Abstract
Methods for in-depth genome-wide characterization of transcriptomes and quantification of transcript levels using various microarray and next-generation sequencing technologies have emerged as valuable tools for understanding cellular physiology and human disease biology and have begun to be utilized in various clinical diagnostic applications. Current methods, however, typically require RNA to be converted to complementary DNA prior to measurements. This step has been shown to introduce many biases and artifacts. In order to best characterize the ‘true’ transcriptome, the single-molecule direct RNA sequencing (DRS) technology was developed. This review focuses on the underlying principles behind the DRS, sample preparation steps, and the current and novel avenues of research and applications DRS offers.
INTRODUCTION
The emergence of microarray technologies during the 1990s1–3 and their wide-spread application to understand biological processes and human disease has resulted in numerous ‘mysteries’ in genomics, epigenomics, and transcriptomics to be resolved, and revolutionized the way we perform biomedical research. The impact on transcriptomics was particularly noteworthy, as a simplistic view of transcriptomes was replaced with a larger, more complicated view of genome-wide transcription, where a large fraction of transcripts emanate from unannotated parts of the genome.4,5 The recent emergence of high-throughput next-generation DNA sequencing technologies6 and their application to transcriptomics by various complementary DNA (cDNA) sequencing (RNA-Seq) technologies7,8 have resulted in an even more complicated view of the transcriptome and its regulation, demonstrating our limited knowledge in this area. RNA-Seq technologies have eliminated some of the technical challenges posed by hybridization-based microarray strategies, such as limited dynamic range of detection and high background due to cross-hybridization,8 but several fundamental shortcomings still remain, including their requirement for cDNA synthesis and other sample manipulation and amplification steps which introduce various biases and artifacts (discussed below). Consequently, there is an ever-growing need for more accurate and efficient molecular and computational tools for transcriptomics applications.
To address the limitations of current RNA-Seq strategies and to simplify and reduce the costs of the genome-wide transcriptome surveys, we recently developed the direct RNA sequencing (DRS) technology.9 DRS involves sequencing of natural RNA molecules without their prior conversion to cDNA. The technology offers the highest potential to eliminate artifacts in RNA measurements and allow us to understand the ‘true’ nature of normal and disease transcriptomes and pathways in an unbiased manner. It also opens new avenues of research and is particularly advantageous for emerging clinical applications. After briefly summarizing the challenges we face with RNA-Seq methods in use today, we will review the basics of DRS and the biomedical research applications it offers.
DIFFICULTIES ASSOCIATED WITH CDNA SEQUENCING METHODS
The conversion of RNA into cDNA is the common step in microarray- and sequencing-based RNA analysis approaches today. This step is necessary because the technologies in use require amplified molecules for detection. This is in contrast to the currently available gold standard methods, such as RNase protection assays and northern blots, which measure RNA abundance directly without cDNA synthesis. Even though the conversion of RNA into cDNA is known to be associated with multiple technical issues (discussed below), ‘cDNA-free’ assays are not as widely used as cDNA-based expression measurement tools, because they are laborious, semi-quantitative, not easily scalable, and usually do not perform well in detecting very low quantities of RNA species.
One limitation of cDNA-based approaches is the tendency of various reverse transcriptases (RTs) to generate spurious second-strand cDNA due to their DNA-dependent DNA polymerase activities,10,11 which presents difficulties in sense versus antisense transcript determination.12 This artifact is thought to occur by either a hairpin loop at the 3′ end of the first-strand cDNA or by re-priming, from either RNA fragments or primers used for the first-strand synthesis. Although strategies to alleviate this artifact has been proposed,13 their success and whether or not they introduce additional artifacts have not been fully examined. Strand-specific libraries can be prepared through various approaches including RNA–RNA ligations and bisulfite treatment,14–16 but these methods are laborious and highly inefficient. Another limitation is template switching.17–19 During the process of reverse transcription, the nascent cDNA being synthesized can sometimes dissociate from the template RNA and re-anneal to a different stretch of RNA with a sequence similar to the initial template, generating artifactual cDNAs. In addition to causing difficulties in RNA quantification, template-switching causes problems in the identification of exon–intron boundaries and true chimeric transcripts. RTs can also synthesize cDNA in a primer-independent manner, which is thought to be caused by self-priming due to RNA secondary structure and results in the generation of random cDNA. Furthermore, RTs are error-prone due to their lack of proofreading mechanisms20 and yield low quantities of cDNA, necessitating the use of large quantities of input RNA and relatively high levels of amplification.
In addition to the limitations of cDNA synthesis, RNA-seq approaches available today are limited by coverage nonuniformity, which may be a result of biases introduced during fragmentation, priming with random hexamers,21,22 cDNA synthesis (discussed above), ligation,23,24 amplification,25 and sequencing.25–27 In addition, the commonly used RNA-seq strategies result in transcript length bias because of multiple fragmentation and RNA or cDNA size selection steps they employ. These biases result in one’s ability to identify differentially expressed genes dependent on transcript length,28 thus resulting in complications for downstream analyses such as gene ontology analyses.29
HOW DOES DRS WORK?
The ability to sequence RNA molecules directly without cDNA conversion has long been desired. The initial attempts in the 1970s on determining RNA sequence content were in fact without cDNA synthesis, attempted by Helen Donnis-Keller, Alan M. Maxam, and Walter Gilbert, using the tendency of various RNases to cut RNA molecules at certain nucleotides.30 However, the ability to sequence RNA fully, and also in massively parallel manner, was not possible until recently.9 This is because DRS ability may strictly require single-molecule sequencing capabilities. The commonly available sequencing platforms from Illumina, Life Technologies, and Roche are amplified molecule technologies, which sequence combined clonal populations of molecules generated by methods such as bridge or emulsion polymerase chain reaction. Since amplifying RNA molecules directly without cDNA conversion has not been examined in detail previously, the extent to which DRS capability can be achieved with amplified molecule sequencing technologies is unknown.
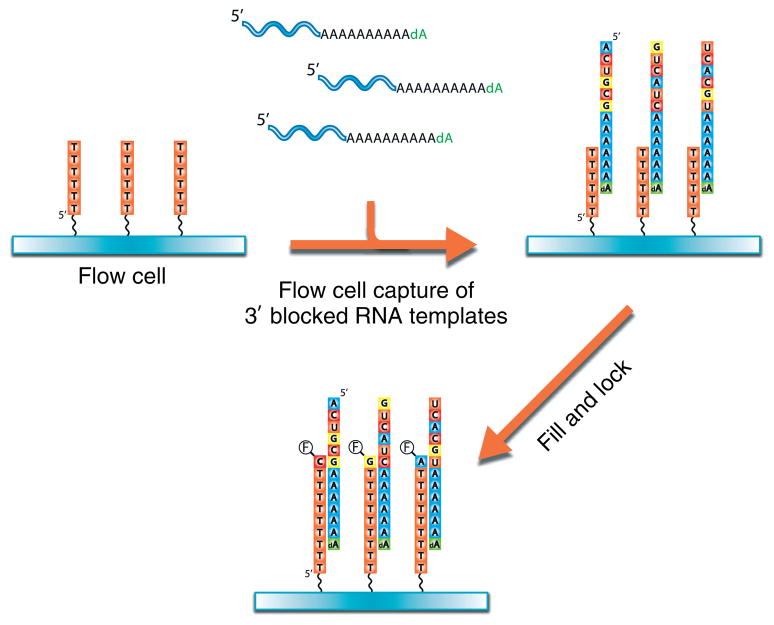
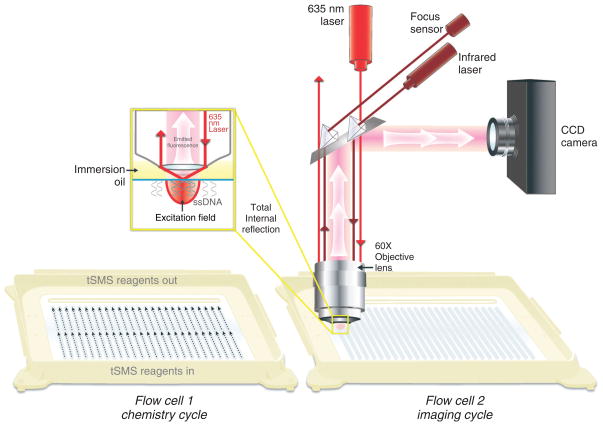
DRS was developed using the single-molecule sequencer commercially available from the Helicos BioSciences Corporation.9 The sequencing flow cell surfaces are composed of ultraclean glass containing poly(dT) oligonucleotides covalently attached at their 5′ ends (Figure 1). These oligonucleotides serve two purposes: (1) the capture of 3′ poly(A)-tail containing nucleic acids onto surfaces by hybridization and (2) priming and initiation of sequencing steps. The current requirement for RNA preparation is the presence of 3′ poly(A) tail that is ‘blocked’ at its 3′ end with a terminal 3′ deoxy nucleotide. 3′ polyadenylation and blocking of RNA templates are performed using Escherichia coli or yeast poly-A polymerases with rATP and 3′dATP, respectively, although the characterization of RNA species that naturally contain a poly(A) tail, such polyadenylation, is not required and direct hybridization of poly(A)+ RNAs to surfaces can be performed after the 3′ blocking step. After hybridization of RNA templates to the poly(dT) capture-primers, in order to begin sequencing at the unique template region adjacent to the polyA tail, each primer–template duplex molecule is ‘filled’ in with excess dTTP by DNA polymerase and then ‘locked’ in position with A-, C-, and G-Virtual Terminator (VT) nucleotide analogs. VTs are nucleotides used for sequencing, containing a fluorescent dye and chemically cleavable groups that prevent the addition of another nucleotide.31 After washing away the excess, unincorporated dye-labeled nucleotides, the surface is irradiated with a laser at an angle that allows total internal reflection at the surface. In such a situation, an evanescent field is generated so that only molecules very close to the surface are able to be excited by the laser. This reduces the background level of fluorescence such that single molecules can be detected by a highly sensitive charge-coupled device camera (Figure 2). After image acquisition across desired number of positions per channel, the liquid in each channel is replaced with a mixture that cleaves the fluorescent dye and VT off the incorporated nucleotide, rendering the strands suitable for another round of incorporation. The sequencing-by-synthesis reaction continues with the addition of the next VT nucleotide (A, C, G, or T) followed by rinsing, imaging, and cleavage. Repeating this cycle many times provides a large set of images from which the base incorporations are detected and then used to generate sequence information for each individual molecule.
FIGURE 1.
Illustration of direct RNA sequencing sequencing preparation procedure. Poly-adenylated and 3′ blocked RNA is captured on surfaces containing covalently bound poly(dT) oligonucleotide (3′ end of the poly(dT) oligonucleotide faces ‘up’). A ‘fill and lock’ step is performed, where the ‘fill’ step is performed with natural thymidine and polymerase, and a ‘lock’ step is performed with fluorescently labeled A-, C-, and G-Virtual-Terminator nucleotides and polymerase. These steps correct any misalignments that may be present in polyA and polyT duplexes and ensure that the sequencing starts in the RNA template rather than the poly-adenylated tail. Imaging is performed to locate the positions of the templates.
FIGURE 2.
Diagram of single-molecule sequencing instrument optics. A 635-nm laser is used to illuminate the surface through the objective lens using total internal reflection. This generates an evanescent wave that results in a restricted excitation field, important for the reduction of background fluorescence. Fluorescent single molecules within the excitation field on the flow cell surface emit light, which is captured by the objective lens and detected by the charge-coupled device camera.
Improvements since the initial publication allowed genome-wide analyses to be performed with DRS. Each Helicos DRS run now contains up to 50 independent channels and produce between 800,000 and 12,000,000 aligned reads [25–55 nucleotides (nts) in length, median 33 nts] per channel depending on the requested run time (2–4 days) and throughput (e.g., imaging quantity per channel). Error rates are in the range of 4%, dominated by missing base errors (~2–3%), whereas insertion (~1%) and substitution (~0.5%) errors are lower. Loading of each channel at the optimal levels generally requires 2–3 femtomoles of 3′ polyadenylated RNA templates (~300 picograms, with an average size of 300 nts).
APPLICATIONS OF DRS
The DRS technology has the potential to alleviate many limitations inherent in the transcriptomics methods in use today and provide new avenues of research. The DRS not only eliminates the reverse transcription step, but also the other sample manipulation steps such as ligation and amplification, thus resulting in minimal distortion in the representation of RNA templates. The natural strand-specificity of DRS and its requirement for only femtomole quantities of RNA are advantageous for all aspects of RNA research.
Perhaps the natural initial application of DRS is the comprehensive mapping of polyadenylation sites and gene expression determination. Much attention has been devoted during the past several years to understand the mechanisms operating at the promoters of genes and directing transcription initiation. This was a result of technological advances, allowing the adaptation of methods such as chromatin immunoprecipitation and nucleosome mapping for genome-wide analyses using microarray and sequencing technologies. On the other hand, the genome-wide localization and characteristics of polyadenylation sites have not been investigated in detail due to technical limitations. Our knowledge in this area primarily originates from expressed sequence tag databases and predictions relying on polyadenylation-associated motif elements,32–34 which does not offer resolution at the individual nucleotide level of resolution. Attempts to map polyadenylation sites with cDNA sequencing35 and microarray data36 were also mostly unsuccessful, because of the additive nature of these data sets. In other words, for each nucleotide locations across the genome, these approaches rely on signals accumulating from multiple transcripts, leading to the loss of resolution for determining the location of the polyadenylation site at nucleotide resolution.
DRS offers a simple route for polyadenylation site mapping, as demonstrated recently.37 Given its nature of capturing polyadenylated RNAs on poly(dT)-coated surfaces and sequencing after a ‘fill and lock’ step, DRS reads emerge immediately upstream of the polyA-tail. Thus, 5′ end of DRS reads signify polyadenylation cleavage locations. DRS procedure is capable of capturing polyA+ mRNA population from total RNAs or cell lysates directly without additional polyadenylation. The data generated are quantitative, thus for the first time allows genome-wide study of alternative polyadenylation states in both quantitative and qualitative manner across biological settings of interest. The data can also be used to generate gene expression profiles of polyA+ mRNAs within cells. Furthermore, custom flow cell surfaces, such as poly(dG), poly(dC), or sequence-specific capture primers, can be developed to enable unique applications for DRS.
DRS can also be adapted for the vast majority of RNA analyses being performed today. Although current DRS sequence reads offer the researcher a read length of 25 to upward of 55–60 nts, the ability to accurately detect the full compendium of transcript isoforms will be limited. Yet, whole transcriptome profiling for quantitation, mutation detection, and quantitation of individual splice junctions can be done with RNA fragmentation using standard methods, followed by polyadenylation of the RNAs. One advantage of DRS is the universality of sample preparation steps for different applications. In other words, unlike cDNA methods which require different cDNA synthesis and sample manipulation steps for short and long RNAs, DRS requires only 3′ polyadenylated templates. Thus, both short and long RNAs can be sequenced together in a single experiment.
CONCLUSIONS
DRS offers an attractive option for transcriptome studies and emerging applications in diagnostics. Future efforts will focus on several areas to expand DRS’ capabilities: (1) The current DRS chemistry relies on the addition of one nucleotide per cycle. Being able to add two or four nucleotides per cycle may improve read lengths and decrease sequencing time. (2) Alternative polymerases that can add nucleotides in a more efficient and higher fidelity manner can be generated through targeted mutation or enzyme evolution. Such improvements may improve sequencing error rates and read lengths. (3) Although the current read lengths up to 55 nts may be sufficient for fused transcript and alternative splicing detection, paired read capabilities may also be needed in order to increase coverage per sequencing experiment. With the application of DRS to biomedical studies, we expect DRS to stimulate novel advances in many areas of genomics.
References
- 1.Fodor• SP, et al. Light-directed, spatially addressable parallel chemical synthesis. Science. 1991;251:767–773. doi: 10.1126/science.1990438. [DOI] [PubMed] [Google Scholar]
- 2.Lennon GG, Lehrach H. Hybridization analyses of arrayed cDNA libraries. Trends Genet. 1991;7:314–317. doi: 10.1016/0168-9525(91)90420-u. [DOI] [PubMed] [Google Scholar]
- 3.Southern EM, Maskos U, Elder JK. Analyzing and comparing nucleic acid sequences by hybridization to arrays of oligonucleotides: evaluation using experimental models. Genomics. 1992;13:1008–1017. doi: 10.1016/0888-7543(92)90014-j. [DOI] [PubMed] [Google Scholar]
- 4.Berretta J, Morillon A. Pervasive transcription constitutes a new level of eukaryotic genome regulation. EMBO Rep. 2009;10:973–982. doi: 10.1038/embor.2009.181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kapranov P, Willingham AT, Gingeras TR. Genome-wide transcription and the implications for genomic organization. Nat Rev Genet. 2007;8:413–423. doi: 10.1038/nrg2083. [DOI] [PubMed] [Google Scholar]
- 6.Metzker ML. Sequencing technologies—the next generation. Nat Rev Genet. 2010;11:31–46. doi: 10.1038/nrg2626. [DOI] [PubMed] [Google Scholar]
- 7.van Vliet AH. Next generation sequencing of microbial transcriptomes: challenges and opportunities. FEMS Microbiol Lett. 2010;302:1–7. doi: 10.1111/j.1574-6968.2009.01767.x. [DOI] [PubMed] [Google Scholar]
- 8.Wang Z, Gerstein M, Snyder M. RNA-Seq: a revolutionary tool for transcriptomics. Nat Rev Genet. 2009;10:57–63. doi: 10.1038/nrg2484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ozsolak F, et al. Direct RNA sequencing. Nature. 2009;461:814–818. doi: 10.1038/nature08390. [DOI] [PubMed] [Google Scholar]
- 10.Gubler U. Second-strand cDNA synthesis: mRNA fragments as primers. Methods Enzymol. 1987;152:330–335. doi: 10.1016/0076-6879(87)52038-9. [DOI] [PubMed] [Google Scholar]
- 11.Spiegelman S, et al. DNA-directed DNA polymerase activity in oncogenic RNA viruses. Nature. 1970;227:1029–1031. doi: 10.1038/2271029a0. [DOI] [PubMed] [Google Scholar]
- 12.Wu JQ, et al. Systematic analysis of transcribed loci in ENCODE regions using RACE sequencing reveals extensive transcription in the human genome. Genome Biol. 2008;9:R3. doi: 10.1186/gb-2008-9-1-r3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Perocchi F, Xu Z, Clauder-Munster S, Steinmetz LM. Antisense artifacts in transcriptome microarray experiments are resolved by actinomycin D. Nucleic Acids Res. 2007;35:e128. doi: 10.1093/nar/gkm683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.He Y, Vogelstein B, Velculescu VE, Papadopoulos N, Kinzler KW. The antisense transcriptomes of human cells. Science. 2008;322:1855–1857. doi: 10.1126/science.1163853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Mamanova L, et al. FRT-seq: amplification-free, strand-specific transcriptome sequencing. Nat Methods. 2010;7:130–132. doi: 10.1038/nmeth.1417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Parkhomchuk• D, et al. Transcriptome analysis by strand-specific sequencing of complementary DNA. Nucleic Acids Res. 2009 doi: 10.1093/nar/gkp596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Cocquet J, Chong A, Zhang G, Veitia RA. Reverse transcriptase template switching and false alternative transcripts. Genomics. 2006;88:127–131. doi: 10.1016/j.ygeno.2005.12.013. [DOI] [PubMed] [Google Scholar]
- 18.Mader RM, et al. Reverse transcriptase template switching during reverse transcriptase-polymerase chain reaction: artificial generation of deletions in ribonucleotide reductase mRNA. J Lab Clin Med. 2001;137:422–428. doi: 10.1067/mlc.2001.115452. [DOI] [PubMed] [Google Scholar]
- 19.Roy SW, Irimia M. When good transcripts go bad: artifactual RT-PCR ‘splicing’ and genome analysis. Bioessays. 2008;30:601–605. doi: 10.1002/bies.20749. [DOI] [PubMed] [Google Scholar]
- 20.Roberts JD, et al. Fidelity of two retroviral reverse transcriptases during DNA-dependent DNA synthesis in vitro. Mol Cell Biol. 1989;9:469–476. doi: 10.1128/mcb.9.2.469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Armour CD, et al. Digital transcriptome profiling using selective hexamer priming for cDNA synthesis. Nat Methods. 2009;6:647–649. doi: 10.1038/nmeth.1360. [DOI] [PubMed] [Google Scholar]
- 22.Hansen KD, Brenner SE, Dudoit S. Biases in Illumina transcriptome sequencing caused by random hexamer priming. Nucleic Acids Res. 2010 doi: 10.1093/nar/gkq224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Faulhammer D, Lipton RJ, Landweber LF. Fidelity of enzymatic ligation for DNA computing. J Comput Biol. 2000;7:839–848. doi: 10.1089/10665270050514963. [DOI] [PubMed] [Google Scholar]
- 24.Housby JN, Southern EM. Fidelity of DNA ligation: a novel experimental approach based on the polymerisation of libraries of oligonucleotides. Nucleic Acids Res. 1998;26:4259–4266. doi: 10.1093/nar/26.18.4259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kozarewa I, et al. Amplification-free Illumina sequencing-library preparation facilitates improved mapping and assembly of (G+C)-biased genomes. Nat Methods. 2009;6:291–295. doi: 10.1038/nmeth.1311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Dohm JC, Lottaz C, Borodina T, Himmelbauer H. Substantial biases in ultra-short read data sets from high-throughput DNA sequencing. Nucleic Acids Res. 2008;36:e105. doi: 10.1093/nar/gkn425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Goren A, et al. Chromatin profiling by directly sequencing small quantities of immunoprecipitated DNA. Nat Methods. 2010;7:47–49. doi: 10.1038/nmeth.1404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Oshlack A, Wakefield MJ. Transcript length bias in RNA-seq data confounds systems biology. Biol Direct. 2009;4:14. doi: 10.1186/1745-6150-4-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Young MD, Wakefield MJ, Smyth GK, Oshlack A. Gene ontology analysis for RNA-seq: accounting for selection bias. Genome Biol. 2010;11:R14. doi: 10.1186/gb-2010-11-2-r14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Donis-Keller H, Maxam AM, Gilbert W. Mapping adenines, guanines, and pyrimidines in RNA. Nucleic Acids Res. 1977;4:2527–2538. doi: 10.1093/nar/4.8.2527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Bowers J, et al. Virtual terminator nucleotides for next-generation DNA sequencing. Nat Methods. 2009;6:593–595. doi: 10.1038/nmeth.1354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Graber JH, McAllister GD, Smith TF. Probabilistic prediction of Saccharomyces cerevisiae mRNA 3′-processing sites. Nucleic Acids Res. 2002;30:1851–1858. doi: 10.1093/nar/30.8.1851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lutz CS. Alternative polyadenylation: a twist on mRNA 3′ end formation. ACS Chem Biol. 2008;3:609–617. doi: 10.1021/cb800138w. [DOI] [PubMed] [Google Scholar]
- 34.Tian B, Hu J, Zhang H, Lutz CS. A large-scale analysis of mRNA polyadenylation of human and mouse genes. Nucleic Acids Res. 2005;33:201–212. doi: 10.1093/nar/gki158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Nagalakshmi U, et al. The transcriptional landscape of the yeast genome defined by RNA sequencing. Science. 2008;320:1344–1349. doi: 10.1126/science.1158441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.David L, et al. A high-resolution map of transcription in the yeast genome. Proc Natl Acad Sci U S A. 2006;103:5320–5325. doi: 10.1073/pnas.0601091103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ozsolak F, et al. Comprehensive polyadenylation site maps in yeast and human reveal pervasive alternative polyadenylation. Cell. 2010;143:1018–1029. doi: 10.1016/j.cell.2010.11.020. [DOI] [PMC free article] [PubMed] [Google Scholar]