Abstract
Understanding the mechanisms that promote aberrant tumour cell survival is critical for the determination of novel strategies to combat colorectal cancer. We have recently shown that the anti-apoptotic protein BAG-1, highly expressed in pre-malignant and colorectal cancer tissue, can potentiate cell survival through regulating NF-κB transcriptional activity. In this study, we identify a novel complex between BAG-1 and the p50-p50 NF-κB homodimers, implicating BAG-1 as a co-regulator of an atypical NF-κB pathway. Importantly, the BAG-1-p50 complex was detected at gene regulatory sequences including the epidermal growth factor receptor [EGFR] and COX-2 [PTGS2] genes. Suppression of BAG-1 expression using siRNA was shown to increase EGFR and suppress COX-2 expression in colorectal cancer cells. Furthermore, mouse embryonic fibroblasts derived from the NF-κB1 [p105/p50] knockout mouse were used to demonstrate that p50 expression was required for BAG-1 to suppress EGFR expression. This was shown to be functionally relevant as attenuation of BAG-1 expression increased ligand activated phosphorylation of EGFR in colorectal cancer cells. In summary, this paper identifies a novel role for BAG-1 in modulating gene expression through interaction with the p50-p50 NF-κB complexes. Data presented led us to propose that BAG-1 can act as a selective regulator of p50-p50 NF-κB responsive genes in colorectal tumour cells, potentially important for the promotion of cell survival in the context of the fluctuating tumour microenvironment. As BAG-1 expression is increased in the developing adenoma through to metastatic lesions, understanding the function of the BAG-1-p50 NF-κB complexes may aid in identifying strategies for both the prevention and treatment of colorectal cancer.
Introduction
Colorectal cancer (CRC) is the second leading cause of cancer death in many industrialised countries (Cancer Research UK, 2007); understanding the mechanisms that promote colorectal tumour cell survival is critical for the identification of novel strategies for the prevention and treatment of this important cancer.
BAG-1 was first discovered when identifying novel Bcl-2 and glucocorticoid receptor interacting proteins (Takayama et al., 1995; Zeiner and Gehring, 1995 respectively). BAG-1 has since been shown to interact with a wide variety of molecular targets and consequently has been shown to regulate a number of key cellular processes such as proliferation, cell signalling, transcription, differentiation and apoptosis (reviewed in Townsend et al., 2005). Characterised by the BAG domain (BD), BAG-1 is a member of a family of related proteins of which there are at least six in humans (reviewed in Takayama and Reed, 2001). The BAG-1 gene encodes multiple isoforms, generated from alternate translation of a single mRNA (Packham et al., 1997; reviewed in Sharp et al., 2004). The BAG-1L isoform is found to be localised within the nucleus, BAG-1M is both cytoplasmic and nuclear. BAG-1S, although reported to be located predominantly within the cytoplasm (Brimmell et al., 1999; Packham et al., 1997; Takayama et al., 1998; Yang et al., 1998), is both nuclear and cytoplasmic in colorectal epithelial cells (Barnes et al 2005). These differences in subcellular localisation are thought to be important for biological function, for example the cytoplasmic BAG-1S interacts with Raf-1 to promote cell survival (Song et al., 2001), whereas, the nuclear BAG-1L plays a role in transcription through interacting with DNA (Niyaz et al., 2001) and transcription factors (Cutress et al., 2003; Froesch et al., 1998, reviewed in Gehring et al., 2009).
BAG-1 is over-expressed in a number of cancers, with studies focusing on breast (Brimmell et al., 1999), lung (Rorke et al., 2001), colon (Clemo et al., 2008) and oral squamous cell carcinoma (Shindoh et al., 2000; Wood et al., 2009). In the colon, nuclear BAG-1 has been associated with shorter overall patient survival (Kikuchi et al., 2002). It is therefore of interest that, when studying BAG-1 expression in colorectal tumour tissue, we recently demonstrated that BAG-1 increased NF-κB transcriptional activity (Clemo et al., 2008); activation of NF-κB having been associated with resistance to therapy (Arlt et al., 2002; Weldon et al., 2001).
NF-κB was first discovered in 1986 as a nuclear factor binding to the intronic kappa-light-chain enhancer element in B cells (Sen and Baltimore, 1986). NF-κB is a family of transcription factors that regulate a number of biological processes including inflammation, apoptosis and cell proliferation (reviewed in Chen and Greene, 2004). In mammals there are five members in the NF-κB family; RelA (p65), c-Rel, RelB, NF-κB1 (p105/p50) and NF-κB2 (p100/p52)(reviewed in Ghosh et al., 1998; Xiao et al., 2006). The NF-κB family share a conserved rel homology domain in their N-terminus, which is required for dimer formation, DNA binding and interaction with the inhibitor of NF-κB (IκB) (Gilmore and Temin, 1988; Kieran et al., 1990; reviewed in Perkins, 2007). p65, RelB and c-Rel contain a transactivation domain (TAD) which is necessary for the activation of transcription (Kamens et al., 1990; reviewed in Perkins, 2007; Ryseck et al., 1992; Schmitz and Baeuerle, 1991). NF-κB1 (p105/p50) and NF-κB2 (p100/p52) both lack this TAD but have been shown to induce transcription when bound to other co-activating proteins, for example Bcl-3 (Franzoso et al., 1992, reviewed in Beinke and Ley, 2004; Schmitz and Baeuerle, 1991).
In unstimulated cells, NF-κB is predominantly cytoplasmic through interaction with inhibitors of NF-κB (IκBs) (Baeuerle and Baltimore, 1988; reviewed in Lin et al., 2010). Upon stimulation (such as by cytokines, bacterial products and growth factors) (reviewed in Pahl, 1999), NF-κB can be activated by two main pathways. These are the canonical and non-canonical pathways (otherwise referred to as the ‘classical’ and ‘non-classical’). In response to stimulation, NF-κB dimers translocate into the nucleus and bind to DNA at specific κB sites (reviewed in Lin et al., 2010). In addition to the canonical and non-canonical pathways, p50-p50 NF-κB homodimers are generated as part of an atypical NF-κB pathway (reviewed in Pereira and Oakley 2008). Although not as extensively studied as some of the other NF-κB dimers, evidence from the literature shows that the p50-p50 homodimers are localised to the nucleus in unstimulated or “resting” cells where they bind to DNA and repress transcription (Hou et al., 2003; Zhong et al., 2002). In stimulated cells, the ability of the p50-p50 homodimers to act as a transcriptional activators or repressors is dependent on the co-factors that interact with the NF-κB complex at the DNA (reviewed in Glass and Rosenfeld, 2000). As the p50-p50 NF-κB homodimeric complexes can bind κB elements with high affinity, it is suggested that the p50-p50 homodimers may impact on the function of the transcriptionally active heterodimeric NF-κB complexes (reviewed in Pereira and Oakley 2008).
Previously, in colorectal tumour cells, BAG-1 has been shown to regulate NF-κB transcriptional activity (Clemo et al., 2008), although the mechanism has not been described. In the current study we establish that BAG-1 can interact with p50-p50 NF-κB homodimers at a consensus κB site and importantly, using the EGFR gene as an example, we reveal that the BAG-1-p50 complex can regulate gene expression. Hence this paper identifies a novel role for BAG-1 as a co-regulator of gene expression through interaction with the p50-p50 NF-κB complexes, and suggests a potentially important role for this complex in colorectal carcinogenesis.
Materials and methods
Cell line and cell culture conditions
The human colorectal carcinoma-derived cell line HCT116 was obtained from the American Type Culture Collection (Rockville, USA). The human colorectal carcinoma-derived HCA7 colony 29 cell line (herein referred to as HCA7) was a gift from Dr. S. Kirkland (Imperial College London, UK). The NF-κB+/+ and −/− MEF cell lines were a gift from J. Caamano (Birmingham University, UK).
RNAi
Cells were reverse transfected using Lipofectamine 2000 (Invitrogen, Paisley, UK) with small interfering RNAs (siRNAs) from Applied Biosystems (Warrington, UK) targeting BAG-1 or a negative control sequence (50nM) as described previously (Clemo et al., 2008) or from Dharmacon, (Lafayette, USA) targeting NF-κB1, murine BAG-1 or a negative control siRNA (25nM siGENOME SMARTpool).
DNA transfection
Cells were transiently transfected using Lipofectamine 2000 (Invitrogen, Paisley, UK) with pIRESneo2 expression plasmids encoding BAG-1L or BAG-1S, or a pRSV NF-κBp50 expression plasmid. The empty pIRES neo2 or pRSV plasmids were used as the negative controls.
BAG-1SNLS and BAG-1SNES fusion proteins
The BAG-1S isoform was cloned into a pIRESneo2 fused to either a nuclear localisation signal (NLS) or a nuclear exit signal (NES) using primers 5′-GTAGCTAGCGAAGAGATGGTGGACCTCCAAAAGAAGCTGGAGGAGCTGGAGCTGAATCGGAGCCAGGAGGTG for BAG-1SNES and GGTAGCTAGCGAAGAGATGCCAAAAAAGAAGAGAAAGGTAAATCGGAGCCAGGAGGTG for BAG-1NLS; common reverse primer was 5′-ATGAGGATCCTCACTCGGCCGAGGGCAAAGT.
NF-κB reporter assays
Growing cells were transiently transfected with either the NF-κB reporter plasmid pNF-κB-TA-luc or with the control plasmid pTA-luc (Clontech, Oxford, UK) including the pRL-SV40 renilla plasmid (Promega, Southampton, UK) using Lipofectamine 2000 following the manufacturer’s protocol. Following lysis, luciferase activities were measured using the Dual-Luciferase Reporter Assay System (Promega, Southampton, UK) according to the manufacturer’s instructions.
Immunoblotting
Whole cell lysates were prepared and subjected to immunoblotting as previously described (Williams et al., 1993) using antibodies against BAG-1 (G3E2; kind gift from G. Packham, Southampton University, UK) and NF-κB1 (E10: sc-8414; Santa Cruz Biotechnology, Ca, USA).
Immunofluorescence
Proteins were visualised as previously described (Barnes et al., 2005). BAG-1 was visualised using the polyclonal anti-BAG-1 (TB3) antibody which recognises all three BAG-1 isoforms (kind gift from G. Packham, Southampton University, UK).
Immunohistochemistry
Immunohistochemistry was carried out as previously described (Clemo et al., 2008); Formalin-fixed, paraffin embedded normal human large intestinal sections were obtained from the Department of Histopathology, Bristol Royal Infirmary, Bristol, UK with local Ethics Committee approval. NF-κB1 was detected using the E10 antibody (sc-8414; Santa Cruz Biotechnology, Ca, USA), BAG-1 was detected using the TB3 antibody (G. Packham, Southampton University, UK).
Preparation of nuclear protein extracts
A Nuclear Extraction Kit (Active Motif, Rixensart, Belgium) was used as per manufacturer’s instructions. The protein concentration of the nuclear fractions was determined using the Bio-Rad DC Protein assay kit (Bio-Rad, Hertfordshire, UK).
Oligonucleotide-pulldown Assay
The assay was essentially carried out following manufacturer’s instructions (Santa Cruz Biotechnology, Ca, USA), using NF-κB binding oligonucleotide sequences (wild-type: 5′-AGTTGAGGGGACTTTCCCAGGC-3; mutant: 5′-AGTTGAGGCGACTTTCCCAGGC-3′). However, the binding buffer used was 8.5mM HEPES pH7.9; 1mM KCl, 1mM MgCl2, 1mM DTT, 7.5% Ficoll, 1mg/ml BSA and 1-4μg [dIdC].
Electrophoretic Mobility Shift Assay (EMSA)
The EMSA was carried out using standard techniques as previously described (Smartt et al., 2003). For supershift assays, 1μl of BAG-1 (G3E2; Kind gift from G. Packham, Southampton University, UK) or NF-κB antibody (NF-κB1; sc-114; p65: sc-372 and NF-κB2: sc-298; Santa Cruz Biotechnology, Ca, USA) was used.
Quantitative Reverse Transcription Polymerase Chain Reaction (Q-RT-PCR)
Total RNA extraction and comparative quantitative real-time polymerase chain reaction (Q-RT-PCR) was performed as previously described (Clemo et al., 2008). QuantiTect Primer Assays and primers for EGFR and PTGS2 were obtained from Qiagen Ltd, (Crowley, West Sussex, UK).
Co-immunopreciptation
Preparation of immunoprecipitating antibody
All immunoprecipitating antibodies were obtained from Santa Cruz Biotechnology (Ca, USA); BAG-1 antibody (C16: sc-939), NF-κB antibodies (NF-κB1: E10; sc-8414x; p65: F6; sc-8008x) and the IgG antibody (sc-2027) as an irrelevant control. 5μg of antibody was suspended in 25% Immunoprecipitation Matrix slurry (IP Matrix; Santa Cruz Biotechnology, Santa Cruz, California, USA)
Preparation of cell lysate
Cell pellets were suspended in immunopreciptation lysis buffer supplemented with protease inhibitors (Complete Mini Protease Inhibitors, Roche Diagnostics Ltd, W.Sussex, UK), sonicated in a Bioruptor UCD-200 Waterbath Sonicator (Diagebode Inc, Sparta, New Jersey, USA) and then resuspended in the lysis buffer with protease inhibitors. Immunopreciptiation was carried out essentially as previously described (Arhel et al., 2003).
Chromatin immunoprecipitation (ChIP)
Chromatin Immunoprecipitation (ChIP) was carried out as previously described (Kaidi et al., 2007) using BAG-1 antibody (C16; sc-939) and NF-κB1 antibody (H119; sc-7179) or a species matched IgG control (Santa Cruz Biotechnology (Ca, USA ). Primers flanking the NF-κB1 binding sites at the EGFR promoter (Thornburg et al., 2003) were (forward, 5-GGGGACCCGAATAAAGGAGCAGTT-3′; reverse, 5-CTGAGGAGTTAATTTCCGAGAGGGG-3′) and the PTGS2 promoter (forward,5′-CTGTTGAAAGCAACTTAGCT-3′; reverse, 5′-AGACTGAAAACCAAGCCCAT-3′, Deng et al., 2003). Re-ChiP: For the Re-ChIP assay, the complexes were eluted from the primary immunoprecipitation followed by re-immunoprecipitation with a second antibody.
Statistical analysis
Statistical analyses were carried out using a Student’s t-test in Microsoft Excel and represented as: * = P < 0.05; ** = P < 0.01; *** = P < 0.001
Results
Nuclear localisation of BAG-1 is important for the regulation of TNF-α-induced NF-κB transcriptional activity
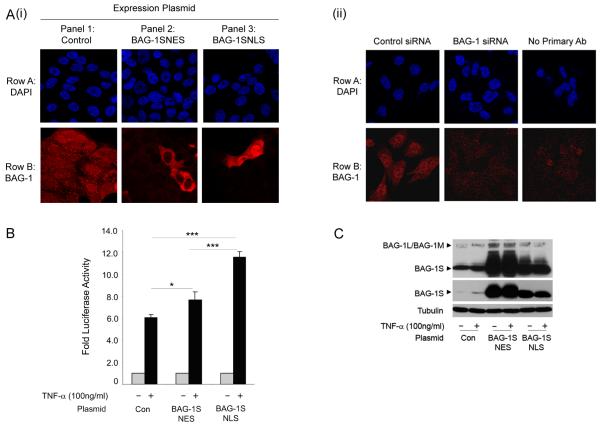
As the BAG-1L isoform had been shown to significantly enhance TNF-α-induced NF-κB transcriptional activity (in colorectal carcinoma cells, Clemo et al., 2008, as well as adenoma and human embryonic kidney cell lines, data not shown), initial experiments were carried out to investigate the importance of the nuclear localisation of BAG-1 for regulating NF-κB activity. Using the BAG-1S isoform, (common to the C terminus of BAG-1L containing the BAG domain but does not have an endogenous nuclear localisation signal, Brimmell et al., 1999), fusion proteins were made introducing either a nuclear localisation (BAG-1SNLS) or nuclear exit signal (BAG-1SNES) (kind gift, G. Packham, Southampton University, UK) thus limiting the expression of the exogenous protein to either the nucleus or the cytoplasm respectively. The localisation of the fusion proteins was confirmed in HCA7 cells (as these cells have an increased cytoplasmic volume compared to HCT116 cells, facilitating the visualisation of subcellular compartments, figure 1Ai). Expression of endogenous BAG-1 protein was found to be both nuclear and cytoplasmic, whereas the BAG-1SNES fusion protein was concentrated in the cytoplasm and the BAG-1SNLS protein in the nucleus. The ability of the fusion proteins to potentiate TNF-α-induced NF-κB transcriptional activity was tested using a luciferase reporter assay, the results are summarised in figure 1B. A highly significant increase in TNF-α-induced NF-κB transcriptional activity was detected in HCT116 cells when BAG-1S was fused to a NLS when compared to BAG-1S fused to a NES (figure 1B), despite the fact that expression of the BAG-1SNES protein was higher than the BAG-1SNLS protein (figure 1C). (Of note, it remains possible that the small but significant increase in NF-κB transcriptional activity observed in the TNF-α treated BAG-1SNES transfected cells is due to the induction of the endogenous nuclear BAG-1L and 1M isoforms in the transfected cells and previously reported, Hinnit et al., 2010). Taken together, these results demonstrate that TNF-α-induced NF-κB activity is enhanced by the nuclear localisation of the BAG-1 protein and justifies further investigation into the mechanism by which BAG-1 protein can potentiate NF-κB transcriptional activity in the nucleus.
Figure 1. Enforced nuclear localisation of the BAG-1S isoform increases TNF-α induced NF-κB transcriptional activity.
(A) (i) Confocal immunofluorescence microscopy was used to show the localisation of BAG-1 in HCA7 cells transfected with the control IRES plasmid, BAG-1SNES or BAG-1SNLS expression plasmids. Row A: DAPI staining showing the cell nuclei; Row B: Alexa Fluor-546 TRITC fluorescence showing BAG-1 staining. It is important to note that endogenous BAG-1 expression could not be detected in panel 2 or panel 3 as the intensity of the confocal microscope was decreased in order to visualise the localisation of the exogenous fusion proteins. (ii) The specificity of the antibody was confirmed in cells transfected with the control negative / BAG-1 siRNA , as previously reported (Clemo et al., 2008) Row A: DAPI staining showing the cell nuclei; Row B: Alexa Fluor-546 TRITC fluorescence showing BAG-1 staining. (B) Transcriptional activation of the NF-κB luciferase reporter in HCT116 cells transfected with control plasmid, BAG-1SNES or BAG-1SNLS expression plasmids and treated with TNF-α (100ng/ml) for 16 hours. The results shown are representative of three separate experiments done in triplicate (± standard deviation). Statistical analysis was carried out using a Student’s t-test * = P < 0.05; ** = P < 0.01; *** = P < 0.001. (C) Western analysis confirmed over-expression of the BAG-1SNES and BAG-1SNLS fusion proteins (middle panel shows lower exposure to demonstrate expression of the exogenous BAG-1S fusion proteins). With TNF-α treatment, an increase in endogenous BAG-1 expression was detected. Equal loading was confirmed by α-tubulin. Of note, expression of exogenous BAG-1SNES or BAG-1SNLS proteins increased expression of the endogenous BAG-1L/BAG-1M proteins (as previously reported, Hinnit et al., 2010).
BAG-1 interacts with the p50-p50 homodimeric NF-κB complex
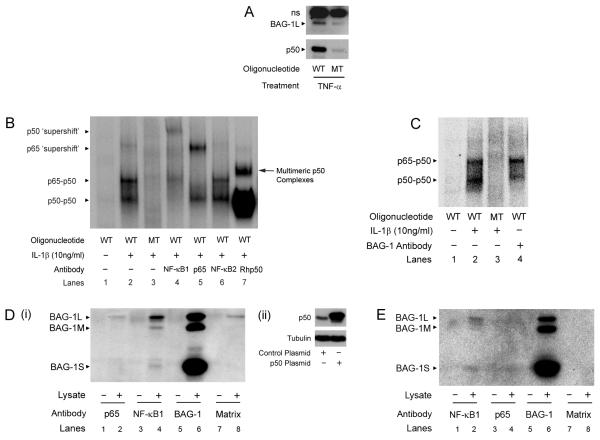
Previously it has been established that the nuclear localised BAG-1 is able to bind DNA and regulate transcription of reporter constructs (Zeiner et al., 1999; Niyaz et al., 2001). Furthermore, in prostate and breast cancer cells BAG-1 has been shown to interact with nuclear hormone receptors and regulate their transcriptional activity (Froesch et al., 1998; Liu et al., 1998). Therefore, because of the ability to bind DNA, we investigated whether BAG-1 could be isolated in association with an NF-κB-DNA binding motif in colorectal cancer cells. Initially, we performed an oligopulldown assay using the consensus NF-κB binding sequence GGGGACTTTCCC, and were able to demonstrate that BAG-1 could be detected at the κB binding site in HCT116 cells treated with TNF-α (100ng/ml) (figure 2A). To investigate whether BAG-1 was associated with the NF-κB complexes themselves, we initially treated HCT116 cells with a number of different NF-κB activators to maximise NF-κB signalling in the cells. HCT116 cells treated with IL-1β (10ng/ml) resulted in an approximate 10-fold induction of the NF-κB reporter construct as compared to four-fold induction by TNF-α (100ng/ml) and less than two-fold induction with LPS (10ng/ml, data not shown). Therefore treatment with IL-1β (10ng/ml) was used to stimulate NF-κB DNA binding in subsequent experiments. Within the literature, cytokine stimulation of colonic epithelial cells has predominantly been associated with increased binding of the p65-p50 heterodimer and p50-p50 homodimer NF-κB complexes using an electrophoretic mobility shift assay (EMSA) (Inan et al., 2000). Importantly both p65-p50 heterodimers and p50-p50 homodimers have high affinity for the consensus κB sequence used (Duckett et al., 1993). Addition of specific antibodies raised against individual NF-κB subunits allowed the complexes in the stimulated cells to be identified (figure 2B, lanes 4-6). A recombinant p50 protein (Rhp50) was added to indicate the position of the p50-p50 homodimers (lane 7). Addition of the NF-κB2 [p100/p52] antibody (which acts as a negative control as p52 NF-κB complexes weakly associate with the GGGGACTTTCCC binding motif) did not supershift any of the bands (lane 6).
Figure 2. BAG-1 interacts with the p50-p50 homodimeric NF-κB complex.
(A) BAG-1 interacts with a consensus NF-κB DNA binding site as shown by an oligonucleotide pulldown assay. ns denotes a non-specific band. HCT116 cells were transfected with the BAG-1L expression plasmid and treated with TNF-α (100ng/ml) for 16 hours. After cellular fractionation, the nuclear lysate was incubated with either the wild-type (GGGGACTTTCCC, WT) or mutant (GGCGACTTTCCC, MT) oligonucleotide. (B) IL-1β (10ng/ml) treated HCT116 nuclear fractions were analysed by EMSA. Nuclear lysates were incubated with the mutant (MT) oligonucleotide to control for non-specific interactions (Lane 3). 1μg of NF-κB1, p65 or NF-κB2 antibody were incubated with nuclear lysate and wild-type (WT) oligonucleotide (Lanes 4, 5, 6). 0.5μg of recombinant human p50 (Rhp50) was incubated with the nuclear lysate and wild-type oligonucleotide to show the position of the p50-p50 homodimeric NF-κB complex (Lane 7), and other multimeric complexes (Duckett et al. 1993). (C) HCT116 nuclear fractions were treated with IL-1β (10ng/ml) for 16 hours. G3E2 BAG-1 antibody was added to the HCT116 nuclear lysate, followed by incubation with the wild-type (WT) radiolabelled oligonucleotide (Lane 4). Results are representative of three independent experiments. BAG-1 interacts with the p50 protein. (D) (i) HCT116 cells were seeded for 72 hours and transfected with the control or p50 expression plasmid (shown by Western analysis, ii). 48 hours following transfection, cells were lysed and proteins immunoprecipitated using antibodies against p65 (Lane 2), NF-κB1 (Lane 4) and BAG-1 (Lane 6); (Lane 8 is the no antibody control). Western analysis identified BAG-1 protein interactions within the whole cell lysate. Results are representative of three separate experiments. (E) BAG-1 interacts with the p50 protein in untransfected HCT116 cells. Endogenous protein was immunoprecipitated as before. Western analysis was carried out to determine endogenous BAG-1 protein interactions within the whole cell lysate. Proteins were immunoprecipitated using antibodies against NF-κB1 (Lane 2), p65 (Lane 4) and BAG-1 (Lane 6). Results are representative of three independent experiments.
To identify whether BAG-1 interacted with an NF-κB transcriptional complex in colorectal epithelial cells, the BAG-1 antibody G3E2 was incubated with the nuclear lysate and radiolabelled oligonucleotide (figure 2C). Addition of the antibody resulted in a consistent decrease of the p50-p50 homodimer band and no loss of the p65-p50 heterodimeric band. Of note, contrary to expectation, this suggested that BAG-1 associated with the p50-p50 homodimeric NF-κB transcriptional complex and not the canonical p65-p50 NF-κB heterodimeric complex, raising the interesting possibility that BAG-1 could be regulating NF-κB activity through interaction with the p50-p50 homodimer NF-κB complex.
To confirm the interaction between the BAG-1 and p50 proteins, a co-immunoprecipitation assay in HCT116 cells transfected with a p50 expression plasmid was performed. The p50 expressing HCT116 cell lysates (figure 2D ii) were precipitated with anti-p65, NF-κB1 or BAG-1 antibodies and interrogated by Western analysis using the mouse anti-BAG-1 antibody (figure 2Di). As expected, the anti-BAG-1 antibody precipitated all BAG-1 isoforms as indicated by strong bands at 36, 46 and 50kDa (lane 6). The nuclear localised BAG-1 isoforms (predominantly BAG-1L) were also precipitated with the anti-NF-κB1 antibody (lane 4) but not with the anti-p65 antibody (lane 2), further implicating BAG-1 as a p50 binding protein. In addition, the reverse precipitation was carried out; p50 was precipitated by the anti-p65 and BAG-1 antibodies in HCT116 cells transfected with a BAG-1 expression plasmid (data not shown). Importantly, the BAG-1-p50 interaction was confirmed when the immunoprecipitation was repeated with endogenous protein expression (in untransfected HCT116 cells; figure 2E). Using the anti-BAG-1 antibody to interrogate bound protein, the anti-NF-κB1 antibody precipitated the nuclear BAG-1L isoform (lane 2), whereas BAG-1 protein was not precipitated by the antibody raised against p65 (lane 4). These results confirm that BAG-1 interacts with the p50 but not the p65 NF-κB protein in the HCT116 colorectal carcinoma derived cell line.
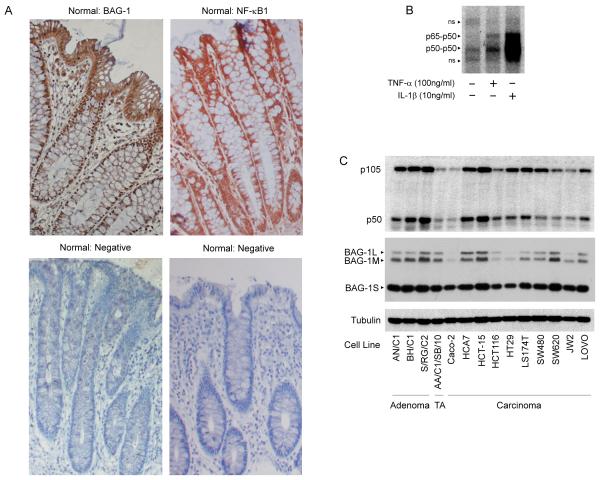
The potential significance of an interaction with NF-κB1 (p50) in the colon is emphasised by the work of the Giardina group. They showed that p50-p50 homodimers were differentially expressed in the normal crypt, being the predominant complex in the non-proliferating cells (Inan et al., 2000). In accordance with these findings, we have found that NF-κB1 is highly expressed at the top of normal colonic crypt but is also expressed at the bottom of the crypt coincident with nuclear BAG-1 expression (figure 3A). NF-κB1 represents the predominant NF-κB complex in stimulated colorectal cancer cells (figure 3B) and both BAG-1 and NF-κB1 , are also expressed in all cell lines investigated in vitro (figure 3C). This data supports a role for NF-κB1 in tissue homeostasis in the colon, and emphasises the potential significance of the novel finding that BAG-1 interacts with NF-κB1 in colorectal tumour cells.
Figure 3. Expression of BAG-1 and NF-κB1 in normal colorectal epithelium and cell lines.
(A) BAG-1 and NF-κB1 expression in paraffin-embedded normal colorectal epithelial tissue. BAG-1 expression was detected using the BAG-1 (TB3; Brimmel et al., 1999) antibody, NF-κB1 expression was detected using the anti-NF-κB1 (E10) antibody. Both were visualised using DAB (brown staining) and counter-stained with haematoxylin (blue staining), objective x10. (The negative control for staining is normal colonic epithelium without primary BAG-1 or NF-κB1 antibody; objective x10). (B) DNA bound NF-κB complexes in cytokine stimulated HCT116 cells. 3μg of HCT116 nuclear fractions were treated with TNF-α (100ng/ml) or IL-1β (10ng/ml) for 16 hours and analysed by EMSA using the wild type [WT, GGGGACTTTCCG] labelled oligonucleotide to detect active NF-κB DNA binding complexes. Non-specific bands are denoted with ns. (C) Western analysis to determine NF-κB1 and BAG-1 protein expression in a panel of colorectal cell lines (TA denotes transformed adenoma). Equal loading was confirmed by α-tubulin. Results are representative of three independent experiments.
Suppression of NF-κB1 signalling increases apoptosis in HCT116 cells
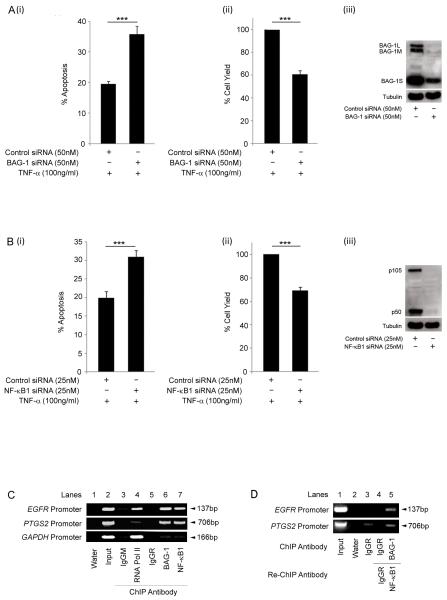
As BAG-1 has been previously been shown to promote tumour cell survival through regulation of NF-κB activity (Clemo et al., 2008), we wished to investigate whether NF-κB1 expression was implicated in colorectal tumour cell survival. HCT116 cells were transfected with either BAG-1 or NF-κB1 siRNA and apoptosis measured 48 h after treatment with TNF-α (100ng/ml), results are summarised in figure 4. Data shows that not only does loss of BAG-1 increase apoptosis in TNF-α treated cells consistent with the previous report (figure 4A, Clemo et al., 2008), but that suppression of NF-κB1 also promotes cell death in TNF-α treated cells (figure 4B). Taken together, this data implicates a role for both BAG-1 and NF-κB1 in promoting tumour cell survival.
Figure 4. BAG-1 and NF-κB1 promote cell survival in HCT116 cells.
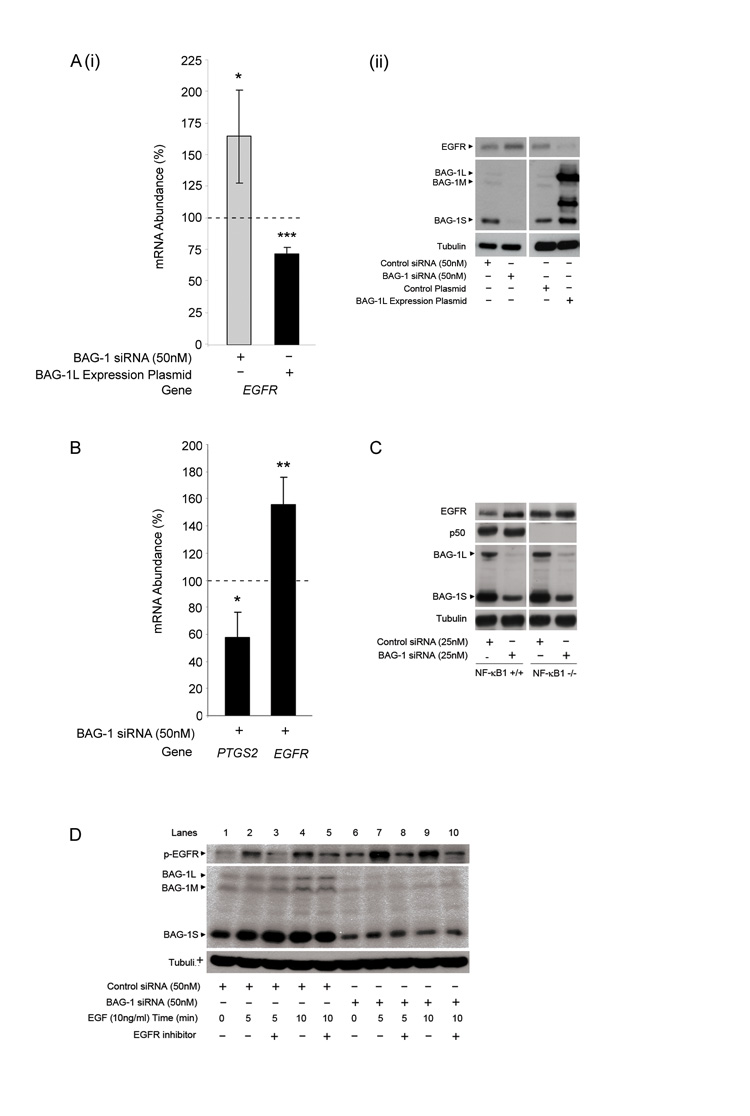
HCT116 cells were transfected with (A) control negative / BAG-1 or (B) control negative / NF-κB1 siRNA; suppression of protein expression was confirmed by Western analysis (iii). Cells were treated with TNF-α (100ng/ml) for 16h and apoptosis (i) and attached cell yield (ii) assessed. The results shown are the mean of three independent experiments done in triplicate (± standard deviation). Statistical analysis was carried out using a Student’s t-test * = P < 0.05; ** = P < 0.01; *** = P < 0.001. BAG-1 and p50 are present at the same binding site on the EGFR and PTGS2 promoters. (C) ChIP analysis of the EGFR, PTGS2 and GAPDH promoters in HCT116 cells using antibodies to immunoprecipitate the indicated proteins. The IgGM and IgGR antbodies are used as negative controls for non-specific binding of the antibody to the chromatin fragments. Results are representative of four independent experiments. p50 and BAG-1 are part of the same DNA complex at gene promoters. (D) Re-ChIP analysis of the EGFR and PTGS2 promoters in the HCT116 cell line using the indicated combination of antibodies.
The BAG-1-p50 complex can be detected at the promoter of the EGFR and PTGS2 genes
The previous EMSA allowed the detection of the BAG-1-p50-p50 homodimeric NF-κB complex at a κB consensus sequence in colorectal carcinoma derived cells. However to establish whether the complex was functional in the cells, the potential of BAG-1 to modulate p50-p50 regulated gene expression was investigated. A focused miniarray performed on mRNA obtained from HCT116 cells transfected with BAG-1 siRNA and treated with TNF-α (100ng/ml) or IL-1β (10ng/ml) had identified EGFR as possible BAG-1 regulated genes (data not shown). Furthermore, as the expression of the EGFR gene had previously been described as regulated by p50-p50 homodimeric NF-κB complexes (Thornburg et al., 2003), EGFR was chosen for further study. To investigate whether the BAG-1-p50 complex could be detected at the EGFR promoter, we carried out a chromatin immunoprecipitation [ChIP] assay using antibodies against both BAG-1 and NF-κB1 (figure 4C). Recovered DNA underwent PCR analysis using primers flanking the κB sites in the EGFR promoter (Thornburg et al., 2003, refer to methods section); the PCR products were detected by gel electrophoresis. RNA pol II was precipitated as a positive control for binding to the EGFR promoter (lane 4). DNA complexes were precipitated with the control mouse and rabbit IgG antibodies to account for non-specific interaction of the antibodies (lanes 3 and 5). Importantly, a positive interaction between both BAG-1 and NF-κB1 within the same region of the EGFR promoter was detected in HCT116 cells (figure 4C).
To determine whether BAG-1 and p50 were part of the same DNA complex at the EGFR promoter, a re-ChIP was carried out (figure 4D). In HCT116 cells, DNA complexes were firstly immunoprecipitated with the NF-κB1 antibody and then underwent a second immunoprecipitation using the BAG-1 antibody. DNA was recovered and underwent PCR analysis using the EGFR promoter primers; PCR products were identified using gel electrophoresis (figure 4D). Protein DNA complexes were not detected using the anti-IgGR antibody ruling out non-specific binding of the antibodies (lane 3 and 4). Detection of a PCR product (lane 5) demonstrated that BAG-1 and p50 are indeed part of the same DNA complex at the EGFR promoter. In addition, as results from the focused microarray also identified the COX-2 gene (PTGS2) as a possible BAG-1 regulated gene, it was of interest to show that the BAG-1-p50 complex could also be detected at the PTGS2 promoter (figure 4C and D) providing further evidence for the binding of the BAG-1-p50 homodimeric NF-κB complex to gene regulatory sequences.
BAG-1 regulates EGFR expression in colorectal cancer cells
To investigate the consequence of the interaction of BAG-1 with the p50-p50 homodimers at the gene promoter, we were able to show that EGFR is a transcriptional target of BAG-1 by studying the mRNA abundance of EGFR by Q-RT-PCR in cells with increased or suppressed BAG-1 expression (figure 5Ai). Importantly, although the potentially repressive BAG-1-p50 complex was detected at the EGFR promoter in unstimulated HCT116 cells (figure 4), basal expression of EGFR was detected (figure 5Aii) suggesting the complex may be involved in limiting expression of the protein, rather than completely inhibiting it. Results show that suppression of BAG-1 protein expression using RNAi resulted in an increase in EGFR mRNA abundance (~1.7 fold; figure 5Ai) and protein (figure 5Aii). Conversely, expression of exogenous BAG-1L suppressed EGFR mRNA (figure 5Ai) and protein (figure 5Aii). These data establish BAG-1 as a repressor of EGFR expression in colorectal cancer cells.
Figure 5. BAG-1 regulation of EGFR expression and function is dependent on NF-κB1 expression.
(A) (i) HCT116 cells were transfected with control negative / BAG-1 siRNA or control plasmid / BAG-1L expression plasmid for 48 hours. Q-RT-PCR was carried out to show EGFR mRNA abundance; all mRNA values were normalised to the housekeeping gene, TBP. Data is presented as a percentage change of levels in control transfected cells, represented by the dotted line. Data points are of three independent experiments carried out in triplicate (± standard deviation). Statistical analysis was carried out using a Student’s t-test * = P < 0.05; ** = P < 0.01; *** = P < 0.001. (ii) Western analysis to show regulation of EGFR protein +/− BAG-1 expression. Equal loading was confirmed by α-tubulin. Data is representative of three independent experiments. (B) HT29 cells (positive for COX-2 expression) were transfected with control negative or BAG-1 siRNA for 48 hours. Q-RT-PCR was carried out to show PTGS2 and EGFR mRNA abundance; all mRNA values were normalised to the housekeeping gene, TBP. Data is presented as a percentage change of levels in control transfected cells, represented by the dotted line. Data points are of three independent experiments carried out in triplicate (± standard deviation). Statistical analysis was carried out using a Student’s t-test * = P < 0.05; ** = P < 0.01; *** = P < 0.001. (C) NF-κB1+/+ and NF-κB1−/− MEF cells were transfected with 25nM of control siRNA or mBAG-1 siRNA. Western analysis was carried out to show the regulation of EGFR protein expression following suppression of the BAG-1 protein in NF-κB1+/+ and −/− MEF cells. Equal loading was confirmed by α-tubulin. Results are representative of three independent experiments. (D) HCT116 cells treated with EGF (10ng/ml) for 72 hr after siRNA transfection either in the presence or absence of an EGFR inhibitor CL-387.785 (10μM) added 1h prior to EGF treatment. Activation of the EGFR upon EGF treatment was shown by the phosphorylation status of the receptor, specificity confirmed by the blockade with the EGFR inhibitor. Western blotting was used to confirm suppression of BAG-1 expression by siRNA. Equal loading was confirmed by α-tubulin.
BAG-1 can promote PTGS2 gene expression in colorectal cancer cells
To determine whether the BAG-1:p50 complex is commonly associated with gene repression we also investigated the effect of loss of BAG-1 expression on PTGS2 expression (shown to have BAG-1-p50 complexes at its promoter, figure 4C and D). PTGS2 mRNA abundance was investigated by Q-RT-PCR in HT29 (COX-2 expressing) cells transfected with BAG-1 siRNA (figure 5B). Interestingly, data show that suppression of BAG-1 protein expression resulted in a decrease in PTGS2 mRNA abundance (~0.6 fold), suggesting that BAG-1 actually potentiates expression of the PTGS2 gene. This is in contrast to the increased EGFR expression on suppression of BAG-1 in the HT29 cells (~1.5 fold, figure 5B). Taken together, this study suggests that BAG-1 can either repress or enhance gene expression, dependent on the target gene.
BAG-1 fails to regulate EGFR expression in NF-κB1 knock out mouse embryonic fibroblasts
To establish whether this regulation of EGFR expression by BAG-1 is dependent on the interaction with the p50-p50 homodimeric NF-κB complex, EGFR protein expression was studied in NF-κB1 knock out [NF-κB1−/−] mouse embryonic fibroblasts (MEFs) and compared to the response in age matched wild type [NF-κB1+/+] MEFs (a kind gift from J. Caamamo Birmingham University, UK). NF-κB1+/+ and NF-κB1−/− MEFs were transfected with 25nM of control scrambled or murine BAG-1 [mBAG-1] siRNA and the level of EGFR expression determined by Western analysis (figure 5C). Results show that suppression of BAG-1 expression in NF-κB1+/+ MEFs lead to an increase in EGFR protein expression (a 60% decrease in BAG-1 expression lead to a 66% increase in EGFR protein expression). In contrast, this response was attenuated in the NF-κB1−/− MEF cells resulting in no detectable up-regulation of the EGFR protein expression (a 57% suppression of BAG-1 expression caused a less than 1% increase in the EGFR protein expression). These results further demonstrate that suppression of BAG-1 expression increases EGFR protein levels and demonstrate that the regulation of EGFR expression by BAG-1 is mediated via a p50 dependent mechanism.
The BAG-1-p50 complex suppresses epidermal growth factor signalling in colorectal cancer cells
To determine whether the regulation of EGFR expression by BAG-1 is sufficient to modulate signalling through the EGF pathway, HCT116 cells were transfected with 50nM of control scrambled or BAG-1 siRNA and treated with EGF (10ng/ml), results are summarised in figure 5D. Western analysis using a phosphor-specific antibody was used to demonstrate activation of the EGFR, which could be blocked by addition of the EGFR inhibitor [10μM CL-387.785] confirming specificity of the assay (lanes 3,5,8 and 10). Activation of the EGFR was detected on addition of 10ng/ml EGF to the cultures 72h after siRNA transfection (5 min (lane 2) and 10 min (lane 4) after addition of the EGF ligand) which was blocked by addition of the EGFR inhibitor (lanes 3 and 5). Strikingly, when BAG-1 expression is suppressed by siRNA (lanes 6-10) there is increase phosphorylation of the EGFR on addition of EGF (5 min (lane 7) and 10 min (lane 9) after addition of 10ng/ml EGF). These data reveal an increase in the phosphorylation, and hence activation, of the EGFR in the presence of the ligand when BAG-1 protein expression is suppressed by siRNA. This is reflected in an increase in phospho-ERK (a downstream target of the EGF signalling pathway) when BAG-1 expression is suppressed (data not shown), confirming that the regulation of EGFR expression by BAG-1 is sufficient to significantly attenuate signalling through the pathway.
Taken together this study has identified for the first time that BAG-1 interacts with the p50-p50 homodimeric NF-κB complex, and that these complexes can be detected on the promoters of genes known to be important in colorectal carcinogenesis, including EGFR and PTGS2. Of note, although the role of both EGF and COX-2 signalling are well established in colorectal carcinogenesis, we propose that it is the combined effect of the BAG-1-p50 complex on the expression of a number of genes that is important for promoting colorectal tumour survival. This finding has implications not only for normal tissue homeostasis but suggest that BAG-1-p50 NF-κB complexes could play a fundamental role in the de-regulation of key signalling pathways in colorectal carcinogenesis.
Discussion
This investigation has established that nuclear localisation of BAG-1 is important in the regulation of NF-κB activity, identifying BAG-1 as a novel binding partner of the p50-p50 homodimeric NF-κB complex. The function of BAG-1 as a regulator of gene expression through interaction with p50-p50 NF-κB complexes has potentially significant implications for colorectal carcinogenesis. Although not as extensively studied as some of the other NF-κB dimers, p50-p50 homodimers are important signalling complexes generally recognised as repressors of transcription due to the lack of a transactivation domain (reviewed in Piera and Oakley, 2008). There have been several reports of p50-p50 homodimers repressing key NF-κB associated genes, including TNF-α and IL-8 (Baer et al., 1998; Tong et al., 2004). However, there are also reports of p50-p50 homodimers acting as transcriptional activators. Studies have shown p50-p50 homodimers to increase the expression of the NF-κB target genes such as IL-6 in melanoma cells and IL-10 in macrophages (Cao et al., 2006; Karst et al., 2009). It appears that the ability of the p50-p50 homodimeric NF-κB complex to act as a transcriptional activator or repressor is dependent on the co-factors that interact with the NF-κB complex at the DNA (reviewed in Glass and Rosenfeld, 2000). For example, a study by Zhong et al used a co-immunoprecipitation assay to demonstrate that p50 interacted with HDAC-1 in unstimulated murine pre-B cells, resulting in transcriptional silencing of the IL-8 and TNF-α genes (Zhong et al., 2002). On the other hand, the p50-p50 homodimers have been shown to become transcriptional activators when interacting with the nuclear BCL-3 protein (Bours et al., 1993; Fujita et al., 1993). As a co-chaperone, it is of interest to hypothesise that either by stabilising the p50-DNA complexes or influencing the recruitment of other co-factors, BAG-1 may be able to either repress and/or trans-activate gene expression.
In the colon, consistent with previous reports (Inan et al., 2000), we found NF-κB1 to be expressed throughout the normal colonic crypt. Interestingly, BAG-1 has been described as predominantly nuclear at the bottom of the crypt and associated with colorectal epithelial cell survival (Clemo et al., 2008; shown in figure 3; Barnes et al., 2005). Taken together with evidence from the literature, we would propose that the nuclear localisation of BAG-1 in association with p50-p50 homodimeric NF-κB complexes is able to regulate gene expression potentially involved in preserving the proliferating compartment of the colonic crypt. Consistent with this hypothesis, the increased expression of BAG-1 in colonic tumour cells would promote the pro-survival function of the p50-p50 homodimeric NF-κB complexes, identified as the predominant NF-κB complexes in stimulated colorectal tumour cells (figure 3B) and as such high BAG-1 expression would contribute to cell survival and hence be associated with poor therapeutic response in colorectal cancer patients (as reported by Kikuchi et al., 2003). As NF-κB signalling, which is up-regulated in response to cytotoxic treatments, is accountable for many tumours being resistant to current cancer therapies (reviewed in Nakanishi and Toi, 2005), the emerging role of BAG-1 in the regulation of NF-κB function has potentially significant therapeutic implications. Although NF-κB is a crucial player in many cellular processes, targeting the BAG-1-p50 complex may provide a mechanism for inhibiting the tumour-promoting/survival function of NF-κB whilst still retaining many of the normal physiological processes. Interestingly, a previous study reported that another member of the BAG family (BAG-3) could promote DNA binding of NF-κB subunits mediated via the conserved BAG domain (Rosati et al., 2007). Furthermore, a recent study in HeLa cells reported suppression of BAG-1 to attenuate NF-κB activation, although in this report they found BAG-1 to promote NF-κB activity through down-regulation of IκBα leading to the nuclear accumulation of NF-κB (Maier et al., 2010). Therefore, it is of interest to speculate that the BAG proteins may have a multi-factorial mode of action, able to modulate the function of NF-κB at more than one stage in the signalling pathway, whether dependent on cell type or stimulus remains to be determined. However, because of the potential role of p50-p50 homodimeric complexes in colorectal tissue homeostasis (Inan et al. 2000), we would propose that the recruitment of BAG-1 to the p50-p50 homodimeric NF-κB complex may have a particularly important selective advantage for the survival of colorectal tumour cells.
Through interacting with the EGFR promoter, the BAG-1-p50 complex was shown to repress signalling through the EGF pathway. However, EGF signalling has been reported to promote cell survival and proliferation through activating the RAS-MAPK and PI3K-AKT pathways (reviewed in Yarden, 2001). These reports suggest that high levels of EGF signalling would be beneficial for promoting colorectal carcinogenesis, and hence an apparent paradox exists as BAG-1 is up-regulated in colorectal cancer tissue and can inhibit EGFR gene and protein expression. However, there are conditions where, by driving proliferation, EGFR expression could be detrimental to cancer cells (for example when attempting to adapt and survive in response to a changing microenvironment within the developing tumour, Kaidi et al., 2007). However, it is equally important to note that in the colon, activating mutations of K-ras commonly occur in approximately 50% of colorectal cancers (Bos et al., 1987), and as a consequence tumour cells have constitutive MAPK activation and may not require EGF signalling for cell survival (reviewed in Jiang et al., 2009). Furthermore, there are situations where high EGFR expression may be unfavourable to the cells. For example, EGFR has been shown to sequester the insulin receptor substrate-1 (IRS-1), which is an essential component of the insulin-like growth factor type 1 receptor (IGF-1R), implying that high EGFR expression could inhibit the IGF-II survival signalling pathway (Knowlden et al., 2008). Inhibition of EGFR expression by BAG-1 could therefore actually promote cell survival through the IGF-II pathway. We suggest that many of the stress induced signalling pathways stimulated by the microenvironment in a developing tumour, such hypoxia or energy deprivation, could increase the formation of BAG-1-p50 complexes resulting in suppression of genes involved in proliferation (such as EGFR) and promoting genes involved in survival and adaptation (such as the COX2/prostaglandin pathway).
In conclusion, this paper identifies a novel role for BAG-1 as a co-regulator of gene expression through interaction with the p50-p50 NF-κB complexes. Data presented have led us to propose that BAG-1 can act as a selective regulator of p50-p50 NF-κB responsive genes in colorectal tumour cells, important for promoting cell survival in the context of the fluctuating tumour microenvironment. As BAG-1 expression is increased in the developing adenoma through to metastatic lesions, understanding the function of the BAG-1 p50-NF-κB complexes may aid in identifying novel strategies for both the prevention and treatment of colorectal cancer.
Acknowledgements
This work was funded by a Cancer Research UK programme grant, the Citrina Foundation, The Wellcome Trust and by the John James Bristol Foundation.
We thank the Medical Research Council for providing an Infrastructure Award to establish the School of Medical Sciences Cell Imaging Facility at the University of Bristol.
Footnotes
Conflict of interest: The authors declare no conflict of interest.
References
- Arhel NJ, Packham G, Townsend PA, Collard TJ, H-Zadeh AM, Sharp A, et al. The retinoblastoma protein interacts with BAG-1 in human colonic adenoma and carcinoma derived cell lines. Int J Cancer. 2003;106:364–71. doi: 10.1002/ijc.11257. [DOI] [PubMed] [Google Scholar]
- Arlt A, Vorndamm J, Muerkoster S, Yu H, Schmidt WE, Folsch UR, et al. Autocrine production of interleukin 1beta confers constitutive nuclear factor kappaB activity and chemoresistance in pancreatic carcinoma cell lines. Cancer Res. 2002;62:910–916. [PubMed] [Google Scholar]
- Baer M, Dillner A, Schwartz RC, Sedon C, Nedospasov S, Johnson PF. Tumor necrosis factor alpha transcription in macrophages is attenuated by an autocrine factor that preferentially induces NF-kappaB p50. Mol Cell Biol. 1998;18:5678–5689. doi: 10.1128/mcb.18.10.5678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baeuerle PA, Baltimore D. I kappa B: a specific inhibitor of the NF-kappa B transcription factor. Science. 1988;242:540–546. doi: 10.1126/science.3140380. [DOI] [PubMed] [Google Scholar]
- Barnes JD, Arhel NJ, Lee SS, Sharp A, Al-Okail M, Packham G, et al. Nuclear BAG-1 expression inhibits apoptosis in colorectal adenoma-derived epithelial cells. Apoptosis. 2005;10:301–11. doi: 10.1007/s10495-005-0804-8. [DOI] [PubMed] [Google Scholar]
- Beinke S, Ley SC. Functions of NF-kappaB1 and NF-kappaB2 in immune cell biology. Biochem J. 2004;382:393–409. doi: 10.1042/BJ20040544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bos JL, Fearon ER, Hamilton SR, Verlaan-de Vries M, van Boom JH, van der Eb AJ, et al. Prevalence of ras gene mutations in human colorectal cancers. Nature. 1987;327:293–297. doi: 10.1038/327293a0. [DOI] [PubMed] [Google Scholar]
- Bours V, Franzoso G, Azarenko V, Park S, Kanno T, Brown K, et al. The oncoprotein Bcl-3 directly transactivates through kappa B motifs via association with DNA-binding p50B homodimers. Cell. 1993;72:729–739. doi: 10.1016/0092-8674(93)90401-b. [DOI] [PubMed] [Google Scholar]
- Brimmell M, Burns JS, Munson P, McDonald L, O’Hare MJ, Lakhani SR, et al. High level expression of differentially localized BAG-1 isoforms in some oestrogen receptor-positive human breast cancers. Br J Cancer. 1999;81:1042–1051. doi: 10.1038/sj.bjc.6690805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cancer Research UK . Bowel Cancer Factsheet. Cancer Research UK; 2007. Website. [Google Scholar]
- Cao S, Zhang X, Edwards JP, Mosser DM. NF-kappaB1 (p50) homodimers differentially regulate pro- and anti-inflammatory cytokines in macrophages. J Biol Chem. 2006;281:26041–26050. doi: 10.1074/jbc.M602222200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen LF, Greene WC. Shaping the nuclear action of NF-kappaB. Nat Rev Mol Cell Biol. 2004;5:392–401. doi: 10.1038/nrm1368. [DOI] [PubMed] [Google Scholar]
- Clemo NK, Collard TJ, Southern SL, Edwards KD, Moorghen M, Packham G, et al. BAG-1 is up-regulated in colorectal tumour progression and promotes colorectal tumour cell survival through increased NF-kappaB activity. Carcinogenesis. 2008;29:849–857. doi: 10.1093/carcin/bgn004. [DOI] [PubMed] [Google Scholar]
- Cutress RI, Townsend PA, Sharp A, Maison A, Wood L, Lee R, et al. The nuclear BAG-1 isoform, BAG-1L, enhances oestrogen-dependent transcription. Oncogene. 2003;22:4973–4982. doi: 10.1038/sj.onc.1206688. [DOI] [PubMed] [Google Scholar]
- Deng WG, Zhu Y, Wu KK. Up-regulation of p300 binding and p50 acetylation in tumor necrosis factor-alpha-induced cyclooxygenase-2 promoter activation. J Biol Chem. 2003;278:4770–4777. doi: 10.1074/jbc.M209286200. [DOI] [PubMed] [Google Scholar]
- Duckett CS, Perkins ND, Kowalik TF, Schmid RM, Huang ES, Baldwin AS, Jr, et al. Dimerization of NF-κB2 with RelA(p65) regulates DNA binding, transcriptional activation, and inhibition by an I kappa B-alpha (MAD-3) Mol Cell Biol. 1993;13:1315–1322. doi: 10.1128/mcb.13.3.1315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Franzoso G, Bours V, Park S, Tomita-Yamaguchi M, Kelly K, Siebenlist U. The candidate oncoprotein Bcl-3 is an antagonist of p50/NF-kappa B-mediated inhibition. Nature. 1992;359:339–342. doi: 10.1038/359339a0. [DOI] [PubMed] [Google Scholar]
- Froesch BA, Takayama S, Reed JC. BAG-1L protein enhances androgen receptor function. J Biol Chem. 1998;273:11660–11666. doi: 10.1074/jbc.273.19.11660. [DOI] [PubMed] [Google Scholar]
- Fujita T, Nolan GP, Liou HC, Scott ML, Baltimore D. The candidate proto-oncogene bcl-3 encodes a transcriptional coactivator that activates through NF-kappa B p50 homodimers. Genes Dev. 1993;7:1354–1363. doi: 10.1101/gad.7.7b.1354. [DOI] [PubMed] [Google Scholar]
- Gehring U. Multiple, but Concerted Cellular Activities of the Human Protein Hap46/BAG-1M and Isoforms. Int J Mol Sci. 2009;10:906–928. doi: 10.3390/ijms10030906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghosh S, May MJ, Kopp EB. NF-kappa B and Rel proteins: evolutionarily conserved mediators of immune responses. Annu Rev Immunol. 1998;16:225–260. doi: 10.1146/annurev.immunol.16.1.225. [DOI] [PubMed] [Google Scholar]
- Gilmore TD, Temin HM. v-rel oncoproteins in the nucleus and in the cytoplasm transform chicken spleen cells. J Virol. 1988;62:703–714. doi: 10.1128/jvi.62.3.703-714.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glass CK, Rosenfeld MG. The coregulator exchange in transcriptional functions of nuclear receptors. Genes Dev. 2000;14:121–141. [PubMed] [Google Scholar]
- Hinitt CAM, Wood J, Lee SS, Williams AC, Howarth JL, Glover CP, Uney JB, Hague A. BAG-1 enhances cell-cell adhesion, reduces proliferation and induces chaperone-independent suppression of hepatocyte growth factor-induced epidermal keratinocyte migration. Experimental Cell Research. 2010;316:2042–60. doi: 10.1016/j.yexcr.2010.04.016. [DOI] [PubMed] [Google Scholar]
- Hou S, Guan H, Ricciardi RP. Phosphorylation of serine 337 of NF-kappaB p50 is critical for DNA binding. J Biol Chem. 2003;278:45994–45998. doi: 10.1074/jbc.M307971200. [DOI] [PubMed] [Google Scholar]
- Inan MS, Tolmacheva V, Wang QS, Rosenberg DW, Giardina C. Transcription factor NF-kappaB participates in regulation of epithelial cell turnover in the colon. Am J Physiol Gastrointest Liver Physiol. 2000;279:G1282–1291. doi: 10.1152/ajpgi.2000.279.6.G1282. [DOI] [PubMed] [Google Scholar]
- Jiang Y, Kimchi ET, Staveley-O’Carroll KF, Cheng H, Ajani JA. Assessment of K-ras mutation: a step toward personalized medicine for patients with colorectal cancer. Cancer. 2009;115:3609–3617. doi: 10.1002/cncr.24434. [DOI] [PubMed] [Google Scholar]
- Kaidi A, Williams AC, Paraskeva C. Interaction between beta-catenin and HIF-1 promotes cellular adaptation to hypoxia. Nat Cell Biol. 2007;9:210–7. doi: 10.1038/ncb1534. [DOI] [PubMed] [Google Scholar]
- Kamens J, Richardson P, Mosialos G, Brent R, Gilmore T. Oncogenic transformation by vrel requires an amino-terminal activation domain. Mol Cell Biol. 1990;10:2840–2847. doi: 10.1128/mcb.10.6.2840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karst AM, Gao K, Nelson CC, Li G. Nuclear factor kappa B subunit p50 promotes melanoma angiogenesis by upregulating interleukin-6 expression. Int J Cancer. 2009;124:494–501. doi: 10.1002/ijc.23973. [DOI] [PubMed] [Google Scholar]
- Kieran M, Blank V, Logeat F, Vandekerckhove J, Lottspeich F, Le Bail O, et al. The DNA binding subunit of NF-kappa B is identical to factor KBF1 and homologous to the rel oncogene product. Cell. 1990;62:1007–1018. doi: 10.1016/0092-8674(90)90275-j. [DOI] [PubMed] [Google Scholar]
- Kikuchi R, Noguchi T, Takeno S, Funada Y, Moriyama H, Uchida Y. Nuclear BAG-1 expression reflects malignant potential in colorectal carcinomas. Br J Cancer. 2002;87:1136–1139. doi: 10.1038/sj.bjc.6600579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knowlden JM, Jones HE, Barrow D, Gee JM, Nicholson RI, Hutcheson IR. Insulin receptor substrate-1 involvement in epidermal growth factor receptor and insulin-like growth factor receptor signalling: implication for Gefitinib (‘Iressa’) response and resistance. Breast Cancer Res Treat. 2008;111:79–91. doi: 10.1007/s10549-007-9763-9. [DOI] [PubMed] [Google Scholar]
- Lin Y, Bai L, Chen W, Xu S. The NF-kappaB activation pathways, emerging molecular targets for cancer prevention and therapy. Expert Opin Ther Targets. 2010;14:45–55. doi: 10.1517/14728220903431069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu R, Takayama S, Zheng Y, Froesch B, Chen GQ, Zhang X, et al. Interaction of BAG-1 with retinoic acid receptor and its inhibition of retinoic acid-induced apoptosis in cancer cells. J Biol Chem. 1998;273:16985–16992. doi: 10.1074/jbc.273.27.16985. [DOI] [PubMed] [Google Scholar]
- Maier JV, Volz Y, Berger C, Schneider S, Cato AC. Depletion of the cellular levels of BAG-1 proteins attenuates phorbol ester-induced downregulation of IκBα and nuclear accumulation of NF-κB. Biochem Biophys Res Commun. 2010;401:406–411. doi: 10.1016/j.bbrc.2010.09.067. [DOI] [PubMed] [Google Scholar]
- Nakanishi C, Toi M. Nuclear factor-kappaB inhibitors as sensitizers to anticancer drugs. Nature Reviews Cancer. 2005;5:297–309. doi: 10.1038/nrc1588. [DOI] [PubMed] [Google Scholar]
- Niyaz Y, Zeiner M, Gehring U. Transcriptional activation by the human Hsp70-associating protein Hap50. J Cell Sci. 2001;114:1839–1845. doi: 10.1242/jcs.114.10.1839. [DOI] [PubMed] [Google Scholar]
- Packham G, Brimmell M, Cleveland JL. Mammalian cells express two differently localized BAG-1 isoforms generated by alternative translation initiation. Biochem J. 1997;328:807–813. doi: 10.1042/bj3280807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pahl HL. Activators and target genes of Rel/NF-kappaB transcription factors. Oncogene. 1999;18:6853–6866. doi: 10.1038/sj.onc.1203239. [DOI] [PubMed] [Google Scholar]
- Pereira SG, Oakley F. Nuclear factor-kappaB1: regulation and function. Int J Biochem Cell Biol. 2008;40:1425–1430. doi: 10.1016/j.biocel.2007.05.004. [DOI] [PubMed] [Google Scholar]
- Perkins ND. Integrating cell-signalling pathways with NF-kappaB and IKK function. Nat Rev Mol Cell Biol. 2007;8:49–62. doi: 10.1038/nrm2083. [DOI] [PubMed] [Google Scholar]
- Rorke S, Murphy S, Khalifa M, Chernenko G, Tang SC. Prognostic significance of BAG-1 expression in nonsmall cell lung cancer. Int J Cancer. 2001;95:317–322. doi: 10.1002/1097-0215(20010920)95:5<317::aid-ijc1055>3.0.co;2-j. [DOI] [PubMed] [Google Scholar]
- Rosati A, Leone A, Del Valle L, Amini S, Khalili K, Turco MC. Evidence for BAG3 modulation of HIV-1 gene transcription. J Cell Physiol. 2007;210:676–683. doi: 10.1002/jcp.20865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ryseck RP, Bull P, Takamiya M, Bours V, Siebenlist U, Dobrzanski P, et al. RelB, a new Rel family transcription activator that can interact with p50-NF-kappa B. Mol Cell Biol. 1992;12:674–684. doi: 10.1128/mcb.12.2.674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmitz ML, Baeuerle PA. The p65 subunit is responsible for the strong transcription activating potential of NF-kappa B. Embo J. 1991;10:3805–3817. doi: 10.1002/j.1460-2075.1991.tb04950.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sen R, Baltimore D. Multiple nuclear factors interact with the immunoglobulin enhancer sequences. Cell. 1986;46:705–716. [PubMed] [Google Scholar]
- Sharp A, Crabb SJ, Townsend PA, Cutress RI, Brimmell M, Wang XH, et al. BAG-1 in carcinogenesis. Expert Rev Mol Med. 2004;6:1–15. doi: 10.1017/S1462399404007537. [DOI] [PubMed] [Google Scholar]
- Shindoh M, Adachi M, Higashino F, Yasuda M, Hida K, Nishioka T, et al. BAG-1 expression correlates highly with the malignant potential in early lesions (T1 and T2) of oral squamous cell carcinoma. Oral Oncol. 2000;36:444–449. doi: 10.1016/s1368-8375(00)00025-7. [DOI] [PubMed] [Google Scholar]
- Smartt HJ, Elder DJ, Hicks DJ, Williams NA, Paraskeva C. Increased NF-kappaB DNA binding but not transcriptional activity during apoptosis induced by the COX-2-selective inhibitor NS-398 in colorectal carcinoma cells. Br J Cancer. 2003;89:1358–1365. doi: 10.1038/sj.bjc.6601266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song J, Takeda M, Morimoto RI. BAG-1-Hsp70 mediates a physiological stress signalling pathway that regulates Raf-1/ERK and cell growth. Nat Cell Biol. 2001;3:276–282. doi: 10.1038/35060068. [DOI] [PubMed] [Google Scholar]
- Takayama S, Reed JC. Molecular chaperone targeting and regulation by BAG family proteins. Nat Cell Biol. 2001;3:E237–241. doi: 10.1038/ncb1001-e237. [DOI] [PubMed] [Google Scholar]
- Takayama S, Krajewski S, Krajewska M, Kitada S, Zapata JM, Kochel K, et al. Expression and location of Hsp70/Hsc-binding anti-apoptotic protein BAG-1 and its variants in normal tissues and tumor cell lines. Cancer Res. 1998;58:3116–3131. [PubMed] [Google Scholar]
- Takayama S, Sato T, Krajewski S, Kochel K, Irie S, Millan JA, et al. Cloning and functional analysis of BAG-1: a novel Bcl-2-binding protein with anti-cell death activity. Cell. 1995;80:279–284. doi: 10.1016/0092-8674(95)90410-7. [DOI] [PubMed] [Google Scholar]
- Thornburg NJ, Pathmanathan R, Raab-Traub N. Activation of nuclear factor-kappaB p50 homodimer/Bcl-3 complexes in nasopharyngeal carcinoma. Cancer Res. 2003;63:8293–8301. [PubMed] [Google Scholar]
- Tong X, Yin L, Washington R, Rosenberg DW, Giardina C. The p50-p50 NF-kappaB complex as a stimulus-specific repressor of gene activation. Mol Cell Biochem. 2004;265:171–183. doi: 10.1023/b:mcbi.0000044394.66951.4d. [DOI] [PubMed] [Google Scholar]
- Townsend PA, Stephanou A, Packham G, Latchman DS. BAG-1: a multi-functional pro-survival molecule. Int J Biochem Cell Biol. 2005;37:251–259. doi: 10.1016/j.biocel.2004.03.016. [DOI] [PubMed] [Google Scholar]
- Weldon CB, Burow ME, Rolfe KW, Clayton JL, Jaffe BM, Beckman BS. NF-kappa B-mediated chemoresistance in breast cancer cells. Surgery. 2001;130:143–150. doi: 10.1067/msy.2001.115512. [DOI] [PubMed] [Google Scholar]
- Williams AC, Browne SJ, Yeudal WA, Paterson IC, Marshall CJ, Lane DP, Paraskeva C. Molecular events including p53 and k-ras alterations in the in vitro progression of a human colorectal adenoma cell line to an adenocarcinoma. Oncogene. 1993;8:3063–3072. [PubMed] [Google Scholar]
- Wood J, Lee SS, Hague A. BAG-1 proteins in oral squamous cell carcinoma. Oral Oncol. 2009;45:94–102. doi: 10.1016/j.oraloncology.2008.07.013. [DOI] [PubMed] [Google Scholar]
- Xiao G, Rabson AB, Young W, Qing G, Qu Z. Alternative pathways of NF-kappaB activation: a double-edged sword in health and disease. Cytokine Growth Factor Rev. 2006;17:281–293. doi: 10.1016/j.cytogfr.2006.04.005. [DOI] [PubMed] [Google Scholar]
- Yang X, Chernenko G, Hao Y, Ding Z, Pater MM, Pater A, et al. Human BAG-1/RAP46 protein is generated as four isoforms by alternative translation initiation and overexpressed in cancer cells. Oncogene. 1998;17:981–989. doi: 10.1038/sj.onc.1202032. [DOI] [PubMed] [Google Scholar]
- Yarden Y. The EGFR family and its ligands in human cancer. signalling mechanisms and therapeutic opportunities. Eur J Cancer. 2001;37(Suppl 4):S3–8. doi: 10.1016/s0959-8049(01)00230-1. [DOI] [PubMed] [Google Scholar]
- Zeiner M, Gehring U. A protein that interacts with members of the nuclear hormone receptor family: identification and cDNA cloning. Proc Natl Acad Sci U S A. 1995;92:11465–11469. doi: 10.1073/pnas.92.25.11465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeiner M, Niyaz Y, Gehring U. The hsp70-associating protein Hap46 binds to DNA and stimulates transcription. Proc Natl Acad Sci U S A. 1999;96:10194–10199. doi: 10.1073/pnas.96.18.10194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhong H, May MJ, Jimi E, Ghosh S. The phosphorylation status of nuclear NF-kappa B determines its association with CBP/p300 or HDAC-1. Mol Cell. 2002;9:625–636. doi: 10.1016/s1097-2765(02)00477-x. [DOI] [PubMed] [Google Scholar]