Abstract
In a genetic association study, it is often desirable to perform an overall test of whether any or all single-nucleotide polymorphisms (SNPs) in a gene are associated with a phenotype. Several such tests exist, but most of them are powerful only under very specific assumptions about the genetic effects of the individual SNPs. In addition, some of the existing tests assume that the direction of the effect of each SNP is known, which is a highly unlikely scenario. Here we propose a new kernel-based association test (KBAT) of joint association of several SNPs. Our test is non-parametric and robust, and does not make any assumption about the directions of individual SNP effects. It can be used to test multiple correlated SNPs within a gene and can also be used to test independent SNPs or genes in a biological pathway. Our test uses an analysis of variance (ANOVA) paradigm to compare variation between cases and controls to the variation within the groups. The variation is measured using kernel functions for each marker, and then a composite statistic is constructed to combine the markers into a single test. We present simulation results comparing our statistic to the U-statistic based method by Schaid et al. and another statistic by Wessel and Schork. We consider a variety of different disease models and assumptions about how many SNPs within the gene are actually associated with disease. Our results indicate that our statistic has higher power than other statistics under most realistic conditions.
Keywords: genetic similarity, association study, multilocus association
INTRODUCTION
In a genome-wide association study or a large-scale candidate gene study, the fundamental scientific goal is to identify genes that are associated with a phenotype. Yet for the most part in such studies, statistical testing is done at the individual marker level rather than at the gene level. Testing at the gene level may be scientifically more sensible, and could also reduce multiple comparison problems. There are several approaches to gene-level testing in the literature, but they are not entirely satisfactory for a variety of reasons. These statistics usually perform well only under very specific genetic conditions, thus restricting their use in real applications.
In a case-control design (binary trait), one possible test for joint association of multiple single-nucleotide polymorphisms (SNPs) within a gene is the maximum of the single SNP chi-square statistics, known as Max-Single [Schaid, et al. 2005]. The alternative hypothesis for such an approach is that at least one of the SNPs tested is associated with the disease. The p-value of the Max-Single statistic can be found using a permutation procedure. Another well known procedure is the multivariate Hotelling’s T2 statistic proposed by Fan and Knapp [2003]. Here each marker genotype is scored using a numerical dosage scheme. For the joint test of association of all SNPs, the mean genotype score vectors between the cases and controls are compared using the T2 statistic. Under a specific genotype scoring, this method is equivalent to using logistic regression with the SNP genotypes as covariates [Schaid, et al. 2005]. Roeder et al. [2005] compared the power of Max-Single (with p-values by permutation), T2, and a test procedure based on the maximum of a smooth curve fitted to the single marker statistics. They showed that in general the Max-Single statistic has higher power than the Hotelling’s T2 statistic. Their alternative method of using the mode of the fitted values in a non-parametric regression of single chi-square statistics depends on the choice of the regression function. The gain in power of such a method over the Max-Single is not substantial and in some cases its power is less than that of the Max-Single statistic. However if there are multiple risk variants within the gene, the Max-Single may not be powerful [Schaid, et al. 2005]. Similarly the mode statistic may not be a powerful strategy when there are multiple risk variants within the gene.
An approach that is more powerful when there are multiple associated SNPs was developed by Schaid et al. [2005]. It is a non-parametric test based on U-statistics. The U-statistic is computed as the average of kernel function scores for a pair of individuals with several different possible kernel functions. The Schaid et al. statistic, known as the ‘Zglobal’ statistic, has higher power than the Hotelling’s T2 statistic or the Max-Single statistic whenever there is more than one associated SNP within a gene. The T2 statistic loses power because its degrees of freedom are proportional to the number of markers tested. Schaid et al. [2005] were able to reduce the degrees of freedom by taking a linear combination of the marker statistics using a weight vector, which depends on the data matrix. However, the U-statistic approach requires the knowledge of the risk allele for each SNP, information that is not typically available. Wang and Elston [2007] developed a variant of the Schaid et al. statistic that uses a different set of weights based on the Fourier transform. Their simulation results show that their statistic is more powerful than the Max-Single and the logistic regression (equivalently the T2 statistic). Note that the powers of the statistics based on linear combination of the marker scores as proposed in Schaid et al. [2005] and Wang and Elston [2007] are affected by the direction of the genotype scores at each SNP [Wang and Abbott 2008].
Another approach to gene-level testing is to assume that affected individuals will share the same genetic material around the causative locus. This genetic similarity of the affected individuals has been exploited in several association studies [Beckmann, et al. 2005; Kwee, et al. 2008; Tzeng, et al. 2003; Wessel and Schork 2006]. Wessel and Schork [2006] developed a test for phenotype-genotype associations using the matrix of genetic similarity among individuals using an idea proposed by McArdle and Anderson [2001]. This method, called, Multivariate Distance Matrix Regression (MDMR), can be used in association studies for both discrete and continuous multivariate phenotypes with multi-marker genotypes, gene-expression data, and even sequence data [Schork, et al. 2008]. MDMR is expected to be more powerful for genetic markers within a small genomic region than for a set of independent SNPs across the genome. Lin and Schaid [2008] have shown that MDMR has higher power compared to seven other methods for multilocus association.
To reduce the number of degrees of freedom associated with the multimarker joint association tests, another possibility is to work with a few principal components instead of using all the markers [Gauderman, et al. 2007; Wang and Abbott 2008]. Another approach that works in a reduced dimensional space is proposed in a recent paper by Kwee et al. [2008]. They used genotype similarity measured through non-parametric functions in a semiparametric-regression model for quantitative trait association; as outlined by the authors, this approach is extendable to binary traits (case-control association studies). The non-parametric function in the model is in turn defined in a reduced dimensional space through kernel functions that score the genotype similarity. The test of association of marker genotypes is developed by using the relationship between the least squares kernel machines estimates of the non-parametric function in their semi-parametric model and the variance components analysis in a mixed linear model. The authors however have not tested the power of this approach compared to other dimension reduction approaches such as principal components regression [Gauderman, et al. 2007; Wang and Abbott 2008] and the MDMR [Wessel and Schork 2006]. It is important to note that the marker genotypes in MDMR, through the similarity matrix, are used as dependent variables whereas in the usual regression analysis, such as the one discussed above, they are used as independent variables.
We propose a new test procedure based on genotype similarity measures obtained as scores of kernel functions. Our goal is to develop a kernel-based association test (KBAT) statistic to test association between a phenotype and a set of SNPs, without making assumptions about the directions of the SNP effects, and ideally, with high power even if several SNPs are associated with the phenotype. Our KBAT statistic does not use information on correlation between the SNPs, but it is valid even when such correlation is present. This method can be used both for a set of markers in linkage disequilibrium (LD) with each other within a small genomic region, as well as for a set of independent markers. For each marker we first score the genotype similarity between individuals within the case group and within the control group using a symmetric kernel function. We then compare the average similarity scores between cases and controls in an ANOVA-like context. The final test combines the statistics for each marker. We use simulation to compare the power of our KBAT statistic with that of the Zglobal statistic by Schaid et al. [2005] and the MDMR statistic by Wessel and Schork [2006].
METHODS
Suppose n1 cases and n2 controls are genotyped at K SNP markers. There could be linkage disequilibrium (LD) among these K markers or they may be a set of independent markers. The KBAT statistic proposed here globally tests for joint association of the markers with the disease. The test developed here is based on genotype similarities between individuals within a group (case or control). These similarities are measured using one of several possible kernel functions, some of which are discussed below. The similarity scores are then considered as the observations for the ANOVA model, based on which we develop the test statistic for testing the joint association of multiple SNPs. Throughout the rest of the paper we use the term 'similarity score' to refer to the score used to measure the degree of similarity between two individuals using the chosen kernel function.
CHOICE OF KERNELS
For scoring genetic similarity between two individuals, we have considered eight different kernels, which are defined below. The first kernel described below, the “AM” kernel, does not require knowledge of the risk allele for each SNP; the other kernels do require such knowledge.
Allele match (AM) kernel
The Allele Match kernel score assays the number of alleles common between the genotypes gi and gj of two individuals i and j. The score is 4 if gi and gj are the same; 2 if one is a heterozygote and the other is a homozygote; 0 if they don’t share any common alleles.
Allele share (AS) kernel
The Allele Share kernel score assays the number of risk alleles shared between gi and gj. The scores are 0, 1, 2 according as they share 0, 1, or 2 risk alleles. Other kernels considered here are additive or product kernels.
Additive kernels
Additive kernel similarity score between two individuals is defined as h(gi, gj) = wi + wj, wi being the score corresponding to gi. Depending on the choice of wi we get the following three kernel scores which fall under the additive kernels:
Linear dosage (LIN) kernel
The linear dosage kernel score wi is the number of risk alleles that genotype gi contains.
Recessive (REC) kernel
Here, wi takes the value 1 if gi is homozygous for the risk allele, otherwise it is 0.
Quadratic (QUAD) kernel
For this kernel, wi is 1 if gi is homozygous for the non-risk allele, otherwise it is twice the number of risk alleles that gi has.
Product (Prod) kernel
We have introduced the product kernels to examine the multiplicative effect of genotype dosage values to model the similarity score, i.e., h(gi, gj) = wiwj. We have considered three product kernels of the type “Prod-l.m.n” where “l.m.n” denotes the way wi scores for the three genotypes a/a, a/b or b/b respectively; for example, if ‘b’ is the risk allele,
We thus get three different product kernels “Prod-0.1.2”, “Prod-1.2.3”, and “Prod-1.2.4” for suitable choices of l, m, and n. The similarity scores corresponding to additive and product kernels are given in Table I.
TABLE I.
Similarity scores associated with the pair of genotypes gi and gj using all the 8 kernels.
| Additive kernels | |||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| REC | LIN | QUAD | AM | ||||||||||||
| wi(gi) | 0 (a/a) |
0 (a/b) |
1 (b/b) |
wi(gi) | 0 (a/a) |
1 (a/b) |
2 (b/b) |
wi(gi) | 1 (a/a) |
2 (a/b) |
4 (b/b) |
wi(gi) | (a/a) | (a/b) | (b/b) |
| wj(gj) | wj(gj) | wj(gj) | wj(gj) | ||||||||||||
| 0 (a/a) | 0 | 0 | 1 | 0 (a/a) | 0 | 1 | 2 | 1 (a/a) | 2 | 3 | 5 | (a/a) | 4 | 2 | 0 |
| 0 (a/b) | 0 | 0 | 1 | 1 (a/b) | 1 | 2 | 3 | 2 (a/b) | 3 | 4 | 6 | (a/b) | 2 | 4 | 2 |
| 1 (b/b) | 1 | 1 | 2 | 2 (b/b) | 2 | 3 | 4 | 4 (b/b) | 5 | 6 | 8 | (b/b) | 0 | 2 | 4 |
| Product kernels | |||||||||||||||
| Prod-0.1.2 | Prod-1.2.3 | Prod-1.2.4 | AS | ||||||||||||
| wi(gi) | 0 (a/a) |
1 (a/b) |
2 (b/b) |
wi(gi) | 1 (a/a) |
2 (a/b) |
3 (b/b) |
wi(gi) | 1 (a/a) |
2 (a/b) |
4 (b/b) |
wi(gi) | 0 (a/a) |
1 (a/b) |
2 (b/b) |
| wj(gj) | wj(gj) | wj(gj) | wj(gj) | ||||||||||||
| 0 (a/a) | 0 | 0 | 0 | 1 (a/a) | 1 | 2 | 3 | 1 (a/a) | 1 | 2 | 4 | 0 (a/a) | 0 | 0 | 0 |
| 1 (a/b) | 0 | 1 | 2 | 2 (a/b) | 2 | 4 | 6 | 2 (a/b) | 2 | 4 | 8 | 1 (a/b) | 0 | 1 | 1 |
| 2 (b/b) | 0 | 2 | 4 | 3 (b/b) | 3 | 6 | 9 | 4 (b/b) | 4 | 8 | 16 | 2 (b/b) | 0 | 1 | 2 |
Two of the kernels defined above, Allele Match (AM) and Allele Share (AS) are based on sharing of any specific alleles between the two individuals and no numerical scheme for scoring similarities is assumed whereas in the other six kernels a dosage scheme for scoring the alleles in the genotypes is assumed. Further, we also assume an additive function for scoring the similarities for the three kernels, Linear dosage (LIN), Recessive (REC) and Quadratic (QUAD). For the rest of the three kernels Prod-0.1.2, Prod-1.2.3 and Prod-1.2.4 we use a product function for scoring similarities. Actual similarity scores using the above kernel functions are given in the Table I. We do not consider the dominant kernel as Schaid et al. [2005] clearly discussed its limitations.
DEVELOPMENT OF TEST STATISTIC
Let be a measure of genotype similarity for the two individuals i and j with genotypes , respectively, for the marker k (k = 1,2,…,K) in group l. We assume both individuals belong to the same group l (l=1 denotes case and l=2 denotes control). Within each group l there are ml(= nl(nl −1) / 2) distinct similarity scores corresponding to the ml possible pairs of the nl individuals. We first develop the statistic for a single marker and then extend it to multiple markers. So, for the sake of simplicity we drop k from our notation for a while.
Let yl(ij) = hl(gi, gj) denote the similarity score for the (i,j)-th pair in the l-th group (l = 1,2). We model the similarity score values for pairs of individuals in cases and controls using a one-way ANOVA model as:
| (1) |
Here μ denotes the overall grand mean or the general effect for pairs of individuals, αl is the group specific treatment effect for similarity scores over the general effect, and el(ij) are the error components. The similarity scores from each pair (i,j) form the within group observations for the above one-way ANOVA model. Although the average similarity scores among the two groups could be compared using t-statistics, for generality we have decided to use the ANOVA model. This gives us the option of easily extending this approach to work with diseases which are categorized into several groups instead of simply cases and controls.
We assume,
It is important to note that for a given l, yl(ij) (= hl(gi, gj)) values are correlated and this dependence leads to the above covariance structure. Here, ρ is the correlation coefficient between the similarity scores of two pairs of individuals when one individual appears in both pairs within a case or control group.
If the similarity scores across the cases and controls were similar, we would expect α1 = α2, so we can test for disease marker association by testing the hypothesis H0 : α1 = α2 = 0.
Let denote the U-statistic for the kth marker (k = 1,2,…,K) in group l. Define the within group sum of squares (SSW) corresponding to kth marker as , and between group sum of squares (SSB) for the marker as . Therefore for testing association of disease with the kth marker, we can use the following statistic, which is analogous to the well-known ANOVA statistic, .
For testing the disease association using all K markers, we propose to use the following statistic, which is based on between and within sum of squares of all markers:
Thus we obtain a kernel-based association test (KBAT) statistic where the similarity scores are used as dependent observations as in MDMR.
It is possible to construct several other alternatives to the KBAT statistic given above. For example, instead of the ratio of the sum of individual marker SSB to sum of SSW, we could consider the sum of ratios of between to within sums of squares of individual markers, i.e., , as suggested by a reviewer. This alternative statistic is called here KBAT-A and the performances of these two statistics are compared. Further, it also possible to construct weighted linear combinations of both the KBAT and KBAT-A statistics with several choices for the weights such as the single SNP p-values and Hardy-Weinberg p-values [Hoh, et al. 2001]. Here we have not attempted to evaluate such weighted statistics.
Two important points to note: (1) the similarity scores are not normally distributed and also the yl(ij) are not all independent. Hence the statistic 𝔍 may not follow an F distribution, thus requiring either simulation or permutation to calculate the p-value for the statistic; and (2) the test is one-sided to the right, because the ratio of expectations of the numerator to the denominator in 𝔍 is greater than its value under the null hypothesis (See Appendix B in the Supplement for a mathematical justification). However, this one-sided test is valid only if the correlation coefficient between yl(ij) values is positive, which is true for the type of kernels we used for our proposed method (Appendix A in the Supplement). Thus, we propose to reject H0 if the observed 𝔍 > Fα, where the significance threshold Fα at the level α is obtained empirically. If the null hypothesis is rejected we conclude that at least one of the SNPs used in the joint test is associated with the disease. Hence if the joint test is rejected, the risk-associated SNPs may be identified using single SNP tests.
Note that in the usual ANOVA method we take the ratio of the mean squares (MS) where the mean square is just the sum of squares divided by the corresponding degrees of freedom. But in this case, we use sum of squares instead of mean squares because one is just a scalar multiple of the other, and since even if we used the mean square, it would not lead to a closed form of the distribution of the test statistic. Also, since we have to do simulation or permutation to calculate the p-value, it is simpler to use the sum of squares in our KBAT statistic.
Zglobal AND MDMR STATISTICS
We compare our KBAT method of testing multiple SNPs with two other statistics: the Zglobal [Schaid, et al. 2005] and the MDMR [Wessel and Schork 2006]. For the sake of completeness, we briefly review these two statistics.
Schaid et al. [2005] developed their Zglobal method using U-statistics based on similarity scores as defined by a kernel function. As before we define the U-statistic:
Let U̅1 and U̅2 be the vector of U-statistics for all markers for the case and control groups respectively. To test disease marker association, Schaid et al. [2005] proposed the statistic , where where 1 is a vector with each component as 1 and V0 denotes the variance-covariance matrix, which is to be estimated from the data. The distribution of Zglobal can be approximated by a normal distribution. If the differences (U̅1−U̅2) are not in the same direction for all the markers, then the test based on linear combinations for K marker may lose power.
The MDMR statistic [Wessel and Schork 2006] is based on a multivariate distance matrix, where the distance matrix S is defined in terms of similarity score as before. Their choice of similarity score is similar to the allele match kernel, and so does not depend on knowledge of the risk allele. In principle, any other kernel appropriate for defining a similarity between the genotypic profiles of two individuals might also be considered. Based on the similarity score, the similarity matrix for pairs of individuals, whose (i,j)-th element is as follows:
The distance matrix D calculated as D = 11′ − S, where 1 is a vector with each component as 1. Let where dij is the (i,j)-th element in D and let X be an N×M matrix of data on M phenotypic variables corresponding to N individuals. Define a centered matrix using A as and H as H = X(X′X)−1 X′. The F-statistic for testing trait marker association as used by Wessel and Schork [2006] is given by
Note that this statistic can also be used for testing association between a multivariate quantitative phenotype and the genotypes. For numerical stability, the sums of squares in the numerator and the denominator are usually calculated using QR decomposition of the H matrix. In some cases it is possible to get a negative FMDMR statistic because some of the eigenvalues of the G matrix might be negative. This can be avoided by computing the Total Sums of Squares and Error Sums of Squares using only the positive eigenvalues [McArdle and Anderson 2001]. In fact for some similarity scores, we observe that the F-statistic computed using only the positive eigenvalues (denoted here as FMDMR+) has increased power compared to the MDMR statistic calculated based on all the eigenvalues (see “Results” section).
SIMULATIONS
We have performed extensive simulations to evaluate the performance of the methods under two different scenarios: one with no LD among the simulated SNPs (called simulation I) and the other with LD (Simulation II). Simulation I examines the effect of the number of risk variants as compared to the total number of variants tested. In simulation II, our focus is on understanding the effect of LD on the test.
For simulation I, we simulate ten independent markers, which are unlinked and in linkage equilibrium with each other. Two allele frequency patterns of minor allele frequency (MAF) for each of the SNPs are considered, i.e. one with 0.05 and the other 0.1. Genotypes of 500 population controls are generated assuming Hardy-Weinberg equilibrium (HWE). To ascertain the cases, we vary the number of causative SNPs from 1 to 5 among the total of 10 SNPs. The cases are ascertained on the basis of the genotypes at the causal loci and using an appropriate penetrance function. For this simulation we assume that the Relative Risk (RR) at each causative locus is 1.5 or 1.25 and hence the combined relative risk for r causative loci is (1.5)r or (1.25)r. The details of ascertainment of cases using multiple causative SNPs are described in the Appendix C in the Supplement. In another simulation scenario I-A, we fixed the combined relative risk to be either 1.5 or 1.25 and varied the number of causative loci. For example, if the combined relative risk for two causative SNPs is 1.5, then risk of each causative SNP is taken to be (1.5)1/2.
For simulation II (LD among markers) we have simulated data sets using the observed haplotype frequencies for two genes, gene I and II, published in previous studies [Sha, et al. 2007; 2005]. Gene I has exactly 10 SNPs (called as gene III in Sha et al [2005]), which helps us to compare with simulation I which is based on the same number of SNPs without LD. We first generate a large number of haplotypes with probabilities proportional to the observed haplotype frequencies for the genes. We then select two haplotypes at random and pair them to form two haplotypes for an individual. Finally the unphased SNP genotypes are formed by taking the corresponding alleles from the haplotype pairs. We then select a single SNP as a causative locus (for gene I SNP 6 is set as the causative locus which has MAF approximately 0.108 and for gene II SNP 3 is set as the causative one, which has a MAF around 0.424) and generate the case-control data sets as described above for simulation I. Further, to understand the power gain with more than one causal SNP in the case of correlated SNPs, we have also considered more than one causal SNP within these two genes. Ascertainment of cases using multiple causative SNPs were done as in simulation I.
Under the null, the exact distribution of the proposed KBAT statistic is very difficult to obtain and therefore to compute the p-values we may have to use a permutation based procedure. Note that for this statistic, a permutation procedure can easily be adopted because the genotype similarity scores between individuals need to be computed only once and a permuted dataset can be generated by shuffling case and control labels. The present simulation study has several choices for the parameters such as the MAF, RR, several different models for the RRs and several different choices for the kernel functions. Therefore it is very time consuming to compute the power of the test using permutation-based p-values for the present simulation since we would need to obtain the p-value for each of the 1000 data sets generated under each of these specific combinations of parameters and models for RRs. Hence we have decided to generate the empirical percentile points of the statistic using the datasets generated under the null distribution. For simulation I, we have used 10,000 datasets and for simulation II, we have used 5,000 datasets. The test hypothesis is rejected at levelα if the computed statistics exceeds the upper αth percentile point of the empirical distribution.
To have a uniform comparison we have also used the empirical distribution for the MDMR and Zglobal statistics. We observed that for simulation I (independent markers), the distribution of Zglobal is well approximated by a normal distribution for all kernel functions, while for simulation II (markers in LD) we observed that percentile points of Zglobal's empirical null distribution are not well approximated by the normal percentile points for some kernel functions. Further, exact distributions of the MDMR statistics FMDMR and FMDMR+ are also not known under the null distribution which led us to consider the empirical distribution for the simulation study.
For both simulations I and II, the power is calculated based on 1,000 datasets, each with 500 cases and 500 controls, simulated under the alternative model. Power, for a given level of significance α, is defined as the proportion of test statistics for datasets generated under the alternative model exceeding the upper αth percentile point of the null distribution of that statistic. The datasets under alternative hypothesis were generated for three different models of relative risks, namely, additive, multiplicative, and recessive models. We considered all eight kernel functions described above, and furthermore considered scenarios in which the risk alleles at each locus were either known or unknown.
RESULTS
In all our simulations the type I error rate is fixed at 5% because we used the empirical percentile points of the null distribution of each test statistic. However to estimate the empirical type I error rate of the KBAT statistic with different kernels, we first randomly selected 9,000 replicates out of the 10,000 replicates generated under the null distribution. The empirical 95th percentile point based on the 9,000 null replicates was first obtained. The proportion of test statistics for the remaining 1,000 null replicates exceeding the empirical 95th percentile point gives the type I error rate, which varies from 0.047 to 0.053 for our simulation I for various choices of the kernel function.
To restrict our discussions to either one of the two alternative statistic i.e. KBAT and KBAT-A, we first present a comparison between the two. Under the additive model, in simulation I, KBAT-A performs better than KBAT. But for other models the power largely depends on the choice of the kernel function (Supplemental Table IV-A). For correlated SNPs there is no clear pattern favoring either KBAT or KBAT-A except under the AM kernel, for which KBAT performs better than KBATA. For most of the kernel functions under the multiplicative and additive models KBAT performs better than KBAT-A (Supplemental Table XIV). We recommend the use of KBAT because it performs well for correlated SNPs with the AM kernel for which knowledge of the risk allele is not needed.
In the remainder of this section we discuss the relative power of four statistics KBAT, Zglobal, FMDMR and FMDMR+ only based on the allele match kernel (AM) because the AM kernel does not require the knowledge of risk allele and it is easy to use this kernel function in real applications. A comparison of other kernel functions is presented in the following sub-section.
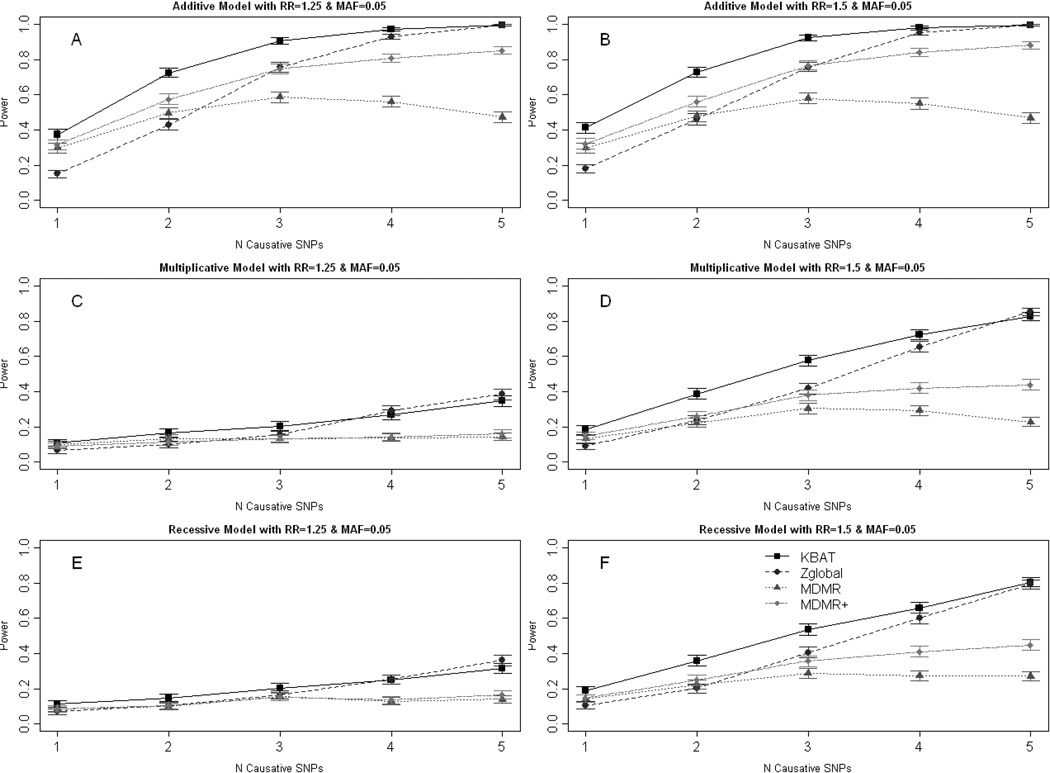
Evaluation of power through extensive simulations reveals that our proposed KBAT statistic performs well. First, we discuss the results for simulation I, i.e., for the set of simulations with independent SNPs. In this simulation the combined relative risk increases as the number of causative SNPs increases. The powers of the four statistics using the AM kernel are summarized in Figure 1 for selected genetic models. Note that, for the Zglobal statistic, the simulations assume that the risk allele is known, so our results represent the best possible performance for that statistic.
Figure 1.
Empirical power at α = 0.05 for the three methods based on 1,000 replicates using allele match kernel under various models for genotype relative risk. (A) Additive model with RR=1.25 and MAF = 0.05 (B) Additive model with RR=1.5 and MAF = 0.05 (C) Multiplicative model with RR=1.25 and MAF = 0.1 (D) Multiplicative Model with RR=1.5 and MAF = 0.1 (E) Recessive model with RR = 1.25 and MAF=0.05 (F) Recessive model with RR = 1.5 and MAF=0.05
The KBAT statistic has higher power than Zglobal under most of the simulation models we considered. In the extreme case of low effect sizes, small MAF, and more than 40% causative SNPs among the total SNPs, Zglobal does have higher power than the KBAT statistic (Figure 1C and 1E). Under this scenario the overall power for both of the methods is low, however. Moreover the mean genotype difference between the cases and the controls for each SNP must be in the same direction for the Zglobal statistic to work well [Schaid, et al. 2005]. That is, we need the knowledge of the risk allele for each SNP, which is not known in advance. Note that in our simulation I and I-A, we have assumed that the risk allele to be the same among all the 10 SNPs, thus providing the knowledge of risk allele required by Zglobal. In real applications we would probably expect only a few causative SNPs among all the tested SNPs and in this situation the KBAT statistic is more powerful than the Zglobal test. The MDMR procedure is not as powerful as KBAT or Zglobal, although it is very general and can be used for both continuous and discrete traits. It is interesting to note that in some cases FMDMR+ has higher power than FMDMR.
Results under the simulation scenario I-A are summarized in Table II. In this simulation scenario, the combined relative risk of is 1.5 irrespective of the number of causative SNPs. When two causative SNPs are assumed, under our multiplicative model (see simulations section) each has a relative risk of (1.5)1/2 ≈ 1.22, and similarly each has a relative risk of (1.5)1/3 ≈ 1.15 when three causative SNPs are assumed, etc. We find that the KBAT has higher power compared to Zglobal and MDMR in this case also. It is interesting to note that for some models, even if the total effect size is fixed, the power increases as the number of causative SNPs increases. This indicates the fact that the joint test of association will have higher power compared to single SNP association when there are multiple SNPs with moderate effect sizes.
TABLE II.
Empirical power of KBAT, Zglobal and MDMR for simulation I-A at the 5% significance level based on 1,000 replicates.
| Additive Model | Multiplicative Model | Recessive Model | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| #LL* | KBAT | Zglobal | FMDMR | FMDMR+ | KBAT | Zglobal | FMDMR | FMDMR+ | KBAT | Zglobal | FMDMR | FMDMR+ |
| 1 | 0.412 | 0.178 | 0.296 | 0.322 | 0.182 | 0.089 | 0.132 | 0.148 | 0.188 | 0.107 | 0.143 | 0.146 |
| 2 | 0.690 | 0.410 | 0.449 | 0.534 | 0.132 | 0.094 | 0.092 | 0.104 | 0.146 | 0.103 | 0.110 | 0.110 |
| 3 | 0.905 | 0.745 | 0.548 | 0.729 | 0.129 | 0.109 | 0.075 | 0.075 | 0.120 | 0.092 | 0.076 | 0.080 |
| 4 | 0.969 | 0.916 | 0.521 | 0.800 | 0.114 | 0.085 | 0.067 | 0.073 | 0.101 | 0.089 | 0.075 | 0.081 |
| 5 | 0.993 | 0.983 | 0.461 | 0.859 | 0.105 | 0.100 | 0.068 | 0.063 | 0.119 | 0.091 | 0.081 | 0.093 |
Power computed using the allele match kernel with the combined relative risk of 1.5, minor allele frequency of 0.05 and population prevalence of 0.02; #LL indicates number of liability loci. Best power within each model is highlighted in bold.
Table III presents the results of simulation II (LD among markers). This simulation assumes a single causative locus with relative risk of 1.25 and population prevalence 0.02. The results for gene I are given in table III and the results for gene II are given in Supplemental Table XII. In simulation II, knowledge of risk allele was not made available to Zglobal and because of LD among the markers Zglobal may also suffer from the directionality of the risk allele. Hence we find that Zglobal performs very poorly compared to both KBAT and FMDMR. In general, KBAT statistic is more powerful than the others. However, in presence of relatively common causal allele, MDMR seems to be more powerful for gene-based association (simulation II). Our simulations show that Zglobal probably will not perform very well in a gene-based association unless the risk alleles across all the SNPs within the gene are known.
TABLE III.
Empirical power of KBAT, Zglobal and MDMR for gene I at the 5% significance level based on 1,000 replicates.
| Additive Model | Multiplicative Model | Recessive Model | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Kernel | KBAT | Zglobal | FMDMR | FMDMR+ | KBAT | Zglobal | FMDMR | FMDMR+ | KBAT | Zglobal | FMDMR | FMDMR+ |
| AM | 0.971 | 0.060 | 0.913 | 0.918 | 0.323 | 0.049 | 0.214 | 0.228 | 0.274 | 0.060 | 0.197 | 0.200 |
| AS | 0.958 | 0.160 | 0.891 | 0.891 | 0.292 | 0.081 | 0.216 | 0.216 | 0.238 | 0.064 | 0.208 | 0.208 |
| REC | 0.951 | 0.227 | 0.882 | 0.014 | 0.265 | 0.063 | 0.228 | 0.036 | 0.265 | 0.077 | 0.183 | 0.039 |
| LIN | 0.825 | 0.496 | 0.900 | 0.158 | 0.171 | 0.125 | 0.207 | 0.063 | 0.160 | 0.092 | 0.192 | 0.075 |
| QUAD | 0.916 | 0.345 | 0.933 | 0.935 | 0.231 | 0.091 | 0.247 | 0.269 | 0.225 | 0.088 | 0.225 | 0.240 |
| Prod-0.1.2 | 0.960 | 0.108 | 0.746 | 0.157 | 0.295 | 0.060 | 0.149 | 0.077 | 0.253 | 0.066 | 0.131 | 0.054 |
| Prod-1.2.3 | 0.942 | 0.225 | 0.834 | 0.036 | 0.275 | 0.086 | 0.178 | 0.046 | 0.247 | 0.073 | 0.154 | 0.046 |
| Prod-1.2.4 | 0.965 | 0.128 | 0.889 | 0.039 | 0.298 | 0.076 | 0.212 | 0.038 | 0.260 | 0.085 | 0.196 | 0.042 |
Single causative locus with RR of 1.25 and population prevalence is assumed to be 0.02. Best power within each model is highlighted in bold
We have also attempted to compare the power of KBAT and MDMR in the case of correlated SNPs with more than one causal locus (Supplemental Table XIII). For gene I, with more than one causal SNP, KBAT is more powerful than FMDMR and FMDMR+. In the case of gene II, KBAT sometimes has slightly reduced power compared to MDMR. Note that in gene II the MAF of the causal loci are relatively common (more than 33%; see Table II in Sha et al [2005])). We may conclude that if the risk allele is common then MDMR has slightly increased power compared to KBAT.
Comparison of Kernel Functions
As we have discussed, in most realistic situations, risk alleles for each marker will be unknown and so the AM kernel is the only realistic choice. But in some situations investigators may know the risk alleles, and could in theory gain power by taking advantage of that knowledge by using a different kernel. We have considered several kernel functions for genotype similarity but we find that the powers for both KBAT and Zglobal test statistics do not vary too much across the kernels when risk alleles are known. In our simulations we find that of powers for different models using the KBAT statistic do not favor any particular choice of kernel, although in some situations the allele match kernel actually has slightly increased power compared to other kernel functions. In most of the situations the product kernel “Prod-1.2.4” has slightly increased power compared to the other two product kernels (see Supplemental Tables).
For gene I, under every model KBAT with AM kernel has the maximum power when compared across different statistics and kernel functions (Table III). Within each kernel, KBAT has higher power in most of the cases but for linear and quadratic kernels either FMDMR or FMDMR+ has higher power than KBAT. In general, across many kernel functions KBAT has higher power than the MDMR. For gene II this pattern is not very clear. Maximum power under each model is still obtained by KBAT statistics but with different kernel functions (supplement Table XII). In the case of more than one causal SNPs, with the AM kernel the MDMR statistic has higher power compared to KBAT but with the QUAD kernel KBAT has comparable power to MDMR.
Overall, if risk alleles are unknown, we recommend the use of the KBAT statistic with the allele match kernel. If there are several high frequency SNPs we recommend other kernel functions such as the QUAD or product kernel.
DISCUSSION
Here we have proposed a novel procedure for multi-locus genotype-phenotype association testing. We use an analysis of variance formulation and assign scores to genotypes of cases and controls based on different choices of kernels for measuring genotype similarity between pairs of individuals. We use the kernels as given in Schaid et al. [2005] and also propose a few new ones, but we emphasize the use of the AM kernel because it does not require knowledge of the risk allele at each locus. We find that our KBAT statistic is more powerful than the Zglobal and MDMR statistics in most of the simulation scenarios considered here. The only exception is the situation when the causal allele is relatively common. So in presence of rare causal SNPs, KBAT is more powerful than other methods. Our test can also be used even when there are more than two categories for the phenotypes, for example, mild and severe disease status with a set of controls. Our statistic could be further modified to incorporate prior knowledge (such as functional or molecular information) by using weights to take a linear combination of marker-wise statistics instead of a simple sum.
The power gain for KBAT over MDMR in most cases may be due to several reasons. In the calculation of KBAT, the similarity computation is restricted within case groups and controls groups and there is no comparison of case-control similarity. The statistic is based on comparison of the average similarity between these two groups. In contrast, the ideal distance matrix in MDMR statistic would be a matrix with zero distances between pairs of cases and between pairs of controls and non-zero distances for case-control pairs. In general, such distance matrices are seldom obtained, which may reduce the power of MDMR. Furthermore, the MDMR similarity score is computed as the average of similarity scores based on individual markers. We have observed that a KBAT statistic using such an average similarity score did not perform very well compared to the current KBAT statistic which uses the combination of individual marker statistics. This use of an average similarity score may be another reason for loss of power of MDMR.
We also investigated a variation of the MDMR statistic proposed by Wessel and Schork [2006]. The MDMR statistic uses genetic distances between the individuals derived from the similarity scores. Except for the distance measures derived through the AM kernel, all others will result in nonstandard distance i.e., the genetic distance between two individuals with identical genotypes may not be zero. For example, with the LIN kernel (Table I) the diagonal entries are not the same and therefore the distance obtained using this measure will result in non-zero distance for the same individual, i.e., the diagonal entries of the distance matrix will not be zero. Our simulation studies show that such nonstandard distance measures can also be used in this statistic. In some cases, we have also observed that the statistic FMDMR+, which is computed by restricting to positive eigenvalues for the matrices involved in the calculation of within and between sums of squares, has higher power compared to FMDMR. The power for FMDMR+ increases as a function of the number of risk alleles for additive and multiplicative models (Figure 1), which is similar to KBAT and Zglobal. If there are multiple risk variants within the set of variants tested and we use the allele match kernel, FMDMR+ is better than FMDMR. For some other kernels, such as allele share, recessive etc, FMDMR+ does not work well as it is based on a nonstandard distance matrix (see results in Table III and Supplemental Table XII under simulation II).
There are at least two proposals in the literature for reducing the degrees of freedom in joint association tests by using linear combinations of marker statistics (Schaid et al. [2005]; Wang and Elston [2007]). However, such test statistics inherently require knowledge of the risk alleles. We find that in the absence of knowledge of the risk alleles, the weighted linear combination didn’t help to improve the power of the Zglobal statistic. Similarly the statistic proposed by Wang and Elson [2007] is also found to be not very powerful in some cases (see also [Chapman and Whittaker 2008; Wang and Abbott 2008]).
Global association tests discussed here are used to detect the joint association of multiple SNPs with the disease. Rejection of the null hypothesis only indicates that there might be some SNPs in the whole set that are associated with disease risk. However, the problem of identifying the particular SNPs which are associated with disease is not addressed here. Of course, one could attempt to use the single SNP statistics to identify the subset of SNPs associated with the trait. Also an approach similar to the sum statistics proposed in Wille et al. [2003] for correlated SNPs can be used to identify subsets of associated SNPs. However, the power of such approaches using the statistics proposed here needs further investigation.
In our simulation study, we have considered scenarios where the markers are independent and also where the markers are in LD. The KBAT can also be used for joint association of a set of SNPs in a biological pathway. We are currently using this method for a pathway-based association study. Furthermore, our method can be potentially extended for testing associations using sequence similarities or similarities defined using copy numbers.
The KBAT statistic, as presented here, assumes no missing genotypes. If the SNPS are in LD we can impute missing genotypes using software such as PLINK [Purcell, et al. 2007] or other imputation procedures [Marchini, et al. 2007]. If the SNPs are independent, genotype similarity between individuals with missing genotypes can be computed by taking the average similarity of all possible combinations for the missing genotypes. We plan to implement this approach in our extension and study the power loss due to imputing missing genotypes. The KBAT statistics have been implemented in an R program that can be obtained from the corresponding author.
Supplementary Material
ACKNOWLEDGMENTS
This research was funded by the Fogarty International Center (FIC) grant 5D43TW006180 "India-US Research Training Program in Genetics" (PI: Weeks) and the University of Pittsburgh, USA.
REFERENCES
- Beckmann L, Thomas DC, Fischer C, Chang-Claude J. Haplotype sharing analysis using mantel statistics. Hum Hered. 2005;59(2):67–78. doi: 10.1159/000085221. [DOI] [PubMed] [Google Scholar]
- Chapman J, Whittaker J. Analysis of multiple SNPs in a candidate gene or region. Genet Epidemiol. 2008;32(6):560–566. doi: 10.1002/gepi.20330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fan R, Knapp M. Genome association studies of complex diseases by case-control designs. Am J Hum Genet. 2003;72(4):850–868. doi: 10.1086/373966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gauderman WJ, Murcray C, Gilliland F, Conti DV. Testing association between disease and multiple SNPs in a candidate gene. Genet Epidemiol. 2007;31(5):383–395. doi: 10.1002/gepi.20219. [DOI] [PubMed] [Google Scholar]
- Hoh J, Wille A, Ott J. Trimming, Weighting, and Grouping SNPs in Human Case-Control Association Studies. Genome Res. 2001;11(12):2115–2119. doi: 10.1101/gr.204001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwee LC, Liu D, Lin X, Ghosh D, Epstein MP. A powerful and flexible multilocus association test for quantitative traits. Am J Hum Genet. 2008;82(2):386–397. doi: 10.1016/j.ajhg.2007.10.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin WY, Schaid DJ. Power comparisons between similarity-based multilocus association methods, logistic regression, and score tests for haplotypes. Genet Epidemiol. 2008 doi: 10.1002/gepi.20364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marchini J, Howie B, Myers S, McVean G, Donnelly P. A new multipoint method for genome-wide association studies by imputation of genotypes. Nat Genet. 2007;39(7):906–913. doi: 10.1038/ng2088. [DOI] [PubMed] [Google Scholar]
- McArdle BH, Anderson MJ. Fitting multivariate models to community data: a comment on distance-based redundancy analysis. Ecology. 2001;82(1):290–297. [Google Scholar]
- Purcell S, Neale B, Todd-Brown K, Thomas L, Ferreira MA, Bender D, Maller J, Sklar P, de Bakker PI, Daly MJ, et al. PLINK: a tool set for whole-genome association and population-based linkage analyses. Am J Hum Genet. 2007;81(3):559–575. doi: 10.1086/519795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roeder K, Bacanu S-A, Sonpar V, Zhang X, Devlin B. Analysis of single-locus tests to detect gene/disease associations. Genet Epidemiol. 2005;28(3):207–219. doi: 10.1002/gepi.20050. [DOI] [PubMed] [Google Scholar]
- Schaid DJ, McDonnell SK, Hebbring SJ, Cunningham JM, Thibodeau SN. Nonparametric tests of association of multiple genes with human disease. Am J Hum Genet. 2005;76(5):780–793. doi: 10.1086/429838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schork NJ, Wessel J, Malo N. DNA sequence-based phenotypic association analysis. Adv Genet. 2008;60:195–217. doi: 10.1016/S0065-2660(07)00409-9. [DOI] [PubMed] [Google Scholar]
- Sha Q, Chen HS, Zhang S. A new association test using haplotype similarity. Genet Epidemiol. 2007;31(6):577–593. doi: 10.1002/gepi.20230. [DOI] [PubMed] [Google Scholar]
- Sha Q, Dong J, Jiang R, Zhang S. Tests of association between quantitative traits and haplotypes in a reduced-dimensional space. Annals of Human Genetics. 2005;69(Pt 6):715–732. doi: 10.1111/j.1529-8817.2005.00216.x. [DOI] [PubMed] [Google Scholar]
- Tzeng J-Y, Devlin B, Wasserman L, Roeder K. On the identification of disease mutations by the analysis of haplotype similarity and goodness of fit. Am J Hum Genet. 2003;72(4):891–902. doi: 10.1086/373881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang K, Abbott D. A principal components regression approach to multilocus genetic association studies. Genet Epidemiol. 2008;32(2):108–118. doi: 10.1002/gepi.20266. [DOI] [PubMed] [Google Scholar]
- Wang T, Elston RC. Improved power by use of a weighted score test for linkage disequilibrium mapping. Am J Hum Genet. 2007;80(2):353–360. doi: 10.1086/511312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wessel J, Schork NJ. Generalized genomic distance-based regression methodology for multilocus association analysis. Am J Hum Genet. 2006;79(5):792–806. doi: 10.1086/508346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wille A, Hoh J, Ott J. Sum statistics for the joint detection of multiple disease loci in case-control association studies with SNP markers. Genet Epidemiol. 2003;25(4):350–359. doi: 10.1002/gepi.10263. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.