Abstract
We demonstrate a localization microscopy analysis method that is able to extract results in live cells using standard fluorescent proteins and Xenon arc lamp illumination. Our Bayesian analysis of blinking and bleaching (3B analysis) method models the entire dataset simultaneously as being generated by a number of fluorophores which may or may not be emitting light at any given time. The resulting technique allows many overlapping fluorophores in each frame, and unifies the analysis of localization from blinking and bleaching events. By modeling the entire dataset we are able to use each reappearance of a fluorophore to improve the localization accuracy. The high performance of this technique allows us to reveal the nanoscale dynamics of podosome formation and dissociation with a resolution of 50 nm on a four second timescale.
Introduction
High resolution optical microscopy methods have pushed the resolution of a microscope system below the Abbe limit by utilizing a non-linear sample response to illumination light 1. This is often achieved by switching fluorophores between a dark state and a bright state 2. Stimulated emission depletion (STED) can be used to shrink the effective size of the scanning beam in a confocal system 3; saturated structured illumination (SSIM) can extract information hidden in the Moiré patterns produced when a grating is projected onto the sample 4; and localization microscopy techniques, such as photoactivatable localization microscopy (PALM) 5 and stochastic optical reconstruction microscopy (STORM) 6, build up a high resolution image from the localized positions of many single fluorophores. The application of these techniques to live cell imaging promises dynamic information on complex protein structures with nanoscale resolution 7,8,9,10. The ideal technique would be experimentally simple, fast, and the switching between fluorophore states would not damage the sample. However, several factors still limit the utility of high resolution microscopy techniques for live cell imaging applications.
STED requires a specialized microscope and a complex alignment procedure. On live cells STED has achieved 28 frames per second at 62 nm resolution with low photon numbers, over a field of view of 2.5 × 1.8 μm 9. As STED is a scanning technique, increasing the field of view decreases the frame rate. SSIM is not limited in this way since it is a widefield technique; however, it also requires a specialized microscope, can be prone to deconvolution artifacts 4, and biological high resolution images have not been published. Non-saturated structured illumination (SIM) has been carried out at frame rates up to 11 Hz in live cells 11, but can at most provide around 100 nm resolution.
Current localization techniques require images in which fluorescence emission from individual fluorophores does not overlap. This limits the number of fluorophores which can be localized in a given frame and thus limits the timescale at which useful images of complex structures can be built up. Achieving the non-overlapping fluorophore emission necessary for conventional localization microscopy analysis requires switching a large fraction of probes into a non-emitting state. This is done by either activating small populations of fluorophores (usually using near-ultraviolet light) while imaging with longer wavelengths 5,6,12 or by keeping a large fraction of probes in a non-emitting state using relatively high-intensity illumination (kW/cm2) at a single imaging wavelength under suitable chemical conditions 13,14. These wavelengths and intensities have been shown to damage live samples15. Current localization techniques have, however, allowed localization-based imaging of simple structures in live cells at a temporal resolution of 10 s without near-ultraviolet activation light 7 and a temporal resolution of 0.5 s with activation 10.
Another method which utilises fluorescence blinking to boost the achievable resolution is called super-resolution optical fluctuation imaging (SOFI), which has been shown to improve images of samples labelled with quantum dots and organic dyes 16,17. This method assumes that the blinking of neighbouring fluorophores is uncorrelated, and uses the temporal correlation between pixels of the image to sharpen the effective point spread function. Analysing 1000-frame datasets of biological samples leads to a 1.4 to 2-fold improvement in resolution 17. There are two other image analysis methods which are able to deal with relatively dense localisation data, DAOSTORM 18 and simultaneous multiple emitter fitting 19. These methods can analyse localization microscopy data with some overlapping fluorophores in each image, but do not use fluorophore reappearance to improve localisation.
Podosomes are cytoskeletal structures which are associated with cell adhesion, migration and degradation of the extracellular matrix 20,21. They consist of an actin core surrounded by a ring of integrin-associated proteins such as talin and vinculin. This ring was thought to be roughly round, and the podosomes were thought to form and dissociate over a period of about 5–10 minutes. In widefield images the process of formation and dissociation appears as a fading out of the structure 22, though we have also seen cases where the podosome appears to slowly unwind.
Here we present a Bayesian localization microscopy method that allows localization data to be extracted from widefield images of live cells labeled with a standard fluorescent protein. Our method allows the use of data from overlapping fluorophores, and the use of information from bleaching events, blinking events, and changes due to fluorophores being added or removed by the cell. We carried out high frame rate imaging on a standard widefield microscope with xenon arc lamp illumination. We used a Bayesian technique to model the resulting high-density fluorophore image data as arising from a number of fluorophores, each of which can emit light, but which do not necessarily emit light in every frame. By modeling the whole dataset as arising from a number of fluorophores, we could use all fluorophore reappearances, even in non-adjacent frames, and thus use all the photons collected from a fluorophore to improve determination of its position. This Bayesian analysis of bleaching and blinking data (3B analysis) method allowed us to perform localization microscopy with a spatial resolution of 50 nm and a temporal resolution of 4 s on podosomes in living cells expressing an mCherry fusion of a truncated talin construct. Note that here resolution is taken to be the smallest distance at which two talin strands can be separated, and the localization as the apparent full width at half maximum (FWHM) of a strand of talin. We found that podosomes frequently exhibit a polygonal structure and are highly dynamic over a timescale of tens of seconds.
Results
Standard fluorescent proteins have been shown to blink and bleach in a live-cell environment, even under illumination from standard non-laser light sources. We developed the 3B analysis method to model an entire dataset consisting of a sequence of high frame rate images generated from large numbers of fluorescent proteins or other fluorophores undergoing blinking and bleaching processes. We modeled the entire dataset with a factorial Hidden Markov Model (FHMM) 23. In this Bayesian technique, the state of a system at a certain time is determined by a transition matrix and the state of the system at the previous timepoint. The state of the system is statistically linked to the observation (the link is not direct as the process of observation itself is subject to noise). In order to render the calculation both accurate enough and algorithmically tractable, we hybridized two Hidden Markov Model (HMM) inference methods, the forward algorithm 24 and Markov Chain Monte Carlo sampling (MCMC) (see methods section). We calculate many samples of the model parameters and build up a probability map of the positions of fluorophores taken from these samples. Each calculation of a particular set of model parameters uses samples of the state taken using MCMC and generates a set of fluorophore positions using a maximum a posteriori (MAP) calculation.
Note that if there are multiple models which fit the data well, then it is very likely that we will have samples of both models. At the end, we marginalize out the fluorophore state, to give a distribution of fluorophore positions. Any ambiguity will cause the final model to be blurred out, so it will be unlikely to report falsely high resolution where there is ambiguity (where resolution is defined as the ability to image two fluorophores or line structures as separate). While there exists only one model which corresponds to the real world, given the data it is not possible to decide which is the correct one and averaging a multitude of almost correct models seems an effective way to represent the underlying structure. A subtle yet important point is that the analysis is integrating over the parameters (state sequences and continuous parameters). The output is not a single model, it is an ensemble of models for different samplings of the state sequences. For each model in the ensemble, the optimised positions of the fluorophores are displayed, but the positions are also integrated out when making the decisions about the number of fluorophores in the image. So the outcome of the analysis does not include a specific state sequence; the outcome has integrated over a sampling of different possible state sequences.
Correlative results
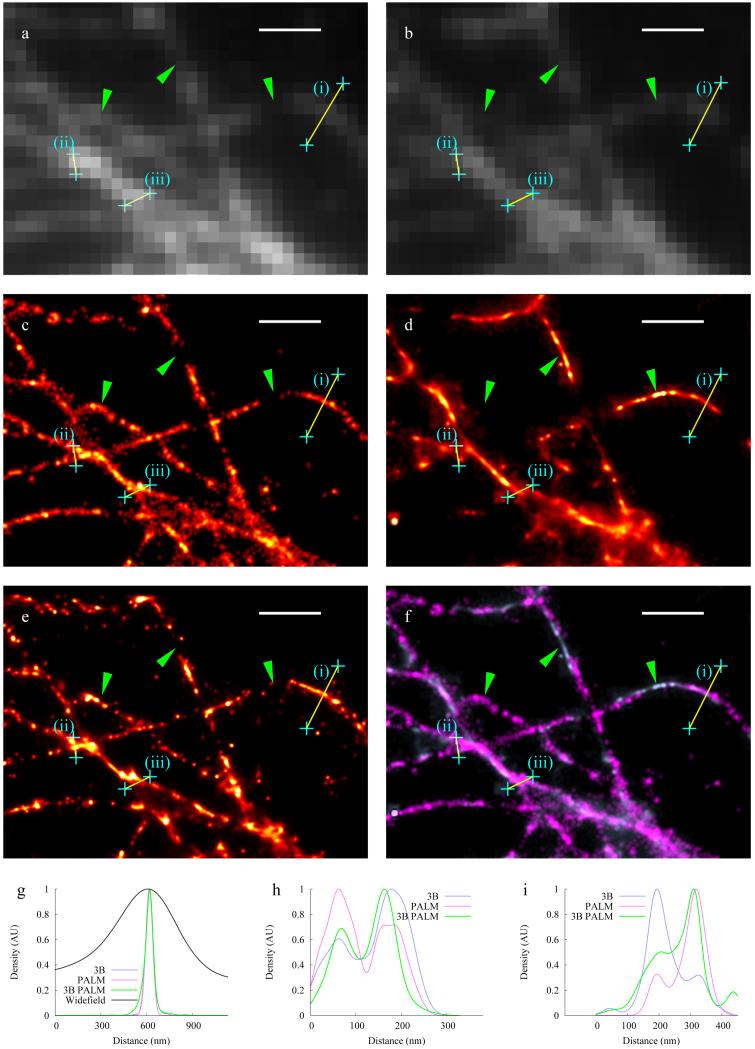
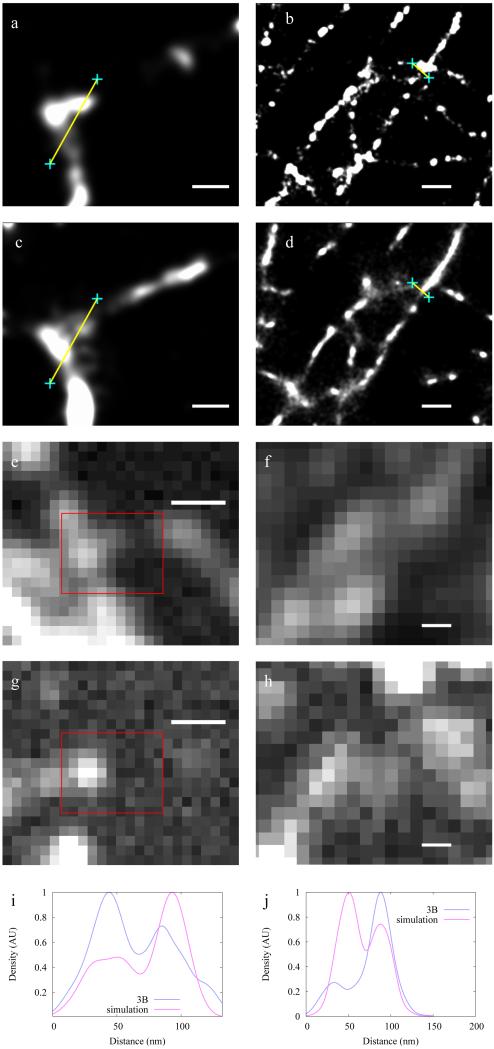
In order to verify that 3B analysis produces a result which reflects the underlying structure when used on experimental (rather than simulated) data, correlative experiments were carried out. We chose to label tubulin, since the network of tubulin strands gives rise to strands crossing at many different distances and angles, allowing the resolution to be assessed by determining when the strands can be distinguished. Tubulin was labeled with PAGFP for PALM imaging and mCherry for Bayesian localization imaging using 3B analysis (Figure 1). Widefield images created by averaging the frames in the two datasets (Figs. 1a,b) show that not all features visible in one dataset are visible in another since the incorporation efficiencies of mCherry and PA-GFP into the microtubule vary. The incorporation of these proteins into the microtubule is at a low level so some areas have more mCherry-Tubulin molecules and other areas more PA-GFP-tubulin. Some areas that show particularly clear widefield discrepancy are indicated with green arrows.
Figure 1.
Correlative measurements using PALM imaging and Bayesian localization imaging on tubulin. (a, b) Widefield images created by averaging all frames of the PALM image dataset of tubulin-PAGFP (a), and the Bayesian localization image dataset of tubulin-mCherry (b). (c-d) Super-resolution images generated by analyzing the PALM tubulin-PAGFP dataset from a using standard PALM analysis 5 (c) and analyzing the tubulin-mCherry dataset from b using 3B analysis (d). (e) Image generated from the 3B tubulin-mCherry dataset from a using 3B analysis. (f) Overlay of c and e. Green arrows indicate regions with differences in apparent structure that arise from labeling differences. Linescans corresponding to lines (i-iii) are shown in panels (g-i) respectively, with 3B data shown in blue, PALM data shown in pink, 3B PALM shown in green, and widefield data shown in black. Scale bars are 1 μm.
Using 3B analysis we created a probability map by building up many MAP positions obtained from different samplings of state sequences (Fig. 1d). Due to the high level of fluorophore overlap in all frames, this mCherry data is not analyzable with standard localization microscopy analysis techniques. With the exception of the labeling discrepancies noted above, the 3B analysis results (Fig. 1d) show good agreement with the PALM data (Fig. 1c), with features separated by distances down to 100 nm visible in both data sets, and where features are present in both datasets they agree to high resolution as shown in the overlay of the PALM and 3B images (Fig. 1f). Additionally, applying 3B analysis to the PALM dataset yields very similar structure to the original PALM analysis (Fig. 1e).
Experiments on fixed podosomes
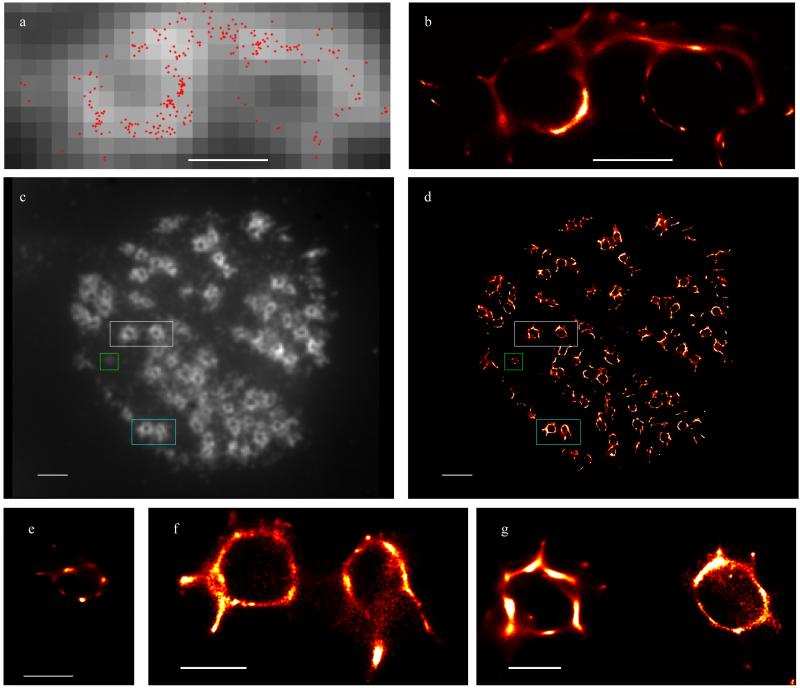
Vinculin in fixed podosome samples was immunolabeled with Alexa 488 and samples were mounted in PBS with 100 mM mercaptoethanol added as a reducing agent to induce blinking 14. (Supplementary Fig. 1). The sample was illuminated using a laser at 488nm with a nominal power of 1 kW/cm2. Series of 300 images were collected, with collection taking around 6 s. A video of the raw data is shown in Supplementary Video 1. Note that there are many overlapping emitting fluorophores in the majority of the frames. This prevents us from using the standard thresholding and fitting image analysis techniques normally used to reconstruct PALM or STORM images. A typical widefield image obtained by averaging over all 300 images is shown as the background in Figure 2a. An example of MAP positions created from one sampling of 3B analysis is shown in red in Figure 2a (many MAP positions are built up to create the final probability map).
Figure 2.
3B analysis of vinculin in fixed cells with podosomes, labeled with Alexa 488. (a) An example maximum likelihood estimate for one set of MCMC samples superimposed on a widefield image created by averaging all 300 images. (b) Probability map created by building up MAP positions created using different sets of MCMC samples. Scale bars correspond to 500 nm. (c,d) Whole cell (widefield and 3B analysis respectively) with areas displayed in (e),(f) and (g) identified by green, turquoise and white rectangles respectively. Scale bars correspond to (a,b) 500 nm, (c,d) 2 μm, (e,f,g) 500 nm.
The apparent thickness of the vinculin strands varies between 6nm and 60nm, with the variation probably arising from variation in the number of fluorophore reappearances in different areas, the number of photons detected in one appearance, and variations in the distribution of vinculin. The structure of the podosome is clearly seen to be geometrical, and this is in agreement with recent high resolution microscopy observations using STED and SIM (at about 120 nm resolution) (M. Walde, J.M., G.E.J., R.H., S.C., Manuscript in preparation). The higher resolution reveals a small structure joining the two podosomes.
We applied 3B analysis to an entire cell, which is displayed in Figure 2c-d. This revealed a small number of podosomes of diameter 300 nm, well below the standard diameter of around 500 nm (see for example Figure 2e), which only appear as a blob of brightness in the widefield image. As in SIM and STED studies of podosomes (M. Walde, J.M., G.E.J., R.H., S.C., Manuscript in preparation), we also see that vinculin strands tend to bind at angles of 120–130° (see Supplementary Fig. 2).
Experiments on podosomes in living cells
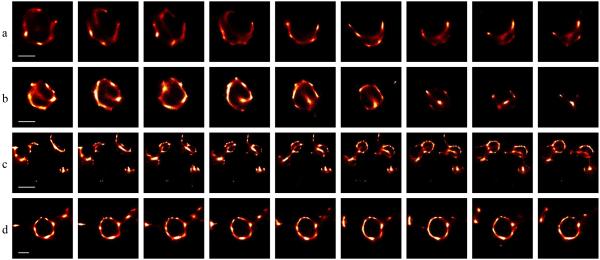
THP-1 cells stably expressing an mCherry-tagged, truncated talin construct (amino acids 1974-2541) were generated by lentiviral gene-transduction. This talin mutant comprises the second integrin binding domain of talin and has previously been shown to be an excellent marker of podosome rings in living cells 25. We illuminated live samples (maintained at 37°C) with a mercury arc lamp supplying a nominal power (measured before the objective) of 12 W/cm2 at the sample in the wavelength range 615-687 nm and acquired a series of 5000 images at 50 fps. An example of the data is shown in Supplementary Video 2. 3B analysis was applied to sequences of 200 frames, corresponding to an acquisition time of 4 s. In Figure 3, data is shown for selected timepoints from the reconstructed image sequence. Complete reconstructed datasets are shown in Supplementary Videos 3-6, with the timeshift between video frames being 50 frames or 0.5 seconds (though the temporal resolution is still 4 s).
Figure 3.
Podosomes with an mCherry-tagged truncated talin construct forming and dissociating in a live cell. (a, b) A podosome being dissociated (scalebar 400 nm). (c) Podosomes being formed (scalebar 1 μm). (d) A steady state podosome (scalebar 400 nm). Each reconstructed frame uses 200 frames (4 s) and frames are spaced 600 frames (12 s) apart. Videos of the podosomes shown in (a)–(d) are given in Supplementary Videos 3-6 respectively.
We determined the localization precision from the FWHM of a linescan perpendicular to a talin strand. This gave values as low as 18 nm. We defined the resolution as the distance at which two talin strands could be separated, as measured by a linescan. This was determined to be 50 nm (Supplementary Fig. 3).
Note that movement of the fluorophores will cause the analysis to produce an image which is smeared in the direction of movement. This limits the resolution achievable if structures are moving by distances larger than the resolution of the system, on timescales smaller than the acquisition time for a single reconstructed high resolution image. Podosomes were chosen as a suitable test system for imaging as they form and dissociate over a period of several minutes, and most do not appear to move around the cell during this time (if movement is observed, it is generally restricted to a few hundred nm).
Podosomes can clearly be observed undergoing assembly and two different modes of breakdown. In the first mode of podosome breakdown, the podosome exhibits a small break and then one end of the break gradually retracts, producing an ‘unwinding’ effect (Fig. 3a). This retraction appears to be associated with the formation of small (250 nm diameter) struts in the region of the cell where the podosome is being dissociated. In the second mode of podosome dissociation, struts (around 450 nm in length) repeatedly form across the podosome, drawing the talin in to a central point until it has all been removed (Fig. 3b). We also observe a podosome formation in which the strut appears to have an important function, with the podosome nucleating from a strut and then expanding on either side of it (Fig. 3c). Once the podosome has formed and the strut has served its purpose, it appears to be broken down. Some podosomes exhibit no apparent changes in the widefield image, and are relatively stable at the nanoscale (Fig. 3d).
More complex structures can also be observed, composed of a number of joined podosomes and struts. Imaging of a motile cell reveals highly dynamic behavior of these complex structures, with podosome structures changing on a timescale of tens of seconds. Figure 4a reveals one example of such behavior, where two groups of podosomes become joined after each group extends a strut. Where the two struts join, a miniature podosome-like structure forms, and the two groups of podosomes are pulled further together. Similar behaviour is seen across the whole cell (Supplementary Video 7).
Figure 4.
Dissociation and formation of groups of podosomes in a motile cell. (a,b) Dissociation and formation of linked podosomes. (c) Separated podosomes joining together. A video containing (a)–(c) along with the rest of the cell is given in Supplementary Video 7. Each reconstructed frame uses 200 frames (4 seconds) and frames are spaced 1000 frames (20 seconds) apart. Scalebars are 800nm.
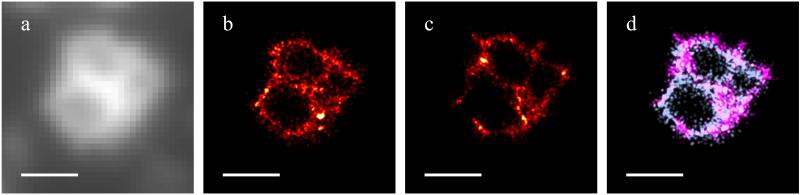
In order to determine whether the truncated talin construct which we used for live cell imaging experiments is representative of the structure of the podosome protein ring, we performed two-color measurements in fixed cells. We fixed cells expressing the mCherry tagged truncated talin construct and immunolabeled vinculin with Alexa 488. The same embedding conditions as for other fixed cell experiments were used. The truncated talin construct and the vinculin showed extremely similar structures (Fig. 5d). The vinculin seemed to be localised slightly more to the periphery of the ring, and the talin more to the center, while the short strands at the edge were more visible in the vinculin. This hints that the localisation of different proteins in the ring are subtly different and demonstrates that the 3B analysis method can be used to build up a map of the spatial organisation of different podosome components.
Figure 5.
3B analysis of fixed cell data to determine colocalisation of vinculin and the truncated talin construct in podosomes. (a) Widefield image of vinculin labelled with Alexa-488. (b,c) The individual 3B analysis images shown in glowscale for talin (b) and vinculin (c). (d) Superposition of images from 3B analysis showing the truncated talin construct (in cyan) and vinculin 3B data (in magenta). Scalebars are 1 μm.
Simulations
To further validate the Bayesian localization imaging results from our 3B analysis method we analyzed simulated datasets. Bayesian fitting methods use wide priors over a large number of parameters to fit real-world data, which tends to have narrow, unknown distributions over these parameters. Simulations created using the fitting distributions may provide extremely poor results so we created simulated datasets by using blinking sequences and fluorophore positions from the PALM correlative data. Each simulated frame was created using the fluorophore positions from sixteen PALM frames (Fig. 6). Average separation of nearest neighbour fluorophores was 112 nm, considerably smaller than the point spread function FWHM (270 nm), leading to a large degree of fluorophore overlap in simulated frames (Fig. 6g and h).
Figure 6.
Simulations demonstrating the performance of the 3B analysis method. Ground truth simulated image data (a,b), 3B analysis reconstructions (c,d) and linescans (i,j) show the 3B analysis method achieving good reproduction of the structure and 50 nm resolution. For the simulations (a,b), the simulated widefield image (e,f) and a typical frame (g,h) is given. Note that (a,c) correspond to the boxed region in (e,g). Scalebars are 50 nm in (a,c) and 200 nm in all other panels.
The simulations showed that the 3B analysis method achieves 50 nm resolution at an intersection of two strands (Figs. 6i and j). The method gave the correct structure but does not pick up every fluorophore. Artificial thinning and thickening of the structures was observed in different areas but the magnitude of all of these effects was below 20 nm. In all cases the intensity between close spots was somewhat enhanced, making the spots appear more ‘connected’ than in the PALM data. This suppresses the resolution along a line of fluorophores compared to that perpendicular to a line of fluorophores. We quantified the resolution perpendicular to lines of fluorophores in all our experiments.
Discussion
The 3B analysis method removes a number of barriers to getting good localization information that exist with other approaches. The experiments are easy to implement: live cell experiments use fluorescent proteins, widefield microscope and arc lamp illumination that are standard in most cell biology labs. With this equipment, it is possible to achieve a 50 nm resolution with data from only a few seconds of acquisition, and it is possible to image for extended periods. The software which we use to perform the analysis in this paper is available in Supplementary Software; updated versions can be obtained from http://3bmicroscopy.com.
The computational effort of our method is linear in the number of fluorophores × the number of pixels. For a cell such as that shown in Figure 2, the data was modeled as arising from 10,000 fluorophores, and 200 sets of MCMC samples were taken to build up the probability map. Note that our method has a natural mechanism for trading off temporal and spatial resolution; analyzing more frames simultaneously raises the spatial resolution but lowers the temporal resolution. Comparing our method to SOFI, both methods can deal with images with overlapping fluorophores, but SOFI requires more data than 3B analysis and delivers a more limited resolution improvement. In 3B analysis, it is possible that structures can be artificially sharpened by including models with fewer fluorophores than the data, but in simulations we have found these effects to be considerably below the resolution limit.
Rather than revealing an improved resolution picture of an apparently smooth process in podosomes, achieving high resolution revealed an entirely new level of complexity. Podosomes were previously thought to smoothly form and dissociate over a period of several minutes 22. In contrast, our results indicate that podosomes are highly dynamic structures. It appears that smaller ring type structures down to 230 nm play an important part in podosome dynamics. Rather than strands simply being parts of partially grown podosomes, our results indicate that they may play a role in seeding new areas of podosomes. In another form, as struts spanning across a podosome, they appear to be associated with podosome formation and dissociation.
Our use of standard fluorescent proteins opens up the use of high spatial and temporal resolution microscopy to a whole new range of samples. Many of them, like podosomes, may turn out to be complex all the way down to the nanoscale.
Supplementary Material
Acknowledgments
We acknowledge helpful discussions with A. Fraser, F. Viola, P. Fox-Roberts, O. Mandula and J. Sleep. We thank M. Kielhorn for assistance in aligning the optical system and K. Glocer for critical reading of the manuscript. We thank M. Parsons (King’s College London) for providing the template plasmid. We acknowledge support from the EU Seventh Framework Programme (FP7 Project GA 215597 (S.C. and R.H.), EU FP7 Project ITN 237946 T3Net (G.E.J.)), the Wellcome Trust (S.C., J.M., G.E.J.), the Medical Research Council (G.E.J.), and the Royal Society (S.C.).
Appendix
Methods
Sample preparation for correlative measurements
B16-F1 cells were seeded on coverslips coated with 25 μg/mL laminin as previously described 26. Cells were then co-transfected with tubulin-PAGFP and tubulin-mCherry with Fugene 6 (Roche) following manufacturers recommendation. Streaming time-lapse images were acquired with an Olympus IX71 TIRF microscope using a 60x 1.45 NA objective (Olympus, Center Valley, PA) and fluorescence emission was detected with an EM-CCD camera (Andor Technology, DV887ECS-BV). PAGFP constructs were imaged with 100 ms integration times (488 nm laser power was 400-800 μW going into the microscope). mCherry constructs were imaged with 50 ms integration times (561 nm laser power was 1 mW going into the microscope). PALM datasets were 5000 frames and 3B datasets were 1000 frames, and both were corrected for drift. For PALM data, single molecules were fitted with theoretical Gaussians and PALM images reconstructed as previously described 5.
Sample preparation for podosome observations
To observe podosomes, the THP1 cell line (Human acute monocytic leukemia cell line) was used, which can be stimulated to differentiate into macrophages. Podosome formation was induced in these cells by seeding them on fibronectin coated cover glasses in the presence of the cytokine TGFβ1 (1 ng/ml). Vinculin staining was conducted using VN-1 anti-vinculin mouse monoclonal antibody (Sigma) conjugated to Alexa 488 anti-mouse secondary antibody. Coverslips were mounted in PBS containing 100 mM mercaptoethanol to induce blinking at a suitable rate for dSTORM imaging 14.
Lentiviral-mediated gene transduction of THP-1
Polymerase-chain reaction (PCR) was used to amplify cDNA encoding residues 1975-2541 of human talin from a template plasmid. The resulting sequence was cloned via the ZERO-BLUNT vector (Invitrogen) into the MCS of the pLNT/Sffv-mCherry-MCS vector generating the mCherry-talin(1975-2541) lentiviral expression construct. VSV-G pseudotyped lentivirus encoding mCherry-talin(1975-2541) were packaged in 293T by transient transfection of cells with the pΔ8.91 and pMD.G accessory plasmids along with the pLNT/Sffv transfer vector encoding the talin construct. Supernatants containing lentivirus were harvested 48 h-post transfection, 0.45 μm-filtered and stored at −80°C. THP-1 were transduced with lentivirus by incubation with lentiviral supernatants for 24 h, subsequently washed by sequential centrifugation and resuspension steps, and finally left for an additional three days to express the fusion protein. 24 h prior to live-cell imaging experiments THP-1 were seeded at a density of 2.5×106 cell/ ml on fibronectin-coated (10 μg/ml) glass coverslips in the presence of 1 ng/ml TGFβ to induce cell attachment and podosome formation. For all imaging experiments coverslips with adherent cells were mounted onto purpose-built glass viewing chambers.
For two colour experiments cell containing the truncated mcherry-talin construct were fixed and stained as described in the previous section.
Microscopy for podosome experiments
For fixed cell experiments a widefield Zeiss Axiovert 200M microscope was used with an oil immersion objective (63×, numerical aperture (NA) 1.4; Zeiss) and a 2.5× Optovar. Illumination was provided by an Argon ion laser (Innova 90 coherent), emitting at 488 nm. Images were recorded using a Cascade II EMCCD camera (Photometrics), with square pixels and a pixel pitch of 16 μm (each recorded pixel corresponds to 102 nm in real space). Frame rate varied between 50 and 60 frames/s.
For live cell experiments a widefield Olympus IX81 was used with an oil immersion objective (100×, NA 1.4; Olympus). Illumination was provided by a Sutter Lambda Ls Xe arc lamp coupled with a liquid light guide, with a Comar GFP-RFP filter set (for RFP, excitation at 537.5-592.5 nm, emission at 615-687 nm, and dichroic at 590-700 nm). Images were recorded using a cascade II EMCCD camera (properties as above). For Figure 3, no Optovar was used, meaning each recorded pixel corresponds to 160 nm in real space. For Figure 4 a 1.6 × Optovar was used, meaning each recorded pixel corresponds to 100 nm on the sample. The drift in these experiments was assessed by monitoring the drift of bead samples over time. The beads were imaged using the same frame rate, and for the same amount of time, as a live cell experiment. The drift was found to be within the localization error, with the mean of the localization position varying by up to 10 nm over 5000 images (acquired over 98 s). We therefore ignore drift effects in our analysis, since the expected drift over the 200 images which we use to reconstruct an image is expected to be 0.4 nm.
Simulations
Simulations were created using fluorophore positions from 4800 out of the 5000 PALM frames, with each simulated frame created from 16 PALM frames, giving 300 simulated frames. The simulated frames were created from two groups of PALM frames such that simulated frame 0 consists of PALM frames 0–7 and 2400–2407, simulated frame 1 consists of PALM frames 8–15 and 2408–2415, and so on. This prevents later frames from becoming unrealistically sparse. The FWHM of the simulated PSF (point spread function) was set to 1.56 pixels which corresponds to a FWHM of 270 nm at 86 nm per pixel (this halves the number of nm per pixel compared to the original PALM dataset, but since the positions of the fluorophores are set relative to pixels, it also decreases the distance between fluorophores by a factor of two). This created datasets with overlapping fluorophores. Simulated images were created using Gaussian shaped spots of average brightness 1200 photons on a background level of average brightness 600 photons, with Poisson noise. The photon counts and therefore the signal to noise ratio was set using photon counts from the background and isolated fluorophores in the fixed cell dataset.
Analysis
The image series was modeled using a Factorial Hidden Markov Model (FHMM) 23 as arising from a number of fluorophores. Each fluorophore is modeled using a Markov Model which has three possible states, emitting (light), not emitting, and bleached. The fluorophore can transfer between the emitting and not emitting states, and can also transfer from the not emitting to the bleached state. Once in the bleached state the fluorophore cannot leave it. The state transition diagram for fluorophores is given in Supplementary Figure 4. From our estimates of the lifetimes and transition probabilities associated with the energy levels 27,14, and the frame time of around 0.02 s, we calculated estimates for all the model probabilities in a given frame. We obtained values of P1=0.16. P2=0.84, P3=P4=0.495, P5=0.01. To calculate these, typical values of the lifetimes and various transition probabilities were taken. The singlet state lifetime was taken to be 10−7s and a fluorophore was taken to be 105 times more likely to transition from the ground to the singlet state than from the ground to the triplet state. The frame rate of the camera was taken to be between 50 Hz and 60 Hz i.e. each frame takes 1.8×10−2 s. So each frame is 18,000 singlet lifetimes, in each of which the fluorophore has a probability of 0.99999 of returning to the singlet state. The values of P3 and P4 were taken with a variety of typical triplet lifetimes (10−3 to 10−2) assuming a monoexponential decay. This led to a values of P4 between 0.1 and 0.8. The value of 0.495 was chosen as being reasonably central to this spread, given the large uncertainties in the input values.
The same state transition probabilities were used for fitting both fixed cell and live cell datasets, as only a broadly correct prior is needed for this type of modelling. The structure observed in the reconstructed image was not found to vary to an extent which altered the observed structure if the state transition probabilities were varied within physically realistic values. We assume that neighboring fluorophore states are statistically completely independent. Note that our results are only weakly dependent on the priors.
Without blinking or bleaching, the method would have the same limits as deconvolution. To enhance the blinking, a switching probe with better dynamics could be employed.
We calculate the relative probability that a fluorophore is present compared to the null hypothesis that the data arises from noise. The model evidence for each hypothesis can be calculated by integrating out over state sequences (blinking and bleaching state in each frame) using the forward algorithm 24, and integrating out over continuous variables using Laplace’s approximation 28. However, the forward algorithm calculation is exponential in the number of fluorophores. An alternative approach is to take a statistical sample of state sequences; but this does not provide sufficiently accurate results. We therefore use a hybrid of the forward algorithm and a state sampling technique. A detailed description of the algorithm is given in Supplementary Note.
The algorithm is then run on user selected areas. The user must specify the pixel size (which is used to calculate the predicted point spread function size), and the starting number of fluorophores in the area The final result of the algorithm is a density map of the positions of fluorophores yielded. Further details on the parameters and reconstruction algorithm are given in Supplementary Note.
In order to analyse a region of around 1.5×1.5 μm in size, 6 hours processing on a single core i7 (3.33 GHz) is required. Larger areas can be analysed, the time required scales with the area. In order to analyse large areas the analysis is broken down into a number of small areas. To analyse large images or video data, a cluster computer is required.
References
- 1.Heintzmann R, Ficz G. Breaking the resolution limit in light microscopy. Methods in Cell Biology. 2007;81:561–580. doi: 10.1016/S0091-679X(06)81026-5. [DOI] [PubMed] [Google Scholar]
- 2.Hell SW. Microscopy and its focal switch. Nat. Methods. 2009;6:24–32. doi: 10.1038/nmeth.1291. [DOI] [PubMed] [Google Scholar]
- 3.Klar TA, Jakobs S, Dyba M, Egner A, Hell SW. Fluorescence microscopy with diffraction resolution barrier broken by stimulated emission. Proc. Natl. Acad. Sci. 2000;97:8206–8210. doi: 10.1073/pnas.97.15.8206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Heintzmann R, Jovin TM, Cremer C. Saturated patterned excitation microscopy - a concept for optical resolution improvement. J. Opt. Soc. Am. A. 2002;19:1599–1609. doi: 10.1364/josaa.19.001599. [DOI] [PubMed] [Google Scholar]
- 5.Betzig E, et al. Imaging intracellular fluorescent proteins at nanometer resolution. Science. 2006;313:1642–1645. doi: 10.1126/science.1127344. [DOI] [PubMed] [Google Scholar]
- 6.Rust MJ, Bates M, Zhuang X. Sub-diffraction-limit imaging by stochastic optical reconstruction microscopy (STORM) Nat. Methods. 2006;3:793–796. doi: 10.1038/nmeth929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wombacher R, et al. Live-cell super-resolution imaging with trimethoprim conjugates. Nat. Methods. 2010;7:717–719. doi: 10.1038/nmeth.1489. [DOI] [PubMed] [Google Scholar]
- 8.Shroff H, Galbraith CG, Galbraith JA, Betzig E. Live-cell photoactivated localization microscopy of nanoscale adhesion dynamics. Nat. Methods. 2008;5:417–423. doi: 10.1038/nmeth.1202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Westphal V, et al. Video-rate far-field optical nanoscopy dissects synaptic vesicle movement. Science. 2008;320:246–249. doi: 10.1126/science.1154228. [DOI] [PubMed] [Google Scholar]
- 10.Jones SA, Shim S-H, He J, Zhuang X. Fast, three-dimensional super-resolution imaging of live cells. Nature Methods. 2011;8:499–505. doi: 10.1038/nmeth.1605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kner P, Chhun BB, Griffis ER, Winoto L, Gustafsson MGL. Super-resolution video microscopy of live cells by structured illumination. Nat. Methods. 2009;6:339–342. doi: 10.1038/nmeth.1324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Heilemann M, Dedecker P, Hofkens J, Sauer M. Photoswitches: Key molecules for subdiffraction-resolution fluorescence imaging and molecular quantification. Laser and Photonics Reviews. 2009;3:180–202. [Google Scholar]
- 13.Zhuang X. Nano-imaging with STORM. Nature Photonics. 2009;3:365–367. doi: 10.1038/nphoton.2009.101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Heilemann M, van de Linde S, Mukherjee A, Sauer M. Super-resolution imaging with small organic fluorophores. Angew. Chem. Int. Ed. 2009;48:6903–6908. doi: 10.1002/anie.200902073. [DOI] [PubMed] [Google Scholar]
- 15.Lippincott-Schwartz J, Manley S. Putting super-resolution fluorescence microscopy to work. Nat. Methods. 2009;6:21–23. doi: 10.1038/nmeth.f.233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Dertinger T, Colyera R, Iyera G, Weissa S, Enderleind J. Fast, background-free, 3D super-resolution opticalfluctuation imaging (SOFI) Proc. Natl. Acad. Sci. 2009;106:22287–22292. doi: 10.1073/pnas.0907866106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Dertinger T, Heilemann M, Vogel R, Sauer M, Weiss S. Superresolution optical fluctuation imaging with organic dyes. Angewandte Chemie. 2010;122:9631–9633. doi: 10.1002/anie.201004138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Holden S, Uphoff S, Kapanidis A. DaoSTORM: an algorithm for high-density super-resolution microscopy. Nature Methods. 2011;8:279–280. doi: 10.1038/nmeth0411-279. [DOI] [PubMed] [Google Scholar]
- 19.Huang F, Schwartz SL, Byars JM, Lidke KA. Simultaneous multiple-emitter fitting for single molecule super-resolution imaging. Biomedical Optics Express. 2011;2:1377–1393. doi: 10.1364/BOE.2.001377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Linder S, Aepfelbacher M. Podosomes: adhesion hot-spots of invasive cells. Trends Cell Biol. 2003;13:376–385. doi: 10.1016/s0962-8924(03)00128-4. [DOI] [PubMed] [Google Scholar]
- 21.Linder S, Kopp P. Podosomes at a glance. J. Cell Sci. 2005;118:2079–2082. doi: 10.1242/jcs.02390. [DOI] [PubMed] [Google Scholar]
- 22.Monypenny J, et al. Role of WASP in cell polarity and podosome dynamics of myeloid cells. Eur. J. Cell Biol. 2011;90:198–204. doi: 10.1016/j.ejcb.2010.05.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ghahramani Z, Jordan MI. Factorial hidden markov models. Mach. Learn. 1997;29(2–3):245–273. [Google Scholar]
- 24.Rabiner LR. A tutorial on hidden markov models and selected applications in speech recognition. Proceedings of the IEEE. 1989;77(2):257–286. [Google Scholar]
- 25.Wiesner C, Faix J, Himmel M, Bentzien F, Linder S. KIF5B and KIF3A/KIF3B kinesins drive MT1-MMP surface exposure, CD44 shedding, and extracellular matrix degradation in primary macrophages. Blood. 2010;116:1559–1569. doi: 10.1182/blood-2009-12-257089. [DOI] [PubMed] [Google Scholar]
- 26.Burnette DT, et al. A role for actin arcs in the leading-edge advance of migrating cells. Nat. Cell Bio. 2011;13:371–382. doi: 10.1038/ncb2205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Xie XS. Optical studies of single molecules at room temperature. Annu. Rev. Phys. Chem. 1998;49:441–480. doi: 10.1146/annurev.physchem.49.1.441. [DOI] [PubMed] [Google Scholar]
- 28.MacKay DJC. Information theory, inference, and learning algorithms. Cambridge University Press; 2003. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.