Abstract
The type I and III interferon (IFN) families consist of cytokines rapidly induced during viral infection that confer antiviral protection on target cells and are critical components of innate immune responses and the transition to effective adaptive immunity. The regulation of their expression involves an intricate and stringently regulated signaling cascade, initiated by recognition most often of viral nucleic acid in cytoplasmic and endosomal compartments and involving a series of protein conformational rearrangements and interactions regulated by helicase action, ubiquitin modification, and protein aggregation, culminating in kinase activation and phosphorylation of critical transcription factors and their regulators. The many IFN subtypes induced by viruses confer amplification, diversification, and cell-type specificity to the host response to infection, providing fertile ground for development of antiviral therapeutics and vaccines.
Introduction
Type I and type III interferon (IFN) are a diverse family of cytokines, related by structure, regulation, and function. In humans and most mammals, the classical type I IFN proteins are encoded by a single IFN-β gene, a dozen or so IFN-α genes, plus various more distantly related genes and pseudogenes for IFN-ε, κ, τ, δ, ζ, ω, and v, depending on species. All these genes share clear sequence homology and chromosomal location, indicative of derivation from a single ancestral gene through duplication and diversification. In contrast, the more distantly related type III IFN family (3 genes for IFN-λ1, λ2, and λ3, also called IL28α/β and IL29) is encoded on a different chromosome and is more closely related in structure and sequence to the cytokine IL10 [1]. However, a majority of these cytokines is induced in response to viral infection and in turn induce resistance to viral replication in target cells, thereby justifying their classification as IFNs [2]. As would be expected for such a diverse group of cytokines that function in related pathways, they display both significant commonalities as well as important functional differences, which will be the focus of this review. Since this represents well-trodden ground, we will attempt to concentrate on recent developments in the field.
While conferring necessary and beneficial physiologic attributes, IFNs produce powerful effects that include numerous modulations of cell physiology, including effects on cell proliferation, survival, differentiation, protein translation, and metabolism, in addition to inhibition of viral replication at numerous stages of the viral lifecycle [3]. Thus it is unsurprising that their production is tightly regulated, the result of an acutely activated signaling pathway and tightly regulated gene expression, achieved through stringent regulation of transcription as well as of posttranscriptional control of mRNA stability, and translation [4]. In addition, many elements of the regulatory pathways governing both the signaling pathway and IFN gene expression are themselves regulated by IFN, providing an intricate network of overlapping feed-forward and feedback regulatory loops, overlaid on homeostatic control of basal expression levels controlled by autocrine/paracrine cytokine signaling [5,6].
Transcriptional regulation of IFN gene expression
A major level of control of IFN production depends on transcriptional control. IFN gene expression is maintained at near silent levels in the absence of stimulus, through a combination of the absence of activated transcription factors and the constitutive presence of repressive machinery. Gene repression is provided through both a repressed chromatin configuration involving an occluding nucleosome and the recruitment of gene-specific transcriptional repressors [7]. A number of transcriptional repressors have been implicated in negative regulation, including IRF2 [8], a factor that binds to one of the positive regulatory elements in the IFN-α and –β promoters, both competing for binding by positive regulators and actively repressing transcription. Following stimulation, IRF2 is replaced by an activating IRF protein, most often IRF3 or IRF7, although under some circumstances/cell types, IRF1 and IRF5 may play positive roles [9]. In addition to being replaced, there is evidence for degradation of IRF2 in virus-infected cells. BLIMP or PRDI-BFI is another negatively acting transcriptional repressor associated with the IFN-β promoter, although in most circumstances the expression of this protein is induced along with the IFN-β gene, and it acts as a post-induction repressor to attenuate IFN-β gene expression. Interestingly, degradation of IRF3 may also play a role in post-induction repression [10].
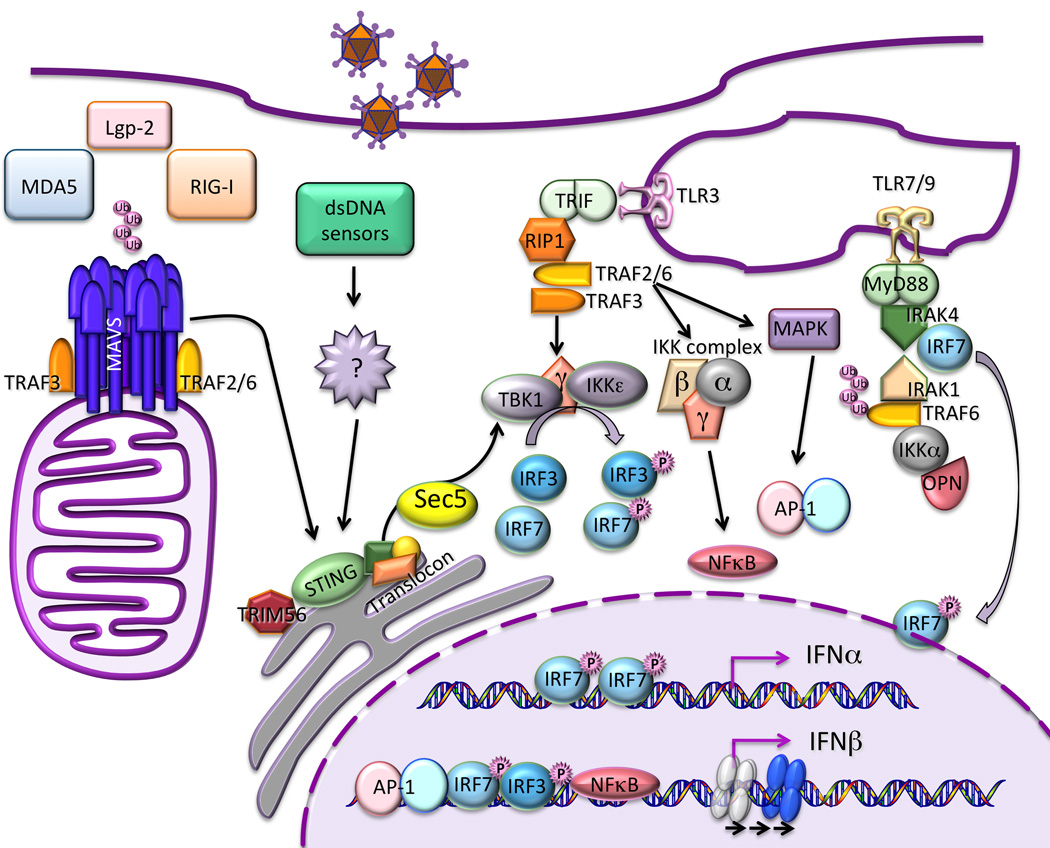
The general paradigm for type I IFN gene induction involves recruitment of sequence-specific transcription factors that are activated by phosphorylation in response to signaling cascades stimulated during viral infection (see below). The IFNβ promoter contains four positive regulatory domains (PRDI-IV), which are occupied by overlapping transcription factor complexes [11]. IRF-3 (early during infection, due to its constitutive synthesis) and IRF-7 (with delayed kinetics, due to its inducible expression through a positive feedback loop) bind PRDI and III; the ATF-2/c-Jun AP-1 complex binds PRDIV; and the p50/RelA NF-κB complex binds PRDII (Fig. 1). In addition, roles for the architectural protein HMGA1 and for a positioned nucleosome have been defined [12]. The binding of each of these components in the correct orientation and location as well as in an orchestrated temporal sequence results in activation of the IFNβ promoter in response to viral infection [13]. Requirement for a higher order assembly of multiple transcription factor components for effective transcription of the IFN-β gene triggers its expression in a stochastic and possibly monoallelic fashion in a minority of virus-infected cells [14], through a mechanism involving long range interchromosomal interactions with multiple independent loci [15]. In the context of virus-induced activation of the IFNβ promoter, NF-κB, IRF3 and IRF7 appear to be the most important transcription factors that play essential and non-overlapping roles [16,17].
Figure 1. Induction IFN by viruses.
Virus infection triggers a complex signaling cascade involving ubiquitin modification and protein aggregation, leading to kinase activation and transcription factor phosphorylation. Signaling is initiated by the detection of viral nucleic acid by both cytoplasmic helicase receptors and endosomal transmembrane Toll-like receptors, activating distinct signaling pathways that converge on transcription factor phosphorylation. Kinase activation is controlled by signaling protein aggregates that assemble as scaffolds on organellar surfaces, including mitochondria, endoplasmic reticulum, and possibly peroxisomes. Phosphorylated IRF3 and IRF7 and activated NF-κB translocate to the cell nucleus, where they bind to enhancer/promoter regions of IFN genes. IFN-β (and probably some IFN-λ subtypes) requires assembly of a multiprotein enhanceosome that alters chromatin structure, sliding an occluding nucleosome that would otherwise prevent polymerase recruitment. The multiple type I and III IFN genes are largely regulated in analogous fashions, although differential utilization of multiple IRF isoforms, some of which are subject to feedforward and feedback regulation, produces distinct patterns of induction of the individual genes under different circumstances.
An interesting recent insight into negative regulation of IFN gene expression documented the role of a positioned nucleosome that occludes access to the IFN-β promoter start site in the absence of virus infection [18]. In resting cells, although the enhancer region upstream of the IFN-β promoter is relatively nucleosome-free and therefore potentially available for binding of activator complexes, the transcriptional start site is blocked by a repressive nucleosome, preventing recruitment of Pol II and the general transcriptional machinery. This barrier to transcription is overcome through the action of the SWI/SNF remodeling complex, which is recruited to promoter-bound acetylated histones due to bromodomain interactions of its BRG1 and BRM subunits [19]. ATP-dependent remodeling results in repositioning of the nucleosome downstream of the start site, allowing transcriptional initiation. Mutational analysis resulting in removal of the positioned nucleosome greatly relaxed the regulation of IFN-β expression, allowing it to be induced in response to cytokines such as TNF-α or IFN-γ as well as viral infection [20]. Given that these cytokines activate only individual transcription factors (NF-κB and IRF-1, respectively), this result suggests that relieving the barrier of the occluding nucleosome requires the coordinated activity of multiple, independent transcription factors.
In contrast to IFN-β, the IFN-α genes display a simpler although similar regulatory architecture, and individual members of the gene family can be subject to distinct regulation through incompletely understood mechanisms. Specifically, there appears to be no direct role for NF-κB or AP-1 complexes binding IFN-α promoters, but members of the IRF transcription factor family are essential [21]. Roles for IRF1, IRF3, IRF4, IRF5, and IRF8 have been documented as inducers of IFN-α genes [22], with IRF3 and IRF7 playing largely universal roles while the other family members regulate IFN production in a highly cell type-specific manner. The multiplicity of IRF isoforms may provide a degree of gene-specific regulation to the IFN-α family, since their promoters display differential affinities for distinct IRF proteins [23,24]. However, why IFN-α genes can be regulated by a single type of transcription factor while the IFN-β gene requires several cooperating complexes, and why different cells require distinct members of the IRF family to accomplish IFN-α gene induction are intriguing but as yet unanswered questions.
Although type I IFN genes are tightly regulated in response to viral infection, it has become clear that IFN-β is also constitutively produced by uninfected cells at exceedingly low but physiologically significant levels [25]. In contrast to viral induction of IFN-β, constitutive secretion appears to be independent of IRF3 and 7; instead, there is a switch to dependence on the AP-1 subunit c-Jun. Constitutive IFN appears to function by maintaining homeostatic levels of important signaling and response factors that are themselves the products of IFN-stimulated target genes, thereby insuring an available pool of critical proteins, such as STAT1 [25]. The absence of constitutive IFN results in a number of aberrations, such as deregulation of the hematopoietic stem cell niche, bone resorption, leukocyte homeostasis and activity, anti-viral responses, anti-tumor immunity and autoimmune diseases [26]. Induction of the IFN-λ gene family reflects another twist to the story. These genes display IRF and NF-κB binding sites at their promoters, and early studies suggested that IFN-λ1 was regulated similarly to IFN-β, dependent on IRF3 and NF-κB, while IFNλ2/3 regulation resembled IFN-α, dependent on IRF7 [27,28]. Nonetheless, they can be regulated differently than the type I IFN genes, at least in some cell types infected with certain viruses [2,29], and there is evidence that IRF and NF-κB proteins function independently on IFN-λ promoters, rather than in concert [30]. Additional evidence for distinct regulation of IFN-λ synthesis comes from genetic studies of viral susceptibility. Polymorphism of the IFN-λ3 locus correlates strongly with resolution of HCV infection and sustained response to treatment [31–33]. While the molecular mechanism underlying the action of this polymorphism remains undetermined, it suggests that subtle changes in IFN-λ gene expression or function can have profound effect on viral infection.
Virus-induced signaling pathways regulating IFN gene expression
Sensing microbial pathogens is the first step required to initiate and modulate the IFN-dependent innate immune response. Cellular sensors responsible for pathogen recognition can be classified into 4 groups: the membrane bound Toll-like receptors (TLR), the cytoplasmic RIG-like receptors (RLR) mainly responsible for sensing RNA viruses, the nucleotide binding oligomerization domain (NOD)-like receptors, and a disparate family of cytoplasmic and possibly nuclear DNA sensors [34]. The signaling cascades downstream of pathogen recognition that lead to type I and III interferon promoter stimulation have been the subject of intense research in the past decade. A remarkable paradigm for signal propagation has emerged from this research, documenting critical roles for sensing nucleic acids by helicases, protein modification by ubiquitin ligases, and induced protein interaction and aggregation that creates platforms for kinase/substrate assembly and phosphorylation (Fig. 1).
Signaling events downstream of cytoplasmic RLR
Three related DExD/H box containing RNA helicases called retinoic acid inducible gene (RIG-I), melanoma differentiation-associated gene 5 (MDA5), and LGP2 function as sensors of RNA virus infection. RIG-I and MDA5 signal the presence of primarily 5’-phosphorylated RNA and long double-stranded RNA, respectively, while LGP2 appears to play an auxiliary role in RNA recognition [35,36]. RNA binding induces a conformational change and exposes N-terminal caspase activation and recruitment domains (CARD) that bind to the CARD of the mitochondrially-localized adaptor molecule MAVS (mitochondrial antiviral signaling), also known as IPS-1 (IFNβ promoter stimulator 1), VISA (virus induced signaling adaptor), and Cardif (CARD adaptor inducing IFNβ). Activation of RIG-I by short double-stranded and 5’-phosphorylated RNA also requires interaction with unanchored polyubiquitin chains of Lys 63 linkage [37]. Recent advances demonstrated a remarkable activation mechanism for MAVS: in the presence of both viral RNA and K63 polyubiquitin chains, RIG-I promotes the formation of large prion-like MAVS aggregates on the mitochondrial membrane [38]. This auto-propagatory conformational switch may be one of the features that confers exceptionally high sensitivity and amplification to antiviral responses.
Ubiquitin modification appears to be a common component of antiviral signaling. Whether MDA5 triggers MAVS aggregation has not yet been reported, but this mechanism is likely since MDA5 CARDs also have the ability to bind K63 polyubiquitin chains. Adaptors of the TRAF (tumor necrosis factor receptor associated factor) family [39], in particular TRAF2, TRAF3, and TRAF6, serve as ubiquitin ligases for signaling and are recruited by MAVS aggregates [38]. TRAF proteins are in turn essential for the recruitment/activation of the MAP (mitogen activated protein) kinase, the IKK complex, and IKK related kinases TBK1/IKKε. These kinases respectively activate activator protein (AP)-1, NF-κB and IRF3 and 7 that are required for IFN gene induction [40].
A number of modulators have been reported to positively or negatively modify the function of these primary components and add to the complexity of this tightly regulated pathway. An unusual one is STING (Stimulator of Interferon Genes), a transmembrane protein localized in the endoplasmic reticulum, which was recently shown to play a critical role in RIG-I but not MDA-5 mediated signaling [41], thereby possibly allowing diversification of signal propagation. However, its precise mode of action is still under scrutiny and remains controversial [42]. Given its association with the translocon, it has been suggested that STING participates directly in TBK1 recruitment through physical interaction with the exocyst complex. Indeed, Sec5, a component of the exocyst complex, has been shown to directly recruit and activate TBK1, also a component of the exocyst [43]. Recruitment of TBK1 by STING [44] appears to be another process that is again regulated by ubiquitin modification [45].
Although the RLR proteins are considered primary sensors of viral nucleic acid, there are additional signaling steps regulated by helicases, presumably triggered following binding foreign nucleic acid. Roles for DDX3 and DDX60 in viral induction of IFN have recently been documented, although the exact mechanism of their action remains unclear. Interestingly, in addition to being required for augmenting IFN production, these helicases are also targeted by viruses to evade immune recognition [46–49].
Signaling events downstream of TLRs
TLRs are predominant receptors in innate immune cells such as dendritic cells, and they serve as the primary viral sensors for IFN production in plasmacytoid dendritic cells (pDC), cells that are capable of producing very high levels of IFN and are therefore critical for antiviral defense [50]. pDCs express only 2 types of TLRs, 7 and 9, that bind single-stranded RNA and CpG DNA, respectively, and employ a unique signal transduction pathway [51]. After binding their respective ligands, the adaptor protein myeloid differentiation primary response gene 88 (MyD88) is recruited to these receptors and in turn recruits IRAK (IL-1R associated kinase) 1 and 4 and IRF7. TRAF3 and 6 are subsequently recruited to this activation complex. IRF7 is thought to be directly phosphorylated by IRAK1 following a PIN1-dependent isomerization reaction [52], although IKKα may be an additional activating kinase. IKKα was shown to be capable of directly phosphorylating IRF7 following activation by NIK [53] and to be essential for IFNα production by TLR7 [54]. Osteopontin has been reported to be yet another critical player for IRF7 activation, though its role remains ill defined [55].
TLR3, a sensor for extracellular and endosomal double-stranded RNA, signals through an alternative pathway by recruiting the adaptor TRIF, also known as TICAM-1 [56]. TRAF3-TRIF interaction serves as a link to the recruitment of the IRF kinases, TBK1 and IKKε. NAP (NAK associated protein) 1 and TRAF proteins also play a role in the formation of an active IRF kinase complex [57,58]. The other arm of the pathway involves the recruitment of RIP1 and TRAF6, even though the involvement of TRAF6 is still controversial and might be cell type specific, with this arm culminating in the activation of NF-κB and AP-1.
Signaling events downstream of DNA sensors
Unraveling the identity of sensors for foreign DNA has been an area of intense research these past years, resulting in the identification of a growing number of potential players. Among the identified candidates are: DAI, AIM2, IFI16, pol III and most recently DDX41. The signaling events downstream of these receptors are still largely uncharacterized, as is the issues of specificity and redundancy, but a number of known players of the TLR and/or RLR pathways are involved in the cellular response to DNA. A strict requirement for STING, TBK1 and IRF3 has been clearly demonstrated in the majority of studies. However, much detail of the signaling pathways still needs to be worked out, such as the potential for individual specificity or activity of the different sensors or the existence of a unique versus multiply redundant DNA sensing pathways [59,60].
The Antiviral Response
Once type I (IFN-α and IFN-β) and type III (IFN-λ) IFNs are induced by infection or other stimuli, they are secreted by infected cells or stimulated pDCs and bind distinct surface receptors on target cells. Although the signaling pathways triggered downstream of type I and type III receptor engagement lead to similar transcriptional responses, the receptors themselves are distinct. IFN-α and -β bind to a ubiquitously expressed receptor (IFNAR) made up of IFNAR1 and IFNAR2 chains [61]. IFN-λ binds an unrelated receptor consisting of IFN-λ receptor 1 (IFNLR1) and a second subunit shared with the IL-10 receptor, IL10R2. Binding of any of these IFNs to their respective receptors leads to activation of the transcription factor ISGF3 [62] composed of STAT1, STAT2, and IRF9 [50], and the transcriptional activation of a common set of interferon stimulated genes (ISGs) [63–65]. It is the proteins encoded by the ISGs that mediate the antiviral, immunostimulatory, and antiproliferative effects of these cytokines [66]. The importance of this pathway for our survival cannot be overstated. Only three patients lacking IFN-α/β responsiveness have ever been identified, and none has survived infancy [67,68]. Each of these patients carried a stat1 gene mutation; two died with overwhelming virus infection, the third of complications of bone marrow transplantation. Thus, the recent discovery of IFN-λ explained the heretofore puzzling observation that mice lacking IFNAR were not uniformly susceptible to viruses thought to be controlled by this pathway [69–71]. Therefore, IFN-α/β and IFN-λ are redundant in some settings, in that both can activate ISGF3 formation, but a Stat1 mutation would inactivate both of these pathways.
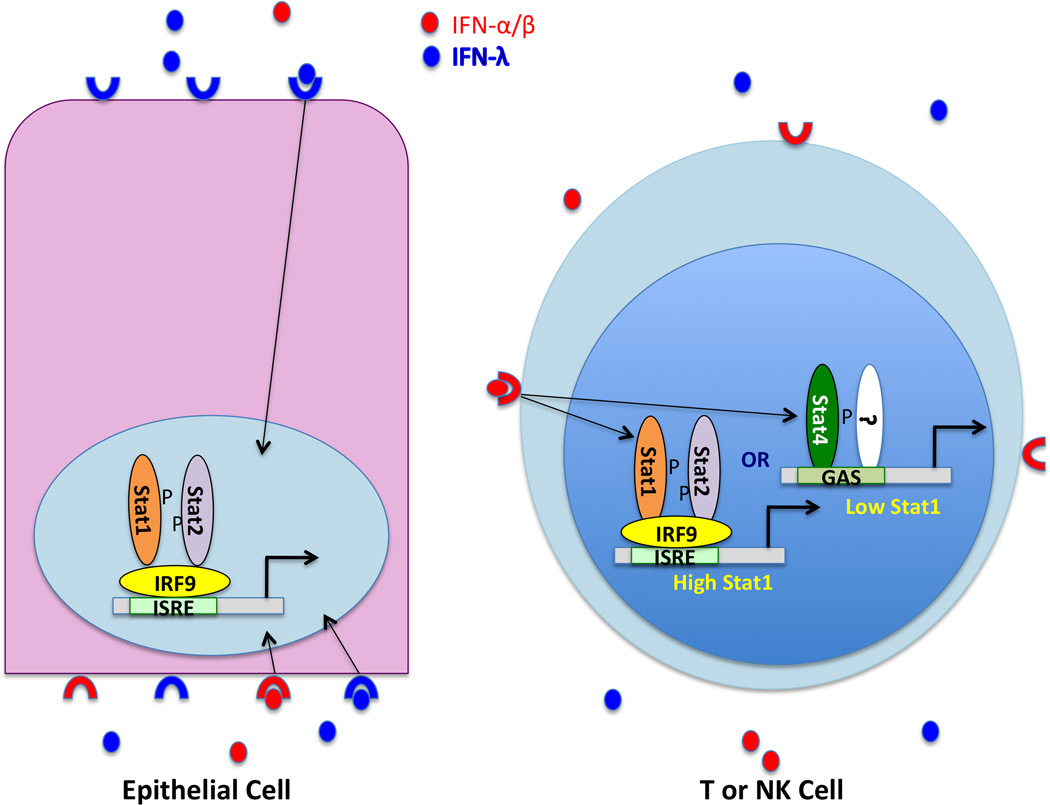
The most striking difference between IFN-α/β and IFN-λ action is due to receptor distribution (Fig.2). While the IFN-α/β receptor is present on all cells, and all nucleated cells can produce and respond to IFN-α/β, expression of the IFNLR chain of the IFN-λ receptor is thought to be limited primarily to epithelial cells [72]. Several studies have shown that the epithelium of the intestine, lung, and vagina can be protected from viral infection by IFN-λ treatment [73,74], and it is preferentially induced by influenza A virus and respiratory syncytial virus infection [75,76]. Likewise, hepatocytes express IFNLR1 [65], and clinical trials of IFN-λ treatment for HCV, a hepatotropic virus, are showing promise [101]. In addition to differential induction patterns, there are also indications that IFN-λ signaling may be resistant to feedback mechanisms targeting IFN-α [77], allowing it to be a more effective antiviral, at least under some circumstances. In a mouse model of rotavirus infection, IFN-λ is more protective than IFN-α, but the basis for this enhanced efficacy has yet to be determined [74]. These accumulated data suggest that these distinct but convergent pathways allow a nuanced response to virus infection, with the diversity of IFN types allowing enhanced IFN production and prolonged action in the anatomic compartments that are open to the outside of the body and therefore serve as major portals for pathogen entry.
Figure 2. Tissue-specific biological effects of type I and III IFN.
IFN-α, -β and -λ are induced by virus infection and secreted by both infected cells and plasmacytoid DCs. Their relative ratio may vary with the pathogen, but IFN-λ is the predominant cytokine secreted by either cell type in the course of influenza virus infection. IFNs can be detected in the blood and in bronchoalveolar lavage fluid during respiratory infection. Respiratory and intestinal epithelial cells are polarized, and there is evidence that IFN receptor expression on intestinal epithelia is polarized as well, with IFNAR localized to basolateral surfaces and IFNLR present on both basolateral and apical surfaces. Receptors on the basolateral surface can detect IFNs present in blood, while apical receptors will see IFNs present at the luminal surface. Signaling through IFNAR or the IFN-λ receptor leads to Stat1 and Stat2 tyrosine phosphorylation, and formation of the ISGF3 complex consisting of phospho-Stat1, phospho-Stat2 and IRF9, which activates target gene expression following nuclear translocation. T and NK cells lack the IFNLR chain, and therefore cannot respond to IFN-λ. However, the response to signaling through IFNAR is conditional (see also Fig. 3). In naïve mice, both Stat1 and Stat4 are present in NK and T cells, with constitutive levels of Stat1 much lower than Stat4. In this state, IFN-α/β signaling results predominantly in Stat4 phosphorylation, which occurs in a Stat2 independent manner and triggers IFN-γ production. In the course of viral infection, Stat1 levels are induced by IFN signaling, altering the ratio of Stat1 to Stat4 in stimulated cells. The consequences of this shift are only partly understood, but serve to explain seemingly contradictory responses of T and NK cells to IFN-α treatment. For example, despite the known antiproliferative effect of IFN-α, virus specific CD8 T cell expansion can occur at a time when IFN levels are high. However, low levels of Stat1 in actively dividing CD8 T cells from infected mice render this population resistant to the anti-proliferative effects of IFN.
Immunoregulation by IFN
In addition to their antiviral action, IFN-α and -β play a major role in orchestrating the development of the adaptive immune response to infection. They do this indirectly, via upregulation of cytokines, chemokines, and intermediate signaling molecules that affect immune cell activation, growth and trafficking, and also via direct effects on dendritic cells (DCs), NK cells and lymphocytes. DCs act both as sensors of infection and antigen presenting cells, and IFNs are one of the signals that signify viral infection. Treatment of immature DCs with IFN-α/β results in upregulation of maturation markers and an enhanced ability to stimulate B and T cells [78–80]. Along with maturation, IFN-α/β treatment results in secretion of chemokines and cytokines including IL-10, IL-15, BAFF and APRIL production by DCs [81–83]. Many different effects of IFN-α/β on IL-12 production by DCs have been reported, suggesting that this may be context dependent. In combination, the IFN induced DC maturation and cytokine secretion promote antibody production and class switching by B cells, and cross priming of CD8+ T cells [80,84].
It has also become clear that IFN-α/β signaling in T, B and NK cells has important cell-autonomous consequences, and that responses to IFN-α/β by these cell types occur by both canonical and non-canonical IFN signaling pathways [85–88]. In the absence of infection, the abundance of the Stat1 protein is low, but it is rapidly induced by rising IFN-α/β levels in response to infection. In NK cells, early IFN-α stimulation drives primarily Stat4 rather than Stat1 phosphorylation, leading to enhanced IFN-γ production. However, as Stat1 levels rise, IFNAR stimulation will preferentially activate Stat1, resulting in enhanced NK cell killing and a loss of IFN-γ production. Thus, it is the ratio of Stat1 and Stat4, influenced by the cytokine milieu, that dictates the behavior of this cell type in response to changing conditions [89]. Although NK cells have a remarkably high basal level of Stat4, similar events have also been demonstrated for T cells. To carry out the viral clearance necessary for host defense, virus-specific CD8+ T cells must proliferate early in infection. This would appear to present a conundrum, because activated Stat1 responses are antiproliferative and access to Stat4 is critical for IFN-γ production. However, antigen-specific, CD8+ T cells actively degrade Stat1, promoting Stat4 activation; while Stat1 activation in non specific T cells inhibits their expansion [86].
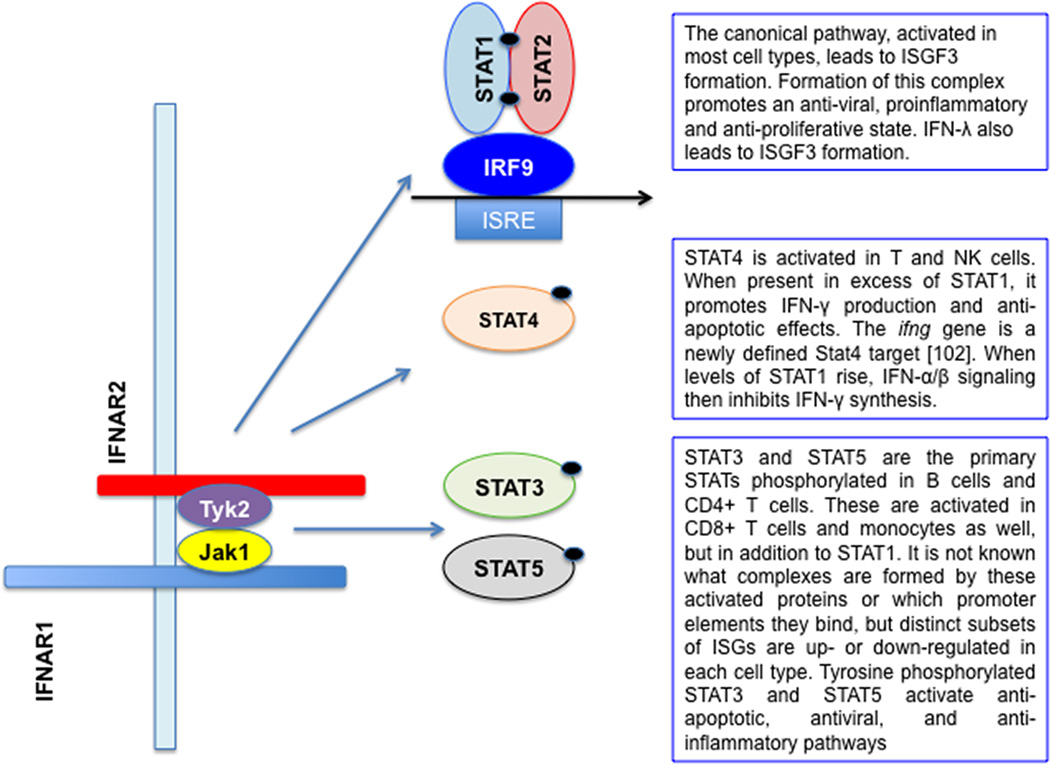
This nuanced response to IFNAR activation by immune cells has now been demonstrated for B and CD4+ T cells as well, and taken together these studies create a new paradigm regarding the role of IFN-α/β in anti-viral immunity (Fig.3). Not only are these cytokines essential for rapid establishment of the antiviral state, they are also required for development of a robust lymphocyte response to clear virus and mount an anti-viral antibody response. Recent work with PBMCs from healthy donors demonstrated differential Stat1, Stat3 and Stat5 activation by IFN-β in monocytes, T cells and B cells [88], with very low levels of Stat1 activation in B and CD4+ T cells. Therefore, in some cells types, Stat1 activation by IFNAR is inefficient, and alternative sets of genes are transcriptionally up or down-regulated by IFN-αβ [88]. The effects of Stat activation are broadly understood [90]. Stat1 activation is proinflammatory and proapoptotic, Stat3 activation antagonizes transcriptional activation by Stat1 [91], both Stat3 and Stat5 activation promote cell proliferation, Stat4 activation is anti-apoptotic and essential for Th1 differentiation [85,92]. The mechanisms that determine which Stats will be activated in a given cell type are not yet known. A comprehensive review of cell type specific type I IFN signaling outlines the various possible mechanisms, including altered receptor expression, differential ligand binding (β versus α-subtypes), variable activation of kinases, presence/activation of cell type specific transcription factors and previous cytokine exposure[93].
Figure 3. Cell type-specific biological effects of type I IFN during virus infection.
IFN-α, and -β are induced by virus infection and secreted by both infected cells and plasmacytoid DCs. In most cell types, signaling through IFNAR receptor leads to Stat1 and Stat2 tyrosine phosphorylation, and formation of the canonical ISGF3 complex consisting of phospho-Stat1, phospho-Stat2 and IRF9, which activates target gene expression following nuclear translocation. However, the response to signaling through IFNAR is both cell specific and context dependent. In most cells studied Stat3 is also activated by IFN-αβ, and acts as a brake on Stat1-mediated, antiviral, proinflammatory, proapoptotic gene expression. Both Stat1 and Stat4 are activated in NK and CD4+ T cells, but Stat4-mediated IFN-γ production by these cells requires low levels of Stat1. Stats1, 3 and 5 are activated by IFN-β treatment of resting B and T cells, with a cell type specific pattern, with very little Stat1 activation in B cells and CD4+ T cells. Stat5, like Stat3, is anti-apoptotic and promotes cell proliferation. However, even in the absence of pStat1, antiviral ISGs can be expressed through IRF-containing complexes binding to alternative enhancer elements [88]. The extent to which IFN-λ activates Stats other than Stat1 and Stat2 has yet to be determined.
It will be interesting to learn the extent to which IFNLR signaling parallels that of IFNAR. As with IFN-α/β, Stats 1–5 can be activated in cultured cells by IFNLR signaling [63], but this has yet to be explored in vivo. Given that IFN-α/β and IFN-λ use distinct receptors, it would be surprising if their signaling were identical. In addition, the ability of immune cells to respond to IFN-λ remains a matter for debate. Studies based on ISG induction in mouse tissues [72,94] and measurement of full length IFNLR1 mRNA in human hepatocytes and PBMCs [95,96] suggest that this receptor is expressed at very low levels in immune cells; but there are also reports describing IFN-λ signaling and receptor expression in some DC and lymphocyte subsets [97–99]. This is an important issue and further studies correlating IFNLR1 mRNA expression, protein expression at the cell surface and signaling are needed to determine whether, like IFN-α/β, IFN-λ plays a direct role in shaping the adaptive immune response. Clinical trials of IFN-λ for HCV treatment were based on the hypothesis that limited IFN-λ receptor tissue distribution would decrease the viral load in infected hepatocytes [65,100] without the toxicity that accompanies IFN-α treatment. This expectation has so far been met [101], lending support to the emerging paradigm that both IFN-α/β and IFN-λ contribute to host antiviral defense, with IFN-α/β being also a major regulator of immune cell function with both beneficial and detrimental sequelae.
Conclusion
More than 50 years after the discovery of IFN, we are far from fully understanding how this complicated network of signals is regulated and triggered. Recent discoveries in the area of virus sensing and IFN induction, the advent of IFN-λ, and a new focus on Stat1-independent type I IFN responses, have both reinforced and challenged established paradigms, leading to an appreciation that this arm of host defense is more finely tuned than had been previously suspected.
The discovery of IFN-λ, and its differential induction by virus, underscores the fact that there is still much to understand about pathogen-specific interactions with innate host defenses. The possibility that this powerful antiviral cytokine is regulated differently than IFN-α/β adds a new dimension to our thinking about how the antiviral state is initiated and maintained. Relatively little is known about the mechanisms of IFN-λ induction, and the question of how widely the IFNLR is expressed is still debated. We hypothesize that this new family of IFNs has functions distinct from IFN-α/β, but this has yet to be definitively shown.
Another paradigm shift involves our growing appreciation of how IFN-α/β signaling can trigger opposing effects in different cell types. The realization that Stats other than Stat1 and Stat2 are activated through the IFNAR represents a big step forward in our understanding of how adaptive anti-viral responses are orchestrated. In addition to inducing ISGF3-mediated gene regulation, activation of alternative Stats appears to enable rapid differentiation and proliferation of antigen specific cells in the context of high IFN-α/β levels. Rather than opposing the outgrowth of virus specific T and B cells, these cytokines appear to play an important role in promoting this process. It is likely that, as new mechanistic details emerge, an enhanced understanding of these interactions will lead us a step closer to deployment of more effective antiviral vaccines and therapies.
Highlights.
Viral infection induces type I and III IFN through recognition of viral nucleic acid.
Ubiquitin is a key signaling component downstream of nucleic acid receptors.
Aggregation of signaling intermediates on organellar scaffolds activates downstream kinases.
Phosphorylation activates transcription factors that reorganize chromatin and recruit coactivators to induce IFN gene transcription.
Cell type restricted expression of IFN receptors results in tissue specific responses. Differential regulation of STAT protein abundance hones distinct responses across the infection cycle.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Donnelly RP, Kotenko SV. Interferon-lambda. a new addition to an old family. J Interferon Cytokine Res. 2010;30:555–564. doi: 10.1089/jir.2010.0078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Iversen MB, Paludan SR. Mechanisms of type III interferon expression. J Interferon Cytokine Res. 2010;30:573–578. doi: 10.1089/jir.2010.0063. [DOI] [PubMed] [Google Scholar]
- 3.Stetson DB, Medzhitov R. Type I interferons in host defense. Immunity. 2006;25:373–381. doi: 10.1016/j.immuni.2006.08.007. [DOI] [PubMed] [Google Scholar]
- 4. Lin WJ, Zheng X, Lin CC, Tsao J, Zhu X, Cody JJ, Coleman JM, Gherzi R, Luo M, Townes TM, et al. Posttranscriptional control of type I interferon genes by KSRP in the innate immune response against viral infection. Mol Cell Biol. 2011;31:3196–3207. doi: 10.1128/MCB.05073-11. Documents a new mechanism for translational control of type I IFN synthesis.
- 5.Marie I, Durbin JE, Levy DE. Differential viral induction of distinct interferon-alpha genes by positive feedback through interferon regulatory factor-7. EMBO J. 1998;17:6660–6669. doi: 10.1093/emboj/17.22.6660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Levy DE, Marie IJ. How viruses elicit interferon production. Triggering the innate immune response to viral infection. In: Chaugeux J-P, Palese P, editors. Modulation of host gene expression and of innate immunity by viruses. Kluwer Academic Publishers; 2005. [Google Scholar]
- 7. Ford E, Thanos D. The transcriptional code of human IFN-beta gene expression. Biochim Biophys Acta. 2010;1799:328–336. doi: 10.1016/j.bbagrm.2010.01.010. *This paper reviews current models for transcriptional control of IFN-β gene expression.
- 8.Taniguchi T, Ogasawara K, Takaoka A, Tanaka N. IRF family of transcription factors as regulators of host defense. Annu Rev Immunol. 2001;19:623–655. doi: 10.1146/annurev.immunol.19.1.623. [DOI] [PubMed] [Google Scholar]
- 9.Paun A, Pitha PM. The IRF family, revisited. Biochimie. 2007;89:744–753. doi: 10.1016/j.biochi.2007.01.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Ye J, Maniatis T. Negative regulation of interferon-beta gene expression during acute and persistent virus infections. PLoS One. 2011;6:e20681. doi: 10.1371/journal.pone.0020681. **This paper demonstrates that regulation of IRF3 can affect the persistance as well as the induction of IFN gene expression and can be targeted during persistant viral infection to impair innate immunity.
- 11.Goodbourn S. The regulation of beta-interferon gene expression. Semin Cancer Biol. 1990;1:89–95. [PubMed] [Google Scholar]
- 12.Maniatis T, Falvo JV, Kim TH, Kim TK, Lin CH, Parekh BS, Wathelet MG. Structure and function of the interferon-beta enhanceosome. Cold Spring Harb Symp Quant Biol. 1998;63:609–620. doi: 10.1101/sqb.1998.63.609. [DOI] [PubMed] [Google Scholar]
- 13.Lin R, Mustafa A, Nguyen H, Gewert D, Hiscott J. Mutational analysis of interferon (IFN) regulatory factors 1 and 2. Effects on the induction of IFN-beta gene expression. J Biol Chem. 1994;269:17542–17549. [PubMed] [Google Scholar]
- 14. Chen S, Short JA, Young DF, Killip MJ, Schneider M, Goodbourn S, Randall RE. Heterocellular induction of interferon by negative-sense RNA viruses. Virology. 2010;407:247–255. doi: 10.1016/j.virol.2010.08.008. **This paper examines molecular mechanisms that limit type I IFN production during infection to a minority of infected cells.
- 15.Apostolou E, Thanos D. Virus Infection Induces NF-kappaB-dependent interchromosomal associations mediating monoallelic IFN-beta gene expression. Cell. 2008;134:85–96. doi: 10.1016/j.cell.2008.05.052. [DOI] [PubMed] [Google Scholar]
- 16.Sato M, Suemori H, Hata N, Asagiri M, Ogasawara K, Nakao K, Nakaya T, Katsuki M, Noguchi S, Tanaka N, et al. Distinct and essential roles of transcription factors IRF-3 and IRF-7 in response to viruses for IFN-alpha/beta gene induction. Immunity. 2000;13:539–548. doi: 10.1016/s1074-7613(00)00053-4. [DOI] [PubMed] [Google Scholar]
- 17.Hata N, Sato M, Takaoka A, Asagiri M, Tanaka N, Taniguchi T. Constitutive IFN-alpha/ beta signal for efficient IFN-alpha/beta gene induction by virus. Biochem Biophys Res Commun. 2001;285:518–525. doi: 10.1006/bbrc.2001.5159. [DOI] [PubMed] [Google Scholar]
- 18.Agalioti T, Lomvardas S, Parekh B, Yie J, Maniatis T, Thanos D. Ordered recruitment of chromatin modifying and general transcription factors to the IFN-beta promoter. Cell. 2000;103:667–678. doi: 10.1016/s0092-8674(00)00169-0. [DOI] [PubMed] [Google Scholar]
- 19.Horn PJ, Peterson CL. The bromodomain. a regulator of ATP-dependent chromatin remodeling? Front Biosci. 2001;6:D1019–D1023. doi: 10.2741/horn. [DOI] [PubMed] [Google Scholar]
- 20.Lomvardas S, Thanos D. Modifying gene expression programs by altering core promoter chromatin architecture. Cell. 2002;110:261–271. doi: 10.1016/s0092-8674(02)00822-x. [DOI] [PubMed] [Google Scholar]
- 21.Tamura T, Yanai H, Savitsky D, Taniguchi T. The IRF family transcription factors in immunity and oncogenesis. Annu Rev Immunol. 2008;26:535–584. doi: 10.1146/annurev.immunol.26.021607.090400. [DOI] [PubMed] [Google Scholar]
- 22.Tailor P, Tamura T, Kong HJ, Kubota T, Kubota M, Borghi P, Gabriele L, Ozato K. The feedback phase of type I interferon induction in dendritic cells requires interferon regulatory factor 8. Immunity. 2007;27:228–239. doi: 10.1016/j.immuni.2007.06.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Levy DE, Marie I, Prakash A. Ringing the interferon alarm. differential regulation of gene expression at the interface between innate and adaptive immunity. Curr Opin Immunol. 2003;15:52–58. doi: 10.1016/s0952-7915(02)00011-0. [DOI] [PubMed] [Google Scholar]
- 24.Genin P, Vaccaro A, Civas A. The role of differential expression of human interferon--a genes in antiviral immunity. Cytokine Growth Factor Rev. 2009;20:283–295. doi: 10.1016/j.cytogfr.2009.07.005. [DOI] [PubMed] [Google Scholar]
- 25. Gough DJ, Messina NL, Hii L, Gould JA, Sabapathy K, Robertson AP, Trapani JA, Levy DE, Hertzog PJ, Clarke CJ, et al. Functional crosstalk between type I and II interferon through the regulated expression of STAT1. PLoS Biol. 2010;8 doi: 10.1371/journal.pbio.1000361. e1000361. **This paper shows the consequences of modulating Stat protein abundance on subsequent cytokine responses.
- 26.Gough DJ, Messina NL, Clark CPJ, Levy DE, Johnstone RW. Constitutive type I interferon secretion positively and negatively modulates homeostatic balance. 2011 Submitted. [Google Scholar]
- 27.Onoguchi K, Yoneyama M, Takemura A, Akira S, Taniguchi T, Namiki H, Fujita T. Viral infections activate types I and III interferon genes through a common mechanism. J Biol Chem. 2007;282:7576–7581. doi: 10.1074/jbc.M608618200. [DOI] [PubMed] [Google Scholar]
- 28.Osterlund P, Pietila T, Veckman V, Kotenko S, Julkunen I. IFN regulatory factor family members differentially regulate the expression of type III IFN (IFN-lambda) genes. J Immunol. 2007;179:3434–3442. doi: 10.4049/jimmunol.179.6.3434. [DOI] [PubMed] [Google Scholar]
- 29.Iversen MB, Ank N, Melchjorsen J, Paludan SR. Expression of type III interferon (IFN) in the vaginal mucosa is mediated primarily by dendritic cells and displays stronger dependence on NF-kappaB than type I IFNs. J Virol. 2010;84:4579–4586. doi: 10.1128/JVI.02591-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Thomson SJ, Goh FG, Banks H, Krausgruber T, Kotenko SV, Foxwell BM, Udalova IA. The role of transposable elements in the regulation of IFN-lambda1 gene expression. Proc Natl Acad Sci U S A. 2009;106:11564–11569. doi: 10.1073/pnas.0904477106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Thomas DL, Thio CL, Martin MP, Qi Y, Ge D, O'Huigin C, Kidd J, Kidd K, Khakoo SI, Alexander G, et al. Genetic variation in IL28B and spontaneous clearance of hepatitis C virus. Nature. 2009;461:798–801. doi: 10.1038/nature08463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Tanaka Y, Nishida N, Sugiyama M, Kurosaki M, Matsuura K, Sakamoto N, Nakagawa M, Korenaga M, Hino K, Hige S, et al. Genome-wide association of IL28B with response to pegylated interferon-alpha and ribavirin therapy for chronic hepatitis C. Nat Genet. 2009;41:1105–1109. doi: 10.1038/ng.449. [DOI] [PubMed] [Google Scholar]
- 33. Suppiah V, Moldovan M, Ahlenstiel G, Berg T, Weltman M, Abate ML, Bassendine M, Spengler U, Dore GJ, Powell E, et al. IL28B is associated with response to chronic hepatitis C interferon-alpha and ribavirin therapy. Nat Genet. 2009;41:1100–1104. doi: 10.1038/ng.447. **References 31–33 document IFN-λ polymorphisms that impact antiviral responses to HCV.
- 34.O'Neill LA, Bowie AG. Sensing and signaling in antiviral innate immunity. Curr Biol. 2010;20:R328–R333. doi: 10.1016/j.cub.2010.01.044. [DOI] [PubMed] [Google Scholar]
- 35.Satoh T, Kato H, Kumagai Y, Yoneyama M, Sato S, Matsushita K, Tsujimura T, Fujita T, Akira S, Takeuchi O. LGP2 is a positive regulator of RIG-I-and MDA5-mediated antiviral responses. Proc Natl Acad Sci U S A. 2010;107:1512–1517. doi: 10.1073/pnas.0912986107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Pollpeter D, Komuro A, Barber GN, Horvath CM. Impaired cellular responses to cytosolic DNA or infection with Listeria monocytogenes and vaccinia virus in the absence of the murine LGP2 protein. PLoS One. 2011;6:e18842. doi: 10.1371/journal.pone.0018842. *References 35 and 36 report the surprising finding that Lgp2 is a positive rather than a negative regulator of viral signaling.
- 37.Zeng W, Sun L, Jiang X, Chen X, Hou F, Adhikari A, Xu M, Chen ZJ. Reconstitution of the RIG-I pathway reveals a signaling role of unanchored polyubiquitin chains in innate immunity. Cell. 2010;141:315–330. doi: 10.1016/j.cell.2010.03.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Hou F, Sun L, Zeng H, Skaug B, Jiang QX, Chen ZJ. MAVS forms functional prion-like aggregates to activate and propagate antiviral innate immune response. Cell. 2011;146:448–461. doi: 10.1016/j.cell.2011.06.041. **References 37 and 38 document the importance of ubiquitin polymerization and MAVS aggregation in viral signaling.
- 39.Oganesyan G, Saha SK, Guo B, He JQ, Shahangian A, Zarnegar B, Perry A, Cheng G. Critical role of TRAF3 in the Toll-like receptor-dependent and -independent antiviral response. Nature. 2006;439:208–211. doi: 10.1038/nature04374. [DOI] [PubMed] [Google Scholar]
- 40.Wertz IE, Dixit VM. Signaling to NF-kappaB. regulation by ubiquitination. Cold Spring Harb Perspect Biol. 2010;2 doi: 10.1101/cshperspect.a003350. a003350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ishikawa H, Barber GN. STING is an endoplasmic reticulum adaptor that facilitates innate immune signalling. Nature. 2008;455:674–678. doi: 10.1038/nature07317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Zhong B, Yang Y, Li S, Wang YY, Li Y, Diao F, Lei C, He X, Zhang L, Tien P, et al. The adaptor protein MITA links virus-sensing receptors to IRF3 transcription factor activation. Immunity. 2008;29:538–550. doi: 10.1016/j.immuni.2008.09.003. [DOI] [PubMed] [Google Scholar]
- 43.Chien Y, Kim S, Bumeister R, Loo YM, Kwon SW, Johnson CL, Balakireva MG, Romeo Y, Kopelovich L, Gale M, Jr, et al. RalB GTPase-mediated activation of the IkappaB family kinase TBK1 couples innate immune signaling to tumor cell survival. Cell. 2006;127:157–170. doi: 10.1016/j.cell.2006.08.034. [DOI] [PubMed] [Google Scholar]
- 44. Ishikawa H, Ma Z, Barber GN. STING regulates intracellular DNA-mediated, type I interferon-dependent innate immunity. Nature. 2009;461:788–792. doi: 10.1038/nature08476. **This paper demonstrates the centrality of STING as a mediator of virus induction of IFN gene expression.
- 45.Tsuchida T, Zou J, Saitoh T, Kumar H, Abe T, Matsuura Y, Kawai T, Akira S. The ubiquitin ligase TRIM56 regulates innate immune responses to intracellular double-stranded DNA. Immunity. 2010;33:765–776. doi: 10.1016/j.immuni.2010.10.013. [DOI] [PubMed] [Google Scholar]
- 46.Miyashita M, Oshiumi H, Matsumoto M, Seya T. DDX60, a DEXD/H box helicase, is a novel antiviral factor promoting RIG-I-like receptor-mediated signaling. Mol Cell Biol. 2011;31:3802–3819. doi: 10.1128/MCB.01368-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Soulat D, Burckstummer T, Westermayer S, Goncalves A, Bauch A, Stefanovic A, Hantschel O, Bennett KL, Decker T, Superti-Furga G. The DEAD-box helicase DDX3X is a critical component of the TANK-binding kinase 1-dependent innate immune response. EMBO J. 2008;27:2135–2146. doi: 10.1038/emboj.2008.126. *References 45 – 47 document novel components of the signaling pathway leading to virus-induced IFN.
- 48.Schroder M, Baran M, Bowie AG. Viral targeting of DEAD box protein 3 reveals its role in TBK1/IKKepsilon-mediated IRF activation. EMBO J. 2008;27:2147–2157. doi: 10.1038/emboj.2008.143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Mulhern O, Bowie AG. Unexpected roles for DEAD-box protein 3 in viral RNA sensing pathways. Eur J Immunol. 2010;40:933–935. doi: 10.1002/eji.201040447. [DOI] [PubMed] [Google Scholar]
- 50.Levy DE. Whence interferon? Variety in the production of interferon in response to viral infection. J Exp Med. 2002;195:F15–F18. doi: 10.1084/jem.20020075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Blasius AL, Beutler B. Intracellular toll-like receptors. Immunity. 2010;32:305–315. doi: 10.1016/j.immuni.2010.03.012. [DOI] [PubMed] [Google Scholar]
- 52.Tun-Kyi A, Finn G, Greenwood A, Nowak M, Lee TH, Asara JM, Tsokos GC, Fitzgerald K, Israel E, Li X, et al. Essential role for the prolyl isomerase Pin1 in Toll-like receptor signaling and type I interferon-mediated immunity. Nat Immunol. 2011;12:733–741. doi: 10.1038/ni.2069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Wang RP, Zhang M, Li Y, Diao FC, Chen D, Zhai Z, Shu HB. Differential regulation of IKK alpha-mediated activation of IRF3/7 by NIK. Mol Immunol. 2008;45:1926–1934. doi: 10.1016/j.molimm.2007.10.034. [DOI] [PubMed] [Google Scholar]
- 54.Hoshino K, Sugiyama T, Matsumoto M, Tanaka T, Saito M, Hemmi H, Ohara O, Akira S, Kaisho T. IkappaB kinase-alpha is critical for interferon-alpha production induced by Toll-like receptors 7 and 9. Nature. 2006;440:949–953. doi: 10.1038/nature04641. [DOI] [PubMed] [Google Scholar]
- 55.Shinohara ML, Lu L, Bu J, Werneck MB, Kobayashi KS, Glimcher LH, Cantor H. Osteopontin expression is essential for interferon-alpha production by plasmacytoid dendritic cells. Nat Immunol. 2006;7:498–506. doi: 10.1038/ni1327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Seya T, Matsumoto M, Ebihara T, Oshiumi H. Functional evolution of the TICAM-1 pathway for extrinsic RNA sensing. Immunol Rev. 2009;227:44–53. doi: 10.1111/j.1600-065X.2008.00723.x. [DOI] [PubMed] [Google Scholar]
- 57.Sasai M, Shingai M, Funami K, Yoneyama M, Fujita T, Matsumoto M, Seya T. NAK-associated protein 1 participates in both the TLR3 and the cytoplasmic pathways in type I IFN induction. J Immunol. 2006;177:8676–8683. doi: 10.4049/jimmunol.177.12.8676. [DOI] [PubMed] [Google Scholar]
- 58. Sasai M, Tatematsu M, Oshiumi H, Funami K, Matsumoto M, Hatakeyama S, Seya T. Direct binding of TRAF2 and TRAF6 to TICAM-1/TRIF adaptor participates in activation of the Toll-like receptor 3/4 pathway. Mol Immunol. 2010;47:1283–1291. doi: 10.1016/j.molimm.2009.12.002. *This paper documents the importance of TRAF ubiquitin ligases in viral signaling.
- 59.Sharma S, Fitzgerald KA. Innate immune sensing of DNA. PLoS Pathog. 2011;7 doi: 10.1371/journal.ppat.1001310. e1001310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Barber GN. Cytoplasmic DNA innate immune pathways. Immunological Reviews. 2011;243:99–108. doi: 10.1111/j.1600-065X.2011.01051.x. [DOI] [PubMed] [Google Scholar]
- 61.de Weerd NA, Samarajiwa SA, Hertzog PJ. Type I interferon receptors. biochemistry and biological functions. J Biol Chem. 2007;282:20053–20057. doi: 10.1074/jbc.R700006200. [DOI] [PubMed] [Google Scholar]
- 62.Levy DE, Kessler DS, Pine R, Reich N, Darnell JE., Jr Interferon-induced nuclear factors that bind a shared promoter element correlate with positive and negative transcriptional control. Genes Dev. 1988;2:383–393. doi: 10.1101/gad.2.4.383. [DOI] [PubMed] [Google Scholar]
- 63.Kotenko SV, Gallagher G, Baurin VV, Lewis-Antes A, Shen M, Shah NK, Langer JA, Sheikh F, Dickensheets H, Donnelly RP. IFN-lambdas mediate antiviral protection through a distinct class II cytokine receptor complex. Nat Immunol. 2003;4:69–77. doi: 10.1038/ni875. [DOI] [PubMed] [Google Scholar]
- 64.Sheppard P, Kindsvogel W, Xu W, Henderson K, Schlutsmeyer S, Whitmore TE, Kuestner R, Garrigues U, Birks C, Roraback J, et al. IL-28, IL-29 and their class II cytokine receptor IL-28R. Nat Immunol. 2003;4:63–68. doi: 10.1038/ni873. [DOI] [PubMed] [Google Scholar]
- 65.Doyle SE, Schreckhise H, Khuu-Duong K, Henderson K, Rosler R, Storey H, Yao L, Liu H, Barahmand-pour F, Sivakumar P, et al. Interleukin-29 uses a type 1 interferon-like program to promote antiviral responses in human hepatocytes. Hepatology. 2006;44:896–906. doi: 10.1002/hep.21312. [DOI] [PubMed] [Google Scholar]
- 66.Schindler C, Levy DE, Decker T. JAK-STAT signaling. from interferons to cytokines. J Biol Chem. 2007;282:20059–20063. doi: 10.1074/jbc.R700016200. [DOI] [PubMed] [Google Scholar]
- 67.Bustamante J, Boisson-Dupuis S, Jouanguy E, Picard C, Puel A, Abel L, Casanova JL. Novel primary immunodeficiencies revealed by the investigation of pediatric infectious disease. Curr Opin Immunol. 2008;20:39–48. doi: 10.1016/j.coi.2007.10.005. [DOI] [PubMed] [Google Scholar]
- 68.Dupuis S, Jouanguy E, Al-Hajjar S, Fieschi C, Al-Mohsen IZ, Al-Jumaah S, Yang K, Chapgier A, Eidenschenk C, Eid P, et al. Impaired response to interferon-alpha/beta and lethal viral disease in human STAT1 deficiency. Nat Genet. 2003;33:388–391. doi: 10.1038/ng1097. [DOI] [PubMed] [Google Scholar]
- 69.Gil MP, Bohn E, O'Guin AK, Ramana CV, Levine B, Stark GR, Virgin HW, Schreiber RD. Biologic consequences of Stat1-independent IFN signaling. Proc Natl Acad Sci U S A. 2001;98:6680–6685. doi: 10.1073/pnas.111163898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Price G, Gaszewska-Mastarlarz A, Moskophidis D. The role of alpha/beta and gamma interferons in development of immunity to influenza A virus in mice. J Virol. 2000;74:3996–4003. doi: 10.1128/jvi.74.9.3996-4003.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Johnson TR, Mertz SE, Gitiban N, Hammond S, Legallo R, Durbin RK, Durbin JE. Role for innate IFNs in determining respiratory syncytial virus immunopathology. J Immunol. 2005;174:7234–7241. doi: 10.4049/jimmunol.174.11.7234. [DOI] [PubMed] [Google Scholar]
- 72.Sommereyns C, Paul S, Staeheli P, Michiels T. IFN-lambda (IFN-lambda) is expressed in a tissue-dependent fashion and primarily acts on epithelial cells in vivo. PLoS Pathog. 2008;4 doi: 10.1371/journal.ppat.1000017. e1000017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Mordstein M, Neugebauer E, Ditt V, Jessen B, Rieger T, Falcone V, Sorgeloos F, Ehl S, Mayer D, Kochs G, et al. Lambda interferon renders epithelial cells of the respiratory and gastrointestinal tracts resistant to viral infections. J Virol. 2010;84:5670–5677. doi: 10.1128/JVI.00272-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Pott J, Mahlakoiv T, Mordstein M, Duerr CU, Michiels T, Stockinger S, Staeheli P, Hornef MW. IFN-lambda determines the intestinal epithelial antiviral host defense. Proc Natl Acad Sci U S A. 2011;108:7944–7949. doi: 10.1073/pnas.1100552108. **References 73–74 demonstrate the antiviral potency of type III IFN on epithelial tissues. Importantly, Pott et al. demonstrate that IFN-λ is more protective than IFN-α/β in the context of rotavirus infection.
- 75. Jewell NA, Cline T, Mertz SE, Smirnov SV, Flano E, Schindler C, Grieves JL, Durbin RK, Kotenko SV, Durbin JE. Lambda interferon is the predominant interferon induced by influenza A virus infection in vivo. J Virol. 2010;84:11515–11522. doi: 10.1128/JVI.01703-09. ** This is the first report demonstrating that IFN-λ is preferentially induced by virus infection, and that it has potent antiviral activity in lung epithelium.
- 76. Okabayashi T, Kojima T, Masaki T, Yokota S, Imaizumi T, Tsutsumi H, Himi T, Fujii N, Sawada N. Type-III interferon, not type-I, is the predominant interferon induced by respiratory viruses in nasal epithelial cells. Virus Res. 2011;160:360–366. doi: 10.1016/j.virusres.2011.07.011. *This is the first report of an infection where IFN-λ alone is induced by virus.
- 77. Francois-Newton V, Magno de Freitas Almeida G, Payelle-Brogard B, Monneron D, Pichard-Garcia L, Piehler J, Pellegrini S, Uze G. USP18-based negative feedback control is induced by type I and type III interferons and specifically inactivates interferon alpha response. PLoS One. 2011;6:e22200. doi: 10.1371/journal.pone.0022200. *This paper documents negative cross-talk between type I and type III IFN signaling.
- 78.Ito T, Amakawa R, Inaba M, Ikehara S, Inaba K, Fukuhara S. Differential regulation of human blood dendritic cell subsets by IFNs. J Immunol. 2001;166:2961–2969. doi: 10.4049/jimmunol.166.5.2961. [DOI] [PubMed] [Google Scholar]
- 79.Montoya M, Schiavoni G, Mattei F, Gresser I, Belardelli F, Borrow P, Tough DF. Type I interferons produced by dendritic cells promote their phenotypic and functional activation. Blood. 2002;99:3263–3271. doi: 10.1182/blood.v99.9.3263. [DOI] [PubMed] [Google Scholar]
- 80.Le Bon A, Schiavoni G, D'Agostino G, Gresser I, Belardelli F, Tough DF. Type I interferons potently enhance humoral immunity and can promote isotype switching by stimulating dendritic cells in vivo. Immunity. 2001;14:461–470. doi: 10.1016/s1074-7613(01)00126-1. [DOI] [PubMed] [Google Scholar]
- 81.Padovan E, Spagnoli GC, Ferrantini M, Heberer M. IFN-alpha2a induces IP-10/CXCL10 and MIG/CXCL9 production in monocyte-derived dendritic cells and enhances their capacity to attract and stimulate CD8+ effector T cells. J Leukoc Biol. 2002;71:669–676. [PubMed] [Google Scholar]
- 82.Mattei F, Schiavoni G, Belardelli F, Tough DF. IL-15 is expressed by dendritic cells in response to type I IFN, double-stranded RNA, or lipopolysaccharide and promotes dendritic cell activation. J Immunol. 2001;167:1179–1187. doi: 10.4049/jimmunol.167.3.1179. [DOI] [PubMed] [Google Scholar]
- 83.Litinskiy MB, Nardelli B, Hilbert DM, He B, Schaffer A, Casali P, Cerutti A. DCs induce CD40-independent immunoglobulin class switching through BLyS and APRIL. Nat Immunol. 2002;3:822–829. doi: 10.1038/ni829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Le Bon A, Etchart N, Rossmann C, Ashton M, Hou S, Gewert D, Borrow P, Tough DF. Cross-priming of CD8+ T cells stimulated by virus-induced type I interferon. Nat Immunol. 2003;4:1009–1015. doi: 10.1038/ni978. [DOI] [PubMed] [Google Scholar]
- 85.Nguyen KB, Watford WT, Salomon R, Hofmann SR, Pien GC, Morinobu A, Gadina M, O'Shea JJ, Biron CA. Critical role for STAT4 activation by type 1 interferons in the interferon-gamma response to viral infection. Science. 2002;297:2063–2066. doi: 10.1126/science.1074900. [DOI] [PubMed] [Google Scholar]
- 86.Gil MP, Salomon R, Louten J, Biron CA. Modulation of STAT1 protein levels. a mechanism shaping CD8 T-cell responses in vivo. Blood. 2006;107:987–993. doi: 10.1182/blood-2005-07-2834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Miyagi T, Gil MP, Wang X, Louten J, Chu WM, Biron CA. High basal STAT4 balanced by STAT1 induction to control type 1 interferon effects in natural killer cells. J Exp Med. 2007;204:2383–2396. doi: 10.1084/jem.20070401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88. van Boxel-Dezaire AH, Zula JA, Xu Y, Ransohoff RM, Jacobberger JW, Stark GR. Major differences in the responses of primary human leukocyte subsets to IFN-beta. J Immunol. 2010;185:5888–5899. doi: 10.4049/jimmunol.0902314. ** This paper is the first to demonstrate that Stats are differentially activated by type I IFN treatment, and that the pattern of Stat activation and gene induction varies by immune cell type.
- 89. Mack EA, Kallal LE, Demers DA, Biron CA. Type I interferon induction of natural killer cell gamma interferon production for defense during lymphocytic choriomeningitis virus infection. MBio. 2011;2 doi: 10.1128/mBio.00169-11. **This paper demonstrates that IFN-γ can be induced in NK cells through type I IFN activating STAT4 at the initial site of viral infection due to abundant STAT4, followed by induction of a type I IFN antiviral program later in infection after induction of elevated STAT1 levels.
- 90.Adamson AS, Collins K, Laurence A, O'Shea JJ. The Current STATus of lymphocyte signaling. new roles for old players. Curr Opin Immunol. 2009;21:161–166. doi: 10.1016/j.coi.2009.03.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91. Wang WB, Levy DE, Lee CK. STAT3 negatively regulates type I IFN-mediated antiviral response. J Immunol. 2011;187:2578–2585. doi: 10.4049/jimmunol.1004128. **This is the first exploration of the role of Stat3 activation following type I IFN treatment.
- 92.Morinobu A, Gadina M, Strober W, Visconti R, Fornace A, Montagna C, Feldman GM, Nishikomori R, O'Shea JJ. STAT4 serine phosphorylation is critical for IL-12-induced IFN-gamma production but not for cell proliferation. Proc Natl Acad Sci U S A. 2002;99:12281–12286. doi: 10.1073/pnas.182618999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.van Boxel-Dezaire AH, Rani MR, Stark GR. Complex modulation of cell type-specific signaling in response to type I interferons. Immunity. 2006;25:361–372. doi: 10.1016/j.immuni.2006.08.014. [DOI] [PubMed] [Google Scholar]
- 94.Lasfar A, Lewis-Antes A, Smirnov SV, Anantha S, Abushahba W, Tian B, Reuhl K, Dickensheets H, Sheikh F, Donnelly RP, et al. Characterization of the mouse IFN-lambda ligand-receptor system. IFN-lambdas exhibit antitumor activity against B16 melanoma. Cancer Res. 2006;66:4468–4477. doi: 10.1158/0008-5472.CAN-05-3653. [DOI] [PubMed] [Google Scholar]
- 95. Diegelmann J, Beigel F, Zitzmann K, Kaul A, Goke B, Auernhammer CJ, Bartenschlager R, Diepolder HM, Brand S. Comparative analysis of the lambda-interferons IL-28A and IL-29 regarding their transcriptome and their antiviral properties against hepatitis C virus. PLoS One. 2010;5:e15200. doi: 10.1371/journal.pone.0015200. *This paper is a detailed examination of IFNLR expression and IFN-λ signaling in both hepatocytes and leukocytes.
- 96.Witte K, Gruetz G, Volk HD, Looman AC, Asadullah K, Sterry W, Sabat R, Wolk K. Despite IFN-lambda receptor expression, blood immune cells, but not keratinocytes or melanocytes, have an impaired response to type III interferons: implications for therapeutic applications of these cytokines. Genes Immun. 2009;10:702–714. doi: 10.1038/gene.2009.72. [DOI] [PubMed] [Google Scholar]
- 97.Dai J, Megjugorac NJ, Gallagher GE, Yu RY, Gallagher G. IFN-lambda1 (IL-29) inhibits GATA3 expression and suppresses Th2 responses in human naive and memory T cells. Blood. 2009;113:5829–5838. doi: 10.1182/blood-2008-09-179507. [DOI] [PubMed] [Google Scholar]
- 98. Megjugorac NJ, Gallagher GE, Gallagher G. Modulation of human plasmacytoid DC function by IFN-lambda1 (IL-29) J Leukoc Biol. 2009;86:1359–1363. doi: 10.1189/jlb.0509347. *This is the first paper to document the effects of IFN-λ on pDC function.
- 99.Mennechet FJ, Uze G. Interferon-lambda-treated dendritic cells specifically induce proliferation of FOXP3-expressing suppressor T cells. Blood. 2006;107:4417–4423. doi: 10.1182/blood-2005-10-4129. [DOI] [PubMed] [Google Scholar]
- 100.Robek MD, Boyd BS, Chisari FV. Lambda interferon inhibits hepatitis B and C virus replication. J Virol. 2005;79:3851–3854. doi: 10.1128/JVI.79.6.3851-3854.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101. Muir AJ, Shiffman ML, Zaman A, Yoffe B, de la Torre A, Flamm S, Gordon SC, Marotta P, Vierling JM, Lopez-Talavera JC, et al. Phase 1b study of pegylated interferon lambda 1 with or without ribavirin in patients with chronic genotype 1 hepatitis C virus infection. Hepatology. 2010;52:822–832. doi: 10.1002/hep.23743. **This paper demonstrates the potential of type III IFN as an antiviral therapeutic.
- 102. Wei L, Vahedi G, Sun HW, Watford WT, Takatori H, Ramos HL, Takahashi H, Liang J, Gutierrez-Cruz G, Zang C, et al. Discrete role of STAT4 and STAT6 transcription factors in tuning epigenetic modifications and transcription during T helper cell differentiation. Immunity. 2010;32:840–851. doi: 10.1016/j.immuni.2010.06.003. **This paper outlines a direct role for Stat4 in the transcriptional and epigenetic changes that occur during CD4+ T cell differentiation.