Abstract
The reinforcing effects and long-term consequences of cocaine self-administration have been associated with brain regions of the mesolimbic dopamine pathway, namely the nucleus accumbens (NAc). Studies of cocaine-induced biochemical adaptations in rodent models have advanced our knowledge; however, unbiased detailed assessments of intracellular alterations in the primate brain are scarce, yet essential, to develop a comprehensive understanding of cocaine addiction. To this end, two-dimensional difference in gel electrophoresis (2D-DIGE) was used to compare changes in cytosolic protein abundance in the NAc between rhesus monkeys self-administering cocaine and controls. Following image normalization, spots with significantly differential image intensities (P < 0.05) were identified, excised, trypsin digested and analyzed by matrix-assisted laser-desorption ionization time-of-flight time-of-flight (MALDI-TOF-TOF). In total, 1098 spots were subjected to statistical analysis with 22 spots found to be differentially abundant of which 18 proteins were positively identified by mass spectrometry. In addition, approximately 1000 protein spots were constitutively expressed of which 21 proteins were positively identified by mass spectrometry. Increased levels of proteins in the cocaine-exposed monkeys include glial fibrillary acidic protein, syntaxin-binding protein 3, protein kinase C isoform, adenylate kinase isoenzyme 5 and mitochondrial-related proteins, whereas decreased levels of proteins included β-soluble N-ethylmaleimide-sensitive factor attachment protein and neural and non-neural enolase. Using a complimentary proteomics approach, the differential expression of phosphorylated proteins in the cytosolic fraction of these subjects was examined. Two-dimensional gel electrophoresis (2DGE) was followed by gel staining with Pro-Q Diamond phosphoprotein gel stain, enabling differentiation of approximately 150 phosphoprotein spots between the groups. Following excision and trypsin digestions, MALDI-TOF-TOF was used to confirm the identity of 15 cocaine-altered phosphoproteins. Significant increased levels were detected for γ-aminobutyric acid type A receptor-associated protein 1, 14-3-3 γ-protein, glutathione S-transferase and brain-type aldolase, whereas significant decreases were observed for β-actin, Rab GDP-dissociation inhibitor, guanine deaminase, peroxiredoxin 2 isoform b and several mitochondrial proteins. Results from these studies indicate coordinated dysregulation of proteins related to cell structure, signaling, metabolism and mitochondrial function. These data extend and compliment previous studies of cocaine-induced biochemical alterations in human post-mortem brain tissue, using an animal model that closely recapitulates the human condition and provide new insight into the molecular basis of the disease and potential targets for pharmacotherapeutic intervention.
Keywords: cocaine, protein expression, nucleus accumbens, phosphorylation, monkey
Introduction
Cocaine abuse remains a significant health concern in the United States and abroad.1,2 Approximately 2.4 million Americans, 12 years and older, currently use cocaine. This represents approximately 0.97% of the population and a 20% increase in the number of users since 2002.1 The propensity to use cocaine is influenced by both positive (euphoric, pleasurable effects) and negative (withdrawal, depressed mood states and drug cravings) consequences, including the development of neuroadaptive changes in specific brain regions.3 Understanding the neurobiological mechanisms that contribute to cocaine abuse is critical for developing new pharmacotherapies and matching clinical diagnoses with appropriate treatment strategies.
The majority of research designed to understand the addictive properties of cocaine has focused largely on the neural circuit that mediates motivational processes, the mesolimbic dopamine pathway. Projections from the ventral tegmental area (VTA) to the nucleus accumbens (NAc) comprise part of the mesolimbic dopamine pathway and have been identified as a critical substrate in the reinforcing effects of cocaine in humans and animal models. Indeed, cocaine has been shown to increase extracellular dopamine concentrations within this pathway, an effect attributed to the drug’s abuse liability. Furthermore, there is ample evidence that repeated cocaine use leads to biochemical adaptations in mesolimbic brain regions and these adaptations appear to be relevant to the processes of sensitization, craving, withdrawal and relapse.4 Studies in rodent models indicate cocaine-induced biochemical alterations in regions associated with the mesolimbic pathway including upregulation of the cyclic AMP (cAMP) pathway,5–8 activator protein 1 family members9–11 and glutamate signaling.12–14
Genomics-based analyses, such as microarrays, have revealed novel mechanisms of drug-induced neuronal and non-neuronal dysregulation in human postmortem brain tissue15,16 and rodent models.17–21 Although these studies have been highly informative in furthering our understanding of drug-induced transcriptional regulation contributing to long-term changes in cellular function, research determining coordinate changes in the expression of multiple proteins following cocaine exposure has been scarce.22 To comprehend the intricate neuroadaptive machinery implicated in the development and expression of cocaine abuse, it is desirable to complement the global gene expression analyses with studies examining the corresponding proteomes. Current understanding of stimulant-induced neurobiological alterations (including expression and functional genomics/proteomics, epigenetic modifications, and so on) is based predominantly on rodent models of human drug taking; however, the direct determination of protein expression status and factors regulating protein expression in primate brain has been lacking, yet it is essential for understanding the consequential molecular pathology of cocaine addiction in humans.
Recently, we interrogated a portion of the NAc proteome in individuals following cocaine overdose using two-dimensional difference in gel electrophoresis (2D-DIGE) combined with tandem mass spectrometry (MS/MS) analysis.23 The abundances of proteins from several families were found to be significantly altered, including proteins belonging to cell structure, synaptic plasticity/signal transduction, mitochondrial function and metabolic pathways. Interestingly, several of the proteins were functionally associated with N-methyl-D-aspartate (NMDA) and α-amino-3-hydroxy-5-methyl-4-isoxazolepropionic acid (AMPA) receptors, previously shown to be increased in the VTA and NAc of cocaine overdose victims.16,24
In addition to assessing changes in the abundance of native proteins, changes in the abundance of posttranslational modifications represent an important means of activation/deactivation of various proteins and thereby significantly influencing cellular function. Phosphorylation, the most prevalent covalent modification of proteins in eukaryotic cells, affects approximately one-third of all proteins at any given time.25 Phosphorylation is catalyzed by a variety of protein kinases including protein kinase A, one of the most well studied in cocaine abuse,4 which phosphorylates serine and threonine residues on multiple targets including cAMP response element-binding protein.26,27 Detecting changes in post-translationally modified proteins in human postmortem tissue is difficult at best; however, similar assessments in nonhuman primate models of cocaine abuse are possible and have the advantage of recapitulating behavioral aspects of human drug intake and offering neuroanatomical and biochemical similarities compared with other species used as models.28
Historically, research examining the abundance of proteins and post-translational modifications as a function of cocaine abuse has been restricted to the serial analysis of individual proteins. With the advent of high-throughput separation and MS-based analysis strategies, it is possible to provide a broad and unbiased coverage of the proteome to delineate the multitude of neurobiological effects of abused drugs.22,29 In the present study, the proteome and phosphoproteome of the NAc was examined following chronic cocaine self-administration in rhesus monkeys. Proteins contained in the cytosolic fraction were assessed by 2D-DIGE and a phospho-specific dye for native and phosphorylated proteins, respectively. Proteins that were differentially expressed between the cocaine and control groups were isolated and followed by de novo identification by matrix-assisted laser-desorption ionization time-of-flight time-of-flight (MALDI-TOF-TOF) MS. These strategies provide new insight into the neurobiological/neuropathological changes associated with chronic intake and may yield novel targets for future drug development.
Materials and methods
Surgery and self-administration procedures
Eight male adult rhesus monkeys (Macaca mulatta) were singly housed with standard enrichment, including social enrichment, human interaction, variety in diet and age appropriate objects as dictated by the Animal Welfare Act and the Emory University Policy for Environmental Enhancement. Each subject was fed Purina monkey chow (Ralston Purina, St Louis, MO, USA), fruits and vegetables. Food and water were available ad libitum. Animal care procedures strictly followed the National institutes of Health Guide for the Care and Use of Laboratory Animals and were approved by the Institutional Animal Care and Use Committee of Emory University and Wake Forest University Schools of Medicine. Four monkeys were surgically prepared with chronically indwelling venous catheters using procedures described previously. 30,31 Under appropriate anesthesia, either isoflurane alone or ketamine in combination with diazepam, and under aseptic conditions, one end of a silicone catheter was passed by way of a jugular or femoral vein to the level of the right atrium and vena cava, respectively. The distal end of the catheter was passed under the skin and attached to a vascular access port (Access Technologies, Skokie, IL, USA), which remained subcutaneous in the center of the back for easy access. The 0.25 ml unit was accessed during testing with special right-angle Huber needles (Access Technologies) that minimize damage to the port membrane and allow for repeated punctures over a year or more. Catheters were flushed after daily test sessions with heparinized saline (0.9%). Daily experimental sessions were conducted within a ventilated, sound-attenuating chamber with each monkey seated in a standard primate chair of the type commercially available (Primate Products, Redwood City, CA, USA). A panel equipped with a response lever and stimulus lights was mounted on the front of the chair. The vascular access port was connected through polyvinyl-chloride tubing to a motor-drive syringe located outside the test chamber to yield a precise injection volume of 2.0 ml during drug self-administration experiments with dose determined by the concentration of drug solution in the syringe. Experimental procedures were controlled online by a microprocessor and electromechanical programming systems, and data were monitored and recorded during daily sessions.
Four monkeys were trained to self-administer cocaine by pressing a response key while seated in a primate chair. Responding was initiated using a 1-response fixed-ratio schedule (FR 1) so that each response in the presence of a red light produced an intravenous drug injection and the brief illumination of a white light followed by a timeout. The ratio value was increased gradually as responding increased. When the schedule value reached FR 20, drug injection no longer followed completion of each FR and, instead, was arranged to follow an increasing number of FR components. Ultimately, the schedule was a second-order schedule of FR 20 components with drug injection following the first component completed after 10 min had elapsed (fixed-interval (FI) 600-s (FR 20:S)). A 2-s white light was presented upon completion of each FR 20 component. Drug administration was accompanied by a change in the stimulus light from red to white for 15 s, followed by a 1-min timeout. Daily sessions consisted of five consecutive 10-min intervals. The unit dose of cocaine remained constant at 0.1 mg kg−1 per injection. 30,32 Use of this second-order procedure and limiting the daily session to approximately 1 h enabled the standardization of total drug intake to 0.5 mg kg−1 per session.
In total, 18–24 h after the last drug self-administration studies, monkeys were anesthetized with Telazol (4.0 mg kg−1; i.v.), administered intravenous heparin followed by an overdose of intravenous sodium pentobarbital. This time point was selected to ensure that no cocaine was present at the time of killing and to avoid any effects associated with cocaine withdrawal. After the confirmed absence of brain stem reflexes was established, the monkeys were transcardially perfused with ice-cold phosphate-buffered saline (PBS; pH 7.2–7.4), the brain was removed and placed in 4 °C PBS for 5 min. Brains were blocked using a rhesus monkey brain matrix that allows 4 mm coronal blocks at various AP locations (Electron Microscopy Sciences, Ft Washington, PA, USA). Tissue was frozen at −80 °C within 40 min of necropsy and stored at −80 °C until further processing.
Protein isolation and fractionation
The NAc was dissected from the rostral pole to the beginning of the anterior commissure, using the internal capsule and lateral ventricle as landmarks33 (Supplementary Figure). A steel mortar and pestle chilled in dry ice were used to pulverize the frozen brain tissue into a dry homogenate in the presence of liquid nitrogen. Tissue proteins were fractionated as described previously.13,23,24,34 Fractionation enables the enrichment of low abundance proteins in distinct cellular fractions compared to total cell/tissue homogenate and therefore allows increased coverage of the analyzed proteome.
Cytosolic fractions from the NAc were evaluated in the present study to compliment and extend similar analysis conducted in postmortem brain of human cocaine overdose victims.23 It is important to note that the membrane fraction was not included in the current analysis primarily due to the limitations of 2DG for analyzing membrane proteins.35 Most hydrophobic (membrane-bound) proteins are insoluble in nondetergent sample buffer used for isoelectric focusing (IEF) and the ones which are soluble precipitate at their respective isoelectric points (pI). Second, the pIs of hydrophobic proteins are generally alkaline and even with the use of extended pH gradients, the proteins are difficult to resolve well at the basic end. Liquid chromatography (LC)-MS methodologies can overcome issues of hydrophobic membrane protein separation. Studies are currently underway in our lab utilizing multidimensional liquid chromatographic separation along with MS to explore the membrane proteome in these subjects.
Experiment 1: Differential native protein expression using 2D-DIGE
Cyanine dye labeling
Minimal labeling of the lysine residues was achieved by reaction with cyanine dyes, as described previously.23,36,37 Briefly, a normalization (pooled) sample was prepared by combining 50 μg from each sample. The labeling of 50 μg of protein sample (each sample) was optimized by labeling with 200 pmol of appropriate dye (suspended in > 99.5% pure dimethylformamide). On each gel, electrophoresis was carried out on a pooled sample labeled with Cy2 and also individuals samples from the cocaine and control groups labeled with either Cy3 or 5.
Two-dimensional polyacrylamide gel electrophoresis
Two-dimensional polyacrylamide gel electrophoresis (2D-PAGE) was executed as described previously.23,36,37 Aliquots of cytosolic proteins were diluted in 400 μl of rehydration buffer and increased to a final volume of 450 μl with DeStreak rehydration buffer (GE Healthcare, Piscataway, NJ, USA). Isoelectric focusing was performed using Immobiline DryStrips (240×3×0.5 mm, pH 4–7 linear; GE Healthcare) on an Ettan IPGphor apparatus (GE Healthcare).23,36,37
Gel image analysis
Image analysis was conducted as described previously,23,36,37 using a Typhoon 9400 scanner (GE Healthcare) to scan all gels at 100 μm resolution. The photomultiplier tube was set to ensure maximum pixel intensity of 85 000–95 000 for all the images in every gel. Image analysis was performed using DeCyder 5.01 software (GE Healthcare). The Difference In-gel Analysis (DIA) mode of DeCyder was employed for protein spot detection and also for normalization of cocaine and control gel images to the pooled sample gel image. The photomultiplier tube was set to ensure maximum pixel intensity of 85 000–95 000 for all the images in every gel. After spot detection, the abundance changes are represented by the normalized volume ratio of cocaine or control to the pooled sample. The following parameters were used for spot filtering; slope > 1.0, area < 350, peak height < 350 and volume < 100 000. The spots were authenticated manually for all the gels. Spot maps from all the gels were first matched by manual landmarks and then in automatic mode by DeCyder Biological Variation Analysis (BVA). Protein spot matches were confirmed manually for all the gels. The average ratio and also the corresponding student’s t-test value for each protein spot was calculated based on all gel images in the DeCyder BVA mode.37 Selected spots were isolated using the Ettan Spot Handling Workstation (GE Healthcare) and picked proteins were prepared for MS analysis as described in the section below entitled in-gel trypsin digestion.
Experiment 2: Phosphoproteome gel staining and image analysis
Aliquots of cytosolic protein fractions were separated by 2D gels in the same manner as the samples for experiment 1 and as described above in the subsection 2D-PAGE. Following, gels were stained for proteins containing phosphorylated moieties according to the instructions provided by the manufacturer (Molecular Probes, Eugene, OR, USA). Gels were fixed in 500 ml of 50% methanol and 10% trichloroacetic acid for 15 h and then sequentially washed thrice with 500 ml distilled water for 15 min, followed by incubation with 500 ml Pro-Q Diamond phospho-protein stain for 2 h in the dark. Gels were then destained with 20% acetonitrile, 50 mM sodium acetate (pH = 4) for 90 min and washed twice for 5 min each in distilled water. All gels were scanned at 100 μm resolution of Typhoon 9400 scanner (GE Healthcare), using the green laser (532 nm) for excitation and 610 nM band pass emission filter for visualization of proteins with phospho-specific moieties. The photo-multiplier tube was set to ensure maximum pixel intensity of 80 000–90 000 for all the gels. ImageQuant V5.2 and DeCyder 5.01 (GE Healthcare) were used to remove extraneous areas to the scanned gel images and for performing image analysis, respectively. Spots were authenticated manually for all the gels during the differential in-gel analysis (DeCyder DIA). Spot maps from all gels were analyzed by manually assigning landmarks and then by automatic mode by DeCyder BVA, which enables the matching of multiple 2DGE gels for comparison and statistical analysis of protein abundance changes. The protein spot matches performed in the automatic mode were also confirmed manually for all the gels.37 To visualize the proteome from this specific pH and mass range, gels were stained with Sypro Ruby stain overnight. The excess stain was removed by 10% methanol and 6% glacial acetic acid for 20 min. The gels were imaged and analyzed, in a similar fashion to the phospho-protein stain using Typhoon 9400 scanner and DeCyder image analysis software, respectively. Student’s t-test was used to determine differential abundance of the phosphoproteins between groups. Individual protein spots from the 2D gels were excised with 1.5 mm diameter gel cutter (The Gel Company, San Francisco, CA, USA).
In-gel trypsin digestion
The excised gel spots from experiments 1 and 2 were washed for 20 min, twice in 100 μl of solution of 50 mM ammonium bicarbonate, 50% methanol (v/v) in distilled water and once in 75% acetonitrile in distilled water for 30 min or until the gel plugs turned opaque. Lyophilized trypsin (20 μg) (883 pmol; Promega, Madison, WI, USA) was reconstituted in 1 ml of 20 mM ammonium bicarbonate and incubated for 15 min at 37 °C. The gel fragments were dried by vacuum centrifugation and then incubated overnight with 10 μl (200 ng) of trypsin at 37 °C. The supernatant from trypsin digest was transferred to a low retention 96-well plate. Peptides from the gel pieces were sequentially extracted twice in 100 μl of extraction buffer (50% (v/v) acetonitrile, 0.1% (v/v) trifluoroacetic acid in distilled water). The original tryptic supernatant and the supernatants from two sequential extractions were combined and dried in a vacuum centrifuge. The dried peptides from each gel plug were dissolved in 5 μl of 50% (v/v) acetonitrile, 0.1% trifluoroacetic acid in distilled water and 0.5 μl deposited on the stainless-steel MALDI target plate. After drying, the spot residue was mixed with 0.5 μl of 5 mg ml−1 of α-cyano-4-hydroxy-cinnamic acid (Sigma-Aldrich, St Louis, MO, USA) in 50% (v/v) acetonitrile, 0.1% trifluoroacetic acid in distilled water.23,37
MALDI-TOF-TOF
Mass spectrometry analyses were performed using the Applied Biosystems 4700 Proteomics Analyzer (MALDI-TOF-TOF; Foster City, CA, USA) in reflector mode for positive ion detection. The laser wavelength and the repetition rate were 355 nm and 200 Hz, respectively. All the MS spectra resulted from accumulation of at least 2000 laser shots. The peak detection criteria used were: minimum S/N of 8, local noise window width mass/charge (m/z) of 200 and minimum full-width half-maximum (bins) of 2.9. The mass spectra were calibrated using the three trypsin auto digest products: fragment 100–107 ([M+H]+ = 842.51 Da), fragment 90–99 ([M+H]+ = 1045.556 Da) and fragment 50–69 ([M+H]+ = 2211.105 Da). A maximum of the 10 strongest precursor ions per sample were chosen for MS/MS analysis. The following mono-isotopic precursor selection criteria were used for the MS/MS: minimum S/N filter of 10, excluding the most commonly observed peptide peaks for trypsin and keratin, and excluding the precursors within 150 resolution. In the TOF1 stage, all ions were accelerated to 1 kV under conditions promoting metastable fragmentation. The peak detection criteria used were: S/N of 8 and local noise window width of 250 (m/z).23
Protein identification
The peak lists generated by the 4000 Series Explorer software (version 3.6) were submitted to GPS Explorer (Applied Biosystems) to search against the National Center for Biotechnology Information nonredundant for protein characterization using the limited M. mulatta database.23,38 The following parameters were used: one allowed missed cleavage, ±50 p.p.m. for m/z error for MS and 0.1 Da (m/z) error for MS/MS, partial modification of cysteine (carbamidomethyl-cysteine), methionine (oxidized) and phosphorylation of serine, threonine and tyrosine. Peptide mass fingerprint (PMF) and MS/MS spectra were interpreted with the MASCOT software (Matrix Science Ltd, London, UK).39,40 Database searches, through MASCOT, using combined PMF and MS/MS datasets were performed through GPS Explorer software. The criterion for identification was a MASCOT confidence interval greater than 95%.
In order to reduce false-positive identification of proteins, an open access (http://www.matrixscience.com/help/decoy_help.html) Perl script ‘decoy.pl.gz.’ was used to randomize the database entries from the forward database (target database) and create a new database (decoy database).41 Randomization, instead of reversal, of peptide sequence was used to establish the decoy database as it is more suitable for PMF incorporated with the MS/MS searches in the current study and also the fact that a reverse decoy database will have at least half of the tryptic peptide mass values unchanged. The decoy database is comprised of random sequences of the same length and has the same average amino-acid composition as the forward database. As no matches are expected to be generated against the decoy database, the number of matches found provides a good estimate of the number of false positives present from the real database. The original search was repeated using identical search parameters against the decoy database (randomized sequences).
De novo sequencing
The method for the de novo peptide sequencing for M. mulatta has been published recently by our group and was adapted for the current study,42 using the PEAKS Studio 4.0 (Bioinformatics Solutions, Waterloo, ON, Canada) de novo sequencing software. One potential obstacle to comprehensive assessment of protein alterations in rhesus monkey is the relative paucity of available protein annotations for this species making it difficult to identify proteins. The use of standard database search engines (for example, MASCOT) has a limitation in that ‘broad species database’ searches are needed resulting in less than optimal protein annotation. This limitation can be overcome in some respects using a de novo sequencing strategy, in which partial or complete amino-acid sequence information is obtained using either manual or automated de novo peptide sequence analysis using PEAKS software. Briefly, when positive characterization was not obtained for all spectra, spectra were subjected to PEAKS de novo analysis. The de novo sequencing parameters used were as follows: parent- and fragment-mass error tolerance of 0.08 U; trypsin as the protease with one maximum missed cleavage allowed; deconvolution of the charge state in the spectra to generate a spectra in which each monoisotopic peak becomes singly charged; partial modification of cysteine (carbamidomethyl-cysteine) and methionine (oxidized). The most abundant peptide fragments ‘b- and y-ions’; the less abundant peptide fragments ‘a-ions’; the neutral losses of water and ammonia for b- and y-ions; and also the immonium ions were utilized to develop confident and complete peptide sequences de novo from MS/MS spectra. The sequences generated from each spectrum were used for protein identification by sequence homology in the mammalian database using either the PEAKS or SPIDER software (Software Protein Identifier). Protein identification was confirmed by protein mass and pI accuracy.
Results
Behavioral data
Three of four subjects readily acquired drug self-administration during the first month of training. The fourth subject required approximately 4 months establishing reliable drug self-administration and drug intake was very erratic during this extended training period. Once stable self-administration behavior was established, all subjects reliably received all scheduled injections during daily sessions. Total drug intake for the group of four subjects over the 18-month period was 37.9±4.6 mg kg−1. Previously, we have reported significant alterations in ionotropic glutamate receptor subunits in the NAc from the subjects used in the present study.24
2D-DIGE
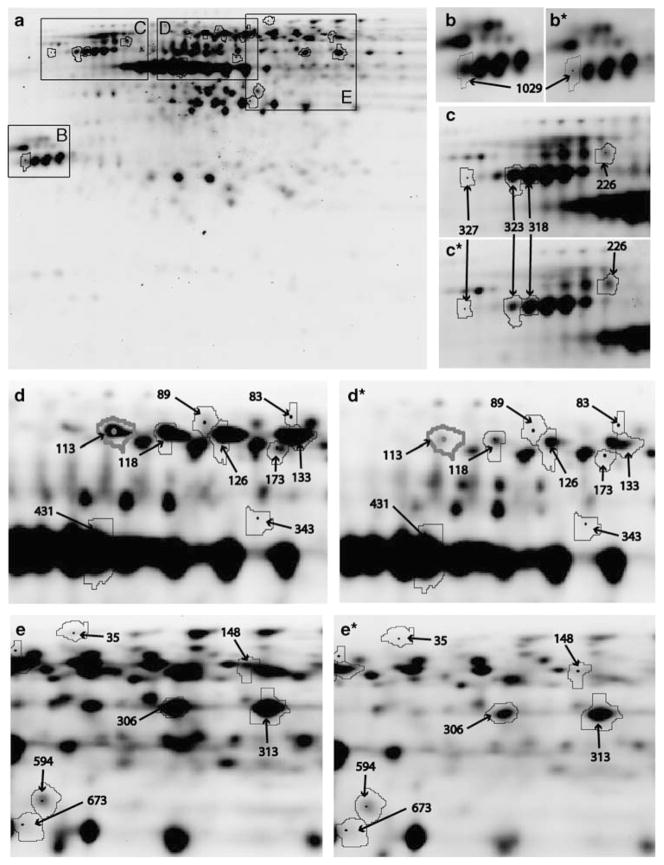
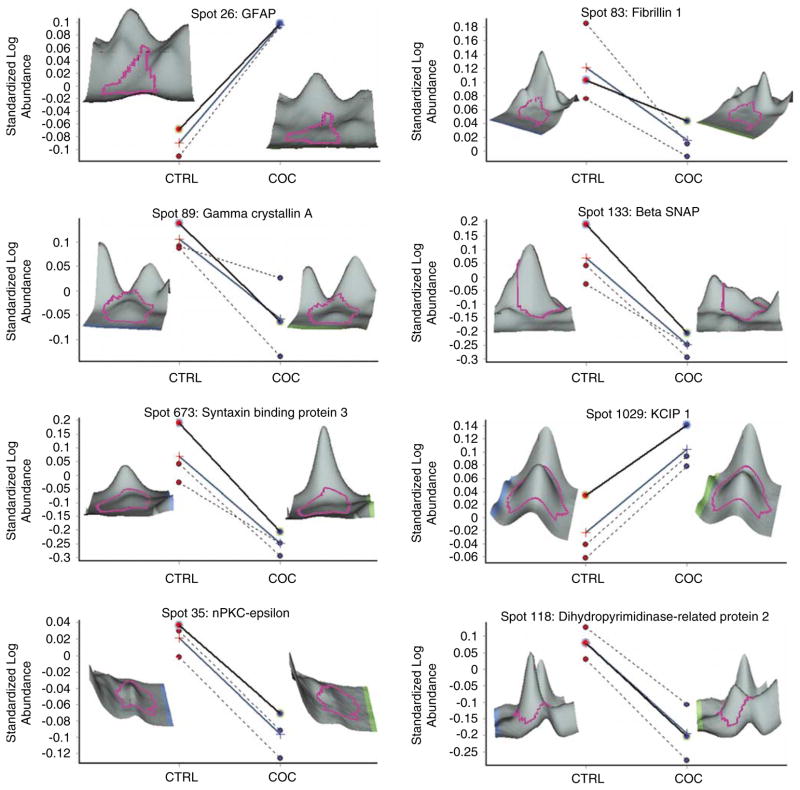
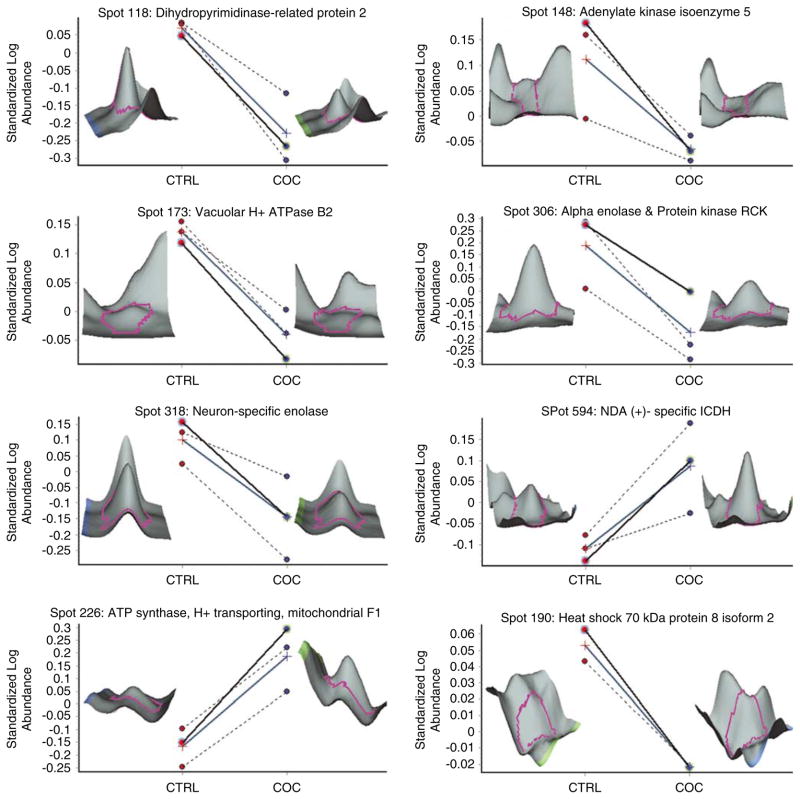
Cytosolic fractions of NAc from CSA rhesus monkeys and controls were compared using the DIGE proteomics strategy to determine differences in the abundance levels of proteins in a pH range of 4–7 to elucidate the molecular mechanisms by which previous exposure to cocaine alters the cytosolic proteome of NAc (Figure 1 and Table 1). The total numbers of protein spots subjected to statistical analysis were 1098. The DeCyder image analysis of the images representing CSA rhesus monkeys and control NAc cytosol proteome elucidated the putative CSA-specific protein spots. Quantification of the individual protein spots revealed that the vast majority of ~1000 gene products relegated to structural and housekeeping functions were quantitatively similar. Image analysis (Figure 1) of fluorescently labeled NAc cytosol lysates loaded on 2D gels identified differential distributions of 22 protein spots (t-test: P < 0.05) characterized by 18 proteins in CSA rhesus monkeys (Figures 2 and 3). Notably, the expression of glial fibrillary acidic protein (GFAP, spot 26) was increased significantly in human NAc cytosol after chronic cocaine exposure as shown in Figure 2. Even though there have been many studies reporting the cocaine-induced alterations in gene expression, this is the first follow-up study which utilizes a noncandidate approach to decipher the involvement of novel proteins and pathways.
Figure 1.
Two-dimensional difference in gel electrophoresis (2D-DIGE)-based comparison of control and cocaine groups. A representative 2D-DIGE image of cytosolic proteins from the nucleus accumbens (NAc) (a). Proteins were fractionated in the first dimension by a 4–7 linear pH gradient (isoelectric points, pI) and in the second dimension by an 8–15% gradient SDS–polyacrylamide gel electrophoresis (PAGE) gel (molecular weight, kDa). Areas of the gel containing spots of interest are marked with boxes and shown separately for control (b–e) and cocaine groups (b*, c*, d*, e*). The numbered protein spots correspond with numbers in Figures 2 and 3, and Table 1.
Table 1.
Proteins identified by MALDI-TOF-TOF following 2D-DIGE analysis
| Spot nos. | Protein ID (PMF) | Accession nos. | Mr (kDa) | pI | Peptides | MASCOT score
|
Target confidence index (%) | Protein quantitation
|
||
|---|---|---|---|---|---|---|---|---|---|---|
| Target | Decoy | T-test (P-value) | Average value | |||||||
| Structural | ||||||||||
| 26 | Glial fibrillary acidic protein, astrocyte | gi|121135 | 49.85 | 5.42 | 8 | 66 | 38 | 98.35 | 0.013 | 1.53 |
| 83 | Fibrillin 1 | gi|24430141 | 31.2 | 4.81 | 23 | 66 | 38 | 98.4 | 0.042 | 1.28 |
| 343 | Actin, cytoplasmic 2 (γ) | gi|15012187 | 41.76 | 5.31 | 7 | 150 | 39 | 100 | 0.032 | −1.50 |
| 413 | Actin, cytoplasmic 2 (γ) | gi|15012187 | 41.76 | 5.31 | 9 | 113 | 33 | 100 | 0.045 | 2.45 |
| 431 | Actin, cytoplasmic 2 (γ) | gi|15012187 | 41.76 | 5.31 | 7 | 75 | 37 | 99.7 | 0.023 | 1.56 |
| 160 | α-Tubulin | gi|10881132 | 49.03 | 4.9 | 6 | 92 | 35 | 100 | 0.27 | −2.27 |
| 89 | γ-Crystallin A | gi|117458 | 21.13 | 7.53 | 6 | 63 | 40 | 96.85 | 0.03 | 1.44 |
| 250 | Unnamed protein product; Homology-filament, intermediate filament protein | gi|34536332 | 49.47 | 5.84 | 13 | 185 | 49 | 100 | 0.079 | −1.74 |
| 257 | Unnamed protein product; Homology-filament, intermediate filament protein | gi|34536332 | 49.47 | 5.84 | 16 | 211 | 43 | 100 | 0.19 | −1.50 |
| 254 | Unnamed protein product; Homology-filament, intermediate filament protein | gi|34536332 | 49.47 | 5.84 | 19 | 261 | 52 | 100 | 0.11 | −1.62 |
| Cell signaling | ||||||||||
| 133 | β-Soluble NSF attachment protein | gi|18202933 | 33.53 | 5.32 | 6 | 56 | 37 | 85.6 | 0.01 | −2.12 |
| 673 | Syntaxin-binding protein 3 | gi|27881593 | 67.79 | 8.1 | 7 | 59 | 25 | 95.15 | 0.022 | 1.34 |
| 1029 | 14-3-3 protein ζ/Δ (protein kinase C inhibitor protein-1) | gi|52000886 | 27.72 | 4.73 | 5 | 51 | 39 | 97.5 | 0.0041 | −1.31 |
| 35 | PKC, epsilon type (nPKC- ε) | gi|125556 | 83.46 | 6.62 | 12 | 66 | 43 | 98.5 | 0.021 | 1.32 |
| 612 | DDAH1 protein | gi|34783629 | 27.11 | 5.23 | 3 | 82 | 38 | 99.9 | 0.36 | −1.19 |
| Metabolism | ||||||||||
| 113 | Dihydropyrimidinase-related protein 2 | gi|33991634 | 62.25 | 5.95 | 12 | 83 | 36 | 99.9 | 0.0085 | −1.86 |
| 118 | Dihydropyrimidinase-related protein 2 | gi|33991634 | 62.25 | 5.95 | 11 | 93 | 36 | 100 | 0.0072 | −1.96 |
| 126 | Dihydropyrimidinase-related protein 2 | gi|33991634 | 62.25 | 5.95 | 11 | 84 | 43 | 100 | 0.03 | −1.65 |
| 148 | Adenylate kinase isoenzyme 5 | gi|9296996 | 22.07 | 5.38 | 9 | 65 | 98.19 | 0.044 | 1.53 | |
| 173 | Vacuolar ATP-synthase subunit B, brain isoform 56/58 kDa, V1 subunit B, isoform 2 | gi|19913428 | 56.46 | 5.57 | 16 | 159 | 41 | 100 | 0.0027 | −1.50 |
| 306 | α-Enolase (non-neural enolase) | gi|119339 | 47.13 | 7.01 | 9 | 84 | 27 | 99.9 | 0.045 | −2.30 |
| Serine/threonine-protein kinase MAK (protein kinase RCK) | gi|462461 | 70 | 9.67 | 8 | 63 | 27 | 96.92 | |||
| 313 | α-Enolase (non-neural enolase) | gi|119339 | 47.13 | 7.01 | 6 | 44 | 35 | 87.8 | 0.033 | −1.75 |
| 323 | γ-Enolase (neural enolase) (NSE) | gi|119348 | 47.26 | 4.99 | 5 | 46 | 37 | 93.1 | 0.046 | −1.73 |
| 318 | NSE | gi|930063 | 47.12 | 4.94 | 7 | 53 | 36 | 97.7 | 0.05 | −1.39 |
| 319 | NSE | gi|930063 | 47.12 | 4.94 | 6 | 72 | 44 | 99.5 | 0.59 | 1.09 |
| 315 | Guanine deaminase | gi|4758426 | 50.97 | 5.44 | 5 | 71 | 30 | 99.2 | 0.70 | −1.05 |
| 689 | Lactate dehydrogenase B | gi|4557032 | 36.61 | 5.71 | 5 | 78 | 40 | 99.8 | 0.52 | −1.15 |
| Mitochondrial | ||||||||||
| 226 | ATP synthase, H+ transporting, mitochondrial F1 complex, β-subunit precursor | gi|32189394 | 56.52 | 5.26 | 12 | 122 | 35 | 100 | 0.014 | 2.30 |
| 139 | Succinyl-CoA:3-ketoacid CoA transferase 1 | gi|10280560 | 56.12 | 7.14 | 2 | 92 | 38 | 100 | 0.21 | 1.41 |
| Miscellaneous | ||||||||||
| 190 | Heat shock 70 kDa protein 8 isoform 2 | gi|24234686 | 53.48 | 5.62 | 7 | 75 | 37 | 98.82 | 0.017 | 1.19 |
| 327 | Brain creatine kinase | gi|17939433 | 42.62 | 5.34 | 6 | 88 | 38 | 99.9 | 0.047 | −1.54 |
| 434 | Brain creatine kinase | gi|21536286 | 42.61 | 5.34 | 9 | 201 | 40 | 100 | 0.43 | −1.16 |
| 668 | T-cell receptor β-chain precursor V region (8.3) | gi|88712 | 12.98 | 6.07 | 6 | 59 | 35 | 86.9 | 0.75 | 1.06 |
Abbreviations: ATP, adenosinetriphosphate; DDAH1, dimethylarginine dimethylaminohydrolase 1; nos., numbers; NSE, neuron-specific enolase; NSF, N-ethylmaleimide-sensitive factor; pI, isoelectric points; PKC, protein kinase C; PMF, peptide mass fingerprint.
Statistically significant differences in protein abundance between the groups are shaded. MASCOT score is shown for the searches against the target and also the decoy databases. Average ratios are provided as cocaine:control. Positive and negative ratios indicate that the protein abundance is greater in the cocaine group and control group, respectively. COC/0 refers to proteins present in three or four cocaine subjects and none of the control subjects. 0/CTR refers to proteins present in three or four control subjects and none of the cocaine subjects.
Figure 2.
Differentially expressed proteins between the control and cocaine groups. Each panel depicts differentially expressed proteins as determined by two-dimensional difference in gel electrophoresis (2D-DIGE). Panels show individual data from the control (CTRL: red circles) and cocaine (COC: blue circles) groups and mean standardized log abundance of the two groups (cross symbols with blue line) for differentially expressed proteins. A three-dimensional image of each protein spot is provided from a representative control and COC subject. All proteins shown are significantly different between the two groups (P < 0.05).
Figure 3.
Differentially expressed proteins between the control and cocaine groups. Each panel depicts differentially expressed proteins as determined by two-dimensional difference in gel electrophoresis (2D-DIGE). Panels show individual data from the control (CTRL: red circles) and cocaine (COC: blue circles) groups and mean standardized log abundance of the two groups (cross symbols with blue line) for differentially expressed proteins. A three-dimensional image of each protein spot is provided from a representative control and cocaine subject. All proteins shown are significantly different between the two groups (P < 0.05).
A number of proteins extracted from 2D gels were constitutively expressed in the control and CSA groups and identified by MALDI-TOF-MS and -MS/MS. The proteins which were constitutively expressed belonged to the following groups: inter- and intracellular signaling, cell morphology, protein folding and stability, cell proliferation and apoptosis fully realizing that such a classification is somewhat arbitrary as a number of proteins could be assigned to more than one functional category. It should be noted that although some of the differentially expressed proteins seen here are already known to be either directly or indirectly involved in cocaine intake, a number of gene products (for example, syntaxin-binding protein 3 (Figure 2)) with unprecedented involvement in cocaine intake were also identified in the current study. The more conservative strategy of comparing results from the decoy and target databases resulted in three false-positive protein identifications: actin, cytoplasmic 2 (γ)—spots 291 and 320, and isocitrate dehydrogenase subunit-α, mitochondrial precursor-spot 594. The proteins identified from the respective spots were not included for subsequent analysis.
Pro-Q diamond for staining phosphoproteins
The specificity, quantitative assessment and the linear dynamic range of the Pro-Q Diamond staining for phosphoproteins has been rigorously established previously.43–47 The specificity of Pro-Q Diamond was re-validated in the current study by examining the differential staining of known phosphorylated and nonphosphorylated proteins. Standard proteins and NAc cytosolic proteins from control and cocaine-treated monkeys were electrophoresed and the gels stained with Pro-Q Diamond and Sypro-Ruby. The staining of specific proteins, ovalbumin and β-casein, with documented phosphorylation sites was observed in the protein standards stained with Pro-Q Diamond. As anticipated bovine serum albumin, avidin and lysozyme were not stained by Pro-Q Diamond due to the lack of phosphorylation sites on these proteins. Specific protein band staining from cocaine and control samples was also observed with Pro-Q Diamond stain.
NAc phosphoproteomes in cytosolic fractions
To determine changes in protein phosphorylation in NAc with cocaine self-administration, 2D gels were stained with the Pro-Q Diamond. DeCyder software-based comparisons of gels revealed that 123 and 152 spots were resolved in the control and cocaine groups, respectively, and spots were consistently matched in at least three of four subjects in each group. DeCyder-based image analysis was performed to quantify staining intensities of the putative phosphoproteins. Representative cytosolic protein spots with significant differences in phosphorylation abundance between control and cocaine groups are shown in the Figure 4. Targeted phosphoprotein spots were identified by MALDI-TOF-TOF (Tables 2 and 3). Although the specificity of the Pro-Q Diamond for staining phosphoproteins has been rigorously established, 43–47 a note of caution in the interpretation of these data. The identification of a spot, stained with Pro-Q Diamond, as a phosphoprotein by MS may be biased if an abundant protein (which may or may not be a phosphoprotein) co-migrates with this particular spot.
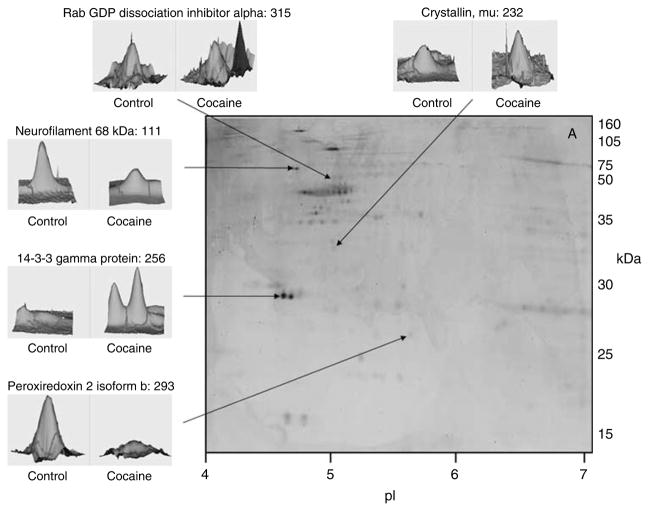
Figure 4.
Comparison of the phosphoproteins between control and cocaine groups. Representative gel of spots visualized following Pro-Q Diamond staining identified as differentially abundant between the two groups. Proteins were fractionated and electrophoresed by a linear 4–7 pH gradient in first dimension and by 12.5% SDS–polyacrylamide gel electrophoresis (PAGE) in second dimension. The identified protein name is followed by the spot number. The numbered protein spots correspond with numbers in Tables 2 and 3.
Table 2.
Phospho-proteins identified by MALDI-TOF-TOF
| Spot nos. | Protein ID (PMF) | Accession nos. | Mr (kDa) | pI | Peptides | MASCOT score
|
CI (%) | Protein quantitation | ||
|---|---|---|---|---|---|---|---|---|---|---|
| Target | Decoy | T-test (P-value) | Average ratio | |||||||
| Structural | ||||||||||
| 204 | Actin, β | Q96HG5 | 40.98 | 5.5 | 21 | 243 | 41 | 100 | 0.019 | −2.08 |
| 205 | ACTG1 protein (γ-actin) | P63261 | 41.76 | 5.3 | 18 | 185 | 49 | 100 | 0.995 | 1.21 |
| 183 | Actin, cytoplasmic 1 | P60709 | 41.71 | 5.3 | 20 | 72 | 43 | 100 | 0.315 | −1.30 |
| 191 | Actin, cytoplasmic 1 | P60709 | 41.7 | 5.3 | 23 | 116 | 46 | 100 | 0.122 | 5.88 |
| 190 | Actin, cytoplasmic 1 | P60709 | 41.7 | 5.3 | 21 | 96 | 35 | 100 | 0.916 | −1.56 |
| 186 | Actin, cytoplasmic 1 | P60709 | 41.7 | 5.3 | 16 | 102 | 39 | 100 | 0.315 | −1.30 |
| 193 | Actin, cytoplasmic 1 | P60709 | 41.7 | 5.3 | 16 | 66 | 37 | 100 | 0.495 | −1.04 |
| 111 | Neurofilament, light polypeptide 68 kDa | P07196 | 61.74 | 4.7 | 16 | 66 | 39 | 100 | 0.032 | −3.57 |
| 7 | Neurofilament triplet M protein 160 kDa | P07197 | 102.38 | 4.9 | 25 | 91 | 38 | 100 | 0.728 | −1.58 |
| Cell signaling | ||||||||||
| 274 | GABAA receptor-associated protein 1 (GEC1) | Q9H0R8 | 14.03 | 8.7 | 5 | 55 | 39 | 100 | N/A | COC/0 |
| 315 | Rab GDP dissociation inhibitor-α | P31150 | 50.55 | 5 | 17 | 54 | 41 | 100 | 0.0008 | −1.54 |
| 119 | Rab GDP dissociation inhibitor-α | P31150 | 50.55 | 5 | 17 | 69 | 38 | 100 | 0.192 | −1.72 |
| 256 | 14-3-3 γ-protein | P61981 | 28.36 | 4.7 | 13 | 92 | 46 | 99.99 | 0.057 | 4.79 |
| 255 | 14-3-3 γ-protein | P61981 | 28.36 | 4.7 | 11 | 86 | 37 | 99.99 | N/A | COC/0 |
| 264 | ρ-GDP dissociation inhibitor-α | P52565 | 23.2 | 5.0 | 6 | 69 | 40 | 100 | 0.241 | 2.22 |
| Metabolism | ||||||||||
| 286 | Glutathione S-transferase pi—Macaca mulatta | Q28514 | 23.42 | 5.9 | 11 | 107 | 46 | 100 | N/A | COC/0 |
| 267 | Glutathione S-transferase pi—human | P09211 | 23.37 | 5.4 | 7 | 54 | 39 | 100 | N/A | COC/0 |
| 293 | Peroxiredoxin 2 isoform b | P32119 | 159.79 | 6.1 | 10 | 141 | 44 | 100 | 0.014 | −3.37 |
| 219 | Brain-type aldolase | P09972 | 39.42 | 6.4 | 17 | 92 | 46 | 100 | N/A | COC/0 |
| 115 | Brain-type aldolase | P09972 | 39.43 | 6.4 | 16 | 93 | 44 | 100 | N/A | COC/0 |
| 245 | Dimethylarginine dimethylaminohydrolase 2 | Q5SRR8 | 29.63 | 5.7 | 8 | 59 | 38 | 100 | N/A | COC/0 |
| 172 | Guanine deaminase | Q9Y2T3 | 52.8 | 5.6 | 22 | 95 | 44 | 100 | N/A | 0/CTR |
| 217 | Fructose-bisphosphate aldolase C | P09972 | 39.43 | 6.4 | 15 | 82 | 39 | 100 | 0.596 | −2.38 |
| Mitochondrial | ||||||||||
| 166 | ATP-synthase β-chain, mitochondrial precursor | P06576 | 56.52 | 5.3 | 20 | 103 | 51 | 100 | 0.569 | −3.69 |
| 164 | ATP-synthase β-chain, mitochondrial precursor | P06576 | 56.52 | 5.3 | 15 | 86 | 41 | 100 | 0.165 | −1.76 |
| 325 | Ubiquitin C-terminal hydroxylase 12 | O75317 | 24.73 | 5.3 | 12 | 120 | 44 | 100 | 0.355 | −1.19 |
| Miscellaneous | ||||||||||
| 232 | Crystallin, μ | Q14894 | 33.75 | 5.1 | 8 | 140 | 43 | 100 | 0.0012 | 1.68 |
| 173 | KIAA1258 protein | Q9ULG2 | 53.4 | 5.5 | 17 | 91 | 41 | 100 | 0.444 | −1.15 |
| 214 | GLUL protein | P15104 | 42.11 | 6.4 | 15 | 55 | 100 | 0.351 | −1.44 | |
| 197 | Creatine kinase B-type | P12277 | 42.59 | 5.3 | 25 | 149 | 39 | 100 | 0.187 | −2.11 |
| 196 | Creatine kinase, brain | Q6FG40 | 42.61 | 5.3 | 14 | 113 | 43 | 100 | 0.753 | 1.43 |
| 114 | Heat-shock 60 kD protein 1 | P10809 | 61.01 | 5.7 | 25 | 142 | 46 | 100 | 0.192 | −1.72 |
| 331 | 60 kDa heat-shock protein, mitochondrial precursor (Hsp60) | P10809 | 61.01 | 5.7 | 17 | 66 | 35 | 100 | 0.730 | 1.26 |
| 101 | Heat-shock 70 kDa protein 8 isoform 1 | Q53GZ6 | 70.85 | 5.4 | 28 | 118 | 53 | 100 | 0.249 | −1.48 |
Abbreviations: ACTG1, γ-actin 1; CI, confidence interval; GABAA, γ-aminobutyric-acid type A; nos., numbers; pI, isoelectric points; PMF, peptide mass fingerprint.
Statistically significant differences in protein abundance between the groups are shaded. MASCOT score is shown for the searches against the target and also the decoy databases. Average ratios are provided as cocaine:control. Positive and negative ratios indicate that the protein abundance is greater in the cocaine group and control group, respectively. COC/0 refers to proteins present in three or four cocaine subjects and none of the control subjects. 0/CTR refers to proteins present in three or four control subjects and none of the cocaine subjects.
Table 3.
Phospho-proteins identified by amino acid sequencing using MALDI-TOF-MS/MS
| Spot # | Protein ID (MSMS) | Accession # | Peptide sequence | Precursor mass | Mass error (ppm) | MASCOT score | Confidence interval (%) |
|---|---|---|---|---|---|---|---|
| 101 | Heat shock 70 kDa protein 8 isoform 1 | Q53GZ6 | FEELNADLFR | 1253.6162 | 0 | 29 | 97.95 |
| TVTNAVVTVPAYFNDSQR | 1981.9982 | 0 | 35 | 99.49 | |||
| 114 | Heat shock 60 kD protein 1 | P10809 | AAVEEGIVLGGGCALLR | 1684.9053 | 0 | 85 | 100 |
| LVQDVANNTNEEAGDGTTTATVLAR | 2560.2588 | 4 | 29 | 98.49 | |||
| 166 | ATP synthase beta chain, mitochondrial precursor | P06576 | AIAELGIYPAVDPLDSTSR | 1988.0354 | 1 | 42 | 99.91 |
| 172 | Guanine deaminase | Q9Y2T3 | FQNIDFAEEVYTR | 1631.7726 | 2 | 48 | 99.98 |
| 196 | Brain creatine kinase | Q6FG40 | DLFDPIIEDR | 1232.6084 | 6 | 38 | 99.66 |
| VLTPELYAELR | 1303.7258 | 0 | 34 | 99.26 | |||
| 197 | Brain creatine kinase | P12277 | VLTPELYAELR | 1303.7256 | 0 | 54 | 99.98 |
| LAVEALSSLDGDLAGR | 1586.8346 | 2 | 33 | 98.69 | |||
| 204 | Actin, beta | Q96HG5 | QEYDESGPSIVHR | 1516.6962 | 4 | 73 | 100 |
| SYELPDGQVITIGNER | 1790.8821 | 5 | 64 | 99.99 | |||
| VAPEEHPVLLTEAPLNPK | 1954.0564 | 4 | 49 | 99.96 | |||
| 205 | ACTG1 protein | P63261 | QEYDESGPSIVHR | 1516.6975 | 3 | 48 | 99.97 |
| SYELPDGQVITIGNER | 1790.889 | 2 | 72 | 100 | |||
| 232 | Crystallin, mu | Q14894 | LVTFYEDR | 1042.5265 | 6 | 35 | 99.31 |
| VPAFLSAAEVEEHLR | 1667.8766 | 1 | 97 | 100 | |||
| 255 | 14-3-3 gamma protein | P61981 | NVTELNEPLSNEER | 1643.7914 | 3 | 57 | 100 |
| 264 | Rho GDI alpha | P52565 | SIQEIQELDKDDESLR | 1917.9427 | 1 | 35 | 99.17 |
| 267 | Glutathione S-transferase pi Human | P09211 | PPYTVVYFPVR | 1337.7222 | 2 | 66 | 100 |
| 286 | Glutathione S-transferase pi macaca mulatta | Q28514 | PPYTVVYFPVR | 1337.7238 | 1 | 31 | 98.63 |
| FQDGDLTLYQSNTFLR | 1917.9314 | 1 | 29 | 97.88 | |||
| 293 | Peroxiredoxin 2 isoform b | P32119 | QITVNDLPVGR | 1211.6733 | 1 | 49 | 99.97 |
| KEGGLGPLNIPLLADVTR | 1863.067 | 2 | 29 | 97.78 | |||
| 325 | Ubiquitin carboxyl-terminal esterase L1 | O75317 | NEAIQAAHDAVAQEGQCR | 1967.8951 | 3 | 101 | 100 |
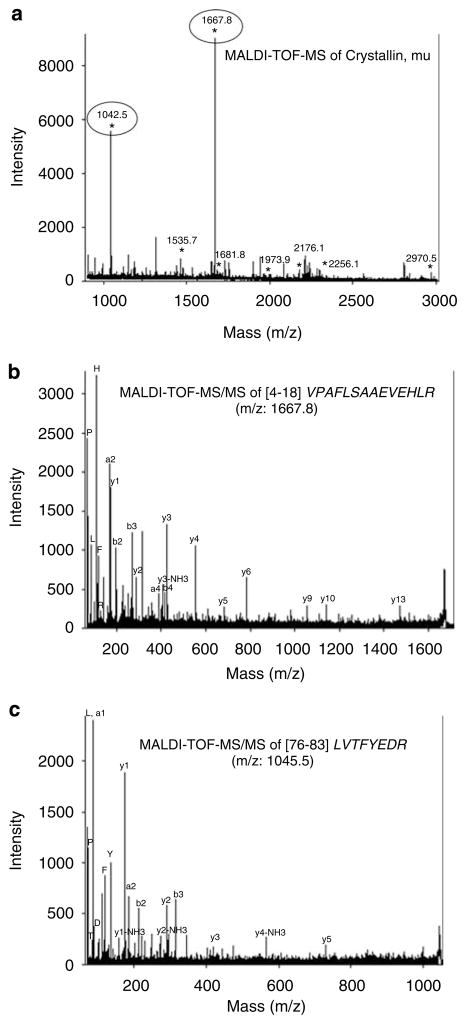
Thirty-six proteins, matched in at least three of four subjects in each group, which were identified by Pro-Q Diamond as phosphorylated proteins between control and cocaine groups. The proteins were identified by MALDI-TOF MS and were corroborated by peptide sequencing using MALDI-TOF-TOF for select peptides. For example, mu crystallin protein was identified by MALDI-MS (Figure 5a) initially and confirmed by amino-acid sequence analysis of its tryptic peptides by MALDI-MS/MS (Figures 5b and c), unequivocally determining the protein’s identity.
Figure 5.
Mass spectrometry identification of mu crystallin. (a) Peptide mass fingerprint of mu crystallin with peptides marked with ‘*’ matched by MASCOT search against the National Center for Biotechnology Information (NCBI) primate database. The x and y axes show the mass/charge (m/z) ratio and the % abundance of the tryptic peptide fragments, respectively. (b) Matrix-assisted laserdesorption ionization time-of-flight tandem mass spectrometry (MALDI-TOF-MS/MS) analysis of peptide fragment from mu crystallin with m/z of 1667.8 was sequenced as VPAFLSAAEVEEHLR. The corresponding y- and b-series ions and also the immonium ions are shown. (c) A similar MALDI-TOF-MS/MS analysis of peptide fragment with m/z 1042.5 was sequenced as LVTFYEDR.
The MS analysis of tryptic digest identified KIAA 1258 protein from protein spot 173 and a BLAST search of the sequence returned guanine deaminase. The identification was confirmed by the following returning parameters for the search: CD-length = 429 residues, 100.0% aligned score = 524 bits (1351), expect = 9e-150. The list of 34 phospho-proteins identified by 2D-PAGE, MS and bioinformatics methods are presented in Table 2 along with an elaboration of the protein identification parameters, MASCOT scores (independent searches against target and decoy databases), spot intensity ratios of all the phospho-proteins and the corresponding P-value. Of the 34 proteins identified by PMF, the identities of 15 proteins was further confirmed by MALDI-MS/MS. We have attempted to organize the differentially expressed proteins into functional categories, fully realizing that such a classification is somewhat arbitrary as proteins could be assigned to more than one functional category. Comparison of results from the decoy and target databases resulted in false-positive protein identification of γ-enolase (spot 180). As noted previously, such proteins were excluded from subsequent analysis.
Discussion
The present study was undertaken to interrogate the cytosolic proteome of the NAc in primate brain as a function of cocaine intake. Previous studies in rodents and human postmortem tissue have demonstrated that cocaine administration can involve transitory and also enduring biochemical alterations in specific brain regions. Furthermore, these changes may be the seat of persistent drug-seeking behavior, craving and also relapse.3,4 This is the first study to provide an unbiased simultaneous assessment of multiple protein abundances and also the post-translational modifications in an animal model of drug abuse and compliments a previous study investigating the cytosolic proteome of the NAc in cocaine overdose victims.23 The use of proteomic technology provides a unique format to query native and post-translationally modified proteomes and determine the proteomic phenotype of cocaine abuse in the primate brain. As in our previous study, subcellular fractionation enabled the quantification of low abundance proteins that would not have been possible by analysis of whole-cell protein homogenates. In the present study, DIGE analysis revealed the differential expression of 18 polypeptides from several protein families including cell structure, synaptic plasticity/signal transduction, metabolism and mitochondrial function. Concurrent analysis of the phosphorylation modifications to the NAc cytosolic proteome of the same subjects revealed significant alterations in complimentary proteins of the aforementioned categories. Notwithstanding, the results of this study provide the first proteomic analysis of chronic cocaine exposure in nonhuman primates and a significant extension of our knowledge of the effects of cocaine in the primate brain.
Structural
The detected isoform of GFAP was significantly increased (53%) in monkeys self-administering cocaine compared to controls. The data compliment a previous report from this laboratory in which GFAP levels in the NAc of cocaine overdose victims were increased on average 40 and 60% above controls levels,23,24 suggesting that such an effect in humans is most likely attributable to the effects of cocaine exposure. GFAP is an intermediate filament protein specific to mature astrocytes and elevated levels of this protein are indicative of astrocytic activation or astrogliosis. Whereas astrocytic activation is known to contribute to neuronal plasticity,48 several studies have suggested that astrocytes may contribute to synaptic plasticity associated with abused drugs.49,50 For example, previous studies have demonstrated elevated GFAP levels in the NAc, ventral tegmental area and hippocampus following various withdrawal periods from cocaine.11,50,51 Alternatively, elevated GFAP levels may reflect a compensatory response, such as stabilization of astrocytic processes52,53 to the effects of cocaine administration. Acute and chronic cocaine administration in mice resulted in increased GFAP immunoreactivity in the dentate gyrus accompanied by changes in astrocyte proliferation and cell morphology.54 The biochemical mechanisms involved in reactive astrocytic gliosis remain to be determined; however, one possible means is activation of dopamine and glutamate receptors on astrocytes leading to stimulation of adenylyl cyclase activity55 and subsequent phosphorylation of transcription factors that bind to the cAMP response elements located on the GFAP promoter region.56 By means of this interaction between astrocytes and neurons,52,57 it is likely that astrocytic cells actively contribute to long-term neuronal changes in response to psychostimulant drugs. Whether these changes are indicative of neurotoxicity or neuroplasticity remains to be determined; however, further understanding of the mechanisms involved in reactive astrocytic gliosis may provide insight into whether such changes are associated with repair and reversal of neuronal damage associated with cocaine abuse.
In addition to increased levels of GFAP, cocaine self-administration also resulted in reduced phosphorylation states of neurofilament (NF) (light polypeptide 68 kDa; −3.57) and β-actin (−2.08). The NF, neuron-specific intermediate filaments (L, M and H corresponding to 68, 160 and 200 kDa) regulate neuronal plasticity by affecting the dynamics and function of cytoskeletal elements such as microtubules and actin thereby regulating neurite outgrowth, axonal caliber and transport. Studies have shown that NR1 and NF-L are colocalized and NF-L appears to be involved in NMDA receptor anchoring and localization.58 More recently, NF-L was demonstrated to increase NMDA receptor expression and prevent NMDA receptor ubiquination.59 Although the role of phosphorylated NF-L on NMDA receptor expression has not been studied, NF-L phosphorylation prevents subsequent NF assembly and induces disassembly in existing NF-L formed filaments.60–62 Nixon and Shiag63 suggest that in postmitotic cells, NF-L phosphorylation prevents polymerization immediately after synthesis (that is increased turnover)—allowing proper integration of NF-M and -H subunits. Therefore, decreased NF-L phosphorylation may be expected to increase NF assembly and stabilize existing filaments—thereby stabilizing NF-L/NR1 interactions. Such an effect would be parsimonious with our recent study demonstrating increased NR1 subunit protein levels in the NAc of cocaine overdose victims and in monkeys self-administering cocaine and increased NR1 subunit phosphorylation in these monkeys as well.24 The lack of detectable change in NF-H phosphorylation is not surprising given that this isoform is the most extensively phosphorylated protein in brain.64 Nonetheless, decreased phosphorylation of NF-L and its known interaction with NMDA receptors provides a mechanism for increased membrane stabilization of NMDA receptors that we have observed following chronic cocaine administration in the NAc of human and nonhuman primate brain.24
Metabolism
Several metabolic proteins were identified as differentially expressed in the NAc following cocaine self-administration subjects in the present study, of particular interest are the proteins involved in the glycolytic pathway and those involved in oxidative regulation (Table 1). Glucose is the principal source of energy for the brain and the oxidation of glucose to pyruvic acid through the glycolytic pathway generates adenosinetriphosphate (ATP) and nicotinamide adenine dinucleotide, sources of energy for cellular function and culminates in the production of pyruvate, which is used in the citric acid cycle for aerobic respiration. Brain-type aldolase C (ALDOC) is involved in the preparatory stage of glycolysis converting fructose 1,6-biphosphate to glyceraldehyde-3 phosphate. Increased levels of phospho-ALDOC observed in the present study suggest a means of enzyme activation that would catalyze the above reaction to produce increased amounts of glyceraldehyde-3 phosphate for the energy-yielding phase of the glycolytic pathway. Interestingly, we found significant downregulation of two members of the enolase family, the family of enzymes involved in the catalysis of phosphoglycerate to phosphoenolpyruvate in the energy-yielding phase of the glycolytic pathway. Levels of γ-enolase, also known as neuron-specific enolase (NSE) and α-enolase, ubiquitously expressed in cell cytoplasm, were significantly decreased in the NAc—in contrast to previous findings in human cocaine overdose victims.23 NSE forms homodimers or heterodimers with α-enolase. The reductions in NSE and α-enolase levels may represent a compensatory mechanism to elevated ALDOC to control glucose utilization in this brain region. These results support the results of a previous study demonstrating decreased glucose utilization in the NAc of rhesus monkeys self-administering cocaine65 and may provide a potential mechanism of this effect. Although not statistically significant, the phosphorylation status of NSE was decreased 1.9-fold in the same monkeys, further suggesting reduced glycolysis in this region following cocaine self-administration.
The phosphorylation status of two proteins involved in oxidative metabolism and identified as differentially expressed in the NAc of monkeys self-administering cocaine were PRDX2 (−3.37) and glutathione S-transferase pi (GSTP1, exclusively expressed in cocaine group). PRDX2 is a highly abundant cytosolic protein primarily involved in redox regulation, which is accomplished by decreasing intracellular peroxides with reducing equivalents from the thioredoxin system. PRDX2 is expressed in neurons, specifically those vulnerable to oxidative stress,66 and functions as the primary regulator of H2O2 generated by cell surface receptors,67 thereby protecting protein and lipids against oxidative stress68 and regulating apoptosis through peroxide elimination. 69 Interestingly, cocaine administration in rodents increases lipid peroxidation,70 and also the presence of reactive oxygen species in striatum and frontal cortex71—both of which indicate the likelihood of H2O2 accumulation and resultant oxidative stress. The present finding of decreased phosphorylation of PRDX2 β-isoform in monkeys corroborates our previous report of decreased protein expression of the PRDX2 α-isoform in the NAc of human cocaine overdose victims.23
Increased levels of phosphorylated GSTP1 isoform may compensate for the reduced phosphorylation of PRDX2. GST activation is one of the principal mechanisms of cellular detoxification catalyzing the conjugation of hydrophobic electrophilic compounds with reduced glutathione and thereby countering the effect of reactive oxygen metabolites.72 Thus, increased phosphorylation of GSTP1 in the NAc of monkeys self-administering cocaine may be a means of protecting cells against potential oxidative damage induced by cocaine. These data compliment a recent study demonstrating increased frequency of a splice variant of GSTP1, which confers increased enzymatic activity and is correlated with cocaine dependence.73 Together, these results indicate that glutathione conjugation is likely a predominant mechanism to attenuate oxidative stress in the NAc induced by cocaine.
Cell signaling
In the present study, four proteins were identified as significantly differentially expressed following cocaine self-administration: β-soluble N-ethylmaleimide-sensitive factor (NSF) attachment protein (β-SNAP; −2.12), syntaxin-binding protein 3 (+1.34), 14-3-3 protein ζ/Δ (−1.31) and protein kinase C epsilon isoform (+1.32). β-SNAP is one of three soluble SNAPs, the β-form of which is brain specific. 74 Targets of NSF-SNAP complexes include SNAP receptors such as synaptobrevin 2, syntaxin, SNAP25, and so on. Disassembly of SNARE complexes on the membrane surface is driven by an ATP-dependent process that requires the NSF-SNAP complex.75 In addition to this more classical function, two additional targets of NSF-SNAP include one in which targets bind directly to NSF (for example, GluR2) and also targets which appear to be isoform specific such as the binding of β-SNAP to synaptotagmin76 or α- and β-SNAP to Pick1 complexed with GluR2.77 The interaction of β-SNAP with GluR2 is particularly intriguing given that previously we have demonstrated increased GluR2 protein levels in membrane fractions from the NAc of monkeys self-administering cocaine.24 Hanley et al.24 have proposed a model of NSF-SNAP regulation of Pick1-AMPA receptor interactions in AMPA receptor cycling. GluR2 subunits that are bound to Pick1 are contained in a receptor pool in which low levels of α/β-SNAP binding disassemble the GluR2/Pick1 complex rendering a portion of GluR2 subunits available for anchoring to the membrane by ABP/GRIP and conversely; high levels of β-SNAP inhibit the NSF-mediated disruption of GluR2/Pick1 interactions leading to internalization of the receptor. Thus, the present findings of downregulation of β-SNAP suggests reduced GluR2 endocytosis in the cocaine monkeys, which may lead to increased levels of the GluR2 subunit at the membrane surface, as reported previously.24
These data are complimented by the identification of two proteins whose phosphorylation status was altered following cocaine self-administration. γ-Aminobutyric-acid type A (GABAA) receptor-associated protein-like 1 (GABARAPL1/GEC1) was recently identified as a member of the protein family that includes GABARAP—sharing 87% identity and 96% similarity. Like GABARAP, GEC1 binds tubulin and exhibits similar subcellular localization.78 GABARAP and GEC1 are biochemical linkers between the GABAA receptor and microtubules and contribute to the intracellular trafficking and membrane insertion of GABAA receptors thereby important in synaptic plasticity.78,79 Given the prevalence of GABAergic synapses in the NAc and their role in the behavioral effects of cocaine, further studies evaluating the mechanisms of GABAA subunit trafficking following cocaine administration are warranted.
Rab proteins are a group of membrane-associated proteins that localize to discrete subcellular compartments and are found in two conformations: bound to GTP in association with membrane and bound to GDP in association with guanine-dissociation inhibitor (GDI). Rab proteins exert their effects on the secretory pathway in which proteins bind Rab proteins specifically in the GTP-bound state.80 In the current study, the phosphorylated form of Rab GDIα was found to be decreased in NAc following cocaine self-administration. Rab GDIα is found predominantly in neural and sensory tissues and acts as a general regulator of all Rab small G proteins implicated in intracellular vesicle trafficking.81,82 Rab GDIα regulates the GDP/GTP exchange reaction of most Rab proteins by inhibiting the dissociation of GDP and the subsequent binding of GTP, implicated in neurotransmission.83 In addition, Rab GDIα has an important in Rab3A recycling to suppress hyperexcitability through modulation of presynaptic forms of plasticity in the hippocampus.84 In the NAc, increased phosphorylation of Rab GDIα may serve as a compensatory mechanism to offset hyperexcitability induced by ionotropic glutamate receptors and increased phosphorylation of NR1 following cocaine self-administration in primates.24
Conclusion
In our continuing effort to understand the neurobiology of cocaine addiction, the current study delineates a preliminary functional proteome of the NAc in rhesus monkeys following chronic self-administration of cocaine and compliments a recent proteomic study of the NAc in human cocaine overdose victims.23 The current study documents quantitative differences in expression and differential phosphorylation of several proteins in the NAc following cocaine self-administration including those involved in cytoskeleton structure, mitochondrial function, energy metabolism and cell signaling. The use of a nonhuman primate model of human cocaine intake to study proteomic alterations associated with long-term cocaine use and abuse offers new perspectives on the associated neuropathology and may provide targets for functional assessments in other models. Determination of the orchestrated changes in proteins, related post-translational modifications and biochemical pathways altered by chronic cocaine intake is essential for providing a comprehensive understanding of the neuropathophysiology associated with drug abuse and necessary for the development of novel pharmacotherapies targeting these proteins.
Supplementary Material
Acknowledgments
This work was supported by National Institutes of Health grants P50 DA006634 (SEH), DA003628 (SEH), DA012498 (SEH), DA016589 (LLH), DA000517 (LLH) and RR000165. We are thankful to Dr Jim Smith for editorial suggestions.
Footnotes
Supplementary Information accompanies the paper on the Molecular Psychiatry website (http://www.nature.com/mp)
References
- 1.SAMHSA. Results from the 2006 National Survey on Drug Use and Health: National Findings. Substance Abuse and Mental Health Services Administration; Rockville, MD: 2007. [Google Scholar]
- 2.WHO. Neuroscience of Psychoactive Substance Use and Dependence. World Health Organization; Geneva: 2004. [Google Scholar]
- 3.Koob GF, Le Moal M. Drug addiction, dysregulation of reward, and allostasis. Neuropsychopharmacology. 2001;24:97–129. doi: 10.1016/S0893-133X(00)00195-0. [DOI] [PubMed] [Google Scholar]
- 4.Nestler EJ. Molecular basis of long-term plasticity underlying addiction. Nat Neurosci Rev. 2001;2:119–128. doi: 10.1038/35053570. [DOI] [PubMed] [Google Scholar]
- 5.Miserendino MJ, Nestler EJ. Behavioral sensitization to cocaine: modulation by the cyclic AMP system in the nucleus accumbens. Brain Res. 1995;674:299–306. doi: 10.1016/0006-8993(95)00030-t. [DOI] [PubMed] [Google Scholar]
- 6.Self DW, Genova LM, Hope BT, Barnhart WJ, Spencer JJ, Nestler EJ. Involvement of cAMP-dependent protein kinase in the nucleus accumbens in cocaine self-administration and relapse of cocaineseeking behavior. J Neurosci. 1998;18:1848–1859. doi: 10.1523/JNEUROSCI.18-05-01848.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Pliakas AM, Carlson RR, Neve RL, Konradi C, Nestler EJ, Carlezon WA., Jr Altered responsiveness to cocaine and increased immobility in the forced swim test associated with elevated cAMP response element-binding protein expression in nucleus accumbens. J Neurosci. 2001;21:7397–7403. doi: 10.1523/JNEUROSCI.21-18-07397.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Carlezon WA, Jr, Thome J, Olson VG, Lane-Ladd SB, Brodkin ES, Hiroi N, et al. Regulation of cocaine reward by CREB. Science. 1998;282:2272–2275. doi: 10.1126/science.282.5397.2272. [DOI] [PubMed] [Google Scholar]
- 9.Hope B, Kosofsky B, Hyman SE, Nestler EJ. Regulation of immediate early gene expression and AP-1 binding in the rat nucleus accumbens by chronic cocaine. Proc Natl Acad Sci USA. 1992;89:5764–5768. doi: 10.1073/pnas.89.13.5764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hiroi N, Brown JR, Haile CN, Ye H, Greenberg ME, Nestler EJ. FosB mutant mice: loss of chronic cocaine induction of Fos-related proteins and heightened sensitivity to cocaine’s psychomotor and rewarding effects. Proc Natl Acad Sci USA. 1997;94:10397–10402. doi: 10.1073/pnas.94.19.10397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Haile CN, Hiroi N, Nestler EJ, Kosten TA. Differential behavioral responses to cocaine are associated with dynamics of mesolimbic dopamine proteins in Lewis and Fischer 344 rats. Synapse. 2001;41:179–190. doi: 10.1002/syn.1073. [DOI] [PubMed] [Google Scholar]
- 12.White FJ, Hu XT, Zhang XF, Wolf ME. Repeated administration of cocaine or amphetamine alters neuronal responses to glutamate in the mesoaccumbens dopamine system. J Pharmacol Exp Ther. 1995;273:445–454. [PubMed] [Google Scholar]
- 13.Hemby SE, Horman B, Tang W. Differential regulation of ionotropic glutamate receptor subunits following cocaine self-administration. Brain Res. 2005;1064:75–82. doi: 10.1016/j.brainres.2005.09.051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Tang W, Wesley M, Freeman WM, Liang B, Hemby SE. Alterations in ionotropic glutamate receptor subunits during binge cocaine self-administration and withdrawal in rats. J Neurochem. 2004;89:1021–1033. doi: 10.1111/j.1471-4159.2004.02392.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Albertson DN, Pruetz B, Schmidt CJ, Kuhn DM, Kapatos G, Bannon MJ. Gene expression profile of the nucleus accumbens of human cocaine abusers: evidence for dysregulation of myelin. J Neurochem. 2004;88:1211–1219. doi: 10.1046/j.1471-4159.2003.02247.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Tang WX, Fasulo WH, Mash DC, Hemby SE. Molecular profiling of midbrain dopamine regions in cocaine overdose victims. J Neurochem. 2003;85:911–924. doi: 10.1046/j.1471-4159.2003.01740.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hemby SE. Morphine-induced alterations in gene expression of calbindin immunopositive neurons in nucleus accumbens shell and core. Neuroscience. 2004;126:689–703. doi: 10.1016/j.neuroscience.2004.01.056. [DOI] [PubMed] [Google Scholar]
- 18.Ang E, Chen J, Zagouras P, Magna H, Holland J, Schaeffer E, et al. Induction of nuclear factor-kappaB in nucleus accumbens by chronic cocaine administration. J Neurochem. 2001;79:221–224. doi: 10.1046/j.1471-4159.2001.00563.x. [DOI] [PubMed] [Google Scholar]
- 19.Yuferov V, Nielsen D, Butelman E, Kreek MJ. Microarray studies of psychostimulant-induced changes in gene expression. Addict Biol. 2005;10:101–118. doi: 10.1080/13556210412331308976. [DOI] [PubMed] [Google Scholar]
- 20.Ahmed SH, Lutjens R, van der Stap LD, Lekic D, Romano-Spica V, Morales M, et al. Gene expression evidence for remodeling of lateral hypothalamic circuitry in cocaine addiction. Proc Natl Acad Sci USA. 2005;102:11533–11538. doi: 10.1073/pnas.0504438102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Backes E, Hemby SE. Discrete cell gene profiling of ventral tegmental dopamine neurons after acute and chronic cocaine self-administration. J Pharmacol Exp Ther. 2003;307:450–459. doi: 10.1124/jpet.103.054965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hemby SE. Assessment of genome and proteome profiles in cocaine abuse. Prog Brain Res. 2006;158:173–195. doi: 10.1016/S0079-6123(06)58009-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Tannu N, Mash DC, Hemby SE. Cytosolic proteomic alterations in the nucleus accumbens of cocaine overdose victims. Mol Psychiatry. 2007;12:55–73. doi: 10.1038/sj.mp.4001914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hemby SE, Tang W, Muly EC, Kuhar MJ, Howell L, Mash DC. Cocaine-induced alterations in nucleus accumbens ionotropic glutamate receptor subunits in human and non-human primates. J Neurochem. 2005;95:1785–1793. doi: 10.1111/j.1471-4159.2005.03517.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zolnierowicz S, Bollen M. Protein phosphorylation and protein phosphatases. De Panne, Belgium, September 19–24, 1999. EMBO J. 2000;19:483–488. doi: 10.1093/emboj/19.4.483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Yamamoto KK, Gonzalez GA, Biggs WH, III, Montminy MR. Phosphorylation-induced binding and transcriptional efficacy of nuclear factor CREB. Nature. 1988;334:494–498. doi: 10.1038/334494a0. [DOI] [PubMed] [Google Scholar]
- 27.Gonzalez GA, Yamamoto KK, Fischer WH, Karr D, Menzel P, Biggs W, III, et al. A cluster of phosphorylation sites on the cyclic AMP-regulated nuclear factor CREB predicted by its sequence. Nature. 1989;337:749–752. doi: 10.1038/337749a0. [DOI] [PubMed] [Google Scholar]
- 28.Creely H, Khaitovich P. Human brain evolution. In: Hemby SE, Bahn S, editors. Progress in Brain Research: Functional Genomics and Proteomics in the Clinical Neurosciences. Elsevier; Amsterdam: 2006. pp. 295–309. [DOI] [PubMed] [Google Scholar]
- 29.Matsumoto I, Alexander-Kaufman K, Iwazaki T, Kashem MA, Matsuda-Matsumoto H. CNS proteomes in alcohol and drug abuse and dependence. Expert Rev Proteomics. 2007;4:539–552. doi: 10.1586/14789450.4.4.539. [DOI] [PubMed] [Google Scholar]
- 30.Howell LL, Wilcox KM. Intravenous drug self-administration in nonhuman primates. In: JJB, editor. Methods of Behavior Analysis in Neuroscience. CRC Press; Boca Raton, FL: 2001. pp. 91–110. [Google Scholar]
- 31.Byrd LD. The behavioral effects of cocaine: rate dependency or rate constancy. Eur J Pharmacol. 1979;56:355–362. doi: 10.1016/0014-2999(79)90266-8. [DOI] [PubMed] [Google Scholar]
- 32.Lindsey KP, Wilcox KM, Votaw JR, Goodman MM, Plisson C, Carroll FI, et al. Effects of dopamine transporter inhibitors on cocaine self-administration in rhesus monkeys: relationship to transporter occupancy determined by positron emission tomography neuroimaging. J Pharmacol Exp Ther. 2004;309:959–969. doi: 10.1124/jpet.103.060293. [DOI] [PubMed] [Google Scholar]
- 33.Paxinos G, Huang X-F, Toga A. The RhesusMonkey Brain in Stereotaxic Coordinates. 1. Academic Press; San Diego, CA: 1999. [Google Scholar]
- 34.Tang W-X, Fasulo WH, Mash DC, Hemby SE. Molecular profiling of midbrain dopamine regions in cocaine overdose victims. J Neurochem. 2003;85:911–924. doi: 10.1046/j.1471-4159.2003.01740.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Churchward MA, Butt RH, Lang JC, Hsu KK, Coorssen JR. Enhanced detergent extraction for analysis of membrane proteomes by two-dimensional gel electrophoresis. Proteome Sci. 2005;3:5. doi: 10.1186/1477-5956-3-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Tannu NS, Hemby SE. Methods for proteomics in neuroscience. In: Hemby SE, Bahn S, editors. Progress in Brain Research: Functional Genomics and Proteomics in the Clinical Neurosciences. Elsevier; Amsterdam: 2006. pp. 41–82. [DOI] [PubMed] [Google Scholar]
- 37.Tannu N, Hemby SE. Two dimensional fluorescence difference gel electrophoresis (2D-DIGE) for comparative proteomics profiling. Nat Protoc. 2006;1:1732–1742. doi: 10.1038/nprot.2006.256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Henzel WJ, Billeci TM, Stults JT, Wong SC, Grimley C, Watanabe C. Identifying proteins from two-dimensional gels by molecular mass searching of peptide fragments in protein sequence databases. Proc Natl Acad Sci USA. 1993;90:5011–5015. doi: 10.1073/pnas.90.11.5011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Mann M, Wilm M. Error-tolerant identification of peptides in sequence databases by peptide sequence tags. Anal Chem. 1994;66:4390–4399. doi: 10.1021/ac00096a002. [DOI] [PubMed] [Google Scholar]
- 40.Yates III., JR Database searching using mass spectrometry data. Electrophoresis. 1998;19:893–900. doi: 10.1002/elps.1150190604. [DOI] [PubMed] [Google Scholar]
- 41.Elias JE, Gygi SP. Target-decoy search strategy for increased confidence in large-scale protein identifications by mass spectrometry. Nat Methods. 2007;4:207–214. doi: 10.1038/nmeth1019. [DOI] [PubMed] [Google Scholar]
- 42.Tannu NS, Hemby SE. De novo protein sequence analysis of Macaca mulatta. BMC Genomics. 2007;8:270. doi: 10.1186/1471-2164-8-270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Martin K, Steinberg TH, Cooley LA, Gee KR, Beechem JM, Patton WF. Quantitative analysis of protein phosphorylation status and protein kinase activity on microarrays using a novel fluorescent phosphorylation sensor dye. Proteomics. 2003;3:1244–1255. doi: 10.1002/pmic.200300445. [DOI] [PubMed] [Google Scholar]
- 44.Steinberg TH, Agnew BJ, Gee KR, Leung WY, Goodman T, Schulenberg B, et al. Global quantitative phosphoprotein analysis using multiplexed proteomics technology. Proteomics. 2003;3:1128–1144. doi: 10.1002/pmic.200300434. [DOI] [PubMed] [Google Scholar]
- 45.Schulenberg B, Goodman TN, Aggeler R, Capaldi RA, Patton WF. Characterization of dynamic and steady-state protein phosphorylation using a fluorescent phosphoprotein gel stain and mass spectrometry. Electrophoresis. 2004;25:2526–2532. doi: 10.1002/elps.200406007. [DOI] [PubMed] [Google Scholar]
- 46.Tannu NS, Rao VK, Chaudhary RM, Giorgianni F, Saeed AE, Gao Y, et al. Comparative proteomes of the proliferating C(2)C(12) myoblasts and fully differentiated myotubes reveal the complexity of the skeletal muscle differentiation program. Mol Cell Proteomics. 2004;3:1065–1082. doi: 10.1074/mcp.M400020-MCP200. [DOI] [PubMed] [Google Scholar]
- 47.Steinberg TH, Agnew BJ, Gee KR, Leung WY, Goodman T, Schulenberg B, et al. Global quantitative phosphoprotein analysis using multiplexed proteomics technology. Proteomics. 2003;3:1128–1144. doi: 10.1002/pmic.200300434. [DOI] [PubMed] [Google Scholar]
- 48.Ullian EM, Christopherson KS, Barres BA. Role for glia in synaptogenesis. Glia. 2004;47:209–216. doi: 10.1002/glia.20082. [DOI] [PubMed] [Google Scholar]
- 49.Narita M, Miyatake M, Shibasaki M, Shindo K, Nakamura A, Kuzumaki N, et al. Direct evidence of astrocytic modulation in the development of rewarding effects induced by drugs of abuse. Neuropsychopharmacology. 2006;31:2476–2488. doi: 10.1038/sj.npp.1301007. [DOI] [PubMed] [Google Scholar]
- 50.Bowers MS, Kalivas PW. Forebrain astroglial plasticity is induced following withdrawal from repeated cocaine administration. Eur J Neurosci. 2003;17:1273–1278. doi: 10.1046/j.1460-9568.2003.02537.x. [DOI] [PubMed] [Google Scholar]
- 51.Beitner-Johnson D, Guitart X, Nestler EJ. Glial fibrillary acidic protein and the mesolimbic dopamine system: regulation by chronic morphine and Lewis-Fischer strain differences in the rat ventral tegmental area. J Neurochem. 1993;61:1766–1773. doi: 10.1111/j.1471-4159.1993.tb09814.x. [DOI] [PubMed] [Google Scholar]
- 52.Haydon PG. Glia: listening and talking to the synapse. Nat Rev Neurosci. 2001;2:185–193. doi: 10.1038/35058528. [DOI] [PubMed] [Google Scholar]
- 53.Weinstein DE, Shelanski ML, Liem RK. Suppression by antisense mRNA demonstrates a requirement for the glial fibrillary acidic protein in the formation of stable astrocytic processes in response to neurons. J Cell Biol. 1991;112:1205–1213. doi: 10.1083/jcb.112.6.1205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Fattore L, Puddu MC, Picciau S, Cappai A, Fratta W, Serra GP, et al. Astroglial in vivo response to cocaine in mouse dentate gyrus: a quantitative and qualitative analysis by confocal microscopy. Neuroscience. 2002;110:1–6. doi: 10.1016/s0306-4522(01)00598-x. [DOI] [PubMed] [Google Scholar]
- 55.Porter JT, McCarthy KD. Astrocytic neurotransmitter receptors in situ and in vivo. Prog Neurobiol. 1997;51:439–455. doi: 10.1016/s0301-0082(96)00068-8. [DOI] [PubMed] [Google Scholar]
- 56.Laping NJ, Teter B, Nichols NR, Rozovsky I, Finch CE. Glial fibrillary acidic protein: regulation by hormones, cytokines, and growth factors. Brain Pathol. 1994;4:259–275. doi: 10.1111/j.1750-3639.1994.tb00841.x. [DOI] [PubMed] [Google Scholar]
- 57.Fields RD, Stevens-Graham B. New insights into neuron-glia communication. Science. 2002;298:556–562. doi: 10.1126/science.298.5593.556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Ehlers MD, Fung ET, O’Brien RJ, Huganir RL. Splice variant-specific interaction of the NMDA receptor subunit NR1 with neuronal intermediate filaments. J Neurosci. 1998;18:720–730. doi: 10.1523/JNEUROSCI.18-02-00720.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Ratnam J, Teichberg VI. Neurofilament-light increases the cell surface expression of the N-methyl-D-aspartate receptor and prevents its ubiquitination. J Neurochem. 2005;92:878–885. doi: 10.1111/j.1471-4159.2004.02936.x. [DOI] [PubMed] [Google Scholar]
- 60.Nakamura Y, Takeda M, Angelides KJ, Tanaka T, Tada K, Nishimura T. Effect of phosphorylation on 68 KDa neurofilament subunit protein assembly by the cyclic AMP dependent protein kinase in vitro. Biochem Biophys Res Commun. 1990;169:744–750. doi: 10.1016/0006-291x(90)90394-3. [DOI] [PubMed] [Google Scholar]
- 61.Gill SR, Wong PC, Monteiro MJ, Cleveland DW. Assembly properties of dominant and recessive mutations in the small mouse neurofilament (NF-L) subunit. J Cell Biol. 1990;111:2005–2019. doi: 10.1083/jcb.111.5.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Sihag RK, Nixon RA. Identification of Ser-55 as a major protein kinase A phosphorylation site on the 70-kDa subunit of neuro-filaments. Early turnover during axonal transport. J Biol Chem. 1991;266:18861–18867. [PubMed] [Google Scholar]
- 63.Nixon RA, Sihag RK. Neurofilament phosphorylation: a new look at regulation and function. Trends Neurosci. 1991;14:501–506. doi: 10.1016/0166-2236(91)90062-y. [DOI] [PubMed] [Google Scholar]
- 64.Petzold A. Neurofilament phosphoforms: surrogate markers for axonal injury, degeneration and loss. J Neurol Sci. 2005;233:183–198. doi: 10.1016/j.jns.2005.03.015. [DOI] [PubMed] [Google Scholar]
- 65.Porrino LJ, Lyons D, Smith HR, Daunais JB, Nader MA. Cocaine self-administration produces a progressive involvement of limbic, association, and sensorimotor striatal domains. J Neurosci. 2004;24:3554–3562. doi: 10.1523/JNEUROSCI.5578-03.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Sarafian TA, Verity MA, Vinters HV, Shih CC, Shi L, Ji XD, et al. Differential expression of peroxiredoxin subtypes in human brain cell types. J Neurosci Res. 1999;56:206–212. [PubMed] [Google Scholar]
- 67.Rhee SG, Chae HZ, Kim K. Peroxiredoxins: a historical overviewand speculative preview of novel mechanisms and emerging concepts in cell signaling. Free Radic Biol Med. 2005;38:1543–1552. doi: 10.1016/j.freeradbiomed.2005.02.026. [DOI] [PubMed] [Google Scholar]
- 68.Yim MB, Chae HZ, Rhee SG, Chock PB, Stadtman ER. On the protective mechanism of the thiol-specific antioxidant enzyme against the oxidative damage of biomacromolecules. J Biol Chem. 1994;269:1621–1626. [PubMed] [Google Scholar]
- 69.Kim H, Lee TH, Park ES, Suh JM, Park SJ, Chung HK, et al. Role of peroxiredoxins in regulating intracellular hydrogen peroxide and hydrogen peroxide-induced apoptosis in thyroid cells. J Biol Chem. 2000;275:18266–18270. doi: 10.1074/jbc.275.24.18266. [DOI] [PubMed] [Google Scholar]
- 70.Kloss MW, Rosen GM, Rauckman EJ. Biotransformation of norcocaine to norcocaine nitroxide by rat brain microsomes. Psychopharmacology (Berl) 1984;84:221–224. doi: 10.1007/BF00427449. [DOI] [PubMed] [Google Scholar]
- 71.Dietrich JB, Mangeol A, Revel MO, Burgun C, Aunis D, Zwiller J. Acute or repeated cocaine administration generates reactive oxygen species and induces antioxidant enzyme activity in dopaminergic rat brain structures. Neuropharmacology. 2005;48:965–974. doi: 10.1016/j.neuropharm.2005.01.018. [DOI] [PubMed] [Google Scholar]
- 72.Landi S. Mammalian class theta GST and differential susceptibility to carcinogens: a review. Mutat Res. 2000;463:247–283. doi: 10.1016/s1383-5742(00)00050-8. [DOI] [PubMed] [Google Scholar]
- 73.Guindalini C, O’Gara C, Laranjeira R, Collier D, Castelo A, Vallada H, et al. A GSTP1 functional variant associated with cocaine dependence in a Brazilian population. Pharmacogenet Genomics. 2005;15:891–893. doi: 10.1097/01213011-200512000-00007. [DOI] [PubMed] [Google Scholar]
- 74.Whiteheart SW, Griff IC, Brunner M, Clary DO, Mayer T, Buhrow SA, et al. SNAP family of NSF attachment proteins includes a brain-specific isoform. Nature. 1993;362:353–355. doi: 10.1038/362353a0. [DOI] [PubMed] [Google Scholar]
- 75.Jahn R, Lang T, Sudhof TC. Membrane fusion. Cell. 2003;112:519–533. doi: 10.1016/s0092-8674(03)00112-0. [DOI] [PubMed] [Google Scholar]
- 76.Schiavo G, Gmachl MJ, Stenbeck G, Sollner TH, Rothman JE. A possible docking and fusion particle for synaptic transmission. Nature. 1995;378:733–736. doi: 10.1038/378733a0. [DOI] [PubMed] [Google Scholar]
- 77.Hanley JG, Khatri L, Hanson PI, Ziff EB. NSF ATPase and alpha-/beta-SNAPs disassemble the AMPA receptor-PICK1 complex. Neuron. 2002;34:53–67. doi: 10.1016/s0896-6273(02)00638-4. [DOI] [PubMed] [Google Scholar]
- 78.Mansuy V, Boireau W, Fraichard A, Schlick JL, Jouvenot M, Delage-Mourroux R. GEC1, a protein related to GABARAP, interacts with tubulin and GABA(A) receptor. Biochem Biophys Res Commun. 2004;325:639–648. doi: 10.1016/j.bbrc.2004.10.072. [DOI] [PubMed] [Google Scholar]
- 79.Wang H, Olsen RW. Binding of the GABA(A) receptor-associated protein (GABARAP) to microtubules and microfilaments suggests involvement of the cytoskeleton in GABARAPGABA(A) receptor interaction. J Neurochem. 2000;75:644–655. doi: 10.1046/j.1471-4159.2000.0750644.x. [DOI] [PubMed] [Google Scholar]
- 80.Stenmark H, Olkkonen VM. The Rab GTPase family. Genome Biol. 2001;2:REVIEWS3007. doi: 10.1186/gb-2001-2-5-reviews3007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Pfeffer SR, Dirac-Svejstrup AB, Soldati T. Rab GDP dissociation inhibitor: putting rab GTPases in the right place. J Biol Chem. 1995;270:17057–17059. doi: 10.1074/jbc.270.29.17057. [DOI] [PubMed] [Google Scholar]
- 82.Novick P, Zerial M. The diversity of Rab proteins in vesicle transport. Curr Opin Cell Biol. 1997;9:496–504. doi: 10.1016/s0955-0674(97)80025-7. [DOI] [PubMed] [Google Scholar]
- 83.Takai Y, Sasaki T, Shirataki H, Nakanishi H. Rab3A small GTP-binding protein in Ca(2+)-dependent exocytosis. Genes Cells. 1996;1:615–632. doi: 10.1046/j.1365-2443.1996.00257.x. [DOI] [PubMed] [Google Scholar]
- 84.Ishizaki H, Miyoshi J, Kamiya H, Togawa A, Tanaka M, Sasaki T, et al. Role of rab GDP dissociation inhibitor alpha in regulating plasticity of hippocampal neurotransmission. Proc Natl Acad Sci USA. 2000;97:11587–11592. doi: 10.1073/pnas.97.21.11587. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.