Abstract
The elongation cycle of protein synthesis involves the delivery of aminoacyl-tRNAs to the A-site of the ribosome, followed by peptide-bond formation and translocation of the tRNAs through the ribosome to reopen the A-site1,2. The translocation reaction is catalyzed by elongation factor G (EF-G) in a GTP-dependent fashion3. Despite the availability of structures of various EF-G-ribosome complexes, the precise mechanism by which tRNAs move through the ribosome still remains unclear. Here we use multiparticle cryo-EM analysis to resolve two previously unseen subpopulations within EF-G-ribosome complexes at sub-nanometer resolution, one of them with a partially translocated tRNA. Comparison of these sub-states reveals that translocation of tRNA on the 30S subunit parallels the swiveling of the 30S-head and is coupled to un-ratcheting of the 30S-body. Since the tRNA maintains contact with the P-site on the 30S-head and simultaneously establishes interaction with the E-site on the 30S-platform, a novel intra-subunit pe/E hybrid state is formed. This state is stabilized by domain IV of EF-G, which interacts with the swiveled 30S-head conformation. These findings provide direct structural and mechanistic insight into the “missing link” in terms of tRNA intermediates involved in the universally conserved translocation process.
Following peptide-bond formation, pre-translocational (PRE) ribosomes carry a peptidyl-tRNA at the A-site and a deacylated-tRNA at the P-site1,3,4. This is a highly dynamic state of the ribosome, which fluctuates between classical states with A- and P-tRNAs and hybrid states with A/P- (A/P denotes that the tRNA is in the A-site on the 30S and the P-site on the 50S subunit) and P/E-tRNAs5-8. Hybrid state formation is coupled to spontaneous rotation of the 30S subunit relative to the 50S subunit9-11 and is stabilized by binding of EF-G12-14. The EF-G/eEF2 induced ratchet-like subunit rearrangement also includes a swivel movement of the head that is approximately orthogonal to the intersubunit rotation of the ribosomal subunits14-16. EF-G catalyses translocation of the hybrid state tRNAs on the 30S subunit to form a post-translocational (POST) state ribosome with tRNAs located at classical P- and E-sites. The translocation process is accelerated by the GTPase activity of EF-G stimulated by the ribosome17,18. However, how tRNAs are translocated with respect to the 30S subunit and how the mRNA is advanced by one codon remains unclear.
Structural snapshots of the translocation process come from cryo-EM and X-ray analysis of EF-G bound to ribosome complexes12-14,19. Despite considerable effort12,20-22, no direct structural information is available for ribosomal PRE complexes containing simultaneously EF-G and an A-tRNA. It appears that this state is too dynamic and transient to be captured, resulting in either a POST state containing EF-G or a PRE state without EF-G bound20,22. Indeed, the stable EF-G bound POST state determined by X-ray crystallography reveals a non-ratcheted ribosome with tRNAs in classical P/P- and E/E-sites19. Therefore, structural insights into intermediate states of translocation, i.e. ratcheted ribosomal EF-G complexes, have utilised complexes without an A-site peptidyl-tRNA12-14.
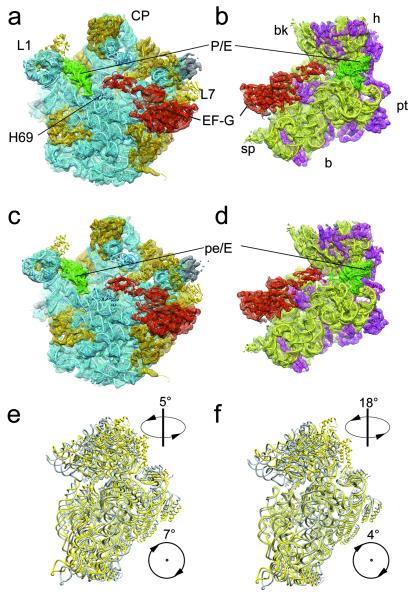
Here we used the antibiotic fusidic acid (FA) to stall EF-G on the 70S ribosome. FA allows GTP hydrolysis by EF-G, but prevents the associated changes in EF-G that normally accompany hydrolysis19. After complex formation and cryo-EM data collection, we employed multiparticle refinement in order to resolve the heterogeneity of the data (supp. Fig. S1). In the first phase of multiparticle refinement a major population of particle images of ribosomes containing EF-G was obtained that yielded a structure in a ratcheted conformation. As refinement progressed to higher resolution the presence of intrinsic conformational heterogeneity necessitated a second phase of multiparticle refinement resulting in two final reconstructions of the 70S•EF-G•GDP•FA complex (Fig. 1), at resolutions of 7.6 Å and 7.8 Å (0.5 FSC criterion), respectively (supp. Fig. S2). Both maps have density attributed to EF-G, but show significant conformational differences (Supp. Mov 1,2): Specifically, the sub-states are distinguished by different degrees of subunit ratcheting, positioning of the L1 protuberance, as well as by the swivel movement of the head of the 30S subunit relative to the body/platform (Fig. 1e, f; supp. Figs. S3, S4; supp. Table 1). Unlike previous cryo-EM and X-ray structures of ribosome•EF-G complexes, movement of the head and body/platform of the 30S subunit is uncoupled: In sub-state I, the 30S subunit is ratcheted by ~7° relative to the 50S subunit, but there is only a modest ~5° swiveling of the 30S head. In contrast, the 30S in sub-state II is only ratcheted by ~4°, but there is a very large ~18° swivel of the head (supp. Table 1).
Fig. 1. Sub-states I (TIPRE) and II (TIPOST) of the 70S•EF-G•GDP•FA complex.
a, b, c, d, The cryo-EM maps of sub-state I (TIPRE) (a, b) and sub-state II (TIPOST) (c , d) of the 70S•EF-G•GDP•FA complex are shown as mesh with docked models in ribbons representation: EF-G (red), tRNA (green), 23S/5S rRNA (blue), 50S ribosomal proteins (orange), 16S rRNA (yellow), and the 30S ribosomal proteins (magenta). The maps are shown from the 30S side with the 30S subunit computationally removed (a, c) and from the 50S side with the 50S subunit computationally removed (b, d). e, f, The 30S subunit of TIPRE (e) and TIPOST (f) (yellow) is compared with the 30S subunit of the POST state19 (grey) by aligning the respective 50S subunits. Arrows with numbers indicate the direction and magnitude (supp. Table 1) of the inter-subunit rotation and the head-swivel from the un-rotated state to TIPRE or TIPOST, respectively.
The identification of two different ratcheted sub-states within the 70S•EF-G•GDP•FA complex prompted us to investigate whether such intrinsic heterogeneity also exists in our previous 70S•EF-G•GMPPNP cryo-EM dataset14. Additional multiparticle refinement indeed revealed that the 70S•EF-G•GMPPNP complex could be subdivided into two sub-states that appeared to be equivalent to those identified in the 70S•EF-G•GDP•FA complex (supp. Fig. S5; supp. Table 1). Our findings here provide supportive evidence for the emerging energy landscape model that allows sampling of several metastable conformations for a defined ribosomal complex8. The sub-nanometer resolution of the 70S•EF-G•GDP•FA subpopulations enabled visualization of secondary structure and thus molecular models to be generated (Fig. 1) by applying our newly developed MDFIT algorithm (see methods). Comparison with available structures reveals that sub-state I is similar in conformation to the ratcheted sub-state of the PRE complex9,10 (supp. Table 1) and also that the tRNA is bound in a hybrid P/E site (Fig. 2a). Therefore we consider sub-state I to be related to a pre-translocational intermediate (TIPRE).
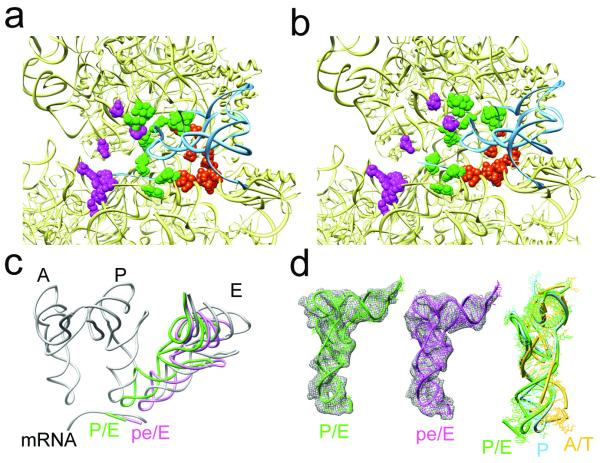
Fig. 2. Localization and conformation of the tRNA of sub-states I (TIPRE) and II (TIPOST).
a, b, Close-up of the tRNA binding regions of the 30S subunit of TIPRE (a) and TIPOST (b). The 30S and tRNAs are shown as yellow and blue ribbons, respectively, whereas ribosomal residues that contact A-, P- and E-tRNAs (magenta, green and orange) are shown as spheres. c, In a common 50S alignment, the P/E-tRNA (green) of TIPRE and the pe/E-tRNA (magenta) of TIPOST together with their respective mRNA codons are compared to mRNA and classical A-, P- and E-tRNA positions (grey). d, Density for the tRNAs (wire-mesh) with molecular models for the P/E-tRNA of TIPRE (left, green) and the pe/E tRNA of TIPOST (middle, magenta). On the right, the model for the P/E-tRNA (green), that is essentially the same as the pe/E-tRNA model (RMSD = 1.5 Å), is compared to a classical P-tRNA (blue) and a A/T-tRNA (yellow) by aligning the acceptor stem, D- and T-stem loops.
In contrast, sub-state II of the 70S•EF-G•GDP•FA complex represents a novel conformational state of the 70S ribosome. Interestingly, the anticodon stem-loop (ASL) of the tRNA has moved by 8 - 10 Å compared to the P/E position of the TIPRE as it maintains strong association with the P-site components of the head and follows the large 18° swivel movement of the head (Fig. 2b; Supp. Mov 3). Because the tRNA interacts simultaneously with P-site components of the head as well as E-site components of the platform, it can be thought of as a 30S intra-subunit hybrid site (Fig. 2b). Moreover, since the contacts of the CCA-end of the tRNA with the E-site on the 50S subunit remain unaffected, we extend the previous nomenclature of hybrid sites5 and refer to this newly identified state as a pe/E hybrid state (P-site on head and E-site on platform of 30S / E-site on 50S).
Interestingly, the ASL of the pe/E-tRNA together with the bound mRNA codon is very close to the position of a fully translocated E/E-tRNA. (Fig. 2c; Supp. Mov 4). Apparently, head-swiveling coupled with partial un-ratcheting of the body/platform of the 30S subunit leads to tRNA translocation, suggesting that sub-state II of the 70S•EF-G•GDP•FA complex is related to a post-translocational intermediate state (TIPOST). We note that although the intermediate states visualized here contain only one tRNA, a second tRNA (ap/P) can be superimposed into the TIPOST state without steric interference with the binding position of EF-G (supp. Fig. S6). Thus the structures presented here appear to be valid models for translocation intermediates (see also supplemental Information for further discussion) but structures of translocation intermediates with two tRNAs will be necessary to validate these predictions. The presence of the RSR in the yeast 80S●eEF2●sordarin complex15 may hint that translocation in pro- and eukaryotes utilize related intermediate conformations. This structure showed a strong head swivel comparable to the bacterial TIPOST combined with a strong intersubunit rotation of the bacterial TIPRE. Thus, the conformation of the 80S●eEF2●sordarin complex15, although obtained with classical single particle methods, may present a further intermediate in between the TIPRE and the TIPOST.
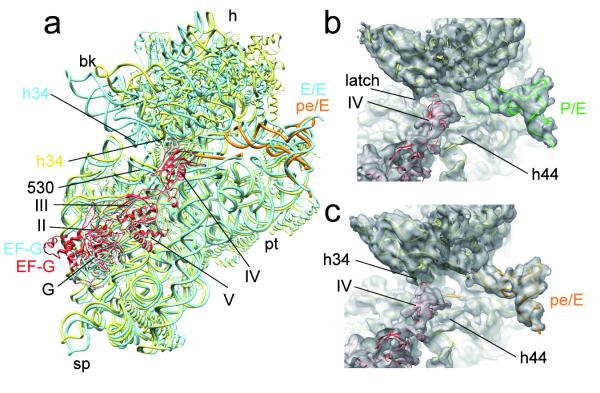
Whereas the P/E- and pe/E-tRNA are in a twisted conformation (Fig. 2d), the overall conformation of EF-G in the TIPRE and TIPOST is remarkably similar to each other (Fig. 1) and to that observed in the cryo-EM reconstruction of the 70S●EF-G●GMPPNP14 as well as in the recent X-ray structure of EF-G•GDP•FA bound to a POST state ribosome19 (Fig. 3a). However, an interesting difference between the two EF-G•GDP•FA sub-states relates to the interaction patterns of domain IV of EF-G: In the TIPRE state, domain IV does not appear to interact significantly with the ribosome (Fig. 3b), whereas a small shift in the binding position of EF-G and the large swivel of the head facilitate a more extensive interaction of domain IV of EF-G with h34 of the 16S rRNA in the TIPOST state (Fig. 3c). h34, together with the 530 region of 16S rRNA, form the so-called latch of the mRNA entry channel. Because of the head-swivel in the TIPOST state, helix h34 has moved ~12 Å away from the 530 region, leading to an opening of the latch (Fig. 3c) similar to that observed previously when eEF2 is trapped on the yeast 80S ribosome with sordarin15. This may facilitate movement of the mRNA-tRNA2 complex. Consistent with this observation, transient protection of h34 by EF-G against chemical modification during the translocation reaction has been reported previously23. Therefore, the direct interaction of domain IV of EF-G with h34 may bias the energy landscape of the ribosome towards the TIPOST.
Fig. 3. EF-G stabilizes the swiveled head movement in the TIPOST state.
a, Comparison of the position of FA-stalled EF-G and the 30S subunit between TIPOST and the POST state 70S•EF-G•GDP•FA19. All shown components of the POST state 70S•EF-G•GDP•FA19 are depicted as blue ribbons. The 30S, EF-G and pe/E-tRNA of the TIPOST are represented by yellow, red and orange ribbons, respectively. b, c, Close-up on the decoding region and domain IV of EF-G in the same orientation as in (a). The surfaces of TIPRE (b) and TIPOST (c) are transparent with molecular models in ribbons representation (30S subunit, yellow; EF-G, red, P/E-tRNA, green and pe/E-tRNA, orange). The arrows mark the closed latch between h34 and the 530 region of TIPRE (b) and the interaction between h34 and domain IV of EF-G within TIPOST.
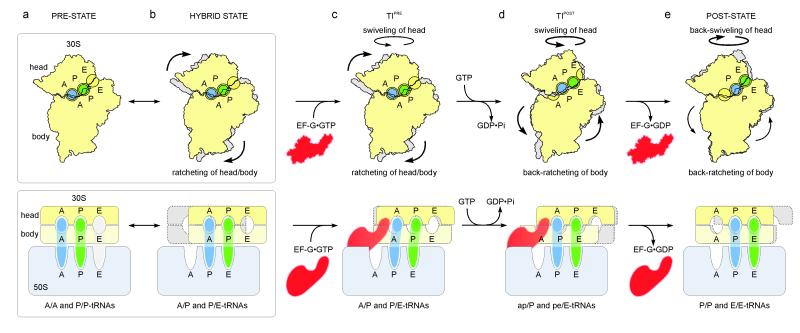
So far, intermediate states of inter-subunit rotation were considered to be intermediates on the pathway to the fully rotated state16. The present findings implicate un-ratcheting (in combination with the large swivel of the 30S head), rather than ratcheting, as being coupled to the translocation movement of the tRNAs and the mRNA with respect to the 30S subunit. Collectively, the insights gained from the structures of the TIPRE and TIPOST states enable us to provide a structural explanation for the process of translocation in a model where tRNA movements are facilitated by head-swivel, ratcheting and un-ratcheting motions of the ribosome (Fig. 4). These motions may be influenced by the GTPase reaction on EF-G as a network of interactions involving domain III and the ordered switch I region of EF-G, and the γ-phosphate of GTP was proposed to stabilize the rotated state of the 30S subunit 14. Accordingly, fast GTP hydrolysis by EF-G17 could destabilizes the direct and indirect interactions of the switch I of EF-G with the maximally rotated 30S subunit14,27, and therefore increases the propensity of the 30S subunit to rotate backwards. The un-ratcheting motion produces an on-coming movement of the body/platform, thereby reducing the distance that the tRNAs have to travel during translocation. Intra-subunit hybrid states allow the 30S subunit to maintain partial contacts with the tRNAs at any time of the translocation reaction. In the context of the ribosome functioning during translocation as a Brownian ratchet machine24, our model suggests that EF-G acts as a dynamic pawl, decoupling the un-ratcheting motions of the ribosome from the transition of hybrid state tRNAs back into classical states. Thereby, EF-G provides directionality and accelerates translocation of the tRNAs via several intermediate inter- and intra-subunit hybrid states into the classical P/P and E/E sites of the POST state.
Fig. 4. Model for translocation.
a, b, The PRE ribosome exists in a dynamic equilibrium between (a) base states with classical A/A- and P/P-tRNAs and (b) rotated states with hybrid A/P- and P/E-tRNAs6,7,9-11. c, Binding of EF-G•GTP to (a) PRE- or (b) hybrid-states stabilizes the ratcheted state12 as observed in the TIPRE. d, Fast GTP hydrolysis by EF-G17 accelerates translocation via an unlocking step on the 30S subunit18. Domain IV of EF-G uncouples un-ratcheting from the reverse movement of the A/P- and P/E-tRNAs back into classical states i.e. a doorstop function. Through head-swiveling and un-ratcheting motion, the tRNAs move from aa/P and pp/E into the 30S intra-subunit ap/P and pe/E hybrid states. e, Complete un-ratcheting of the 30S subunit leads to the POST state 70S•EF-G complex19. Back-swiveling of the 30S-head re-establishes tRNAs in the classical (pp/P) P- and E- (ee/E) states. Translocation is completed by dissociation of EF-G•GDP.
METHODS SUMMARY
Tight-couple 70S ribosomes from T. thermophilus were isolated by sucrose gradient centrifugation and incubated with EF-G in the presence of GTP and FA. Resulting complexes were flash frozen and imaged under low-dose conditions using an FEI – POLARA G2 electron microscope. The collected data were digitized and processed using multiparticle refinement protocols implemented in SPIDER25. In order to interpret the resulting cryo-EM maps in molecular terms, a newly developed algorithm (MDFIT)26 that integrates molecular simulation with experimental maps was employed.
METHODS
Formation of the EF-G•70S•GDP•FA complex
The fusA gene, encoding EF-G, was cloned from Thermus thermophilus HB8 genomic DNA into pET-46 Ek/LIC vector using primers (TthEFG_ for 5′ GCG CGC CCG GTG GTG ATG CAG CTC TTC CTG GGC TCC GCC CTG AAG AAC 3′; TthEFG_ rev 5′ GTT CTT CAG GGC GGA GCC CAG GAA GAG CTG CAT CAC CAC CGG GCG CGC 3′) according to manufacturer’s instructions (Novagen) and expressed in BL21 (DE3) cells. Recombinant EF-G protein was then purified using Ni-NTA affinity column, followed by gel filtration in a buffer containing 10 mM Tris pH 7.8, 100 mM NaCl, 10 mM β-mercaptoethanol. Tight-coupled 70S ribosomes were purified from log-phase T. thermophilus cells using sucrose density gradient centrifugation, as described previously for 30S subunits28. As observed previously, the ribosomes contained an co-purified tRNA14,29. Binding of EF-G to 70S ribosomes was done by incubating 20 μM of purified EF-G protein with 5 μM T. thermophilus 70S ribosomes, 500 μM GTP, 500 μM fusidic acid (FA), for 15 min at 65°C, in a buffer containing 10 mM HEPES-KOH (pH 7.8), 30 mM MgCl2, 75 mM NH4Cl. The occupancy of EF-G in the complexes was about 60-70 %, as judged by centrifugal binding assay28.
Cryo-EM and image processing
Ribosomal complexes were diluted to a concentration of 30 nM and subsequently frozen onto Quantifoil grids using a Vitrobot (FEI) device. Micrographs where collected on a Tecnai G2 Polara (FEI) at 300 kV and 39,000x magnification under low dose conditions (19 e−/Å2) and scanned on a D8200 Primscan drum scanner (Heidelberger Druckmaschinen) with a step size of 4.758 μm corresponding to 1.26 Å on the specimen scale.
The Contrast Transfer Function (CTF) defocus values for the micrographs were determined with CTFind30. Ribosomal projection images were automatically identified using the program Signature31 and were subsequently screened visually or automatically. From the selected projections, a reconstruction was generated by projection matching procedures and refined using the SPIDER software package32. The complete data set comprised 586,848 projection images collected from 677 micrographs at a 1.3 – 4.8 μm defocus range. During the later refinement rounds, positivity of the reference volumes was enforced, the power spectrum of the cryo-EM map was scaled to the power spectrum of a model density derived from the atomic coordinates of the X-ray structure of the 70S ribosome33 and the map was subsequently low-pass filtered according to the current resolution estimate.
After first phase of multiparticle refinement 20,25,34,35 which was performed with 3-times or 2-times decimated pictures, we obtained a major subpopulation (52%; 303,665) that had strong EF-G density (supp. Fig. S1). However, as refinement progressed and the resolution reached the sub-nanometer range the data set was deemed heterogeneous. Parts of the 30S subunit, especially the head domain, became partially disordered. Therefore, a second phase of multiparticle refinement was employed, leading to the subdivision of the data into two further sub-states having EF-G (supp. Fig. S1). Both data sub-sets were further refined individually at full image size. The final reconstructions of sub-state I (113,214 particle images) and sub-state II (156,332 particle images) reached 7.8 Å and 7.6 Å resolution, respectively (Supp. Fig. S2).
Using a similar strategy we revisited the previous data set (362,361 particle images; 371 micrographs) of the 70S•EF-G•GMPPNP complex14. In first phase of multiparticle refinement a major population (118,991 particle images) was further sorted by a second phase of multiparticle refinement resulting into substate I (58,911 particle images) and substate II (38,055 particle images). The resolution of the maps was 9.6 Å and 10.5 Å, respectively. As the resolution for the reconstructions of the 70S•EF-G•GMPPNP complex was significant lower (due to the smaller size dataset) than the resolutions obtained for the 70S•EF-G•GDP•FA complex, we restricted the comparison to a dissection of only the global conformational changes, such as ratcheting and head swiveling.
Nevertheless, this analysis suggests that the 70S•EF-G•GMPPNP complex co-exists in two sub-states that resemble TIPRE and TIPOST of the 70S•EF-G•GDP•FA complex (supp. Fig. S5; supp. Table 1). Interestingly, the ratio of particles within each of the two sub-states of the 70S•EF-G•GMPPNP complex is inverted with respect to 70S•EF-G•GDP•FA complex, i.e. in the 70S•EF-G•GMPPNP complex, the particle ratio of TIPRE and TIPOST is ~3:2 (58,911:38,055), whereas in the 70S•EF-G•GDP•FA complex, the ratio is ~2:3 (113,214:156,332). This means that the majority of ribosomes in the 70S•EF-G complex stalled using a non-hydrolyzable GTP analogue are in TIPRE (sub-state I), having a fully ratcheted 30S subunit, but only a modest head swivel. However, in the 70S•EF-G complex stalled using GTP and fusidic acid (FA), TIPOST (sub-state II) dominates and here an intermediate inter-subunit rotation is coupled with a large head swivel instead (supp. Fig. S4, S5; supp. Table 1).
Structure-based simulation fitting (MDFIT)
To determine atomic models that are consistent with the cryo-EM densities, we employed structure-based molecular simulation26,36,37 together with an energetic term developed by Orzechowski and Tama38, which incorporates the correlation between the simulated and experimental electron density throughout the simulation. Tama and co-workers developed a similar method, which used a standard explicit solvent force field, as opposed to the structure-based force field. The advantage of the structure-based force field is that, because the potential energy function is defined by the x-ray structure, MDFIT retains tertiary contacts present in the x-ray structure without special constraints. Furthermore, since MDFIT explicitly includes all non-hydrogen atoms, there are no atomic clashes and proper stereochemistry is maintained in all fits.
We began the MDFIT procedure with a structure-based potential energy function defined by the classical unratcheted conformation. To induce hybrid-state formation and subunit pivoting we introduced the energetic term based on the correlation between the simulated cryo-EM map and the experimentally determined cryo-EM map. Specifically, the potential energy function is:
| (1) |
where W is the energetic weight of the map, and are the normalized experimental and simulated electron densities at voxel (i,j,k), respectively. The quantity VSB is the structure-based potential energy function. To calculate the simulated map, each atom is described by a Gaussian function of width 5 Å with the tail truncated at 1 % of the peak value. Here, since the structure-based forcefield has 1 unit (all calculations were in reduced units) of stabilizing energy per atom (by construction), we set W to a comparable value of 150,000. The contributions to the force due to Vmap were updated every 200 timesteps. Fitting simulations employed Langevin dynamics. All simulations were performed using code based on Gromacs version 4.0.539,40. Calculations were performed on the Encanto Supercomputer. The structure-based force field is freely available online (smog.ucsd.edu).
Structural Models
The crystallographic structure of the 70S ribosome in complex with EF-G19 (PDB ID: 2WRI/J) was used as an initial structure for MDFIT. Proteins without side chains in the x-ray structure were removed. The C-terminal domain of ribosomal protein L7 from PDB entry 1RQU41 was inserted by hand, prior to fitting. The E-site tRNA was also removed from the initial x-ray structure in accordance with the cryo-EM map. The P-site tRNA was included in the fitting process. To facilitate fitting of the tRNA molecule into a P/E conformation, stabilizing interactions between the tRNA and the ribosome were removed. Stabilizing interactions between EF-G and the ribosome (due to their proximity in the crystal structure) were also excluded from the calculations, as were stabilizing interactions between L7 and all other components in the system. Further, crystallographic interactions found between the 3′-CCA end of the E-site tRNA with the E-site of the 50S subunit were re-introduced as short-ranged (of the type 6-12, see refs 26,36,37) attractive interactions between the 3′-CCA end of the fitted-tRNA and the 50S E-site. Introducing these interactions ensured that the 3′-CCA end of the P/E tRNA was in a conformation identical to a classically-bound E-site tRNA. Though, since these interactions were short-ranged, they only affected the process once the major rearrangements in the tRNA were already achieved. Codon-anti-codon interactions were restrained by harmonic interactions with minima corresponding to the classical configuration.
The tRNA-ribosome-EF-G crystal structure was first manually aligned in VMD42, as a single rigid unit, to the map of the sub-state I (TIPRE). The first round of fitting was performed using the sub-state I (TIPRE) map after subjecting it to a 4 Å Gaussian low-pass filter. This filter reduced noise, effectively smoothing the energetic profile associated with V map, which allowed for more rapid fits. After 106 integration steps, the fit was continued for an additional 106 steps with V map based on the sub-state I (TIPRE) map filtered at 2 Å. The sub-state II (TIPOST) map, filtered at 2 Å, was then fit using the sub-state I (TIPRE) 4 Å fitted structure as the initial structure.
Supplementary Material
Acknowledgments
The present work was supported by grants from the Deutsche Forschungsgemeinschaft DFG (SFB 740 TP A3 and TP Z1, SP 1130/2-1 to C.M.T.S., FU579 1-3 to P.F., HA 1672/7-5 to R.K.H. and WI3285/1-1 to D.N.W.), the European Union 3D-EM Network of Excellence (to CMTS), the European Union and Senatsverwaltung für Wissenschaft, Forschung und Kultur Berlin (UltraStructureNetwork, Anwenderzentrum) and US NIH Grant GM 60635 (to PAP), the Cluster of excellence “Macromolecular complexes” at the Goethe University Frankfurt (DFG Project EXC 115 to P.F. and S.C.), and the HFSP Young Investigators Award HFSP67/07 (to P.F.). We thank the New Mexico Computing Application Center for generous time on the Encanto Supercomputer. PCW is currently funded by a LANL Director’s Fellowship. This work was also supported by the Center for Theoretical Biological Physics sponsored by the NSF (Grant PHY-0822283) with additional support from NSF- MCB-0543906, the LANL LDRD program and NIH Grant R01-GM072686.
Footnotes
Full Methods and any associated references are available in the online version of the paper at www.nature.com/nature
Supplementary Information accompanies the paper on www.nature.com/nature.
Author Contributions A.M., A.L.S. and A.D. prepared the complexes. A.H.R. and T.M. collected the cryo-EM data. A.H.R, J.L., M.B., S.R.C. and C.M.T.S. did the image processing. P.C.W., Y.Y., J.O. and K.Y.S. developed and employed the MDFit method. P.W.H. participated in docking and analysed the FA binding site. A.H.R., R.K.H., S.R.C., P.F., P.A.P. D.N.W. and C.M.T.S. discussed the results and wrote the paper.
Author Information The electron density maps and models of the TIPRE and the TIPOST complexes have been deposited in the 3D-EM and PDB databases with the accession numbers EMD-1798 and EMD-1799, and PDBID 2xsi, 2sxl, 2sxm, and 2xsn.
Reprints and permissions information is available at www.nature.com/reprints.
The authors declare no competing financial interests.
REFERENCES
- 1.Frank J, Spahn CM. The ribosome and the mechanism of protein synthesis. Rep Prog Phys. 2006;69:1383. [Google Scholar]
- 2.Schmeing TM, Ramakrishnan V. What recent ribosome structures have revealed about the mechanism of translation. Nature. 2009;461:1234. doi: 10.1038/nature08403. [DOI] [PubMed] [Google Scholar]
- 3.Shoji S, Walker SE, Fredrick K. Ribosomal Translocation: One Step Closer to the Molecular Mechanism. ACS Chem Biol. 2009 doi: 10.1021/cb8002946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ramakrishnan V. Ribosome structure and the mechanism of translation. Cell. 2002;108:557. doi: 10.1016/s0092-8674(02)00619-0. [DOI] [PubMed] [Google Scholar]
- 5.Moazed D, Noller HF. Intermediate states in the movement of transfer RNA in the ribosome. Nature. 1989;342:142. doi: 10.1038/342142a0. [DOI] [PubMed] [Google Scholar]
- 6.Munro JB, Altman RB, O’Connor N, Blanchard SC. Identification of two distinct hybrid state intermediates on the ribosome. Mol Cell. 2007;25:505. doi: 10.1016/j.molcel.2007.01.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Blanchard SC, et al. tRNA dynamics on the ribosome during translation. Proc Natl Acad Sci U S A. 2004;101:12893. doi: 10.1073/pnas.0403884101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Munro JB, Sanbonmatsu KY, Spahn CM, Blanchard SC. Navigating the ribosome’s metastable energy landscape. Trends Biochem Sci. 2009;34:390. doi: 10.1016/j.tibs.2009.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Agirrezabala X, et al. Visualization of the hybrid state of tRNA binding promoted by spontaneous ratcheting of the ribosome. Mol Cell. 2008;32:190. doi: 10.1016/j.molcel.2008.10.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Julian P, et al. Structure of ratcheted ribosomes with tRNAs in hybrid states. Proc Natl Acad Sci U S A. 2008;105:16924. doi: 10.1073/pnas.0809587105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Fischer N, et al. Ribosome dynamics and tRNA movement by time-resolved electron cryomicroscopy. Nature. 2010;466:329. doi: 10.1038/nature09206. [DOI] [PubMed] [Google Scholar]
- 12.Valle M, et al. Locking and unlocking of ribosomal motions. Cell. 2003;114:123. doi: 10.1016/s0092-8674(03)00476-8. [DOI] [PubMed] [Google Scholar]
- 13.Frank J, Agrawal RK. A ratchet-like inter-subunit reorganization of the ribosome during translocation. Nature. 2000;406:318. doi: 10.1038/35018597. [DOI] [PubMed] [Google Scholar]
- 14.Connell SR, et al. Structural basis for interaction of the ribosome with the switch regions of GTP-bound elongation factors. Mol Cell. 2007;25:751. doi: 10.1016/j.molcel.2007.01.027. [DOI] [PubMed] [Google Scholar]
- 15.Spahn CM, et al. Domain movements of elongation factor eEF2 and the eukaryotic 80S ribosome facilitate tRNA translocation. EMBO J. 2004;23:1008. doi: 10.1038/sj.emboj.7600102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zhang W, Dunkle JA, Cate JH. Structures of the ribosome in intermediate states of ratcheting. Science. 2009;325:1014. doi: 10.1126/science.1175275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Rodnina M, Savelsbergh A, Katunin VI, Wintermeyer W. Hydrolysis of GTP by elongation factor G drives tRNA movement on the ribosome. Nature. 1997;385:37. doi: 10.1038/385037a0. [DOI] [PubMed] [Google Scholar]
- 18.Savelsbergh A, et al. An elongation factor G-induced ribosome rearrangement precedes tRNA-mRNA translocation. Mol Cell. 2003;11:1517. doi: 10.1016/s1097-2765(03)00230-2. [DOI] [PubMed] [Google Scholar]
- 19.Gao YG, et al. The structure of the ribosome with elongation factor G trapped in the posttranslocational state. Science. 2009;326:694. doi: 10.1126/science.1179709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Penczek PA, Frank J, Spahn CM. A method of focused classification, based on the bootstrap 3D variance analysis, and its application to EF-G-dependent translocation. J Struct Biol. 2006;154:184. doi: 10.1016/j.jsb.2005.12.013. [DOI] [PubMed] [Google Scholar]
- 21.Agrawal RK, et al. EF-G-dependent GTP hydrolysis induces translocation accompanied by large conformational changes in the 70S ribosome. Nature Struct.Biology. 1999;6:643. doi: 10.1038/10695. [DOI] [PubMed] [Google Scholar]
- 22.Scheres SH, et al. Disentangling conformational states of macromolecules in 3D-EM through likelihood optimization. Nat Methods. 2007;4:27. doi: 10.1038/nmeth992. [DOI] [PubMed] [Google Scholar]
- 23.Matassova AB, Rodnina MV, Wintermeyer W. Elongation factor G-induced structural change in helix 34 of 16S rRNA related to translocation on the ribosome. Rna. 2001;7:1879. [PMC free article] [PubMed] [Google Scholar]
- 24.Spirin AS. The ribosome as a conveying thermal ratchet machine. J Biol Chem. 2009;284:21103. doi: 10.1074/jbc.X109.001552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Schuette JC, et al. GTPase activation of elongation factor EF-Tu by the ribosome during decoding. EMBO J. 2009;28:755. doi: 10.1038/emboj.2009.26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Whitford PC, et al. Accommodation of aminoacyl-tRNA into the ribosome involves reversible excursions along multiple pathways. RNA. 2010 doi: 10.1261/rna.2035410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ticu C, et al. Conformational changes in switch I of EF-G drive its directional cycling on and off the ribosome. EMBO J. 2009;28:2053. doi: 10.1038/emboj.2009.169. [DOI] [PMC free article] [PubMed] [Google Scholar]
REFERENCES
- 28.Sharma MR, et al. Interaction of Era with the 30S ribosomal subunit implications for 30S subunit assembly. Mol Cell. 2005;18:319. doi: 10.1016/j.molcel.2005.03.028. [DOI] [PubMed] [Google Scholar]
- 29.Yusupov MM, et al. Crystal structure of the ribosome at 5.5 A resolution. Science. 2001;292:883. doi: 10.1126/science.1060089. [DOI] [PubMed] [Google Scholar]
- 30.Mindell JA, Grigorieff N. Accurate determination of local defocus and specimen tilt in electron microscopy. J Struct Biol. 2003;142:334. doi: 10.1016/s1047-8477(03)00069-8. [DOI] [PubMed] [Google Scholar]
- 31.Chen JZ, Grigorieff N. SIGNATURE: A single-particle selection system for molecular electron microscopy. J Struct Biol. 2006 doi: 10.1016/j.jsb.2006.06.001. [DOI] [PubMed] [Google Scholar]
- 32.Frank J, et al. SPIDER and WEB: processing and visualization of images in 3D electron microscopy and related fields. Journal of Structural Biology. 1996;116:190. doi: 10.1006/jsbi.1996.0030. [DOI] [PubMed] [Google Scholar]
- 33.Selmer M, et al. Structure of the 70S ribosome complexed with mRNA and tRNA. Science. 2006;313:1935. doi: 10.1126/science.1131127. [DOI] [PubMed] [Google Scholar]
- 34.Spahn CM, Penczek PA. Exploring conformational modes of macromolecular assemblies by multiparticle cryo-EM. Curr Opin Struct Biol. 2009;19:623. doi: 10.1016/j.sbi.2009.08.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Connell SR, et al. A new tRNA intermediate revealed on the ribosome during EF4-mediated back-translocation. Nat Struct Mol Biol. 2008 doi: 10.1038/nsmb.1469. [DOI] [PubMed] [Google Scholar]
- 36.Whitford PC, et al. An all-atom structure-based potential for proteins: bridging minimal models with all-atom empirical forcefields. Proteins. 2009;75:430. doi: 10.1002/prot.22253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Whitford PC, et al. Nonlocal helix formation is key to understanding S-adenosylmethionine-1 riboswitch function. Biophys J. 2009;96:L7. doi: 10.1016/j.bpj.2008.10.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Orzechowski M, Tama F. Flexible fitting of high-resolution x-ray structures into cryoelectron microscopy maps using biased molecular dynamics simulations. Biophys J. 2008;95:5692. doi: 10.1529/biophysj.108.139451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lindahl E, Hess B, van der Spoel DJ. GROMACS 3.0: A package for molecular simulation and trajectory analysis. J Mol Model. 2001;7:306. [Google Scholar]
- 40.Berendsen HJC, van der Spoel DJ, van Drunen R. GROMACS-a message-passing parallel molecular-dynamics implementation. Comput Phys Commun. 1995;4:435. [Google Scholar]
- 41.Bocharov EV, et al. From structure and dynamics of protein L7/L12 to molecular switching in ribosome. J Biol Chem. 2004;279:17697. doi: 10.1074/jbc.M313384200. [DOI] [PubMed] [Google Scholar]
- 42.Humphrey W, Dalke A, Schulten K. VMD: visual molecular dynamics. J Mol Graph. 1996;14:33. doi: 10.1016/0263-7855(96)00018-5. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.