOI] [PubMed] [Google Scholar]
- [25].Huang YC, Colman RF. Arch. Biochem. Biophys. 2002;401:81–90. doi: 10.1016/S0003-9861(02)00041-3. [DOI] [PubMed] [Google Scholar]
- [26].Taylor AB, Hu G, Hart PJ, McAlister-Henn L. J. Biol. Chem. 2008;283:10872–10880. doi: 10.1074/jbc.M708719200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Panisko EA, McAlister-Henn L. J. Biol. Chem. 2001;276:1204–1210. doi: 10.1074/jbc.M005056200. [DOI] [PubMed] [Google Scholar]
- [28].Lin AP, McCammon MT, McAlister-Henn L. Biochemistry. 2001;40:14291–14301. doi: 10.1021/bi0111707. [DOI] [PubMed] [Google Scholar]
- [29].McCammon MT, McAlister-Henn L. Arch. Biochem. Biophys. 2003;419:222–233. doi: 10.1016/j.abb.2003.08.022. [DOI] [PubMed] [Google Scholar]
- [30].Hu G, Lin AP, McAlister-Henn L. J. Biol. Chem. 2006;281:16935–16942. doi: 10.1074/jbc.M512281200. [DOI] [PubMed] [Google Scholar]
- [31].Przybyla-Zawislak B, Gadde DM, Ducharme K, McCammon MT. Genetics. 1999;152:153–166. doi: 10.1093/genetics/152.1.153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [32].McCammon MT, Epstein CB, Przybyla-Zawislak B, McAlister-Henn L, Butow RA. Mol. Biol. Cell. 2003;14:958–972. doi: 10.1091/mbc.E02-07-0422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [33].Lin AP, Hakala KW, Wein traub ST, McAlister-Henn L. Arch. Biochem. Biophys. 2008;474:205–212. doi: 10.1016/j.abb.2008.03.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [34].Gadde DM, McCammon MT. Arch. Biochem. Biophys. 1997;344:139–149. doi: 10.1006/abbi.1997.0191. [DOI] [PubMed] [Google Scholar]
- [35].Lee ME, Dyer DH, Klein OD, Bolduc JM, Stoddard BL, Koshland DE., Jr. Biochemistry. 1995;34:378–384. doi: 10.1021/bi00001a046. [DOI] [PubMed] [Google Scholar]
- [36].Hu G, McAlister-Henn L. Arch. Biochem. Biophys. 2006;453:207–216. doi: 10.1016/j.abb.2006.06.022. [DOI] [PubMed] [Google Scholar]
- [37].Lin AP, Demeler B, Minard KI, Anderson SL, Schirf V, Galaleldeen A, McAlister-Henn L. Biochemistry. 2011;50:230–239. doi: 10.1021/bi101401h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [38].Garcia JA, Minard KI, Lin AP, McAlister-Henn L. Biochemistry. 2009;48:8869–8878. doi: 10.1021/bi900968a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [39].Kuehn GD, Barnes LD, Atkinson DE. Biochemistry. 1971;10:3945–3951. doi: 10.1021/bi00797a023. [DOI] [PubMed] [Google Scholar]
- [40].Lin AP, McAlister-Henn L. Biochemistry. 2011 [Epub ahead of print] PMID: 21861471. [Google Scholar]
- [41].Dean AM, Lee MH, Koshland DE., Jr. J. Biol. Chem. 1989;264:20482–20486. [PubMed] [Google Scholar]
- [42].LaPorte DC, Chung T. J. Biol. Chem. 1985;260:15291–15297. [PubMed] [Google Scholar]
- [43].LaPorte DC, Walsh K, Koshland DE., Jr. J. Biol. Chem. 1984;259:14068–14075. [PubMed] [Google Scholar]
- [44].Walsh K, Koshland DE., Jr. J. Biol. Chem. 1984;259:9646–9654. [PubMed] [Google Scholar]
- [45].Walsh K, Koshland DE., Jr. J. Biol. Chem. 1985;260:8430–8437. [PubMed] [Google Scholar]
- [46].Lancien M, Gadal P, Hodges M. Plant J. 1998;16:325–333. doi: 10.1046/j.1365-313x.1998.00305.x. [DOI] [PubMed] [Google Scholar]
- [47].Lemaitre T, Hodges M. Plant Cell Physiol. 2006;47:634–643. doi: 10.1093/pcp/pcj030. [DOI] [PubMed] [Google Scholar]
- [48].Ramachandran N, Colman RF. J. Biol. Chem. 1980;255:8859–8864. [PubMed] [Google Scholar]
- [49].Soundar S, Park JH, Huh TL, Colman RF. J. Biol. Chem. 2003;278:52146–52153. doi: 10.1074/jbc.M306178200. [DOI] [PubMed] [Google Scholar]
- [50].Kim YO, Koh HJ, Kim SH, Jo SH, Huh JW, Jeong KS, Lee IJ, Song BJ, Huh TL. J. Biol. Chem. 1999;274:36866–36875. doi: 10.1074/jbc.274.52.36866. [DOI] [PubMed] [Google Scholar]
- [51].Nichols BJ, Hall L, Perry AC, Denton RM. Biochem. J. 1993;295(Pt 2):347–350. doi: 10.1042/bj2950347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [52].Nichols BJ, Perry AC, Hall L, Denton RM. Biochem. J. 1995;310(Pt 3):917–922. doi: 10.1042/bj3100917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [53].Dange M, Colman RF. J. Biol. Chem. 2010;285:20520–20525. doi: 10.1074/jbc.M110.115386. [DOI] [PMC free article] [PubMed] [Google Scholar]