Summary
Many bacteria conditionally express proteinaceous organelles referred to here as microcompartments (Fig. 1). These microcompartments are thought to be involved in a least seven different metabolic processes and the number is growing. Microcompartments are very large and structurally sophisticated. They are usually about 100–150 nm in cross section and consist of 10,000–20,000 polypeptides of 10–20 types. Their unifying feature is a solid shell constructed from proteins having bacterial microcompartment (BMC) domains. In the examples that have been studied, the microcompartment shell encases sequentially acting metabolic enzymes that catalyze a reaction sequence having a toxic or volatile intermediate product. It is thought that the shell of the microcompartment confines such intermediates, thereby enhancing metabolic efficiency and/or protecting cytoplasmic components. Mechanistically, however, this creates a paradox. How do microcompartments allow enzyme substrates, products and cofactors to pass while confining metabolic intermediates in the absence of a selectively permeable membrane? We suggest that the answer to this paradox may have broad implications with respect to our understanding of the fundamental properties of biological protein sheets including microcompartment shells, S-layers and viral capsids.
Introduction
Recent studies indicate that proteinaceous microcompartments are widely used by Bacteria to optimize particular metabolic processes.(1–3) As yet, such compartments have not been found in the Archaea or Eukarya, although new types may await discovery. The microcompartments that have been studied consist of a solid protein shell that encapsulates sequential metabolic enzymes.(4–6) Studies indicate that the general role of the protein shell is to sequester toxic or volatile metabolic intermediates; however, it must also allow enzyme substrates, products and cofactors to pass.(1,7) Current models for microcompartment function propose that a selectively permeable shell and a highly defined architecture mediates the necessary molecular routing.(5,7) This review summarizes current knowledge of the structure, function, genetics, regulation, molecular principles and ecological aspects of bacterial microcompartments focusing on carboxysomes, which are involved in CO2 fixation, and on the microcompartments involved in B12-dependent 1,2-propanediol degradation. In addition, we suggest that studies of microcompartments are focusing attention in a way that will lead to new insights into the functional properties of biological protein sheets.
Carboxysomes
Discovery
Carboxysomes were the first bacterial microcompartments to be discovered. They were observed more than 50 years ago as polyhedral bodies in the cytoplasm of cyanobacteria.(8) Initially, carboxysomes were mistaken for viruses, but further work showed that they play a key role in enhancing autotrophic carbon fixation via the Calvin cycle, and that they are widely distributed among chemoautotrophs and cyanobacteria.(3,7,9,10) Carboxysomes are very large and sophisticated multi-protein complexes. They are about 80–150 nm in cross section and are bounded by a 3–4 nm thick protein shell (Fig. 1). Their mass is about 300 MDa and they are composed of several thousand polypeptides of 10–15 different types.(11,12) There is no indication of associated lipids. The lumen or interior of the carboxysome contains the sequential metabolic enzymes carbonic anhydrase (CA) and ribulose bisphosphate carboxylase monooxygenase (RuBisCO).(6,13) CA converts HCO3− to CO2 within the carboxysome, then RuBisCO converts CO2 and ribulose bisphosphate (RuBP) to two molecules of 3-phosphoglycerate (3-PGA). The shell of the carboxysome is proposed to retain CO2 in the immediate vicinity of RuBisCO by acting as a diffusion barrier.(14,15) In vivo, the carboxysome works in conjunction with C1 transport systems as part of a bacterial CO2-concentrating mechanism (CCM) that is used to increase the level of CO2 in the local vicinity of RuBisCO thereby enhancing carbon fixation and autotrophic growth.(7)
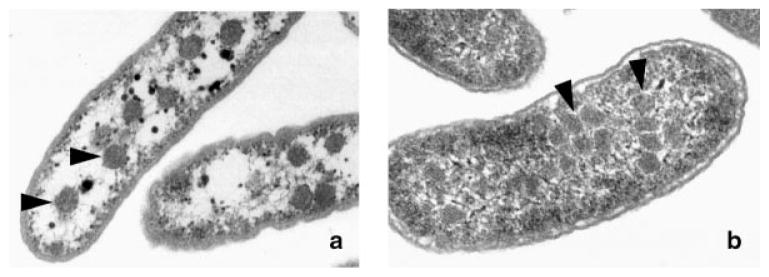
Figure 1.
Electron micrograph of bacterial microcompartments: a: The carboxysomes of H. neapolitanus; b: The organelles formed during growth of Salmonella enterica on 1,2-propanediol. Triangles point to the microcompartments.
Carbon dioxide concentrating mechanisms (CCMs)
CCMs are common in organisms that use the Calvin cycle of autotrophic CO2 fixation including higher plants and bacteria. CCMs improve the efficiency of RuBisCO which is the rate-limiting enzyme of the Calvin cycle. Kcat values for RuBisCO are relatively low ranging from about 1–13 sec−1 depending on the organism.(16) In addition, RuBisCO often uses O2 in place of CO2 in a nonproductive process known as photorespiration which can drain away up to 50% of the carbon fixed by the Calvin cycle. Several types of CCMs are known. The carboxysome is part of a bacterial CCM that appears to be absent from Eukarya and Archaea with possible rare exceptions.(7,17,18) The bacterial CCM begins with active transport processes that concentrate HCO3− within the cellular cytoplasm (Fig. 2A).(7) Within this compartment, HCO3− does not reach equilibrium with CO2 due to the absence of cytoplasmic CA.(19) A carboxysome-associated CA converts HCO3− to CO2 and releases it within the carboxysome. The protein shell of the carboxysome acts as a barrier to CO2 diffusion. This allows CO2 to accumulate proximal to RuBisCO with reduced diffusive loss. Elevated CO2 both increases the rate of CO2 fixation by RuBisCO and suppresses photorespiration. Optimal efficiency requires that CA activity and CO2 fixation are matched.(14,15)
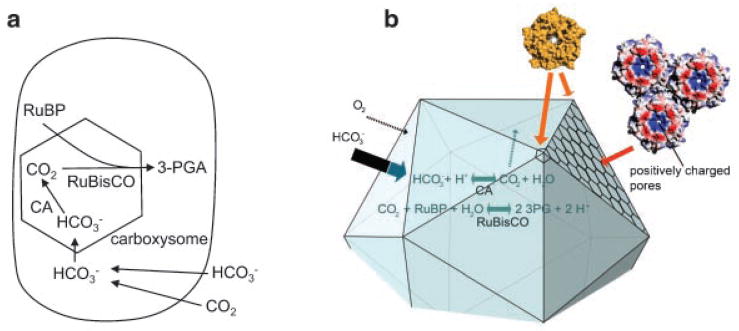
Figure 2.
The bacterial carbon dioxide concentrating mechanism. a: The bacterial CCM starts with transport of inorganic carbon into the cell as HCO3−. Equilibrium between HCO3− and CO2 is not reached due to a lack of carbonic anhydrase in the cytoplasm. HCO3− is converted to CO2 by carboxysomal carbonic anhydrase within the microcompartment lumen. The protein shell of the carboxysome impedes CO2 diffusion. Consequently, CO2 can accumulate in the immediate vicinity of RuBisCO while diffusive loss of CO2 through the cell membrane is minimized. Elevated CO2 proximal to RuBisCO increases carbon fixation and suppresses photorespiration (a nonproductive process in which O2 replaces CO2 as a substrate for RuBisCO). b: A preliminary atomic model of the carboxysome shell based on crystal structures of the component shell proteins.(12) The positively charged pores through the sheet of hexagonal proteins are indicated. These have been postulated to enhance the diffusion of negatively charged molecules such as bicarbonate (thick arrow) across the shell, compared to uncharged molecules such as CO2 and O2 (thin arrows).
The idea that the carboxysome is part of a bacterial CCM was predicted by mathematical models(14,15,20) and is now well-supported experimentally.(7) Genetic investigations showed that mutants of Synechococcus, Synechocystis and Halothiobacillus unable to form carboxysomes were incapable of autotrophic growth at ambient CO2 levels (0.03%) but were able to grow with elevated CO2, (1–5%).(21–25) This phenotype, which is referred to as a high-CO2-requiring (HCR) phenotype, connects carboxysomes to the efficiency of autotrophic CO2 fixation in vivo. A telling property of carboxysome mutants is that their RuBisCO has normal activity and kinetic parameters in vitro.(26,27) This argues against the simplistic idea that the primary function of carboxysomes is to modulate the kinetic properties RuBisCO through direct protein–protein interactions. It has also been shown that localization of CA to the carboxysome is essential for efficient CO2 fixation as expected for a CCM.(13,28) Mutants defective in carboxysomal CA have an HCR phenotype,(29,30) and ectopic expression of human CA in the cytoplasm of Synechococcus causes massive loss of CO2 as well as an HCR phenotype.(19) Moreover, recent in vitro studies support the idea that the shell of the carboxysome acts as a barrier to CO2 diffusion.(31) Carboxysomes were purified from wild-type H. neapolitanus and a strain carrying a deletion of carboxysomal CA (ΔcsoS3). Kinetic studies showed that RuBisCO associated with CA-deficient carboxysomes had a 2.8-fold higher apparent Km for CO2 and that this defect was not corrected by addition of exogenous CA.(31)
Oxygen exclusion by the carboxysome
Increasing the concentration of CO2 within the lumen of the carboxysome via a CCM should be sufficient per se to enhance the carboxylase activity of RuBisCO with the added benefit of reducing competitive inhibition by oxygen. However, it has been suggested that the carboxysome shell might also act as a barrier to molecular oxygen further decreasing photorespiration.(3) In vitro studies with Halothiobacillus neapolitanus and Synechocystis PCC 6803 show that carboxysomal RuBisCO is less sensitive to inhibition by O2.(32) These findings are consistent with oxygen exclusion, but to our knowledge direct evidence is lacking.
Regulation of carboxysome expression
Prior studies showed that carboxysomes are induced by CO2 limitation, providing additional evidence for their role in enhancing carbon fixation.(10) However, relatively little has been reported on the regulatory factors that control carboxysome production. In Nitrobacter winogradskyi, carboxysome genes are proximal to a gene for the transcriptional regulator cbbR. In the β-cyanobacteria, transcriptional regulators CcmR (NdhR) and CmpR regulate Ci transporters involved in the CCM, but to our knowledge have not been shown to regulate carboxysome expression.(7) Transcription of the carboxysome (cso) gene cluster of H. neapolitanus was recently investigated by quantitative PCR.(33) The levels of mRNA for individual Cso proteins generally corresponded to their relative abundance in the carboxysome. However, the mechanism by which transcript levels were modulated and the effects of varied CO2 concentrations were not investigated.(33)
Carboxysomes are composed completely of protein subunits
To better understand the function of carboxysomes, substantial effort has gone into studies of their composition. Investigations conducted thus far indicate that carboxysomes (and other microcompartments) are made completely of protein subunits.(4,26,34) There are no confirmed reports of RNA, DNA, or lipids associated with carboxysomes or other microcompartments. No lipid bilayer or monolayer is visible by electron microscopy under conditions where such structures would be readily apparent. However, the possibility of associated lipids is an open question. It has been reported that two carboxysome proteins (CsoS2A and CsoS2B) are glycosylated,(35) but other occurrences of carbohydrates are unknown.
α- and β-carboxysomes
A variety of studies indicate two main classes of carboxysomes, α and β, which vary slightly in composition. Each class is thought to have co-evolved with a different phylogenetic group of RuBisCO.(36,37) The α-carboxysome is associated with form 1A RuBisCO which is found in chemoautotrophs and α-cyanobacteria while the β-carboxysome is associated with form 1B RuBisCO which is found in the β-cyanobacteria.(37) Overall, α- and β-carboxysomes are thought to have similar architectural and mechanistic principles. The gene names differ between α- and β-carboxysomes. However, many of their protein components are conserved with the exceptions that the CsoS2 and CsoS3 proteins are specific to type α while the CcmM, CcmN and CcaA proteins are characteristic of type β as further described below.(36,38)
Protein composition of the α-carboxysome
The α-carboxysome of H. neapolitanus was purified and a number of its components were identified.(26,39) It is composed of about 12–15 polypeptides including CbbL, CbbS, CsoS2A, CsoS2B, CsoS3, CsoS1A, CsoS1B, CsoS1C, CsoS4A and CsoS4B. CbbL and CbbS are the large and small subunits of form 1A RuBisCO. CsoS1A, CsoS1B and CsoS1C, which are closely related in sequence, form the bulk of the shell. CsoS1 and related proteins contain a conserved motif known as a bacterial microcompartment (BMC) domain defined by the ProSite pattern D-x(0,1)-M-x-K-[SAG](2)-x-[IV]-x-[LIVM]-[LIVMA]-[GCSY]-x(4)-[GDE]-[SGPDR]-[GA] and by Pfam alignments. CsoS2A and CsoS2B are also associated with the shell.(35) They are encoded by the same gene, but differ in their degree of glycosylation.(35) The CsoS3 protein was recently shown to be a novel type of carbonic anhydrase that is tightly associated with the carboxysome shell.(28) Originally, it was thought to be a new class of CA, but a recent crystallographic study showed that it is distantly related to the βclass of CA enzymes.(40) The CsoS4A protein (formerly OrfA) was crystallized and found to be a pentamer, suggesting it may form the vertices of the protein shell.(12) CsoS4A was not identified as a component of purified carboxysomes. It was probably missed due to low abundance, since the ratio of vertex proteins to major shell proteins is estimated to be one per several hundred.
Protein composition of the β-carboxysome
As mentioned above, the β-type carboxysome is thought to have co-evolved with form 1B RuBisCO.(37) Although β-carboxysomes have not been purified to homogeneity, genetic, proteomic and sequence analyses have provided insights into their composition. The polypeptides of β-carboxysomes are designated Ccm for their role in a carbon dioxide concentrating mechanism. β-type carboxysomes commonly include RbcL and RbcS, CcmK CcmO, CcmL, CcmM, and CcmN and CcaA (IcfA).(9,36–38) RbcL and RbcS are the large and small subunits of form 1B RuBisCO. CcaA (IcfA) is a carboxysome-associated carbonic anhydrase.(13,30) No sequence similarity can be detected between CcaA of the β-carboxysome and CsoS3 of the α-carboxysome. CcmK and CcmO are BMC-domain proteins that have homology to CsoS1 which is a major shell protein of the α-carboxysome.(41) CcmL is related in sequence to the CsoS4A and CsoS4B proteins which are likely pentamers that form the vertices of the α-carboxysome shell.(12) CcmM and CcmN are restricted to β-carboxysomes, and may function as organizing factors or in enzyme regulation.(42,43)
Carboxysome genomics
The number and organization of carboxysome genes varies somewhat among different organisms reflecting the compositional variation between types α and β and also suggesting some minor variations within type. The best-studied α-carboxysome is that of H. neapolitanus. This organism has the following carboxysome gene cluster: cbbL-cbbS-csoS2-csoS3-csos4A-csos4B-csoS1C-csoS1A-csoS1B.(38) In all, nine polypeptides are encoded by the H. neapolitanus carboxysome gene cluster and most are known to be components of purified carboxysomes. However, purified carboxysomes from H. neapolitanus were estimated to have 12–15 different polypeptides;(26) hence, some carboxysome genes may still be unidentified. In general, α-carboxysome gene clusters are conserved with the exception that one copy of CsoS1 is sometimes found at the start of the cluster adjacent to the ccbL gene. In addition, a few such clusters appear to include bacterioferritin- and pterin-4 alpha-carbinolamine dehydratase-like genes.(44)
The β-carboxysome has been studied in several cyanobacteria. In Synechocystis PCC 6803, the β-carboxysome genes are arranged as follows: ccmN-ccmM-ccmL-ccmK-ccmK//ccmK-ccmK//ccaA//rbcL-rbcX-rbcS//ccmO.(38) The arrangement in Synechococcus elongatus PCC 7942 is ccmK-ccmK//ccmK-ccmL-ccmM-ccmN-ccmO-rbcL-rbcS//ccaA//rbcX. However, β-carboxysomes have not been purified so there is some ambiguity about their protein content and corresponding genes. As is the case for the α-carboxysome, there is the possibility of additional β-carboxysome genes yet to be identified.
Carboxysomes are icosahedral
For many years, there was uncertainty about the overall shape of carboxysomes. Electron microscopy suggested they were either icosahedra or pentagonal dodecahedra.(45,46) However, recent electron cryotomography of carboxysomes from Synechococcus strain WH8102 (β-type) and H. neapolitanus (α-type) indicate an icosahedral structure.(47,48) The shells of these carboxysomes appear 3–4 nm thick and their sizes range from 114–137 nm for Synechococcus and from 88–108 nm for H. neapolitanus.(48) The molecular mass range is estimated to be 115–355 MDa for H. neapolitanus carboxysomes.(48) About 250 RuBisCO molecules could be visualized per carboxysome arranged in 3–4 concentric layers, but well-defined interactions between the RuBisCO subunits and the shell were not evident.(48,49)
Atomic-level structure of the carboxysome
The electron microscopy described above together with recent x-ray crystallography studies allows the carboxysome shell to be modeled at the atomic level.(5,12) The model is partly based on analogy with icosahedral viral capsids, some of which are constructed from a combination of hexamers and pentamers that occupy the 12 vertices.(50,51) The most-abundant proteins in the shell of the Synechocystis PCC 6803 and H. neapolitanus carboxysomes are the BMC-domain proteins, CcmK and CsoS1, respectively.(3) Both of these proteins are hexamers that form extended protein sheets.(5,52) Hence, CcmK and CsoS1 are proposed to form the 20 triangular faces of the icosahedral shell. Recent studies have also shown that carboxysome proteins CcmL and CsoS4A from Synechocystis PCC 6803 and H. neapolitanus are pentamers, suggesting that these proteins form the 12 vertices.(12) Overall, the shell is thought to be formed from several hundred hexamers and 12 pentamers that pack very tightly into a nearly solid protein shell. Interestingly, structural studies also indicate that specific pores through the shell might serve as molecular conduits that are critical to carboxysome function (Fig. 2B).(5)
Molecular conduits for small molecules span the carboxysome shell
Current models for carboxysome function require that its shell is permeable to RuBP, HCO3− and 3-phosphoglycerate, but restricts the diffusion of CO2.(7) Crystallography of its protein subunits suggests that the carboxysome shell is essentially solid except for small pores and gaps.(5,12,52) The CcmK1, CcmK2 and CcmK4 hexamers have central pores that range in diameter from 4 or 6 Å taking van der Waals radii into account, and the boundaries where these hexamers join to form sheets have gaps 4–6 Å wide.(5,53) The central pores have a large net positive electrostatic potential and the boundary gaps also have conserved positively charged amino acid residues. These findings suggest that the pores and gaps could act as molecular conduits that preferentially allow negatively charged molecules such as RuBP, HCO3− and 3-phosphoglycerate to pass while restricting the movement of uncharged molecules such as CO2 and possibly O2. Similar openings are found in the corresponding shell protein (CsoS1A) from H. neapolitanus further supporting the importance of these features.(52) In the carboxysomes of Synechocystis PCC6803 and H. neapolitanus, the pentameric proteins thought to occupy the vertices of the shell also have narrow pores.(12) However, the small number of pentamers compared to hexamers in the shell suggests that transport across these pores might not be significant.
Higher order structure of carboxysomes
Many of the components of the carboxysome are known, and a rough atomic level model of the shell is available, but a great deal remains to be learned about its internal organization and the interactions that guide its assembly. Recent protein–protein interaction studies investigated protein subcomplexes of the carboxysome.(42,43,54) Two-hybrid analyses with carboxysome proteins from Synechococcus WH8102 indicate a stable binary complex between CsoS2 (unknown function) and CsoS4A (a pentamer proposed to form the vertice of the carboxysome shell).(54) In Synechococcus PCC7942, co-purification by nickel-affinity chromatography indicates a stable trinary complex among carboxysomal carbonic anhydrase (CcaA), putative non-BMC shell protein CcmM (58 kDa form) and RuBisCO, as well as a binary complex between CcmM (35 kDa form) and RuBisCO.(43) CcmM is thought to be a nucleation point for the formation of these complexes. Its N-terminal region has a left-handed parallel beta-helix (LHH) domain similar to that of some carbonic anhydrases and acetyltransferases. Depending on the species, the C-terminal domain of CcmM (residues 250–687) has three or four repeats of a sequence with similarity to the small subunit of RuBisCO, potentially providing the protein–protein interaction sites needed for complex formation. It was proposed that carboxysome subcomplexes described above are critical to recruit CA to the carboxysome and provide a scaffold upon which the shell is assembled.(43) In Synechocystis PCC6803, protein–protein interaction studies also support a key role for CcmM in carboxysome organization.(42) In this organism, CcmM is part of a proposed trinary bicarbonate dehydration complex (BDC) consisting of CcmN, CcaA, and CcmM. Formation of this complex requires the LHH domain of CcmM which also connects the BDC to the carboxysome shell. Interestingly, CcmM of Synechocystis PCC6803 lacks CA activity, but binds CO2/bicarbonate suggesting a possible role in regulation of CA activity.(42) Thus, recent studies suggest CcmM plays a key role in guiding the internal organization of the carboxysome. However, the functional details of carboxysome subcomplexes remain to be elucidated.
Other microcompartments
Microcompartments for B12-dependent 1,2-propanediol utilization (Pdu microcompartments)
For many years, carboxysomes were the only known bacterial microcompartment. Then, in 1994, genetic studies indicated that a BMC-domain protein was involved in B12-dependent 1, 2-propanediol degradation by S. enterica.(55) A few years later, electron microscopy showed that S. enterica conditionally formed microcompartments during B12-dependent growth on 1,2-propanediol and further studies established that microcompartments function in this process.(2) The Pdu microcompartments are very large multi-protein complexes that are generally similar to carboxysomes in size and appearance (Fig. 1). They are 100–150 nm in cross section with a 3–4 nm protein shell. Their shape is roughly polyhedral, but they are more irregular than carboxysomes. Their mass is about 600 MDa and they are composed of about 18,000 individual polypeptides of about 14–18 different types.(44) Like carboxysomes, the Pdu microcompartments are thought to be composed completely from protein subunits.(4) The proposed function of the Pdu microcompartments is to sequester an intermediate of 1,2-propanediol degradation (propionaldehyde) in order to prevent toxicity and diffusive loss.(56,57) Thus, there may be functional analogy between the Pdu microcompartments and carboxysomes in that both are used to confine metabolic intermediates that are not well-retained by lipid bilayers.
Model for the Pdu microcompartment
The current model for the Pdu microcompartment proposes that the first two steps of 1,2-propanediol degradation are confined to the lumen of a microcompartment so that propionaldehyde can be sequestered (see model in Fig. 3).(4) In the model, 1,2-propanediol traverses the microcompartment shell and enters the lumen where it is converted to propionaldehyde by B12-dependent diol dehydratase. Propionaldehyde dehydrogenase then catalyzes the conversion of propionaldehyde +HS-CoA +NAD+→propionyl-CoA +NADH +H+.(4,58) Propionyl-CoA exits the microcompartment and continues through the 1,2-propanediol degradative pathway to propionate and 1-propanol, or enters the methylcitrate pathway where it is converted to pyruvate and succinate.(59–61) These processes allow S. enterica to grow efficiently on 1,2-propanediol as a sole carbon and energy source while at the same time sequestering propionaldehyde. Confinement of propionaldehyde allows its rate of production and consumption to be matched, mitigating toxicity and DNA damage and reducing diffusive loss to the environment since propionaldehyde is poorly retained by lipid bilayers.(62) In the model, two additional enzymes are placed in the lumen of the Pdu microcompartment: diol dehydratase reactivase (PduGH) and adenosyltransferase (PduO). These enzymes support the activity of diol dehydratase by ensuring an adequate supply of coenzyme B12.(63,64)
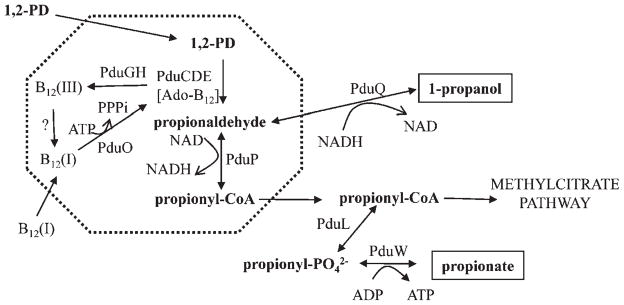
Figure 3.
Model for 1,2-propanediol degradation by S. enterica. The dashed line indicates the shell of the microcompartment which is composed of seven BMC-domain proteins (PduABB′JKTU). The first two steps of 1,2-propanediol degradation (conversion of 1,2-propanediol to propionyl-CoA) occur in the lumen of the microcompartment and the remaining steps in the cytoplasm. The proposed function of this microcompartment is sequestration of propionaldehyde to minimize its toxicity. The Ado-B12 cofactor is sometimes converted to B12(III) in a catalytic by-reaction inactivating diol dehydratase. B12(III) is released from diol dehydratase (PduCDE) by a reactivase (PduGH), and reduced to B12(I) by an unknown enzyme. Then, ATP:cob(I)alamin adenosyltransferase (PduO) converts B12(I) to active cofactor (Ado-B12) which associateswith diol dehydratase to form active holoenzyme. Abbreviations: 1,2-PD, 1,2-propanediol; PduCDE, coenzyme B12-dependent diol dehydratase; PduP, propionaldehyde dehydrogenase; PduL, phosphotransacylase; PduW, propionate kinase; PduQ, 1-propanol dehydrogenase.
Proteomics of the Pdu microcompartments
The protein content of the Pdu microcompartment supports the functional model described above. The Pdu microcompartments were purified intact (Fig. 4).(4) They consist of 14 different major polypeptides (PduABB′CDEGHJKOPTU).(4) Four of the components of the Pdu microcompartment are enzymes: (i) B12-dependent diol dehydratase (PduCDE), (i) diol dehydratase reactivase (PduGH), (iii) adenosyltransferase (PduO), and (iv) propionaldehyde dehydrogenase (PduP). This complement of enzymes supports the idea that the first two steps of 1,2-propanediol degradation occur within the microcompartment, and this is confirmed by enzyme assays that show purified Pdu microcompartments have high levels of diol dehydratase and propionaldehyde dehydrogenase activity.(4,58) The remaining enzymes of the 1,2-propanediol degradative pathway (phosphotransacylase, propionate kinase, and 1-propanol dehydrogenase) are not detected in purified Pdu microcompartments suggesting that these enzymes localize to the cytoplasm of the cell(4). The protein content of the Pdu microcompartments also supports the idea of a shell consisting of seven different BMC-domain proteins (PduABB′JKTU), a somewhat more complex arrangement than the shell of the H. neapolitanus carboxysome, which is composed of three different BMC domain proteins.(4) Thus, protein content underpins many aspects of the current model for the Pdu microcompartment. A caveat of these studies is that low-abundance components of the Pdu microcompartment may have escaped detection, as is likely the case for the protein that is suggested to form the vertice of the carboxysome shell.(12)
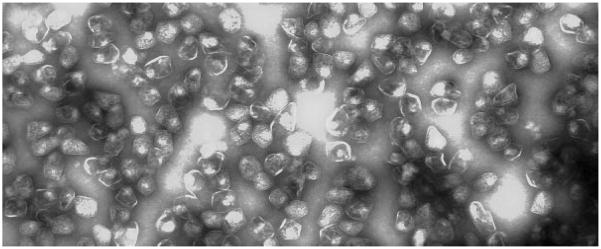
Figure 4.
Purified Pdu microcompartments.
The pathway of 1,2-propanediol degradation
The pathway of 1,2-propanediol degradation presented in model 1 is well-established by genetic and biochemical studies. The overall pathway was first outlined by biochemical studies of whole cells and crude cell extracts.(60,61) B12-dependent diol dehydratase, propionaldehyde dehydrogenase, phosphotransacylase, and propionate kinase have been studied in vitro, and their substrates, products and cofactor requirements have been defined.(58,65–67) Their encoding genes have been identified.(58,65–68) Knockout mutations have established their role in 1,2-propanediol degradation in vivo.(58,65–69) The 1-propanol dehydrogenase has not yet been studied, but its encoding gene has been identified and 1-propanol has been established as a product of 1,2-propanediol degradation.(62,68) The role of diol dehydratase reactivase (PduGH) and adenosyltransferase in 1,2-propanediol degradation has been established by genetic tests.(44,63) Adenosyltransferase has been characterized biochemically(44,63,70) and the properties of reactivase are inferred from studies on a closely related enzyme from Klebsiella.(64,71,72)
The genetics of 1,2-propanediol degradation
The genetics of 1,2-propanediol degradation further supports model 1 and suggests some additional components may be involved in microcompartment formation. In S. enterica, the genes involved in 1,2-propanediol degradation are organized as a contiguous, twenty-three-gene cluster with the following structure: pocR, pduF, pduABCDEGHJKLMNOPQSTUVWX.(68) This locus is one of the largest clusters of functionally related genes found in S. enterica. It encodes a transcriptional regulator (PocR), a 1,2-propanediol facilitator (PduF), three enzymes for B12 metabolism (PduGH, PduO, and PduS), one enzyme used for the de novo synthesis of B12 (pduX), five catabolic enzymes that mediate 1,2-propanediol degradation as described above (PduCDE, PduL, PduP, PduQ, PduW), seven BMC-domain proteins (PduA, PduB, PduBme; PduJ, PduK, PduTand PduU), a protein with homology to the pentamer proposed to form the carboxysome vertices (PduN) and two polypeptides of unknown function (PduM and PduV).(55,63,67,68,73–75) S. enterica strains with deletions of the pduM gene form abnormal microcompartments suggesting that the PduM protein may play a structural role.(44) Pfam analyses indicated that the PduV protein is related to the AAA-family of ATPases, but its role in 1,2-propanediol degradation is unknown.
Mitigation of aldehyde toxicity and diffusive loss
The first evidence that microcompartments might have a role in mitigating aldehyde toxicity was based on comparative genetics.(57) Sequence analyses showed that both 1,2-propanediol and ethanolamine degradation involve BMC-domain proteins.(55,57) A common feature of the 1,2-propanediol and ethanolamine degradative pathways is that both proceed through aldehyde intermediates (propionaldehyde and acetaldehyde, respectively); hence, it was proposed that microcompartments might be formed to sequester aldehydes which were known cytotoxins.(57) Later studies showed that mutants unable to form the Pdu microcompartments accumulated large amounts of propionaldehyde and underwent a temporary period of growth arrest due to propionaldehyde toxicity.(56,62) In addition, propionaldehyde was found to be a mutagen and mutation frequencies were increased in microcompartment mutants during growth on 1,2-propanediol.(62) The aldehyde toxicity model is further supported by the findings that polA (DNA repair polymerase) and gsh (glutathione biosynthesis) mutants were unable to grow on ethanolamine.(76,77) However, other studies have indicated that the main role of the ethanolamine microcompartment might be to prevent the loss of a volatile metabolic intermediate, acetaldehyde.(78) In the case of propionaldehyde (boiling point = 48°C), loss due to volatility appears less important than for acetaldehyde (boiling point = 20°C).(62)
Structure of the Pdu microcompartment
The structure of the Pdu microcompartment is a key to resolving its functional principles. At this time, its structure can be tentatively modeled based on its protein content and analogy with the carboxysome. Carboxysomes are proposed to be icosahedra with triangular faces made from hexamers of BMC-domain proteins and vertices formed from pentamers with homology to the CcmL and CsoS4 proteins.(12) Purified Pdu microcompartments contain two major BMC-domain proteins (PduA and PduJ) that are closely related in sequence to the hexamers proposed to form the faces of the carboxysome.(4) The pdu operon also encodes a homolog of the pentamer proposed to from the vertices of the carboxysome (PduN).(12) Thus, analogy suggests that the shell of the Pdu microcompartment may have faces made from PduA and PduJ hexamers and vertices made from PduN pentamers. However, electron microscopy shows that the Pdu microcompartments are more irregular in shape than carboxysomes suggesting they vary somewhat from a regular icosahedron and may include distinctive structural elements.(4,56,79) In addition, the Pdu microcompartment includes seven BMC-domain proteins whereas carboxysomes generally have 3–5. The crystal structure of one of these shell proteins, PduU, has been determined and shown to differ substantially from the homologous carboxysome shell proteins.(80) This suggests that the shell of the Pdu microcompartments may be more sophisticated than that of the carboxysome which would not be surprising considering its complement of enzymes and their cofactor requirements (Model 1).
An electron conduit through the shell of the Pdu microcompartment
Recent studies of the PduT protein from Citrobacter freundii indicate it has a [4Fe-4S](1)+ iron–sulfur cluster with a midpoint potential of +99mV.(81) The PduT protein was shown to be a component of the Pdu microcompartment in S. enterica and it includes two BMC domains suggesting it may be a shell protein.(4,68) This opens the possibility that PduT might be involved in redox sensing or single electron transfer from the cytoplasm to the lumen of the microcompartment. Alternatively, the iron–sulfur cluster might simply be a structural element. Further studies will be needed to determine whether PduT (and other BMC-domain protein) have specialized roles in microcompartment function.
Microcompartments for B12-dependent ethanolamine utilization (Eut microcompartments)
Studies with S. enterica have demonstrated that a microcompartment is involved in the B12-dependent degradation of ethanolamine.(78,82) Ethanolamine utilization begins with the conversion of ethanolamine to acetaldehyde by B12-dependent ethanolamine ammonia lyase after which acetaldehyde is degraded to acetate and ethanol by a pathway analogous to 1,2-propanediol degradation (model 1).(83,84) Some of the acetyl-CoA formed during ethanolamine degradation is further metabolized via the citric acid cycle and glyoxylate shunt to provide biosynthetic building blocks and additional energy.(59,85) Thus, overall, the degradation of 1,2-propanediol and ethanolamine share many features with the main difference being that 1,2-propanediol is a C3 compound and ethanolamine is a C2 compound. Accordingly, it is thought that the microcompartments involved in these processes have analogous functions. As mentioned above, polA (DNA repair polymerase) and gsh (glutathione biosynthesis) mutants were unable to grow on ethanolamine supporting an aldehyde toxicity model.(76,77) However, other studies have shown that Eut microcompartment mutants lose acetaldehyde (which is quite volatile) to the environment and are unable to grow due to carbon loss.(78) Mechanistically, both functions are met by sequestration of acetaldehyde. It has also been suggested that the Eut microcompartment concentrates substrates and enzymes either to increase metabolic efficiency or to regulate metabolite levels.(86)
General questions
Evolution of the carboxysome and the bacterial CCM
Changes in the Earth’s atmosphere are thought to underlie the evolution of the carboxysome and the bacterial CCM.(36,37) When cyanobacteria first appeared, the Earth’s atmosphere lacked oxygen and contained a high percentage of CO2. Under these conditions, a CCM may have been unnecessary. The planetary decline in atmospheric CO2 and increase in O2 may have provided the selective pressure for evolution of the CCMs.(36,37) The carboxysome is an essential component of a CCM that spread though the bacterial lineage. However, the complexity of the carboxysome is somewhat surprising to us. For example, why has CO2-channeling via a simpler CA-RuBisCO complex not evolved in any known systems, and why hasn’t sequence variation produced a more efficient RuBisCO. An explanation to the latter question is that RuBisCO is limited by its reaction mechanism. It has been proposed that because CO2 is relatively featureless there is an inverse relationship between the ability of RuBisCO to discriminate between O2 and CO2, and its catalytic rate.(16) Given this tradeoff, extant RuBisCOs may be operating at near maximal efficiency. This idea provides a possible partial explanation for the apparent inefficiency of RuBisCO. The reason why a simple channeling mechanism is not competitive with the carboxysome for increasing the efficiency of RuBisCO is unclear and will require a better understanding of the molecular basis of carboxysome function.
Ecological niches of organism having carboxysomes
The RuBisCO reaction is the first step of the Calvin cycle, which is the most widespread pathway of autotrophic CO2 fixation in nature. It is found in plants, algae, cyanobacteria and proteobacteria. Genome analysis indicates that carboxysomes are widely distributed among Bacteria, but are absent from the Eukarya and the Archaea.(36–38) All cyanobacteria that have been examined and many chemoautotrophs have carboxysomes.(7,10) α-carboxysomes are found primarily in oceanic cyanobacteria and chemoautotrophs while β-carboxysomes are found in the β-cyanobacteria, which typically inhabit freshwater and estuarine environments.(7,10) Overall, carboxysomes are widely distributed among bacteria, which play a major role in the global carbon cycle, thus underscoring the importance of the carboxysome and the bacterial CCM.(7,87)
Ecology of organisms having Pdu microcompartments
Genes for B12-dependent 1,2-propanediol degradation are found in a number of bacterial genera including Salmonella, Klebsiella, Shigella, Yersinia, Listeria, Lactobacillus, Lactococcus and at least one species of Escherichia coli (E24377A). In all cases examined, pdu gene clusters are highly conserved indicating that microcompartments are essential to the physiology of 1,2-propanediol degradation. In the environment, 1,2-propanediol is formed as a major product of the fermentation of rhamnose and fucose, which are common sugars in plant cell walls, bacterial capsules, and the glycoconjugates of eukaryotic cells.(60) Hence, 1,2-propanediol metabolism is thought to have particular importance in anaerobic environments such as the large intestines of higher animals, sediments and the depths of soils. This is supported by gene expression studies in S. enterica, which show the pdu operon is maximally induced by 1,2-propanediol under anaerobic conditions.(68,73,75) Induction by 1,2-propanediol is mediated by the positive transcriptional regulator PocR (which also induces the adjacent cob operon, which is required for the de novo synthesis of coenzyme B12) and global regulation is mediated by cAMP/CRP, ArcAB and CsrA.(73,88–90)
1,2-propanediol degradation is implicated in pathogenesis
The capacity for 1,2-propanediol degradation is found among a number of genera that inhabit the large intestines of higher animal including some that are pathogens. Studies conducted with two intracellular human pathogens, Salmonella and Listeria, have implicated 1,2-propanediol degradation in pathogenesis. In Salmonella, pdu genes are induced in host tissues and co-regulated with invasion genes by CsrA.(90,91) In addition, pdu mutants are impaired for growth in macrophages and show a virulence defect in competitive index assayswith mice.(92,93) In Listeria, comparative genomics indicates a link between 1, 2-propanediol degradation and pathogenesis, and several pdu genes are induced during intracellular growth in host cells.(94,95)
Microcompartment diversity
Genomic analyses indicate that bacterial microcompartments are widespread and functionally diverse. Over 40 different genera of Bacteria encode homologues of BMC-domain proteins, although none have been detected in the Archaea or Eukarya.(1) Multiple genes for BMC-domain proteins are often found in gene clusters that also encode microcompartment-associated enzymes or putative enzymes. Variation in such gene clusters suggests bacterial microcompartments have diverse functions. Established microcompartments include carboxysomes and the Pdu and Eut microcompartments (described above). Several putative microcompartments are inferred from gene clustering analyses. (1) A microcompartment associated with a pyruvate-formate lyase homolog is proposed to be involved in the production of ethanol from pyruvate.(96) (2) A microcompartment was suggested to be involved in the oxidation of ethanol by Clostridium Kluyveri.(97) (3) A putative microcompartment in Rhodopirellula baltica SH 1 is associated with a lactate dehydrogenase homologue. (4) A putative microcompartment in Carboxydothermus hydrogenoformans Z-2901 is associated with an isochorismatase-family protein. (5) A putative microcompartment Solibacter usitatus Ellin6076 associated with a dihydrodipicolinate synthase homologue, and the list is expected to grow as more genome sequences become available.(1) Certainly, this diversity is one of the most-intriguing aspects of bacterial microcompartments. It raises important questions about their functional principles, the extent to which those principles are conserved, and the degree to which they can be adapted and modified (or engineered) to fulfill the needs of a particular metabolic system.
The bacterial microcompartment paradox
Our model for the Pdu microcompartment places four enzymes in the lumen: diol dehydratase, propionaldehyde dehydrogenase, adenosyltransferase and reactivase. The substrates of these enzymes are 1,2-propanediol, HS-CoA, NAD+, ATP, and B12(I), and their products are propionyl-CoA, NADH +H+, ADP, inorganic triphosphate and coenzyme B12. For the Pdu microcompartment to function in the manner that we propose, these substrates and products must cross the microcompartment shell, with the exception of coenzyme B12 which can be produced and used within the microcompartment. Not only must these substrates and products pass, our model requires the propionaldehyde is retained. Analogy with carboxysomes suggests that the Pdu microcompartments have a solid protein shell perforated by discreet pores that serve as molecular conduits for metabolites.(5,11,12,52) Hence, a paradox: how can molecular pores retain propionaldehyde while allowing much larger molecules such as B12, NAD+, etc. to pass? The solution is a system that controls movement of small molecules. One possibility is that substrate-specific pores mediate the transit of small molecules across the microcompartment shell. Another possibility is architectural control of molecular routing. For example, lumen enzymes could reside in close association with pores that span the shell such that molecules moving from the cytoplasm into the microcompartment are channeled to active sites. Thus, molecular routing and pore specificity models provide possible explanations to the paradox.
The Pdu microcompartment paradox also applies to the carboxysome. In this case, the question is how CO2 is retained in the lumen while the substrate (RuBP) and product (3-PGA) readily traverse the shell. Based on structural studies, it was proposed that positively charged trans-shell pores act as semi-specific conduits that allow negatively charged molecules, such as the substrates and products of RuBisCO (RuBP and 3-PGA) to pass, but restrict the diffusion of uncharged molecules such as CO2 and O2.(5) It was also suggested that CA might sit on the interior surface of the shell in close proximity with a trans-shell pore which could direct HCO3− to the active site of CA while forbidding the passage of other small molecules.(7,33)
Studies of bacterial microcompartments may reveal new properties of biological protein sheets
As discussed above, a number of studies support the idea that microcompartments have a nearly solid protein shell that encases metabolic enzyme.(5,11,12,52) Both in vitro and in vivo investigations show that the lumen enzymes of microcompartments are catalytically active.(4,26) Thus, it can be concluded that enzyme substrates, products and cofactors must traverse the microcompartment shell in some manner. We know of no other biological system that so clearly focuses attention on the permeability properties of a protein sheet. Thus, bacterial microcompartments may prove to be uniquely important in providing the necessary impetus for a detailed exploration of principles of biological protein sheets. We believe that the principles of microcompartment shells will have broad application to bacteriology, virology, cell biology and biotechnology.
Conclusions
Bacterial microcompartments are large and sophisticated multi-protein complexes that are functionally diverse and widely used by bacteria to optimize metabolic processes. They are unique in that they consist of metabolic enzymes encased within a protein shell. Growing evidence suggests that microcompartments optimize metabolic processes by spatially organizing the encapsulated enzymes and by regulating the movement substrates, cofactors and products. The precise manner in which microcompartments control molecular routing is unknown but elucidating these mechanisms may illuminate new properties of biological protein sheets.
Acknowledgments
Funding agencies: This work was supported by grant MCB0616008 from the National Science Foundation (TAB) and by the BER program of the US Department of Energy Office of Science (TOY).
Abbreviations
- BMC
bacterial microcompartment domain
- CA
carbonic anhydrase
- RuBisCO
ribulose bisphosphate carboxylase monooxygenase
- RuBP
ribulose bisphosphate
- 3-PGA
3-phosphoglycerate
- CCM
CO2 concentrating mechanism
- HCR
high-CO2-requiring
- LHH
left-handed parallel beta-helix
- BDC
bicarbonate dehydration complex
- Pdu
1,2-propanediol utilization
- PduCDE
B12-dependent diol dehydratase
- PduGH
diol dehydratase reactivase
- PduO
adenosyltransferase
- PduP
propionaldehyde dehydrogenase
References
- 1.Bobik TA. Polyhedral organelles compartmenting bacterial metabolic processes. Appl Microbiol Biotechnol. 2006;70:517–525. doi: 10.1007/s00253-005-0295-0. [DOI] [PubMed] [Google Scholar]
- 2.Bobik TA. Bacterial microcompartments. Microbe. 2007;2:25–31. [Google Scholar]
- 3.Cannon GC, Bradburne CE, Aldrich HC, Baker SH, Heinhorst S, et al. Microcompartments in prokaryotes: carboxysomes and related polyhedra. Appl Environ Microbiol. 2001;67:5351–6361. doi: 10.1128/AEM.67.12.5351-5361.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Havemann GD, Bobik TA. Protein content of polyhedral organelles involved in coenzyme B12-dependent degradation of 1,2-propanediol in Salmonella enterica serovar Typhimurium LT2. J Bacteriol. 2003;185:5086–5095. doi: 10.1128/JB.185.17.5086-5095.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kerfeld CA, Sawaya MR, Tanaka S, Nguyen CV, Phillips M, et al. Protein structures forming the shell of primitive bacterial organelles. Science. 2005;309:936–938. doi: 10.1126/science.1113397. [DOI] [PubMed] [Google Scholar]
- 6.Shively JM, Ball F, Brown DH, Saunders RE. Functional organelles in prokaryotes: polyhedral inclusions (carboxysomes) of Thiobacillus neapolitanus. Science. 1973;182:584–586. doi: 10.1126/science.182.4112.584. [DOI] [PubMed] [Google Scholar]
- 7.Price GD, Badger MR, Woodger FJ, Long BM. Advances in understanding the cyanobacterial CO2-concentrating-mechanism (CCM): functional components, Ci transporters, diversity, genetic regulation and prospects for engineering into plants. J Exper Bot Advance accesss. 2007 doi: 10.1093/jxb/erm112. [DOI] [PubMed] [Google Scholar]
- 8.Drews G, Niklowitz W. Beiträge zur Cytologie der Blaualgen. II. Zentroplasma und18 granulare Einschlüsse von Phormidium uncinatum. Arch Mikrobiol. 1956;24:147–162. [PubMed] [Google Scholar]
- 9.Kaplan A, Reinhold L. CO2 concentrating mechanisms in photosynthetic microorganisms. Ann Rev of Plant Physiol and Plant Mol Biol. 1999;50:539–570. doi: 10.1146/annurev.arplant.50.1.539. [DOI] [PubMed] [Google Scholar]
- 10.Shively JM, van Keulen G, Meijer WG. Something from almost nothing: carbon dioxide fixation in chemoautotrophs. Ann Rev Microbiol. 1998;52:191–230. doi: 10.1146/annurev.micro.52.1.191. [DOI] [PubMed] [Google Scholar]
- 11.Yeates TO, Tsai Y, Tanaka S, Sawaya MR, Kerfeld CA. Self-assembly in the carboxysome: a viral capsid-like protein shell in bacterial cells. Biochem Soc Trans. 2007;35:508–511. doi: 10.1042/BST0350508. [DOI] [PubMed] [Google Scholar]
- 12.Tanaka S, Kerfeld CA, Sawaya MR, Cai F, Heinhorst S, et al. Atomic-level models of the bacterial carboxysome shell. Science. 2008;319:1083–1086. doi: 10.1126/science.1151458. [DOI] [PubMed] [Google Scholar]
- 13.Fukuzawa H, Suzuki E, Komukai Y, Miyachi S. A gene homologous to chloroplast carbonic anhydrase (icfA) is essential to photosynthetic carbon dioxide fixation by Synechococcus PCC7942. Proc Natl Acad Sci USA. 1992;89:4437–4441. doi: 10.1073/pnas.89.10.4437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Reinhold L, Kosloff R, Kaplan A. A model for inorganic carbon fluxes and photosynthesis in cyanobacterial carboxysomes. Canadian Journal of Botany-Revue Canadienne De Botanique. 1991;69:984–988. [Google Scholar]
- 15.Reinhold L, Zviman M, Kaplan A, editors. Inorganic carbon fluxes and photosynthesis in cyanobacteria: a quantitative model. Dordrecht: Martinus Nijhoff; 1987. pp. 289–296. [Google Scholar]
- 16.Tcherkez GG, Farquhar GD, Andrews TJ. Despite slow catalysis and confused substrate specificity, all ribulose bisphosphate carboxylases may be nearly perfectly optimized. Proc Natl Acad Sci USA. 2006;103:7246–7251. doi: 10.1073/pnas.0600605103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Giordano M, Beardall J, Raven JA. CO2 concentrating mechanisms in algae: mechanisms, environmental modulation, and evolution. Annu Rev Plant Biol. 2005;56:99–131. doi: 10.1146/annurev.arplant.56.032604.144052. [DOI] [PubMed] [Google Scholar]
- 18.Fathinejad S, Steiner JM, Reipert S, Marchetti M, Allmaier G, et al. A carboxysomal carbon-concentrating mechanism in the cyanelles of the ‘coelacanth’ of the algal world, Cyanophora paradoxa? Physiologia Plantarum. 2008;133:27–32. doi: 10.1111/j.1399-3054.2007.01030.x. [DOI] [PubMed] [Google Scholar]
- 19.Price GD, Badger MR. Expression of human carbonic-anhydrase in the cyanobacterium Synechococcus PCC7942 creates a high CO2-requiring phenotype-evidence for a central role for carboxysomes in the CO2 concentrating mechanism. Plant Physiology. 1989;91:505–513. doi: 10.1104/pp.91.2.505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Fridlyand L, Kaplan A, Reinhold L. Quantitative evaluation of the role of a putative CO2-scavenging entity in the cyanobacterial CO2-concentrating mechanism. Biosystems. 1996;37:229–238. doi: 10.1016/0303-2647(95)01561-2. [DOI] [PubMed] [Google Scholar]
- 21.English RS, Jin S, Shively JM. Use of electroporation to generate a Thiobacillus neapolitanus carboxysome mutant. Applied and Environ Microbiol. 1995;61:3256–3260. doi: 10.1128/aem.61.9.3256-3260.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kaplan A. Analysis of high CO2 requiring mutants indicates a central role for the 5′ flanking region of Rbc and for the carboxysomes in cyanobacterial photosynthesis. Canadian Journal of Botany-Revue Canadienne De Botanique. 1990;68:1303–1310. [Google Scholar]
- 23.Friedberg D, Kaplan A, Ariel R, Kessel M, Seijffers J. The 5′-flanking region of the gene encoding the large subunit of ribulose-1,5-biosphosphate carboxylase/oxygenase is crucial for growth of the cyanobacterium Synechococcus sp. strain PCC 7942 at the level of CO2 in air. J Bacteriol. 1989;171:6069–6076. doi: 10.1128/jb.171.11.6069-6076.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Marcus Y, Schwarz R, Friedberg D, Kaplan A. High CO2 requiring mutant of Anacystis nidulans R2. Plant Physiology. 1986;82:610–612. doi: 10.1104/pp.82.2.610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Price GD, Badger MR. Isolation and characterization of high CO2-requiring-mutants of the cyanobacterium Synechococcus PCC 7942. Plant Physiol. 1989;91:514–525. doi: 10.1104/pp.91.2.514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Cannon GC, Shively JM. Characterization of a homogenous preparation of carboxysomes from Thiobacillus neapolitanus. Arch Microbiol. 1983;134:52–59. [Google Scholar]
- 27.Holthuijzen YA, Kuenen JG, Konings WN. Activity of ribulose-1,5-bisphosphate carboxylase in intact and disrupted carboxysomes of Thiobacillus neapolitanus. FEMS Microbiol Lett. 1987;42:121–124. [Google Scholar]
- 28.So AK, Espie GS, Williams EB, Shively JM, Heinhorst S, et al. A novel evolutionary lineage of carbonic anhydrase (epsilon class) is a component of the carboxysome shell. J Bacteriol. 2004;186:623–630. doi: 10.1128/JB.186.3.623-630.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.So AK, John-McKay M, Espie GS. Characterization of a mutant lacking carboxysomal carbonic anhydrase from the cyanobacterium Synechocystis PCC6803. Planta. 2002;214:456–467. doi: 10.1007/s004250100638. [DOI] [PubMed] [Google Scholar]
- 30.Yu JW, Price GD, Song L, Badger MR. Isolation of a putative carboxysomal carbonic-anhydrase gene from the cyanobacterium Synechococcus Pcc7942. Plant Physiology. 1992;100:794–800. doi: 10.1104/pp.100.2.794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Dou Z, Heinhorst S, Williams EB, Murin CD, Shively JM, et al. CO2 fixation kinetics of Halothiobacillus neapolitanus mutant carboxysomes lacking carbonic anhydrase suggest the shell acts as a diffusional barrier for CO2. J Biol Chem. 2008;283:10377–10384. doi: 10.1074/jbc.M709285200. [DOI] [PubMed] [Google Scholar]
- 32.Marcus Y, Berry JA, Pierce J. Photosynthesis and photorespiration in a mutant of the cyanobacterium Synechocystis PCC-6803 lacking carboxysomes. Planta. 1992;187:511–516. doi: 10.1007/BF00199970. [DOI] [PubMed] [Google Scholar]
- 33.Cai F, Heinhorst S, Shively JM, Cannon GC. Transcript analysis of the Halothiobacillus neapolitanus cso operon. Arch Microbiol. 2008;189:141–150. doi: 10.1007/s00203-007-0305-y. [DOI] [PubMed] [Google Scholar]
- 34.Holthuijzen YA, Breemen JFL, Kuenen GJ, Konings WN. Protein composition of the carboxysomes of Thiobacillus neapolitanis. Arch Microbiol. 1986;144:398–404. [Google Scholar]
- 35.Baker SH, Lorbach SC, Rodriguez-Buey M, Williams DS, Aldrich HC, et al. The correlation of the gene csoS2 of the carboxysome operon with two polypeptides of the carboxysome in Thiobacillus neapolitanus. Arch Microbiol. 1999;172:233–239. doi: 10.1007/s002030050765. [DOI] [PubMed] [Google Scholar]
- 36.Badger MR, Hanson D, Price GD. Evolution and diversity of CO2 concentrating mechanisms in cyanobacteria. Functional Plant Biology. 2002;29:161–173. doi: 10.1071/PP01213. [DOI] [PubMed] [Google Scholar]
- 37.Badger MR, Price GD. CO2 concentrating mechanisms in cyanobacteria: molecular components, their diversity and evolution. J Exp Bot. 2003;54:609–622. doi: 10.1093/jxb/erg076. [DOI] [PubMed] [Google Scholar]
- 38.Cannon GC, Heinhorst S, Bradburne CE, Shively JM. Carboxysome genomics: a status report. Functional Plant Biology. 2002;29:175–182. doi: 10.1071/PP01200. [DOI] [PubMed] [Google Scholar]
- 39.Holthuijzen YA, van Breemen JFL, Kurnen JG, Konings WN. Protein compostion of the carboxysomes of Thiobacillus neapolitanus. Arch Microbiol. 1986;144:398–404. [Google Scholar]
- 40.Sawaya MR, Cannon GC, Heinhorst S, Tanaka S, Williams EB, et al. The structure of beta-carbonic anhydrase from the carboxysomal shell reveals a distinct subclass with one active site for the price of two. J Biol Chem. 2006;281:7546–7555. doi: 10.1074/jbc.M510464200. [DOI] [PubMed] [Google Scholar]
- 41.English RS, Lorbach SC, Qin X, Shively JM. Isolation and characterization of a carboxysome shell gene from Thiobacillus neapolitanus. Mol Microbiol. 1994;12:647–654. doi: 10.1111/j.1365-2958.1994.tb01052.x. [DOI] [PubMed] [Google Scholar]
- 42.Cot SS, So AK, Espie GS. A multiprotein bicarbonate dehydration complex essential to carboxysome function in cyanobacteria. J Bacteriol. 2008;190:936–945. doi: 10.1128/JB.01283-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Long BM, Badger MR, Whitney SM, Price GD. Analysis of carboxysomes from Synechococcus PCC7942 reveals multiple Rubisco complexes with carboxysomal proteins CcmM and CcaA. J Biol Chem. 2007;282:29323–29335. doi: 10.1074/jbc.M703896200. [DOI] [PubMed] [Google Scholar]
- 44.Bobik TA. 2008 Unpublished. [Google Scholar]
- 45.Holthuijzen YA, Vanbreemen JFL, Konings WN, Vanbruggen EFJ. Electron microscopic studies of carboxysomes of Thiobacillus neapolitanus. Arch Microbiol. 1986;144:258–262. [Google Scholar]
- 46.Peters KR. Characterization of a phage-like particle from cells of Nitrobacter. II. Structure and size (author’s transl) Arch Microbiol. 1974;97:129–140. doi: 10.1007/BF00403052. [DOI] [PubMed] [Google Scholar]
- 47.Iancu CV, Ding HJ, Morris DM, Dias DP, Gonzales AD, et al. The structure of isolated Synechococcus strain WH8102 carboxysomes as revealed by electron cryotomography. J Mol Biol. 2007;372:764–773. doi: 10.1016/j.jmb.2007.06.059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Schmid MF, Paredes AM, Khant HA, Soyer F, Aldrich HC, et al. Structure of Halothiobacillus neapolitanus carboxysomes by cryoelectron tomography. J Mol Biol. 2006;364:526–535. doi: 10.1016/j.jmb.2006.09.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Kaneko Y, Danev R, Nagayama K, Nakamoto H. Intact carboxysomes in a cyanobacterial cell visualized by hilbert differential contrast transmission electron microscopy. J Bacteriol. 2006;188:805–818. doi: 10.1128/JB.188.2.805-808.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Caspar DL, Klug A. Physical principles in the construction of regular viruses. Cold Spring Harb Symp Quant Biol. 1962;27:1–24. doi: 10.1101/sqb.1962.027.001.005. [DOI] [PubMed] [Google Scholar]
- 51.Johnson JE, Speir JA. Quasi-equivalent viruses: a paradigm for protein assemblies. J Mol Biol. 1997;269:665–675. doi: 10.1006/jmbi.1997.1068. [DOI] [PubMed] [Google Scholar]
- 52.Tsai Y, Sawaya MR, Cannon GC, Cai F, Williams EB, et al. Structural analysis of CsoS1A and the protein shell of the Halothiobacillus neapolitanus carboxysome. PLoS Biol. 2007;5:e144. doi: 10.1371/journal.pbio.0050144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Tanaka S, Yeates TO. unpublished results. [Google Scholar]
- 54.Gonzales AD, Light YK, Zhang ZD, Iqbal T, Lane TW, et al. Proteomic analysis of the CO2-concentrating mechanism in the open-ocean cyanobacterium Synechococcus WH8102. Canadian Journal Of Botany-Revue Canadienne De Botanique. 2005;83:735–745. [Google Scholar]
- 55.Chen P, Andersson D, Roth JR. The control region of the pdu/cob regulon in Salmonella typhimurium. J Bacteriol. 1994;176:5474–5482. doi: 10.1128/jb.176.17.5474-5482.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Havemann GD, Sampson EM, Bobik TA. PduA is a shell protein of polyhedral organelles involved in the coenzyme B12-dependent degradation of 1,2-propanediol in Salmonella enterica serovar Typhimurium LT2. J Bacteriol. 2002;184:1253–1261. doi: 10.1128/JB.184.5.1253-1261.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Stojiljkovic I, Baeumler AJ, Heffron F. Ethanolamine utilization in Salmonella typhimurium: nucleotide sequence, protein expression, and mutational analysis of the cchA cchB eutE eutJ eutG eutH gene cluster. J Bacteriol. 1995;177:1357–1366. doi: 10.1128/jb.177.5.1357-1366.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Leal NA, Havemann GD, Bobik TA. PduP is a coenzyme-A-acylating propionaldehyde dehydrogenase associated with the polyhedral bodies involved in B12-dependent 1,2-propanediol degradation by Salmonella enterica serovar Typhimurium LT2. Arch Microbiol. 2003;180:353–361. doi: 10.1007/s00203-003-0601-0. [DOI] [PubMed] [Google Scholar]
- 59.Horswill AR, Escalante-Semerena JC. Salmonella typhimurium LT2 catabolizes propionate via the 2-methylcitric acid cycle. J Bacteriol. 1999;181:5615–5623. doi: 10.1128/jb.181.18.5615-5623.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Obradors N, Badia J, Baldoma L, Aguilar J. Anaerobic metabolism of the L-rhamnose fermentation product 1,2-propanediol in Salmonella typhimurium. J Bacteriol. 1988;170:2159–2162. doi: 10.1128/jb.170.5.2159-2162.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Toraya T, Honda S, Fukui S. Fermentation of 1,2-propanediol and 1,2-ethanediol by some genera of Enterobacteriaceae, involving coenzyme B12-dependent diol dehydratase. J Bacteriol. 1979;139:39–47. doi: 10.1128/jb.139.1.39-47.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Sampson EM, Bobik TA. Microcompartments for B12-dependent 1,2-propanediol degradation provide protection from DNA and cellular damage by a reactive metabolic intermediate. J Bacteriol. 2008;190:2966–2971. doi: 10.1128/JB.01925-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Johnson CLVJ, Pechonick E, Park SD, Havemann GD, Leal NA, et al. Functional genomic, biochemical, and genetic characterization of the Salmonella pduO gene, an ATP:cob(I)alamin adenosyltransferase gene. J Bacteriol. 2001;183:1577–1584. doi: 10.1128/JB.183.5.1577-1584.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Mori K, Tobimatsu T, Hara T, Toraya T. Characterization, sequencing, and expression of the genes encoding a reactivating factor for glycerol-inactivated adenosylcobalamin-dependent diol dehydratase. J Biol Chem. 1997;272:32034–32041. doi: 10.1074/jbc.272.51.32034. [DOI] [PubMed] [Google Scholar]
- 65.Bobik TA, Xu Y, Jeter RM, Otto KE, Roth JR. Propanediol utilization genes (pdu) of Salmonella typhimurium: three genes for the propanediol dehydratase. J Bacteriol. 1997;179:6633–6639. doi: 10.1128/jb.179.21.6633-6639.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Liu Y, Leal NA, Sampson EM, Johnson CL, Havemann GD, et al. PduL is an evolutionarily distinct phosphotransacylase involved in B12-dependent 1,2-propanediol degradation by Salmonella enterica serovar Typhimurium LT2. J Bacteriol. 2007;189:1589–1596. doi: 10.1128/JB.01151-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Palacios S, Starai VJ, Escalante-Semerena JC. Propionyl coenzyme A is a common intermediate in the 1,2-propanediol and propionate catabolic pathways needed for expression of the prpBCDE operon during growth of Salmonella enterica on 1,2-propanediol. J Bacteriol. 2003;185:2802–2810. doi: 10.1128/JB.185.9.2802-2810.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Bobik TA, Havemann GD, Busch RJ, Williams DS, Aldrich HC. The propanediol utilization (pdu) operon of Salmonella enterica Serovar Typhimurium LT2 includes genes necessary for the formation of polyhedral organelles involved in coenzyme B12-dependent 1,2-propanediol degradation. J Bacteriol. 1999;181:5967–5975. doi: 10.1128/jb.181.19.5967-5975.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Jeter RM. Cobalamin-dependent 1,2-propanediol utilization by Salmonella typhimurium. J Gen Microbiol. 1990;136:887–896. doi: 10.1099/00221287-136-5-887. [DOI] [PubMed] [Google Scholar]
- 70.Johnson CL, Buszko ML, Bobik TA. Purification and initial characterization of the Salmonella enterica PduO ATP:Cob(I)alamin adenosyltransferase. J Bacteriol. 2004;186:7881–7887. doi: 10.1128/JB.186.23.7881-7887.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Mori K, Tobimatsu T, Toraya T. A protein factor is essential for in situ reactivation of glycerol-inactivated adenosylcobalamin-dependent diol dehydratase. Biosci Biotechnol Biochem. 1997;61:1729–1733. doi: 10.1271/bbb.61.1729. [DOI] [PubMed] [Google Scholar]
- 72.Mori K, Toraya T. Mechanism of reactivation of coenzyme B12-dependent diol dehydratase by a molecular chaperone-like reactivating factor. Biochemistry. 1999;38:13170–13178. doi: 10.1021/bi9911738. [DOI] [PubMed] [Google Scholar]
- 73.Bobik TA, Ailion ME, Roth JR. A single regulatory gene integrates control of vitamin B12 synthesis and propanediol degradation. J Bacteriol. 1992;174:2253–2266. doi: 10.1128/jb.174.7.2253-2266.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Sampson EM, Johnson CL, Bobik TA. Biochemical evidence that the pduS gene encodes a bifunctional cobalamin reductase. Microbiology. 2005;151:1169–1177. doi: 10.1099/mic.0.27755-0. [DOI] [PubMed] [Google Scholar]
- 75.Rondon MR, Escalante-Semerena JC. The poc locus is required for 1,2-propanediol-dependent transcription of the cobalamin biosynthetic (cob) and propanediol utilization (pdu) genes of Salmonella typhimurium. J Bacteriol. 1992;174:2267–2272. doi: 10.1128/jb.174.7.2267-2272.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Rondon MR, Horswill RA, Escalante-Semerena KC. DNA polymerase I function is required for the utilization of ethanolamine, 1,2-propanediol, and propionate by Salmonella typhimurium LT2. J Bacteriol. 1995;177:7119–7124. doi: 10.1128/jb.177.24.7119-7124.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Rondon MR, Kazmierczak R, Escalante-Semerena JC. Glutathione is required for maximal transcription of the cobalamin biosynthetic and 1,2-propanediol utilization (cob/pdu) regulon and for the catabolism of ethanolamine, 1,2-propanediol, and propionate in Salmonella typhimurium. J Bacteriol. 1995;177:5434–5439. doi: 10.1128/jb.177.19.5434-5439.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Penrod JT, Roth JR. Conserving a volatile metabolite: a role for carboxysome-like organelles in Salmonella enterica. J Bacteriol. 2006;188:2865–2874. doi: 10.1128/JB.188.8.2865-2874.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Hacking AJ, Aguilar J, Lin ECC. Evolution of propanediol utilization in Escherichia coli mutants with improved substrate scavenging power. J Bacteriol. 1978;136:522–530. doi: 10.1128/jb.136.2.522-530.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Crowley CS, Sawaya MR, Bobik TA, Yeates TO. 2008 unpublished results. [Google Scholar]
- 81.Parsons JP, Dinesh SD, Deery E, Leech HK, Brindley AA, et al. Biochemical and structural insights into bacterial organelle form and biogenesis. J Biol Chem. 2008 doi: 10.1074/jbc.M709214200. [DOI] [PubMed] [Google Scholar]
- 82.Kofoid E, Rappleye C, Stojiljkovic I, Roth J. The 17-gene ethanolamine (eut) operon of Salmonella typhimurium encodes five homologues of carboxysome shell proteins. J Bacteriol. 1999;181:5317–5329. doi: 10.1128/jb.181.17.5317-5329.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Chang GW, Chang JT. Evidence for the B12-dependent enzyme ethanolamine deaminase in Salmonella. Nature (London) 1975;254:150–151. doi: 10.1038/254150a0. [DOI] [PubMed] [Google Scholar]
- 84.Roof DM, Roth JR. Ethanolamine utilization in Salmonella typhimurium. J Bacteriol. 1988;170:3855–3863. doi: 10.1128/jb.170.9.3855-3863.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Price-Carter M, Tingey J, Bobik TA, Roth JR. The alternative electron acceptor tetrathionate supports B12-dependent anaerobic growth of Salmonella enterica serovar Typhimurium on ethanolamine or 1,2-propanediol. J Bacteriol. 2001;183:2463–2475. doi: 10.1128/JB.183.8.2463-2475.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Brinsmade SR, Paldon T, Escalante-Semerena JC. Minimal functions and physiological conditions required for growth of Salmonella enterica on ethanolamine in the absence of the metabolosome. J Bacteriol. 2005;187:8039–8046. doi: 10.1128/JB.187.23.8039-8046.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Heffelfinger GS, Martino A, Gorin A, Xu Y, Rintoul MD, 3rd, et al. Carbon sequestration in Synechococcus Sp.: from molecular machines to hierarchical modeling. Omics. 2002;6:305–330. doi: 10.1089/153623102321112746. [DOI] [PubMed] [Google Scholar]
- 88.Ailion M, Bobik TA, Roth JR. Two global regulatory systems (Crp and Arc) control the cobalamin/propanediol regulon of Salmonella typhimurium. J Bacteriol. 1993;175:7200–7208. doi: 10.1128/jb.175.22.7200-7208.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Andersson DI, Roth JR. Redox regulation of the genes for cobinamide biosynthesis in Salmonella typhimurium. J Bacteriol. 1989;171:6734–6739. doi: 10.1128/jb.171.12.6734-6739.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Lawhon SD, Frye JG, Suyemoto M, Porwollik S, McClelland M, et al. Global regulation by CsrA in Salmonella typhimurium. Mol Microbiol. 2003;48:1633–1645. doi: 10.1046/j.1365-2958.2003.03535.x. [DOI] [PubMed] [Google Scholar]
- 91.Conner CP, Heithoff DM, Julio SM, Sinsheimer RL, Mahan MJ. Differential patterns of acquired virulence genes distinguish Salmonella strains. Proc Natl Acad Sci USA. 1998;95:4641–4645. doi: 10.1073/pnas.95.8.4641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Heithoff DM, Conner CP, Hentschel U, Govantes F, Hanna PC, et al. Coordinate intracellular expression of Salmonella genes induced during infection. J Bacteriol. 1999;181:799–807. doi: 10.1128/jb.181.3.799-807.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Klumpp J, Fuchs TM. Identification of novel genes in genomic islands that contribute to Salmonella typhimurium replication in macrophages. Microbiology. 2007;153:1207–1220. doi: 10.1099/mic.0.2006/004747-0. [DOI] [PubMed] [Google Scholar]
- 94.Buchrieser C, Rusniok C, Kunst F, Cossart P, Glaser P. Comparison of the genome sequences of Listeria monocytogenes and Listeria innocua: clues for evolution and pathogenicity. FEMS Immunol Med Microbiol. 2003;35:207–213. doi: 10.1016/S0928-8244(02)00448-0. [DOI] [PubMed] [Google Scholar]
- 95.Joseph B, Przybilla K, Stuhler C, Schauer K, Slaghuis J, et al. Identification of Listeria monocytogenes genes contributing to intracellular replication by expression profiling and mutant screening. J Bacteriol. 2006;188:556–568. doi: 10.1128/JB.188.2.556-568.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Wackett LP, Frias JA, Seffernick JL, Sukovich DJ, Cameron SM. Genomic and biochemical studies demonstrating the absence of an alkane-producing phenotype in Vibrio furnissii M1. Appl Environ Microbiol. 2007;73:7192–7198. doi: 10.1128/AEM.01785-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Seedorf H, Fricke WF, Veith B, Bruggemann H, Liesegang H, et al. The genome of Clostridium kluyveri, a strict anaerobe with unique metabolic features. Proc Natl Acad Sci USA. 2008;105:2128–2133. doi: 10.1073/pnas.0711093105. [DOI] [PMC free article] [PubMed] [Google Scholar]