Abstract
The cytokine tumor necrosis factor (TNF) is the primary trigger of inflammation. Like many extracellular signaling proteins, TNF is synthesized as a transmembrane protein; the active signal is its ectodomain, which is shed from cells after cleavage by an ADAM family metalloprotease, TACE/ADAM17. We report that iRhom2/RHBDF2, a proteolytically inactive member of the rhomboid family, is required for TNF release in mice. iRhom2 binds TACE and promotes exit from the endoplasmic reticulum. The failure of TACE ER exit in the absence of iRhom2 prevents furin-mediated maturation and its trafficking to the cell surface, the site of TNF cleavage. Given the role of TNF in autoimmune and inflammatory diseases, iRhom2 may represent an attractive therapeutic target.
Proteolytic release of the extracellular domain of transmembrane proteins is an important mechanism for generating signals that regulate major aspects of animal development, physiology, immunity and pathology (1, 2). An important example of regulated ectodomain shedding is the cytokine TNF, the primary trigger of inflammatory responses. TNF is associated with many human diseases including rheumatoid arthritis, Crohn’s disease, atherosclerosis, psoriasis, sepsis, diabetes, and obesity. Its blockade is licensed as a therapy for a number of conditions, and is being assessed for others (3). Soluble, active TNF is shed from the plasma membrane by the ADAM family metalloprotease TACE (TNFα converting enzyme; also known as ADAM17) (4). Despite the medical importance of TNF and other transmembrane signaling proteins, the regulation of ectodomain shedding remains poorly understood. Both the transmembrane forms of the signaling proteins themselves, and the shedding proteases, are subject to control by posttranslational modification, interaction with specific partners, and regulated intracellular trafficking and compartmentalization (5-9). The relative physiological importance, however, of these different modes of regulation is unclear.
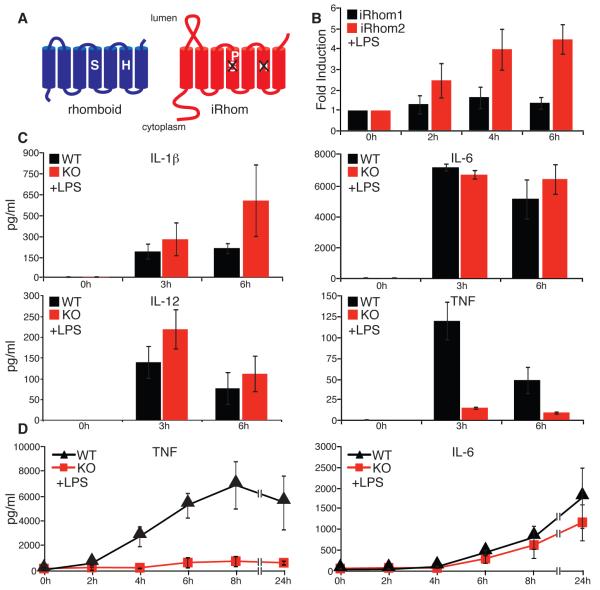
We have investigated the regulation of ectodomain shedding by genetic and cellular approaches, both in Drosophila and mammalian cells. This has led to the recent discovery of a new class of polytopic endoplasmic reticulum (ER) proteins, the iRhoms, which are non-catalytic relatives of rhomboid intramembrane proteases (Fig. 1A) (10). Drosophila iRhom regulates epidermal growth factor (EGF) receptor signaling by interacting with EGF family ligands in the ER, shunting them into the ER-associated degradation (ERAD) pathway (11). iRhoms are conserved in all metazoans, and in cell culture assays their mammalian counterparts, iRhom1 and iRhom2, can also promote ERAD of EGF. In mammals, however, their physiological role is unknown. We therefore generated a null mutation in the gene that encodes iRhom2/RHBDF2 in mice (Fig. S1A). iRhom2−/− mice appeared normal: they were viable and fertile, with no morphological defects. Unlike iRhom1, which is widely expressed, iRhom2 is predominantly expressed in immune cells, particularly macrophages (12, 13), where its expression was specifically upregulated in response to LPS stimulation (Fig 1B). We therefore challenged iRhom2 mutant mice with LPS, to mimic bacterial infection, and measured the serum concentration of pro-inflammatory cytokines. Interleukin (IL)-1β, IL-6 and IL-12 were induced normally but, remarkably, TNF induction was almost completely abolished in mutant mice (Fig. 1C). Macrophages are the most abundant source of TNF; consistent with this, in vitro differentiated bone marrow-derived macrophages (BMDMs) from iRhom2 mutant mice failed to secrete TNF in response to LPS (Fig 1D).
Fig. 1.
iRhom2 is essential for LPS-induced TNF shedding. (A) Comparison of an active rhomboid protease and iRhom. (B) Fold induction of iRhom2 mRNA in response to LPS in WT macrophages (mean of 3 experiments ±SD), as measured by qPCR. mRNA levels were normalized to endogenous actin. (C) Serum concentrations of the indicated cytokines in WT and iRhom2−/− mice (KO) in response to LPS (mean of 5 experiments ±SD), as measured by multi-analyte cytokine profiling. (D) TNF and IL-6 production by LPS-stimulated BMDMs (WT and iRhom2−/− (KO); mean of 3 experiments ±SD), as measured by ELISA. All graphs show mean±SD.
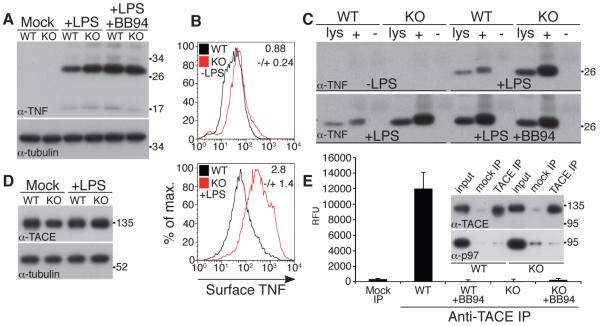
The failure of TNF secretion in iRhom2−/− mice is not caused by its loss of expression: RNA levels were unaffected (Fig. S2A), and protein levels were actually elevated in iRhom2−/− macrophages (Fig. 2A and B). This result was confirmed by cell surface labeling followed by affinity precipitation, which showed an increase in the accumulation of the full length, membrane tethered form of TNF on the plasma membrane of mutant cells (Fig. 2C). Furthermore, inhibition of the shedding protease TACE in wild-type cells by the matrix metalloproteinase inhibitor BB94 (Fig. 2A,C) resulted in a phenotype similar to iRhom2−/− macrophages. Together, these data imply that loss of iRhom2 does not interfere with the induction or intracellular trafficking of TNF, but instead suggests a shedding defect. We tested this hypothesis directly: although TACE levels were normal in both wild-type and mutant macrophages (Fig. 2D), loss of iRhom2 led to a complete abolition of activity of immunoprecipitated TACE (Fig. 2E, S2B). This result explains the loss of TNF shedding in mutant mice and macrophages.
Fig. 2.
TNF biogenesis is normal but TACE proteolytic activity is defective in iRhom2−/− cells. (A) Immunoblots for TNF of extracts from mock, LPS, or LPS/BB94-treated macrophages. Anti-tubulin was used to determine equal loading throughout. (B) Flow cytometric analysis of surface TNF on CD11b+ WT and iRhom2−/− (KO) macrophages 3 hours after LPS treatment. The fold-change in mean fluorescent intensity (KO/WT) is indicated in each graph (mean of 3 experiments ±SD). (C) Whole cell lysate (lys) versus cell surface biotinylated (+) or mock-treated (−) proteins bound to neutravidin resin from WT or KO macrophages were immunoblotted as indicated. (D) Immunoblot of TACE expression in BMDMs. (E) Immunoprecipitates from iRhom2−/− macrophages subjected to a fluorogenic cleavage assay (mean of 3 experiments ±SD). Specificity of the TACE antibody is demonstrated by probing IPs with TACE and p97 antibodies (inset in E). Levels of TACE in the activity assay were equal in WT and KO macrophages (inset E and Fig. S2B).
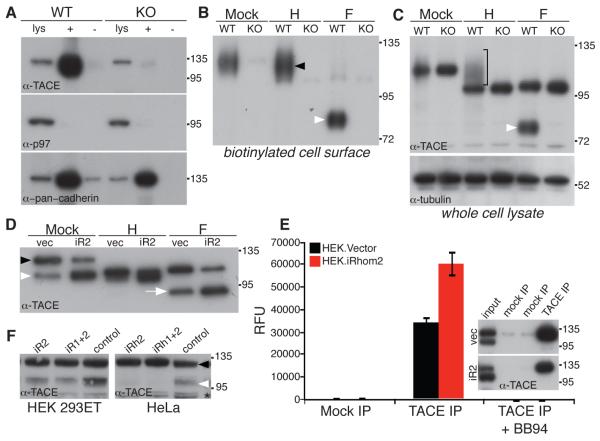
Like TNF, TACE has been the focus of major pharmaceutical interest: not only does it regulate inflammation via TNF, but it is also implicated in cancer by activating EGF receptor ligands (14); furthermore, it is a point of integration for a wide range of incoming signals (15). Despite this intense spotlight, TACE regulation remains poorly understood. Several lines of evidence suggest the trafficking of active enzyme to the plasma membrane is important, but other work implies that at least in some contexts the rate limiting step may be activation of molecules already at the cell surface (5-7, 16, 17). Because most evidence agrees that the plasma membrane is the primary site of TACE activity, we tested whether TACE was present at the cell surface of iRhom2−/− cells. In contrast to wild-type cells, no TACE was found on the surface of iRhom2−/− macrophages (Fig. 3A and B), implying that the cause of TACE inactivity is a fundamental defect in its intracellular trafficking.
Fig. 3.
iRhom2 is essential for ER exit and activation of TACE. (A) Cell surface biotinylation was used to detect TACE at the plasma membrane in WT and iRhom2−/− (KO) macrophages: whole cell lysates (lys); neutravidin captured cell surface proteins (+); mock-treated (−). Cytoplasmic protein p97 was used as a negative control for cell surface biotinylation; surface levels of cadherin are comparable in WT and KO cells. (B) Biotinylated cell surface proteins were deglycosylated with endoglycosidase-H (H), which cleaves ER but not Golgi N-glycans, or PNGase F (F) which removes both. Cell surface TACE is endo-H resistant (black arrow). TACE matures by acquiring Golgi N-glycans, and furin-catalyzed removal of the prodomain. PNGase F treatment of cell surface TACE generates a lower molecular weight polypeptide corresponding to the furin-cleaved species (white arrowhead; see also Fig. S2B,C). (C) TACE immunoblot of whole cells lysates treated with endo-H (H) or PNGase F (F) Bracket: endo-H resistant mature TACE. For PNGase F-treated lysates: the upper band is ER resident, immature TACE, and the lower band is furin-cleaved TACE (white arrow, see also Fig. 3B, S2C,D). (D) Cell extracts from HEK cells stably expressing mouse iRhom2 were deglycosylated with endo-H (H) or PNGase F (F) and immunoblotted for TACE. Unlike mouse BMDM-derived TACE, human TACE can be detected as two species in lysates without prior deglycosylation (immature, black arrowhead; mature, white arrowhead). (E) Activity of TACE immunoprecipitates from HEK cells expressing iRhom2 (mean of 3 experiments ±SD); specificity of the IP was confirmed by western blot (inset). (F) The effect of siRNA knockdown of human iRhom2 (and combined iRhom1/2) on the expression of endogenous mature TACE (white arrowhead; black arrowhead indicates immature TACE) in human HEK 293T and HeLa cells. As a control, an unrelated member of the rhomboid family (RHBDL2) was knocked down. The asterisk denotes a non-specific band.
Endoglycosidase-H (endo-H) sensitive N-glycans are added to TACE in the ER. The sugars are then elaborated in the Golgi apparatus, becoming resistant to endo-H; later in the trans-Golgi network (TGN) the inhibitory N-terminal prodomain is removed by furin (18). These modifications can be used to track the progress of TACE from the ER to the plasma membrane (see Fig. S2C,D for an overview). Consistent with previous reports (7, 18), we found the majority of endogenous TACE to be endo-H sensitive, and therefore located in the ER, in both wild-type and mutant cells (Fig. 3C). In the absence of iRhom2, however, TACE never became endo-H resistant, demonstrating that, unlike in wild-type cells, it was unable to reach the site of addition of Golgi-specific glycans (Fig 3C, bracket). Furthermore, furin cleavage of the prodomain was abolished in the knockout cells (Fig 3C, white arrowhead). We conclude that trafficking from the ER is a limiting step for TACE maturation, and that iRhom2 is an essential component of the mechanism that releases it to the Golgi apparatus. Without iRhom2, therefore, TACE cannot reach the TGN to be activated.
Our results are consistent with iRhom2 being a rate-limiting factor in releasing TACE from the ER. To examine this directly, we expressed mouse iRhom2 in HEK cells and assayed its effect on endogenous human TACE. Overexpression of iRhom2 caused excess TACE to leave the ER, resulting in more furin-processed TACE (Fig 3D, white arrow), which translated into an overall increase in TACE enzyme activity (Fig 3E). This result demonstrates that iRhom2 is rate limiting for TACE activity, and also indicates conservation of human and mouse iRhom2 function. This was confirmed by showing that RNAi knockdown of human iRhom2 inhibited the maturation of endogenous TACE in human cells (Fig. 3F).
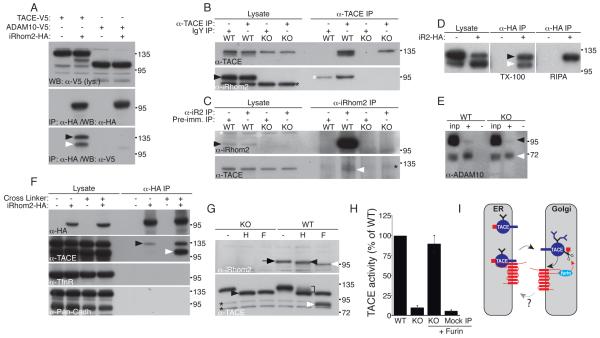
How might iRhom2 control trafficking of TACE? Because most TACE resides in the ER, which is also where the bulk of iRhom2 is detected in COS cells (11), we asked whether they can bind to each other directly. Indeed, when coexpressed in HEK cells, mouse iRhom2 efficiently immunoprecipitated tagged mouse TACE (Fig 4A and S3A), whereas multiple control transmembrane proteins showed no interaction with iRhom2 (Fig 4A,F; Fig. S3A,B). Conversely, tagged iRhom2 was immunoprecipitated by endogenous TACE (Fig S3C). Moreover, we found that both endogenous TACE and iRhom2 proteins could co-immunoprecipitate one another in macrophages, (Fig. 4B,C). iRhom2 interacts with both endogenous full-length TACE and the mature form (Fig. 4A; black and white arrowheads, respectively), although by increasing the stringency of the binding and washing conditions, we found that the interaction is weaker once the TACE prodomain is removed (Fig. 4D; immature and mature TACE indicated by black and white arrowhead, respectively). We also tested whether iRhom2 interacts with ADAM10, TACE’s closest relative (14). In contrast to TACE, no interaction (Fig. 4A) or effect on ADAM10 trafficking (Fig. 4E) was detected, indicating a high level of specificity of iRhom2 for potential client proteins.
Fig. 4.
iRhom2 interacts with TACE and leaves the ER. (A) Immunoprecipitates from HEK cells or HEK cells stably expressing iRhom2-HA, transfected with TACE-V5 or ADAM10-V5, were immunoblotted for iRhom2. Co-immunoprecipitation of immature and mature TACE is indicated by black and white arrowheads, respectively. (B) Macrophage lysates were immunoprecipitated with anti-TACE and immunoblotted for iRhom2 or TACE. Arrowhead: endogenous iRhom2. Black asterisk: a non-specific band detected in both KO and WT. White asterisk: background binding of iRhom2 to the resin. (C) Macrophage lysates were immunoprecipitated with anti-iRhom2 and immunoblotted for iRhom2 or TACE. White arrowhead: endogenous TACE. Black arrowhead: endogenous iRhom. White asterisk: original TACE signal detected following reprobing. Black asterisk: a non-specific band in all IP lanes. (D) HEK cells expressing iRhom2-HA were immunoprecipitated with anti-HA and immunoblotted for TACE. IPs were performed under mild (Triton X-100 detergent), and more stringent (RIPA buffer) conditions. (E) Complete input extracts (inp) and surface biotinylated proteins (+) from WT and iRhom2 KO macrophages immunoblotted for ADAM10. Black arrow: immature ADAM10; white arrow: mature ADAM10. (F) Immunoprecipitates (in RIPA buffer) of iRhom2-HA from HEK cells; precipitates were done +/− the crosslinker DSP. Transferrin receptor and pan-cadherin immunoblots were used as negative controls. (G) Lysates from LPS-treated WT or KO macrophages were treated with endo-H (H) or PNGase-F (F), and immunoblotted for iRhom2. Arrow: fully glycosylated iRhom2. Black and white arrowheads: endo-H-resistant single glycan form and unmodified polypeptide, respectively (iRhom2 has two predicted N-linked glycosylation sites; only one is modified in the Golgi). Lower panel: TACE in the same lysates as control. Black arrowhead: endo-H-sensitive TACE. Bracket: endo-H-resistant TACE. White arrowhead: mature TACE. Asterisks: non-specific bands. (H) TACE immunoprecipitates were preincubated −/+ 100 nM recombinant furin prior to measuring TACE activity. (I) Model of iRhom2 function in macrophages.
The membrane permeable chemical crosslinker DSP could crosslink iRhom2 to both full-length and mature TACE (Fig. 4 F; black and white arrowheads, respectively) in living HEK cells, implying that the interaction occurs in vivo and that the two proteins are less than 12Å apart, and therefore that the binding is probably direct. Together, these data make a strong case that iRhom2 binds directly to TACE, thereby regulating its trafficking from the ER. We envisage two alternative models to explain these results. Either iRhom2 is required in the ER for the folding/maturation of TACE, or it acts as a type of cargo receptor to assist the onward trafficking of TACE. In support of the latter model, we find that iRhom2 in macrophages carries endo-H resistant, Golgi-specific, glycans (Fig 4G), which implies that at least some iRhom2 is trafficked beyond the ER. This is consistent with the observed binding in vivo of iRhom2 to both full-length and mature TACE. The reduction in affinity once the prodomain is removed suggests possible release of TACE from iRhom2 once activated by furin (although we emphasize that this last interpretation is speculative). In further support of our model, and arguing against substantially disrupted folding of TACE, catalytically inactive TACE immunoprecipitated from iRhom2 knockout cells (Fig 2E) can be made active by incubation with recombinant furin (Fig. 4H), indicating that lack of access to furin is the primary cause of TACE inactivity.
These data, along with our previous results (11), suggest that iRhoms are polytopic membrane proteins that bind to single-pass transmembrane proteins in the ER, thereby regulating their subsequent trafficking (Fig. 4I). Depending on the specific iRhom, the client, and/or the cellular context, iRhoms can promote exit from ER, or degradation (11). The ER exit of some Toll-like receptors, which also have a single TMD, depends on an unrelated polytopic membrane protein, Unc93b1 (19). Perhaps trafficking assistance for proteins with single TMDs is common: they may require specific cargo reception machinery, or alternatively, TMD chaperones may prevent non-specific TMD interactions. Strikingly, there is a broad family of other non-proteolytic rhomboid-like proteins that also lack defining features of iRhoms, including, very distantly, derlins (10, 20). It is possible that this wider group of rhomboid-like membrane proteins may also interact with, and regulate, the fate of single-pass transmembrane proteins.
This work provides a mechanistic explanation for why TNF production is abolished in iRhom2 knockout mice. Interestingly, iRhom2 expression is itself upregulated by TNF signaling (21), implying a positive feedback loop: TNF upregulates the mechanism that promotes its own activation. Positive feedback can sharpen physiological signaling responses, but can also promote hyperactivity if normal regulation is disrupted, and underlies a variety of inflammatory and oncogenic pathologies (22). The further medical implication of this work is the potential of interfering with iRhom2 or its binding to TACE as a strategy to block TNF signaling. Although TNF blockade is already licensed as a therapeutic strategy, its use is hampered by side effects. The specificity, expression, and the mouse phenotype we report implies that pharmacological disruption of the interaction between iRhom2 and TACE should block specifically macrophage release of TNF.
Supplementary Material
One sentence summary.
Release of the cytokine TNF is controlled by the rhomboid-like pseudoprotease iRhom2, which binds to and assists the ER exit of TACE, enabling its activation.
Acknowledgements
We thank C. Blobel, R. Hegde, G. Murphy, S. Munro, V. Lastun, C. Luginsland and K. Strisovsky for their advice and generously sharing unpublished reagents. We are grateful to R. Pannell for help with the mouse knockout, and to members of the animal facility for their assistance. The data presented in this paper are reported in the main paper and in the Supporting Online Material. C.A. was supported by a long-term fellowship from The International Human Frontier Science Program Organization and was the recipient of an EMBO Long-Term Fellowship. M.Z. was supported by Erwin-Schrödinger fellowship from the Austrian FWF, and an APART fellowship from the Austrian Academy of Science.
Footnotes
Materials and methods are presented as Supplementary Online Material.
References
- 1.Gooz M. Crit. Rev. Biochem. Mol. Biol. 2010;45:146. doi: 10.3109/10409231003628015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Peschon JJ, et al. Science. 1998;282:1281. doi: 10.1126/science.282.5392.1281. [DOI] [PubMed] [Google Scholar]
- 3.Tracey D, Klareskog L, Sasso EH, Salfeld JG, Tak PP. Pharmacol. Ther. 2008;117:244. doi: 10.1016/j.pharmthera.2007.10.001. [DOI] [PubMed] [Google Scholar]
- 4.Black RA, et al. Nature. 1997;385:729. doi: 10.1038/385729a0. [DOI] [PubMed] [Google Scholar]
- 5.Le Gall SM, Maretzky T, Issuree PD, Niu XD, Reiss K, Saftig P, Khokha R, Lundell D, Blobel CP. J. Cell Sci. 2010;123:3913. doi: 10.1242/jcs.069997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Xu P, Derynck R. Mol. Cell. 2010;37:551. doi: 10.1016/j.molcel.2010.01.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Soond SM, Everson B, Riches DW, Murphy G. J. Cell Sci. 2005;118:2371. doi: 10.1242/jcs.02357. [DOI] [PubMed] [Google Scholar]
- 8.Ha SD, Martins A, Khazaie K, Han J, Chan BM, Kim SO. J. Immunol. 2008;181:690. doi: 10.4049/jimmunol.181.1.690. [DOI] [PubMed] [Google Scholar]
- 9.Lieu ZZ, Lock JG, Hammond LA, La Gruta NL, Stow JL, Gleeson PA. Proc. Natl. Acad. Sci. U S A. 2008;105:3351. doi: 10.1073/pnas.0800137105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lemberg MK, Freeman M. Genome Res. 2007;17:1634. doi: 10.1101/gr.6425307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zettl M, Adrain C, Strisovsky K, Lastun V, Freeman M. Cell. 2011;145:79. doi: 10.1016/j.cell.2011.02.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Su AI, et al. Proc. Natl. Acad. Sci. U S A. 2004;101:6062. doi: 10.1073/pnas.0400782101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Barrett T, et al. Nucleic Acids Res. 2009;37:D885. doi: 10.1093/nar/gkn764. http://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE1133 and http://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE10246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Saftig P, Reiss K. Eur. J. Cell Biol. 2010 doi: 10.1016/j.ejcb.2010.11.005. [DOI] [PubMed] [Google Scholar]
- 15.Murphy G. Semin. Cell. Dev. Biol. 2009;20:138. doi: 10.1016/j.semcdb.2008.09.004. [DOI] [PubMed] [Google Scholar]
- 16.Le Gall SM, Bobe P, Reiss K, Horiuchi K, Niu XD, Lundell D, Gibb DR, Conrad D, Saftig P, Blobel CP. Mol. Biol. Cell. 2009;20:1785. doi: 10.1091/mbc.E08-11-1135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Rousseau S, Papoutsopoulou M, Symons A, Cook D, Lucocq JM, Prescott AR, O’Garra A, Ley SC, Cohen P. J. Cell Sci. 2008;121:149. doi: 10.1242/jcs.018671. [DOI] [PubMed] [Google Scholar]
- 18.Schlondorff J, Becherer JD, Blobel CP. Biochem. J. 2000;347:131. [PMC free article] [PubMed] [Google Scholar]
- 19.Kim YM, Brinkmann MM, Paquet ME, Ploegh HL. Nature. 2008;452:234. doi: 10.1038/nature06726. [DOI] [PubMed] [Google Scholar]
- 20.Greenblatt EJ, Olzmann JA, Kopito RR. Nat. Struct. Mol. Biol. 2011 doi: 10.1038/nsmb.2111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Barrett T, et al. Nucleic Acids Res. 2009;37:D885. doi: 10.1093/nar/gkn764. http://www.ncbi.nlm.nih.gov/geoprofiles?term=GDS1543[ACCN]+nm_024599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Freeman M. Nature. 2000;408:313. doi: 10.1038/35042500. [DOI] [PubMed] [Google Scholar]
- 23.Liu P, Jenkins NA, Copeland NG. Genome Res. 2003;13:476. doi: 10.1101/gr.749203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Adrain C, Strisovsky K, Zettl M, Hu L, Lemberg MK, Freeman M. EMBO Rep. 2011;12:421. doi: 10.1038/embor.2011.50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lammich S, Buell D, Zilow S, Ludwig AK, Nuscher B, Lichtenthaler SF, Prinzen C, Fahrenholz F, Haass C. J. Biol. Chem. 2010;285:15753. doi: 10.1074/jbc.M110.110742. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.