Introduction
In the clinical setting, pathogenic bacteria form biofilms on a variety of medical implants, such as indwelling catheters, artificial heart valves, and pacemakers (Darouiche, 2001; O'Gara and Humphreys, 2001; Wilson, 2001). Biofilms are bacterial communities encased in a hydrated extracellular matrix, which may consist of proteins, polysaccharides, nucleic acids, or combinations of these molecules (Branda et al., 2005). The development of biofilms on medical devices presents significant medical problems because bacteria in the biofilm mode of growth are highly resistant to treatment with antibiotics and clearance by the host’s immune system. Therefore, once these bacterial communities form they are extremely difficult to eradicate with conventional therapies. The biofilm mode of growth has been recognized as an important virulence factor for many pathogens, including Staphylococcus epidermidis and Staphylococcus aureus. Consequently, bacterial biofilms have become important targets for drug discovery and for testing conventional antibacterial compounds for anti-biofilm activity.
Here, we present an in vitro assay for culturing staphylococcal biofilms and biofilms of non-motile gram-positive bacteria under static conditions in microtiter assay plates, and for the quantification of biofilm growth using a simple staining procedure that measures the amounts of bacterial cells and extracellular matrix. This protocol is based on the assay first described by Chistensen et al. (Christensen et al., 1985), and was brought to the attention of wider group of biofilm researchers when it was adapted for genetic screens by O’Toole and Kolter (O'Toole and Kolter, 1998). The protocols described in this unit include improvements described by other groups (Christensen et al., 1995; Cramton et al., 2001a; Deighton et al., 2001; Mack et al., 2001; Merritt et al., 2005) and those developed in our laboratory. While the basic assay is simple to perform and requires a minimum of equipment, it is very powerful and versatile, and with some ingenuity it can be adapted to examine many aspects of bacterial biofilm biology.
In the following pages, we describe a basic assay to measure biofilm formation of several Gram-positive pathogens (Protocol 1) in standard microtiter assay plates. This basic assay can be adapted readily to study several aspects of biofilm formation. For example, we present an alternative protocol for measuring biofilm formation on medically relevant materials, such as silicone. In addition, the basic assay can be adapted for high-throughput screening to identify small molecule inhibitors of biofilm formation or biofilm-defective mutants. In Basic Protocol 2, we describe how to identify the optimal media conditions and incubation times for a high throughput screen described in Basic Protocol 3. Finally, we describe how to adapt the basic assay that you have optimized in basic protocol 2 for the evaluation of the anti-biofilm treatments by measuring the Minimal Biofilm Inhibitory Concentration (MBIC). While these methods are deceptively simple, they require careful attention to detail in order to obtain meaningful results. While we have attempted to highlight the most likely sources of variability in the assay protocols, we encourage you to optimize the assays for the conditions that exist in your laboratory.
Basic Protocol 1. Basic assay for biofilm formation of non-motile Gram-positive bacteria
Here we describe a versatile method for growing and quantifying biofilms cultures in 96-well assay plates. This standard protocol can be used for a variety of non-motile Gram-positive bacterial species, such as Staphylococcus aureus, S. epidermidis, Enterococcus faecalis, and E. faecium. However, you may find that you will need to manipulate the assay conditions to optimize biofilm formation for a particular strain of interest. Guidelines for the variables that can be manipulated to optimize biofilm growth are given in Basic Protocol 2.
Materials list
Sterile solid growth media—Trytpic Soy Agar (TSA) (Difco™, Becton, Dickenson and Co. (BD))
Sterile growth media—Trytpic Soy Broth (TSB) (Bacto™ (BD))
Sterile 80% glycerol (autoclaved)
Culture tubes (e.g. 16 × 150 mm)
40% glucose (filter sterile)
96 well assay plates with lids (Costar 3595-flat bottomed, polystyrene, tissue culture treated or equivalent)
Adhesive foil lids (Costar 6569, optional)
Incubator set at 37 °C
Plate washer, e.g. BioTek ELx405 or equivalent (optional)
Multichannel pipetor (optional)
Pipetting reservoirs
Two 2 L beakers (optional)
Washing solution e.g. Deionized water (dH20), phosphate buffered saline (PBS), or 0.9% saline
Baking oven set at 60 °C
95% ethanol
0.06% crystal violet (w/v) dissolved in deionized water
Microtiter plate reader (Molecular Devices, optional)
Camera (optional)
Preparation of freezer stocks
-
1.
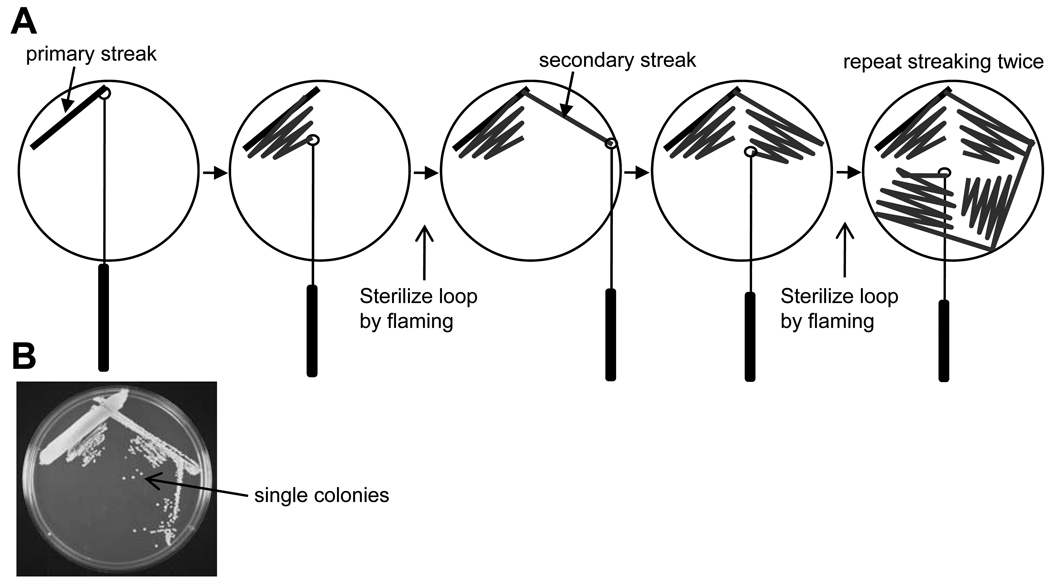
Streak the strain(s) of interest on an appropriate solid growth medium using the streaking technique shown in Fig. 1 to produce single, well-isolated colonies. We routinely use Tryptic Soy Agar (TSA; BD) for cultivating staphylococci and enterococci. However, other solid media, such as Brain Heart Infusion (BHI) agar (BD) or blood agar plates (TSA II + 5% defibrinated sheep blood) will support the growth of several Gram-positive bacterial species.
-
2.
Incubate plates overnight at 37 °C.
-
3.
Pick 1–2 isolated colonies, inoculate a sterile culture tube containing 3 ml Tryptic Soy Broth (TSB), and grow cultures aerobically (shaker or tube roller) overnight at 37 °C.
-
4.
Create a freezer stock of each isolate by mixing 0.2 ml sterile 80% glycerol with 0.8 ml of the overnight culture (final concentration of glycerol is equal to 16%) in a 2 ml cryogenic storage vial (e.g. Nalgene cryogenic vials, catalog number 5000-0020). Place freezer stocks at −80 °C.
Note: 80% glycerol can be readily pipetted at room temperature.
Figure 1.
Streaking bacteria on solid agar to obtain single, well-isolated colonies. A) Streaking plates. Plates can be streaked using a standard wire inoculating loop, a disposable plastic loop, a toothpick, or a long “picking stick”. The wire loop can be sterilized in a flame, but it must be cooled by touching the agar surface prior to making contact with bacteria. Single colonies can be isolated by repeated streaking as illustrated in the figure. B) An example of the results obtained using this method. Single, well-isolated colonies are indicated.
Preparation of inocula
-
5.
Strains. Several readily available biofilm-proficient staphylococcal and enterococcal strains are listed in Table 1.
-
6.
Grow an overnight culture for each strain to be tested. Using a sterile tooth pick, inoculating loop, or applicator stick remove a small portion of the frozen stock culture and transfer it to a sterile culture tube containing 3 ml TSB.
-
7.
Grow the culture(s) overnight at 37 °C under aerobic conditions i.e. in a rotary shaker or in a tube roller.
-
8.
Prepare inocula by diluting the overnight culture(s) 1:100 in the amount of an appropriate assay media required for the experiment. Mix thoroughly. We routinely use TSB or TSB supplemented with additional glucose to a final concentration of 1%. It is important to use a standardized inoculum as biofilm density varies with inoculum size.
Media. An all-purpose media that is appropriate for biofilm assays using many low G+C Gram-positive organisms, such as S. epidermidis, S. aureus, E. faecalis, and E. faecium, is TSB supplemented with glucose to a final concentration of 1%. Note: TSB contains 0.25% glucose; add 1.88 ml of filter sterilized 40% glucose to 100 ml TSB to achieve a final concentration of 1%.
Several strains of S. epidermidis, such as ATCC 35984 (RP62A), form robust biofilms in TSB media containing 0.25% glucose.
Other media, such as Brain Heart Infusion (BHI) or CYGP broth supplemented with 1% glucose have been used for growing Gram-positive biofilms (Balaban et al., 2003; O'Neill et al., 2008). However, depending on the biofilm forming ability of the strain, it may be desirable to adjust the media conditions so that the amount to biofilm formation falls within the linear range of the detection method to be used. See protocol below.
Table 1.
Biofilm-forming staphylococcal and enterococcal strains.
| Species | Strain | Description | Source/Reference |
|---|---|---|---|
| S. aureus | ATCC 35556 (SA113) | Lab strain. Biofilm proficient | ATCC (Iordanescu and Surdeanu, 1976) |
| NCTC 8325 (ATCC 12600, NRS-77) | Lab strain. Biofilm proficient. Parent of SA113 | ATCC or NARSA (Novick, 1967) | |
| UAMS-1 (ATCC 49230) | Clinical isolate. Biofilm proficient | ATCC (Gillaspy et al., 1995) | |
| MW2 (NRS-123) | MRSA | NARSA (Baba et al., 2002) | |
| ATCC 25923 | Reference strain. Biofilm negative | ATCC | |
| S. epidermidis | ATCC 35984 (RP62A) | Clinical isolate. Biofilm proficient | ATCC (Christensen et al., 1982) |
| 1457 | Clinical isolate. Biofilm proficient | (Mack et al., 1992) | |
| O-47 | Clinical isolate. Biofilm proficient | (Heilmann et al., 1996) | |
| 18972 | Clinical isolate. Biofilm proficient | Oscient Pharmaceuticals, Waltham, MA | |
| ATCC 14990 | Reference strain. Biofilm deficient, but is biofilm proficient under oxygen limited conditions. | ATCC | |
| ATCC 12228 | Reference strain. Biofilm negative. | ATCC | |
| E. faecalis | ATCC 51299 | VRE. Biofilm proficient | ATCC |
| ATCC 29212 | Reference strain. Biofilm proficient | ATCC |
Abbreviations: ATCC, American Type Culture Collection (Manassas, VA); NARSA; Network on Antibiotic Resistance in Staphylococcus aureus (Chantilly, VA); MRSA, Methicillin Resistant Staphylococcus aureus; VRE, Vancomycin Resistant Enterococcus.
Biofilm growth
-
9.
Transfer 200 µl of the inoculum to the assay wells of a sterile 96-well assay plate, which corresponds to an inoculum of approximately 5 × 106 cells/well. We generally use tissue-culture treated polystyrene 96-well assay plates with flat bottoms (e.g. Costar 3595) for biofilm assays using staphylococci and enterococci. To compensate for the considerable variability that is inherent in these assays, we recommend using at least 3–4 replicates for each strain/condition, from which we can calculate statistically significant values for average biofilm formation and the standard deviation. We routinely inoculate an entire column (8 wells) of a 96-well assay plate with a single strain or assay condition, which provides enough data points to achieve statistical significance. Each plate should include a media only control (no biofilm formation) and a positive control strain (e.g. see Table 1)
There is considerable variation between tissue culture treated plates produced by different manufacturers, and there is considerable variation between plates from different lots produced by the same manufacturer. These variations in assay plates will influence the results of the assays. Therefore, we recommend trying plates from different manufacturers to identify those that are most suitable for your assay and then using plates from the same lot number for a series of related experiments.
To speed up the inoculation step, a multichannel pipettor can be used to transfer the inocula to the assay plates. If the entire plate is to be inoculated with the same bacterial strain, then a liquid handling device, such as a Multidrop DW (Thermo Scientific) or Well-Mate (Thermo Scientific) can be used to inoculate plates.
-
10.
Cover each plate with the lid supplied by the manufacturer. Alternatively, the assay plates can be covered with an adhesive foil lid (e.g. Costar 6569), which increases biofilm formation by creating an environment with reduced oxygen tension.
Biofilm formation of staphylococci is stimulated by low oxygen conditions (Cramton et al., 2001b), and the foil lid produces an environment with reduced oxygen tension.
-
11.
Transfer inoculated assay plates to an incubator set at 37 °C and incubate them for an appropriate time (16–18 h) without shaking (static culture). Non-motile Gram-positive bacteria, such as staphylococci and enterococci, form biofilms on the bottom of each assay well.
The time of incubation required for robust biofilm formation may vary considerably in a strain-dependent manner. This assay can be used to generate a biofilm growth curve that can be used to identify the optimal time of growth for media/temperature conditions (see Protocol 2)
Processing plates
-
12.
Remove lids from assay plates.
-
13.
After incubation, measure the optical density at 600 nm (OD600) of each well using a multiwell plate reader (e.g. Molecular Devices) to quantify overall growth (biofilm and planktonic). This step is used to identify strains that are defective in overall growth or conditions that inhibit overall growth, resulting in decreased biofilm growth.
-
14.
Remove liquid culture from each well, and remove non-adherent bacteria by washing each well 3–4 times with deionized water (dH20) while taking care to preserve the structure of the biofilm located on the bottom of each assay well. This step can be accomplished in many ways. In our experience, the most reliable method is to use a commercially available plate washer, such as a BioTek ELx-405 (BioTek Instruments Inc, Winooski, VT). This machine will remove the liquid culture and wash the bottom of the well in a consistent, automated fashion. The washing parameters, such as dispensing speed, distance from bottom of aspiration, can be optimized for biofilms formed by different bacterial species. Because many researchers may not have access to a plate washer, two alternative washing methods are described below.
Manual methods for plate washing.
The first method involves removing liquid culture by inverting the assay plate and decanting the liquid into a waste receptacle (a 2 L beaker will suffice) with a gentle flick of the wrist. The entire plate is then “dunked” into a large beaker (e.g. a 2 L beaker) containing dH20 by inserting the entire plate at an angle (approximately 45 °) into the water starting with one end and continuing until the entire plate is submerged. The dH20 in each well is then decanted into a waste beaker as described above. Repeat this process 1–3 times. Invert the washed plates on a paper towel to dry. Because the dunking container will contain bacteria, care should be taken to avoid contact with the dunking solution by wearing gloves and by using a large container of dH20. All contaminated dH20 should be decontaminated by addition of bleach to a final concentration of 10% and contaminated paper towels should be autoclaved. Gloves should be worn while handling assay plates after washing.
-
The second method involves decanting liquid by inverting the plate as above, but a multichannel pipettor is used to carefully transfer 200 µl wash solution (dH20) to each well. Sufficient care must be taken when pipetting liquid into the wells to avoid disrupting the structure of the biofilm. We have found that it is best to tilt the assay plate and touch the side of the wells with the pipet tips while pipetting slowly so the liquid flows down side of well. The wash solution can be decanted as described above. Alternatively, wash solutions can be removed using a multichannel pipet while taking care to avoid disrupting the biofilm. Major sources of variability are pipetting strength and decanting.
The wash solution and method may be varied depending on the assay endpoint. For example, if you need to measure the number of viable cells in each well or if you need to culture bacteria from biofilms, then a sterile buffer, such as phosphate-buffered saline (PBS), Tris-buffered saline (TBS), or 0.9% saline, should be used to wash non-adherent cells from the biofilm cultures.
-
15.
Fixing biofilms. To fix adherent cells prior to staining, incubate washed plates at 60 °C for at least 60 min. This step reduces variability caused by loss of biofilm during the staining process. In our experience, heat fixation is the most reliable method. Staining time is quicker with this method
Alternative fixation methods. If a suitable incubator is not available, robust biofilms can be fixed by the addition of 50 µl 95% ethanol to each well followed by incubation for 30 min. Decant ethanol after fixation and air-dry plates. Because this fixation method can dislodge the biofilms of some strains (e.g. S. aureus), it must be tested on each strain prior to use. In addition, biofilms fixed with ethanol require increased staining times (see below). Althernatively, biofilm cells can be fixed by adding 150 µl/well of Bouin's fxative [0.5 g of picric acid is solubilized in 37.5 ml of distilled water; 12.5 ml of 37% (v/v) formaldehyde and 2.5 ml of concentrated acetic acid are added] for 15 min (Mack et al., 2001). Fixed cells are washed once with PBS and stained as described below.
-
16.
Staining. Biofilms can be detected and quantified using various stains. For most assays we find that staining with a 0.06% (w/v) solution of crystal violet dissolved in dH2O as previously described (Christensen et al., 1995; Christensen et al., 1985) produces excellent results. The amount of CV bound in each well is proportional to the amount of biofilm. This stain is readily available and inexpensive, and it can be evaluated visually, photographed, or quantified spectrophotmetrically using a microtiter plate reader. To stain biofilms add 50 µl 0.06% crystal violet dissolved in dH2O to each well and allow at least 5 minutes for staining if biofilms were heat-fixed (60 °C). If biofilms were fixed with 95% ethanol, allow staining to proceed for at least 60 min. After the staining reaction has been completed, remove excess stain by repeated washing (3–4 washes) with dH2O as described above. The wash solution should be clear after final last washing step.
-
17.
Quantification. The amount of crystal violet bound in each well can be directly quantified spectrophotometrically by measuring the OD600 using an appropriate microplate reader (e.g. Molecular Devices).
Most plate readers measure absorbance in a small area of the assay well. Therefore, the results obtained using this method will be affected when the integrity of the stained biofilms has been disrupted. In addition, this method will not measure biofilms that may form on the sides of the assay well.
Alternatively, the crystal violet can be eluted from stained biofilms by adding 200 µl 30% acetic acid to each well, transferring the eluted dye to a fresh assay plate, and measuring the OD600. Calculate average OD600 values and standard deviation for replicate wells. This method yields results that are more quantitative than those obtained by directly measuring OD600 of crystal violet bound to the bottom of the well. If OD600 readings are ≥ 2.5 (see Fig. 3), then dilute eluted crystal violet 1:10 in 30% acetic acid (add 20 µl CV to 180 µl 30% acetic acid) and measure OD600. Multiply readings by a factor of 10 to obtain the actual OD600 values. This method has the advantage that it measures the amount of crystal violet bound to the entire well, and not just the middle of the well. One disadvantage of this method is that it adds 1–2 more steps to the process and uses 1–2 additional microtiter assay plates, making it less practical for high-throughput screens (see below).
-
18.
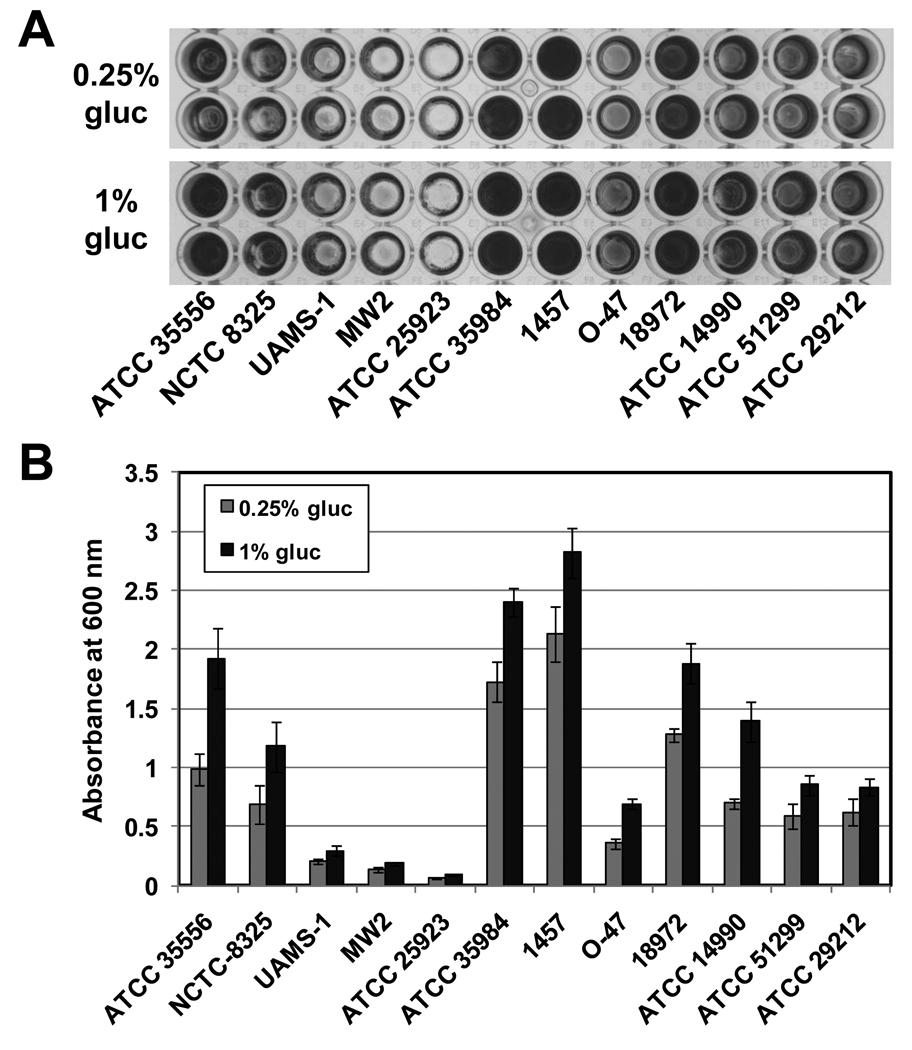
Representative results of this assay using the strains listed in Table 1 are shown in Figure 2.
Figure 3.
Setting up assay plates for biofilm formation on alternative substrates
Figure 2.
Results of a biofilm assay using the strains listed in Table 1 demonstrates the range of phenotypes of individual strains and the stimulatory effect of glucose. Biofilm cultures were grown in 0.5X TSB supplemented with 0.25% or 1% glucose (gluc) at 37 °C for 18 h. A) Photographs of two representative rows from each tissue culture treated 96-well assay that was stained with crystal violet. B) The amount of crystal violet bound was quantified by measuring absorbance at 600 nm and plotted for each strain. Each bar represents the average of 8 individual assay wells, and the error bars represent the standard deviation.
Alternate Protocol 1. Biofilm formation on alternative surfaces, such as medical grade materials
Several medical grade materials, such as silicone or polyurethane, can be purchased in the form of sheeting of various thicknesses. Biofilm formation on these alternative surfaces can be measured by introducing some slight modifications to Basic Protocol 1 as described by Shanks et al. (Shanks et al., 2005).
Materials
Same as Basic Protocol 1, plus:
Silicone sheeting, 0.03” thick (Cardiovascular Instrument Corp, Wakefield, MA)
Polyurethane sheeting (Pellethane 55-D, Specialty Silicone Fabricators, Inc., Paso Robles, CA)
#5 cork borer
24-well assay plate (Costar 3526)
Silicone sealant (e.g. Silicone II, GE Sealants and Adhesives, Huntersville, NC)
Forceps and alcohol for sterilization.
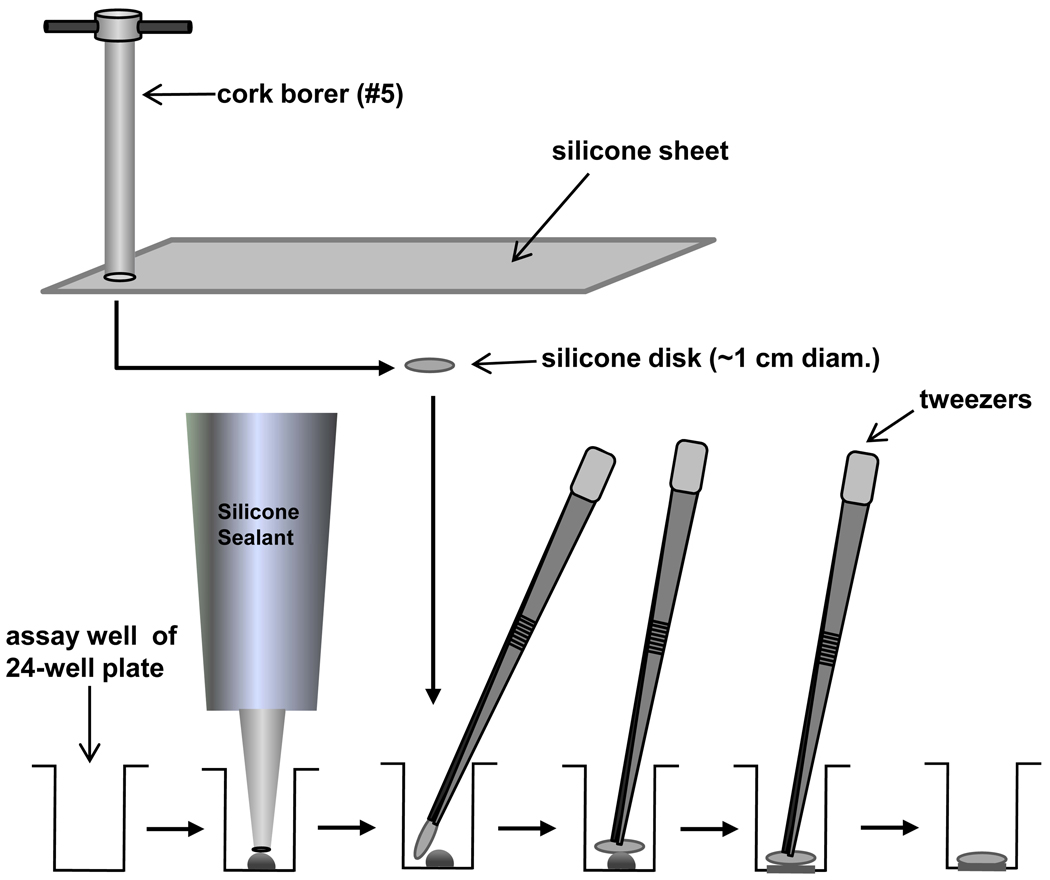
Cut disks from sheets of non-reinforced medical grade silicone or polyurethane sheeting using a #5 cork borer (~1 cm diameter).
Sterilize disks using an autoclave.
-
Attach disks to the bottoms of the wells of a 24-well assay plate (Costar 3526) using silicone sealant (see Fig. 2).
Dispense a small amount of sealant (~100 µl) onto the bottom of the well. Carefully lay a sterile disk on the sealant using forceps that have been sterilized with alcohol and flamed (There should be 1–2 mm space between the disk and the side of the well. If this is not the case, cut smaller disks). Press the disk toward the bottom of the well to evenly distribute the sealant under the disk. Allow the sealant to cure for at least the minimum time recommended by the manufacturer.
Prepare bacterial inocula as described in Basic Protocol 1. You will need 2 ml inocula/assay well.
Inoculate each well with 2 ml and cover the assay plate with the supplied by the manufacturer.
Incubate the assay plates for approximately 18 h at 37 °C.
Remove media and non-adherent cells by repeated manual washing with water using the “dunking” method or by adding and removing wash solutions with a pipet (see basic protocol 1).
Fix adherent cells (biofilms) by heating at 60 °C for 60 min.
Stain the fixed biofilms with 200 µl of 0.06% crystal violet as described above.
Remove excess CV by repeated washing using the dunking method.
To elute the crystal violet, each stained disk must be removed from the bottom of the assay well and transferred to a separate tube. We routinely use 2 ml microcentrifuge tubes for this purpose, however, it is important to ensure that the stained surface is facing the interior of the tube. Add 1 ml 30% acetic acid to each tube, close the tube and vortex.
Transfer eluted crystal violet to 4 separate wells (200 µl/well) of a 96-well assay plate and measure the optical density at 600 nm (OD600) was measured using a microtiter plate reader. Determined the average and standard deviation of the OD600 values obtained for each disk.
Basic Protocol 2. Optimizing assay conditions
Here we describe two experiments designed to optimize two important parameters of the biofilms formation assay: 1) the amount of time for growth of the biofilm cultures, 2) the media conditions that results in robust growth that is within the linear range of detection of the assay set up. Both of these parameters can have profound effects on the outcome of this assay. As you will see, biofilm cultures follow the same pattern of growth phases as do planktonic cultures i.e. a lag phase, an exponential growth phase, and stationary phase. In addition, the biofilm structure in static biofilm cultures begins to fall apart after nutrients have been depleted, resulting in an apparent decrease in biofilm formation. Therefore, it is critical to identify and standardize the time of growth required to reach the desired growth phase. Finally, if the assay is to be used to screen for biofilm-defective mutants or treatments that inhibit biofilm formation, it is advisable to optimize the biofilm growth media to ensure that the amount of crystal violet staining is within the linear range of detection of your microplate reader. This will increase the sensitivity of the screen and the likelihood of identifying mutants or treatments that produce subtle defects in biofilm formation.
Materials
Same as Basic Protocol 1
Biofilm growth curve to optimize time of growth
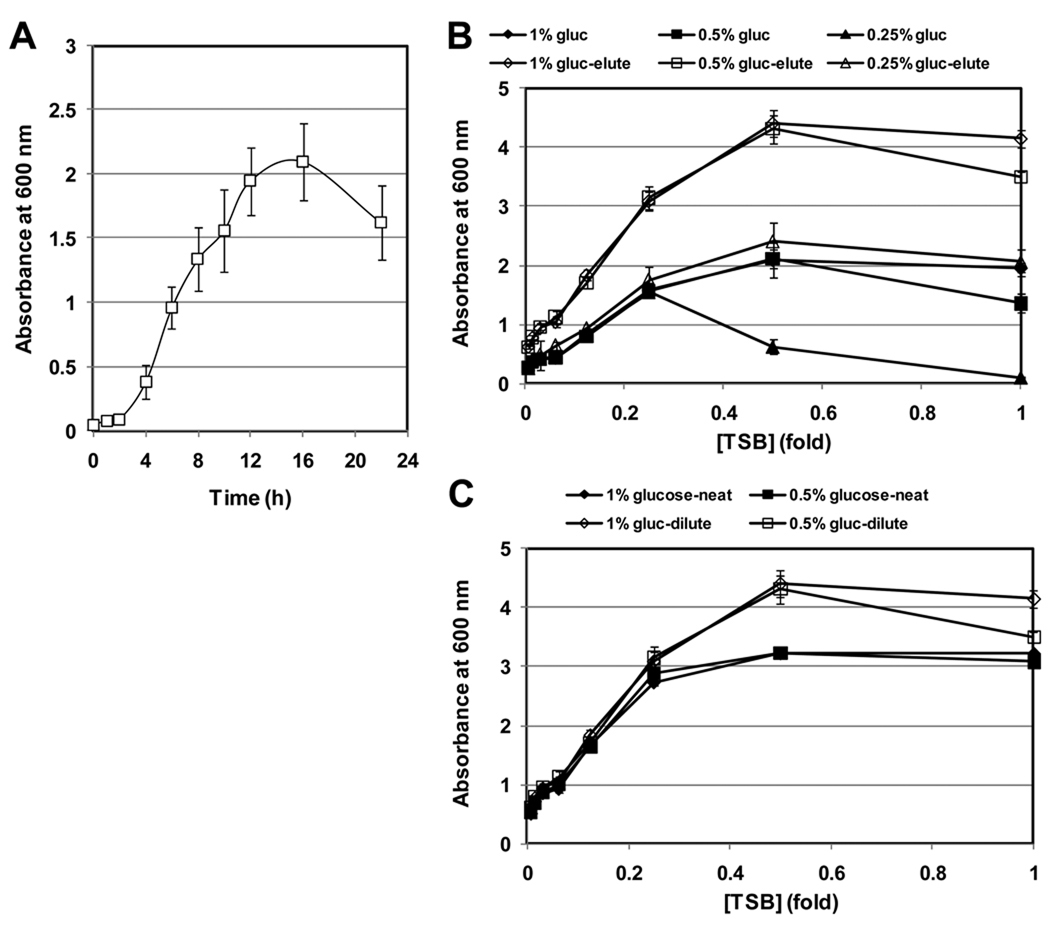
A simple biofilm growth curve can be carried out to identify the optimal time of growth for your biofilm assay. The biofilm growth curve is conceptually identical to a classic planktonic growth curve analysis, except biofilm growth will be measured using the detection method described above. A representative biofilm growth curve is shown in Fig. 3A. Because the biofilm cultures are killed by the detection method, several identical assay plates must be set up, and one plate will be used for each time point that will be analyzed.
Prepare inocula of the strain(s) of interest as described in Basic protocol 1.
-
Transfer 200 µl of the inoculum to the assay wells of several sterile 96-well assay plates as described in Basic protocol 1. Several strains and/or media conditions (3–4) can be analyzed in the same assay. Because the biofilm cultures will be killed by the detection method, inoculate one plate for each time point of the biofilm growth curve. It is not necessary to inoculate the entire plate. We recommend that you inoculate 24–48 wells/plate/strain, or 3–6 columns of a 96-well assay plate. If you measure biofilm formation at 9 time points (see below), you will need a total of 9 assay plates.
We recommend measuring biofilm growth at the following time points: 0, 2, 4, 6, 8, 12, 16, 20, and 24 h.
By inoculating ≥ 24 wells/plate, the number of data points will ensure the average values obtained will be less susceptible to large errors caused by well-to-well and plate-to-plate variations.
S. aureus forms a mature biofilm in 12–16 h in 0.5X TSB supplemented with 1% glucose (see Fig 3).
Cover assay plates and place all assay plates in an incubator set at 37 °C as described above.
Immediately remove one plate (t = 0 h), and process it as described in basic Protocol 1: Measure OD600, wash the plate, fix, and stain biofilms with crystal violet.
Repeat this process for the remaining time points of the biofilm growth curve.
Read the OD600 of the stained biofilms using an appropriate microtiter plate reader.
Calculate the average OD600 of crystal violet bound and the standard deviation for each time point.
Plot the average OD600 of crystal violet bound vs. time of biofilm growth.
Identify optimal time of growth for the assay. Based on the results shown in Fig. 3A, we measure biofilm growth after 16–18 h growth at 37 °C in 0.5X TSB supplemented with 1% glucose.
Optimization of media conditions
We have found that the amount of biofilm formed is directly proportional to the concentration of media when the concentration of glucose is held constant. For example, when various concentrations of TSB are supplemented with 1% glucose biofilm formation is proportional to the concentration of TSB. To produce a series of TSB concentrations, we have found that it is convenient to generate a two-fold dilution series of TSB and supplement each dilution with glucose to the desired concentration. The amount of biofilm formed in each media condition can then be measured using the standard biofilm assay described above. The optimal media condition is defined as the media that produces robust biofilm growth that is within the linear range of detection of your microtiter plate reader.
Generate a two-fold dilution series of TSB (1X to 0.0078X). Add 10 ml 1X TSB to 10 ml sterile dH2O in a sterile tube or flask and mixing thoroughly. Remove 10 ml of the resulting 20 ml 0.5X TSB media and to 10 ml sterile dH2O, resulting in 20 ml of 0.25X TSB. Perform five more 2-fold serial dilutions to generate TSB concentrations ranging from 1X to 0.0078X.
Supplement each TSB dilution with glucose to the desired concentration using Table 2 as a guide.
Prepare inoculum of desired strain in each of the media conditions you have made as described above.
Inoculate assay plates as described above. Inoculate 8 assay wells (1 column) of a 96-well assay plate.
Incubate assay plates at 37 °C for 16–18 h, or the optimal time of growth as determined above.
Measure the OD600 to quantify total growth.
Process assay plates as described above.
Read the OD600 of the stained biofilms using an appropriate microtiter plate reader.
Calculate the average OD600 of crystal violet bound and the standard deviation for each time point.
Plot the average OD600 of crystal violet bound for each glucose concentration vs. TSB concentration. A representative assay result is presented in Fig. 3B.
-
Identify optimal media conditions.
Based on the results shown in Fig. 3B, we routinely use 0.5X TSB supplemented with 1% glucose for biofilm assays. This media condition produces OD600 readings of stained biofilms that are at the upper-most limit of the linear range of detection for both detection methods tested i.e. direct OD600 measurement of crystal violet bound to the well and measurement of eluted crystal violet.
Table 2.
Optimizing media conditions by varying the concentrations of media (TSB) and glucose.
| µl 40% glucose/10 ml media for: | |||
|---|---|---|---|
| [TSB] | 1% gluc | 0.5%gluc | 0.25% gluc |
| 1× | 187 | 62 | 0 |
| 0.5× | 219 | 94 | 31 |
| 0.25× | 234 | 109 | 47 |
| 0.125× | 242 | 117 | 55 |
| 0.0625× | 246 | 121 | 58 |
| 0.0312× | 248 | 123 | 60 |
| 0.0156× | 249 | 124 | 62 |
| 0.0078× | 249 | 124 | 62 |
Basic Protocol 3. A Screening assay for mutants or compounds that inhibit biofilm formation
Materials
Same as Basic Protocol 1, plus:
96-pin replicator
Library of transposon-insertion mutants stored in 96-well plates
Library of small molecules stored in 96-well plates
Dimethyl Sulfoxide (DMSO)
- For compound screening,
- Prepare an appropriate amount of inoculum as described above. You will need approximately 20 ml inoculum/assay plate.
-
Plate set up. Add test compounds to wells in columns 2–11 of a flat-bottomed tissue culture-treated 96-well assay plates (Costar 3595, Corning Life Sciences). Each of the assay wells in columns 2–11 will contain a unique small molecule from the screening library (MSL). The compounds in most screening libraries are stored in DMSO. Add 2–8 µl compounds to each assay well to achieve the desired concentration. We screen compounds at concentrations ranging from 50–100 µM.Generally, the assay will tolerate concentrations of DMSO up to 4%; however, it is important to confirm that biofilm formation is not affected by DMSO using the assay described in Basic Protocol 1.
-
In each assay plate, columns 1 and 12 will contain control cultures, which will serve as negative (0% biofilm inhibition) and positive controls (100% biofilm inhibition), respectively. The negative control wells contain the assay strain treated with the same concentration of DMSO present in the assay wells. The positive control wells will contain either uninoculated media, or the assay strain treated with an antibiotic that will result in complete inhibition of growth making sure that all wells contain the same final concentration of DMSO.For high-throughput screening we find that it is more practical to use an antibiotic that completely inhibits growth as the positive control because of the likelihood of cross-contamination of any uninoculated wells during the inoculation process. Chloramphenicol (100 µg/ml) is an antibiotic that can be used for all of the listed in Table 1.
-
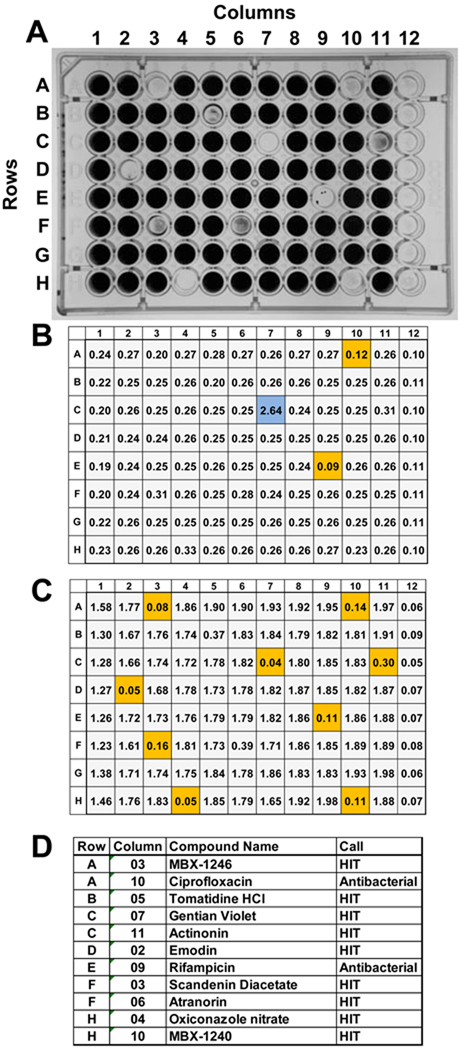
Inoculate assay plates with 200 µl/well of the assay strain diluted in the optimal medium as described above.Screening results obtained using S. epidermidis 18972 and a mock screening plate are presented in Fig. 5A. The mock screening plate contained nine compounds that are inhibitors of biofilm formation and two antibiotics (see Fig. 5D), and the remainder of the wells in columns 2–11 contained DMSO. Columns 1 and 12 contain the 0% INH (DMSO only) and 100% INH (100 µg chloramphenicol/ml; Cm100) controls, respectively. The OD600 values obtained for the planktonic cultures and for crystal violet bound to biofilms are shown in Fig. 5B and 5C, respectively. Compounds that produced ≥ 40% inhibition planktonic growth or ≥ 80% biofilm growth inhibition are highlighted in Fig 5B and 5C, respectively. Eight of the compounds that produced % biofilm inhibition ≥80% and % planktonic growth inhibition ≤ 40% were analyzed in a dose response assay (see below). Seven of the antibiofilm compounds used in the mock screening plate were identified previously in a pilot screen using a compound library comprised of 2000 known bioactive chemicals (Spectrum library; MicroSource Discovery Systems, Gaylordsville, CT). MBX-1240 and MBX-1246 were identified in screen of compounds purchased from Chembridge Corp. (San Diego, CA) (Opperman et al., 2009)
- For mutant screening:
- Transfer 200 µl media to each assay well.
- Plate set up. Inoculate columns 2–11 of a flat-bottomed tissue culture-treated 96-well assay plates (Costar 3595, Corning Life Sciences) with mutant cultures. Inoculate columns 1 and 12 with control cultures, which will serve as negative (0% biofilm inhibition) and positive controls (100% biofilm inhibition), respectively. The negative control wells contain the wild-type strain and the positive control wells contain a biofilm-negative strain (either a different strain or an isogenic biofilm-defective mutant).
- Inoculate plates from a frozen mutant library stored as glycerol stocks in 96-well storage plates using a 96-pin replicator. Sterilize the 96-pin replicator by dipping the pins in 95% ethanol and then flaming it. Press the pins into the frozen cultures, transfer to the assay plate, and move pins up and down to mix.
Seal the assay plates with foil tape or plate lids and incubate them at 37°C for the optimal time as determined above.
Process assay plates as described above.
Calculate the percent inhibition of overall growth (% INH-Growth) caused by each compound was calculated using the formula: (1-(OD600 (compound)/average OD600 (negative control))) × 100.
Calculate the percent inhibition of biofilm growth produced by each compound was calculated using the formula: (1-(CV OD600 (compound)/average CV OD600 (negative control))) × 100.
Compounds or mutants that produced ≥80% biofilm inhibition and ≤40% overall growth inhibition were scored as primary hits.
Retest primary hits in triplicate using the assay described above. Primary hits that produce an average biofilm inhibition of ≥80% and overall growth inhibition of ≤40% are scored as confirmed hits.
Figure 5.
An example of the screening results obtained using S. epidermidis 18972 and a mock screening plate. A) A photograph of the crystal violet stained mock screening plate that contained nine compounds that are inhibitors of biofilm formation and two antibiotics. The remainder of the wells in columns 2–11 contained DMSO. Columns 1 and 12 contain the 0% INH (DMSO only) and 100% INH (100 µg chloramphenicol/ml; Cm100) controls, respectively. The wells containing anti-biofilm and antibacterial compounds exhibited significant reductions in the intensity of crystal violet staining. B) The OD600 values obtained for the planktonic cultures. Compounds that produced ≥ 40% inhibition planktonic growth are highlighted in orange. The compound in well number C07, highlighted in blue, produced an OD600 of 2.64, which is significantly higher than those produced by the 0% INH (DMSO only) controls. This well contained gentian violet, an intensely colored compound that absorbs light at 600 nm. Beware of colored compounds that absorb at 600 nm, as they can mask antibacterial activity. C) The OD600 values obtained for the crystal violet stained biofilm cultures. Compounds that produced ≥ 80% inhibition planktonic growth are highlighted in orange. D) A table listing the compounds used in the mock screening plate, their location on the plate (row, column), and the screening call (HIT or antibacterial). A compound that produced % ≥80% biofilm inhibition and ≤ 40% planktonic growth inhibition were designated anti-biofilm “HITS”. Compounds that produced ≥ 40% planktonic growth inhibition were designated as “Antibacterial” compounds. Seven of the antibiofilm compounds used in the mock screening plate were identified previously in a pilot screen using a compound library comprised of 2000 known bioactive chemicals (Spectrum library; MicroSource Discovery Systems, Gaylordsville, CT). MBX-1240 and MBX-1246 were identified in screen of compounds purchased from Chembridge Corp. (San Diego, CA) (Opperman et al., 2009)
Basic Protocol 4. A quantitative assay for anti-biofilm activity: the Minimal Biofilm Inhibitory Concentration (MBIC) assay
The biofilm assay described above can be used as a quantitative assay for anti-biofilm activity of chemical treatments that is conceptually similar to the microbroth dilution assay described in the CLSI document M7-A7 (CLSI, 2006) with several modifications (described below). While the microbroth dilution assay is used to measure the minimal inhibitory concentration (MIC) an antibacterial compound, this assay measures the Minimal Biofilm Inhibitory Concentration (MBIC, which is defined as concentration of a compound in a serial two-fold dilution series that inhibits biofilm formation by ≥80% as compared to an untreated control.
Materials
Same as Basic Protocol 1, plus:
Dimethyl sulfoxide (DMSO)
-
Prepare a two-fold dilution series of each compound consisting of 10 compound concentrations (e.g. final concentration range: 0.2–100 µM).
The compound dilutions can be prepared by serially diluting each compound 2-fold in DMSO at concentrations that were 50-fold greater than the final concentration (e.g. 10–5000 µM).
-
Add 4 µl of each compound dilution in the dilution series and DMSO only to the assay wells for the untreated controls. The final concentration of DMSO in each well will be 2%.
We have found that the biofilm assay is unaffected by DMSO concentrations up to 4%.
It is convenient to add a dilution series and the untreated control for a single compound to one row (12 wells) of a 96-well assay plate.
Inoculate each assay plate containing compound dilution series with 200 µl of bacterial culture as described above.
Cover the assay plates with adhesive foil lids (Costar 3904, Corning) and incubate the plates for 18 h at 37 °C.
Measure the optical density at 600nm for each well was measured using a multiplate reader to quantify overall bacterial growth.
Wash, fix, and stain assay plates as described above.
Measure the optical density at 600nm for each well was measured using a multiplate reader to quantify bacterial growth.
Calculate the percent inhibition of overall growth (% INH-Growth) caused by each compound was calculated using the formula: (1-(OD600 (compound)/average OD600 (negative control))) × 100.
Calculate the percent inhibition of biofilm growth produced by each compound was calculated using the formula: (1-(CV OD600 (compound)/ average CV OD600 (negative control))) × 100.
Determine the MIC for each compound by identifying the lowest compound concentration that inhibited overall growth by ≥ 80%.
-
Determine the MBIC for each compound by identifying the lowest compound concentration that inhibited biofilm growth by ≥ 80%. Alternatively, the MBIC can be determined visually.
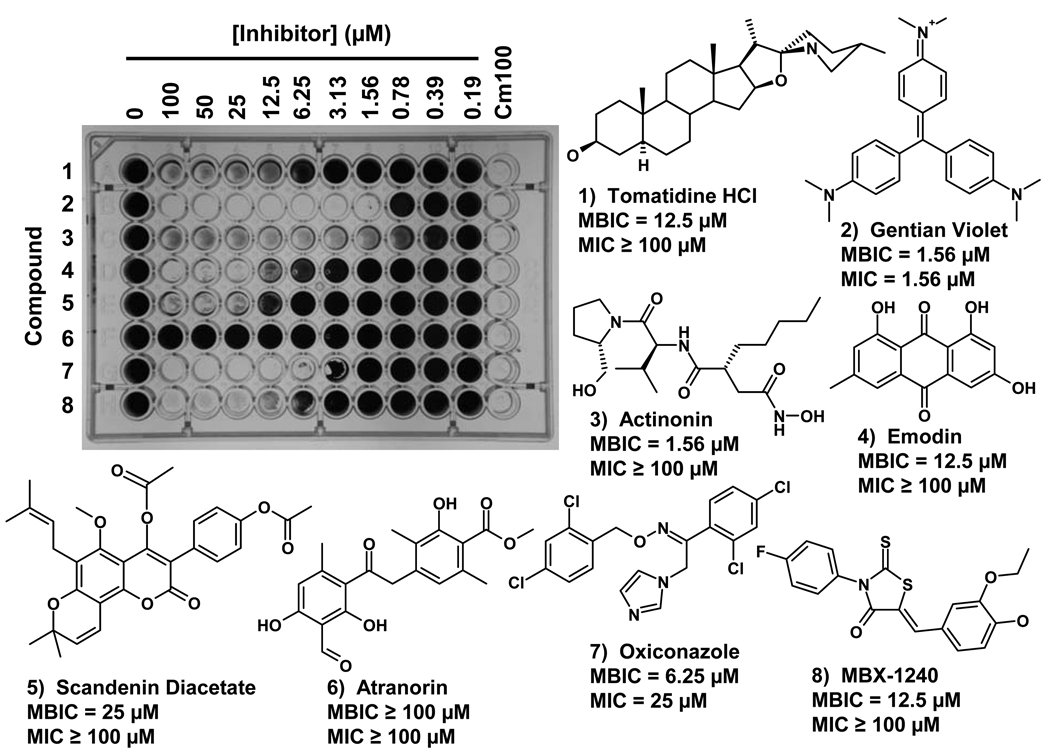
An example of a typical MBIC assay using eight of the anti-biofilm compounds described above against S. epidermidis 18972 is shown in Fig. 6. The compounds used in the assay, their chemical names and structures, and the MBIC and MIC values obtained in the assay are also shown in Fig. 6. These compounds are commercially available from MicroSource Discovery Systems (. Alternatively, three of these compounds (Tomatidine, Actinonin, and Emodin) can be purchased from Sigma-Aldrich (St. Louis, MO).
Figure 6.
An example of a Minimal Biofilm Inhibitory Concentration (MBIC) assay for eight anti-biofilm compounds that were tested against S. epidermidis 18972. The assay plate in which biofilm cultures have been stained with crystal violet is shown in the photograph. The concentrations of each compound tested are indicated above the plate, and the compound tested in each row is indicated on the left side of the plate. The MBIC and MIC are defined as the lowest compound concentration that produces a ≥ 80% inhibition of biofilm growth or planktonic growth, respectively. The compounds used in this assay (Compounds 1–8), their chemical names and structures, and the MBIC and MIC values obtained as a result of the assay are also shown. Note that Gentian violet produced an MIC that was equal to the MBIC. Because this compound strongly absorbs light at 600 nm, the antibacterial activity of this compound became apparent only when it was diluted in the dose response assay. The compounds used in this assay are commercially available from MicroSource Discovery Systems, Gaylordsville, CT). Alternatively, three of these compounds (Tomatidine, Actinonin, and Emodin) can be purchased from Sigma-Aldrich (St. Louis, MO). MBX-1240 can be purchased from ChemBridge Corp. (San Diego, CA).
Reagents and Solutions
Trytpic Soy Agar (TSA)
For Difco™ TSA (Becton, Dickenson and Co. (BD)), add 40 g premixed powder to 1 L dH2O. Mix thoroughly. Dissolve the powder by heating the suspension with frequent mixing until it is boiling. Continue boiling solution for 1 min. Autoclave to sterilize (15 min, 121°C, liquid cycle). Alternatively, add 10–15 g Bacto Agar to 1 L TSB (see below), dissolve agar by heating and autoclave to sterilize. Store at room temperature.
Trytpic Soy Broth (TSB)
For Bacto™ TSB (BD), add 30 g premixed powder to 1 L dH2O. Mix thoroughly. Autoclave to sterilize (15 min, 121°C, liquid cycle). Autoclave this media for the minimal time required for sterilization to prevent degradation of the glucose. Store at room temperature.
Sterile 80% glycerol (v/v)
Add 80 ml glycerol to 20 ml dH2O. Mix well. Autoclave to sterilize (20 min, 121°C, liquid cycle). Store at room temperature.
40% glucose
Add 40 g D-glucose to 100 ml dH2O. Filter sterilize (0.22 µm filter). Store at room temperature
Phosphate Buffered Saline (PBS), pH 7.4 (Sambrook et al., 1989)
Dissolve the following in 800 ml of dH2O: 8 g NaCl, 0.2 g KCl, 1.44 g Na2HPO4, 0.27 g KH2PO4. Adjust the pH to 7.4 with HCl or NaOH. Adjust final volume to 1L by adding dH2O. Autoclave to sterilize (20 min, 121°C, liquid cycle). Store at room temperature
0.9% (w/v) saline
Add 9 g NaCl to 1 L dH2O. Autoclave (20 min, 121°C, liquid cycle) or filter sterilize (0.22 µm filter). Store at room temperature.
0.06% crystal violet (w/v)
Add 0.6 g crystal violet to 1 L dH2O. Mix well. Store at room temperature.
Commentary
a. Background Information
The basic biofilm assay was designed to measure the ability of clinical strains to form biofilms in microtiter assay plates using a dye (crystal violet) that binds to bacterial cells and the extracellular matrix materials. The basic assay is simple to perform and requires minimal equipment; however, it is very powerful and versatile. The basic assay has been adapted to examine many aspects of bacterial biofilm biology. For example, the basic assay protocol has been used to measure biofilm forming ability of clinical isolates (Chokr et al., 2006; Rohde et al., 2001), examine effects of genetic mutations on biofilms [e.g. (Cramton et al., 1999; Rice et al., 2007; Seidl et al., 2008)], the effect of environmental conditions on biofilm formation (Rachid et al., 2000a; Rachid et al., 2000b), to screen for transposon-insertion mutations that affect biofilm formation (Mack et al., 1994; Tu Quoc et al., 2007), and to screen for small molecule inhibitors of biofilm formation and to measure their anti-biofilm activity (Opperman et al., 2009). In addition, the type of stain used can be varied to measure different components of the biofilm. For example, the numbers of bacterial cells can be measured using intercalating dyes that enter living cells, such as Syto 9. However, these dyes will also bind to DNA in the extracellular biofilm matrix. This problem can be solved by using propidium iodide stain together with syto 9, which does not enter viable cells and will quench the fluorescence signal of syto9 bound to extracellular DNA when emission at 530 nm is measured (excitation 485 nm). These dyes can be purchased together in the BacLight Live/Dead kit (Molecular probes, Eugene, OR). Alternatively, propidium iodide can be used to detect extracellular DNA in biofilms (Yarwood et al., 2004). Finally, the fluorescent dye thioflavin T has been used to detect amyloid-like structures formed by fimbrae in biofilms formed by diverse range of Gram-positive and –negative bacterial species (Jordal et al., 2009; Larsen et al., 2007).
The advantages of this assay system are numerous. It is relatively inexpensive to perform, rapid, quantitative, adaptable, and is readily scalable from small scale assays to high-throughput screens. In addition, several studies have demonstrated that mutations that block biofilm formation in the static microtiter assay also dramatically decrease colonization of foreign body infections in animal models (Rupp et al., 2001; Rupp et al., 1999a; Rupp et al., 1999b), which demonstrate the relevance of this assay system. However, the fact that these assays are carried out under static conditions and biofilm cultures are not supplied with a continuous supply of fresh nutrients is a potential drawback of this system, which may result in an inability to generate mature biofilms or to perform experiments lasting more that 18–20 h. In some cases, this drawback can be remedied by replacing the growth media every 12 h during the course of the experiment, enabling biofilms to continue growing until another factor becomes limiting for growth. However, replacing growth media every 12 h becomes impractical for large numbers of assay plates or for labs that have limited hours of operation. Another drawback of this assay is that it is designed to measure biofilm formation and is not suited for measuring effects of compounds/mutations on the viability of biofilm bacteria, as the crystal violet stain measures total biomass (cells and extracellular matrix) and not numbers of viable cells. However, the microtiter format can be adapted for this purpose by using detection methods that quantify viable cells. We have routinely used the BacLight Live/Dead stain (Molecular probes, Eugene, OR) according to the manufacturers’ instructions to measure the relative viability of mature biofilms treated with antibiotics compared to an untreated control. Other research groups have measured ATP levels (Monzon et al., 2001) or have used tetrazolium dyes (Pitts et al., 2003) to measure the relative viability of mature biofilms after various treatments.
It is important to establish a secondary assay or biofilm detection method to confirm the results obtained using the microtiter assay and crystal violet staining. For example, a flow cell system used in conjunction with confocal microscopy could be used to verify biofilm-defective phenotypes of mutants. An alternative biofilm detection method, such as Baclight Live/Dead or determination of viable cfu/well could be used to verify the anti-biofilm effects of small molecule compounds.
b. Critical Parameters and Troubleshooting
The critical parameters for this assay are as follows:
Standardization of media, preparation of inocula, and incubation time. Variations in these parameters can result in variable results.
Assay plates. Variations in tissue culture treated assay plates between manufacturers and between different lots from the same manufacturer can affect results. Attempt to use assay plates from the same manufacturers’ lot for an entire series of experiments.
Because of well-to-well variability, assay at least 4 replicate wells for a given strain and/or condition. For high-throughput screens, confirm each hit in triplicate.
Because of plate-to-plate variability, include positive and negative controls on each plate. A list of some of the difficulties commonly encountered with the techniques described in this unit, their possible cause, and potential solution is shown in Table 3.
Table 3.
Critical parameters and troubleshooting.
| Problem | Possible Cause | Solution |
|---|---|---|
| Poor or no Biofilm forms on bottom of plate | Plates are not tissue-culture treated | Use tissue-culture treated plates |
| Medium may not be adjusted with optimal glucose concentration | Repeat with medium that is properly adjusted with glucose | |
| Bad plate lot | Try a different lot from the same manufacturer, or a different manufacturer | |
| Incubation period too long, biofilms falling apart | Optimize length of incubation | |
| Biofilms robust after washing, fall apart with the addition of ethanol | Heat fixation may be required | Heat fix biofilms at 60 °C for at least 60 min |
| Biofilms robust after heat fixation, mostly gone after staining step | Not heat-fixed long enough | Increase time for heat fixation |
| Stain incubated in wells too long | Decrease staining time | |
| Large hole taken out of biofilms when washed with a plate washer | Aspiration tips may be too low in well, or aspiration rate too high | Raise height of aspiration tips, or decrease the rate of aspiration |
| Decrease number of aspiration steps | ||
| Biofilms on edges of the plate are less dense than biofilms on the interior of the plate | Possible edge effect taking place, incubation parameters not consistent across entire plate | May require an incubation atmosphere that will allow entire plate to grow at the same rate; i.e., anaerobic conditions |
c. Anticipated Results
Representative results for the basic assay, assay optimization, and the MBIC assay are presented in Figs. 2, 4 and 5, respectively.
Figure 4.
Optimization of assay conditions. A) A biofilm growth curve of S. aureus ATCC 35556 grown in 0.5X TSB supplemented with 1% glucose. B) Biofilms were grown in varying media conditions to identify optimal growth conditions and two methods of biofilm detection are compared. Biofilms were grown at 37 °C for 18 h. Biofilms were stained as described above and the amount of crystal violet bound to the bottom of each assay well was measured directly as absorbance at 600 nm, or crystal violet was eluted from each well, diluted 1:10, and the absorbance at 600 nm was measured. C) Comparison of absorbance at 600 nm readings of neat vs. a 1:10 dilution of eluted crystal violet demonstrates the linear range of the assay.
d. Time Considerations
The basic assay can be completed in two days, including growth of the overnight culture, preparation of inocula, and inoculating, incubating, staining and quantifying assay plates. However, the amount of time required for setting up and processing assay plates increases dramatically as the number of plates and/or the number of conditions to be tested is increased. The use of multichannel pipets and/or laboratory automation can reduce the time required for these processes.
LITERATURE CITED
- Baba T, Takeuchi F, Kuroda M, Yuzawa H, Aoki K, Oguchi A, Nagai Y, Iwama N, Asano K, Naimi T, Kuroda H, Cui L, Yamamoto K, Hiramatsu K. Genome and virulence determinants of high virulence community-acquired MRSA. Lancet. 2002;359:1819–1827. doi: 10.1016/s0140-6736(02)08713-5. [DOI] [PubMed] [Google Scholar]
- Balaban N, Giacometti A, Cirioni O, Gov Y, Ghiselli R, Mocchegiani F, Viticchi C, Del Prete MS, Saba V, Scalise G, Dell'Acqua G. Use of the quorum-sensing inhibitor RNAIII-inhibiting peptide to prevent biofilm formation in vivo by drug-resistant Staphylococcus epidermidis. J Infect Dis. 2003;187:625–630. doi: 10.1086/345879. [DOI] [PubMed] [Google Scholar]
- Branda SS, Vik S, Friedman L, Kolter R. Biofilms: the matrix revisited. Trends Microbiol. 2005;13:20–26. doi: 10.1016/j.tim.2004.11.006. [DOI] [PubMed] [Google Scholar]
- Chokr A, Watier D, Eleaume H, Pangon B, Ghnassia JC, Mack D, Jabbouri S. Correlation between biofilm formation and production of polysaccharide intercellular adhesin in clinical isolates of coagulase-negative staphylococci. Int J Med Microbiol. 2006;296:381–388. doi: 10.1016/j.ijmm.2006.02.018. [DOI] [PubMed] [Google Scholar]
- Christensen GD, Baldassarri L, Simpson WA. Methods for studying microbial colonization of plastics. Methods Enzymol. 1995;253:477–500. doi: 10.1016/s0076-6879(95)53040-1. [DOI] [PubMed] [Google Scholar]
- Christensen GD, Bisno AL, Parisi JT, McLaughlin B, Hester MG, Luther RW. Nosocomial septicemia due to multiply antibiotic-resistant Staphylococcus epidermidis. Ann Intern Med. 1982;96:1–10. doi: 10.7326/0003-4819-96-1-1. [DOI] [PubMed] [Google Scholar]
- Christensen GD, Simpson WA, Younger JJ, Baddour LM, Barrett FF, Melton DM, Beachey EH. Adherence of coagulase-negative staphylococci to plastic tissue culture plates: a quantitative model for the adherence of staphylococci to medical devices. J Clin Microbiol. 1985;22:996–1006. doi: 10.1128/jcm.22.6.996-1006.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- CLSI. Methods for Dilution Antimicrobial Susceptibility Tests for Bacteria that Grow Aerobically; Approved Standard-Seventh Edition. 2. Vol. M7-A7 vol. 26. Wayne, PA USA: Clinical and Laboratory Standards Institute; 2006. [Google Scholar]
- Cramton SE, Gerke C, Gotz F. In vitro methods to study staphylococcal biofilm formation. Methods Enzymol. 2001a;336:239–255. doi: 10.1016/s0076-6879(01)36593-x. [DOI] [PubMed] [Google Scholar]
- Cramton SE, Gerke C, Schnell NF, Nichols WW, Gotz F. The intercellular adhesion (ica) locus is present in Staphylococcus aureus and is required for biofilm formation. Infect Immun. 1999;67:5427–5433. doi: 10.1128/iai.67.10.5427-5433.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cramton SE, Ulrich M, Gotz F, Doring G. Anaerobic conditions induce expression of polysaccharide intercellular adhesin in Staphylococcus aureus and Staphylococcus epidermidis. Infect Immun. 2001b;69:4079–4085. doi: 10.1128/IAI.69.6.4079-4085.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Darouiche RO. Device-associated infections: a macroproblem that starts with microadherence. Clin Infect Dis. 2001;33:1567–1572. doi: 10.1086/323130. [DOI] [PubMed] [Google Scholar]
- Deighton MA, Capstick J, Domalewski E, van Nguyen T. Methods for studying biofilms produced by Staphylococcus epidermidis. Methods Enzymol. 2001;336:177–195. doi: 10.1016/s0076-6879(01)36589-8. [DOI] [PubMed] [Google Scholar]
- Gillaspy AF, Hickmon SG, Skinner RA, Thomas JR, Nelson CL, Smeltzer MS. Role of the accessory gene regulator (agr) in pathogenesis of staphylococcal osteomyelitis. Infect Immun. 1995;63:3373–3380. doi: 10.1128/iai.63.9.3373-3380.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heilmann C, Gerke C, Perdreau-Remington F, Gotz F. Characterization of Tn917 insertion mutants of Staphylococcus epidermidis affected in biofilm formation. Infect Immun. 1996;64:277–282. doi: 10.1128/iai.64.1.277-282.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iordanescu S, Surdeanu M. Two restriction and modification systems in Staphylococcus aureus NCTC8325. J Gen Microbiol. 1976;96:277–281. doi: 10.1099/00221287-96-2-277. [DOI] [PubMed] [Google Scholar]
- Jordal PB, Dueholm MS, Larsen P, Petersen SV, Enghild JJ, Christiansen G, Hojrup P, Nielsen PH, Otzen DE. Widespread abundance of functional bacterial amyloid in mycolata and other gram-positive bacteria. Appl Environ Microbiol. 2009;75:4101–4110. doi: 10.1128/AEM.02107-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larsen P, Nielsen JL, Dueholm MS, Wetzel R, Otzen D, Nielsen PH. Amyloid adhesins are abundant in natural biofilms. Environ Microbiol. 2007;9:3077–3090. doi: 10.1111/j.1462-2920.2007.01418.x. [DOI] [PubMed] [Google Scholar]
- Mack D, Bartscht K, Fischer C, Rohde H, de Grahl C, Dobinsky S, Horstkotte MA, Kiel K, Knobloch JK. Genetic and biochemical analysis of Staphylococcus epidermidis biofilm accumulation. Methods Enzymol. 2001;336:215–239. doi: 10.1016/s0076-6879(01)36592-8. [DOI] [PubMed] [Google Scholar]
- Mack D, Nedelmann M, Krokotsch A, Schwarzkopf A, Heesemann J, Laufs R. Characterization of transposon mutants of biofilm-producing Staphylococcus epidermidis impaired in the accumulative phase of biofilm production: genetic identification of a hexosamine-containing polysaccharide intercellular adhesin. Infect Immun. 1994;62:3244–3253. doi: 10.1128/iai.62.8.3244-3253.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mack D, Siemssen N, Laufs R. Parallel induction by glucose of adherence and a polysaccharide antigen specific for plastic-adherent Staphylococcus epidermidis: evidence for functional relation to intercellular adhesion. Infect Immun. 1992;60:2048–2057. doi: 10.1128/iai.60.5.2048-2057.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merritt JH, Kadouri DE, O'Toole GA. Growing and analyzing static biofilms. Curr Protoc Microbiol. 2005 doi: 10.1002/9780471729259.mc01b01s00. Chapter 1, Unit 1B 1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Monzon M, Oteiza C, Leiva J, Amorena B. Synergy of different antibiotic combinations in biofilms of Staphylococcus epidermidis. J Antimicrob Chemother. 2001;48:793–801. doi: 10.1093/jac/48.6.793. [DOI] [PubMed] [Google Scholar]
- Novick R. Properties of a cryptic high-frequency transducing phage in Staphylococcus aureus. Virology. 1967;33:155–166. doi: 10.1016/0042-6822(67)90105-5. [DOI] [PubMed] [Google Scholar]
- O'Gara JP, Humphreys H. Staphylococcus epidermidis biofilms: importance and implications. J Med Microbiol. 2001;50:582–587. doi: 10.1099/0022-1317-50-7-582. [DOI] [PubMed] [Google Scholar]
- O'Neill E, Pozzi C, Houston P, Humphreys H, Robinson DA, Loughman A, Foster TJ, O'Gara JP. A novel Staphylococcus aureus biofilm phenotype mediated by the fibronectin-binding proteins, FnBPA and FnBPB. J Bacteriol. 2008;190:3835–3850. doi: 10.1128/JB.00167-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Toole GA, Kolter R. Initiation of biofilm formation in Pseudomonas fluorescens WCS365 proceeds via multiple, convergent signalling pathways: a genetic analysis. Mol Microbiol. 1998;28:449–461. doi: 10.1046/j.1365-2958.1998.00797.x. [DOI] [PubMed] [Google Scholar]
- Opperman TJ, Kwasny SM, Williams JD, Khan AR, Peet NP, Moir DT, Bowlin TL. Aryl rhodanines specifically inhibit staphylococcal and enterococcal biofilm formation. Antimicrob Agents Chemother. 2009;53:4357–4367. doi: 10.1128/AAC.00077-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pitts B, Hamilton MA, Zelver N, Stewart PS. A microtiter-plate screening method for biofilm disinfection and removal. J Microbiol Methods. 2003;54:269–276. doi: 10.1016/s0167-7012(03)00034-4. [DOI] [PubMed] [Google Scholar]
- Rachid S, Cho S, Ohlsen K, Hacker J, Ziebuhr W. Induction of Staphylococcus epidermidis biofilm formation by environmental factors: the possible involvement of the alternative transcription factor sigB. Adv Exp Med Biol. 2000a;485:159–166. doi: 10.1007/0-306-46840-9_22. [DOI] [PubMed] [Google Scholar]
- Rachid S, Ohlsen K, Witte W, Hacker J, Ziebuhr W. Effect of subinhibitory antibiotic concentrations on polysaccharide intercellular adhesin expression in biofilm-forming Staphylococcus epidermidis. Antimicrob Agents Chemother. 2000b;44:3357–3363. doi: 10.1128/aac.44.12.3357-3363.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rice KC, Mann EE, Endres JL, Weiss EC, Cassat JE, Smeltzer MS, Bayles KW. The cidA murein hydrolase regulator contributes to DNA release and biofilm development in Staphylococcus aureus. Proc Natl Acad Sci U S A. 2007;104:8113–8118. doi: 10.1073/pnas.0610226104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rohde H, Knobloch JK, Horstkotte MA, Mack D. Correlation of biofilm expression types of Staphylococcus epidermidis with polysaccharide intercellular adhesin synthesis: evidence for involvement of icaADBC genotype-independent factors. Med Microbiol Immunol (Berl) 2001;190:105–112. doi: 10.1007/s00430-001-0099-5. [DOI] [PubMed] [Google Scholar]
- Rupp ME, Fey PD, Heilmann C, Gotz F. Characterization of the importance of Staphylococcus epidermidis autolysin and polysaccharide intercellular adhesin in the pathogenesis of intravascular catheter-associated infection in a rat model. J Infect Dis. 2001;183:1038–1042. doi: 10.1086/319279. [DOI] [PubMed] [Google Scholar]
- Rupp ME, Ulphani JS, Fey PD, Bartscht K, Mack D. Characterization of the importance of polysaccharide intercellular adhesin/hemagglutinin of Staphylococcus epidermidis in the pathogenesis of biomaterial-based infection in a mouse foreign body infection model. Infect Immun. 1999a;67:2627–2632. doi: 10.1128/iai.67.5.2627-2632.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rupp ME, Ulphani JS, Fey PD, Mack D. Characterization of Staphylococcus epidermidis polysaccharide intercellular adhesin/hemagglutinin in the pathogenesis of intravascular catheter-associated infection in a rat model. Infect Immun. 1999b;67:2656–2659. doi: 10.1128/iai.67.5.2656-2659.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sambrook J, Fritsch EF, Maniatis T. Molecular Cloning: A Laboratory Manual. Plainville, NY: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- Seidl K, Goerke C, Wolz C, Mack D, Berger-Bachi B, Bischoff M. Staphylococcus aureus CcpA affects biofilm formation. Infect Immun. 2008;76:2044–2050. doi: 10.1128/IAI.00035-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shanks RM, Donegan NP, Graber ML, Buckingham SE, Zegans ME, Cheung AL, O'Toole GA. Heparin stimulates Staphylococcus aureus biofilm formation. Infect Immun. 2005;73:4596–4606. doi: 10.1128/IAI.73.8.4596-4606.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tu Quoc PH, Genevaux P, Pajunen M, Savilahti H, Georgopoulos C, Schrenzel J, Kelley WL. Isolation and characterization of biofilm formation-defective mutants of Staphylococcus aureus. Infect Immun. 2007;75:1079–1088. doi: 10.1128/IAI.01143-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson M. Bacterial biofilms and human disease. Sci Prog. 2001;84:235–254. doi: 10.3184/003685001783238998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yarwood JM, Bartels DJ, Volper EM, Greenberg EP. Quorum sensing in Staphylococcus aureus biofilms. J Bacteriol. 2004;186:1838–1850. doi: 10.1128/JB.186.6.1838-1850.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]