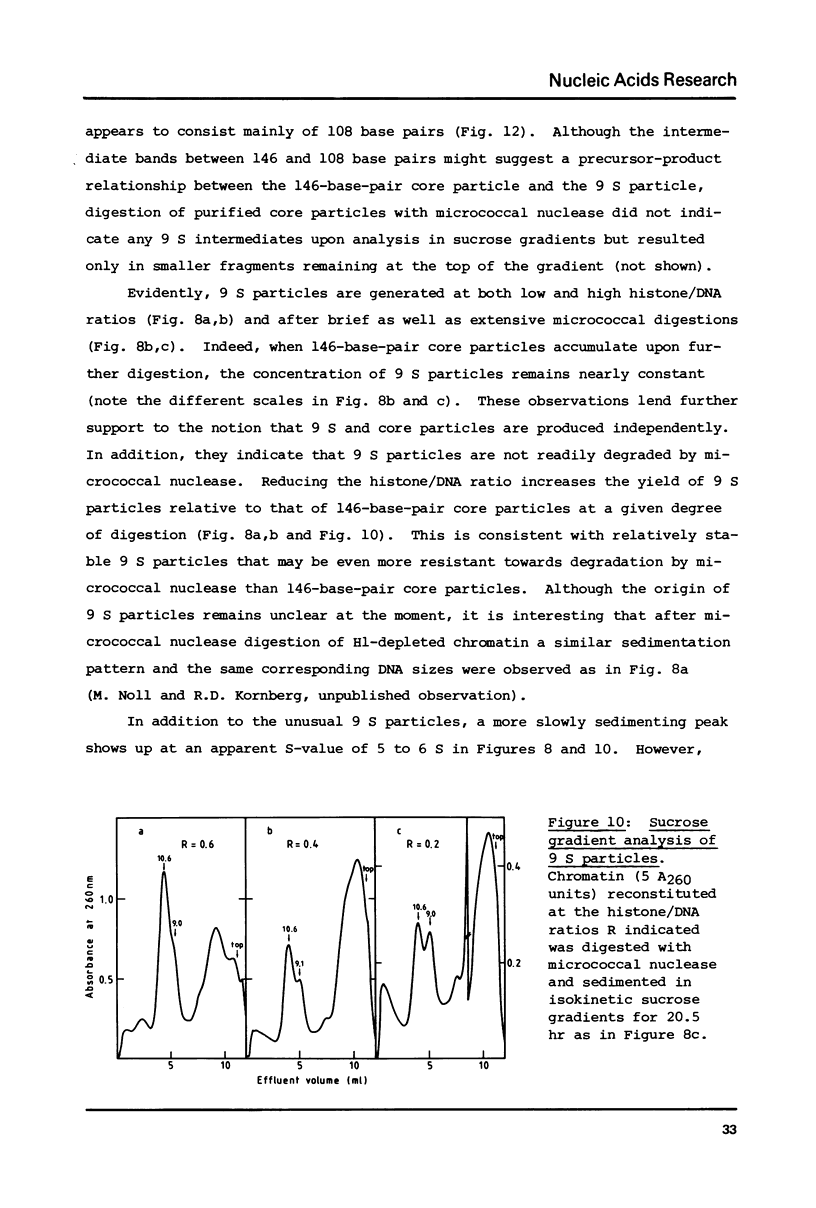
Abstract
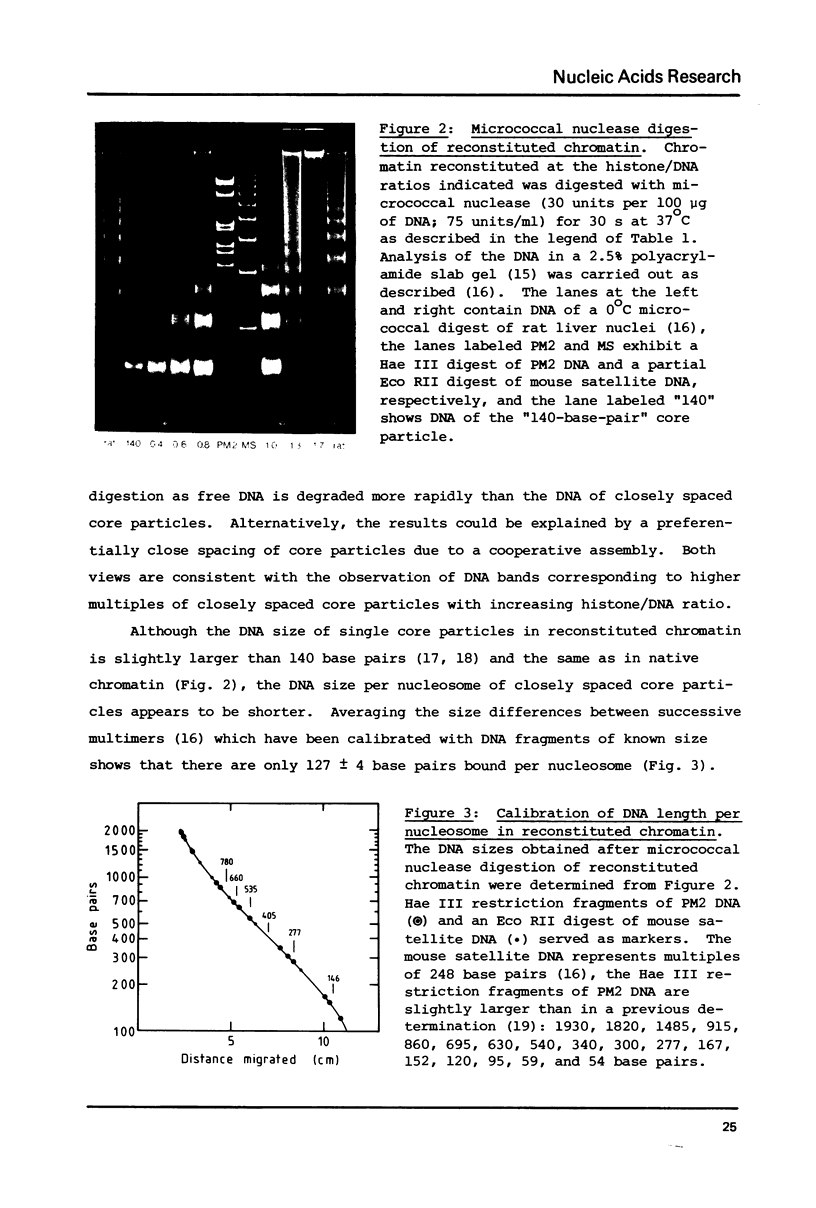
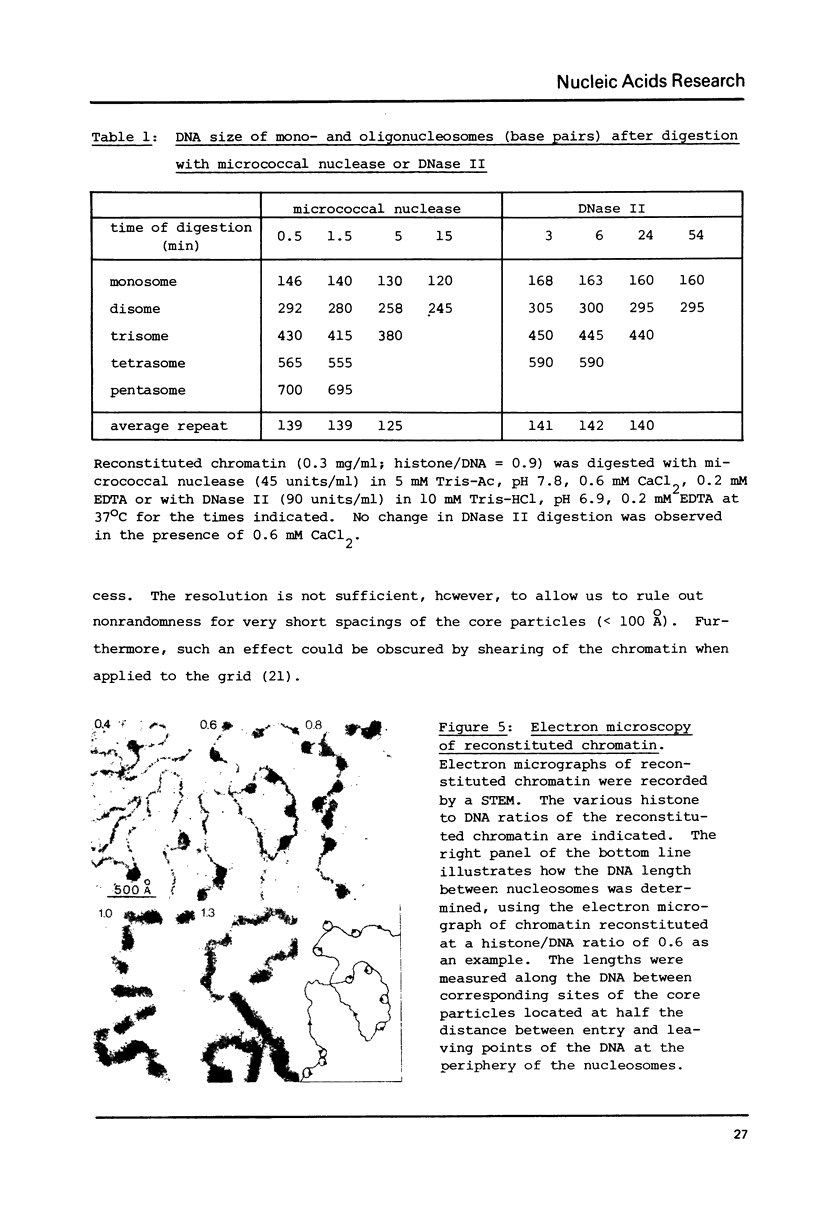
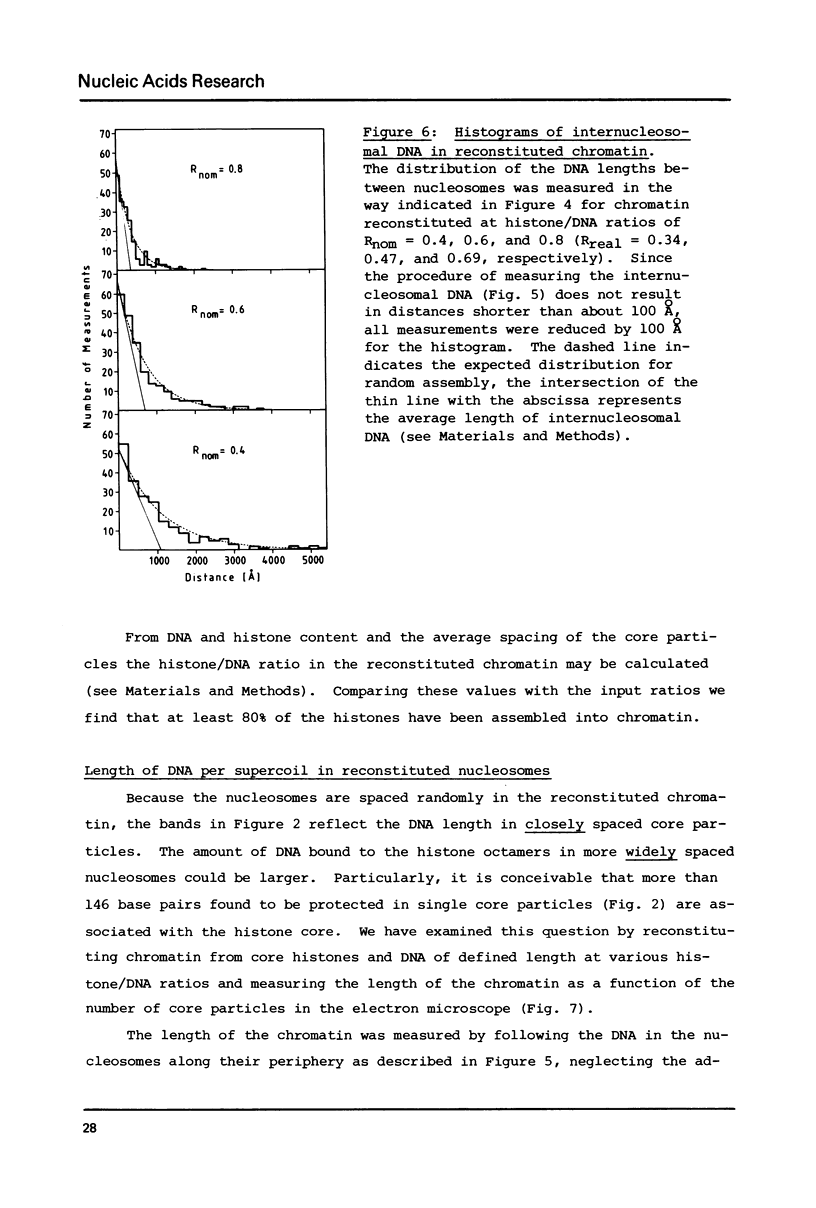
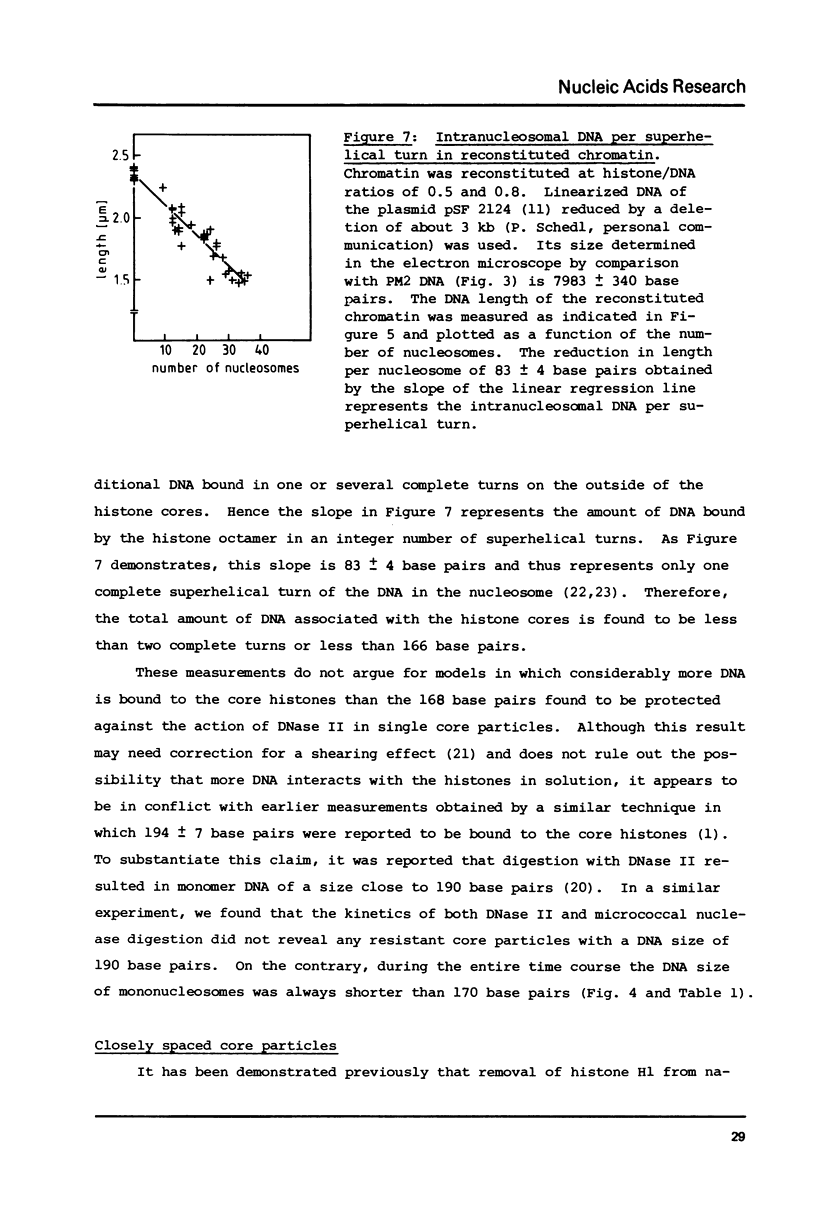
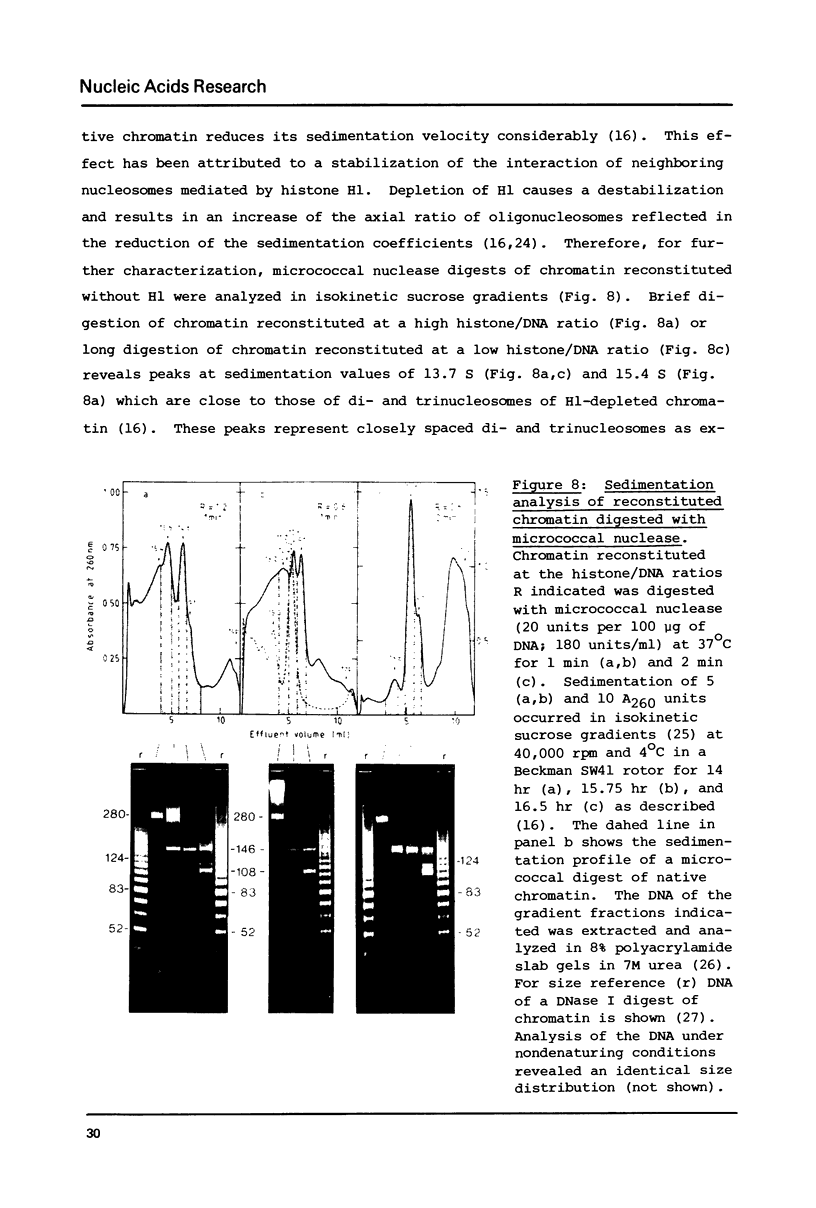
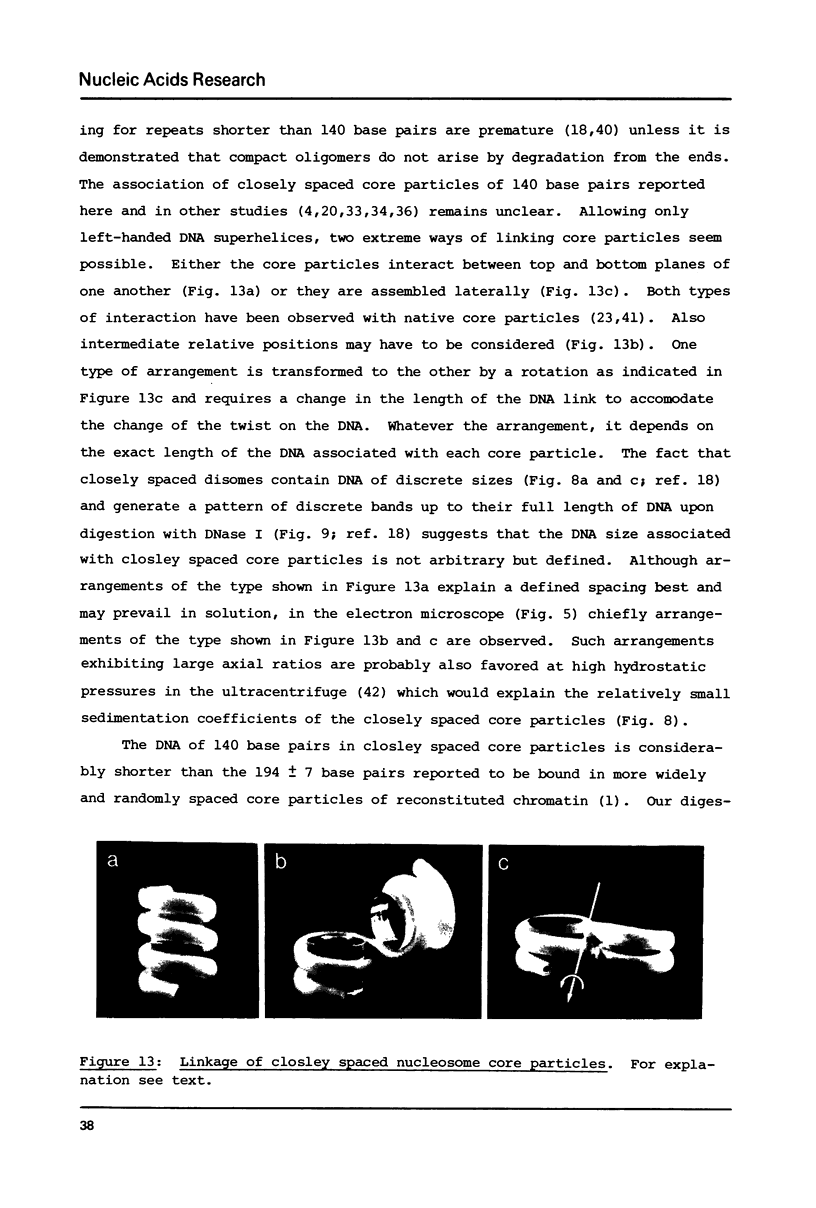
Self-assembly of DNA with the four core histones but in the absence of H1 generates nucleosome core particles which are spaced randomly over large distances. Closely spaced core particles, however, exhibit a preferred short linkage which is not a multiple of 10 base pairs. They bind about 140 base pairs whereas apparently shorter DNA lengths per nucleosome observed after digestion with micrococcal nuclease are the result of degradation from the ends. The DNA length of one superhelical turn in the core particle is 83 +/- 4 base pairs. Single core particles may bind more DNA than closely spaced core particles but probably less than two full turns of 168 base pairs. The internal structures of single and of native core particles are very similar as judged by their amount of DNA, sedimentation coefficient, appearance in the electron microscope, and digestion with DNase I. In addition to core particles, a particle is described which sediments at 9 S and consists of 108 base pairs of DNA bound to the histone octamer. It appears to be the smallest stable "core particle" but it is not a degradation product of the 146-base-pair core particle. Digestion of end-labeled 9 S and nucleosome core particles with DNase I shows distinct differences.
Full text
PDF














Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Bakayev V. V., Bakayeva T. G., Varshavsky A. J. Nucleosomes and subnucleosomes: heterogeneity and composition. Cell. 1977 Jul;11(3):619–629. doi: 10.1016/0092-8674(77)90079-4. [DOI] [PubMed] [Google Scholar]
- Dubochet J., Ducommun M., Zollinger M., Kellenberger E. A new preparation method for dark-field electron microscopy of biomacromolecules. J Ultrastruct Res. 1971 Apr;35(1):147–167. doi: 10.1016/s0022-5320(71)80148-x. [DOI] [PubMed] [Google Scholar]
- Dubochet J., Noll M. Nucleosome arcs and helices. Science. 1978 Oct 20;202(4365):280–286. doi: 10.1126/science.694532. [DOI] [PubMed] [Google Scholar]
- Engel A., Dubochet J., Kellenberger E. Some progress in the use of a scanning transmission electron microscope for the observation of biomacromolecules. J Ultrastruct Res. 1976 Dec;57(3):322–330. doi: 10.1016/s0022-5320(76)80120-7. [DOI] [PubMed] [Google Scholar]
- Finch J. T., Lutter L. C., Rhodes D., Brown R. S., Rushton B., Levitt M., Klug A. Structure of nucleosome core particles of chromatin. Nature. 1977 Sep 1;269(5623):29–36. doi: 10.1038/269029a0. [DOI] [PubMed] [Google Scholar]
- Finch J. T., Noll M., Kornberg R. D. Electron microscopy of defined lengths of chromatin. Proc Natl Acad Sci U S A. 1975 Sep;72(9):3320–3322. doi: 10.1073/pnas.72.9.3320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fulmer A. W., Fasman G. D. Analysis of chromatin reconstitutiion. Biochemistry. 1979 Feb 20;18(4):659–668. doi: 10.1021/bi00571a017. [DOI] [PubMed] [Google Scholar]
- Germond J. E., Bellard M., Oudet P., Chambon P. Stability of nucleosomes in native and reconstituted chromatins. Nucleic Acids Res. 1976 Nov;3(11):3173–3192. doi: 10.1093/nar/3.11.3173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Germond J. E., Hirt B., Oudet P., Gross-Bellark M., Chambon P. Folding of the DNA double helix in chromatin-like structures from simian virus 40. Proc Natl Acad Sci U S A. 1975 May;72(5):1843–1847. doi: 10.1073/pnas.72.5.1843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klevan L., Crothers D. M. Isolation and characterization of a spacerless dinucleosome from H1-deleted chromatin. Nucleic Acids Res. 1977 Dec;4(12):4077–4089. doi: 10.1093/nar/4.12.4077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klevan L., Hogan M., Dattagupta N., Crothers D. M. Electric dichroism studies of the size and shape of nucleosomal particles. Cold Spring Harb Symp Quant Biol. 1978;42(Pt 1):207–214. doi: 10.1101/sqb.1978.042.01.023. [DOI] [PubMed] [Google Scholar]
- Langmore J. P., Wooley J. C. Chromatin architecture: investigation of a subunit of chromatin by dark field electron microscopy. Proc Natl Acad Sci U S A. 1975 Jul;72(7):2691–2695. doi: 10.1073/pnas.72.7.2691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laskey R. A., Honda B. M., Mills A. D., Finch J. T. Nucleosomes are assembled by an acidic protein which binds histones and transfers them to DNA. Nature. 1978 Oct 5;275(5679):416–420. doi: 10.1038/275416a0. [DOI] [PubMed] [Google Scholar]
- Laskey R. A., Mills A. D., Morris N. R. Assembly of SV40 chromatin in a cell-free system from Xenopus eggs. Cell. 1977 Feb;10(2):237–243. doi: 10.1016/0092-8674(77)90217-3. [DOI] [PubMed] [Google Scholar]
- Levitt M. How many base-pairs per turn does DNA have in solution and in chromatin? Some theoretical calculations. Proc Natl Acad Sci U S A. 1978 Feb;75(2):640–644. doi: 10.1073/pnas.75.2.640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levy A., Jakob K. M. Nascent DNA in nucleosome like structures from chromatin. Cell. 1978 Jun;14(2):259–267. doi: 10.1016/0092-8674(78)90112-5. [DOI] [PubMed] [Google Scholar]
- Loening U. E. The fractionation of high-molecular-weight ribonucleic acid by polyacrylamide-gel electrophoresis. Biochem J. 1967 Jan;102(1):251–257. doi: 10.1042/bj1020251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lutter L. C. Kinetic analysis of deoxyribonuclease I cleavages in the nucleosome core: evidence for a DNA superhelix. J Mol Biol. 1978 Sep 15;124(2):391–420. doi: 10.1016/0022-2836(78)90306-6. [DOI] [PubMed] [Google Scholar]
- Maniatis T., Jeffrey A., van deSande H. Chain length determination of small double- and single-stranded DNA molecules by polyacrylamide gel electrophoresis. Biochemistry. 1975 Aug 26;14(17):3787–3794. doi: 10.1021/bi00688a010. [DOI] [PubMed] [Google Scholar]
- Morris N. R. Nucleosome structure in Aspergillus nidulans. Cell. 1976 Jul;8(3):357–363. doi: 10.1016/0092-8674(76)90147-1. [DOI] [PubMed] [Google Scholar]
- Noll H. Characterization of macromolecules by constant velocity sedimentation. Nature. 1967 Jul 22;215(5099):360–363. doi: 10.1038/215360a0. [DOI] [PubMed] [Google Scholar]
- Noll M. DNA folding in the nucleosome. J Mol Biol. 1977 Oct 15;116(1):49–71. doi: 10.1016/0022-2836(77)90118-8. [DOI] [PubMed] [Google Scholar]
- Noll M. Differences and similarities in chromatin structure of Neurospora crassa and higher eucaryotes. Cell. 1976 Jul;8(3):349–355. doi: 10.1016/0092-8674(76)90146-x. [DOI] [PubMed] [Google Scholar]
- Noll M., Hapke B., Schreier M. H., Noll H. Structural dynamics of bacterial ribosomes. I. Characterization of vacant couples and their relation to complexed ribosomes. J Mol Biol. 1973 Apr 5;75(2):281–294. doi: 10.1016/0022-2836(73)90021-1. [DOI] [PubMed] [Google Scholar]
- Noll M. Internal structure of the chromatin subunit. Nucleic Acids Res. 1974 Nov;1(11):1573–1578. doi: 10.1093/nar/1.11.1573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noll M., Kornberg R. D. Action of micrococcal nuclease on chromatin and the location of histone H1. J Mol Biol. 1977 Jan 25;109(3):393–404. doi: 10.1016/s0022-2836(77)80019-3. [DOI] [PubMed] [Google Scholar]
- Noll M., Thomas J. O., Kornberg R. D. Preparation of native chromatin and damage caused by shearing. Science. 1975 Mar 28;187(4182):1203–1206. doi: 10.1126/science.187.4182.1203. [DOI] [PubMed] [Google Scholar]
- Ohlenbusch H. H., Olivera B. M., Tuan D., Davidson N. Selective dissociation of histones from calf thymus nucleoprotein. J Mol Biol. 1967 Apr 28;25(2):299–315. doi: 10.1016/0022-2836(67)90143-x. [DOI] [PubMed] [Google Scholar]
- Oudet P., Gross-Bellard M., Chambon P. Electron microscopic and biochemical evidence that chromatin structure is a repeating unit. Cell. 1975 Apr;4(4):281–300. doi: 10.1016/0092-8674(75)90149-x. [DOI] [PubMed] [Google Scholar]
- Pardon J. F., Worcester D. L., Wooley J. C., Cotter R. I., Lilley D. M., Richards R. M. The structure of the chromatin core particle in solution. Nucleic Acids Res. 1977 Sep;4(9):3199–3214. doi: 10.1093/nar/4.9.3199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prunell A., Kornberg R. D., Lutter L., Klug A., Levitt M., Crick F. H. Periodicity of deoxyribonuclease I digestion of chromatin. Science. 1979 May 25;204(4395):855–858. doi: 10.1126/science.441739. [DOI] [PubMed] [Google Scholar]
- Ruiz-Carrillo A., Jorcano J. L., Eder G., Lurz R. In vitro core particle and nucleosome assembly at physiological ionic strength. Proc Natl Acad Sci U S A. 1979 Jul;76(7):3284–3288. doi: 10.1073/pnas.76.7.3284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simpson R. T., Whitlock J. P. Mapping DNAase l-susceptible sites in nucleosomes labeled at the 5' ends. Cell. 1976 Oct;9(2):347–353. doi: 10.1016/0092-8674(76)90124-0. [DOI] [PubMed] [Google Scholar]
- So M., Gill R., Falkow S. The generation of a ColE1-Apr cloning vehicle which allows detection of inserted DNA. Mol Gen Genet. 1975 Dec 30;142(3):239–249. doi: 10.1007/BF00425649. [DOI] [PubMed] [Google Scholar]
- Spadafora C., Oudet P., Chambon P. The same amount of DNA is organized in in vitro-assembled nucleosomes irrespective of the origin of the histones. Nucleic Acids Res. 1978 Oct;5(10):3479–3489. doi: 10.1093/nar/5.10.3479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steinmetz M., Streeck R. E., Zachau H. G. Closely spaced nucleosome cores in reconstituted histone.DNA complexes and histone-H1-depleted chromatin. Eur J Biochem. 1978 Feb;83(2):615–628. doi: 10.1111/j.1432-1033.1978.tb12131.x. [DOI] [PubMed] [Google Scholar]
- Sussman J. L., Trifonov E. N. Possibility of nonkinked packing of DNA in chromatin. Proc Natl Acad Sci U S A. 1978 Jan;75(1):103–107. doi: 10.1073/pnas.75.1.103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tatchell K., Van Holde K. E. Compact oligomers and nucleosome phasing. Proc Natl Acad Sci U S A. 1978 Aug;75(8):3583–3587. doi: 10.1073/pnas.75.8.3583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tatchell K., Van Holde K. E. Reconstitution of chromatin core particles. Biochemistry. 1977 Nov 29;16(24):5295–5303. doi: 10.1021/bi00643a021. [DOI] [PubMed] [Google Scholar]
- Thomas J. O., Furber V. Yeast chromatin structure. FEBS Lett. 1976 Jul 15;66(2):274–280. doi: 10.1016/0014-5793(76)80521-2. [DOI] [PubMed] [Google Scholar]
- Thomas J. O., Kornberg R. D. An octamer of histones in chromatin and free in solution. Proc Natl Acad Sci U S A. 1975 Jul;72(7):2626–2630. doi: 10.1073/pnas.72.7.2626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yaneva M., Tasheva B., Dessev G. Nuclease digestion of reconstituted chromatin. FEBS Lett. 1976 Nov;70(1):67–70. doi: 10.1016/0014-5793(76)80727-2. [DOI] [PubMed] [Google Scholar]