Abstract
Exposures to sodium metam (NaM) within the developmental period of somitogenesis (10- to 18-h postfertilization [hpf]) results in easily detectable distortions of the notochord by 24 hpf in the developing zebrafish. We hypothesized that NaM-induced transcriptional changes during somitogenesis would reveal the major molecular targets in the zebrafish embryo. Embryos were exposed to NaM beginning at 4 hpf (1000 cells) and total RNA was isolated from embryos at the 3 somite (11 hpf), 10 somite (14 hpf), 18 somite (18 hpf), and larval (24 hpf) stages of development. Using the Affymetrix zebrafish gene array we observed relatively few mRNAs differentially regulated at least twofold at each time point (11 hpf, 101 genes; 14 hpf, 151; 18 hpf, 154; 24 hpf, 33). The transcriptional profiles reveal neurodevelopment and myogenesis as the two primary targets of NaM developmental exposure. Quantitative PCR of several muscle and neuronal genes confirmed the array response. We also followed the structural development of the peripheral nervous system under NaM exposure using antibodies against neuronal structural proteins. Although there was no change in the onset of antibody staining, profound alterations became apparent during the period in which the notochord becomes distorted (> 18 hpf). Motor neuron development observed with the Tg(NBT:MAPT-GFP)zc1 transgenic zebrafish and a primary motor neuron specific antibody showed similar timing in the structural alterations observed in these cell types. Further study of the interactions of dithiocarbamates with the regulatory elements of fast muscle development and neurodevelopment is warranted.
Keywords: developmental toxicity, dithiocarbamate, copper, pesticide
The dithiocarbamate (DTC) chemical class has many important uses as chemical precursors, effluent additives, agricultural pesticides and in experimental and clinical medicine ( WHO, 1998). Sodium metam (NaM) is unique to the DTC chemical class because under the appropriate conditions it readily hydrolyzes into methylisocyanate (MITC) and dissipates into the surrounding atmosphere ( U.S. EPA, 2005). In order to effectively utilize NaM as an agricultural pesticide, the pesticide label requires a firm understanding of many conditions across the entire application area including weather, wind and soil moisture content. Ground and surface water in areas of NaM use have the potential to receive concentrations which could pose a developmental risk to aquatic organisms ( U.S. EPA, 2005) as well as other unintended exposures.
The molecular toxicology of NaM is not well understood despite significant effort studying the mechanisms of neuropathy and immunotoxicity in adult mammals ( Pruett et al., 2006; Thompson et al., 2002). Other subclasses of DTCs have been suspected in chronic disease states and birth defects ( Calvert et al., 2007; Cory-Slechta et al., 2005; Garry, 2004; Garry et al., 2002a, b). DTCs, such as NaM, are classified as low in toxicity to humans and considered potential developmental toxicants ( U.S. EPA, 2005; WHO, 1998). Although little is known about the mechanisms of NaM developmental toxicity, there is a conserved developmental toxicity shared among the DTC class of compounds including NaM and the degradation products, MITC and carbon disulfide ( Tilton et al., 2006; Van Leeuwen et al., 1986) Furthermore, for reasons that are not clear from standard toxicity tests, aquatic organisms are more susceptible to acute and developmental DTC exposure relative to mammals ( U.S. EPA, 2005; WHO, 1998).
The distorted notochord phenotype of DTC developmental exposure in zebrafish coincided with altered expression of established mRNA markers for notochord and muscle development ( Haendel et al., 2004; Tilton et al., 2006). The notochord distortions were phenocopied with neocuproine, a copper specific chelating agent and some DTC-induced distortions were protected with the addition of copper ( Tilton et al., 2006). This was supported by the identification of DTCs in a zebrafish chemical genetic screen which was designed to identify novel compounds that modulate copper homeostasis ( Mendelsohn et al., 2006).
The conserved developmental period of somitogenesis (10–18 hpf, zebrafish and 20–40 days post fertilization, humans) was established as the only period in which DTC exposure resulted in a distorted notochord ( Haendel et al., 2004). This period of development is so named for the sweeping formation and development of the somites which carry many of the cellular precursors of the muscular and skeletal system and the accompanying peripheral neurons. Given the importance of somitogenesis in establishing a proper body form, there are numerous possible targets that include immediate and latent outcomes from toxicant exposure during this period of development. Utilizing AChE-deficient mutant zebrafish and neuronal channel blockers it was determined that neuromuscular function beginning at 18 h postfertilization (hpf) physically distorts and reveals this phenotype of exposure during somitogenesis ( Teraoka et al., 2005). However, it has not been determined whether neuronal structure or function is perturbed by DTC exposure.
The objective of the current study was to identify and investigate the major targets of NaM developmental exposure. To accomplish this we evaluated the transcriptional response in control and NaM-exposed embryos during incremental periods of development using the Affymetrix zebrafish array. Antibodies directed at surface proteins expressed on cells of the peripheral nervous system were utilized to monitor peripheral neuron development from 18 to 24 hpf. A primary motor neuron specific antibody and motor neuron specific transgenic animal allowed the isolated observation of the impacts of NaM on motor neuron development. From these data, the transcriptional signatures during somitogenesis are predictive of the ensuing outcomes which include the profound structural alterations to neurodevelopment.
MATERIALS AND METHODS
Zebrafish maintenance and collection of embryos.
Adult AB strain zebrafish (Danio rerio) and Tg(NBT:MAPT-GFP)zc1 (neural specific beta tubulin promoter) transgenics were raised and kept at standard laboratory conditions of 28°C on a 14-h light/10-h dark photoperiod ( Westerfield, 1995). Fish were maintained in reverse osmosis water supplemented with a commercially available salt solution (0.6% Instant Ocean, Atlanta, GA) at a pH and conductivity range of 6.8–7.0 and 450–520 μS, respectively. Embryos were collected from group spawns and staged as previously described ( Westerfield, 1995). All immunohistochemistry images were acquired from intact animals using Axiovision software, Axiocam HR (Zeiss, Thornwood, NY) mounted to a Zeiss Axiovert 200M motorized inverted microscope. All animal protocols were performed in accordance with Oregon State University Institutional Animal Care and Use Committee guidelines.
Embryonic exposures.
Embryos showing proper and sequential development in the first 3 hpf were selected for exposures and placed in Teflon sealed clear glass vials (25 ml capacity) when they reached 4 hpf. The concentration of NaM utilized in these studies was 1.0μM. NaM was prepared and stored as previously described (standard grade ≥ 98% purity, Chem Service, Inc., West Chester PA) ( Haendel et al., 2004). The NaM dose-response in the zebrafish embryo is well characterized and 1.0μM will reliably produce 100% notochord distortions in embryos by 24 hpf under these conditions. We have also established that there is no tank effect with NaM and no increased mortality in this system with 100 embryos/20 ml (data not shown). For the immunohistochemistry and transgenic exposures our standard protocol of 4–24 hpf at 1μM NaM beginning at 4 hpf with 10–20 embryos per vial for each control or treated vial until embryos were sampled and prepared for analysis. For the array experiments 100 embryos per exposure and developmental time-matched embryo controls were placed in individual treatment vials for each of the 4 developmental time points sampled (11, 14, 18, 24 hpf). Each treatment was then processed with its time-matched control from exposure though array hybridization. The pooling of embryos provided a population level response at each time point and was necessary to obtain enough total RNA for the array and subsequent real-time quantitative PCR (qPCR). Sampling of this population every 3–6 h provides a response across the developmental time span of somitogenesis. Further this captures for study, the period before, during and after the notochord distortions become visibly distorted. Animals were removed from their exposure vials at the appropriate time, washed, and 80 embryos were selected by counting the number of visible somites. The remaining 20 embryos were incubated in Petri dishes to confirm the presence of notochord distortions after 24 hpf (with the exception of 11 hpf which is not a sufficient period of time at this concentration to elicit notochord distortions).
Total RNA isolation.
The pools of embryos with their chorions intact were rinsed and placed in polypropylene tubes. The surrounding water was completely removed with a narrow glass pipette gently worked to the bottom. TRIzol Reagent (500 μl; Invitrogen, Carlsbad, CA) was placed on the embryos and they were quickly homogenized with a hand-held tissue tearer. An additional aliquot of TRIzol (Invitrogen) was added, mixed and the samples were snap frozen in liquid nitrogen followed by storage at −80°C. Total mRNA was isolated from whole emybro homogenates using TRIzol (Invitrogen) followed by cleanup with Rneasy Mini Kits (Qiagen, Valencia, CA) according to manufacturer's instructions. Total RNA quantity was determined using a Nanodrop spectrophotometer and 280/260 ratios were between 1.8 and 2.0. Total RNA provided to the Center for Gene Research and Biotechnology at Oregon State University for array processing was further analyzed using the Agilent 2100 Bioanalyzer (Santa Clara, CA). All total RNA samples tested were devoid of contamination and RNA degradation as measured by the ratio of 28S to 18S RNA peaks.
Affymetrix microarray.
Total RNA from each treatment and control was hybridized to a single array in two separate experiments (exp #1: 11/14/18/24 hpf; exp #2: 11/14 hpf duplicate) for a total of 12 arrays. Hybridizations were conducted on all time points, treatment and control, concurrently in each experiment to control for technical variation. Probe synthesis, hybridization and scanning were conducted by the CGRB using Affymetrix protocols (Santa Clara, CA). Total RNA (770 ng) was used to generate biotinylated complementary RNA (cRNA) for each treatment group using the One-Cycle Target Labeling kit (Affymetrix). Briefly, RNA was reverse transcribed using a T7-(dT)24 6primer and Superscript II reverse transcriptase (Invitrogen) and double stranded cDNA was synthesized and purified. Biotinylated cRNA was synthesized from the double stranded cDNA using T7 RNA polymerase and a biotin-conjugated pseudouridine containing nucleotide mixture provided in the IVT Labeling Kit (Affymetrix). Prior to hybridization, the cRNA was purified, fragmented and 10 μg from each experimental sample was hybridized to zebrafish genome arrays (Zebrafish430_2) according to the Affymetrix GeneChip Expression Analysis Technical Manual (701021Rev. 5). Affymetrix fluidics station 400 was used to wash the arrays and arrays were scanned with an Affymetrix scanner 3000. The Affymetrix eukaryotic hybridization control kit and Poly-A RNA control kit were used to ensure efficiency of hybridization and cDNA amplification. All cRNA for each experiment was synthesized at the same time. Each array image was visualized to discount artifacts, scratches or debris. The Affymetrix zebrafish genome array was designed to detect over 14,900 transcripts. Probe sets comprising 16 different oligonucleotides were designed to detect an individual transcript. Affymetrix .CEL files were imported into GeneSpring 7.1 software (AgilentTechnologies, Palo Alto, CA) and GC-RMA processed to correct for background signal across arrays. Each transcript was normalized to the median signal on the array and all arrays were normalized to the 50th percentile of signal. All statistical analysis of microarray data was conducted using GeneSpring 7.1 software. Significance was determined using the cross gene error model in GeneSpring7.1 for all arrays and all assumptions of variance resulting in a t-test (P < 0.05) identifying signals which were significantly different. Only genes at least 2-fold differentially expressed from control transcript levels from the first experiment were considered in the functional annotations. When comparing the duplicated time points at 11 and 14 hpf a 1.7-fold cut-off was assigned. Sequence similarity to known genes was determined by identifying the full length mRNA sequence for each zebrafish probe set by conducting a BLAST search of each Affymetrix probe set against Genbank (http://www.ncbi.nlm.nih.gov/BLAST/), TIGR (http://tigrblast.tigr.org/tgi/), and Sanger (http://www.sanger.ac.uk/Projects/D_rerio/) databases (Current as of 07.07.2007). The top blast hit (≥ ×10−12) was assigned to the Affymetrix probe for the functional annotations. Unknown transcripts with zebrafish gene consortium (zgc) numbers were also blasted into the expression database maintained by the Zebrafish Information Network.
Quantitative real-time PCR confirmation of array.
We chose to use real-time PCR as a technical confirmation of the array response for certain genes of interest identified in the array study. To do this, we generated gene specific primers using the Affymetrix probe ID sequence as a template using Oligo2 Primer Analysis Software (Cascade, CO). Primers were synthesized by MWG-Biotech (High Point, NC).PCR was conducted using the Opticon 3 real-time PCR detection system (MJ Research, Waltham, MA). We evaluated three independent biological replicates, two from the array experiments and a third from an independent exposure, so that the real-time qPCR assays had an N = 3. Prior to the creation of cDNA, total mRNA was DNase-treated with RQ1 DNase (Promega, Madison, WI) according to the manufacture's protocol. cDNA was prepared from 1 μg RNA per group using Superscript II (Life Technologies, Gaithersburg, MD) and oligo(dT) primers in a final 50 μl volume. Specifically, 1 μl of each cDNA pool was used for each PCR reaction in the presence of SYBR Green, using DyNAmo SYBR Green qPCR kit according to the manufacturer's instructions (Finnzymes, Espoo, Finland). All experimental samples were run in triplicate, unless noted, on the same plate as β-actin. C(t) values for control and NaM-exposed samples were taken by choosing a cross over point across all samples before calculating the relative difference of each sample. Samples were then normalized to the corresponding β-actin value and calculated as ratios of exposed compared with control samples. There was no significant treatment or experimental changes in β-actin C(t) values and was therefore an appropriate reference gene. Negative controls for each experiment consisted of RNA without reverse transcriptase and another without template. Agarose gel electrophoresis and thermal denaturation (melt curve analysis) were conducted to ensure formation of specific products. Primers used in this study are listed in supplemental tables.
Whole-mount neuronal labeling.
Monoclonal antibodies generated against acetylated tubulin (mouse anti-AT, 1:1000; Sigma) label most axons and major peripheral processes in the developing embryo. The zn-12 antibody (mouse anti-Zn12, 1:500; Developmental Studies Hybridoma Bank, University of Iowa) is a monoclonal antibody recognizing L2/HNK-1 tetrasaccharide which is associated with the early “primary” neurons of the zebrafish embryo and also a later stage marker for motor neurons in zebrafish ( Metcalfe et al., 1990). The znp-1 monoclonal antibody (mouse anti-zn-1, 1:500; Developmental Studies Hybridoma Bank, University of Iowa) recognizes the neuropil region and primary motor neuron axons. Animals were fixed overnight in 4% paraformaldehyde in phosphate-buffered saline (PBS) and washed in PBS + 0.1% Tween 20 (PBST). The larvae were permeabilized with 0.005% trypsin (4°C) in PBS on ice for 5 min, rinsed in PBST and postfixed in 4% paraformaldehyde. Permeabilized larvae were blocked in 10% normal goat serum in PBS + 0.5% Triton X-100 for an hour at 22°C and incubated with the primary antibody overnight at 4°C in 1% normal goat serum-PBS + 0.5% Triton X-100. After four 30-min washes in PBST, the larvae were incubated with a secondary antibody (1:1000 Alexa-546 conjugated goat anti-mouse; Invitrogen) for 5 h at 22°C. The larvae were then washed four times for 30 min in PBST and visualized by fluorescence microscopy. All animals were examined and representative pictures were taken. The experiment was repeated for all the developmental time points. Z-stacks (12–30 stacks) were acquired at varied depth increments (10–20 micron) to capture the entire breadth of the embryo.
RESULTS
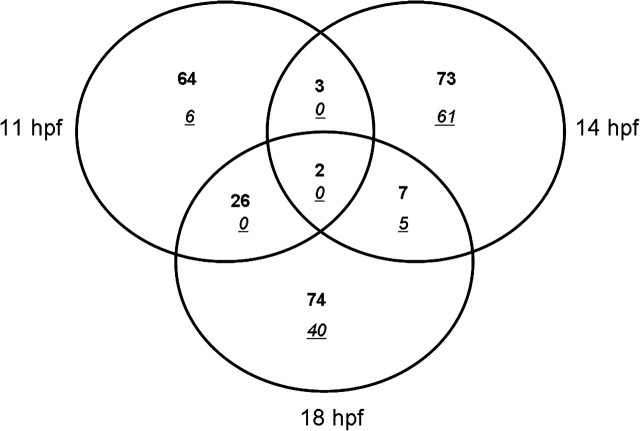
There were fewer than 154 elements differentially regulated greater than 2-fold at any time point (11, 14, 18 hpf) in the first study ( Fig. 1). The majority of the responses during somitogenesis resulted in an increase in transcription relative to the time-matched controls, particularly at 11 hpf in the first study. Only two genes were shared between 11, 14, and 18 hpf; titin that encodes a muscle fiber protein, and an unknown gene. We duplicated the finite response at the 11 and 14 hpf time points in a second study (≤ 84 altered transcripts) but chose to focus most of our attention to the first study (Supplemental Data). Despite the limited number of genes, we were able to observe similar patterns of muscle and neuronal impairment between the studies. For example at 14 hpf, 3 of the 20 shared gene elements were found to be > 1.7-fold (p < 0.05) using a t-test. The two identifiable elements corresponded to the genes, titin Danio rerio (Genbank #AY081167.1) (study 1: + 3.8-fold change; study 2: + 4.2-fold change) and titin, cardiac muscle(Homo sapiens) (study 1: < 2.0-fold; study 2: + 1.9-fold change) (Genbank BI878949) (p = 0.047 and p = 0.039, respectively) between the two studies. Although the small total number of elements differentially regulated during developmental exposure to NaM limits the statistical and pathway analysis tools, we were able to discern the major developmental targets of NaM developmental exposure and exploit this signature to inform our subsequent studies.
FIG. 1.
Venn diagram of twofold differentially regulated genes (+, bold; −, italic and underlined) in study 1 and shared gene elements between 11, 14, 18 hpf from zebrafish embryos exposed to NaM beginning at 4 hpf. Only two elements were shared among all three time points, center, and 3–26 elements shared between any two time points.
Almost immediately apparent from the gene lists was the number of misregulated muscle related transcripts at each time point ( Table 1). The differential regulation of genes encoding fast muscle fibers across the three early time points was noteworthy and suggests target specificity. Several novel and known zebrafish titins were consistently upregulated across all time points. Myosin light and heavy chains are initially down regulated at 14 hpf; however, by 18 hpf the expression returns to normal or is elevated in a pattern consistent with the other muscle related transcripts. This reflects either an overall increase in the amount of fast muscle expression or a shift in the tightly controlled and sequential process of vertebrate muscle development.
TABLE 1.
Genes Related to Myogenesis Differentially Regulated by NaM Exposure during Somitogenesis(Bold Values—Shared Elements between Time Points)
| Affymetrix ID | Fold change | GenBank ID | Annotation | |
| 11 hpf | Dr.1662.1.S1_at | 3.6 | AI601291 | Danio rerio similar to titin isoform N2-B |
| Dr.14676.1.A1_at | 2.6 | BM072351 | Homo sapiens myosin X | |
| Dr.20990.2.S1_at | 3.2 | AY081167.1 | D. rerio titin | |
| Dr.4681.1.A1_at | 2.4 | BI878949 | Mus musculus Ttn protein | |
| Dr.17854.1.S1_at | 2.3 | AL716966 | D. rerio UNC45-related protein | |
| 14 hpf | Dr.20990.2.S1_at | 3.8 (p = 0.046) | AY081167.1 | D. rerio titin |
| Dr.23877.2.S1_at | 2.4 | AL727618 | D. rerio capping protein (actin filament) muscle Z-line, beta | |
| Dr.6099.1.S1_at | 2.1 | CD283621 | Unknown, GO molecular function—actin binding | |
| Dr.344.1.S1_at | −2.5 | BM957948 | D. rerio myosin, light polypeptide 9, regulatory | |
| Dr.4812.1.S1_s_at | −2.7 | NM_152982.1 | D. rerio myosin, heavy polypeptide 2, fast muscle specific (myhz2) | |
| 18 hpf | Dr.4681.1.A1_at | 5.4 | BI878949 | M. musculus Ttn protein |
| Dr.20990.2.S1_at | 4.0 | AY081167.1 | D. rerio titin | |
| Dr.1662.1.S1_at | 3.9 | AI601291 | D. rerio similar to titin isoform N2-B | |
| Dr.10620.1.S1_at | 3.0 | NM_181653.1 | D. rerio fast muscle troponin T isoform TnnT3b | |
| Dr.23405.1.S1_at | 2.9 | BM403462 | Cyprinus carpio myosin heavy chain, fast skeletal muscle | |
| Dr.4723.1.A1_a_at | 2.2 | AI641664 | Predicted: H. sapiens sarcalumenin (SRL) | |
| Dr.17871.2.S1_at | 2.2 | AL723844 | Bos taurus mRNA for nonmuscle myosin heavy chain B | |
| Dr.11242.1.A1_at | 2.1 | BG306206 | Predicted: D. rerio a gene for a novel troponin I | |
| Dr.4723.1.A1_x_at | 2.0 | AI641664 | Predicted: H. sapiens sarcalumenin (SRL) | |
| Dr.5224.1.A1_at | 2.0 | AW116068 | D. rerio similar to Nebulin | |
| Dr.26411.2.S1_s_at | −2.5 | BQ783571 | D. rerio fast muscle troponin I | |
| Dr.18576.1.S1_at | −3.2 | AY113680.1 | D. rerio Arp3 mRNA | |
| Dr.7952.1.S1_at | −3.2 | BC045421.1 | D. rerio pyruvate kinase, muscle |
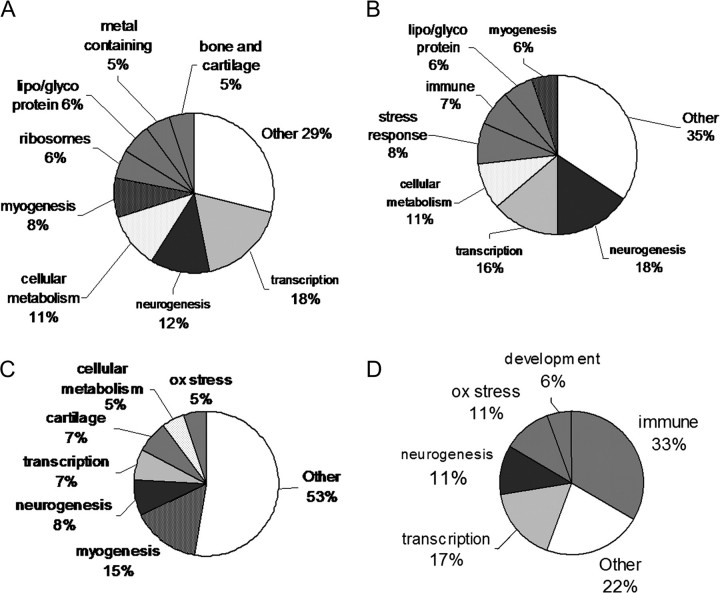
Functional grouping of the differentially regulated transcripts from the first study revealed genes important in myogenesis, neurogenesis, transcription, and cellular metabolism ( Figs. 2A–C, Supplemental Data). Functional groupings representing < 5% of the annotated gene lists were combined and graphed as “other.” Surprisingly, at 24 hpf there were only 33 elements differentially regulated (supplemental data). The response at 24 hpf, albeit small, appears distinct from the earlier time points and perhaps shares more similarity to the transcriptional responses that would be predicted from exposure to the juvenile or adult animal ( Fig. 2D). Considering the notochord is clearly distorted by 24 hpf and revealed beginning at > 18 hpf with the onset of spontaneous muscle contractions, this later time point will not be discussed further.
FIG. 2.
Functional groupings of annotations of the four developmental time points under study (A) 11 hpf, (B) 14 hpf, (C) 18 hpf, (D) 24 hpf.
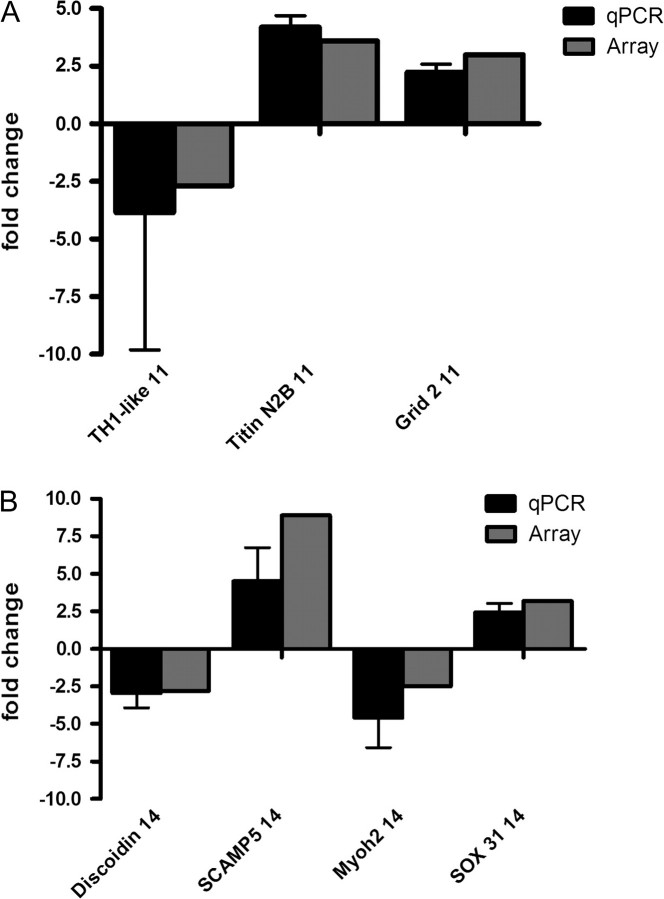
We utilized the transcriptional profiles to obtain a broad picture of the genomic responses within the whole embryo from DTC developmental exposure ( Figs. 1 and 2). The transcriptional responses observed from the arrays were confirmed by real-time qPCR using primers designed to target 4 and 12.5% of the total differentially regulated genes (> 1.7-fold) from the two array experiments, respectively ( Fig. 3). The primers were selected to represent the transcriptional profiles in the functional groupings discussed and presented in Figure 2. We confirmed the array response for the muscle transcript, titin N2B, at 11 hpf, as well as the disruption of the normal sequential and interdependent timing of myogenesis at 14 hpf with the down regulation of myosin heavy 2 (Myoh2) ( Fig. 3). Given the thiol reactive potential of DTCs, we were interested in confirming the up regulation of glutathione S-transferase pi at 14 hpf ( Fig. 3) as well as at 24 hpf (+ 2.1-fold array/+2.6-fold qPCR) and the intervening 18 hpf (+1.1-fold array/+1.7-fold qPCR). Sizzled expression, a gene involved in dorsal-ventral patterning of the body axis and WNT and BMP pathways, was differentially regulated at 24 hpf + 2.9-fold (+ 2.1-fold qPCR).
FIG. 3.
Real-time qPCR of representative genes of interest from NaM-exposed embryos compared with the primary array experimental data for these genes. Samples from 11 hpf (A) and 14 hpf (B) total RNA were run in triplicate (*duplicate) from pools of the two array experiments and a third independent replicate.
Of particular interest was the consistent identification of a number of transcripts differentially regulated at every time point related to neuronal function and development ( Fig. 2). Within this functional group no recognizable pattern could be identified save perhaps a perturbation in synaptic function. One example is secretory carrier membrane protein (SCAMP)-5 which was upregulated in response to NaM at 14 hpf indicating a potential adverse impact to the proper maturation and function of the neuronal synapse ( Fig. 3). The array response for Tryptophan hydroxylase 1, like (TH1-like, 11 hpf), ionotropic glutamate receptor delta 2 (Grid 2, 11 hpf), and SRY-box containing gene 31 (Sox 31, 14 hpf), associated with the proper function and development of the spinal interneuron and central nervous system was also confirmed with qPCR. Using the available databases several unknown differentially regulated transcripts (using zgc annotations) were identified as having possible restricted expression in the brain and spinal chord (ZFIN.org) ( Thisse et al., 2001).
With evidence for impacts to neurodevelopment during somitogenesis we were led to investigate the structural development of the peripheral nervous system before, during, and after the notochord distortion. Using whole-mount immunohistochemistry in fixed embryos and transgenic live animals exposed to NaM, the structural development of peripheral, motor, and early “primary” neurons in control and NaM-exposed embryos could be easily followed after 18 hpf. The ZN-12 antibody, an early marker of primary neurons (and later motor neurons) showed no differences at 18.5 hpf between control and NaM-exposed embryos (A, B, Fig. 4). The timing of the first appearance of labeled cells was unchanged by NaM exposure indicating no significant developmental delay of nervous system development. Furthermore, the structural connections of neurons between the peripheral nervous system and brain show no distinctive structural or developmental defects. This is an important observation considering the muscle contractions which reveal the distortion are spinal-derived reflex contractions which require no brain input.
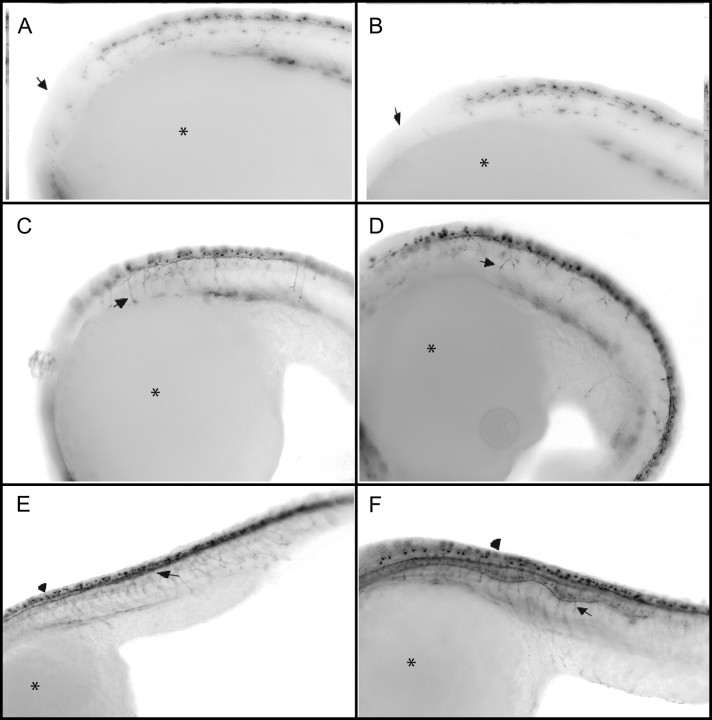
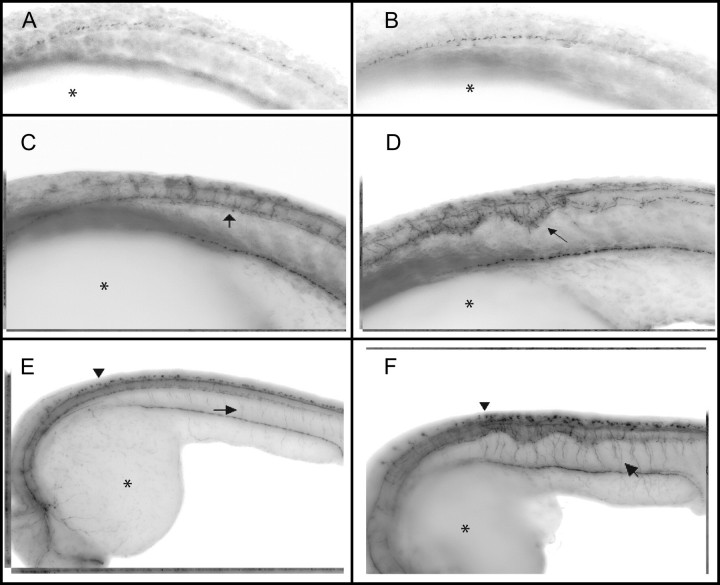
FIG. 4.
Whole-mount immunohistochemistry using the Zn-12 antibody recognizing L2/HNK-1 tetrasaccharide associated with the “primary” neurons in development and later motor neurons. Images are of inverted fluorescent pictures with the embryo wrapping around the yolk (*) with the head to the left. (A) (10×) control animal and (B) (10×) NaM-exposed embryos at 18.5 hpf shows no differences in the first appearance or in the development of the peripheral and central nervous system connections (arrow). In the same orientation (C) (8.5×) control and (D) (8.5×) NaM-exposed embryos at 21 hpf. The prominently stained cell bodies of the Rohon-Beard cells are aligned dorsally to the developing spinal chord. The distorted notochord is apparent by 21 hpf in NaM-exposed embryos (D) and primary neuronal tracks are more highly branched compared with controls C (arrows). (E) (8.5×) Control and (F) (8.5×) NaM-exposed embryos at 24 hpf. The spinal interneuron tracks are clearly distorted in the exposed embryo, however the increased area of staining may reflect the physical spreading of adjacent spinal cord area caused by the notochord distortion and not increases in the number of neurons (E, F—arrow).
By 21 hpf, a clear distortion of the notochord is evident resulting in a noticeable distortion of the “primary” neuronal networks. In the NaM-exposed animals the “primary” neurons appear to extend down in a regular repeating pattern comparable to controls and innervate the myotome (C, D Fig. 4). In NaM-exposed animals there were consistent abnormal neuronal branching which coincided with the magnitude and location of the distorted notochord. By 24 hpf, the normally tightly packed and straight spinal tracts in the animals are deformed along with the notochord in the NaM-exposed animals (E, F, Fig. 4).
The labeling of acetylated alpha tubulin (AAT) in the peripheral nervous system also did not reveal disruptions of the normal cell labeling at 18 hpf( Figs. 5A and 5B). At 19.5 hpf the distortions becomes apparent and AAT labeling shows a clear change in cellular structure and intensified staining in the ventral tract of the spinal neurons in NaM-exposed animals ( Figs. 5C and 5D). This became even more evident by 24 hpf ( Figs. 5E and 5F). Further, the motor neurons appear to reach into the mesoderm to innervate the developing muscle. This can also be seen in Figures 4E and 4F as these antibodies both label motor neurons at 24 hpf. Rohon-Beard cells show some possible irregular patterns in exposed embryos (arrowheads, Figs. 4E and 4F; Figs. 5E and 5F) although there was no obvious change to the number of these cells.
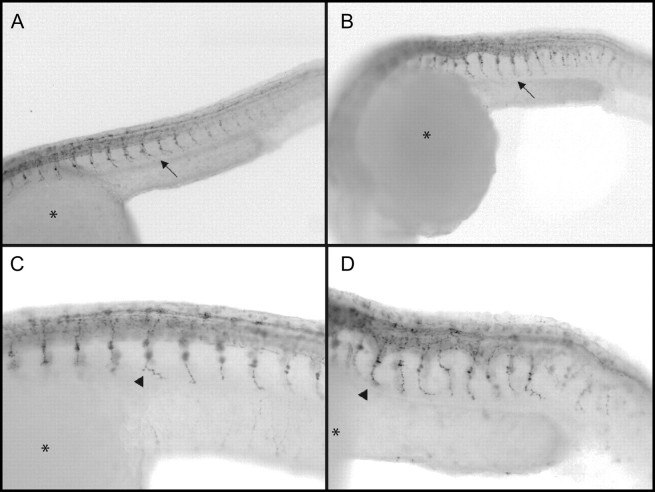
FIG. 5.
Whole-mount immunohistochemistry showing the labeling acetylated alpha tubulin (AAT) in the peripheral nervous system of control and exposed embryos at 18, 19.5, and 24 hpf. Images are of inverted fluorescent pictures with the embryo wrapping around the yolk (*) with the head to the left. Single plain images of control (A) and (B) 1.0μM NaM-exposed embryos at 18 hpf show no change in the first appearance of staining at 18 hpf. By 19.5 hpf the spontaneous muscle contractions have begun and Z-stack images comparing the controls (C) to 1.0μM NaM-exposed embryos (D) there is a clear change in cellular structure and intensified staining in the ventral tract of the spinal neurons of exposed animals (arrows). The motor neurons appear to innervate muscle properly in both control (E) and 1.0μM NaM-exposed embryos at 24 hpf (F), arrows. There was also clusters and possibly more Rohon-Beard cells in the 24 hpf exposed (F) compared with controls (E), arrowhead. *Yolk.
Using the Znp-1 antibody to label primary motor neurons, it is clear these cells reach the developing muscle along largely normal tracts despite the distorted notochord ( Figs. 6A and 6B). Upon closer examination these cells are irregular and slightly malformed, indicating the possible involvement of a structural impairment in the appearance of the distorted notochord > 18 hpf ( Figs. 6C and 6D). Using Tg(NBT:MAPT-GFP)zc1 transgenic animals to label both primary and secondary motor neurons it was evident, as observed with Znp-1, that the primary neurons reach the muscle and cellular irregularities exist in the neuronal cell types ( Figs. 7A and 7B). However, under higher magnification the secondary motor neurons in areas of distorted notochord do not reach the developing muscle and exhibit stunted neuronal tracts and irregular branching compared with controls ( Figs. 7C and 7D). Although there are clear transcriptional indications of neuronal impairment during somitogenesis the ramifications on neuronal structure do not become visibly apparent until the initiation of the muscle contractions which reveal the notochord distortion.
FIG. 6.
ZNP-1 monoclonal antibody recognizing the neuropil region and primary motor neurons of control and 1μM NaM-exposed embryos at 24 hpf. Single plain images are of inverted fluorescent pictures with the embryo wrapping around the yolk (*) with the head to the left. Controls (A) and exposed embryos (B) at 5×. The primary motor neurons which form prior to the notochord distortion appear to reach their intended muscular targets, arrows. However, upon closer examination at 10x there are clear cellular irregularities in the axon and cell bodies of these cell types in NaM-exposed animals (D) relative to controls at 10× (C), arrowheads.
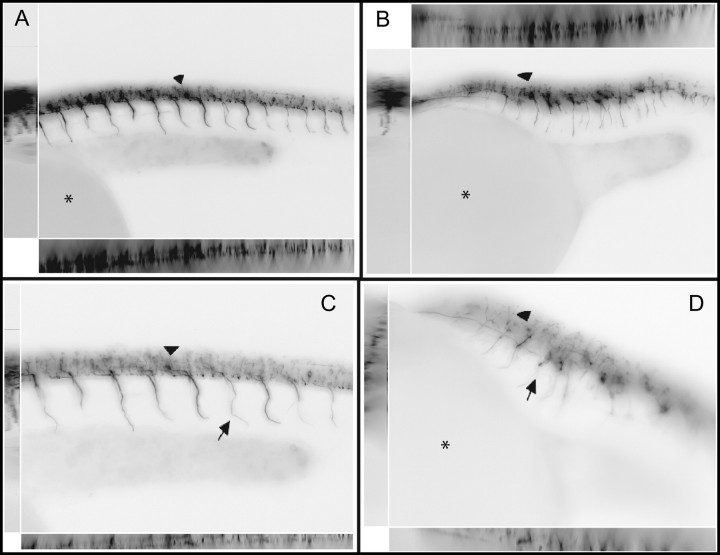
FIG. 7.
Tg(NBT:MAPT-GFP)zc1 transgenic animals with neuronal fluorescence in motor neurons exposed to NaM at 24 hpf. Images are of inverted fluorescent pictures with the embryo wrapping around the yolk (*) with the head to the left. Controls at 8.5× (A) and Tg(NBT:MAPT-GFP)zc1 embryos exposed to NaM from 4 to 24 hpf at 8.5× (B). Controls (C) and NaM exposed embryos (D) at 10×. (D). The primary motor neurons which form prior to the notochord distortion appear to reach their intended muscular targets, arrows. Secondary motor neurons which form during and after the notochord distortion have tracts and cell bodies which are clearly perturbed and stunted, in the exposed embryos, arrowheads.
DISCUSSION
The transcriptional profiles observed in this study suggest that DTCs target the somites during the developmental period of somitogenesis. The cellular fates of the somites make up the precursors of the eventual muscular-skeletal system in adult vertebrates. The normally tightly controlled sequence of vertebrate muscle development was clearly impaired by NaM exposure, appearing to specifically target fast muscle development ( Table 1). There is an interdependence of muscle and neuronal development that is under investigation by many groups ( Drapeau et al., 2002; Eisen and Melancon, 2001). Myogenesis genes are expressed as early as 10 hpf in the zebrafish and, when normalized to developmental stage, compare favorably with the patterns in mouse ( Xu et al., 2000a). A gradient of muscle gene expression closely follows the wave of somitogenesis in an anterior to posterior direction which is preceded and reinforced by several muscle regulatory factors (MRFs) ( Hsiao et al., 2003; Xu et al., 2000a).
Both known and suspected MRFs involved in the determination and differentiation of muscle cells were identified in the gene lists or by utilizing the available expression databases (Zfin) ( Hsiao et al., 2003; Xu et al., 2000b). In the larval animal we presented evidence for the alteration of one of many MRFs (i.e., MyoD), whose orchestrated expression patterns control proper muscle development ( Haendel et al., 2004). Therefore, it was not surprising that muscle transcriptional impairment was observed following NaM exposure during somitogenesis in the whole embryo ( Fig. 2). Although different in etiology and mechanism, exposure to toxicants such as cadmium and fipronil also impact this delicate relationship ( Chow and Cheng, 2003; Stehr et al., 2006). These data reveal that NaM targets fast muscle development and suggests MRFs are as an important molecular target. Regardless, myogenesis as a target of developmental toxicants is in need of more detailed molecular studies.
The consistent identification of a number of transcripts in the array gene profile related to neuronal function was confirmed by monitoring the expression of several of these genes of interest by qPCR ( Figs. 2, 3). For example, NaM led to increased expression of SCAMP-5 ( Figs. 2, 3) which is highly enriched in synaptic vesicles ( Fernandez-Chacon and Sudhof, 2000). Confirmation of the disruption of transcription of neuronal genes with known localized developmental expression in the central nervous system (CNS) and/or spinal interneurons (e.g., Sox31, Grid 2, and TH1) suggest potential for DTCs to cause neurobehavioral impairments ( Girard et al., 2001; Katayama et al., 2004; Teraoka et al., 2004). Although the mechanisms of DTC developmental neurotoxicity need to be explored in greater detail; DTCs impair the proper maturation and function of the nervous system throughout the embryo. Other pesticides exhibiting developmental neurotoxicity include chlorpyrifos and dichlorodiphenyldichloroethylene, also impair neuronal development and function in both the laboratory and in epidemiological studies ( Jameson et al., 2006; Torres-Sanchez et al., 2007). For these reasons the developing nervous system has been the focus of both toxicologists and developmental biologists for some time ( Downes and Granato, 2004; U.S. EPA, 2001). Further study of DTCs during zebrafish development will aid in the identification of relevant indicators of both toxicant exposure and effect.
Teraoka et al. (2005) reported the abolishment of peripheral neuronal function by pharmacological agents and neuromuscular mutants prevented the appearance of the distorted notochord in DTC exposed embryos. However, this study did not determine whether aberrant neuronal structure or function was an underlying cause. Using whole-mount immunohistochemistry in fixed and transgenic live animals exposed to NaM, we followed the structural development of peripheral, motor, and early “primary” neurons in control and exposed embryos after 18 hpf ( Figs. 4, 5, 6). This provided complementary and overlapping observations linking early developmental transcriptional responses to later structural impairment. It is now clear that structural impairment to the peripheral nervous system by NaM became evident as the notochord becomes distorted. The timing of the first appearance of labeled cells was unchanged with treatment indicating there is no discernable developmental delay of nervous system development. Immunohistochemical results also did not reveal improper or ill-timed connections between the CNS and peripheral neurons. Although the cellular patterning was impaired, there was no obvious change in the number of spinal interneurons, Rohon-Beard cells or motor neurons. From these data it is logical to propose that the misregulated transcripts within the CNS and peripheral nervous system could manifest as latent behavioral effects, possibly unrelated to the notochord distortions.
We utilized more specific labeling techniques to explore the observations of motor neuron development made with both zn12 and AAT staining ( Figs. 4E and 4F, Figs. 5E and 5F). The primary motor neuron specific antibody Zn-1 and a motor neuron specific transgenic animal Tg(NBT:MAPT-GFP)zc1 confirmed that NaM exposure did not inhibit the primary motor neurons from forming and innervating the developing muscle tissue along mostly normal tracts. There were, however, clear cellular abnormalities ( Figs. 6, 7) in need of further study. Secondary motor neurons which form during and after the notochord distortions, were clearly impacted during and after the distortion. It remains to be defined whether improper secondary motor neuron structural development plays a direct role in body axis deformation or is simply a secondary consequence.
We previously demonstrated the copper dependence and efficacy of the DTC-induced distorted notochord phenotype ( Tilton et al., 2006). Another laboratory using zebrafish in a genetic screen to identify small molecules which perturb copper homeostasis reported the DTC-distorted notochord phenotype in some of the small molecules identified ( Mendelsohn et al., 2006). Within the list of these small molecules were several DTCs and the copper chelator neocuproine. Injection of ATP7a, a copper transport protein, into mutant and neocuproine exposed animals prevented the development of the notochord distortion. Given the neurodevelopmental impacts reported in this study it is interesting that ATP7a has recently been implicated in improper synaptogenesis and Meake's disease ( Niciu et al., 2006, 2007).
DTCs present a complex class of compounds which have broad potencies and diverse biotransformation and environmental degradation pathways. We chose to utilize the notochord distortion as a phenotypic anchor due to its obvious nature, sensitivity and impact to the developing vertebrate in order to study the mechanisms of DTC developmental toxicity. Control of early muscle and neuronal development are clearly major targets for NaM during somitogenesis resulting in the structural and functional deficits which become apparent with the distorted notochord. Further studies will need to determine the nonteratogenic effects of DTCs. Clearly, the role of copper in the mechanism of action of DTC developmental neuromuscular toxicity is in need of further molecular study.
SUPPLEMENTAL DATA
Supplementary data are available online at http://toxsci.oxfordjournals.org/.
Acknowledgments
We wish to acknowledge Jane LaDue for animal husbandry and maintenance and the rest of the Tanguay laboratory for technical and editorial support.
References
- Calvert GM, Alarcon WA, Chelminski A, Crowley MS, Barrett R, Correa A, Higgins S, Leon HL, Correia J, Becker A, et al. Case report: Three farmworkers who gave birth to infants with birth defects closely grouped in time and place—Florida and North Carolina, 2004-2005. Environ. Health Perspect. 2007;115:787–791. doi: 10.1289/ehp.9647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chow ESH, Cheng SH. Cadmium affects muscle type development and axon growth in zebrafish embryonic somitogenesis. Toxicol. Sci. 2003;73:149–159. doi: 10.1093/toxsci/kfg046. [DOI] [PubMed] [Google Scholar]
- Cory-Slechta DA, Thiruchelvam M, Barlow BK, Richfield EK. Developmental pesticide models of the Parkinson's disease phenotype. Environ. Health Perspect. 2005 doi: 10.1289/ehp.7570. doi: 10.1289/ehp.7570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Downes G, Granato M. Acetylcholinesterase function is dispensable for sensory neurite growth but is critical for neuromuscular synapse stability. Dev. Biol. 2004;270:232–245. doi: 10.1016/j.ydbio.2004.02.027. [DOI] [PubMed] [Google Scholar]
- Drapeau P, Saint-Amant L, Buss RR, Chong M, McDearmid JR, Brustein E. Development of the locomotor network in zebrafish. Prog. Neurobiol. 2002;68:85–111. doi: 10.1016/s0301-0082(02)00075-8. [DOI] [PubMed] [Google Scholar]
- Eisen JS, Melancon E. Interactions with identified muscle cells break motoneuron equivalence in embryonic zebrafish. Nat. Neurosci. 2001;4:1065–1070. doi: 10.1038/nn742. [DOI] [PubMed] [Google Scholar]
- Fernandez-Chacon R, Sudhof TC. Novel SCAMPs lacking NPF repeats: Ubiquitous and synaptic vesicle-specific forms implicate SCAMPs in multiple membrane-trafficking functions. J. Neurosci. 2000;20:7941–7950. doi: 10.1523/JNEUROSCI.20-21-07941.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garry VF. Pesticides and children. Toxicol. Appl. Pharmacol. 2004;198:152–163. doi: 10.1016/j.taap.2003.11.027. [DOI] [PubMed] [Google Scholar]
- Garry VF, Harkins ME, Erickson LL, Long-Simpson LK, Holland SE, Burroughs BL. Birth defects, season of conception, and sex of children birth to pesticide applicators living the Red River Valley of Minnesota, USA. Environ. Health Perspect. 2002b;110(Suppl. 3):441–449. doi: 10.1289/ehp.02110s3441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garry VF, Harkins M, Lyubimov A, Erickson L, Long L. Reproductive outcomes in the women of the Red River Valley of the north. I. The spouses of pesticide applicators: Pregnancy loss, age at menarche, and exposures to pesticides. J. Toxicol. Environ. Health A. 2002a;65 doi: 10.1080/00984100290071333. [DOI] [PubMed] [Google Scholar]
- Girard F, Cremazy F, Berta P, Renucci A. Expression pattern of the Sox31 gene during zebrafish embryonic development. Mech. Dev. 2001;100:71–73. doi: 10.1016/s0925-4773(00)00491-3. [DOI] [PubMed] [Google Scholar]
- Haendel MA, Tilton F, Bailey GS, Tanguay RL. Developmental toxicity of the dithiocarbamate pesticide sodium metam in zebrafish. Toxicol. Sci. 2004;81:390–400. doi: 10.1093/toxsci/kfh202. [DOI] [PubMed] [Google Scholar]
- Hsiao CD, Tsai WY, Horng LS, Tsai HJ. Molecular structure and developmental expression of three muscle-type troponin T genes in zebrafish. Dev. Dyn. 2003;227:266–279. doi: 10.1002/dvdy.10305. [DOI] [PubMed] [Google Scholar]
- Jameson RR, Seidler FJ, Qiao D, Slotkin TA. Chlorpyrifos affects phenotypic outcomes in a model of mammalian neurodevelopment: Critical stages targeting differentiation in PC12 cells. Environ. Health Perspect. 2006;114:667–672. doi: 10.1289/ehp.8750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katayama T, Imaizumi K, Yoneda T, Taniguchi M, Honda A, Manabe T, Hitomi J, Oono K, Baba K, Miyata S, et al. Role of ARF4L in recycling between endosomes and the plasma membrane. Cell Mol. Neurobiol. 2004;24:137–147. doi: 10.1023/B:CEMN.0000012719.12015.ec. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mendelsohn BA, Yin C, Johnson SL, Wilm TP, Solnica-Krezel L, Gitlin JD. Atp7a determines a hierarchy of copper metabolism essential for notochord development. Cell. Metab. 2006;4:155–162. doi: 10.1016/j.cmet.2006.05.001. [DOI] [PubMed] [Google Scholar]
- Metcalfe WK, Myers PZ, Trevarrow B, Bass MB, Kimmel CB. Primary neurons that express the L2/HNK-1 carbohydrate during early development in the zebrafish. Development. 1990;110:41–504. doi: 10.1242/dev.110.2.491. [DOI] [PubMed] [Google Scholar]
- Niciu MJ, Ma XM, El Meskini R, Pachter JS, Mains RE, Eipper BA. Altered ATP7A expression and other compensatory responses in a murine model of Menkes disease. Neurobiol. Dis. 2007;27:278–291. doi: 10.1016/j.nbd.2007.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niciu MJ, Ma XM, El Meskini R, Ronnett GV, Mains RE, Eipper BA. Developmental changes in the expression of ATP7A during a critical period in postnatal neurodevelopment. Neuroscience. 2006;139:947–964. doi: 10.1016/j.neuroscience.2006.01.044. [DOI] [PubMed] [Google Scholar]
- Pruett SB, Fan R, Zheng Q. Involvement of three mechanisms in the alteration of cytokine responses by sodium methyldithiocarbamate. Toxicol. Appl. Pharmacol. 2006;213:172–178. doi: 10.1016/j.taap.2005.10.004. [DOI] [PubMed] [Google Scholar]
- Stehr CM, Linbo TL, Incardona JP, Scholz NL. The developmental neurotoxicity of fipronil: Notochord degeneration and locomotor defects in zebrafish embryos and larvae. Toxicol. Sci. 2006;92:270–278. doi: 10.1093/toxsci/kfj185. [DOI] [PubMed] [Google Scholar]
- Teraoka H, Russell C, Regan J, Chandrasekhar A, Concha ML, Yokoyama R, Higashi K, Take-Uchi M, Dong W, Hiraga T, et al. Hedgehog and Fgf signaling pathways regulate the development of tphR-expressing serotonergic raphe neurons in zebrafish embryos. J. Neurobiol. 2004;60:275–288. doi: 10.1002/neu.20023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teraoka H, Urakawa S, Nanba S, Nagai Y, Dong W, Imagawa T, Tanguay RL, Svoboda KR, Handley-Goldstonem HM, Stegeman JJ, et al. Muscular contractions in the zebrafish embryo are necessary to reveal thiuram-induced notochord distortions. Toxicol. Appl. Pharmacol. 2005 doi: 10.1016/j.taap.2005.06.016. doi: 10.1016/j.taap.2005.06.016. [DOI] [PubMed] [Google Scholar]
- Thisse B, Degrave A, Thisse C. Expression of zebrafish genome during embryogenesis. ZFIN Database. 2001 [Google Scholar]
- Thompson RW, Valentine HL, Valentine WM. In vivo and in vitro hepatotoxicity and glutathione interactions of N-methyldithiocarbamate and N,N-dimethyldithiocarbamate in the rat. Toxicol. Sci. 2002;70:269–280. doi: 10.1093/toxsci/70.2.269. [DOI] [PubMed] [Google Scholar]
- Tilton F, La Du JK, Vue M, Alzarban N, Tanguay RL. Dithiocarbamates have a common toxic effect on zebrafish body axis formation. Toxicol. Appl. Pharmacol. 2006;216:55–68. doi: 10.1016/j.taap.2006.04.014. [DOI] [PubMed] [Google Scholar]
- Torres-Sanchez L, Rothenberg SJ, Schnaas L, Cebrian ME, Osorio E, Del Carmen Hernandez M, Garcia-Hernandez RM, Del Rio-Garcia C, Wolff MS, Lopez-Carrillo L. In utero p,p'-DDE exposure and infant neurodevelopment: A perinatal cohort in Mexico. Environ. Health Perspect. 2007;115:435–439. doi: 10.1289/ehp.9566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- U.S. EPA. Health Effects Division. Vol. 2005. Washington, D.C: Office of Pesticide Programs; 2001. The determination of whether dithiocarbamate pesticides share a common mechanism of toxicity. [Google Scholar]
- U.S. EPA. Environmental Fate and Ecological Risk Assessment for the Existing Uses of Metam-Sodium. 2005;Vol. 2005 EPA-HQ-OPP-2004-0159-0019. Office of Prevention, Pesticides, and Toxic Substances. [Google Scholar]
- Van Leeuwen CJ, Espeldoorn A, Mol F. Aquatic toxicological aspects of dithiocarbamates and related compounds. III. Embryolarval studies with rainbow trout (Salmo Gairdneri) Aquat. Toxicol. 1986;9:129–145. [Google Scholar]
- Westerfield M. The Zebrafish Book. Eugene, OR: University of Oregon Press; 1995. [Google Scholar]
- WHO. Vol. 1998. Geneva, Switzerland: International Program On Chemical Safety; 1998. Dithiocarbamate Pesticides, Ethylenethiourea and Propylenethiourea: A General Introduction. [Google Scholar]
- Xu Y, He J, Wang X, Lim TM, Gong Z. Asynchronous activation of 10 muscle-specific protein (MSP) genes during zebrafish somitogenesis. Dev. Dyn. 2000b;219:201–215. doi: 10.1002/1097-0177(2000)9999:9999<::aid-dvdy1043>3.3.co;2-9. [DOI] [PubMed] [Google Scholar]
- Xu L, Li AP, Kaminski DL, Ruh MF. 2,3,7,8 Tetrachlorodibenzo-p-dioxin induction of cytochrome P4501A in cultured rat and human hepatocytes. Chem. Biol. Interact. 2000a;124:173–189. doi: 10.1016/s0009-2797(99)00149-0. [DOI] [PubMed] [Google Scholar]