Abstract
Cerebellar ataxia is a prominent clinical symptom in patients with mitochondrial DNA (mtDNA) disease. Often this is progressive with onset in young adulthood. Here, we performed a detailed neuropathological investigation of the olivary-cerebellum in 14 genetically and clinically well-defined patients with mtDNA disease. Quantitative neuropathological investigation showed varying levels of loss of Purkinje cells, and neurons of the dentate nucleus and inferior olivary nuclei. Typically, focal Purkinje cell loss was present in patients with the m.3243A>G mutation due to the presence of microinfarcts with relative preservation of neuronal cell populations in olivary and dentate nuclei. In contrast, patients with the m.8344A>G mutation or recessive POLG mutations showed extensive and global neuronal cell loss in all 3 olivary-cerebellum areas examined. Molecular analysis of mutated mtDNA heteroplasmy levels revealed that neuronal cell loss occurred independently of the level of mutated mtDNA present within surviving neurons. High levels of neuronal respiratory chain deficiency, particularly of complex I, were detected in surviving cells; levels of deficiency were greater in regions with extensive cell loss. We found a relationship between respiratory deficiency and neuronal cell density, indicating that neuronal cell death correlates with respiratory deficiency. These findings highlight the vulnerability of the olivary-cerebellum to mtDNA defects.
Keywords: Cerebellar ataxia, Mitochondrial DNA, Mitochondrial disease, Neurodegeneration, Olivo-Cerebellum
INTRODUCTION
Neurodegeneration is an important cause of neurological disability in patients with mitochondrial DNA (mtDNA) disease (1). These diseases arise either from primary defects of mtDNA, such as point mutations or large-scale mtDNA deletions, or from secondary defects such as mtDNA depletion or deletions due to defects in nuclear genes involved in mtDNA maintenance (2). The CNS has a high requirement for energy and is, therefore, dependent on mitochondria for adenosine triphosphate (ATP) generation (3-5). ATP generation occurs via oxidative phosphorylation through a series of 5 respiratory chain complexes embedded in the inner mitochondrial membrane. These complexes are partly encoded by the mitochondrial genome. It is not surprising, therefore, that in patients with mtDNA disease, neurons are especially vulnerable to degeneration.
Although neurological deficits in mtDNA disease may be manifold, cerebellar ataxia is a frequently reported symptom in patients with mtDNA defects (6-12). A recent UK-based MRC-funded cohort study estimates that the frequency of ataxia is high, with 225 out of 345 patients with mitochondrial disease showing symptoms consistent with ataxia (Nesbitt, McFarland et al unpublished observation). Cerebellar ataxia is often progressive in these patients and is a major cause of disability. Because cerebellar ataxia is commonly described in patients with mtDNA disease, there have been a number of attempts to characterize neurodegeneration using neuroimaging techniques. These are often useful as a non-invasive assessment of CNS degeneration that assists in the diagnosis of mtDNA disorders and enables documentation of progression. Typically, reduced cerebellar volume is reported and may be attributed to atrophy (13, 14). Indeed, a large retrospective study involving 113 patients showed that cerebellar atrophy might even be the primary neuroradiological finding in patients with mtDNA defects (15). However, imaging studies give only a limited insight into an ongoing neurodegenerative process for which understanding of the precise mechanisms remains undetermined.
Neuropathological studies are limited in mtDNA disease due to the challenges associated with obtaining postmortem material and published studies tend to focus on single cases. Despite this, there is evidence of cerebellar involvement in the pathogenesis of mtDNA disease. Typically, studies have revealed cerebellar atrophy, Purkinje cell loss, abnormal dendritic arborization and the presence of ischemic-like lesions (16-18). However, there have been no attempts to correlate the severity of clinical manifestations with neuropathological changes associated with specific molecular defects.
Despite the known involvement of the cerebellar dysfunction in the genesis of ataxia in mtDNA disease, very little is understood about the molecular mechanisms underpinning the neurodegeneration. This study aims to increase our understanding of the relationship between clinical symptoms of ataxia, neuropathological changes and the molecular attributes of neurons within the olivo-cerebellum pathway in a large group of clinically and genetically well-defined cases with mtDNA defects. This work will provide an important platform from which we can begin to explore the mechanisms of neurodegeneration due to mitochondrial dysfunction and could prove important for the development of novel therapeutic strategies for the treatment of ataxia. It could also have further implications for the understanding the role of mitochondrial dysfunction in other neurodegenerative disorders such as Alzheimer disease and Parkinson disease.
MATERIALS AND METHODS
Clinical Assessment of Patients
Patients with clinically and genetically defined mitochondrial DNA disease were selected from the Newcastle Brain Tissue Resource (Table 1). Adult patients with a known molecular genetic defect were selected. The participants had received repeated clinical assessments annually until death and had agreed to donate their brain tissue for research purposes. Patients were rated using the Newcastle Mitochondrial Disease Adult Scale (NMDAS) to monitor disease progression (19). The rating scales given for cerebellar ataxia at final clinical assessment are included in Table 1, thereby confirming that all the included patients had ataxia.
Table 1.
Clinical Summary of Patients
| Pt 1 | Pt 2 | Pt 3 | Pt 4 | Pt 5 | Pt 6 | Pt 7 | Pt 8 | Pt 9 | Pt 10 | Pt 11 | Pt 12 | Pt 13 | Pt 14 | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Age (y) | 60 | 57 | 45 | 36 | 42 | 20 | 30 | 42 | 55 | 34 | 40 | 24 | 59 | 50 |
| Gender | F | F | M | F | F | F | M | F | M | F | F | F | M | M |
| Genotype | m.3243A>G | m.3243A>G | m.3243A>G | m.3243A>G | m.3243A>G | m.3243A>G | m.3243A>G | m.8344A>G | m.14709T>C | m.13094T>C | Single, large- scale mtDNA deletion |
POLG (p.Ala467Thr, p.Trp748Ser) and multiple mtDNA deletions |
POLG (p.Gly848Ser, p.Ser1104Cys) and multiple mtDNA deletions |
POLG (p.Ala467Thr, p.X1240Cys) and multiple mtDNA deletions |
| Disease duration (y) | 33 | 32 | 8 | 15 | 22 | 10 | 11 | 37 | 21 | 2 | 37 | 4 | 37 | 35 |
| Degree of cell loss | ||||||||||||||
| Inferior olivary neurons |
50% | 42% | ND | 42% | 0% | ND | ND | 85% | 44% | ND | 52% | 91% | 69% | 81% |
| Purkinje cells | 70% | 59% | 59% | 59% | 0% | ND | ND | 67% | 30% | 40% | 60% | 81% | 65% | 90% |
| Dentate nucleus neurons |
0% | 56% | 26% | 56% | 74% | ND | ND | 75% | 74% | 43% | 23% | 96% | 45% | 53% |
| Clinical Features | ||||||||||||||
| Ataxia (NMDAS rating) |
+ (3/5) | + (1/5) | + (2/5) | + (3/5) | + (2/5) | + (5/5) | + (1/5) | + (5/5) | + (3/5) | + (1/5) | + (4/5) | + (4/5) | + (3/5) | + (4/5) |
| Stroke | + | + | + | + | + | + | ||||||||
| Epilepsy | + | + | + | + | + | + | + | |||||||
| Dementia | + | + | + | + | + | + | + | + | ||||||
| Encephalopathy | + | + | + | + | + | + | + | |||||||
| Migraine | + | + | + | |||||||||||
| Peripheral neuropathy |
+ | + | + | + | + | |||||||||
| Myoclonus | + | + | + | + | + | |||||||||
| Areflexia | + | + | + | + | ||||||||||
| Myopathy | + | + | + | + | + | + | + | + | + | + | ||||
| Myalgia | + | |||||||||||||
| Fatigue | + | + | + | + | + | + | + | + | + | |||||
| Depression | + | + | + | + | + | + | + | + | + | |||||
| Optic atrophy | + | + | ||||||||||||
| Pigmented retinopathy |
+ | + | + | |||||||||||
| Cataracts | + | |||||||||||||
| Ophthalmoplegia | + | + | + | + | + | |||||||||
| Ptosis | + | + | + | + | ||||||||||
| Diplopia | + | + | + | |||||||||||
| Deafness | + | + | + | + | + | + | + | |||||||
| Dysarthria | + | + | + | + | + | + | ||||||||
| Dysphonia | + | + | ||||||||||||
| Dysphagia | + | + | + | + | + | + | ||||||||
| Constipation | + | + | + | + | + | |||||||||
| Irritable bowel | + | + | + | + | + | + | ||||||||
| Vomiting | + | + | + | + | ||||||||||
| Low BMI | + | + | + | + | + | |||||||||
| Cardiac failure | + | + | + | + | ||||||||||
| Heart block | + | + | ||||||||||||
| Arrhythmia | + | + | + | + | ||||||||||
| Cardiomyopathy | + | + | + | + | + | |||||||||
| Resp. failure | + | + | + | + | ||||||||||
| Recurrent asp. pneumonias |
+ | |||||||||||||
| Diabetes | + | + | + | + | + | |||||||||
| Thyroid disease | + | + | ||||||||||||
| Infertility/Miscarriage | + | + | ||||||||||||
| Other | Early menopause |
IHD | PCOS | Renal TP | Renal failure | Paresthesia,PF O |
Dysmenorrhea | IHD | Osteoporosis | Parkinsonism | ||||
| MRI/CT: focal lesions &/or atrophy |
Multiple areas of low attenuation in L frontal lobe; ischemic changes in post cerebellum |
N/A | L occipital infarct |
Basal ganglia calcification; mild generalised atrophy; bi- occipital ischemic changes |
Normal for age |
N/A | Florid symmetrical calcification of basal ganglia and thalami; minor degree of cerebral atrophy/ moderate cerebellar atrophy for age |
N/A | N/A | Infarct in R and L subthalamic, and R medial occipital lobe; further signal change noted in R superior colliculus |
Periventricular hyperintensities and deep white matter increase in signal in posterior frontal and parietal lobes |
Abnormal areas of grey and white matter suggestive of ischemic change |
Multiple tiny foci of signal abnormality in cerebral white matter with some hazy confluence in peritrigonal white matter |
High signal intensity involving hippocampus and para- hippocampal gyrus on the R; patchy high signal intensity within pons; signal change within cerebellar hemispheres & middle cerebral peduncle |
| Clinical diagnosis | MELAS | MELAS | MELAS | MELAS | MELAS | MELAS | MELAS | MERRF | Ataxia, retinopathy |
MELAS/ Leigh disease |
KSS | arPEO | arPEO | arPEO, sensory neuronopathy |
NMDAS cerebellar ataxia: 0 – none, 1 – normal gait but hesitant heel-toe, 2 – gait reasonably steady. Unable to maintain heel-toe walking or mild upper limb dysmetria. 3 – ataxic gait (but walks unaided) or upper limb intention tremor and pointing. Unable to walk heel-toe (falls immediately). 4 – severe ataxia – gait grossly unsteady without support, or upper limb ataxia sufficient to affect feeding. 5 – wheel chair dependent primarily due to ataxia or upper limb ataxia prevents feeding.
arPEO = autosomal recessive, progressive external ophthalmoplegia; F = female; IHD = Ischemic heart disease; KSS = Kearns-Sayre Syndrome; L = left; M = male; MELAS = Mitochondrial encephalopathy with lactic acidosis and stroke-like episodes; MERRF = Myoclonic epilepsy with ragged-red fibres; N/A = not applicable; NMDAS = Newcastle Mitochondrial Disease Adult Scale; PCOS = Polycystic ovary syndrome; PFO = patent foramen ovale; R = right; resp. = respiratory; Renal TP = renal tubulopathy; + = positive finding.
Neuropathology
All the brain tissue used in the current study was obtained from the Newcastle Brain Tissue Resource. The brain tissue collection for both mtDNA disease (n = 14) and control subjects (n = 15) was ethically approved by the Newcastle and North Tyneside Local Research Ethics Committee. For each region of interest investigated, suitable control tissue was acquired (Supplementary Table 1). Patients were between 20 and 68 years of age with a mean age of 42.4 ± 12.8 years (standard deviation). Controls used for the assessment of inferior olivary nucleus (Control: 1-2), Purkinje cells (Control: 3-7), and dentate nucleus (Control: 8-15) had a mean age of 53.5 ± 0.7, 70 ± 7.6 and 52.13 ± 12.5 years, respectively. Whereas patient and control tissue was suitably age-matched for inferior olivary nucleus and dentate nucleus, the controls used to investigate Purkinje cells were somewhat older. No statistical differences were determined for postmortem interval or length of fixation between patient and control tissue.
Histological Stains and Immunohistochemistry
Routine neuropathological histological stains, including cresyl fast violet (CFV), Hematoxylin and eosin (H&E), Loyez and Bielschowsky impregnation, were applied to formalin-fixed, paraffin-embedded (FFPE) tissues. Immunohistochemical staining was performed on 5 μm thick FFPE sections. Sections were deparaffinized and rehydrated through Histoclear (National Diagnostics, Charlotte, NC) and ethanol series. Antigen retrieval was performed using the optimal method for each primary antibody (Supplementary Table 2). Endogenous peroxide was blocked by incubating sections in a 3% H2O2 solution (Sigma-Aldrich, UK). A range of primary antibodies was used to identify various markers, including mitochondrial proteins, presynaptic terminal protein and cytoskeletal components in neuronal and astroglial cells. Primary antibodies were diluted at the optimal concentration and applied to the tissue for 1 hour at room temperature (RT). Sections were washed in 3 changes of Tris-buffered saline (TBS) for 5 minutes and incubated with a Universal probe (Menarini Diagnostics, Wokingham, UK) to recognize the mouse or rabbit epitope of the primary antibody, for 30 minutes. Sections were washed in 3 changes of TBS and a horseradish peroxidase-polymer (Menarini Diagnostics) was applied to the tissue for 30 minutes at RT. The sections were then visualized with 3, 3′-diaminobenzadine (DAB; Sigma-Aldrich, UK), counterstained in Mayer’s hematoxylin and the nuclei were blued in Scott’s tap water.
Two-Dimensional Neuron Counting and Calculation of Neuron Density in Olivary-Cerebellum
Due to limited tissue availability, a 2 -dimensional neuronal cell counting protocol was employed using a stereological workstation with a modified light microscope (Olympus BX51), motorized stage for automatic sampling, CCD color video and stereology software (Stereo Investigator, MBF Bioscience, Williston, VT). The medulla oblongata was sectioned in the horizontal plane and the cerebellum with entire dentate nucleus was cut in the sagittal plane at 20 μm. This was sampled at approximately the same level in each patient and control to allow for direct comparisons. One section was sampled from each patient and control and stained with CFV to allow identification of neuronal populations.
The total counts of Purkinje cells were performed on individual lobules of the cerebellum to calculate Purkinje cell density. The boundaries of the Purkinje cell layer in individual cerebellar lobules were identified at ×10 magnification using the closed contour function of Stereo Investigator. Purkinje cells were defined as neurons if they had a visible nucleolus and clear cytoplasmic profile. Purkinje cells fulfilling these criteria were counted in each lobular area at ×40 magnification and the Purkinje cell densities (number per mm2) were then calculated for each lobule. Because there were no significant changes in cell density in specific lobules from the patients vs. controls, the data were pooled to evaluate overall Purkinje cell density.
The grey matter ribbons of the inferior olivary and dentate nuclei were identified at ×2.5 magnification and the grey matter boundaries were outlined. Dentate nuclei were subdivided into quadrants to gauge the pattern of neuronal loss and provide some evidence of projections from the nuclei, which may be particularly affected by perturbed mitochondrial metabolism (20). This information was stored and dentate nuclear areas calculated. At ×100 magnification, the entire neuronal cell population within the ribbon was counted using the criteria of a clear and visible nucleolus and cytoplasmic profile and densities of olivary and dentate neurons were first calculated (number per mm2) in each quadrant. Because there were no significant differences between quadrants in any of the patients, the data were pooled to identify overall changes in neuron cell density.
Sequential Cytochrome c Oxidase/Succinate Dehydrogenase Histochemistry
A sequential histochemical assay to determine cytochrome c oxidase (COX) and succinate dehydrogenase (SDH) activities was used to identify COX-deficient/SDH-positive neurons in frozen tissue sections as previously described (21). Briefly, 10-μm-thick frozen section mounted on a glass slide were incubated with COX medium at 37°C for 50 minutes, followed by 3 washes with phosphate-buffered saline (PBS), incubation with SDH medium at 37°C for 45 minutes, washed in PBS and dehydrated in a graded ethanol series before being cleared in Histoclear and mounted with DPX (Raymond A. Lamb, UK). For those sections used for molecular analysis, the section were dehydrated in ethanol and allowed to air-dry for 1 hour at RT before being stored at −20°C.
Quantification of Neuronal Respiratory Chain Deficiency
Respiratory deficient neurons were identified either by immunohistochemical (IHC) methods on FFPE sections or by histochemical methods on frozen sections, which reveal protein expression levels or enzyme activity, respectively. A neuron was judged respiratory deficient by an absence of DAB staining in the neuronal cell cytoplasm following IHC or by blue staining of the neuronal cytoplasm following histochemical staining for COX/SDH. Numbers of respiratory deficient cells and total number of cells were then counted and the proportion of respiratory deficient neurons given as a percentage. Because this study has quantified the percentage of neurons harboring a respiratory chain deficiency for a number of oxidative phosphorylation markers, particularly complex I and complex IV, a severity index was devised. This depended on the following scoring system: 0% neurons deficient = 0, 1-9% neurons deficient = 1, 10-24% neurons deficient = 2, 25-49% neurons deficient = 3, 50-74% neurons deficient = 4 and 75-100% neurons deficient = 5. The score was applied to the IHC-stained nuclear-encoded subunits 19, 20 and 30 comprising complex I. The scoring system was also applied to the IHC of the mitochondrially encoded COX-I (complex IV subunit I), and to COX/SDH histochemistry as these provide a close correlation (22). A minimum score of 0, implying an absence of deficiency, and maximum of 15, implying severe deficiency, could be obtained. Because not all markers could be evaluated in some patients, percentage levels could then be derived from the complex I and complex IV score (Table 2).
Table 2.
Neurons in the Olivary-Cerebellum Exhibit Deficiency For Complex I and Complex IV Of the Mitochondrial Respiratory Chain
| Inferior olivary nucleus | ||||||||
|---|---|---|---|---|---|---|---|---|
| CI-19 | CI-20 | CI-30 | Complex I score | COXI | COXIV | COX/SDH | Complex IV score | |
| Pt 1 | 22 | 82 | 74 | 11/15 (73%) | 58 | 40 | N/A | 4/5 (80%) |
| Pt 2 | N/A | N/A | N/A | N/A | N/A | N/A | N/A | N/A |
| Pt 3 | N/A | N/A | N/A | N/A | N/A | N/A | N/A | N/A |
| Pt 4 | 96 | 99 | 100 | 15/15 (100%) | 2 | 2 | N/A | 1/5 (20%) |
| Pt 5 | 75 | 99 | 88 | 15/15 (100%) | 37 | 72 | N/A | 3/5 (60%) |
| Pt 6 | 37 | 13 | 11 | 7/15 (47%) | 0 | 0 | N/A | 0/5 (0%) |
| Pt 7 | 52 | 93 | N/A | 9/10 (90%) | 0 | 0 | N/A | 0/5 (0%) |
| Pt 8 | 50 | 60 | 72 | 12/15 (80%) | 43 | 38 | N/A | 3/5 (60%) |
| Pt 9 | 66 | 100 | 98 | 14/15 (93%) | 81 | 73 | N/A | 5/5 (100%) |
| Pt 11 | 99 | 98 | 100 | 15/15 (100%) | 80 | 82 | N/A | 5/5 (100%) |
| Pt 12 | 55 | 100 | 100 | 14/15 (93%) | 5 | 15 | N/A | 1/5 (20%) |
| Pt 13 | 98 | 76 | 99 | 15/15 (100%) | 62 | 79 | N/A | 4/5 (80%) |
| Pt 14 | 98 | 100 | 100 | 15/15 (100%) | 88 | 64 | N/A | 5/5 (100%) |
| Purkinje cells | ||||||||
| Pt 1 | 10 | 15 | 36 | 7/15 (47%) | 5 | 20 | 0 | 1/10 (10%) |
| Pt 2 | N/A | 3 | 8 | 2/15 (13%) | 3 | 0 | N/A | 1/5 (20%) |
| Pt 3 | N/A | 10 | 36 | 5/10 (50%) | 5 | 0 | N/A | 1/5 (20%) |
| Pt 4 | 5 | N/A | 34 | 4/10 (40%) | 1 | 0 | 0 | 1/10 (10%) |
| Pt 5 | N/A | 45 | 20 | 5/10 (50%) | 0 | 0 | N/A | 0/5 (0%) |
| Pt 6 | 42 | 0 | N/A | 4/10 (40%) | 0 | 0.5 | N/A | 0/5 (0%) |
| Pt 7 | 15 | 65 | N/A | 6/10 (60%) | 29 | N/A | N/A | 3/5 (60%) |
| Pt 8 | 0 | 0 | 2 | 3/15 (20%) | 0 | 0 | 10 | 2/10 (20%) |
| Pt 9 | 51 | 49 | 53 | 11/15 (73%) | 3 | 0 | 10 | 3/10 (30%) |
| Pt 10 | 5 | N/A | 30 | 4/10 (40%) | 0 | 0 | 0 | 0/10 (0%) |
| Pt 11 | 2 | 2 | 10 | 4/15 (27%) | 0 | 0 | 10 | 2/10 (20%) |
| Pt 12 | N/A | 51 | N/A | 4/5 (80%) | 10 | 16 | N/A | 2/5 (40%) |
| Pt 13 | N/A | N/A | 37 | 3/5 (60%) | 5 | 10 | 20 | 3/10 (30%) |
| Pt 14 | N/A | N/A | N/A | N/A | 26 | 5 | 20 | 5/10 (50%) |
| Dentate nucleus | ||||||||
| Pt 1 | 22 | 32 | 9 | 6/15 (40%) | 0 | 0 | 0 | 0/10 (0%) |
| Pt 2 | N/A | 2 | 0.5 | 2/10 (20%) | 2 | 0 | N/A | 1/5 (20%) |
| Pt 3 | N/A | 8 | 11 | 3/10 (30%) | 1 | 0 | N/A | 1/5 (20%) |
| Pt 4 | 5 | 37 | 19 | 6/15 (40%) | 0 | 0 | 0 | 0/10 (0%) |
| Pt 5 | N/A | 9 | 10 | 3/10 (30%) | 0 | 0 | N/A | 0/5 (0%) |
| Pt 6 | 6 | 12 | N/A | 3/10 (30%) | 0 | 0 | N/A | 0/5 (0%) |
| Pt 7 | 11 | 2 | N/A | 3/10 (30%) | 0 | 0 | N/A | 0/5 (0%) |
| Pt 8 | 14 | 40 | 15 | 7/15 (47%) | 3 | 0 | 20 | 3/10 (30%) |
| Pt 9 | 40 | 78 | 63 | 12/15 (80%) | 3 | 0 | 0 | 1/10 (10%) |
| Pt 10 | N/A | N/A | N/A | N/A | N/A | N/A | 0 | 0/5 (0%) |
| Pt 11 | 25 | 43 | 33 | 9/15 (60%) | 2 | 8 | 20 | 3/10 (30%) |
| Pt 12 | N/A | N/A | 100 | 5/5 (100%) | 55 | 48 | N/A | 4/5 (80%) |
| Pt 13 | N/A | 19 | 10 | 4/10 (40%) | 0.5 | 0.2 | 20 | 3/10 (30%) |
| Pt 14 | N/A | N/A | N/A | N/A | N/A | N/A | N/A | N/A |
| Mitochondrial Respiratory Chain | ||||||||
Cl19, 20, 30 = complex 1 subunits,19 kDa, 20 kDa, 30 kDa; COX = cytochrome c oxidase; COX1 = complex 1; COXIV = complex IV; N/A – tissue not available for analysis; SDH = succinate dehydrogenase.
Laser Microdissection and DNA Extraction
For laser microdissection, 20-μm-thick cryostat sections were mounted onto polyethylene naphthalate-membrane slides (Leica Microsystems, UK). Because the levels of neuronal COX-deficiency were low in these patients, COX/SDH histochemistry was performed. Individual neurons with a COX-deficient/SDH-positive cell body were microdissected from the inferior olivary nucleus, Purkinje cells and dentate nucleus using a Leica laser microdissection microscope (Leica Microsystems LMD 6000, UK). DNA extraction was performed overnight using a standard lysis buffer (23).
Determination of Percentage Level Mutated mtDNA to Wild-Type mtDNA
Pyrosequencing for point mutations
The quantitation of mtDNA mutation load was performed using Pyromark Assay Design Software v.2.0 (Qiagen, West Sussex, UK) to design locus specific PCR and pyrosequencing primers that amplified a PCR product spanning the mutation region using a forward primer and a biotinylated reverse primer (Integrated DNA Technologies, Glasgow, UK; Supplementary Table 3). Pyrosequencing was achieved on the Pyromark Q24 platform according to the manufacturer’s protocol, employing a mutation-specific pyrosequencing primer. Pyromark Q24 software was used to quantify mutated mtDNA heteroplasmy levels by directly comparing the relevant peak heights of both wild-type and mutant mtDNA at this site.
Real-time PCR for mtDNA deletions
A multiplex real-time PCR (RT-PCR) MT-ND1/MT-ND4 assay was used to quantify the levels of mtDNA deletions in individual neurons (24-26). The assay provides a deletion (heteroplasmy) level in the neurons. A total of 5 μl of sample was used and run in triplicate in a 96-well plate. A 24-μl master mix consisted of deionized water, TaqMan Universal mastermix (Applied Biosystems, Paisley, UK), 100 nM MT-ND1 VIC probe, 100 nM MT-ND4 FAM probe, 300 nM MT-ND1 FOR, 300 nM MT-ND1 REV, 300 nM MT-ND4 FOR, and 300 nM MT-ND4 REV primers (Eurofins, Wolverhampton, UK; Supplementary Table 3). The PCR program consisted of a 2-minute incubation at 50°C, 10 minutes at 95°C and 40 cycles of amplification; 15-second denaturation at 95°C, and 1 minute at 60°C for hybridization of probes and primers and DNA synthesis. Known deletion-level standards, a blood-positive control and a blood-negative control, all run in triplicate, were added to the assays, as previously described (26).
RESULTS
Patients
Clinical evaluation using the NMDAS rating scale showed that all 14 patients displayed symptoms consistent with cerebellar ataxia. In these patients, the severity of ataxia varied without any relation to the mtDNA defect type (Table 1), with the highest levels of impairment observed in patients with either m.3243A>G (Patient 6), m.8344A>G (Patient 8), single, large-scale mtDNA deletion (Patient 11), or recessive POLG mutations (Patients 12 - 14).
General Neuropathological Findings
The majority of patients had variable degrees of cerebellar abnormalities; there was variable and selective loss of one or more neuronal types among Purkinje cells, olivary and dentate neurons and/or granule cells. Evidence of myelin loss, dendritic abnormalities and axonal degeneration were apparent in patients with a high degree of neuronal cell loss (Fig. 1). In accordance with previous findings in Kearns-Sayre syndrome (KSS), we found myelin loss and spongiform degeneration of the white matter tracts of Patient 11 (Fig. 1Aiii) (18). We recently discussed the mechanisms underpinning loss of myelin in this patient and attribute these changes to oligodendrocyte dysfunction (27). Other patients with different mtDNA defects also showed varying degrees of myelin loss but this was likely secondary to axonal/neuronal loss because, in general, it appeared to occur in regions affected by severe neuron loss. Patients with the m.3243A>G mutation typically exhibited intact myelin with only mild myelin pallor in 1 case (Patient 5; Fig. 1Aii) and selective myelin loss within regions of microinfarcts. Several patients had profound myelin loss of the dentate nucleus outflow tract, which correlated with marked neuron loss from the dentate nucleus (Fig. 1Aiv). These included patients with m.8344A>G, m.14709T>C and recessive POLG mutations.
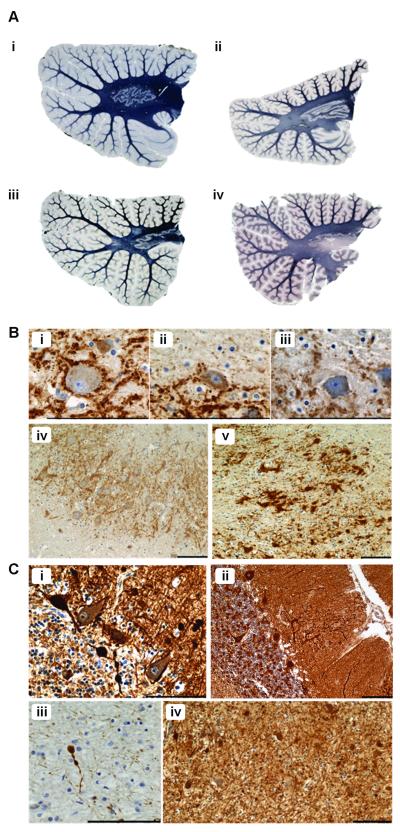
Figure 1.
Myelin loss, axonal and dendritic pathology in patients with mitochondrial DNA (mtDNA) defects are likely to be secondary to neuronal cell loss in all cases except Kearns-Sayre syndrome (KSS). (A) Relative to control (i; Loyez) there is mild myelin pallor in some patients with the m.3243A>G mutation (ii; Loyez, Patient 5); spongiform degeneration and myelin loss are seen in a single mtDNA deletion patient (iii; Loyez, Patient 11 with KSS). There is severe myelin loss of the dentate nucleus outflow tract in patients with POLG mutations (iv; Loyez, Patient 12). (B) There are markedly reduced presynaptic terminals around dentate nucleus neurons in m.3243A>G (ii; synaptophysin, Patient 4) and m.8344A>G (iii; synaptophysin, Patient 8) cases vs. controls (i; synaptophysin). Relative to control (iv; synaptophysin) there are large intensely labeled presynaptic-like structures lacking morphologically distinguishable post-synaptic dentate neurons in a patient with m.14709T>C mutation (v; synaptophysin, Patient 9), indicating relative increase of input from Purkinje cells when the dentate neuron loss exceeds Purkinje cell loss. (C) There are multiple axonal swellings in the granular cell layer of the cerebellum (i; SMI31; single mtDNA deletion, Patient 11) and thickened dendritic trees containing ‘trapped’ mitochondria in the molecular layer (ii; porin; single mtDNA deletion, Patient 11). There are also axonal spheroids in the deep white matter (iii and iv; synaptophysin; single mtDNA deletion, Patient 11). Scale bar: 100 μm.
Synaptic disorganization assessed using synaptophysin immunohistochemistry was prominent in many cases. The main changes were in the dentate nucleus. There was often an absence of presynaptic terminals on neurons in patients with extensive Purkinje cell loss compared to controls (Fig. 1Bi-iii), indicating loss of input to the dentate nucleus. In contrast, Patient 9 was the only case in which there were large, intensely labeled synaptic-like structures lacking morphologically distinguishable post-synaptic neurons, possibly surrounding “ghosts” of degenerating neurons (Fig. 1Biv, v). This suggests relatively increased input to the dentate nucleus because the extent of the dentate neuron loss exceeded the Purkinje cell loss in this patient.
Axonal and dendritic abnormalities were observed in almost all patients and were particular prominent in Purkinje cells. Most notably, axonal ‘torpedoes,’ or swellings, were present in the proximal portion of Purkinje cell axons. These swellings largely consisted of neurofilaments and mitochondria and were localized in the Purkinje cell, granular cell and molecular cell layers (Fig. 1Ci). Axonal spheroids in the deep white matter were often seen in conjunction (Fig. 1Ciii, iv). Dendritic abnormalities consisted of an increase in dendritic arborization, thickening of the dendrites and evidence of ‘trapped’ mitochondria (Fig. 1Cii). Often the dendritic trees became more expansive in those Purkinje cells neighboring regions where cell loss had occurred, suggesting a compensatory mechanism.
There was evidence of cerebellar atrophy with increased inter-folial spaces, mainly due to widespread atrophy of the molecular layer and loss of granule cells. This was typically observed in patients with the m.3243A>G mutations (Patient 5; Fig. 2B,iv). The presence of small, shrunken and eosinophilic “dark” neurons was the most widespread morphological alteration (Patient 8; Fig. 2Aiii).
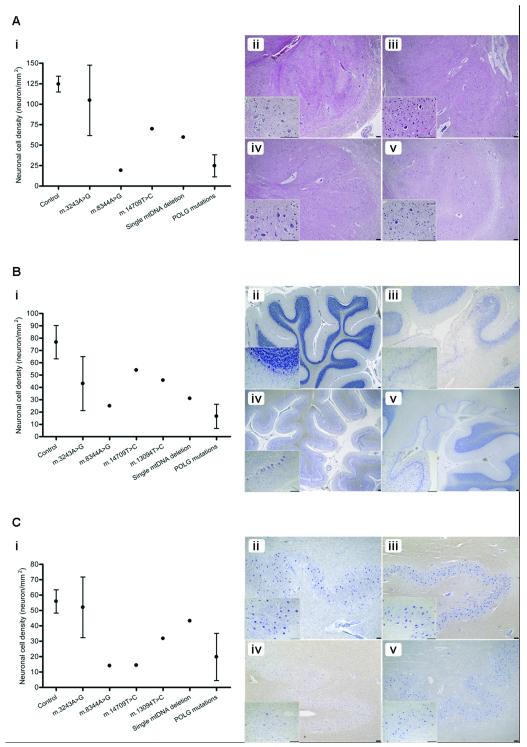
Figure 2.
Variation in neuron loss across the olivo-cerebellum in patients with mitochondrial DNA (mtDNA) mutations. (A) Inferior olivary neuronal densities vary remarkably according to the mtDNA defect vs. control tissues (n = 2) (i). Cell loss is most severe in association with the m.8344A>G (iii; H&E) and recessive POLG (iv; H&E) mutations with only moderate loss associated with the m.14709T>C mutation (v; H&E) vs. control tissues (ii; H&E). (B) Purkinje cell density is reduced in all patients with mtDNA defects vs. controls (n = 5) (i). This is most apparent in a patient with POLG mutations in which neuron loss is global and with the presence of microinfarcts (v; cresyl fast violet [CFV)]). Atrophy of the cerebellar cortex is severe in m.3243A>G (iv; CFV) and Purkinje cell loss often occurs focally in microinfarcts (iii; CFV) when compared to control tissue (ii; CFV). (C) Neuronal cell density in the dentate nucleus also shows wide variation in patients relative to controls (n = 8) (i). Typically, patients with the m.3243A>G mutation show preservation of neurons (iii; CFV) vs. control tissue (ii; CFV); cell loss is severe in m.8344A>G (iv; CFV), and minimal in single mtDNA deletion patients (v; CFV). Scale bar: 100 μm.
Variations in Neuronal Cell Loss in Different Areas of the Olivary-Cerebellum
Quantification of neuronal cell density revealed significant variations in cell loss across the olivary-cerebellum in relation to mtDNA defects (Fig. 2). Percentage levels of cell loss are shown in Table 1. Patient 5 (m.3243A>G) was the only patient that demonstrated an increase in cell density for the inferior olivary and dentate nucleus neurons vs. controls. This was likely due to severe cerebellar atrophy; the percentage values for cell loss are listed as 0% in Table 1. Cell loss was typically most profound in all regions in association with the m.8344A>G and POLG mutations. For example, Patient 12 (p.Ala47Thr and p.Trp748Ser POLG mutations) exhibited 81%, 91% and 96% loss of neurons from Purkinje cells (Fig. 2Bv), inferior olivary and dentate nucleus, respectively. Although patients with the m.3243A>G demonstrated inter-patient variability in cell density, the main finding was of severe Purkinje cell loss (likely due to the presence of ischemic-like lesions), with an average ~50% of cells lost (Table 1; Fig. 2Biii). Cell density was generally reduced in all other patients with different mtDNA defects. It appeared that for Patient 9 (m.14709T>C), neuronal cell loss was most severe in the dentate nucleus with 74% loss whereas the inferior olivary nucleus and Purkinje cells were only moderately affected. Approximately 50% of neurons were lost from the olivo-cerebellum in Patient 10 with an m.13094T>C mutation. Purkinje cells and inferior olivary neurons were predominantly affected in Patient 11 due to the single, large-scale mtDNA deletion where cell loss reached 52% and 60%, respectively. In that case cell loss in the dentate nucleus was minimal (Fig. 2C v).
Microinfarcts
Quantification of neuronal cell density revealed that patients with the common m.3243A>G mutation associated with the mitochondrial encephalopathy with lactic acidosis and stroke-like episodes syndrome (MELAS) displayed microinfarcts or ischemic-like lesions in the cerebellar cortex (Fig. 2 Biv). Typically, these lesions affect not only the Purkinje cells but also the molecular layer, granular cell layer and adjacent white matter. In m.3243A>G patients, neuronal cell loss was predominately due to the microinfarcts. In cases with the myoclonic epilepsy with ragged red fibers-associated mutation, m.8344A>G and recessive p.Ala467Thr and p.Trp748Ser POLG mutations, microinfarcts were also found but there was also more widespread Purkinje cell loss not associated with microinfarcts (Fig. 2 Bv). Furthermore, assessment of the cerebellar lobules suggested that in all cases evaluated microinfarcts typically occurred in the posterior inferior lobules of the cerebellum.
Astrogliosis
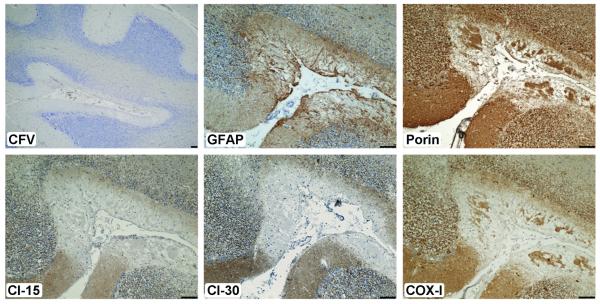
Immunohistochemistry with for glial fibrillary acidic protein, a marker of astrocytes, revealed proliferation of the astroglial cell populations in regions where cell loss was greatest. This was particularly obvious microinfarcts in which proliferation of the Bergmann glia was prominent. In addition, deficiency of components of the mitochondrial respiratory chain in the astroglia was evident in serial sections (Fig. 3). This suggests that astroglia may harbor respiratory-compromised mitochondria.
Figure 3.
Astrogliosis in regions of microinfarcts with evidence of severe respiratory chain deficiency in a patient with m.3243A>G. There are numerous microinfarcts in the cerebellar cortex (cresyl fast violet, [CFV)]); typically, these are accompanied by marked proliferation of glial fibrillary acidic protein (GFAP)-positive astrocytes (in a serial section). Evaluation of the mitochondrial proteins shows that mitochondrial density is reduced due to the necrosis (porin), but expression of complex I (subunits Cl-15 and -30) is completely absent; cytochrome c oxidase-1 (COX-I) is diminished. These observations suggest that astrocytes may also harbor respiratory-deficient mitochondria. Scale bar: 100 μm.
Correlation between Neuronal Cell Loss and a High NMDAS Score for Cerebellar Ataxia
We observed a strong correlation between a high NMDAS score for ataxia and evidence of a low neuronal cell density in all 3 areas of the olivary-cerebellum, suggesting that the greater the impairment the less chance that cellular density is maintained. Our data indicates that a high functional impairment is accompanied by a physical loss of neurons (because low cell density indicates cell loss). This correlation was found to be significant in the inferior olivary nucleus (Pearson’s correlation = −0.902, p = 0.000), the Purkinje cell (Pearson’s correlation = −0.628, p = 0.029) and dentate nucleus (Pearson’s correlation = −0.577, p = 0.049) neuronal populations.
Molecular Genetic Investigation of Levels of Mutated mtDNA
Molecular genetic analysis was performed on individual isolated Purkinje cells and neurons of the inferior olivary and dentate nuclei to determine mutation load. Pyrosequencing of mtDNA point mutations revealed high percentage levels in remaining cells throughout the olivo-cerebellum (Table 3). High levels of point mutation did not correlate with the degree of cell loss. This was particularly evident in Patient 9 where the m.14709T>C mutation levels were near homoplasmy yet only dentate nucleus neurons reveal severe cell loss. Patients with the m.3243A>G mutation generally showed higher percentage levels of mutated mtDNA that are typically greater than 73%. A lack of correlation between high mutation load and cell loss is clearly demonstrated by Patient 4 (m.3243A>G) in which the level of m.3243A>G reached 88% in inferior olivary neurons despite there being only 42% cell loss in this region (Tables 1, 3).
Table 3.
Percentage Level Heteroplasmy for Mutated or Deleted mtDNA
| Patient Number | Inferior olivary neurons | Purkinje cells | Dentate nucleus neurons |
|---|---|---|---|
| 1 | 93 ± 15.7 (n=11) | 90 ± 9.2 (n=15) | 77.5 ± 8.9 (n=6) |
| 4 | 88.3 ± 2.1 (n=4) | 80.8 ± 15.7 (n=11) | 73.3 ± 20.6 (n=12) |
| 6 | ND | 83.8 ± 3.8 (n=13) | 84.9 ± 11.1 (n=13) |
| 7 | ND | 91.7 ± 14.9 (n=13) | 86.4 ± 16.8 (n=20) |
| 8 | 86.1 ± 9.1 (n=8) | 91.9 ± 4 (n=13) | 87.4 ± 18.9 (n=17) |
| 9 | ND | 95 ± 9.6 (n=11) | 95.4 ± 5.7 (n=11) |
| 10 | 44.2 ± 18 (n=13) | 47.2 ± 21.2 (n=13) | 51.3 ± 19.1 (n=11) |
| 11 | 67.9 ± 11.9 (n=7) | 68.1 ± 9.4 (n=7) | 43.9 ± 6.8 (n=7) |
| 12 | ND | 19.9 ± 20 (n=10) | ND |
| 13 | ND | 50.7 ± 24 (n=21) | 24.9 ± 24.1 (n=21) |
| 14 | ND | 46.9 ± 31.3 (n=23) | ND |
Data are mean per cent ± SD; n = number of neurons laser-dissected and examined in each assay; ND = not determined.
In contrast, patients with recessive POLG mutations and accumulated multiple mtDNA deletions showed relatively low levels of mtDNA deletion following RT-PCR. The highest level (50.7%) of mtDNA deletion was seen in the Purkinje cells from Patient 13 and the lowest (19.9%) in Purkinje cells from Patient 12. This was despite the extreme cell loss seen in these patients. Neuron cell loss did not correlate with the level of mutated mtDNA in remaining neurons. Therefore, the level of mutation alone does not appear to determine where neuron cell loss will occur. However, we did not evaluate mtDNA depletion in these patients.
Mitochondrial Protein Changes
Various mitochondrial markers were employed to investigate the selective vulnerability of neurons. Porin immunohistochemistry revealed punctate, uniformly high staining within neuronal cytoplasm in both control and patients, indicating that mitochondrial mass was abundant. However, neurons within the inferior olivary nucleus were the exception showing unusual mitochondrial localization in both patients and controls. Generally, mitochondria within these neurons were distributed around the periphery of the cell body or were perinuclear, whereas the cytoplasm remained devoid of mitochondria.
Quantification of the respiratory chain proteins revealed marked respiratory chain deficiency; there was complex I deficiency within remaining neuronal populations but only low levels of complex IV-deficiency (Table 2). The percentage level neuronal respiratory chain deficiency correlated with the degree of cell loss, particularly in the Purkinje cell and dentate nucleus neuronal populations. Typically patients with extreme cell loss demonstrate high percentage levels of complex I-deficient neurons and moderate levels of complex IV-deficient neurons. The data obtained from Patient 11 (m.14709T>C) support these findings with severe neuron loss from the dentate nucleus at 74% with a complex I and complex IV severity index of 60% and 30%, respectively (Table 2). Conversely, patients with the m.3243A>G mutation where cell loss was moderate, a lower percentage of neurons were deficient for complex I and complex IV was unaffected. For example, Patient 3 (m.3243A>G) demonstrated a 26% reduction in neuron density in the dentate nucleus with remaining cells revealing a complex I and complex IV severity index of 30% and 20%. This was also seen in Patient 10 (m.13094T>C) and Patient 13 (POLG mutations and multiple mtDNA deletions) where Purkinje cell and dentate nucleus cell loss reached 40% to 45% and the complex I and complex IV severity index revealed moderate and mild scores, respectively (Table 2; Fig. 4). Intriguingly, there was a stronger relationship between complex IV deficiency and cell loss, suggesting that development of complex IV deficiency is particularly detrimental to neuronal viability (Purkinje cells; Pearson’s correlation coefficient = 0.736, p = 0.006. Dentate nucleus; Pearson’s correlation coefficient = 0.606, p = 0.048).
Figure 4.
High levels of complex I-deficient neurons can be detected in remaining neuronal populations. The inferior olivary nucleus shows an unusual distribution of mitochondria (m.3243A>G, first column) (porin), whereas mitochondria appear abundant throughout the remainder of the cerebellum in all cases as can be seen in the patient with the m.13094T>C mutation (middle column) and a patient with recessive POLG mutation (third column). High levels of complex I-deficient neurons are observed throughout the olivo-cerebellum, nearing 100% of inferior olivary nucleus in m.3243A>G patient but there is marked deficiency throughout the remainder of the cerebellum. Complex IV-deficiency was rarely detected and low in many cases; cytochrome c oxidase/succinic acid dehydrogenase (COX/SDH) histochemistry often revealed intact neuronal COX activity. Scale bar: 100 μm.
The neurons within the inferior olivary nucleus showed widespread deficiency for complex I in the absence of neuronal cell loss (Fig. 4; Patient 4). Therefore, all patients had a high complex I severity index, often reaching 100% (Table 2).
DISCUSSION
Despite the recognition of cerebellar ataxia as a frequently reported neurological deficit in patients with mtDNA disease, the neurodegenerative changes underpinning this disorder are largely unknown. The present study performs a detailed quantitative clinical, neuropathological and molecular investigation on a large number of patients with a variety of mtDNA abnormalities to determine the mechanisms of neurodegeneration in the olivo-cerebellar pathway.
General Pathology
Generalized cerebellar atrophy and eosinophilic dark neurons are commonly detected throughout the olivo-cerebellum in mtDNA disease. We also document degenerative changes in connectivity throughout these structures. These changes include the formation of axonal spheroids and “torpedoes,” axonal loss, synaptic changes and demyelination. All cases investigated appear to show degenerative changes in axons and dendrites that correlate with the extreme cell loss in these regions. Evidence of Purkinje cell axonal degeneration has been reported in patients with single, large-scale mtDNA deletions and the m.3243A>G mutation (21, 28). The current study extends this observation to other patients with different mtDNA mutations and suggests that axonal abnormalities are more widespread than previously thought in mtDNA disease. It is intriguing to note that similar swellings have been documented in patients with essential tremor, Alzheimer disease and Parkinson disease (29-31). It is uncertain what these changes represent but it is likely that they would result in axonal block in both an anterograde and retrograde direction and would likely lead to Purkinje cell death. The loss of myelin in these regions is likely a secondary consequence of axonal and neuronal demise. This would be true for all cases except KSS in which white matter abnormalities far outweighed grey matter abnormalities, consistent with previous reports (32).
Correlation of Cell Loss with Ataxia
Analyses of neuronal cell density and NMDAS scores revealed a strong association between severe ataxia and reduced neuronal density, though the olivo-cerebellum showed considerable variation in both the pattern and severity of neurodegeneration in the cases with mtDNA defects. The emergence of the infarct-like lesions is an important finding since this is not specific to MELAS patients with the m.3243A>G mutation. In fact, microinfarcts in the cerebellum and other brain areas have also been detected in patients with the m.8344A>G mutation and heterozygous POLG mutations (p.Ala467Thr and p.Trp748Ser; p.Gly848Ser and p.Ser1104Cys) herein and by others (33-35). Intriguingly, clinical assessment of patients with the m.8344A>G and POLG mutations showed stroke-like episodes, which are associated with MELAS but also occur in these patients (36, 37). Whereas these infarct-like lesions resemble true microinfarcts, they do not conform to a particular vascular territory and have a cortical laminar distribution. Patients with the m.3243A>G suffer Purkinje cell loss, which tends to occur focally in regions of microinfarct, suggesting that the main mode of cell loss occurs through these lesions. The mechanisms behind the formation of these lesions are not known but this type of lesion has been documented in cerebral ischemia and hypoxia that arises from localized energy deficiency (35).
This study employed 2-dimensional neuron counting protocol because of limited availability of the patient postmortem CNS tissues and the need to make maximal use of available material. Because there is some debate in the scientific community regarding the usefulness of 3-dimensional stereology (39), and other studies have shown that 2- and 3-dimensional methodologies yield similar results (40), we used the former. Our results show a clear and obvious reduction of neuronal cell density in patient tissues vs. control tissues. This is further strengthened in the assessment of Purkinje cells where the higher age of our control cases was documented where Purkinje cell loss might be a feature (41-43).
Astrocytes in regions of cell loss are readily identified in mtDNA disease (18, 38). However, whether this alleviates or contributes to the pathophysiology of disease remains to be determined. Indeed, it has been postulated that dysfunctional astrocytes might be key players in grey matter pathology in KSS (38). Recent work in embryonic stem cells with genetic mutations in complex I and complex IV-encoded mtDNA subunits differentiated into neurons also reveals the presence of astrocytes in the culture system. Severe mutant cells with complex I-deficiency show an overall increase in the number of astrocytes; however, they exhibit a reduced mitochondrial membrane potential and lowered glutathione (44). It is possible that while mitochondrial dysfunction is evident in astrocytes they are not as sensitive to mtDNA mutations because they can switch to glycolytic metabolism when ATP levels are lowered (45). It may be that the proliferation of astrocytes in mtDNA diseases might be a protective response to try to support neurons with dysfunctional mitochondria.
Heteroplasmy Level of Mutated mtDNA and Respiratory Chain Deficiency in Neurons
Previous investigations of heteroplasmic mutations, including the m.3243A>G mutation, have shown that the proportion of mutated to wild-type mtDNA in patients’ non-CNS tissue is crucial in the development of a biochemical defect and clinical abnormality (46). Therefore, evaluation of mutated mtDNA heteroplasmy levels in neurons from different regions of the CNS might account for the selective vulnerability in patients with mtDNA disease. However, combining our neuropathological data, such as cell loss, with mutated mtDNA heteroplasmy reveals a lack of correlation. Typically, point mutations, including m.3243A>G and m.8344A>G, show high heteroplasmy in remaining neurons, whereas mtDNA deletions show relatively low heteroplasmy and the m.14709T>C mutation reaches near homoplasmy. This lack of correlation between cell loss and heteroplasmy has also been previously reported (21, 47) and suggests that there must be other mechanisms that determine which cells undergo cell death. These could include nuclear modifiers, neuronal mtDNA content, neuron subtype specific neurotransmitter metabolism and other factors.
We found high levels of complex I-deficiency in surviving neurons in the olivo-cerebellum. A number of recent studies have shown elevated levels of neuronal complex I-deficiency in CNS tissues from patients with POLG mutations (10). Here, deficiencies of complex I appeared to be most profound in nuclei with the greatest degree of cell loss. It is surprising that such high levels of complex I-deficient neurons exist without undergoing cell death, suggesting that complex-I deficiency is not critical for determining the fate of neurons but is associated with cell loss. This also might indicate that complex I is affected earlier in disease pathogenesis. In contrast, complex IV-deficiency was only found at very low levels within the neuronal populations and in regions where cell loss was most extensive. We believe the mechanisms underpinning development of respiratory deficiency and neuronal cell loss are likely to be due to the progression of the molecular defect. A high degree of heteroplasmy with associated low levels of wild type mtDNA would initially lead to complex I deficiency and, as the molecular defect progresses, neurons develop complex IV deficiency. Neurons with complex IV deficiency have a higher propensity to undergo cell death. This can be explained by the strong relationship between presence of complex IV deficiency and lowered cell density.
Conclusions
This study highlights the importance of correlating clinical, molecular and neuropathological changes occurring in the CNS tissues from patients with mtDNA defects. Our results show that whereas there is clear variability in the extent of neurodegeneration in the olivo-cerebellar pathway, the proportion of neurons with respiratory deficiency appears to correlate with the degree of cell loss. We also show correlation between the clinical severity of disease and the severity of neuronal cell loss. This suggests that clinical assessments of patients involving the NMDAS is informative for determining the level of functional impairments but might also provide some insight into neurodegenerative changes occurring in the brain.
Supplementary Material
Acknowledgments
This work was supported by the Wellcome Trust (074454/Z/04/Z), the Newcastle University Centre for Brain Ageing and Vitality supported by BBSRC, EPSRC, ESRC and MRC as part of the cross-council Lifelong Health and Wellbeing Initiative (G0700718), UK NIHR Biomedical Research Centre for Ageing and Age-related disease award to the Newcastle upon Tyne Hospitals NHS Foundation Trust and the UK NHS Specialist Commissioners which funds the “Rare Mitochondrial Disorders of Adults and Children” Diagnostic Service in Newcastle upon Tyne (http://www.mitochondrialncg.nhs.uk), MRC Mitochondrial Disease Patient Cohort Study (G800674). Tissue for this study was provided by the Newcastle Brain Tissue Resource which is funded in part by a grant from the UK Medical Research Council (G0400074) and the Alzheimer’s Society and Alzheimer’s Research Trust as part of the Brains for Dementia Research project.
Footnotes
This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
REFERENCES
- 1.McFarland R, Taylor RW, Turnbull DM. A neurological perspective on mitochondrial disease. Lancet Neurol. 2010;9:829–40. doi: 10.1016/S1474-4422(10)70116-2. [DOI] [PubMed] [Google Scholar]
- 2.Taylor RW, Turnbull DM. Mitochondrial DNA mutations in human disease. Nature Rev. 2005;6:389–402. doi: 10.1038/nrg1606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bristow EA, Griffiths PG, Andrews RM, Johnson MA, Turnbull DM. The distribution of mitochondrial activity in relation to optic nerve structure. Arch Ophthalmol. 2002;120:791–6. doi: 10.1001/archopht.120.6.791. [DOI] [PubMed] [Google Scholar]
- 4.Erecinska M, Silver IA. Ions and energy in mammalian brain. Prog Neurobiol. 1994;43:37–71. doi: 10.1016/0301-0082(94)90015-9. [DOI] [PubMed] [Google Scholar]
- 5.Hollenbeck PJ, Saxton WM. The axonal transport of mitochondria. J Cell Sci. 2005;118:5411–9. doi: 10.1242/jcs.02745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Schulte C, Synofzik M, Gasser T, Schols L. Ataxia with ophthalmoplegia or sensory neuropathy is frequently caused by POLG mutations. Neurology. 2009;73:898–900. doi: 10.1212/WNL.0b013e3181b78488. [DOI] [PubMed] [Google Scholar]
- 7.Funakawa I, Kato H, Terao A, et al. Cerebellar ataxia in patients with Leber’s hereditary optic neuropathy. J Neurol. 1995;242:75–7. doi: 10.1007/BF00887819. [DOI] [PubMed] [Google Scholar]
- 8.Casali C, Fabrizi GM, Santorelli FM, et al. Mitochondrial G8363A mutation presenting as cerebellar ataxia and lipomas in an Italian family. Neurology. 1999;52:1103–4. doi: 10.1212/wnl.52.5.1103. [DOI] [PubMed] [Google Scholar]
- 9.Fukuhara N. MERRF: a clinicopathological study. Relationships between myoclonus epilepsies and mitochondrial myopathies. Revue Neurologique. 1991;147:476–9. [PubMed] [Google Scholar]
- 10.Hakonen AH, Goffart S, Marjavaara S, et al. Infantile-onset spinocerebellar ataxia and mitochondrial recessive ataxia syndrome are associated with neuronal complex I defect and mtDNA depletion. Hum Mol Genet. 2008;17:3822–35. doi: 10.1093/hmg/ddn280. [DOI] [PubMed] [Google Scholar]
- 11.Van Goethem G, Luoma P, Rantamaki M, et al. POLG mutations in neurodegenerative disorders with ataxia but no muscle involvement. Neurology. 2004;63:1251–7. doi: 10.1212/01.wnl.0000140494.58732.83. [DOI] [PubMed] [Google Scholar]
- 12.Shoffner JM, Kaufman A, Koontz D, et al. Oxidative phosphorylation diseases and cerebellar ataxia. Clinical Neurosci. 1995;3:43–53. [PubMed] [Google Scholar]
- 13.Wray SH, Provenzale JM, Johns DR, Thulborn KR. MR of the brain in mitochondrial myopathy. AJNR Am J Nueroradiol. 1995;16:1167–73. [PMC free article] [PubMed] [Google Scholar]
- 14.Toyono M, Nakano K, Kiuchi M, et al. A case of MERRF associated with chronic pancreatitis. Neuromuscul Disord. 2001;11:300–4. doi: 10.1016/s0960-8966(00)00176-0. [DOI] [PubMed] [Google Scholar]
- 15.Scaglia F, Wong L-JC, Vladutiu GD, Hunter JV. Predominant cerebellar volume loss as a neuroradiologic feature of pediatric respiratory chain defects. AJNR: Am J Neuroradiol. 2005;26:1675–80. Erratum appears in AJNR Am J Neuroradiol 2005 Sep;26:2165. [PMC free article] [PubMed] [Google Scholar]
- 16.Mori O, Yamazaki M, Ohaki Y, et al. Mitochondrial encephalomyopathy with lactic acidosis and stroke like episodes (MELAS) with prominent degeneration of the intestinal wall and cactus-like cerebellar pathology. Acta Neuropathol (Berl) 2000;100:712–7. doi: 10.1007/s004010000209. [DOI] [PubMed] [Google Scholar]
- 17.Tanahashi C, Nakayama A, Yoshida M, Ito M, Mori N, Hashizume Y. MELAS with the mitochondrial DNA 3243 point mutation: a neuropathological study. Acta Neuropathol (Berl) 2000;99:31–8. doi: 10.1007/pl00007403. [DOI] [PubMed] [Google Scholar]
- 18.Sparaco M, Bonilla E, DiMauro S, Powers JM. Neuropathology of mitochondrial encephalomyopathies due to mitochondrial DNA defects. J Neuropathol Exp Neurol. 1993;52:1–10. doi: 10.1097/00005072-199301000-00001. [DOI] [PubMed] [Google Scholar]
- 19.Schaefer AM, Phoenix C, Elson JL, McFarland R, Chinnery PF, Turnbull DM. Mitochondrial disease in adults: a scale to monitor progression and treatment. Neurology. 2006;66:1932–4. doi: 10.1212/01.wnl.0000219759.72195.41. [DOI] [PubMed] [Google Scholar]
- 20.Su M, Yoshida Y, Hirata Y, Satoh Y, Nagata K. Degeneration of the cerebellar dentate nucleus in corticobasal degeneration: neuropathological and morphometric investigations. Acta Neuropathol. 2000;99:365–70. doi: 10.1007/s004010051137. [DOI] [PubMed] [Google Scholar]
- 21.Betts J, Jaros E, Perry RH, et al. Molecular neuropathology of MELAS: level of heteroplasmy in individual neurones and evidence of extensive vascular involvement. Neuropathol Appl Neurobiol. 2006;32:359–73. doi: 10.1111/j.1365-2990.2006.00731.x. [DOI] [PubMed] [Google Scholar]
- 22.Mahad DJ, Ziabreva I, Campbell G, et al. Detection of cytochrome c oxidase activity and mitochondrial proteins in single cells. J Neurosci Methods. 2009;184:310–9. doi: 10.1016/j.jneumeth.2009.08.020. [DOI] [PubMed] [Google Scholar]
- 23.Taylor RW, Barron MJ, Borthwick GM, et al. Mitochondrial DNA mutations in human colonic crypt stem cells. J Clin Invest. 2003;112:1351–60. doi: 10.1172/JCI19435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bender A, Krishnan KJ, Morris CM, et al. High levels of mitochondrial DNA deletions in substantia nigra neurons in aging and Parkinson disease. Nat Genet. 2006;38:515–7. doi: 10.1038/ng1769. [DOI] [PubMed] [Google Scholar]
- 25.Kraytsberg Y, Kudryavtseva E, McKee AC, Geula C, Kowall NW, Khrapko K. Mitochondrial DNA deletions are abundant and cause functional impairment in aged human substantia nigra neurons. Nat Genet. 2006;38:518–20. doi: 10.1038/ng1778. [DOI] [PubMed] [Google Scholar]
- 26.Krishnan KJ, Bender A, Taylor RW, Turnbull DM. A multiplex real-time PCR method to detect and quantify mitochondrial DNA deletions in individual cells. Anal Biochem. 2007;370:127–9. doi: 10.1016/j.ab.2007.06.024. [DOI] [PubMed] [Google Scholar]
- 27.Lax NZ, Campbell GR, Reeve AK, et al. Loss of Myelin Associated Glycoprotein in Kearns-Sayre Syndrome. Arch Neurol. doi: 10.1001/archneurol.2011.2167. In press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Tanji K, Vu TH, Schon EA, DiMauro S, Bonilla E. Kearns-Sayre syndrome: unusual pattern of expression of subunits of the respiratory chain in the cerebellar system. Ann Neurol. 1999;45:377–83. doi: 10.1002/1531-8249(199903)45:3<377::aid-ana14>3.0.co;2-m. [DOI] [PubMed] [Google Scholar]
- 29.Louis ED, Faust PL, Vonsattel JP, et al. Torpedoes in Parkinson’s disease, Alzheimer’s disease, essential tremor, and control brains. Mov Disord. 2009;24:1600–5. doi: 10.1002/mds.22567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Louis ED, Faust PL, Vonsattel JP, et al. Neuropathological changes in essential tremor: 33 cases compared with 21 controls. Brain. 2007;130:3297–307. doi: 10.1093/brain/awm266. [DOI] [PubMed] [Google Scholar]
- 31.Louis ED, Yi H, Erickson-Davis C, Vonsattel JP, Faust PL. Structural study of Purkinje cell axonal torpedoes in essential tremor. Neurosci Lett. 2009;450:287–91. doi: 10.1016/j.neulet.2008.11.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Oldfors A, Fyhr IM, Holme E, Larsson NG, Tulinius M. Neuropathology in Kearns-Sayre syndrome. Acta Neuropathol. 1990;80:541–6. doi: 10.1007/BF00294616. [DOI] [PubMed] [Google Scholar]
- 33.Berkovic SF, Carpenter S, Evans A, et al. Myoclonus epilepsy and ragged-red fibres (MERRF). 1. A clinical, pathological, biochemical, magnetic resonance spectrographic and positron emission tomographic study. Brain. 1989;112:1231–60. doi: 10.1093/brain/112.5.1231. [DOI] [PubMed] [Google Scholar]
- 34.McKelvie PA, Morley JB, Byrne E, Marzuki S. Mitochondrial encephalomyopathies: a correlation between neuropathological findings and defects in mitochondrial DNA. Journal of the neurological sciences. 1991;102:51–60. doi: 10.1016/0022-510x(91)90093-m. [DOI] [PubMed] [Google Scholar]
- 35.Tzoulis C, Neckelmann G, Mork SJ, et al. Localized cerebral energy failure in DNA polymerase gamma-associated encephalopathy syndromes. Brain. 2010;133:1428–37. doi: 10.1093/brain/awq067. [DOI] [PubMed] [Google Scholar]
- 36.Deschauer M, Tennant S, Rokicka A, et al. MELAS associated with mutations in the POLG1 gene. Neurology. 2007;68:1741–2. doi: 10.1212/01.wnl.0000261929.92478.3e. [DOI] [PubMed] [Google Scholar]
- 37.Tanji K, Gamez J, Cervera C, et al. The A8344G mutation in mitochondrial DNA associated with stroke-like episodes and gastrointestinal dysfunction. Acta Neuropathol. 2003;105:69–75. doi: 10.1007/s00401-002-0604-y. [DOI] [PubMed] [Google Scholar]
- 38.Tanji K, DiMauro S, Bonilla E. Disconnection of cerebellar Purkinje cells in Kearns-Sayre syndrome. J Neurol Sci. 1999;166:64–70. doi: 10.1016/s0022-510x(99)00114-8. [DOI] [PubMed] [Google Scholar]
- 39.Benes FM, Lange N. Two-dimensional versus three-dimensional cell counting: a practical perspective. Trends Neurosci. 2001;24:11–7. doi: 10.1016/s0166-2236(00)01660-x. [DOI] [PubMed] [Google Scholar]
- 40.Todtenkopf MS, Vincent SL, Benes FM. A cross-study meta-analysis and three-dimensional comparison of cell counting in the anterior cingulate cortex of schizophrenic and bipolar brain. Schizophr Res. 2005;73:79–89. doi: 10.1016/j.schres.2004.08.018. [DOI] [PubMed] [Google Scholar]
- 41.Andersen BB, Gundersen HJG, Pakkenberg B. Aging of the human cerebellum: a stereological study. J Comp Neurol. 2003;466:356–65. doi: 10.1002/cne.10884. [DOI] [PubMed] [Google Scholar]
- 42.Luft AR, Skalej M, Schulz JB, et al. Patterns of age-related shrinkage in cerebellum and brainstem observed in vivo using three-dimensional MRI volumetry. Cereb Cortex. 1999;9:712–21. doi: 10.1093/cercor/9.7.712. [DOI] [PubMed] [Google Scholar]
- 43.Torvik A, Torp S, Lindboe CF. Atrophy of the cerebellar vermis in ageing. A morphometric and histologic study. J Neurol Sci. 1986;76:283–94. doi: 10.1016/0022-510x(86)90176-0. [DOI] [PubMed] [Google Scholar]
- 44.Abramov AY, Smulders-Srinivasan TK, Kirby DM, et al. Mechanism of neurodegeneration of neurons with mitochondrial DNA mutations. Brain. 2010;133:797–807. doi: 10.1093/brain/awq015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Hertz L, Peng L, Dienel GA. Energy metabolism in astrocytes: high rate of oxidative metabolism and spatiotemporal dependence on glycolysis/glycogenolysis. J Cereb Blood Flow Metab. 2007;27:219–49. doi: 10.1038/sj.jcbfm.9600343. [DOI] [PubMed] [Google Scholar]
- 46.Chinnery PF, Howell N, Lightowlers RN, Turnbull DM. Molecular pathology of MELAS and MERRF. The relationship between mutation load and clinical phenotypes. Brain. 1997;120:1713–21. doi: 10.1093/brain/120.10.1713. [DOI] [PubMed] [Google Scholar]
- 47.Zhou L, Chomyn A, Attardi G, Miller CA. Myoclonic epilepsy and ragged red fibers (MERRF) syndrome: selective vulnerability of CNS neurons does not correlate with the level of mitochondrial tRNAlys mutation in individual neuronal isolates. J Neurosci. 1997;17:7746–53. doi: 10.1523/JNEUROSCI.17-20-07746.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.