Abstract
Chemokines and inflammatory cytokines are key regulators of immunity and inflammation during viral infections. Hepatitis C virus (HCV) is a hepatotropic RNA virus frequently associated with chronic liver inflammation and hepatocellular carcinoma. Intrahepatic levels of chemokines and cytokines are elevated in chronic HCV infections, but the underlying mechanisms remain unclear. We find that Toll like receptor-3 (TLR3) senses HCV infection in cultured hepatoma cells, leading to NF-κB activation and the production of numerous chemokines and inflammatory cytokines, such as RANTES, MIP-1α, MIP-1β, IP-10 and IL-6. The chemokine/cytokine induction occurred late in HCV infection and was abrogated when HCV was UV-inactivated prior to infection, indicating a dependence on cellular recognition of HCV replication products. Gel shift and chromatin immunoprecipitation assays revealed that NF-κB plays a pivotal role in HCV-induced chemokine/cytokine transcription. Mutations specifically disrupting the dsRNA binding activity of TLR3 ablated the chemokine/cytokine response to HCV infection, indicating that HCV dsRNA was the pathogen associated molecular pattern triggering TLR3 signaling. In vitro synthesized HCV dsRNAs with a minimal length of ~80-100 bp activated TLR3-dependent chemokine expression, regardless of the genome position from which they derive. In contrast, HCV ssRNAs, including those derived from the structured 3′NTR highly potent for RIG-I activation, failed to do so. Moreover, robust production of chemokines and inflammatory cytokines was also observed in primary human hepatocytes following stimulation with extracellular poly-I:C, a TLR3 ligand.
Conclusion
Our data suggest that TLR3-mediated chemokine and inflammatory cytokine responses may play an important role in host immune responses to HCV and the pathogenesis of HCV-associated liver diseases.
Keywords: Hepatitis C Virus, Toll-like receptor 3, NF-κB, chemokine, double-stranded RNA
INTRODUCTION
Infections with the hepatitis C virus (HCV) affect approximately 130 million people worldwide and pose a major threat to human health. HCV is a positive-sense, single-stranded (ss) RNA virus that has a restricted tropism for hepatocytes. Remarkably, HCV persists in ~70% of infected individuals, causing chronic intrahepatic inflammation and putting patients at risk of developing cirrhosis and hepatocellular carcinoma (1). Clearance of HCV infection depends on the development of vigorous and broad CD4 and CD8 T cell responses, which, however, often fail and are replaced with an intermediate cytotoxic T cell response unable to eliminate the infection but strong enough to cause hepatocyte destruction (2). Central to T cell homing to the liver is the induction of a family of small chemotactic cytokines called chemokines that regulate migration of leukocytes and their recruitment to inflammation sites. As such, chemokines play crucial roles in inflammatory processes and the transition from innate to adaptive immunity (3, 4). Intrahepatic chemokines such as the CCR5 ligands, RANTES, MIP-1β and MIP-1α, and the CXCR3 ligand, IP-10, are elevated in hepatitis C patients, and the levels of some of them are reported to correlate with the severity of liver inflammation (3-5). However, the cellular source and the underlying mechanism of chemokine induction during HCV infection remain elusive (2).
Toll-like receptor-3 (TLR3) and the RIG-I-like receptors (RLRs, RIG-I and MDA5) constitute two parallel classes of cellular sensors that recognize viral pathogen associated molecular patterns (PAMP) and initiate innate immune responses. Despite operating via distinct adaptors and mechanisms, both pathways culminate in the activation of IRF3-dependent interferon (IFN) antiviral response and production of NF-κB-dependent proinflammatory mediators. TLR3 and RLRs sense viral dsRNA, a major viral PAMP, in endosomal and cytoplasmic compartments, respectively. RIG-I also recognizes viral RNAs bearing 5′-triphosphates (6). The importance of RLR and TLR3 pathways in innate immunity to HCV is suggested by the fact that HCV encodes a serine protease, NS3/4A, which inactivates both pathways by cleaving two adaptor proteins, MAVS and TRIF (7-10). While much has been learned concerning how TLR3 and RIG-I pathways act to control HCV replication through activating IRF3 and antiviral interferon-stimulated gene (ISG) expression (8, 11-13), little is known about the mechanisms governing the chemokine and proinflammatory cytokine response to HCV infection. A better understanding of the latter is crucial to broadening our knowledge on immune response to and pathogenesis of HCV and to design new effective immunotherapies for hepatitis C. Here, we demonstrate that TLR3 senses HCV infection in cultured hepatocytes, leading to NF-κB activation and production of proinflammatory chemokines/cytokines previously reported in hepatitis C patients. Our study also defines the molecular features of HCV dsRNA that serves as the PAMP for TLR3.
EXPERIMENTAL PROCEDURES
Cells, viruses and miscellaneous reagents
See supplementary text.
Plasmids, HCV ssRNAs and dsRNAs
Conventional PCR and recombinant DNA techniques were used to generate the HCV cDNA plasmids (all from HCV-N strain) for in vitro transcription of HCV ssRNAs and dsRNAs. pBSII-core and pBSII-E1-p7 contained cDNA inserts encoding the full-length HCV core and E1/E2/p7, respectively in pBluscript-II backbone (Stratagene). pBSII-HCV-NS-3′NTR contained a 6.6-kb insert spanning partial NS2 through NS5B and the entire 3′NTR sequences. pCMV-NS5A contained full-length NS5A-coding sequence in pCMV-Tag4a (Stratagene). HCV ssRNAs were synthesized from linearized vectors using T7 (for +strand RNAs of core, E1-p7 and NS-3′NTR origins and −strand of NS5A RNA) and T3 (for −strand RNAs of core, E1-p7 and NS-3′NTR origins and +strand of NS5A RNA) MegaScript kits (Ambion). The cDNA templates for synthesizing HCV dsRNAs with varying lengths (ranging from 50- to 250-bp) were amplified by PCR using pBSII-E1-P7 as the template and the indicated oligonucleotide primers (sequences available online in Table S1) with introduced T7 promoter sequence at 5′ termini. In vitro transcriptions were performed using the MEGAshortScript T7 kit (Ambion). To form dsRNA duplexes, equal molar amounts of +ssRNAs and −ssRNAs were combined in a microcentrifuge tube and incubated at 100°C for 5 min, followed by a slow cool-down to room temperature.
Bio-Plex Multiplex Cytokine Assay
Culture supernatants of cells following the indicated treatments were assessed for cytokines and chemokines by the Bio-Plex Pro™ human cytokine 27-plex immunoassay (Bio-Rad) as per manufacturer’s suggestions.
Quantitative PCR (qPCR)
Total RNA was isolated from cells following the indicated treatments using Trizol (Invitrogen) and subsequently was programmed for synthesis of cDNA. The abundance of RANTES, MIP-1β and 28S (internal control) transcripts, and HCV RNA were analyzed by qPCR using gene-specific primers (sequences available online in Table S2) and GoTaq qPCR Master Mix (Promega) on an iCycler IQ5 real-time PCR system (Bio-Rad).
Reporter gene assay
The promoter activities of RANTES and PRDII in transfected cells mock-infected or infected with HCV for the indicated time periods were determined using luciferase reporter assay as described (12, 14).
Immunoblot Analysis
Detailed methods have been presented elsewhere (7, 12, 15). The following polyclonal (pAb) and monoclonal (mAb) antibodies were used: anti-IκBα (C-21) and anti-p65 (C-20) pAbs and anti-Lamin A/C (636) mAb (Santa Cruz); anti-HCV NS5A (9E10) mAb (a gift from Charles Rice); anti-actin mAb (Sigma); anti-ISG15 (a gift from Arthur Haas) and anti-ISG56 pAbs (12); and peroxidase-conjugated secondary anti-rabbit and anti-mouse pAbs (Southern Biotech). The protein bands were visualized by enhanced chemiluminescence (Millipore), followed by exposure to x-ray films.
Chromatin immunoprecipitation (ChIP) assay
7.5-TLR3 cells (~5×106) were mock-infected or infected with HCV (MOI=0.5) for 48 h, and then were processed for ChIP assay as described previously (14). The antibodies used for ChIP were from Santa Cruz (anti-p65) and Active Motif (control IgG). The ChIP-enriched samples were subjected to qPCR quantification of various chemokine and TFF1 (negative control) promoter sequences using specific primers presented online in Table S3.
RESULTS
HCV induces production of proinflammatory cytokines and chemokines in Huh7.5 cells stably reconstituted with functional TLR3 signaling
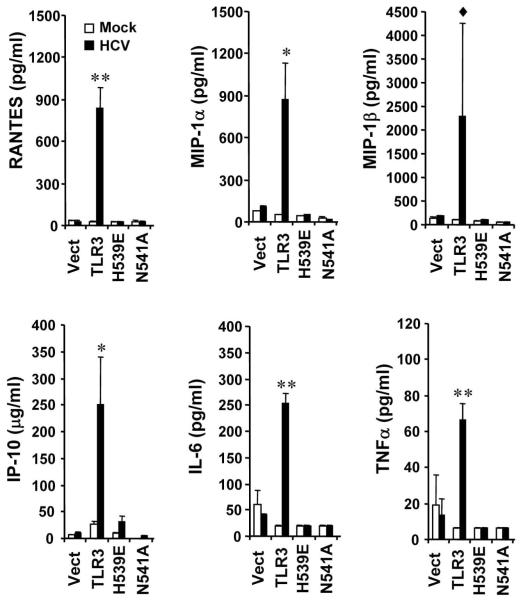
To investigate whether TLR3 plays a role in hepatoceullar proinflammatory response to HCV infection, we determined the production of cytokines/chemokines in HCV-infected 7.5-TLR3 cells, which were derived from Huh7.5 cells by stably reconstituting the expression of human TLR3. As controls, we studied Huh7.5 cells expressing the control vector (Vect) and mutant TLR3s defective for signaling due to incapability of dsRNA binding (H539E and N541A). All these cells are RIG-I defective and the effects observed are due to activation of the TLR3 pathway in the absence of RIG-I. We have previously utilized these cell lines to demonstrate that TLR3 senses HCV infection and triggers a weak ISG response, thereby moderately reducing HCV propagation when cells were infected at low MOIs (12). We infected cells with JFH1 virus at an MOI of 0.2 for 72 h, and then harvested the culture supernatants to measure cytokine/chemokine production by a Bio-Plex Multiplex Cytokine Assay. At this MOI HCV did not replicate differentially among these Huh7.5-derivatives (12) and all the cells were infected at 72 h as determined by immunostaining of NS5A (data not shown). Compared to mock-infected cells, six chemokines/cytokines were strongly induced by HCV infection in 7.5-TLR3 cells, including RANTES (37-fold), MIP-1β (25-fold), MIP-1α (17-fold), IL-6 (13-fold), IP-10 (10-fold) and TNF-α (10-fold) (Fig. 1). In addition, HCV weakly induced the production of IL-17 (5-fold), G-CSF (4-fold), IL-1β (3-fold), GM-CSF (2-fold) and eotaxin (2-fold) (data not shown). Importantly, none of these cytokines/chemokines were induced by HCV in the 7.5-Vect cells or 7.5-H593E and 7.5-N541A cells expressing mutant TLR3 (Fig. 1 and data not shown). These data reveal that TLR3 signaling, but not TLR3 expression per se, confers Huh7.5 cells the ability to produce proinflammatory mediators in response to HCV infection.
Fig. 1. HCV infection induces the production of various chemokines/cytokines in Huh7.5 cells stably reconstituted with functional TLR3 signaling.
Huh7.5 cells expressing the control vector (Vect), TLR3 or the indicated TLR3 mutant (H593E or N541A) were infected with JFH1 virus (MOI=0.2) for 3 days prior to measuring cytokine levels in culture supernatants by bioplex assay. “*” and “**” indicate statistical differences exist between mock- and HCV-infected 7.5-TLR3 cells with a p-value of <0.05 and <0.01, respectively. “◆” denotes p=0.13 between mock- and HCV-infected 7.5-TLR3 cells.
TLR3-mediated upregulation of chemokine transcription following HCV infection depends on viral replication
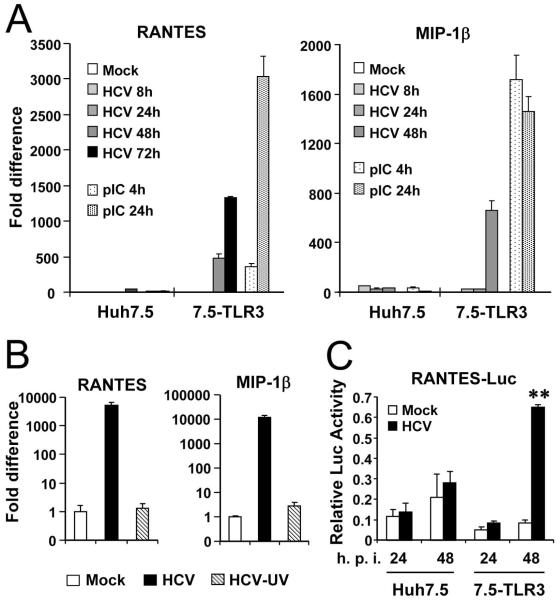
To characterize the mechanism of TLR3-mediated chemokine/cytokine induction in HCV-infected cells, we studied the temporal kinetics of mRNA expression by qPCR for RANTES, one of the most upregulated chemokines. In 7.5-TLR3 cells, RANTES mRNA levels did not increase at 8 h and 24 h postinfection, but were elevated by 477- and 1326-fold at 48 h and 72 h, respectively. In contrast, RANTES mRNA abundance was relatively unaffected in HCV-infected Huh7.5 cells. As a positive control, poly-I:C strongly stimulated RANTES mRNA expression in 7.5-TLR3 cells (by 365- and 3034-fold at 4 h and 24 h post-treatment, respectively), but had little effect in Huh7.5 cells (Fig. 2A, left panel). We obtained similar results when examining MIP-1β mRNA expression (right panel). The delayed kinetics of chemokine induction by HCV in infected 7.5-TLR3 cells implies that viral replication is needed to generate the HCV PAMP to activate TLR3 signaling, while HCV entry and uncoating are insufficient to do so. Consistent with this point, UV-inactivated HCV virions completely lost the chemokine-inducing capacity (Fig. 2B). We next determined whether the RANTES induction by HCV was regulated at transcriptional level. To this end, 7.5-TLR3 and Huh7.5 cells were transfected with a luciferase reporter construct under the control of RANTES promoter, followed by infection with HCV (MOI=1). Consistent with the mRNA quantitation data (Fig. 2A), activation of RANTES promoter was observed only in 7.5-TLR3 cells at 48 h post HCV infection but not at 24 h (Fig. 2C). Collectively, these data suggest that HCV replication product(s) induces chemokine expression by triggering TLR3-dependent activation of chemokine transcription.
Fig. 2. TLR3-mediated chemokine induction by HCV infection depends on viral replication.
Huh7.5 and 7.5-TLR3 cells were infected with JFH1 virus (HCV, MOI=1). Induction of RANTES and MIP-1β transcripts at indicated time points was determined by qPCR, using poly-I:C stimulation in medium as a positive control (A). Reporter assay of HCV activation of the RANTES promoter is shown in (C). “**” indicates statistical difference exists between mock- and HCV-infected 7.5-TLR3 cells with a p-value of <0.01. (B) qPCR quantification of RANTES and MIP-1β transcripts in 7.5-TLR3 cells mock-infected and infected with JFH1 virus (HCV) or UV-inactivated JFH1 virus (HCV-UV) (MOI=1) for 48 h.
NF-κB is activated in HCV-infected 7.5-TLR3 cells in a manner that parallels the kinetics of chemokine induction
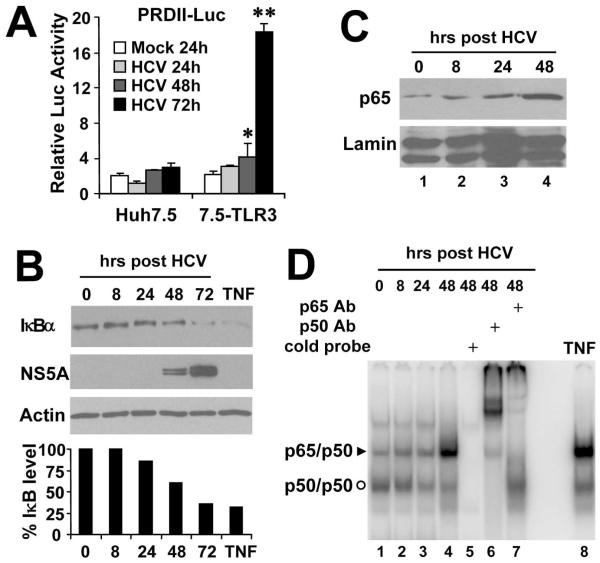
Because NF-κB plays a pivotal role in regulating the expression of proinflammatory cytokines and chemokines, we assessed NF-κB activity in HCV-infected cells. Reporter gene assays demonstrated that the NF-κB-dependent PRDII promoter was activated by 2- and 9-fold, respectively, in 7.5-TLR3 cells at 48 and 72 h post HCV infection but not at earlier times (Fig. 3A). In contrast, the PRDII promoter activity did not change significantly in HCV-infected Huh7.5 cells. The kinetics of TLR3-dependent activation of the PRDII promoter by HCV infection thus closely mirrored that of the chemokine upregulation (Fig. 2). The signal-induced degradation of IκB results in liberation of the p65/p50 heterodimer complexes of NF-κB, followed by their nuclear translocation and binding to target promoters. In HCV-infected 7.5-TLR3 cells, degradation of IκBα was evident at 48 h and further enhanced at 72 h, concomitant with the increase in HCV replication as visualized by the accumulation of viral NS5A protein (Fig. 3B). Consistent with this, substantial nuclear accumulation of p65 NF-κB subunit was observed at 48 h post-infection in 7.5-TLR3 cells (Fig. 3C). Furthermore, at 48 h the nuclear p65/p50 complex was induced by HCV infection as revealed by EMSA with an NF-κB-specific DNA probe (Fig. 3D). This effect resulted specifically from TLR3 signaling, since substantially less p65/p50 complex was formed in HCV-infected Huh7.5 cells and 7.5-N541A cells expressing a TLR3 mutant defective for dsRNA binding than was in 7.5-TLR3 cells (Fig. S1). Taken together, these data demonstrate that HCV replication induces NF-κB activity in a TLR3-dependent fashion with kinetics similar to that of chemokine/cytokine induction, indicating a regulatory role for NF-κB in the latter process.
Fig. 3. HCV infection triggers NF-κB activation with delayed kinetics in 7.5-TLR3 cells.
(A) PRDII promoter activities in Huh7.5 and 7.5-TLR3 cells at various time points post-infection with JFH1 virus (MOI=1). “*” and “**” indicate statistical differences exist between mock- and HCV-infected 7.5-TLR3 cells with a p-value of <0.05 and <0.01, respectively. (B) through (D) 7.5-TLR3 cells were infected with JFH1 virus (MOI=0.5) for the indicated time periods. An uninfected culture treated with TNF (15 ng/ml, 2 h) was included as a positive control for NF-κB activation. (B) Immunoblot analysis was conducted to determine the time-course IκBα degradation, which was quantified by densitometry (lower graph). (C) Immunoblotting of p65 accumulation in the nuclear extracts at various time points post-infection. Immunoblotting of Lamin A/C was shown as a loading control. (D) EMSA analysis of time-course nuclear p65/p50 binding to an NF-κB probe derived from the immunoglobulin gene promoter. Both p65 and p50 antibodies supershifted the NF-κB complex in nuclear extracts of 7.5-TLR3 cells infected with HCV for 48 h (lanes 6 and 7). TNFα treatment (15 ng/ml, 2 h) was included as a positive control for NF-κB activation (lane 8)
NF-κB controls HCV-induced chemokine transcription
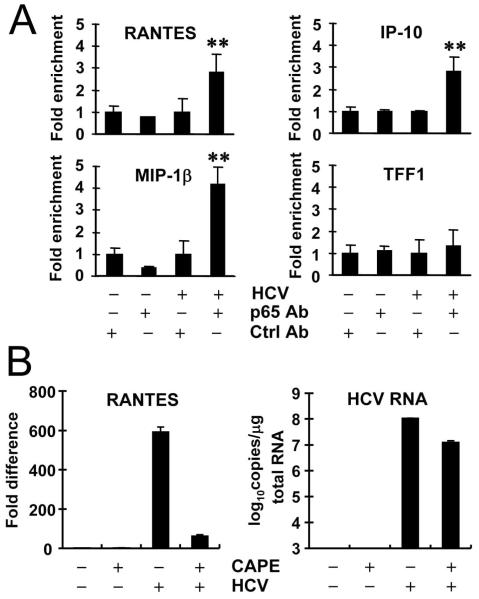
We next performed ChIP experiments to determine whether NF-κB binds to chemokine promoters in HCV-infected 7.5-TLR3 cells. We found that HCV infection substantially augmented p65 NF-κB subunit binding to three chemokine promoters, RANTES, MIP-1β and IP-10, but not to the control TFF1 promoter devoid of NF-κB recognition site (Fig. 4A), indicating that the p65/p50 NF-κB complexes formed in the nucleus of HCV-infected 7.5-TLR3 cells (Fig. 3D) directly controlled the transcription from these chemokine promoters. Confirming this, treatment with CAPE, a potent NF-κB inhibitor (16), abrogated HCV-induced upregulation of transcripts for RANTES (Fig. 4B) and MIP-1β (data not shown). Collectively, these data confirm that NF-κB governs TLR3-mediated chemokine/cytokine induction by HCV infection.
Fig. 4. NF-κB controls chemokine induction in HCV-infected 7.5-TLR3 cells.
(A) qPCR quantification of the p65 NF-κB subunit binding to the indicated promoters after ChIP enrichment of chromatin from cells mock-infected or infected with JFH1 virus (MOI=0.5) for 48 h. The TFF1 promoter devoid of NF-κB binding element was used as a negative control. “**” indicates statistical differences exist between HCV/p65 antibody and mock/p65 antibody groups with a p-value of <0.01. (B) 7.5-TLR3 cells were mock-infected or infected with JFH1 virus (MOI=0.5) for 8 h, and subsequently mock-treated or treated with CAPE for 64 h prior to qPCR analysis of RANTES mRNA and HCV RNA levels.
HCV dsRNA duplex, but not structured viral RNA, is the ligand for activating TLR3
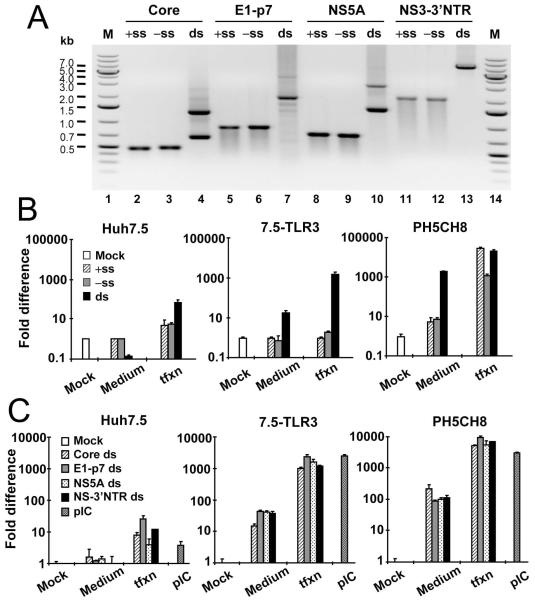
HCV RNA is highly structured and contains double-stranded (ds) regions in various portions of the HCV genome, especially in the 5′- and 3′-nontranslated regions (NTRs) and the core- and NS5B-coding regions (17). Because TLR3 binds dsRNA, the activation of TLR3 signaling during HCV infection may involve TLR3 recognition of structured HCV RNA (of either positive- and negative-strand) or HCV dsRNA intermediates generated during viral RNA replication, or both. To distinguish between these possibilities, we synthesized in vitro a 6.6-kb subgenomic HCV RNA of both positive-sense (+ss) and negative-sense (−ss) encompassing partial NS2 sequence, the entire NS3 to NS5B coding region and the intact 3′NTR (here referred to as NS-3′NTR) (Fig. 5A, lanes 11 and 12). To resemble HCV dsRNA replicative intermediates, the +ss and −ss NS-3′NTR RNAs were annealed to form dsRNA duplexes (lane 13). When added to culture medium of 7.5-TLR3 cells, NS-3′NTR dsRNA duplexes, but not +ss or −ss NS-3′NTR RNA, upregulated RANTES transcripts (by 17-fold) (Fig. 5B). As expected, Huh7.5 cells had no response to either ds or ss NS-3′NTR RNA. When the various NS-3′NTR RNA ligands were complexed with lipofectamine to enhance their intracellular delivery, again only NS-3′NTR dsRNA stimulated RANTES expression. RANTES was upregulated by ~1600-fold in transfected 7.5-TLR3 cells, but was only induced by 70-fold in Huh7.5 cells. The latter phenotype most likely resulted from MDA5 signaling, which is diminished yet weakly detectable in Huh7.5 cells (our unpublished observations). Collectively, these data suggest that HCV dsRNAs generated during viral replication are the HCV PAMP that activates TLR3, while structured HCV RNAs of either strand are not. We further confirmed these findings in a more physiologic setting using the PH5CH8 cells, which are T-antigen immortalized human hepatocytes expressing TLR3 endogenously and containing a robust TLR3 signaling pathway (15). As shown in the right panel of Fig. 5B, NS-3′NTR dsRNA, but not +ss or −ss NS-3′NTR RNA, potently upregulated RANTES transcripts (by 1975-fold) when added to culture medium (to specifically engage TLR3 in these cells). When transfected into PH5CH8 cells, both +ss and −ss NS-3′NTR RNA potently activated RANTES transcription, as did NS-3′NTR dsRNA. This was not unexpected, as PH5CH8 cells contain an intact RIG-I pathway (15) (as opposed to Huh7.5 cells) that recognizes 3′NTR HCV RNA, which is a potent ligand for RIG-I (11).
Fig. 5. dsRNA is the HCV PAMP that activates TLR3 and subsequent chemokine expression.
(A) Gel electrophoresis of in vitro synthesized HCV ssRNAs and dsRNAs derived from the indicated HCV genome region. +ss: positive-sense strand; −ss: negative-sense strand; ds: double-strand RNA duplex; M: GeneRuler 1kb plus DNA ladder (Fermentas). (B) Huh7.5, 7.5-TLR3 and PH5CH8 cells in 24-well plates were mock-treated, incubated in culture medium (5 μg/0.5 ml) or transfected (1 μg/well) with the +ss, −ss or the ds NS-3′NTR RNA for 6 h. Induction of RANTES mRNA was determined by qPCR. (C) Cells were mock-treated, incubated in culture medium or transfected with the indicated HCV dsRNA, or incubated with poly-I:C in culture medium (pIC) for 6 h. Induction of RANTES mRNA was determined by qPCR.
DsRNAs are readily detectable in cells replicating HCV genomes (18). We confirmed this observation in HCV-infected cells using a monoclonal antibody specifically reacting to dsRNA. More importantly, we observed that HCV dsRNAs substantially co-localized with TLR3 as discrete cytoplasmic foci (Fig. S2). This provides further support for the notion that HCV dsRNA replicative intermediates are sensed by TLR3 intracellularly.
Activation of TLR3-dependent chemokine induction by HCV dsRNAs is independent of the genome-position from which HCV dsRNAs derive
RIG-I preferentially senses a segment of HCV RNA derived from 3′ NTR rich in poly-U/UC motifs (11). To determine whether the activation of TLR3 is influenced by genome position and/or nucleotide composition of HCV dsRNA, we compared the chemokine-inducing abilities of various HCV dsRNAs derived from different regions of HCV genome with varying lengths, including core (0.6-kb), E1 to p7 (1.9-kb), NS5A (1.4-kb) and the NS-3′NTR (6.6-kb) (Fig. 5A). Regardless of their length and nucleotide composition, these different HCV dsRNAs stimulated RANTES mRNA expression with comparable efficiency in both 7.5-TLR3 and PH5CH8 cells (Fig. 5C). Therefore, the TLR3 sensing of HCV dsRNA solely relies on the dsRNA structure, but not on the genome position or nucleotide composition of HCV dsRNA. Additionally, the data reveal that the ability to stimulate TLR3-dependent chemokine production reaches a plateau when HCV dsRNA is longer than 0.6 kb.
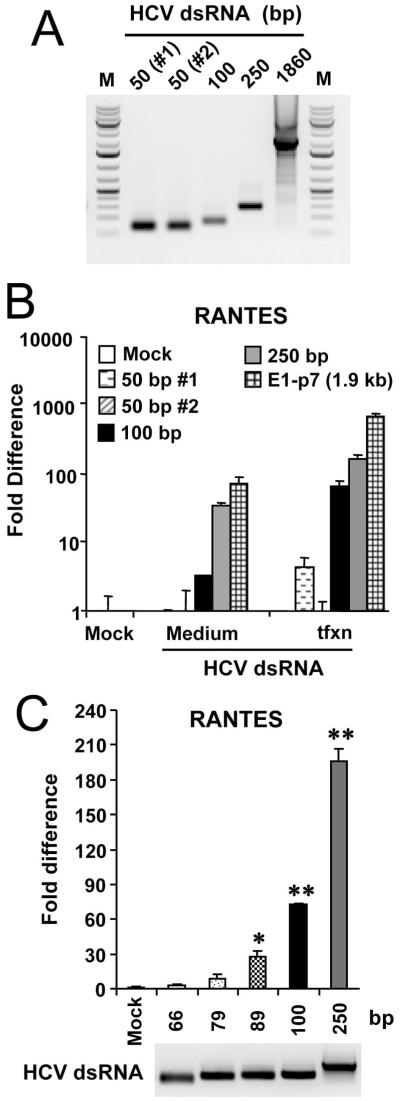
The activation of TLR3 signaling requires HCV dsRNAs with a minimal length of around 80-100 bp
A minimal length of 40-48 bp is required for dsRNA activation of NF-κB in HEK293 cells stably expressing TLR3 (19). To determine the length requirement for HCV dsRNA to activate TLR3 in hepatocytes, we synthesized in vitro a number of HCV dsRNAs derived from the E2/p7-coding region of increasing length, i.e., 50-, 100-, and 250-bp (Fig. 6A), and compared them to the 1.9-kb E1-p7 dsRNA for the capacity to induce RANTES in 7.5-TLR3 cells (Fig. 6B). We found that HCV dsRNAs with a length of ≥ 100 bp all reproducibly upregulated RANTES transcripts when added to culture medium or introduced into cells by transfection. In contrast, there was no reproducible effect on RANTES induction by the two 50-bp HCV dsRNAs, irrespective of the delivery route. Additional refined length mapping experiments revealed that while a 79-bp HCV dsRNA weakly activated RANTES expression, robust activation of TLR3 signaling was achieved when HCV dsRNAs were ≥ 89-bp (Fig. 6C). These data suggest that the efficient activation of TLR3 in hepatocytes requires HCV dsRNA with a minimal length of around 80-100 bp.
Fig. 6. HCV dsRNAs need to have a minimal length of ~80-100 bp to activate TLR3 signaling.
(A) Gel electrophoresis of in vitro synthesized HCV dsRNAs with various length. The 50-, 100- and 250-bp long dsRNAs were all derived from E2/p7-coding region, while the 1860-bp dsRNA was synthesized from the entire E1-p7 coding sequence. (B) 7.5-TLR3 cells were mock-treated, incubated in culture medium or transfected with the indicated HCV dsRNA for 6 h. Induction of RANTES mRNA was determined by qPCR. (C) qPCR analysis of RANTES mRNA induction in 7.5-TLR3 cells transfected with individual HCV dsRNAs of the specified lengths. “*” and “**” denote statistical differences exist when compared with mock-treated cells with a p-value of <0.05 and <0.01, respectively. The lower panel shows gel electrophoresis of the individual HCV dsRNAs used.
Robust production of cytokines and chemokines in primary human hepatocytes (PHHs) following engagement of TLR3
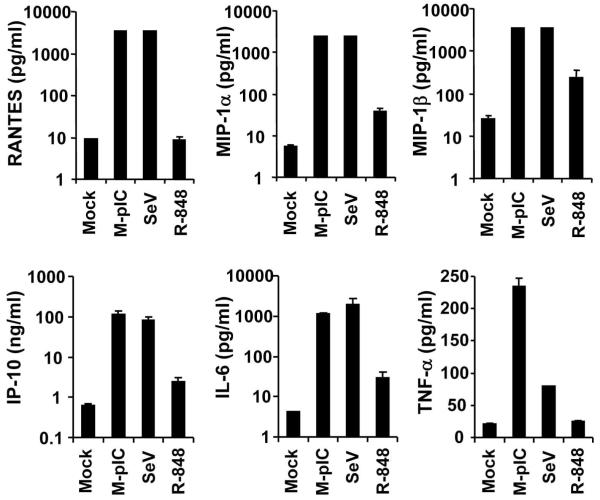
We previously demonstrated that human hepatocytes express TLR3 in situ and isolated PHHs mount a strong ISG response to extracellular poly-I:C stimulation in vitro (12). To determine whether TLR3 signaling in PHHs leads to production of proinflammatory chemokines/cytokines as we observed in HCV-infected 7.5-TLR3 cells, we stimulated PHHs with poly-I:C for 18 h and measured various cytokine/chemokine levels in culture supernatants. It was found that all the cytokines/chemokines induced by HCV in 7.5-TLR3 cells (Fig. 1) were secreted in large quantities from poly-I:C-treated PHHs (Fig. 7). Specifically, production of RANTES, MIP-1α, MIP-1β, IP-10 and IL-6 was upregulated by at least a hundred-fold by poly-I:C, a phenomenon also observed in Sendai virus (SeV)-infected PHHs. Interestingly, TNFα was more efficiently upregulated by poly-I:C (11-fold) than by SeV (4-fold), as was G-CSF (229-fold by poly-I:C vs. 3-fold by SeV, data not shown), indicating that these two cytokines are preferentially induced via the TLR3 pathway over RIG-I in PHHs. When PHHs were treated with the TLR7/8 ligand, R-848, there was weak upregulation (4- to 10-fold) of MIP-1α, MIP-1β, IP-10 and IL-6 but no induction of RANTES, TNFα (Fig. 7) and G-CSF (data not shown), suggesting although engagement of TLR7/8 can moderately induce certain cytokines/chemokines, this pathway plays a minor role in sensing viral infections to produce inflammatory mediators in hepatocytes as compared with the TLR3 and RIG-I pathways. Taken together, the experiments in PHHs demonstrate that TLR3 is a prominent innate immune pathway in human hepatocytes responsible for induction of proinflammatory response to viral infections.
Fig. 7. Engagement of TLR3 by poly-I:C in primary human hepatocyte (PHH) cultures leads to robust production of proinflammatory cytokines/chemokines.
Cells in 12-well plates were mock-treated, stimulated by poly I:C added to culture medium (M-pIC, 25 μg in 0.5 ml medium), infected with SeV (50 HAU), or treated with 10 μM R-848 for 18 h. Culture supernatants were subjected to bioplex assay measuring cytokine/chemokine production. Note both M-pIC- and SeV-treated groups produced high levels of RANTES, MIP-1α and MIP-1β that exceeded the upper detection limit of the assay. The values plotted for these 3 chemokines in these two treatment groups are the upper limit of the assay.
DISCUSSION
Chemokines and cytokines are critical regulators of liver inflammation, and innate and adaptive immunity to HCV, the complex orchestration of which is suggested to determine the outcome of HCV infection (3, 4). The recruitment of T cells to the liver is central in host immune response to HCV infection (2), and is believed to depend mainly on two chemokine receptors expressed on activated T cells, CXCR3 and CCR5 (3, 4). Intrahepatic expression of the ligands for CXCR3 (IP-10, I-TAC and Mig) and CCR5 (RANTES, MIP-1β and MIP-1α) is elevated in hepatits C patients, and the levels of IP-10 and RANTES have been linked to the degree of liver inflammation (3-5). However, the cellular source and mechanism of induction for these chemokines were unclear. We demonstrate in this study that upon infection by HCV, cultured hepatoma cells secret proinflammatory mediators including RANTES, MIP-1β, MIP-1α and IP-10 via TLR3-mediated recognition of HCV dsRNA and activation of NF-κB. Importantly, these observations were not limited to hepatoma Huh7.5 cells reconstituted for TLR3 expression, and we have shown the same repertoire of chemokines and cytokines to be highly upregulated following stimulation by poly-I:C in PHHs (Fig. 7), which contain a robust TLR3 signaling pathway (12). Therefore, not only does the TLR3 pathway mediate the establishment of an antiviral state against HCV infection (12), but it also plays an important role in initiating proinflammatory responses to HCV in hepatocytes and in bridging the innate and adaptive immunity.
The induction of chemokines/cytokines via the TLR3 pathway showed delayed kinetics following HCV infection and did not commence until robust viral replication took place (Fig. 2), implying that HCV replication is needed to produce the PAMP for engagement of TLR3. Consistent with this, UV-inactivated HCV virions were unable to upregulate chemokines (Fig. 2B). The latter result also indicates that HCV entry and virion uncoating processes do not trigger TLR3 activation, neither do the HCV genomic RNAs released upon virion disassembly early after infection. Our finding that HCV dsRNA duplexes, but not structured HCV ssRNAs highly potent for RIG-I activation, are capable of stimulating chemokine expression in 7.5-TLR3 cells (Fig. 5 and see discussions below) explains why TLR3 activation depends on HCV replication, because the latter process yields viral dsRNAs (Fig. S2) (18), the HCV ligands for TLR3. Our results suggest a model in which TLR3 mediates the late phase hepatocellular response to HCV infection by sensing viral dsRNA replicative intermediates, secondary to RIG-I-mediated early response built upon sensing genomic HCV RNA (8, 11, 20). Additionally, TLR3-mediated IFN (12) and cytokine responses may provide a positive feedback to that via RIG-I, whose expression is inducible by IFNs and certain cytokines such as TNFα (21).
The mechanism of TLR3-mediated chemokine/cytokine induction in HCV-infected hepatoma cells revealed in the current study mainly involves the activation of NF-κB-dependent gene transcription, at least for several of the most upregulated chemokines such as RANTES and MIP-1β (Figs. 2-4). Induction of IFNs is not involved in this process, because neither mRNAs for IFN-β and IFN-λs nor secreted antiviral activity was detected at 48 h post HCV infection (data not shown), when active NF-κB complex and chemokine transcription were first observed (Fig. 2-3). Apart from being transcriptionally controlled by NF-κB, a subset of cytokines/chemokines contains AU-rich elements (ARE) in the 3′NTR of their mRNAs that regulate their expression posttranscritionally by controlling mRNA stability and/or translation. Among these cytokines/chemokines are IL-6, TNFα and MIP-1α (22), etc, which were found to be induced in HCV-infected 7.5-TLR3 cells (Fig. 1). Although the TLR3-TRIF pathway is less capable of promoting the stabilization of ARE-containing mRNAs than is the MyD88-dependent pathways (23), further studies are needed to investigate whether ARE-mediated enhancement of mRNA stability/translation also contributes to TLR3-dependent upregulation of specific proinflammatory mediators during HCV infection in hepatocytes.
Out study defines the molecular features of HCV PAMP required for TLR3 activation as HCV dsRNA duplexes ≥ 80-100-bp, regardless of the genome-location or nucleotide composition. Although it is established that TLR3 senses dsRNA (24), whether TLR3 recognizes HCV RNA duplexes or the secondary structures present in HCV RNA (such as the stem-loops in core- and NS5B-coding and 3′NTR regions) has not been determined previously. Our data demonstrated that HCV dsRNA duplexes generated during viral RNA replication are the ligands for TLR3 (Fig. 5), while the secondary structures present in either +ss or −ss HCV RNAs are not, as evidenced by the finding that neither strand of the NS-3′NTR RNA containing complex stem-loop structures in NS5B and 3′NTR sequences activated RANTES expression (Fig. 5B). Further supporting this, the +ss and −ss HCV RNAs derived from HCV core-coding region, which contains two stem-loops (17), also failed to activate TLR3 unless the two RNA strands were annealed to form dsRNA duplexes (data not shown). We speculate that the highly structured HCV ssRNAs may not present the perfect dsRNA structure required for binding to and/or form a stable complex with TLR3 as opposed to HCV dsRNA duplexes.
RIG-I preferentially recognizes a segment of HCV 3′NTR RNA rich in poly-U/UC nucleotides (11). This is not the case for TLR3, since HCV dsRNA duplexes derived from core, E-p7, NS5A and NS-3′NTR regions all activated TLR3 with similar potency (Fig. 5C), regardless of their nucleotide compositions. This suggests that the dsRNA duplex structure is the sole determinant for the HCV dsRNA recognition by TLR3 as long as the dsRNA meets the minimal length requirement (see below). We found that HCV dsRNA has to be at least 80-100-bp to reproducibly activate TLR3 and trigger chemokine induction (Fig. 6). This is consistent with the intracellular localization of TLR3 expressed in reconstituted 7.5-TLR3 cells and of that expressed endogenously in human hepatocytes in situ (12). Although 40-48-bp long dsRNAs bind to TLR3 in vitro and are able to activate TLR3 expressed on the surface of HEK293 cells, only dsRNAs longer than 90-bp can activate TLR3 that is exclusively intracellular (19). A longer stretch of dsRNA may be required to form stable dsRNA-TLR3 receptor dimer complexes in the endosomes (19), where TLR3 appears to be expressed in hepatocytes (Fig. S2) (12).
How dsRNAs generated during HCV RNA replication are delivered to TLR3 in the luminal compartment of endolysosomes is unknown. Possibly, this process is facilitated by autophagy, as demonstrated for recognition of VSV infection in plasmacytoid dendritic cells by TLR7 (25), a viral RNA sensing TLR also residing in endosomes. While this hypothesis requires further investigation, preliminary evidence suggests that inhibition of autophagy by 3-Methyladenine disrupts poly-I:C-induced TLR3 signaling to RANTES and ISG56 induction in both 7.5-TLR3 and PH5CH8 cells (Fig. S3). Furthermore, Bafilomycin A1, which specifically inhibits acidification of the endolysosome (a terminal step in autophagy), blocked poly-I:C-induced ISG expression and NF-κB activation in these hepatocyte cell lines (Fig. S4). The potential dependence on autophagy for HCV activation of TLR3 signaling would contrast with that reported for RIG-I, which is negatively regulated by autophagy (26).
The identification of TLR3 as an active player in mediating proinflammatory cytokine/chemokine responses to HCV in hepatocytes provides novel insights into host immune response to HCV and pathogenesis of HCV-associated liver injury. It remains to be investigated in vivo how much TLR3-mediated signaling contributes to antiviral defense and protective immune responses that culminate in HCV clearance, and how much it is involved in chronic liver inflammation and the progression to fibrosis and ultimately hepatocellular carcinoma. Answers to these questions will yield valuable information to develop novel immunotherapies for hepatitis C.
Supplementary Material
Acknowledgments
Financial support:
This work was supported by National Institutes of Health grants AI069285 (K.L.) and CA133322 (L.M.P.).
List of Abbreviations
- HCV
hepatitis C virus
- TLR3
Toll-like receptor 3
- RLR
RIG-I-like receptors
- PAMP
pathogen associated molecular pattern
- IFN
interferon
- RIG-I
retinoic inducible gene I
- MAVS
mitochondrial antiviral signaling protein
- TRIF
Toll-IL1 receptor homology domain containing adaptor inducing interferon β
- IRF3
interferon regulatory factor-3
- NF-κB
nuclear factor kappa B
- ISG
interferon stimulated gene
- dsRNA
double-stranded RNA
- MOI
multiplicity of infection
- qPCR
quantitative PCR
- ChIP
chromatin immunoprecipitation
- EMSA
electrophoretic mobility shift assay
- NS
nonstructural
- NTR
non-translated region
- CAPE
Caffeic acid phenethyl ester
- PHH
primary human hepatocyte
- TLR7
Toll-like receptor 7
Footnotes
Contact information:
Kui Li, PhD. Department of Microbiology, Immunology and Biochemistry, University of Tennessee Health Science Center, 858 Madison Avenue, Memphis, TN 38163, USA. Tel: 1-901-448-2571; Fax: 1-901-448-7360; kli1@uthsc.edu.
REFERENCES
- 1.Lemon SM. Induction and evasion of innate antiviral responses by hepatitis C virus. J Biol Chem. 2010;285:22741–22747. doi: 10.1074/jbc.R109.099556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bowen DG, Walker CM. Adaptive immune responses in acute and chronic hepatitis C virus infection. Nature. 2005;436:946–952. doi: 10.1038/nature04079. [DOI] [PubMed] [Google Scholar]
- 3.Zeremski M, Petrovic LM, Talal AH. The role of chemokines as inflammatory mediators in chronic hepatitis C virus infection. J Viral Hepat. 2007;14:675–687. doi: 10.1111/j.1365-2893.2006.00838.x. [DOI] [PubMed] [Google Scholar]
- 4.Wald O, Weiss ID, Galun E, Peled A. Chemokines in hepatitis C virus infection: pathogenesis, prognosis and therapeutics. Cytokine. 2007;39:50–62. doi: 10.1016/j.cyto.2007.05.013. [DOI] [PubMed] [Google Scholar]
- 5.Helbig KJ, Ruszkiewicz A, Lanford RE, Berzsenyi MD, Harley HA, McColl SR, Beard MR. Differential expression of the CXCR3 ligands in chronic hepatitis C virus (HCV) infection and their modulation by HCV in vitro. J Virol. 2009;83:836–846. doi: 10.1128/JVI.01388-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kawai T, Akira S. Toll-like receptor and RIG-I-like receptor signaling. Ann N Y Acad Sci. 2008;1143:1–20. doi: 10.1196/annals.1443.020. [DOI] [PubMed] [Google Scholar]
- 7.Li K, Foy E, Ferreon JC, Nakamura M, Ferreon AC, Ikeda M, Ray SC, et al. Immune evasion by hepatitis C virus NS3/4A protease-mediated cleavage of the Toll-like receptor 3 adaptor protein TRIF. Proc Natl Acad Sci U S A. 2005;102:2992–2997. doi: 10.1073/pnas.0408824102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Loo YM, Owen DM, Li K, Erickson AK, Johnson CL, Fish PM, Carney DS, et al. Viral and therapeutic control of IFN-beta promoter stimulator 1 during hepatitis C virus infection. Proc Natl Acad Sci U S A. 2006;103:6001–6006. doi: 10.1073/pnas.0601523103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Meylan E, Curran J, Hofmann K, Moradpour D, Binder M, Bartenschlager R, Tschopp J. Cardif is an adaptor protein in the RIG-I antiviral pathway and is targeted by hepatitis C virus. Nature. 2005;437:1167–1172. doi: 10.1038/nature04193. [DOI] [PubMed] [Google Scholar]
- 10.Li XD, Sun L, Seth RB, Pineda G, Chen ZJ. Hepatitis C virus protease NS3/4A cleaves mitochondrial antiviral signaling protein off the mitochondria to evade innate immunity. Proc Natl Acad Sci U S A. 2005;102:17717–17722. doi: 10.1073/pnas.0508531102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Saito T, Owen DM, Jiang F, Marcotrigiano J, Gale M., Jr. Innate immunity induced by composition-dependent RIG-I recognition of hepatitis C virus RNA. Nature. 2008;454:523–527. doi: 10.1038/nature07106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wang N, Liang Y, Devaraj S, Wang J, Lemon SM, Li K. Toll-like receptor 3 mediates establishment of an antiviral state against hepatitis C virus in hepatoma cells. J Virol. 2009;83:9824–9834. doi: 10.1128/JVI.01125-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sumpter R, Jr., Loo YM, Foy E, Li K, Yoneyama M, Fujita T, Lemon SM, et al. Regulating Intracellular Antiviral Defense and Permissiveness to Hepatitis C Virus RNA Replication through a Cellular RNA Helicase, RIG-I. J Virol. 2005;79:2689–2699. doi: 10.1128/JVI.79.5.2689-2699.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wang N, Dong Q, Li J, Jangra RK, Fan M, Brasier AR, Lemon SM, et al. Viral induction of the zinc finger antiviral protein is IRF3-dependent but NF-kappaB-independent. J Biol Chem. 2010;285:6080–6090. doi: 10.1074/jbc.M109.054486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Li K, Chen Z, Kato N, Gale M, Jr., Lemon SM. Distinct poly(I-C) and virus-activated signaling pathways leading to interferon-beta production in hepatocytes. J Biol Chem. 2005;280:16739–16747. doi: 10.1074/jbc.M414139200. [DOI] [PubMed] [Google Scholar]
- 16.Natarajan K, Singh S, Burke TR, Jr., Grunberger D, Aggarwal BB. Caffeic acid phenethyl ester is a potent and specific inhibitor of activation of nuclear transcription factor NF-kappa B. Proc Natl Acad Sci U S A. 1996;93:9090–9095. doi: 10.1073/pnas.93.17.9090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Tuplin A, Wood J, Evans DJ, Patel AH, Simmonds P. Thermodynamic and phylogenetic prediction of RNA secondary structures in the coding region of hepatitis C virus. RNA. 2002;8:824–841. doi: 10.1017/s1355838202554066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Targett-Adams P, Boulant S, McLauchlan J. Visualization of double-stranded RNA in cells supporting hepatitis C virus RNA replication. J Virol. 2008;82:2182–2195. doi: 10.1128/JVI.01565-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Leonard JN, Ghirlando R, Askins J, Bell JK, Margulies DH, Davies DR, Segal DM. The TLR3 signaling complex forms by cooperative receptor dimerization. Proc Natl Acad Sci U S A. 2008;105:258–263. doi: 10.1073/pnas.0710779105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wagoner J, Austin M, Green J, Imaizumi T, Casola A, Brasier A, Khabar KS, et al. Regulation of CXCL-8 (interleukin-8) induction by double-stranded RNA signaling pathways during hepatitis C virus infection. J Virol. 2007;81:309–318. doi: 10.1128/JVI.01411-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Matsumiya T, Prescott SM, Stafforini DM. IFN-epsilon mediates TNF-alpha-induced STAT1 phosphorylation and induction of retinoic acid-inducible gene-I in human cervical cancer cells. J Immunol. 2007;179:4542–4549. doi: 10.4049/jimmunol.179.7.4542. [DOI] [PubMed] [Google Scholar]
- 22.Khabar KS. Post-transcriptional control during chronic inflammation and cancer: a focus on AU-rich elements. Cell Mol Life Sci. 2010;67:2937–2955. doi: 10.1007/s00018-010-0383-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Datta S, Novotny M, Li X, Tebo J, Hamilton TA. Toll IL-1 receptors differ in their ability to promote the stabilization of adenosine and uridine-rich elements containing mRNA. J Immunol. 2004;173:2755–2761. doi: 10.4049/jimmunol.173.4.2755. [DOI] [PubMed] [Google Scholar]
- 24.Alexopoulou L, Holt AC, Medzhitov R, Flavell RA. Recognition of double-stranded RNA and activation of NF-kappaB by Toll-like receptor 3. Nature. 2001;413:732–738. doi: 10.1038/35099560. [DOI] [PubMed] [Google Scholar]
- 25.Lee HK, Lund JM, Ramanathan B, Mizushima N, Iwasaki A. Autophagy-dependent viral recognition by plasmacytoid dendritic cells. Science. 2007;315:1398–1401. doi: 10.1126/science.1136880. [DOI] [PubMed] [Google Scholar]
- 26.Ke PY, Chen SS. Activation of the unfolded protein response and autophagy after hepatitis C virus infection suppresses innate antiviral immunity in vitro. J Clin Invest. 2011;121:37–56. doi: 10.1172/JCI41474. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.