Highlights
► Animal viruses have many diverse mechanisms to assert translation dominance in cells. ► Viral IRES elements are diverse in structure and sequence but interact with initiation factors and/or ribosomes similarly. ► Viruses use alternate proteins to deliver initiator methionyl tRNA to ribosomes after inducing eIF2a phosphorylation. ► Some viruses may use long range RNA interactions to modulate translation.
Abstract
Viruses have adapted a broad range of unique mechanisms to modulate the cellular translational machinery to ensure viral translation at the expense of cellular protein synthesis. Many of these promote virus-specific translation by use of molecular tags on viral mRNA such as internal ribosome entry sites (IRES) and genome-linked viral proteins (VPg) that bind translation machinery components in unusual ways and promote RNA circularization. This review describes recent advances in understanding some of the mechanisms in which animal virus mRNAs gain an advantage over cellular transcripts, including new structural and biochemical insights into IRES function and novel proteins that function as alternate met-tRNAimet carriers in translation initiation. Comparisons between animal and plant virus mechanisms that promote translation of viral mRNAs are discussed.
Introduction
Viruses use diverse mechanisms to outcompete cellular mRNAs for translation machinery and minimal genome sizes (summarized in Table 1 ). This review will focus on only three of many categories of viral translation control mechanisms, thus readers are referred to a more comprehensive review of the other mechanisms [1•].
Table 1.
Overview of unusual animal virus translation mechanisms
| Virus translation mechanism | Virus | Viral gene product |
|---|---|---|
| Leaky scanning | HIV | Env [83] |
| Human papillomavirus 16 | E7 [84] | |
| SARS | orf 7b [85] | |
| Termination-reinitiation | Influenza B | M2 [86] |
| RSV | M2-2 [87] | |
| Calicivirus | VP2 [88] | |
| Shunting | Adenovirus | Late TL mRNAs[89, 90] |
| Duck Hepatitis Virus | Polymerase [91] | |
| Avian Reovirus | σC [92] | |
| Sendai virus | Y1, Y2 [93] | |
| Cap-independent IRES-mediated translation | Picornaviruses | All proteins [11] |
| HCV | All proteins [94, 95, 96] | |
| Pestivirus | All proteins [97] | |
| HIV 1, 2 | Gag [16, 98] | |
| KSHV | v-FLIP [99] | |
| Herpes Simplex | Thymidine kinase [13] | |
| Dicistrovirus | Orf 2 [18, 46] | |
| Ribosome frame-shifting | Sindbis virus | 6K [100] |
| Coronavirus | Orf1b [101, 102] | |
| HIV | Pol [103] | |
| Astrovirus | Pol [104, 105] | |
| VPg binding initiation factors | FCV, Norovirus | Orf 1 [106, 107, 108] |
Viruses generally exploit the dependence of the host translation machinery on the m7G cap structure at the 5′ end of the mRNA. The 5′ cap is recognized by the eukaryotic initiation factor 4F protein complex (eIF4F; Figure 1 ). The 43S complex is recruited through interactions between the eIF4G moiety of eIF4F and the 40S-bound eIF3 to form the 48S preinitiation complex (Figure 1). The 40S ribosome then scans for the AUG initiator codon where the 60S ribosome subunit joins to initiate translation. Interactions between cap-bound eIF4G and the poly(A)-bound poly(A) binding protein produce a closed loop mRNP that enhances cap-dependent translation [2, 3]. Although the precise explanation for mRNA circularization is unknown, it is hypothesized to increase binding affinity of eIF4F for the cap and the efficiency of ribosome recycling for subsequent translation initiation events [4, 5, 6].
Figure 1.
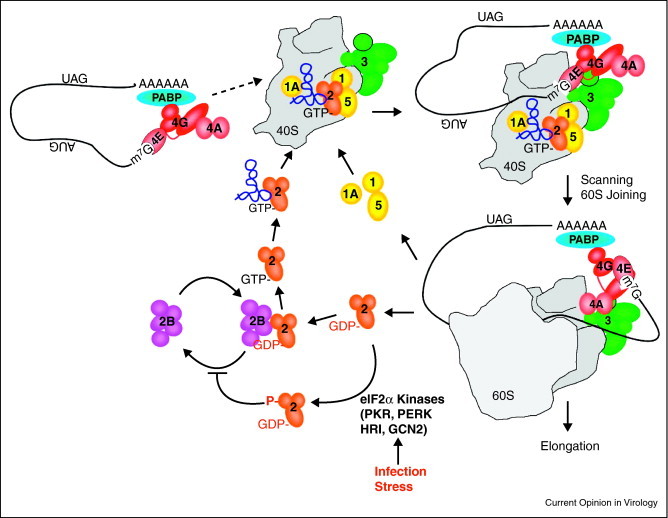
Schematic depiction of translation initiation and the eIF2 nucleotide exchange cycle. Eukaryotic initiation factor 4F, which consists of the cap binding protein (eIF4E), a scaffolding protein (eIF4G) and an RNA helicase (eIF4A), recognizes the m7G cap structure. The mRNA can circularize in accordance with the closed-loop model via interaction between PABP at the 3′ terminus and eIF4G complexed with the 5′ cap. Next, the 43S preinitiation complex, composed of the 40S small ribosomal subunit and initiation factors eIF1, eIF1A, heterotrimeric eIF2(α,β,γ), eIF5 and multisubunit eIF3, can bind the mRNA via interaction between eIF3 and eIF4G to form the 48S complex. eIF2 delivers the initiator methionyl tRNA as a ternary complex, comprised of eIF2·GTP·met-tRNAimet. The 40S ribosomal subunit then scans to locate the AUG codon where the 60S joins, some factors are ejected including eIF2, and the 80S ribosome enters the elongation phase of protein synthesis (reviewed in [109]). eIF2 activity relies on GTP hydrolysis and the guanine nucleotide exchange factor eIF2B must recycle GTP·eIF2 before it can be used in subsequent rounds of translation initiation (depicted by the eIF2:GTP exchange cycle). Several kinases have been identified that act on eIF2 to inhibit met-tRNAimet delivery by phosphorylating eIF2α, which include: PKR, a double-stranded RNA-dependent protein kinase typically activated during infection by RNA viruses in animals, PERK, which is activated in response to ER stress, HRI, a heme-sensing molecule, and GCN2, which senses nutrient availability.
Many plant and animal virus mRNAs are differentiated from host mRNAs by being uncapped, and therefore cannot recruit eIF4F by canonical means. Viruses inhibit cap-dependent translation through many mechanisms, including cleavage of translation initiation factors (reviewed in [7]). This affords virus mRNAs a competitive advantage by increasing availability of translational machinery for cap-independent translation. While many animal viruses cleave initiation factors to promote viral translation, this mechanism is not conserved in plant viruses, as a translation factor-specific protease has not been identified.
New concepts for IRES-mediated translation initiation in animal viruses
A lack of dependence on 5′ cap structures is a major mechanism exploited by many viruses to functionally distinguish and promote virus mRNA translation. IRES elements are RNA sequence or structures that function in lieu of the cap to recruit required translation factors and ribosomal subunits to the vicinity of the start codon. Cap-independent translation using IRESs was first observed in the poliovirus (PV) and EMCV genomic RNAs, and has since been characterized in many viral and cellular mRNAs including all picornaviruses, hepatitis C virus and pestiviruses, c-myc, p53 and the yeast URE2 IRES element [8, 9, 10, 11, 12]. Even DNA viruses such as Kaposi's Sarcoma Associated Herpes Virus, Epstein-Barr Virus and Herpes simplex virus utilize IRES elements, the latter for production of thymidine kinase, which is associated with pathogenicity and drug resistance [13]. IRES RNA structures are typically situated upstream of the initiating AUG codon, however HIV and eIF4GI contain IRES elements within the open reading frame [14, 15, 16]. Animal virus IRES elements are analogous to plant virus 3′ cap-independent translation enhancers (3′CITEs; reviewed in companion article), which function as bipartite pseudo-IRES elements to recruit initiation factors. A crucial feature of IRES-mediated translation allows continued or enhanced expression of virus proteins during cell stress when cap-dependent translation is repressed.
Virus IRES elements are currently grouped into classes based on their requirement for canonical translation initiation factors (as outlined in [17]). Type 1 and 2 IRES elements, in which type 2 lack a scanning step after ribosome binding, require several canonical initiation factors, including the C-terminus of eIF4G (a product of viral proteases) that recruits the 40S subunit via interaction with eIF3 (note similarity to cap-dependent translation; Figure 2 ). Type 1 IRES elements include PV and Hepatitis A Virus IRESs and Type 2 IRES elements include EMCV. Type 3 IRES elements, such as HCV, require only eIF3 and eIF2. Type 4 IRES elements like Cricket paralysis virus intergenic region (CrPV; IGR) and Plautia stali intestine virus IGR require no translation initiation factors, and bypass the need for met-tRNAi met by initiating at an alanine codon [18].
Figure 2.
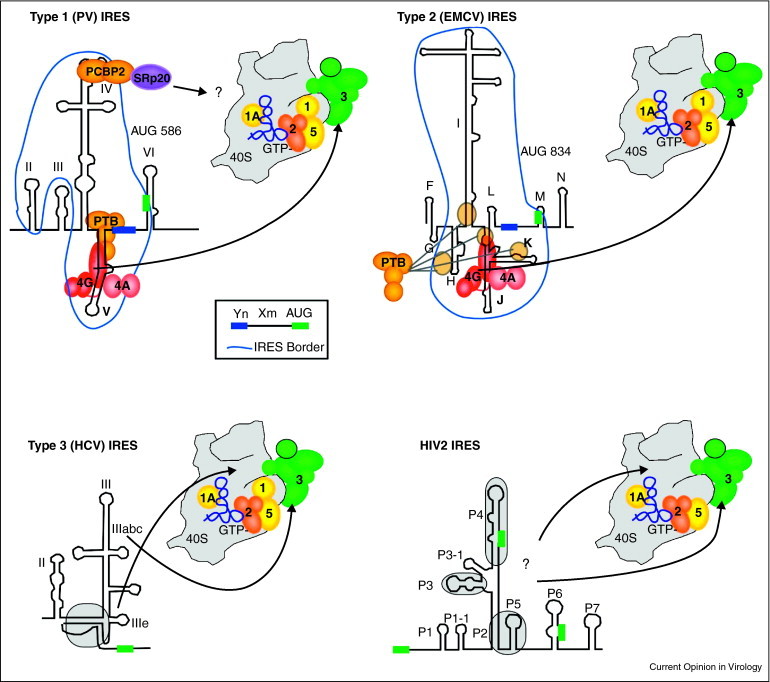
Initiation on virus internal ribosome entry sites (IRES). Virus IRESs use various non-canonical interactions with initiation factors and/or the 40S ribosome subunit. eIF4E is not involved in the binding of viral IRESs shown. Type 1 and 2 IRESs bind the central heat domain of eIF4GI in an analogous position and orientation on stem loop structures adjacent to conserved oligopyrimidine-spacer-AUG motifs (Yn-Xm-AUG) at the 3′ border of the IRES elements. The AUG codon basepairs with the initiator tRNA delivered by eIF2. eIF4GI binding is stimulated by eIF4A and modulated by PTB, which binds the same stem-loop structure as eIF4GI in Type 1 IRESs and binds a more diffuse footprint of RNA domains in Type 2 IRESs [31••, 33]. PV Type 1 IRES requires SRp20 for PCBP2 ITAF function and may play a role in bridging the IRES to the 43S complex [26]. Type 3 IRESs and the unclassified HIV2 IRES utilize interactions between eIF3 and 40S. The HCV pseudoknot (shaded in grey) positions the initiation codon (green box) in the mRNA binding cleft of the 40S subunit [36] and the apical stem loops of domain III interact directly with eIF3 [110, 111, 112]. HIV2 IRES contains 4 domains conserved among primate lentiviruses (shaded, P3, P4, P2–P5). HIV2 IRES can bind 48S preinitiation complexes at three AUGs (green boxes) [16]; however, eIF1 is not found in these complexes [37].
In addition to canonical translation initiation factors, other proteins enhance and regulate activity of many IRES elements (IRES trans-acting factors; ITAFs). ITAFs are thought to provide chaperone function, but may play other roles that are important in overcoming translation restriction during cell stress or innate immune blockades [19]. More tightly folded IRESs are proposed to create less dependence on eIFs and ITAFs for function [20]. Consistent with this observation, structural data indicate the Type 3 (HCV) and Type 4 (CrPV) IRESs are very compact [21•, 22••].
Though many reports catalog dependency of IRESs on ITAFs, little is known about how larger Type 1 or 2 IRESs actually interact with ribosomes and how ITAFs and canonical initiation factors contribute to activity. PCBP2 plays multifunctional and crucial roles for PV translation by binding the 5′ terminal cloverleaf structure, stemloop IV of the IRES, and also PABP on the poly(A) tail to circularize the PV RNA after PABP cleavage by a viral proteinase [23, 24]. Interestingly, PCBP2 requires the cellular splicing factor SRp20 for function as an ITAF on PV RNA [25, 26]. The precise mechanism is unknown, but SRp20 may provide a ribosome recruitment/bridging role (Figure 2). Polypyrimidine tract binding protein (PTB) augments translation of PV, and host Cat-1 and c-myc IRES elements [27, 28, 29, 30]. PTB functions as an RNA chaperone to reorganize the PV IRES RNA structure in a manner that increases the affinity of eIF4G for the IRES element [31••, 32].
Recent reports indicate that diverse IRESs may interact with initiation factors and ribosomes in very similar ways. Despite a lack of sequence relatedness, both Type 1 and Type 2 viral IRESs bind eIF4GI and eIF4A in analogous regions immediately upstream of the same Yn-Xm-AUG stem loop motif (Figure 2; [33]). This induces conformational changes in RNA structure at the 3′ border of the IRES and suggests a model for both IRES types in which eIF4G binds the IRES first, and then recruits eIF4A and eIF4B that promote conformational changes to allow 43S binding at an adjacent site (Figure 2). Similarly, IRESs from diverse groups may interact with the ribosome in comparable unifying mechanisms. Small ribosome subunit protein 25 (Rps25) is a crucial interaction partner for both HCV and CrPV IGR activity [34], without which they cannot bind 40S ribosomes. Since binding of both HCV and IGR induce related conformational remodeling in the 18S RNA, it will be interesting to determine if Rps25p is involved in these conformational changes. CryoEM analysis of WT and Rps25p deleted ribosomes with IGR reveals that Rps25p interacts with IRES RNA near the head domain of the ribosome, and together with neighboring Rps5, constitute the major binding domain on the ribosome. Though other IRES RNA loops interact with the ribosome decoding center, they do not contribute to IRES-ribosome binding affinity [35]. Finally, both the conserved HCV pseudoknot and the P-site binding domain of CrPV IGR fold into tRNA-like structures to mimic tRNA interaction with mRNA [21•, 22••, 36]. These results indicate that though IRESs are diverse, their basic mechanisms for interaction with factors and ribosomes may be quite similar.
Recently a new report on the understudied HIV IRESs indicates they may straddle Type 2 and 3 IRES classifications. A conserved core stem loop structure located in HIV-1, HIV-2 and SIV-IRESs was found to bind eIF3 and 40S ribosomes, similar to Type 3 IRESs (Figure 2). However, analysis of stalled initiation complexes on these IRESs showed they contain all canonical initiation factors except eIF4E and eIF1. The latter is surprising since eIF1 normally binds 40S subunits in conjunction with eIF5, eIF1A, and eIF3. This result suggests the HIV IRES uses pools of 40S subunits containing only eIF3 to assemble initiation complexes [37].
Use of alternative factors for initiator tRNA delivery
Animal RNA virus infection often produces double stranded RNA and cell stress responses that activate eIF2α kinases PKR or HRI, respectively, which phosphorylate the alpha subunit of eIF2 (reviewed in [38]) and result in repression of global translation by depleting the ternary complex (Figure 1; [39]). Some viruses evade the antiviral activation of PKR through interesting antagonistic mechanisms (reviewed in [40, 41]). Viruses also encode proteins to directly deal with limited ternary complexes. For instance, Herpes simplex virus protein ICP34.5 recruits protein phosphatase P1, known to dephosphorylate eIF2α, to reactivate translation after an initial phase of inhibition [42, 43]. This mechanism is recapitulated during coronavirus and African swine fever virus by Gene 7 and DP71L, respectively [44, 45]. Alternatively, several viruses have evolved alternate pathways to avoid the requirement for eIF2 ternary complex binding to 40S ribosomes to support viral translation during eIF2 phosphorylation. The Dicistroviridae class of viruses (including CrPV) contain IGR IRES elements resembling tRNAs, which efficiently bind the P-site of the ribosome and promote initiation often at an alanine codon [18, 46]. This circumvents the need for translation initiation using met-tRNAi met since translation is initiated using elongator tRNAs.
Recent reports have emerged of a resistance strategy unique to animal viruses to cope with eIF2α phosphorylation; the use of alternative proteins for initiator tRNA delivery. These include eukaryotic initiation factor 2A, a single polypeptide unrelated to the heterotrimeric complex eIF2 [47, 48], Ligatin (also known as eIF2D; [49, 50••]), MCT1/DENR [50••], and eukaryotic initiation 5B (eIF5B) (Figure 3 ; [51, 52]). Identification of these proteins indicates a more diverse repertoire of pathways exists for translation initiation than previously thought, and highlights the need to investigate translation initiation of other viruses displaying eIF2-independent translation.
Figure 3.
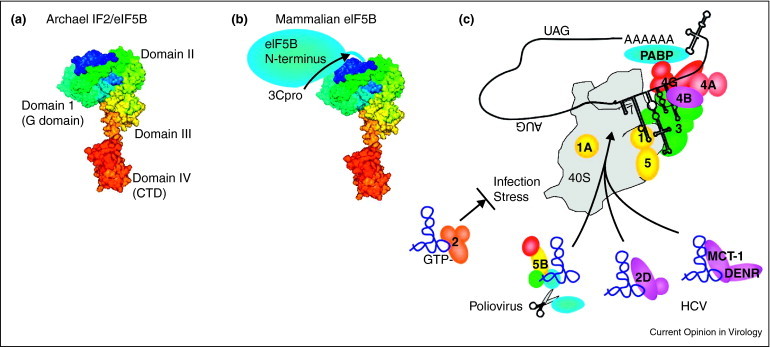
eIF5B and other eIF2-independent mechanisms for translation initiation. (A) The crystal structure of archaeal IF2/eIF5B is organized into 4 highly conserved domains (labeled 1-IV). (B) Mammalian eIF5B contains the C-terminal half that is homologous to archaeal IF2 and a N-terminal regulatory domain whose structure is unknown that is removed by PV 3Cpro cleavage. (C) Infection and stress activate eIF2α kinases that reduce concentrations of eIF2·GTP·met-tRNAimet ternary complex, inhibiting translation from viral IRES elements. PV and HCV may overcome this restriction by recruiting alternate proteins to help deliver met-tRNAimet in GTP-dependent or GTP-independent mechanisms.
eIF2A was first identified based on its ability to direct binding of met-tRNAi met with 40S ribosomal subunits in an AUG codon-dependent manner [48]. In yeast, eIF2A was shown to act as a suppressor of cap-independent initiation on the URE2, PABP and GIC1 IRES elements [53, 54]. The authors hypothesized that kinetic limitations of the eIF2A-dependent pathway for met-tRNAi met delivery inhibit initiation while eIF2 is active (perhaps because of overlapping binding sites on the ribosome). However, when eIF2 is inactivated, translation can still proceed on IRES-containing transcripts at a slower rate. This hypothesis has yet to be directly tested, but positions eIF2A as an important stress-responsive protein for translation of RNA viruses. Consistent with this hypothesis, eIF2A is important for ongoing Sindbis virus translation when high levels of eIF2α phosphorylation are observed [55]. New data indicate that eIF2A can mediate delivery of met-tRNAi met to the 40S subunit to form 48S complexes on the HCV IRES element using purified components [56••]. Furthermore, domain IIId of the HCV IRES was shown to interact with eIF2A directly. These results suggest a mechanism wherein the IRES recruits the eIF2A:met-tRNAi met complex to the ribosome rather than the ribosome recruiting the eIF2A:met-tRNAi met complex, at least on some mRNAs. Kinetics of eIF2A expression and activity during eIF2α phosphorylation, the precise mechanism of action for met-tRNAi met delivery by eIF2A, and validation of additional mRNA targets must be investigated to conclusively demonstrate a role for eIF2A at this step of translation initiation.
In an attempt to further characterize the role of eIF2A, Dmitriev et al. discovered an important role for ligatin in 48S complex formation on the HCV IRES element (Figure 3C; [49]). Interestingly, ligatin, which they termed eIF2D, can promote 48S complex formation using phenylalanine-tRNA on the HCV IRES element when the initiating AUG is mutated to a UUU codon. This result suggests alternative initiation codons may be used to produce proteins with altered functions or half-lives. Skabkin et al. extended these findings to show Ligatin can promote formation of 48S complexes on the CSFV IRES element and the Sindbis virus 26S RNA [50••]. Interestingly, the proteins MCT1 and DENR, which are homologous to the N and C-termini of ligatin, respectively, can work simultaneously to promote 48S complex formation (Figure 3C). Further work is necessary to delineate in vivo activity of ligatin in translation initiation of these viral mRNAs.
Eukaryotic initiation factor 5B is the eukaryotic homolog of bacterial initiation factor 2 (IF2; [57]). The domain architecture of mammalian eIF5B is highly reminiscent of archaeal eIF5B with the exception of a large N-terminal extension on mammalian eIF5B that may regulate met-tRNAi met binding (compare Figure 3A and B; [58]). Indeed, archaeal eIF5B can directly bind met-tRNAi met, although S. cerevisiae eIF5B has a relatively low affinity [59]. These results suggest the N-terminus of mammalian eIF5B must be removed in order for it to function as a met-tRNAi met carrier molecule. Consistent with this hypothesis, poliovirus 3C proteinase cleaves eIF5B during infection liberating the C-terminal fragment and potentiating its use in met-tRNAi met delivery [60]. The C-terminus is then capable of enhancing IRES-mediated translation of PV when ternary complex is depleted [52]. Interestingly, HCV and CSFV have also been shown to act in an eIF2-independent mode using eIF5B to assemble initiation complexes [51, 61]. In the cases of the HCV and CSFV IRES elements, eIF5B can function in the presence of the N-terminal extension. Therefore, it will be of interest to delineate the role of the N-terminus in regulating met-tRNAi met delivery, and determine whether either cleavage or posttranslational modification contribute to this function during viral infection.
Although viruses differ in the route to efficient initiation of protein synthesis, many plus sense RNA viruses can employ similar strategies to enable ongoing protein synthesis during global translation inhibition by eIF2 phosphorylation. Interestingly, plant viruses do not contend with robust eIF2 phosphorylation despite the presence of PKR in plant cells [62]; these viruses clearly have a different set of parameters to enable expression of viral proteins. Perhaps activity of alternative met-tRNAi met delivery proteins is higher on plant virus RNAs thereby making them more competitive with ongoing cellular protein synthesis.
Long-range RNA interactions that increase translation competitiveness
Animal viruses with capped genomes or mRNAs must also compete for translational machinery, but must navigate downregulation of cap-dependent translation that is often associated with infection. Closed loop structures mediated by long range RNA:RNA kissing interactions are a common paradigm in plant viruses that have rarely been described in animal viruses. However, in animal viruses the 3′UTRs can enhance viral IRES-mediated translation, though precise mechanisms are lacking [63, 64, 65, 66, 67, 68, 69, 70].
One example of the influence of the 3′UTR on translation of a 5′ capped, non-polyadenylated animal virus is observed in the Dengue virus (DEN) mRNA. Two conserved dumbbell-shaped RNA structures within the 3′ UTR not only form local pseudoknots important for RNA replication, but also work cooperatively to stimulate translation [71]. DEN can switch from cap-dependent to a non-canonical cap-independent translation mode when eIF4E is limiting. The mechanism does not involve an IRES, but requires a closed loop [72]. The details of this mechanism are unclear but probably involves RNA:RNA hybridization through the cyclization sequence (CS) and the upstream AUG region (UAR) [73, 74•]. PABP may stimulate translation by enhancing closed loop formation by binding the 3′UTR [74•] and eIF4F simultaneously, despite the lack of a poly(A) tail. The dumbbell structures in the 3′ UTR may also help to recruit trans-acting or initiation factors to stimulate translation. The 3′UTR of DEN mRNA has been demonstrated to interact with typical ITAFs PTB, La, and YB-1 [75, 76, 77], which could function to enhance DEN translation in a manner similar to their influence in IRES-mediated translation. Overall, the RNA elements in the DEN 3′ UTR may function similarly to many plant viruses with 3′ UTR CITEs (see companion review in this issue) to stimulate translation when eIF4E becomes limited from stress or innate immunity activation. It will be important to further elucidate host factors that interact with the sequences in the 3′ UTR and identify their precise mechanisms of translational regulation.
Long range RNA kissing interactions have also been described for HCV, where the RNA domain 5BSL3.2 within the 3′ region of the NS5B ORF base pairs with a portion of the HCV IRES. Unlike DEN, this RNA:RNA interaction downregulates HCV IRES translation and may play a role in directing viral RNA to switch from translation to RNA replication [78••]. Further, PCBP2, which functions as an ITAF in PV translation, binds both HCV 5′ and 3 UTR. Electron microscopy demonstrated that PCBP2, probably via dimerization, converted RNA from linear to circular forms. Interaction of PCBP2 with HCV replicons stimulated translation, perhaps via formation of closed loops, though the mechanism was not determined [79]. These results indicate that HCV circularization modulates translation in multiple, possibly temporal ways.
Long range RNA interactions linking the 5′ and 3′ UTR have otherwise only been described in the FMDV viral RNA [80]. Despite a known importance for the 3′UTR of FMDV in stimulating IRES-mediated translation [68], the functional relevance of this interaction is only speculative. More studies dedicated to understanding the functional relevance of interactions between the 5′ and 3′ UTRs, or possibly 3′ coding regions, of animal viruses may highlight important roles in recruiting translation factors, similar to the paradigm in plant CITEs. Studies of plant RNA virus translation systems may identify additional, noncanonical proteins that are important for efficient competition with cellular mRNAs for the translational machinery.
Perspectives and future directions
Animal and plant viruses have evolved cunning mechanisms to ensure competition with cellular mRNAs for the translation machinery. IRESs share key functions with the 3′ CITEs prevalent in plant virus systems, including the emerging theme of tRNA mimicry; however, no animal virus has yet been found with a 3′ CITE. This is despite the recurring animal virus theme of 5′-3′ closed loop mRNA structures that in principle could allow ribosome recruitment to 3′ structures in spatial proximity to nearby 5′ initiator codons. The principle of complex recruitment to one end of the RNA and transfer to the other end exists in enteroviruses, where the negative strand RNA replication complex assembles on a 5′ cloverleaf structure that, within a closed loop, positions the replicase near the 3′ end of the template to initiate replication on the other end of the template (reviewed in [81, 82]). It is unclear whether this paradigm exists for translation in animal cells, but it may partly emerge with further study of flaviviruses such as DEN. Avoidance of innate immunity and stress responses could explain the sharp divergence between plant and animal systems regarding these responses and how viruses must adapt. This is highlighted in the novel responses to stress inhibition of translation where animal viruses employ different cellular proteins to deliver met-tRNAi met to the ribosome. The differences between plant and animal virus translation, and translation of cellular mRNAs indicate that themes are paralleled between the mRNA templates with slight variations to account for organism-specific translational regulation. Additional mechanistic insights could be gained if more attention is devoted towards bridging common translational control mechanisms in the animal and plant virus kingdoms.
References and recommended reading
Papers of particular interest, published within the period of review, have been highlighted as:
• of special interest
•• of outstanding interest
Acknowledgements
We would like to apologize to those researchers whose work was not discussed in this review owing to space constraints. LCR is supported by NIH T32 AI07471 and work in our laboratory is supported by NIH Public Health Service Grant AI50237.
References
- 1•.Lopez-Lastra M., Ramdohr P., Letelier A., Vallejos M., Vera-Otarola J., Valiente-Echeverria F. Translation initiation of viral mRNAs. Rev Med Virol. 2010;20:177–195. doi: 10.1002/rmv.649. [DOI] [PMC free article] [PubMed] [Google Scholar]; An excellent review of viral mechanisms of translational control.
- 2.Gallie D.R. A tale of two termini: a functional interaction between the termini of an mRNA is a prerequisite for efficient translation initiation. Gene. 1998;216:1–11. doi: 10.1016/s0378-1119(98)00318-7. [DOI] [PubMed] [Google Scholar]
- 3.Mazumder B., Seshadri V., Fox P.L. Translational control by the 3′-UTR: the ends specify the means. Trends Biochem Sci. 2003;28:91–98. doi: 10.1016/S0968-0004(03)00002-1. [DOI] [PubMed] [Google Scholar]
- 4.Amrani N., Ghosh S., Mangus D.A., Jacobson A. Translation factors promote the formation of two states of the closed-loop mRNP. Nature. 2008;453:1276–1280. doi: 10.1038/nature06974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bonderoff J.M., Lloyd R.E. Time-dependent increase in ribosome processivity. Nucleic Acids Res. 2010;38:7054–7067. doi: 10.1093/nar/gkq566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kahvejian A., Svitkin Y.V., Sukarieh R., M’Boutchou M.N., Sonenberg N. Mammalian poly(A)-binding protein is a eukaryotic translation initiation factor, which acts via multiple mechanisms. Genes Dev. 2005;19:104–113. doi: 10.1101/gad.1262905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lloyd R.E. Translational control by viral proteinases. Virus Res. 2006;119:76–88. doi: 10.1016/j.virusres.2005.10.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Jang S.K., Krausslich H.G., Nicklin M.J., Duke G.M., Palmenberg A.C., Wimmer E. A segment of the 5′ nontranslated region of encephalomyocarditis virus RNA directs internal entry of ribosomes during in vitro translation. J Virol. 1988;62:2636–2643. doi: 10.1128/jvi.62.8.2636-2643.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Komar A.A., Lesnik T., Cullin C., Merrick W.C., Trachsel H., Altmann M. Internal initiation drives the synthesis of Ure2 protein lacking the prion domain and affects [URE3] propagation in yeast cells. EMBO J. 2003;22:1199–1209. doi: 10.1093/emboj/cdg103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Nanbru C., Lafon I., Audigier S., Gensac M.C., Vagner S., Huez G., Prats A.C. Alternative translation of the proto-oncogene c-myc by an internal ribosome entry site. J Biol Chem. 1997;272:32061–32066. doi: 10.1074/jbc.272.51.32061. [DOI] [PubMed] [Google Scholar]
- 11.Pelletier J., Sonenberg N. Internal initiation of translation of eukaryotic mRNA directed by a sequence derived from poliovirus RNA. Nature. 1988;334:320–325. doi: 10.1038/334320a0. [DOI] [PubMed] [Google Scholar]
- 12.Stoneley M., Paulin F.E., Le Quesne J.P., Chappell S.A., Willis A.E. C-Myc 5′ untranslated region contains an internal ribosome entry segment. Oncogene. 1998;16:423–428. doi: 10.1038/sj.onc.1201763. [DOI] [PubMed] [Google Scholar]
- 13.Griffiths A., Coen D.M. An unusual internal ribosome entry site in the herpes simplex virus thymidine kinase gene. Proc Natl Acad Sci USA. 2005;102:9667–9672. doi: 10.1073/pnas.0504132102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Byrd M.P., Zamora M., Lloyd R.E. Translation of eukaryotic translation initiation factor 4GI (eIF4GI) proceeds from multiple mRNAs containing a novel cap-dependent internal ribosome entry site (IRES) that is active during poliovirus infection. J Biol Chem. 2005;280:18610–18622. doi: 10.1074/jbc.M414014200. [DOI] [PubMed] [Google Scholar]
- 15.Herbreteau C.H., Weill L., Decimo D., Prevot D., Darlix J.L., Sargueil B., Ohlmann T. HIV-2 genomic RNA contains a novel type of IRES located downstream of its initiation codon. Nat Struct Mol Biol. 2005;12:1001–1007. doi: 10.1038/nsmb1011. [DOI] [PubMed] [Google Scholar]
- 16.Weill L., James L., Ulryck N., Chamond N., Herbreteau C.H., Ohlmann T., Sargueil B. A new type of IRES within gag coding region recruits three initiation complexes on HIV-2 genomic RNA. Nucleic Acids Res. 2010;38:1367–1381. doi: 10.1093/nar/gkp1109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hellen C.U. IRES-induced conformational changes in the ribosome and the mechanism of translation initiation by internal ribosomal entry. Biochim Biophys Acta. 2009;1789:558–570. doi: 10.1016/j.bbagrm.2009.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hertz M.I., Thompson S.R. Mechanism of translation initiation by Dicistroviridae IGR IRESs. Virology. 2011;411:355–361. doi: 10.1016/j.virol.2011.01.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Komar A.A., Hatzoglou M. Cellular IRES-mediated translation: the war of ITAFs in pathophysiological states. Cell Cycle. 2011;10:229–240. doi: 10.4161/cc.10.2.14472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Filbin M.E., Kieft J.S. Toward a structural understanding of IRES RNA function. Curr Opin Struct Biol. 2009;19:267–276. doi: 10.1016/j.sbi.2009.03.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21•.Lavender C.A., Ding F., Dokholyan N.V., Weeks K.M. Robust and generic RNA modeling using inferred constraints: a structure for the hepatitis C virus IRES pseudoknot domain. Biochemistry. 2010;49:4931–4933. doi: 10.1021/bi100142y. [DOI] [PMC free article] [PubMed] [Google Scholar]; Computer modeling of the pseudoknot domain of HCV predicts tRNA-like molecular mimicry.
- 22••.Zhu J., Korostelev A., Costantino D.A., Donohue J.P., Noller H.F., Kieft J.S. Crystal structures of complexes containing domains from two viral internal ribosome entry site (IRES) RNAs bound to the 70S ribosome. Proc Natl Acad Sci USA. 2011;108:1839–1844. doi: 10.1073/pnas.1018582108. [DOI] [PMC free article] [PubMed] [Google Scholar]; X-ray crystallography of IGR IRES elements bound to the 70S ribosome resemble tRNA:ribosome interactions.
- 23.Herold J., Andino R. Poliovirus RNA replication requires genome circularization through a protein-protein bridge. Mol Cell. 2001;7:581–591. doi: 10.1016/S1097-2765(01)00205-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sean P., Nguyen J.H., Semler B.L. Altered interactions between stem-loop IV within the 5′ noncoding region of coxsackievirus RNA and poly(rC) binding protein 2: effects on IRES-mediated translation and viral infectivity. Virology. 2009;389:45–58. doi: 10.1016/j.virol.2009.03.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bedard K.M., Daijogo S., Semler B.L. A nucleo-cytoplasmic SR protein functions in viral IRES-mediated translation initiation. EMBO J. 2007;26:459–467. doi: 10.1038/sj.emboj.7601494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Fitzgerald K.D., Semler B.L. Re-localization of cellular protein SRp20 during poliovirus infection: bridging a viral IRES to the host cell translation apparatus. PLoS Pathog. 2011;7:e1002127. doi: 10.1371/journal.ppat.1002127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Cobbold L.C., Spriggs K.A., Haines S.J., Dobbyn H.C., Hayes C., de Moor C.H., Lilley K.S., Bushell M., Willis A.E. Identification of internal ribosome entry segment (IRES)-trans-acting factors for the Myc family of IRESs. Mol Cell Biol. 2008;28:40–49. doi: 10.1128/MCB.01298-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Cobbold L.C., Wilson L.A., Sawicka K., King H.A., Kondrashov A.V., Spriggs K.A., Bushell M., Willis A.E. Upregulated c-myc expression in multiple myeloma by internal ribosome entry results from increased interactions with and expression of PTB-1 and YB-1. Oncogene. 2010;29:2884–2891. doi: 10.1038/onc.2010.31. [DOI] [PubMed] [Google Scholar]
- 29.Hellen C.U., Witherell G.W., Schmid M., Shin S.H., Pestova T.V., Gil A., Wimmer E. A cytoplasmic 57-kDa protein that is required for translation of picornavirus RNA by internal ribosomal entry is identical to the nuclear pyrimidine tract-binding protein. Proc Natl Acad Sci USA. 1993;90:7642–7646. doi: 10.1073/pnas.90.16.7642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Majumder M., Yaman I., Gaccioli F., Zeenko V.V., Wang C., Caprara M.G., Venema R.C., Komar A.A., Snider M.D., Hatzoglou M. The hnRNA-binding proteins hnRNP L and PTB are required for efficient translation of the Cat-1 arginine/lysine transporter mRNA during amino acid starvation. Mol Cell Biol. 2009;29:2899–2912. doi: 10.1128/MCB.01774-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31••.Kafasla P., Morgner N., Robinson C.V., Jackson R.J. Polypyrimidine tract-binding protein stimulates the poliovirus IRES by modulating eIF4G binding. EMBO J. 2010;29:3710–3722. doi: 10.1038/emboj.2010.231. [DOI] [PMC free article] [PubMed] [Google Scholar]; Provides mechanistic insight into the role of PTB in regulating PV IRES-mediated translation.
- 32.Song Y., Tzima E., Ochs K., Bassili G., Trusheim H., Linder M., Preissner K.T., Niepmann M. Evidence for an RNA chaperone function of polypyrimidine tract-binding protein in picornavirus translation. RNA. 2005;11:1809–1824. doi: 10.1261/rna.7430405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.de Breyne S., Yu Y., Unbehaun A., Pestova T.V., Hellen C.U. Direct functional interaction of initiation factor eIF4G with type 1 internal ribosomal entry sites. Proc Natl Acad Sci USA. 2009;106:9197–9202. doi: 10.1073/pnas.0900153106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Landry D.M., Hertz M.I., Thompson S.R. RPS25 is essential for translation initiation by the Dicistroviridae and hepatitis C viral IRESs. Genes Dev. 2009;23:2753–2764. doi: 10.1101/gad.1832209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Muhs M., Yamamoto H., Ismer J., Takaku H., Nashimoto M., Uchiumi T., Nakashima N., Mielke T., Hildebrand P.W., Nierhaus K.H. Structural basis for the binding of IRES RNAs to the head of the ribosomal 40S subunit. Nucleic Acids Res. 2011;39:5264–5275. doi: 10.1093/nar/gkr114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Berry K.E., Waghray S., Doudna J.A. The HCV IRES pseudoknot positions the initiation codon on the 40S ribosomal subunit. RNA. 2010;16:1559–1569. doi: 10.1261/rna.2197210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Locker N., Chamond N., Sargueil B. A conserved structure within the HIV gag open reading frame that controls translation initiation directly recruits the 40S subunit and eIF3. Nucleic Acids Res. 2011;39:2367–2377. doi: 10.1093/nar/gkq1118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Garcia M.A., Meurs E.F., Esteban M. The dsRNA protein kinase PKR: virus and cell control. Biochimie. 2007;89:799–811. doi: 10.1016/j.biochi.2007.03.001. [DOI] [PubMed] [Google Scholar]
- 39.Dever T.E., Yang W., Astrom S., Bystrom A.S., Hinnebusch A.G. Modulation of tRNA(iMet), eIF-2, and eIF-2B expression shows that GCN4 translation is inversely coupled to the level of eIF-2.GTP.Met-tRNA(iMet) ternary complexes. Mol Cell Biol. 1995;15:6351–6363. doi: 10.1128/mcb.15.11.6351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Schneider R.J., Mohr I. Translation initiation and viral tricks. Trends Biochem Sci. 2003;28:130–136. doi: 10.1016/S0968-0004(03)00029-X. [DOI] [PubMed] [Google Scholar]
- 41.Pisarev A.V., Shirokikh N.E., Hellen C.U. Translation initiation by factor-independent binding of eukaryotic ribosomes to internal ribosomal entry sites. C R Biol. 2005;328:589–605. doi: 10.1016/j.crvi.2005.02.004. [DOI] [PubMed] [Google Scholar]
- 42.Li Y., Zhang C., Chen X., Yu J., Wang Y., Yang Y., Du M., Jin H., Ma Y., He B. ICP34.5 of herpes simplex virus facilitates the initiation of protein translation by bridging eIF2{alpha} and protein phosphatase 1. J Biol Chem. 2011;286:24785–24792. doi: 10.1074/jbc.M111.232439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Verpooten D., Feng Z., Valyi-Nagy T., Ma Y., Jin H., Yan Z., Zhang C., Cao Y., He B. Dephosphorylation of eIF2alpha mediated by the gamma134.5 protein of herpes simplex virus 1 facilitates viral neuroinvasion. J Virol. 2009;83:12626–12630. doi: 10.1128/JVI.01431-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Cruz J.L., Sola I., Becares M., Alberca B., Plana J., Enjuanes L., Zuniga S. Coronavirus gene 7 counteracts host defenses and modulates virus virulence. PLoS Pathog. 2011;7:e1002090. doi: 10.1371/journal.ppat.1002090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Zhang F., Moon A., Childs K., Goodbourn S., Dixon L.K. The African swine fever virus DP71L protein recruits the protein phosphatase 1 catalytic subunit to dephosphorylate eIF2alpha and inhibits CHOP induction but is dispensable for these activities during virus infection. J Virol. 2010;84:10681–10689. doi: 10.1128/JVI.01027-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Hertz M.I., Thompson S.R. In vivo functional analysis of the Dicistroviridae intergenic region internal ribosome entry sites. Nucleic Acids Res. 2011;39:7276–7288. doi: 10.1093/nar/gkr427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Adams S.L., Safer B., Anderson W.F., Merrick W.C. Eukaryotic initiation complex formation. Evidence for two distinct pathways. J Biol Chem. 1975;250:9083–9089. [PubMed] [Google Scholar]
- 48.Merrick W.C., Anderson W.F. Purification and characterization of homogeneous protein synthesis initiation factor M1 from rabbit reticulocytes. J Biol Chem. 1975;250:1197–1206. [PubMed] [Google Scholar]
- 49.Dmitriev S.E., Terenin I.M., Andreev D.E., Ivanov P.A., Dunaevsky J.E., Merrick W.C., Shatsky I.N. GTP-independent tRNA delivery to the ribosomal P-site by a novel eukaryotic translation factor. J Biol Chem. 2010;285:26779–26787. doi: 10.1074/jbc.M110.119693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50••.Skabkin M.A., Skabkina O.V., Dhote V., Komar A.A., Hellen C.U., Pestova T.V. Activities of Ligatin and MCT-1/DENR in eukaryotic translation initiation and ribosomal recycling. Genes Dev. 2010;24:1787–1801. doi: 10.1101/gad.1957510. [DOI] [PMC free article] [PubMed] [Google Scholar]; Role for Ligatin and MCT1/DENR in delivery of initiator tRNA to HCV-like IRES elements and SV 26S mRNA.
- 51.Terenin I.M., Dmitriev S.E., Andreev D.E., Shatsky I.N. Eukaryotic translation initiation machinery can operate in a bacterial-like mode without eIF2. Nat Struct Mol Biol. 2008;15:836–841. doi: 10.1038/nsmb.1445. [DOI] [PubMed] [Google Scholar]
- 52.White J.P., Reineke L.C., Lloyd R.E. Poliovirus switches to an eIF2-independent mode of translation during infection. J Virol. 2011;85:8884–8893. doi: 10.1128/JVI.00792-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Komar A.A., Gross S.R., Barth-Baus D., Strachan R., Hensold J.O., Goss Kinzy T., Merrick W.C. Novel characteristics of the biological properties of the yeast Saccharomyces cerevisiae eukaryotic initiation factor 2A. J Biol Chem. 2005;280:15601–15611. doi: 10.1074/jbc.M413728200. [DOI] [PubMed] [Google Scholar]
- 54.Reineke L.C., Merrick W.C. Characterization of the functional role of nucleotides within the URE2 IRES element and the requirements for eIF2A-mediated repression. RNA. 2009;15:2264–2277. doi: 10.1261/rna.1722809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Ventoso I., Sanz M.A., Molina S., Berlanga J.J., Carrasco L., Esteban M. Translational resistance of late alphavirus mRNA to eIF2alpha phosphorylation: a strategy to overcome the antiviral effect of protein kinase PKR. Genes Dev. 2006;20:87–100. doi: 10.1101/gad.357006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56••.Kim J.H., Park S.M., Park J.H., Keum S.J., Jang S.K. eIF2A mediates translation of hepatitis C viral mRNA under stress conditions. EMBO J. 2011;30:2454–2464. doi: 10.1038/emboj.2011.146. [DOI] [PMC free article] [PubMed] [Google Scholar]; Evidence that eIF2A is capable of met-tRNAimet delivery for translation initiation on the HCV IRES element.
- 57.Lee J.H., Choi S.K., Roll-Mecak A., Burley S.K., Dever T.E. Universal conservation in translation initiation revealed by human and archaeal homologs of bacterial translation initiation factor IF2. Proc Natl Acad Sci USA. 1999;96:4342–4347. doi: 10.1073/pnas.96.8.4342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Dever T.E., Roll-Mecak A., Choi S.K., Lee J.H., Cao C., Shin B.S., Burley S.K. Universal translation initiation factor IF2/eIF5B. Cold Spring Harb Symp Quant Biol. 2001;66:417–424. doi: 10.1101/sqb.2001.66.417. [DOI] [PubMed] [Google Scholar]
- 59.Guillon L., Schmitt E., Blanquet S., Mechulam Y. Initiator tRNA binding by e/aIF5B, the eukaryotic/archaeal homologue of bacterial initiation factor IF2. Biochemistry. 2005;44:15594–15601. doi: 10.1021/bi051514j. [DOI] [PubMed] [Google Scholar]
- 60.de Breyne S., Bonderoff J.M., Chumakov K.M., Lloyd R.E., Hellen C.U. Cleavage of eukaryotic initiation factor eIF5B by enterovirus 3C proteases. Virology. 2008;378:118–122. doi: 10.1016/j.virol.2008.05.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Pestova T.V., de Breyne S., Pisarev A.V., Abaeva I.S., Hellen C.U. eIF2-dependent and eIF2-independent modes of initiation on the CSFV IRES: a common role of domain II. EMBO J. 2008;27:1060–1072. doi: 10.1038/emboj.2008.49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Langland J.O., Jin S., Jacobs B.L., Roth D.A. Identification of a plant-encoded analog of PKR, the mammalian double-stranded RNA-dependent protein kinase. Plant Physiol. 1995;108:1259–1267. doi: 10.1104/pp.108.3.1259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Bradrick S.S., Walters R.W., Gromeier M. The hepatitis C virus 3′-untranslated region or a poly(A) tract promote efficient translation subsequent to the initiation phase. Nucleic Acids Res. 2006;34:1293–1303. doi: 10.1093/nar/gkl019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Chattopadhyay M., Shi K., Yuan X., Simon A.E. Long-distance kissing loop interactions between a 3′ proximal Y-shaped structure and apical loops of 5′ hairpins enhance translation of Saguaro cactus virus. Virology. 2011;417:113–125. doi: 10.1016/j.virol.2011.05.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Chiu W.W., Kinney R.M., Dreher T.W. Control of translation by the 5′- and 3′-terminal regions of the dengue virus genome. J Virol. 2005;79:8303–8315. doi: 10.1128/JVI.79.13.8303-8315.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Dobrikova E., Florez P., Bradrick S., Gromeier M. Activity of a type 1 picornavirus internal ribosomal entry site is determined by sequences within the 3′ nontranslated region. Proc Natl Acad Sci USA. 2003;100:15125–15130. doi: 10.1073/pnas.2436464100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Isken O., Grassmann C.W., Yu H., Behrens S.E. Complex signals in the genomic 3′ nontranslated region of bovine viral diarrhea virus coordinate translation and replication of the viral RNA. RNA. 2004;10:1637–1652. doi: 10.1261/rna.7290904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Lopez de Quinto S., Saiz M., de la Morena D., Sobrino F., Martinez-Salas E. IRES-driven translation is stimulated separately by the FMDV 3′-NCR and poly(A) sequences. Nucleic Acids Res. 2002;30:4398–4405. doi: 10.1093/nar/gkf569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Song Y., Friebe P., Tzima E., Junemann C., Bartenschlager R., Niepmann M. The hepatitis C virus RNA 3′-untranslated region strongly enhances translation directed by the internal ribosome entry site. J Virol. 2006;80:11579–11588. doi: 10.1128/JVI.00675-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Weinlich S., Huttelmaier S., Schierhorn A., Behrens S.E., Ostareck-Lederer A., Ostareck D.H. IGF2BP1 enhances HCV IRES-mediated translation initiation via the 3′UTR. RNA. 2009;15:1528–1542. doi: 10.1261/rna.1578409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Manzano M., Reichert E.D., Polo S., Falgout B., Kasprzak W., Shapiro B.A., Padmanabhan R. Identification of cis-acting elements in the 3′-untranslated region of the dengue virus type 2 RNA that modulate translation and replication. J Biol Chem. 2011;286:22521–22534. doi: 10.1074/jbc.M111.234302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Edgil D., Polacek C., Harris E. Dengue virus utilizes a novel strategy for translation initiation when cap-dependent translation is inhibited. J Virol. 2006;80:2976–2986. doi: 10.1128/JVI.80.6.2976-2986.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Alvarez D.E., Filomatori C.V., Gamarnik A.V. Functional analysis of dengue virus cyclization sequences located at the 5′ and 3′UTRs. Virology. 2008;375:223–235. doi: 10.1016/j.virol.2008.01.014. [DOI] [PubMed] [Google Scholar]
- 74•.Polacek C., Friebe P., Harris E. Poly(A)-binding protein binds to the non-polyadenylated 3′ untranslated region of dengue virus and modulates translation efficiency. J Gen Virol. 2009;90:687–692. doi: 10.1099/vir.0.007021-0. [DOI] [PubMed] [Google Scholar]; PABP binds the 3′UTR of DEN mRNA and enhances translation possibly by promoting closed-loop formation.
- 75.Agis-Juarez R.A., Galvan I., Medina F., Daikoku T., Padmanabhan R., Ludert J.E., del Angel R.M. Polypyrimidine tract-binding protein is relocated to the cytoplasm and is required during dengue virus infection in Vero cells. J Gen Virol. 2009;90:2893–2901. doi: 10.1099/vir.0.013433-0. [DOI] [PubMed] [Google Scholar]
- 76.De Nova-Ocampo M., Villegas-Sepulveda N., del Angel R.M. Translation elongation factor-1alpha, La, and PTB interact with the 3′ untranslated region of dengue 4 virus RNA. Virology. 2002;295:337–347. doi: 10.1006/viro.2002.1407. [DOI] [PubMed] [Google Scholar]
- 77.Paranjape S.M., Harris E. Y box-binding protein-1 binds to the dengue virus 3′-untranslated region and mediates antiviral effects. J Biol Chem. 2007;282:30497–30508. doi: 10.1074/jbc.M705755200. [DOI] [PubMed] [Google Scholar]
- 78••.Romero-Lopez C., Berzal-Herranz A. The functional RNA domain 5BSL3.2 within the NS5B coding sequence influences hepatitis C virus IRES-mediated translation. Cell Mol Life Sci. 2011 doi: 10.1007/s00018-011-0729-z. epub ahead of print. [DOI] [PMC free article] [PubMed] [Google Scholar]; Interaction of 3′ RNA domains in the HCV genome negatively regulates HCV IRES-mediated translation initiation, paralleling some plant virus systems.
- 79.Wang L., Jeng K.S., Lai M.M. Poly(C)-binding protein 2 interacts with sequences required for viral replication in the Hepatitis C Virus (HCV) 5′ untranslated region and directs HCV RNA replication through circularizing the viral genome. J Virol. 2011;85:7954–7964. doi: 10.1128/JVI.00339-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Serrano P., Pulido M.R., Saiz M., Martinez-Salas E. The 3′ end of the foot-and-mouth disease virus genome establishes two distinct long-range RNA-RNA interactions with the 5′ end region. J Gen Virol. 2006;87:3013–3022. doi: 10.1099/vir.0.82059-0. [DOI] [PubMed] [Google Scholar]
- 81.Liu Y., Wimmer E., Paul A.V. Cis-acting RNA elements in human and animal plus-strand RNA viruses. Biochim Biophys Acta. 2009;1789:495–517. doi: 10.1016/j.bbagrm.2009.09.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Zoll J., Heus H.A., van Kuppeveld F.J., Melchers W.J. The structure-function relationship of the enterovirus 3′-UTR. Virus Res. 2009;139:209–216. doi: 10.1016/j.virusres.2008.07.014. [DOI] [PubMed] [Google Scholar]
- 83.Schwartz S., Felber B.K., Fenyo E.M., Pavlakis G.N. Env and Vpu proteins of human immunodeficiency virus type 1 are produced from multiple bicistronic mRNAs. J Virol. 1990;64:5448–5456. doi: 10.1128/jvi.64.11.5448-5456.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Stacey S.N., Jordan D., Williamson A.J., Brown M., Coote J.H., Arrand J.R. Leaky scanning is the predominant mechanism for translation of human papillomavirus type 16 E7 oncoprotein from E6/E7 bicistronic mRNA. J Virol. 2000;74:7284–7297. doi: 10.1128/jvi.74.16.7284-7297.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Schaecher S.R., Mackenzie J.M., Pekosz A. The ORF7b protein of severe acute respiratory syndrome coronavirus (SARS-CoV) is expressed in virus-infected cells and incorporated into SARS-CoV particles. J Virol. 2007;81:718–731. doi: 10.1128/JVI.01691-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Horvath C.M., Williams M.A., Lamb R.A. Eukaryotic coupled translation of tandem cistrons: identification of the influenza B virus BM2 polypeptide. EMBO J. 1990;9:2639–2647. doi: 10.1002/j.1460-2075.1990.tb07446.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Ahmadian G., Randhawa J.S., Easton A.J. Expression of the ORF-2 protein of the human respiratory syncytial virus M2 gene is initiated by a ribosomal termination-dependent reinitiation mechanism. EMBO J. 2000;19:2681–2689. doi: 10.1093/emboj/19.11.2681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Napthine S., Lever R.A., Powell M.L., Jackson R.J., Brown T.D., Brierley I. Expression of the VP2 protein of murine norovirus by a translation termination-reinitiation strategy. PLoS ONE. 2009;4:e8390. doi: 10.1371/journal.pone.0008390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Yueh A., Schneider R.J. Selective translation initiation by ribosome jumping in adenovirus-infected and heat-shocked cells. Genes Dev. 1996;10:1557–1567. doi: 10.1101/gad.10.12.1557. [DOI] [PubMed] [Google Scholar]
- 90.Yueh A., Schneider R.J. Translation by ribosome shunting on adenovirus and hsp70 mRNAs facilitated by complementarity to 18S rRNA. Genes Dev. 2000;14:414–421. [PMC free article] [PubMed] [Google Scholar]
- 91.Sen N., Cao F., Tavis J.E. Translation of duck hepatitis B virus reverse transcriptase by ribosomal shunting. J Virol. 2004;78:11751–11757. doi: 10.1128/JVI.78.21.11751-11757.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Racine T., Duncan R. Facilitated leaky scanning and atypical ribosome shunting direct downstream translation initiation on the tricistronic S1 mRNA of avian reovirus. Nucleic Acids Res. 2010;38:7260–7272. doi: 10.1093/nar/gkq611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Latorre P., Kolakofsky D., Curran J. Sendai virus Y proteins are initiated by a ribosomal shunt. Mol Cell Biol. 1998;18:5021–5031. doi: 10.1128/mcb.18.9.5021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Fraser C.S., Doudna J.A. Structural and mechanistic insights into hepatitis C viral translation initiation. Nat Rev Microbiol. 2007;5:29–38. doi: 10.1038/nrmicro1558. [DOI] [PubMed] [Google Scholar]
- 95.Hellen C.U., Pestova T.V. Translation of hepatitis C virus RNA. J Viral Hepat. 1999;6:79–87. doi: 10.1046/j.1365-2893.1999.00150.x. [DOI] [PubMed] [Google Scholar]
- 96.Lukavsky P.J. Structure and function of HCV IRES domains. Virus Res. 2009;139:166–171. doi: 10.1016/j.virusres.2008.06.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Fletcher S.P., Jackson R.J. Pestivirus internal ribosome entry site (IRES) structure and function: elements in the 5′ untranslated region important for IRES function. J Virol. 2002;76:5024–5033. doi: 10.1128/JVI.76.10.5024-5033.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Buck C.B., Shen X., Egan M.A., Pierson T.C., Walker C.M., Siliciano R.F. The human immunodeficiency virus type 1 gag gene encodes an internal ribosome entry site. J Virol. 2001;75:181–191. doi: 10.1128/JVI.75.1.181-191.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Low W., Harries M., Ye H., Du M.Q., Boshoff C., Collins M. Internal ribosome entry site regulates translation of Kaposi's sarcoma-associated herpesvirus FLICE inhibitory protein. J Virol. 2001;75:2938–2945. doi: 10.1128/JVI.75.6.2938-2945.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Firth A.E., Chung B.Y., Fleeton M.N., Atkins J.F. Discovery of frameshifting in Alphavirus 6K resolves a 20-year enigma. Virol J. 2008;5:108. doi: 10.1186/1743-422X-5-108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Baranov P.V., Henderson C.M., Anderson C.B., Gesteland R.F., Atkins J.F., Howard M.T. Programmed ribosomal frameshifting in decoding the SARS-CoV genome. Virology. 2005;332:498–510. doi: 10.1016/j.virol.2004.11.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Park S.J., Kim Y.G., Park H.J. Identification of RNA pseudoknot-binding ligand that inhibits the -1 ribosomal frameshifting of SARS-coronavirus by structure-based virtual screening. J Am Chem Soc. 2011;133:10094–10100. doi: 10.1021/ja1098325. [DOI] [PubMed] [Google Scholar]
- 103.Mazauric M.H., Seol Y., Yoshizawa S., Visscher K., Fourmy D. Interaction of the HIV-1 frameshift signal with the ribosome. Nucleic Acids Res. 2009;37:7654–7664. doi: 10.1093/nar/gkp779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Lewis T.L., Matsui S.M. An astrovirus frameshift signal induces ribosomal frameshifting in vitro. Arch Virol. 1995;140:1127–1135. doi: 10.1007/BF01315421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Lewis T.L., Matsui S.M. Astrovirus ribosomal frameshifting in an infection-transfection transient expression system. J Virol. 1996;70:2869–2875. doi: 10.1128/jvi.70.5.2869-2875.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Daughenbaugh K.F., Fraser C.S., Hershey J.W., Hardy M.E. The genome-linked protein VPg of the Norwalk virus binds eIF3, suggesting its role in translation initiation complex recruitment. EMBO J. 2003;22:2852–2859. doi: 10.1093/emboj/cdg251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Daughenbaugh K.F., Wobus C.E., Hardy M.E. VPg of murine norovirus binds translation initiation factors in infected cells. Virol J. 2006;3:33. doi: 10.1186/1743-422X-3-33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Goodfellow I., Chaudhry Y., Gioldasi I., Gerondopoulos A., Natoni A., Labrie L., Laliberte J.F., Roberts L. Calicivirus translation initiation requires an interaction between VPg and eIF 4 E. EMBO Rep. 2005;6:968–972. doi: 10.1038/sj.embor.7400510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Merrick W.C. Cap-dependent and cap-independent translation in eukaryotic systems. Gene. 2004;332:1–11. doi: 10.1016/j.gene.2004.02.051. [DOI] [PubMed] [Google Scholar]
- 110.Easton L.E., Locker N., Lukavsky P.J. Conserved functional domains and a novel tertiary interaction near the pseudoknot drive translational activity of hepatitis C virus and hepatitis C virus-like internal ribosome entry sites. Nucleic Acids Res. 2009;37:5537–5549. doi: 10.1093/nar/gkp588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Odreman-Macchioli F.E., Tisminetzky S.G., Zotti M., Baralle F.E., Buratti E. Influence of correct secondary and tertiary RNA folding on the binding of cellular factors to the HCV IRES. Nucleic Acids Res. 2000;28:875–885. doi: 10.1093/nar/28.4.875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Sizova D.V., Kolupaeva V.G., Pestova T.V., Shatsky I.N., Hellen C.U. Specific interaction of eukaryotic translation initiation factor 3 with the 5′ nontranslated regions of hepatitis C virus and classical swine fever virus RNAs. J Virol. 1998;72:4775–4782. doi: 10.1128/jvi.72.6.4775-4782.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]


