Abstract
KirBac channels are prokaryotic homologs of mammalian inwardly-rectifying (Kir) potassium channels and recent crystal structures of both Kir and KirBac channels have provided a major insight into their unique structural architecture. However, all of the available structures are closed at the helix bundle-crossing and therefore the structural mechanisms that control opening of their primary activation gate remain unknown. In this study, we engineered the inner pore-lining helix (TM2) of KirBac3.1 to trap the bundle-crossing in an apparently open conformation, and determined the crystal structure of this mutant channel to 3.05 Å resolution. Contrary to previous speculation, this novel structure suggests a mechanistic model in which rotational ‘twist’ of the cytoplasmic domain is coupled to opening of the bundle-crossing gate via a network of inter- and intra-subunit interactions that involves the TM2 C-linker, slide-helix, G-loop and the CD-loop.
INTRODUCTION
Inwardly rectifying potassium (Kir) channels regulate membrane electrical excitability and K+ transport in a wide range of cell types. Their activity controls many diverse processes such as heart rate, vascular tone, insulin secretion and salt/fluid balance, and an increasing number of disease states are now known to be directly associated with abnormal Kir channel function1. Like many other types of ion channels, the activity of Kir channels is controlled by dynamic conformational changes which regulate the flow of K+ ions through the central pore of the channel1,2. This process is known as ‘gating’ and understanding the molecular mechanism of these dynamic changes in Kir channel structure is critical to our understanding of how these channels function in both health and disease.
The primary gating mechanism in most classes of K+ channel is thought to involve an iris-like motion of the pore-lining inner transmembrane helices which constrict the permeation pathway at the helix bundle-crossing3. In voltage-gated K+ channels, this gating motion is physically coupled to the movement of a transmembrane voltage-sensor, whilst in Kir channels conformational changes within the large cytoplasmic domains (CTD) are thought to control movement of the bundle-crossing gate3-6. This provides a mechanism for the action of ligands such as G-proteins, ATP, H+ and PIP2 which all bind to these intracellular domains to regulate Kir channel function1,2. Other structural domains, such as the selectivity filter and the cytoplasmic ‘G-loop’ have also been proposed to act as physical gates in Kir channels7-10. However, their role is secondary to that of the bundle-crossing gate which is regarded as the primary activation gate in all major classes of K+ channels, and which must always be in an open conformation for the channel to be conductive.
Full understanding of these gating processes requires high-resolution structural information of a Kir channel trapped in multiple gating conformations, and over the last few years several different Kir channel structures have been solved by X-ray crystallography; these include a eukaryotic Kir channel11 as well as a number of homologous prokaryotic ‘KirBac’ channels4 and chimeras between them12, and a series of structures of KirBac3.1 with the CTD in multiple orientations13. However, in all of these Kir and KirBac channel structures the bundle-crossing gate is ‘closed’ and despite their enormous value, they therefore provide limited insight into the structural changes which must occur to allow these channels to open. This apparent preference for the bundle-crossing gate to crystallise in the closed state strongly suggests that it must represent a low energy state of the channel, and that novel strategies are therefore required to stabilise the open state of the channel.
In this study we now report the X-ray crystal structure of an open-state KirBac channel at 3.05 Å resolution. This structure not only illustrates how the bundle-crossing opens, but more importantly, suggests how opening of the primary activation gate may be physically coupled to conformational changes in the CTD.
RESULTS
Overall structure of a mutant KirBac3.1 channel
We have previously isolated a number of mutations in KirBac3.1 which directly increase channel activity14. Many of these mutations introduced charged amino acids into TM2 suggesting that steric/charge repulsion between these side-chains may assist channel opening at the bundle crossing. We therefore reasoned that such mutants might have the ability to stabilise the open state of the channel. One such gating mutant is S129R which is located very close to the bundle crossing and in most other Kir/KirBac channels this residue is a glycine15,16.
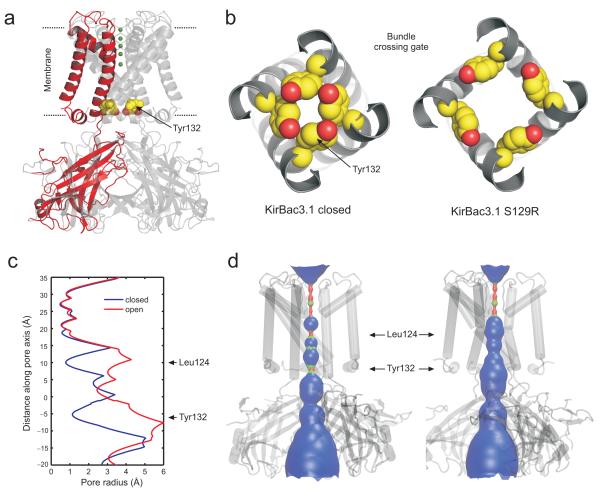
We have previously expressed and purified this mutant channel and shown that it is functionally active14. Crystallisation of this mutant was therefore pursued and crystals belonging to P4212 spacegroup were successfully grown and diffracted up to 3.05 Å resolution. The structure was solved by molecular replacement and a final model containing 280 out of 301 residues of the construct was refined to an R-factor of 22.0% with an RFree of 25.9% with 99.6% of the residues in the final model lying in the most favourable and additionally allowed regions of the Ramachandran plot. The full data collection and refinement statistics are shown in Table 1. Visualisation of this structure and comparison to KirBac3.1 in the closed state revealed major structural changes in the transmembrane helices and also at the bundle-crossing (Fig. 1a-b).
Table 1. Data collection and refinement statistics.
| KirBac3.1(S129R) | |
|---|---|
| Data collection | |
| Space group | P 4 21 2 |
| Cell dimensions | |
| a=b, c (Å) | 106.24 89.80 |
| α=β=γ (°) | 90.00 |
| Resolution (Å) | 106.24 -3.05 (3.21-3.05) * |
| R merge | 0.262 (0.945) |
| I / σI | 5.5 (2.0) |
| Completeness (%) | 99.9 (99.8) |
| Redundancy | 6.8 (6.9) |
| Refinement | |
| Resolution (Å) | 89.80-3.05 |
| No. reflections | 10244 |
| Rwork / Rfree (%) | 22.04 / 25.89 |
| No. atoms | 2218 |
| Protein | 2191 |
| Ligand/ion | 7 |
| Water | 20 |
| B-factors | 54.39 |
| Protein | 54.67 |
| Ligand/ion | 40.77 |
| Water | 29.34 |
| R.m.s. deviations | |
| Bond lengths (Å) | 0.010 |
| Bond angles (°) | 1.08 |
Values in parentheses are for highest-resolution shell.
Figure 1.
Structure of the KirBac3.1 S129R mutant in an apparently open conformation. (a) Overall structure of S129R mutant with one subunit highlighted in red for clarity. Ions within the selectivity filter are shown in green and residue Tyr132 which forms the primary gate at the bundle-crossing is shown as CPK spheres in all four subunits. (b) Bottom up view of the bundle-crossing gate in an example of a closed state KirBac3.1 (PDB 2WLJ) and the S129R mutant channel. Tyr132 side chains are shown as CPK spheres. (c) Pore radius profile of a closed state KirBac3.1 (PDB 2WLJ) (Closed, blue) and the S129R mutant where the Arg129 side chain has been replaced by a serine (Open, red). (d) The pore-lining surface and structure of the open and closed KirBac3.1 channels with the position of the major constrictions Tyr132 and Leu124 marked by arrows.
KirBac3.1 trapped in an apparently open conformation
In all of the previous KirBac3.1 structures the bundle-crossing gates are fully closed13. This is defined by a tight constriction of the pore at Tyr132 in TM2 which occludes the conductive pathway and acts as the gate at this position2,4,17. However, in the structure presented here, the S129R mutation has trapped the bundle-crossing in an apparently open conformation (Fig. 1). In the closed state structures of KirBac3.1, the Cα-Cα distances at Tyr132 range from 12.2 Å to 13.5 Å 13,whereas in the S129R mutant this distance is increased to 17.1 Å (Fig. 1b). The engineered mutant Arg129 side-chains in TM2 create an additional constriction within the pore (Supplementary Fig. 1). However, because Arg129 is a serine in wild-type KirBac3.1, and a glycine in all other Kir/KirBac channels18,19, we therefore analysed the pore-radius of this new structure with a serine at position 129. This reveals that the bundle-crossing gate would be open and conductive when the channel is in this conformation and that there is free access to the selectivity filter from the intracellular side of the channel (Fig. 1c-d). The S129R mutation therefore appears to have stabilised the TM-helices and the bundle-crossing gate in a conformation that mimics the wild-type channel in an open state.
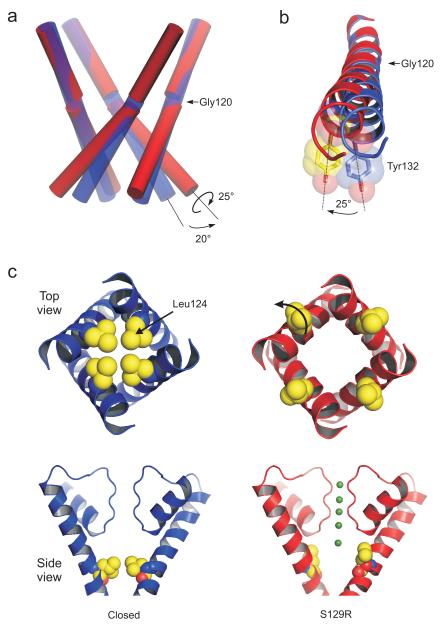
Bending of TM2 at the ‘glycine hinge’
The global rearrangements propagating from these changes at the bundle-crossing are in agreement with earlier modelling studies of TM2 motions in KirBac3.120 as well as the gating motions observed in the open state crystal structures of truncated and full-length KcsA21-23. Compared to the closed state, the lower part of TM2 bends by up to 20° at a highly conserved ‘glycine hinge’ residue18,19 (Gly120 in KirBac3.1) whilst the upper part remains relatively rigid. However, in addition to this bending or ‘kinking’ of TM2, we also observe a rotation, or twisting of the lower section of TM2 along its helical axis by approximately 25° (Fig. 2a,b) and there is good overlay of the TM2 helices of KirBac3.1-S129R with the open 17 Å crystal structure of KcsA (Supplementary Fig. 2)21,22. No twisting is observed in the outer helix (TM1) during opening of the bundle-crossing; however, TM1 is displaced outward and follows the movement of TM2 (see Supplementary Movie 1).
Figure 2.
Bending of the inner TM2 helices during channel opening (a) Conformational changes of the inner helices from the closed state (blue cylinders) to the conformation (red cylinders) seen in the S129R mutant. The lower section of TM2 kinks by up to 20° at the conserved glycine hinge (Gly120) and also rotates around its helical axis by 25° when viewed from below. (b) The clockwise rotation of TM2 is shown viewed from below. Overlay of TM2 helices from the S129R structure (red) and a closed state Kir3.1 (blue, PDB 2WLJ). Tyr132 is shown as a transparent CPK sphere. (c) Opening of the inner cavity constriction formed by Leu124. Top and side views are shown on the S129R structure (red) and a closed state KirBac3.1 (blue, PDB 2WLJ). The position of ions within the filter of the S129R structure is shown in green. The rotation of TM2 moves Leu124 away from the pore. Leu124 is equivalent to the rectification control site in TM2 of eukaryotic Kir channels.
Rotation of TM2 opens a secondary gate within the pore
This rotational movement of TM2 is important because, in addition to the bundle-crossing gate (Tyr132), a secondary constriction exists at Leu124 in the closed state structures (Fig. 1c) 13,17. Rotation of TM2 induced by opening of the bundle-crossing means that Leu124 is now twisted away from the central cavity, thus increasing the effective pore diameter at this position from less than 2 Å to >8 Å (Fig. 2c). Poisson-Boltzmann electrostatic calculations reveal a dramatic reduction in the energetic barrier to K+ permeation at this point (Supplementary Fig. 3) producing an electrostatic profile that becomes progressively more favourable for the movement of K+ towards the selectivity filter in this open conformation. Movement of this side-chain upon channel opening is also of particular interest because Leu124 in KirBac3.1 is equivalent to the ‘TM2 rectification control site’ in eukaryotic Kir channels (e.g. Asp172 in Kir2.1)24.
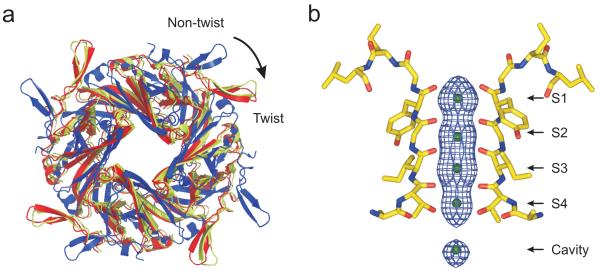
A twisted yet conductive conformation of KirBac3.1
In the recent series of KirBac3.1 structures reported by Clarke et al.13, the CTD displays significant rotational diversity with respect to the pore i.e. the ‘twist and non-twist’ conformations. Comparison of these structures to the S129R structure show that it is rotated and in a ‘twist’ configuration (Fig. 3a). It was also suggested that the twist/non-twist status of the different closed-state structures correlates with the conductive status of the selectivity filter, with only the ‘non-twist’ conformations being conductive13. However, despite being in the twist conformation, analysis of the ion occupancy in the S129R structure shows that all four K+ binding sites (S1-S4) are occupied (Fig. 3b) which is clearly representative of a conductive filter conformation21,22,25,26. This new conformation is therefore in direct contrast to the gating model proposed by Clarke et al.13
Figure 3.
S129R is in a twisted yet conductive conformation. (a) The intracellular assemblies of a non-twist (blue, PDB 2WLJ) and twist (yellow, PDB 2X6C) conformation13 are viewed from the top relative to their superimposed pore domains (not shown). The relative position of the S129R CTD is also overlaid (red) showing that it is in the ‘twist’ conformation. (b) Fo-Fc OMIT map of electron density in the selectivity filter of the S129R mutant channels contoured at ~3σ showing clear density in all four binding sites, as well as the cavity site.
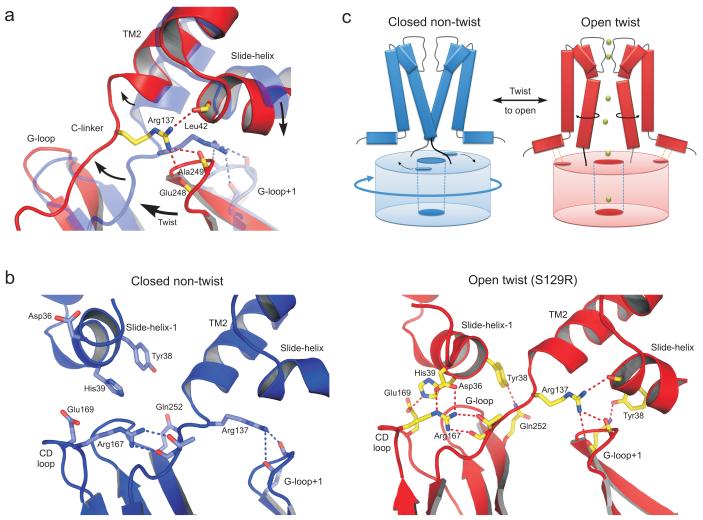
The C-linker couples pore opening to movement of the CTD
A common mechanistic theme in other major classes of tetrameric cation channels is that allosteric changes in domains attached to the TM helices are mechanically coupled to opening and closing of the bundle crossing gate3. A rotational movement or ‘twist’ of the CTD could therefore conceivably promote opening of the bundle-crossing gate4,13. However, this would require tight physical coupling of the TM-helices to the CTD. Therefore, in this new structure it is interesting to note that the ‘C-linker’ between TM2 and the CTD is displaced relative to its position in the ‘non-twist’ conformation (Fig. 4a). This causes the C-linker to become involved in a network of interactions that links the bundle crossing gate with the slide-helix as well as two loops within the CTD (the CD- and the G-loop) that have been implicated in the control of channel gating7,12,27,28. These interactions would stabilise the interface between the twisted CTD and the TM/Pore domain (TMD) in the open state (Fig. 4b).
Figure 4.
Interaction network between the CTD and the TMD. (a) The C-linker in the twist S129R structure (red) is displaced by up to 5Å compared to the closed non-twist structure (PDB 2WLJ, transparent blue). During twisting and opening Arg137 shifts its interaction with backbone carbonyls on the G-loop of the adjacent subunit (G-loop+1) to include an intrasubunit interaction with the slide-helix. The major motions are indicated by arrows. (b) Comparison of the network of interactions formed in the S129R twist open conformation (red) with those in a closed non-twist conformation (PDB 2WLJ). Movement of the slide helix and displacement of the C-linker allow an extensive network of inter and intra-subunit interactions to form (see also Supplementary Movie 2) (c) Cartoon of gating model proposed by the motions observed in the S129R structure. The twist conformation of the CTD pre-tensions the C-linker and also brings the slide-helix into register with the CD-loop (dot shown on CTD), thereby coupling rotational movement of the CTD to opening at the bundle-crossing gate.
One of the residues in this network is the highly-conserved Arg137 within the C-linker itself which provides a direct interaction between the C-linker and the G-loop of the adjacent subunit. A similar interaction is observed in some of the closed state structures13, but local rearrangements caused by opening of the bundle-crossing now displace the C-linker, allowing Arg137 to form an additional intra-subunit contact with a backbone carbonyl of Leu42 on the slide-helix (Fig. 4a).
This network is further expanded by interaction of the C-linker with a highly-conserved arginine on the CD-loop of the CTD (Arg167) thereby providing an additional connection to the slide-helix of the adjacent subunit (Fig. 4b). This connection can only occur in the twist conformations because, when compared to the non-twist structures (Fig. 4b), there is a downward movement of the slide-helix bringing Asp36 into direct contact with both the η- and ε-nitrogens of Arg167 on the CD-loop of the adjacent subunit. This also brings His39 into contact with Glu169 (Fig. 4b and Supplementary Fig. 4). Movement of the slide-helix and its interaction with the CD loop is seen in other twisted structures13, but in this new conformation this extended network is now directly coupled to the opening of the bundle-crossing gate.
Other novel interactions also form upon opening of the bundle-crossing gate. Notably, the highly conserved Tyr38 in the slide-helix now interacts with Gln252 in the G-loop (Fig 4b) and disruption of this interaction (with mutation Y38F) reduces the functional activity of KirBac3.1 consistent with its possible role in stabilisation of the open state (Supplementary Fig. 5). Interestingly, unlike the Asp36/Arg167 interaction which takes place on the outer surface of the slide-helix, the Tyr38/Gln252 interaction occurs on the inner side of the slide-helix and results in a direct intersubunit linkage between the slide-helix of one subunit and both the CD- and G-loops of the adjacent subunit.
DISCUSSION
In this study we present the first high-resolution structure of a KirBac channel with the bundle-crossing gate in an open conformation. This new structure provides a significant extension to the conformational landscape available for this class of dynamic proteins; it demonstrates that TM2 bends at a conserved glycine hinge, and that rotation of TM2 also contributes to opening of a secondary gate within the pore. Furthermore, opening of the bundle-crossing gate involves a network of interactions between the TMD and CTD which illustrate how rotational movement of the CTD may be coupled to Kir/KirBac channel gating.
It has been suggested that the selectivity filter plays an important role in Kir/KirBac channel gating, and indeed several of our recent studies including X-ray footprinting of KirBac3.1, as well as the identification of gating mutations, directly support this notion14,29. However, the filter can only act as gate if and when the bundle-crossing is open, and until now high-resolution structural information about how the primary activation gate opens has been unavailable. Engineering the bundle-crossing gate of KirBac3.1 into a conformation that mimics the open state has therefore allowed us, for the first time, to visualise a potential mechanism of channel opening.
The major structural changes which occur in TM2 as the bundle-crossing opens are consistent with those seen in other open state K+ channels and a similar rotation of TM2 has now been observed in the open-state structure of full-length KcsA23. Importantly, this rotation of TM2 also opens a secondary gate within the inner cavity of KirBac3.1 at Leu124. Although Leu124 will not act as a gate independently of the bundle crossing it may play an important role in ‘sealing’ the pore in the closed state as it is located just below the cavity ion binding site (Fig. 2c). Intriguingly, movement of Leu124 was also detected by our previous X-ray footprinting study of KirBac3.129 and a similar rotation of the equivalent residue in KcsA (Phe103) has recently been observed23. However, of perhaps greater significance is the fact Leu124 is equivalent to the rectification control site in TM2 of eukaryotic Kir channels (e.g. Asp172 in Kir2.124) where the presence of a negatively charged side chain influences the binding of Mg2+ and polyamines and thereby the degree of inward-rectification. Rotational movement of this side-chain during channel opening (Fig. 2) could therefore play a major role in defining how these classical pore blockers interact with this site in the open vs. closed states.
Whether this KirBac3.1 structure represents a ‘fully open’ conformation is not known. Much wider openings of the bundle-crossing (up to 32 Å) have been observed with truncated versions of KcsA 21,22. However, the presence of the C-terminal domain in KcsA appears to restrict these openings to about 21 Å (ref 23) and so the large KirBac CTD is also likely to impose structural constraints on the size of the maximal opening which can occur. Nevertheless, the S129R mutant is functionally active and therefore a wider dilation must be possible to overcome the artificial constriction formed by the mutant Arg129 side-chains (Supplementary Fig. 1). Interestingly, introduction of a negative charge at position 129 also activates KirBac3.1 (Supplementary Fig. 5), and similar charged mutations at the bundle-crossing have been shown to activate both KirBac and Kir channels14,30 suggesting that wider openings must be possible in these mutant channels.
It has previously been suggested that the rotational status of the CTD is directly linked to the conductive state of the selectivity filter, and that this does not require movement of the bundle-crossing gate. In particular, it was proposed that the twist configuration induces a non-conductive selectivity filter13. However, the novel structure we present here is in the twist configuration, yet it is both conductive within the filter and open at the bundle-crossing, thus directly contradicting the gating model proposed by Clarke et al13. At this stage it is not yet known whether any functional form of C-type inactivation occurs within the selectivity filter of KirBac3.1, but it is likely that that any allosteric coupling between the TM-helices and the selectivity filter will require a movement of the bundle crossing gate similar to that seen in the elegant crystallographic studies of KcsA21-23,31.
Instead, this new conformation suggests a model in which rotational movement of the CTD may be directly coupled to channel opening at the bundle-crossing gate, and that the twist configuration is prerequisite for this to occur. Likewise, although the G-loop has been proposed to act as an independent gate7,12 and is clearly an important regulator of Kir/KirBac channel activity8, its primary role may be to couple conformational changes in the CTD to the C-linker, and thereby opening of the bundle-crossing gate.
Importantly, this mechanism also provides a rationale for the action of a number of compounds which are likely to influence these interactions, e.g. cholesterol with the CD loop28, and also PIP2 with the C-linker and CTD32,33. In particular, it is interesting to note that in eukaryotic Kir channels, which are activated by PIP2, the C-linker contains an insertion of three additional charged residues (Supplementary Fig. 6) thought to interact with PIP2 and which would clearly influence this gating mechanism when they are present34. In support of this idea, a structure of chicken Kir2.2 has very recently been solved with PIP2 bound, and although this structure is also closed at the bundle-crossing, it appears to represent a ‘pre-open’ state where PIP2 interacts directly with the C-linker to ‘pre-tension’ this region in preparation for opening of the bundle-crossing35. Given the fundamentally conserved structural basis of K+ channel gating at the bundle crossing, it is perhaps no surprise that the C-linker appears to play such an important role in Kir/KirBac channel gating and is consistent with similar roles for this linker region in the gating of other tetrameric cation channels 36-39.
In conclusion, the gating model suggested by this new conformation is markedly different from that proposed previously13. Instead of being an obstacle to channel opening, we propose that the twist configuration is required for opening of the bundle-crossing gate by allowing a network of interactions to form between the TMD and CTD, and we present a simplified cartoon model summarising this (Fig. 4c, see also Supplementary Movie 2). Clearly further work will be required to determine the causality of the changes we observe in the structure of the bundle-crossing gate and the network of novel interactions that this produces. Likewise, the question of whether the bundle-crossing opens wider than the 17Å opening seen here will also be the subject of future studies. Nevertheless, this novel open-state structure of KirBac3.1 now provides an important extension to the available conformational landscape for this important class of ion channels.
Supplementary Material
ACKNOWLEDGEMENTS
We thank the staff at the I24 beamline at the Diamond Light Source. This work was supported by the BBSRC and the Wellcome Trust. R.DeZ was supported by a Marie Curie Intra-European Fellowship.
APPENDIX
METHODS
Protein expression and crystallisation
The S129R mutant was introduced using site-directed mutagenesis into a synthetic KirBac3.1 in pET30a where the open-reading frame had been codon-optimised for expression in E. coli. The protocol for expression and purification of this mutant channel was identical to that previously described for wild-type KirBac3.129, and following gel-filtration the triDM detergent was exchanged into 14 mM Hega-10 using vivaspin concentrators with a 100 kDa cutoff. Protein crystals were then grown using the sitting drop method in 10% (v/v) Glycerol, 90 mM Hepes 7.2, 20% (v/v) PEG400, 5% (w/v) PEG4k and 2.5% (w/v) PEG8k using ~6 mg ml−1 protein concentration in 1:1 protein-reservoir ratio. Crystals appeared after 3-4 days at 20°C and were cryo-cooled under liquid nitrogen prior to analysis.
Data collection and structure determination
The KirBac3.1 S129R crystals belong to P4212 spacegroup with cell dimensions of a=b=106.2 Å and c=89.8 Å. The asymmetric unit has a solvent content of 69.4% and contains one molecule. Data was collected at 100K using a Pilatus 6M detector at the I-24 beamline at the Diamond Light Source (Harwell, Oxfordshire) at a wavelength of λ= 0.9778 Å to a resolution of up to 3.05 Å. Dataset statistics are provided in Table 1. Data were processed with Mosflm/Scala (CCP4 program)40 and space group confirmed with Pointless41. 5% of the reflections were set aside in the free R set. Molecular replacement was performed with Phaser (CCP4 suite)42, using a search model derived from PDB ID 2X6C processed with Chainsaw (CCP4)40. The model was refined in real space interactively using Coot43 and refined using Buster-TNT44, which included a final round of translation, libration and screw-rotation (TLS) anisotropic refinement as implemented in Buster-TNT. The final model contains 280 residues, out of 301 (residues 12-26, 33-277, 281-300 inclusive); in addition, connectivity between residues 26 and 32 can be established at lower sigma threshold, but they were not included in the final model as it was not possible to build and refine residues in this region at full occupancy with certainty. The model also contains 20 solvent molecules and 7 ions. Ions are positioned on the four-fold axis and are therefore modelled with occupancies of less than 25%. The final model was validated using MolProbity45, and presented very good stereochemistry with over 99.6% of all residues in favoured and additionally allowed regions of the Ramachandran plot. The coordinates have been deposited in the PDB using code 3ZRS.
Structure analysis
The superimpositions of the individual domains were performed using LSQKAB (CCP4 Supported program) and analysed in Coot43. The ribbon diagrams and actual videos were made with PyMol (http://www.pymol.org/). Cavity analysis was carried out using HOLE46 and cylinder representations and cavity visualisations were performed with VMD47. The contribution to the electrostatic potential of a potassium ion along the pore axis of the channel was calculated using the Adaptive Poisson-Boltzmann Solver (APBS) software package48 with a methodology similar to the one previously applied to nanopores49. The potassium ion pathway and axis were defined using the program HOLE, which was also used to calculate pore radii. Charges and radii were assigned using the software PDB2PQR50, which was also used to add missing atoms of non-resolved side chains before calculating electrostatic energies. Energies were calculated using a NaCl bath with an ionic strength of 0.2 M at 298K. The protein (dielectric constant ε = 10) was embedded into a dielectric slab (ε = 2), mimicking the membrane environment with membrane thickness set to 45 Å. The pore itself has been excluded from the membrane and assigned a solvent dielectric (ε = 80). Supplementary movies were animated using PyMol. Starting structures were aligned along the transmembrane domain of 2WLJ (residues Trp46-Gly120) before interpolation. Intermediate structures were calculated using GROMACS451. The consensus secondary structure was modelled after the secondary structure of 3ZRS. Missing extracellular loops in 2X6C and the missing βL-M loop in 3ZRS were built using MODELLERv9.952.
Accession codes
The atomic coordinates of the KirBac3.1 S129R structure have been deposited in the Protein Data Bank under the accession code 3ZRS.
Footnotes
COMPETING FINANCIAL INTERESTS: The authors declare no competing financial interests.
AUTHOR CONTRIBUTIONS
S.J.T and C.V-B conceived and designed the research. R.DeZ and V.N.B. expressed and crystallized the mutant protein. V.N.B, R.DeZ and L.Z. collected the diffraction data. V.N.B and J.R.C.M. determined and refined the structure with contribution from R.DeZ. V.N.B, L.Z and M.R.S analyzed and interpreted the structure. L.Z. performed complementation studies. C.V-B, M.S.P.S and S.J.T supervised the project. V.N.B and S.J.T wrote the manuscript with the help of comments from all authors.
REFERENCES
- 1.Hibino H, et al. Inwardly rectifying potassium channels: their structure, function, and physiological roles. Physiol Rev. 2010;90:291–366. doi: 10.1152/physrev.00021.2009. [DOI] [PubMed] [Google Scholar]
- 2.Bichet D, Haass FA, Jan LY. Merging functional studies with structures of inward-rectifier K+ channels. Nat Rev Neurosci. 2003;4:957–67. doi: 10.1038/nrn1244. [DOI] [PubMed] [Google Scholar]
- 3.Swartz KJ. Towards a structural view of gating in potassium channels. Nat Rev Neurosci. 2004;5:905–16. doi: 10.1038/nrn1559. [DOI] [PubMed] [Google Scholar]
- 4.Kuo A, et al. Crystal structure of the potassium channel KirBac1.1 in the closed state. Science. 2003;300:1922–6. doi: 10.1126/science.1085028. [DOI] [PubMed] [Google Scholar]
- 5.Kuo A, Domene C, Johnson LN, Doyle DA, Venien-Bryan C. Two different conformational states of the KirBac3.1 potassium channel revealed by electron crystallography. Structure. 2005;13:1463–72. doi: 10.1016/j.str.2005.07.011. [DOI] [PubMed] [Google Scholar]
- 6.Domene C, Doyle DA, Venien-Bryan C. Modeling of an ion channel in its open conformation. Biophys J. 2005;89:L01–3. doi: 10.1529/biophysj.105.060855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Pegan S, et al. Cytoplasmic domain structures of Kir2.1 and Kir3.1 show sites for modulating gating and rectification. Nat Neurosci. 2005;8:279–87. doi: 10.1038/nn1411. [DOI] [PubMed] [Google Scholar]
- 8.Proks P, et al. A gating mutation at the internal mouth of the Kir6.2 pore is associated with DEND syndrome. EMBO Rep. 2005;6:470–5. doi: 10.1038/sj.embor.7400393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Proks P, Antcliff JF, Ashcroft FM. The ligand-sensitive gate of a potassium channel lies close to the selectivity filter. EMBO Rep. 2003;4:70–5. doi: 10.1038/sj.embor.embor708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Domene C, Klein ML, Branduardi D, Gervasio FL, Parrinello M. Conformational changes and gating at the selectivity filter of potassium channels. J Am Chem Soc. 2008;130:9474–80. doi: 10.1021/ja801792g. [DOI] [PubMed] [Google Scholar]
- 11.Tao X, Avalos JL, Chen J, MacKinnon R. Crystal structure of the eukaryotic strong inward-rectifier K+ channel Kir2.2 at 3.1 A resolution. Science. 2009;326:1668–74. doi: 10.1126/science.1180310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Nishida M, Cadene M, Chait BT, Mackinnon R. Crystal structure of a Kir3.1-prokaryotic Kir channel chimera. EMBO J. 2007;26:4005–15. doi: 10.1038/sj.emboj.7601828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Clarke OB, et al. Domain reorientation and rotation of an intracellular assembly regulate conduction in Kir potassium channels. Cell. 2010;141:1018–29. doi: 10.1016/j.cell.2010.05.003. [DOI] [PubMed] [Google Scholar]
- 14.Paynter JJ, et al. Functional complementation and genetic deletion studies of KirBac channels: activatory mutations highlight gating-sensitive domains. J Biol Chem. 2011;285:40754–61. doi: 10.1074/jbc.M110.175687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sun S, Gan JH, Paynter JJ, Tucker SJ. Cloning and functional characterization of a superfamily of microbial inwardly rectifying potassium channels. Physiol Genomics. 2006;26:1–7. doi: 10.1152/physiolgenomics.00026.2006. [DOI] [PubMed] [Google Scholar]
- 16.Shang L, Tucker SJ. Non-equivalent role of TM2 gating hinges in heteromeric Kir4.1/Kir5.1 potassium channels. Eur Biophys J. 2008;37:165–71. doi: 10.1007/s00249-007-0206-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Tai K, Haider S, Grottesi A, Sansom MS. Ion channel gates: comparative analysis of energy barriers. Eur Biophys J. 2009;38:347–54. doi: 10.1007/s00249-008-0377-x. [DOI] [PubMed] [Google Scholar]
- 18.Magidovich E, Yifrach O. Conserved gating hinge in ligand- and voltage-dependent K+ channels. Biochemistry. 2004;43:13242–7. doi: 10.1021/bi048377v. [DOI] [PubMed] [Google Scholar]
- 19.Jin T, et al. The βγ subunits of G proteins gate a K+ channel by pivoted bending of a transmembrane segment. Mol Cell. 2002;10:469–81. doi: 10.1016/s1097-2765(02)00659-7. [DOI] [PubMed] [Google Scholar]
- 20.Grottesi A, Domene C, Hall B, Sansom MS. Conformational dynamics of M2 helices in KirBac channels: helix flexibility in relation to gating via molecular dynamics simulations. Biochemistry. 2005;44:14586–94. doi: 10.1021/bi0510429. [DOI] [PubMed] [Google Scholar]
- 21.Cuello LG, et al. Structural basis for the coupling between activation and inactivation gates in K+ channels. Nature. 2010;466:272–5. doi: 10.1038/nature09136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Cuello LG, Jogini V, Cortes DM, Perozo E. Structural mechanism of C-type inactivation in K+ channels. Nature. 2010;466:203–8. doi: 10.1038/nature09153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Uysal S, et al. Mechanism of activation gating in the full-length KcsA K+ channel. Proc Natl Acad Sci USA. 2011;108:11896–9. doi: 10.1073/pnas.1105112108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Stanfield PR, et al. A single aspartate residue is involved in both intrinsic gating and blockage by Mg2+ of the inward rectifier, IRK1. J Physiol. 1994;478:1–6. doi: 10.1113/jphysiol.1994.sp020225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zhou Y, Morais-Cabral JH, Kaufman A, MacKinnon R. Chemistry of ion coordination and hydration revealed by a K+ channel-Fab complex at 2.0 Å resolution. Nature. 2001;414:43–8. doi: 10.1038/35102009. [DOI] [PubMed] [Google Scholar]
- 26.Berneche S, Roux B. Energetics of ion conduction through the K+ channel. Nature. 2001;414:73–7. doi: 10.1038/35102067. [DOI] [PubMed] [Google Scholar]
- 27.Singh DK, Rosenhouse-Dantsker A, Nichols CG, Enkvetchakul D, Levitan I. Direct regulation of prokaryotic Kir channel by cholesterol. J Biol Chem. 2009;284:30727–36. doi: 10.1074/jbc.M109.011221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Rosenhouse-Dantsker A, Leal-Pinto E, Logothetis DE, Levitan I. Comparative analysis of cholesterol sensitivity of Kir channels: role of the CD loop. Channels (Austin) 2010;4:63–6. doi: 10.4161/chan.4.1.10366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Gupta S, et al. Conformational changes during the gating of a potassium channel revealed by structural mass spectrometry. Structure. 2010;18:839–46. doi: 10.1016/j.str.2010.04.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Khurana A, et al. Forced gating motions by a substituted titratable side chain at the bundle crossing of a potassium channel. J Biol Chem. 2011;286:36686–93. doi: 10.1074/jbc.M111.249110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Cordero-Morales JF, et al. Molecular determinants of gating at the potassium-channel selectivity filter. Nat Struct Mol Biol. 2006;13:311–8. doi: 10.1038/nsmb1069. [DOI] [PubMed] [Google Scholar]
- 32.D’Avanzo N, Cheng WW, Wang S, Enkvetchakul D, Nichols CG. Lipids driving protein structure? Evolutionary adaptations in Kir channels. Channels (Austin) 2010;4:139–41. doi: 10.4161/chan.4.3.12129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Tucker SJ, Baukrowitz T. How highly charged anionic lipids bind and regulate ion channels. J Gen Physiol. 2008;131:431–8. doi: 10.1085/jgp.200709936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Leal-Pinto E, et al. Gating of a G protein-sensitive mammalian Kir3.1 prokaryotic Kir channel chimera in planar lipid bilayers. J Biol Chem. 2010;285:39790–800. doi: 10.1074/jbc.M110.151373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hansen SB, Tao X, MacKinnon R. Structural basis of PIP2 activation of the classical inward rectifier K+ channel Kir2.2. Nature. 2011;477:495–8. doi: 10.1038/nature10370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Taraska JW, Zagotta WN. Structural dynamics in the gating ring of cyclic nucleotide-gated ion channels. Nat Struct Mol Biol. 2007;14:854–60. doi: 10.1038/nsmb1281. [DOI] [PubMed] [Google Scholar]
- 37.Schumacher MA, Rivard AF, Bachinger HP, Adelman JP. Structure of the gating domain of a Ca2+-activated K+ channel complexed with Ca2+/calmodulin. Nature. 2001;410:1120–4. doi: 10.1038/35074145. [DOI] [PubMed] [Google Scholar]
- 38.Long SB, Campbell EB, Mackinnon R. Voltage sensor of Kv1.2: structural basis of electromechanical coupling. Science. 2005;309:903–8. doi: 10.1126/science.1116270. [DOI] [PubMed] [Google Scholar]
- 39.Enkvetchakul D, Jeliazkova I, Bhattacharyya J, Nichols CG. Control of inward rectifier K channel activity by lipid tethering of cytoplasmic domains. J Gen Physiol. 2007;130:329–34. doi: 10.1085/jgp.200709764. [DOI] [PMC free article] [PubMed] [Google Scholar]
Additional references for methods section
- 40.Collaborative Computational Project, Number 4 The CCP4 Suite: Programs for Protein Crystallography. Acta Cryst D. 1994;50:760–763. doi: 10.1107/S0907444994003112. [DOI] [PubMed] [Google Scholar]
- 41.Evans PR. Scaling and assessment of data quality. Acta Cryst D. 2006;62:82–82. doi: 10.1107/S0907444905036693. [DOI] [PubMed] [Google Scholar]
- 42.McCoy AJ, et al. Phaser crystallographic software. J Appl Cryst. 2007;40:658–674. doi: 10.1107/S0021889807021206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Emsley P, Lohkamp B, Scott WG, Cowtan K. Features and development of Coot. Acta Cryst D. 2010;66:486–501. doi: 10.1107/S0907444910007493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Blanc E, et al. Refinement of severely incomplete structures with maximum likelihood in BUSTER-TNT. Acta Cryst D. 2004;60:2210–2221. doi: 10.1107/S0907444904016427. [DOI] [PubMed] [Google Scholar]
- 45.Chen VB, et al. MolProbity: all-atom structure validation for macromolecular crystallography. Acta Cryst D. 2010;66:12–21. doi: 10.1107/S0907444909042073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Smart OS, Wang X, Wallace BA, Sansom MS. HOLE: a program for the analysis of the pore dimensions of ion channel structural models. J Mol Graph. 1996;14:354–60. 376. doi: 10.1016/s0263-7855(97)00009-x. [DOI] [PubMed] [Google Scholar]
- 47.Humphrey W, Dalke A, Schulten K. VMD - Visual Molecular Dynamics. J. Mol. Graphics. 1996;14:33–38. doi: 10.1016/0263-7855(96)00018-5. [DOI] [PubMed] [Google Scholar]
- 48.Holst M, M, Saied F. Numerical solution of the nonlinear Poisson-Boltzmann equation: Developing more robust and efficient methods. J Comput Chem. 1995;16:337–364. [Google Scholar]
- 49.Beckstein O, Tai K, Sansom MS. Not ions alone: Barriers to ion permeation in nanopores and channels. J Am Chem Soc. 2004;126:14694–14695. doi: 10.1021/ja045271e. [DOI] [PubMed] [Google Scholar]
- 50.Dolinsky TJ, et al. PDB2PQR: expanding and upgrading automated preparation of biomolecular structures for molecular simulations. Nucleic Acids Res. 2007;35:522–5. doi: 10.1093/nar/gkm276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Van der Spoel D, et al. GROMACS: Fast, flexible and free. J Comput. Chem. 2005;26:1701–1718. doi: 10.1002/jcc.20291. [DOI] [PubMed] [Google Scholar]
- 52.Sali A, A, Blundell TL. Comparative protein modelling by satisfaction of spatial restraints. J Mol Biol. 1993;234:779–815. doi: 10.1006/jmbi.1993.1626. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.