Abstract
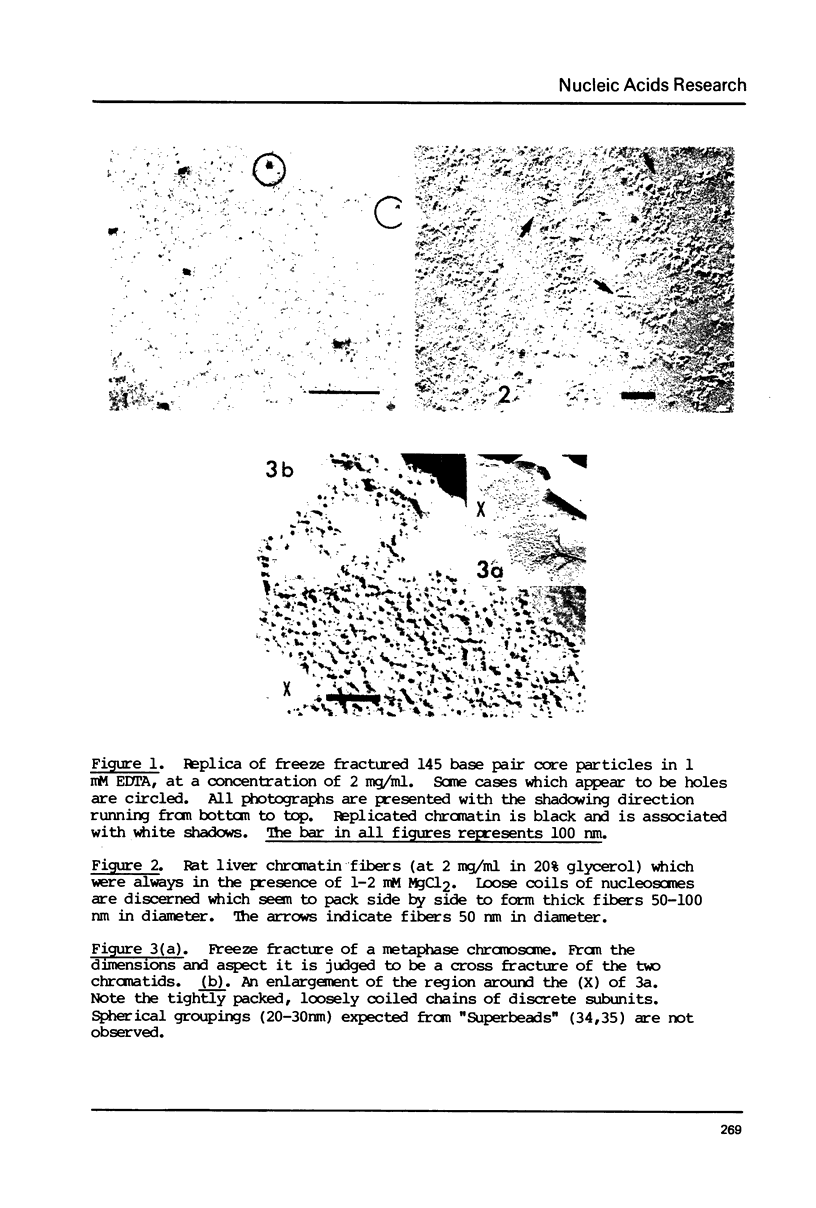
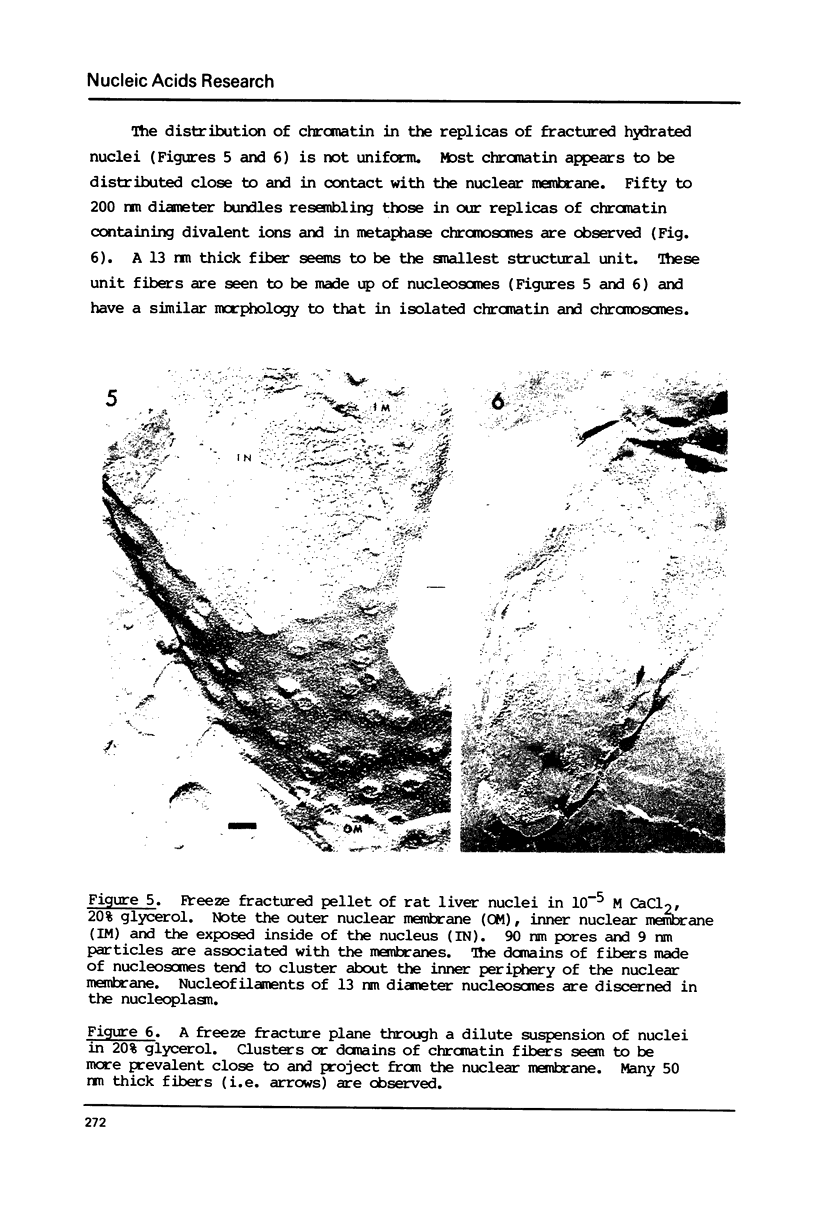
Chromatin gels, metaphase chromosomes, and intact nuclei were studied by freeze fracturing followed by electron microscopy. The results complement and extend those obtained by classical electron microscopy techniques as they are obtained without fixation or dehydration. The freeze fracturing technique permits a determination of the hydrated diameters of nucleosomes in chromatin and in nuclei to be 13 nm by comparing to simultaneously studied test objects. Nucleosomes in chromatin fibers are closely spaced but are discrete particles in all conditions studied. In the presence of divalent ions, most chromatin in solution, chromosomes, and nuclei is organized into fibers whose thickness is larger than 40 nm. The images are not at all compatible with a super bead organization of the nucleofilament. Freeze fractures of intact nuclei provides information on the distribution of chromatin in a hydrated unfixed state. The images suggest that most of the chromatin is localized in large domains in contact with the inner nuclear membrane.
Full text
PDF











Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Baudy P., Bram S., Vastel D., Lepault J. Chromatin subunit small angle neutron scattering: a DNA rich coil surrounds a protein-DNA core. Biochem Biophys Res Commun. 1976 Sep 7;72(1):176–183. doi: 10.1016/0006-291x(76)90976-1. [DOI] [PubMed] [Google Scholar]
- Beams H. W., Mueller S. Effects of ultracentrifugation on the interphase nucleus of somatic cells with special reference to the nuclear envelope-chromatin relationship. Z Zellforsch Mikrosk Anat. 1970;108(3):297–308. doi: 10.1007/BF00336521. [DOI] [PubMed] [Google Scholar]
- Blaustein M. P. The interrelationship between sodium and calcium fluxes across cell membranes. Rev Physiol Biochem Pharmacol. 1974;70:33–82. doi: 10.1007/BFb0034293. [DOI] [PubMed] [Google Scholar]
- Bram S. A double coil chromatin sub-unit model. Biochimie. 1975 Nov-Dec;57(11-12):1301–1306. doi: 10.1016/s0300-9084(76)80542-1. [DOI] [PubMed] [Google Scholar]
- Bram S. A model for the nucleosome core particle subunit. Biochem Biophys Res Commun. 1978 Mar 30;81(2):684–691. doi: 10.1016/0006-291x(78)91591-7. [DOI] [PubMed] [Google Scholar]
- Bram S., Baudy P., Lepault J., Hermann D. Chromatin very small angle neutron scattering: further evidence for a 30 nm diameter super coil in dilute solutions. Nucleic Acids Res. 1977 Jul;4(7):2275–2282. doi: 10.1093/nar/4.7.2275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bram S., Butler-Browne G., Baudy P., Ibel K. Quaternary structure of chromatin. Proc Natl Acad Sci U S A. 1975 Mar;72(3):1043–1045. doi: 10.1073/pnas.72.3.1043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bram S., Kouprach S., Baudy P. DNA structure in chromatin and in solution studied by electron microscopy and neutron and X-ray scattering. Cold Spring Harb Symp Quant Biol. 1978;42(Pt 1):23–29. doi: 10.1101/sqb.1978.042.01.005. [DOI] [PubMed] [Google Scholar]
- Branton D. Fracture faces of frozen membranes. Proc Natl Acad Sci U S A. 1966 May;55(5):1048–1056. doi: 10.1073/pnas.55.5.1048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Compton J. L., Hancock R., Oudet P., Chambon P. Biochemical and electron-microscopic evidence that the subunit structure of Chinese-hamster-ovary interphase chromatin is conserved in mitotic chromosomes. Eur J Biochem. 1976 Nov 15;70(2):555–568. doi: 10.1111/j.1432-1033.1976.tb11047.x. [DOI] [PubMed] [Google Scholar]
- Daskal Y., Mace M. L., Jr, Wray W., Busch H. Use of direct current sputtering for improved visualization of chromosome topology by scanning electron microscopy. Exp Cell Res. 1976 Jun;100(1):204–212. doi: 10.1016/0014-4827(76)90343-8. [DOI] [PubMed] [Google Scholar]
- Daskal Y., Mace M. L., Jr, Wray W., Busch H. Use of direct current sputtering for improved visualization of chromosome topology by scanning electron microscopy. Exp Cell Res. 1976 Jun;100(1):204–212. doi: 10.1016/0014-4827(76)90343-8. [DOI] [PubMed] [Google Scholar]
- DuPraw E. J. Evidence for a 'folded-fibre' organization in human chromosomes. Nature. 1966 Feb 5;209(5023):577–581. doi: 10.1038/209577a0. [DOI] [PubMed] [Google Scholar]
- DuPraw E. J. Evidence for a 'folded-fibre' organization in human chromosomes. Nature. 1966 Feb 5;209(5023):577–581. doi: 10.1038/209577a0. [DOI] [PubMed] [Google Scholar]
- Escaig J., Géraud G., Nicolas G. Congélation rapide de tissus biologiques. Mesure des températures et des vitesses de congélation par thermocouple en couche mince. C R Acad Sci Hebd Seances Acad Sci D. 1977 Jun 13;284(22):2289–2292. [PubMed] [Google Scholar]
- Finch J. T., Klug A. Solenoidal model for superstructure in chromatin. Proc Natl Acad Sci U S A. 1976 Jun;73(6):1897–1901. doi: 10.1073/pnas.73.6.1897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finch J. T., Lutter L. C., Rhodes D., Brown R. S., Rushton B., Levitt M., Klug A. Structure of nucleosome core particles of chromatin. Nature. 1977 Sep 1;269(5623):29–36. doi: 10.1038/269029a0. [DOI] [PubMed] [Google Scholar]
- Franke W. W., Deumling B., Zentgraf H., Falk H., Rae P. M. Nuclear membranes from mammalian liver. IV. Characterization of membrane-attached DNA. Exp Cell Res. 1973 Oct;81(2):365–392. doi: 10.1016/0014-4827(73)90527-2. [DOI] [PubMed] [Google Scholar]
- Gulik-Krzywicki T., Costello M. J. The use of low temperature X-ray diffraction to evaluate freezing methods used in freeze-fracture electron microscopy. J Microsc. 1978 Jan;112(1):103–113. doi: 10.1111/j.1365-2818.1978.tb01158.x. [DOI] [PubMed] [Google Scholar]
- Gulik-Krzywicki T., Costello M. J. The use of low temperature X-ray diffraction to evaluate freezing methods used in freeze-fracture electron microscopy. J Microsc. 1978 Jan;112(1):103–113. doi: 10.1111/j.1365-2818.1978.tb01158.x. [DOI] [PubMed] [Google Scholar]
- Kiryanov G. I., Manamshjan T. A., Polyakov V. Y., Fais D., Chentsov J. S. Levels of granular organization of chromatin fibres. FEBS Lett. 1976 Sep 1;67(3):323–327. doi: 10.1016/0014-5793(76)80557-1. [DOI] [PubMed] [Google Scholar]
- Langmore J. P., Wooley J. C. Chromatin architecture: investigation of a subunit of chromatin by dark field electron microscopy. Proc Natl Acad Sci U S A. 1975 Jul;72(7):2691–2695. doi: 10.1073/pnas.72.7.2691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olins A. L., Olins D. E. Spheroid chromatin units (v bodies). Science. 1974 Jan 25;183(4122):330–332. doi: 10.1126/science.183.4122.330. [DOI] [PubMed] [Google Scholar]
- Oudet P., Gross-Bellard M., Chambon P. Electron microscopic and biochemical evidence that chromatin structure is a repeating unit. Cell. 1975 Apr;4(4):281–300. doi: 10.1016/0092-8674(75)90149-x. [DOI] [PubMed] [Google Scholar]
- Panyim S., Bilek D., Chalkley R. An electrophoretic comparison of vertebrate histones. J Biol Chem. 1971 Jul 10;246(13):4206–4215. [PubMed] [Google Scholar]
- Panyim S., Bilek D., Chalkley R. An electrophoretic comparison of vertebrate histones. J Biol Chem. 1971 Jul 10;246(13):4206–4215. [PubMed] [Google Scholar]
- Paulson J. R., Laemmli U. K. The structure of histone-depleted metaphase chromosomes. Cell. 1977 Nov;12(3):817–828. doi: 10.1016/0092-8674(77)90280-x. [DOI] [PubMed] [Google Scholar]
- Paulson J. R., Laemmli U. K. The structure of histone-depleted metaphase chromosomes. Cell. 1977 Nov;12(3):817–828. doi: 10.1016/0092-8674(77)90280-x. [DOI] [PubMed] [Google Scholar]
- RIS H. Ultrastructure and molecular organization of genetic systems. Can J Genet Cytol. 1961 Jun;3:95–120. doi: 10.1139/g61-015. [DOI] [PubMed] [Google Scholar]
- Renz M., Nehls P., Hozier J. Involvement of histone H1 in the organization of the chromosome fiber. Proc Natl Acad Sci U S A. 1977 May;74(5):1879–1883. doi: 10.1073/pnas.74.5.1879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sedat J., Manuelidis L. A direct approach to the structure of eukaryotic chromosomes. Cold Spring Harb Symp Quant Biol. 1978;42(Pt 1):331–350. doi: 10.1101/sqb.1978.042.01.035. [DOI] [PubMed] [Google Scholar]
- Sedat J., Manuelidis L. A direct approach to the structure of eukaryotic chromosomes. Cold Spring Harb Symp Quant Biol. 1978;42(Pt 1):331–350. doi: 10.1101/sqb.1978.042.01.035. [DOI] [PubMed] [Google Scholar]
- Shaw B. R., Schmitz K. S. Quasielastic light scattering by biopolymers. Conformation of chromatin multimers. Biochem Biophys Res Commun. 1976 Nov 22;73(2):224–232. doi: 10.1016/0006-291x(76)90697-5. [DOI] [PubMed] [Google Scholar]
- Stubblefield E., Wray W. Architecture of the Chinese hamster metaphase chromosome. Chromosoma. 1971;32(3):262–294. doi: 10.1007/BF00284839. [DOI] [PubMed] [Google Scholar]
- Thoma F., Koller T. Influence of histone H1 on chromatin structure. Cell. 1977 Sep;12(1):101–107. doi: 10.1016/0092-8674(77)90188-x. [DOI] [PubMed] [Google Scholar]
- Thoma F., Koller T. Influence of histone H1 on chromatin structure. Cell. 1977 Sep;12(1):101–107. doi: 10.1016/0092-8674(77)90188-x. [DOI] [PubMed] [Google Scholar]
- Wray W. Isolation of metaphase chromosomes, mitotic apparatus, and nuclei. Methods Cell Biol. 1973;6:283–306. doi: 10.1016/s0091-679x(08)60053-9. [DOI] [PubMed] [Google Scholar]
- Wray W., Stubblefield E. A new method for the rapid isolation of chromosomes, mitotic apparatus, or nuclei from mammalian fibroblasts at near neutral pH. Exp Cell Res. 1970 Mar;59(3):469–478. doi: 10.1016/0014-4827(70)90656-7. [DOI] [PubMed] [Google Scholar]







