Abstract
Cancer cells with p53 mutations, in general, grow more aggressively than those with wild-type p53 and show “gain of function”(GOF) phenotypes such as increased growth rate, enhanced resistance to chemotherapeutic drugs, increased cell motility and tumorigenicity; although the mechanism for this function remains unknown. In this communication we report that p53-mediated NF-κB2 up-regulation significantly contributes to the aggressive oncogenic behavior of cancer cells. Lowering the level of mutant p53 in a number of cancer cell lines resulted in a loss of GOF phenotypes directly implicating p53 mutants in the process. RNAi against NF-κB2 in naturally occurring cancer cell lines also lowers GOF activities. In H1299 cells expressing mutant p53, chromatin immunoprecipitation (ChIP) assays indicate that mutant p53 induces histone acetylation at specific sites on the regulatory regions of its target genes. ChIP assays using antibodies against transcription factors putatively capable of interacting with the NF-κB2 promoter show increased interaction of CBP and STAT2 in the presence of mutant p53. Thus, we propose that in H1299 cells, mutant p53 elevates expression of genes capable of enhancing cell proliferation, motility, and tumorigenicity by inducing acetylation of histones via recruitment of CBP and STAT2 on the promoters causing CBP-mediated histone acetylation.
Keywords: Mutant p53, gain of function, transactivation
Introduction
p53 mutants found in cancer cells are largely defective in wild-type (WT) p53 functions such as apoptosis and cell cycle arrest [1–3] mostly as a result of loss of the sequence-specific transactivation functions of WT p53. We and others have shown that mutant p53 can transactivate promoters of cellular growth-related genes [4, 5], independent of the presence of the WT p53 DNA-binding site [6, 7]. We have identified several important cell growth and survival related genes whose expression seems to be regulated by three common p53 mutants (p53-R175H, -R273H and -D281G) [8, 9]. Other laboratories have also reported genes that are influenced by p53 mutants [10–13].
Mutant p53 expression has been shown to result in oncogenic and proliferative processes such as increased tumorigenicity [14, 15], increased metastasis and invasiveness [16], increased growth in soft agar [17], decreased sensitivity to chemotherapeutic drugs [8, 18, 19], increased resistance to γ-irradiation [20], accelerated chemical carcinogenesis [21], disruption of the spindle checkpoint [22, 23], activated topoisomerase I activity [24], increased growth rate [25] and induction of gene amplification [26]. Many of the gain of function (GOF) data come from p53-null systems where the re-expressed mutant p53 levels were comparable to those observed in cancer cells [16]. This suggests the strong possibility that GOF has a true patho-physiological role, which may lead to aggressive cancer development and poorer prognosis. The molecular mechanism of GOF phenotypes and up-regulation of gene expression by mutant p53 has not been clarified yet.
WT p53 induces apoptosis after DNA damage caused by cytotoxic drugs, and it appears that a cell’s p53 mutational status may determine the efficacy of many of these drugs [11, 18, 19]. It has been shown that mutant p53 expression (in cells devoid of WT p53) can lead to decreased sensitivity to drugs such as doxorubicin, etoposide, cisplatin and others [18, 19]. This can be partially explained by an effect of mutant p53 on p73 and p63 [27, 28]. In a previous study, we presented evidence to show that part of this chemoresistance may also arise as a result of up-regulation of p52/p100 NF-κB2 by mutant p53 [8].
The NF-κB family of transcription factors regulates expression of many genes involved in growth, differentiation, survival, development and inflammation [29, 30]. In mammals this group has five members: Rel A (p65), Rel B, c-Rel, p50/p105 (NF-κB1) and p52/p100 (NF-κB2) [31]; the factors function primarily as p50/Rel or p52/Rel heterodimers, although they may also function as different homo- and heterodimers. The NF-κB2 protein is synthesized as p100 precursor that gets processed to p52 for more functional activities upon activation of the pathway. p52 over-expression can lead to lymphocyte hyperplasia and transformation [32]. Constitutive nuclear NF-κB activity has emerged as a hallmark for many human leukemias, lymphomas, and several other cancers [33].
Inhibition of NF-κB sensitizes many tumor cells to death-inducing stimuli, including chemotherapeutic agents [33–37]. Thus, transactivation of NF-κB could be a crucial step by which mutant p53 induces oncogenic progression. Activation of NF-κB appears to protect tumor cells from apoptosis, through induction of anti-apoptotic genes [38, 39], while p52 over-expression has been shown to inhibit pro-apoptotic genes as well [40].
In this study, using a number of cancer cell lines expressing mutant p53 we show that GOF activities are dependent on the p53 level. We also demonstrate that up-regulation of NF-κB2 in H1299 lung cancer cells expressing mutant p53 is caused by changes in chromatin structure on the NF-κB2 promoter, and increasing interaction of crucial transcription factors with the promoter.
Materials and methods
Cell lines
Five human lung cancer cells lines: H23 (p53-M246I, NSCLC), H1048 (p53-R273C, SCLC), H1437 (p53-R267P, NSCLC), KNS-62 (p53-R249S, NSCLC), H1299 (p53-null, NSCLC); two human breast cancer cell lines: MDA-MB-468 (p53-R273H) and SK-BR-3 (p53-R175H); and a human melanoma cell line MDA-MB-435 (p53-G266E) were used in these studies, and grown in media prescribed by ATCC.
Generation of stable cell lines
Stable cell lines were generated after transfection of p53-null H1299 lung carcinoma cells with mutant p53 expression plasmids (or expression vector alone), which contain a neomycin resistance gene as described [15]. Mutant p53 knock down cell lines were generated by using lentivirus expressing short hairpin RNA (shRNA) against p53 utilizing lentivirus systems (Open Biosystems) following the manufacturer protocol. Clones were isolated using puromycin selection at 1–3 μg/ml.
Drug sensitivity assay
Drug sensitivity assays were carried out as described by us earlier [41]. In general, cancer cells expressing mutant p53 were plated at 10,000 cells/10cm dish and treated with a final concentration of 1–6 uM etoposide (Sigma; St. Louis, MO); concentration of the drug used depended on the cell lines used and preliminary earlier experiments. In our hands different cell lines respond differently towards the same concentration of etoposide as far as cell death is concerned as measured by colony formation assays. We have done preliminary experiments where we used different concentrations of the drug to determine the sensitivity for individual cell lines. These experiments dictated what concentration we ultimately chose for the final experiment the data of which we have presented. For control, in order to avoid over-crowding, we plated 1000 cells/10cm dish treated with DMSO vehicle. The number of cells used per assay varies depending on the cell line, its plating efficiency of the cells and sensitivity towards the chemotherapeutic drug under consideration; but for the same cell line and experiment we used identical number of cells.
After drug/DMSO treatment, plates were washed and the media replaced. The surviving cells were allowed to form colonies for 2–3 weeks with periodic changes of media. Colonies were fixed with methanol, stained with methylene blue and counted as described earlier [41]. The percent survival was calculated by dividing the average number of drug-treated colonies by the average number of DMSO-treated colonies and multiplying by 100. This was done for each siRNA treatment, either si-control or si-p53. All experiments were done in triplicate, and repeated multiple times. The error bars represent standard deviation from the average number of colonies counted.
Growth assay
Growth assays were carried out as described by us earlier with slight modifications [8, 9]. Cells were plated at 50,000 cells/6cm dish in triplicate for five time points and harvested after incubation with trypsin and counted using a Coulter Counter (Beckman). Multiple cell clones were used for each assay. All experiments were done in triplicate, and repeated multiple times. The error bars represent standard deviation from the triplicates.
PARP cleavage assay
To determine if p53 knock down results in apoptotic cell death, we performed a transient infection with lentivirus expressing p53 shRNA (or GFP shRNA) to knock down mutant p53 in H1048 lung cancer cells expressing mutant p53 and assess apoptosis without selection. For the PARP Western blot experiments we used H1048 cells treated with etoposide (9 uM) for 48 hours as a positive control for apoptosis. The assay was carried out using an antibody from Cell Signalling.
Xenograft assay
Nu/nu mice were used for the tumorigenicity studies. For all injections, 1x107 cells/250μl media were used. Mice were injected subcutaneously on the flanks and tumors allowed to grow to a maximum size of 1cm, measuring periodically as described before (17, 18). Three different clones of H1048 cells with mutant p53 levels reduced by shRNA were used in comparison to two GFP shRNA control cell lines to rule out clonal variations.
siRNA transfection
Breast or lung cancer cells were nucleofected with siRNA directed against a specific or non-specific gene (luciferase) using a nucleofector and a nucleofector kit following the manufacturer’s protocol (Amaxa Inc.; Gaithersburg, MD). Sequences used were: Control (C): 5’-CAU GUC AUG UGU CAC AUC ACT T -3’ and 5’-GAG AUG UGA CAC AUG ACA UGT T -3’, p53: 5‘-GCA UGA ACC GGA GGC CCA UTT-3‘ and 5‘-AUG GGC CUC CGG UUC AUG CTT-3‘ [42] and NF-κB2: 5’-GCC CUG AGU GCC UGG AUC UTT-3’ and 5’-CGG GAC UCA CGG ACC UAG ATT-3’. Twenty-four hours after nucleofection, cells were trypsinized, counted, plated and then exposed to etoposide or DMSO as a control for 48h and colony formation assays performed as described above.
Chromatin immunoprecipitation
Chromatin immunoprecipitations (ChIP) were performed as described [9, 43]. To crosslink protein and DNA, cell cultures were incubated in 2% formaldehyde for 10min at ambient temperature and then 200mM glycine was added for a further 10min. Cells were washed in cold PBS, scraped and centrifuged. Pellets were resuspended in lysis buffer containing 1% protease inhibitors and then sheared by multiple passages through a 27.5 gauge needle followed by 25min of sonication on ice such that the chromatin was fragmented to 500–2000 bp length. Following centrifugation, the protein content of the supernatants was determined and equal amounts used for immunoprecipitation overnight at 4ºC with gentle tilting with anti-acetylated histone H3 antibody or IgG as a control. Immune complexes were captured using Protein A-agarose, then washed sequentially in RIPA buffer (150mM NaCl, 50mM Tris pH8, 0.1% SDS, 0.5% sodium deoxycholate, 1% NP-40), high salt buffer (500mM NaCl, 50mM Tris pH8, 0.1% SDS, 1% NP-40), twice in LiCl buffer (250mM LiCl, 50mM Tris pH8, 0.5% sodium deoxycholate, 1% NP-40) then twice in TE buffer. Protein was eluted from beads in fresh elution buffer (20% SDS, 10mM DTT, 100mM NaHCO3), crosslinking reversed overnight at 65ºC in the presence of NaCl, and then samples were ethanol-precipitated. Following centrifugation, pellets were resuspended in TE buffer and incubated sequentially with 10mg/ml RNase A (30min) and 20mg/ml proteinase K (1h). Samples were phenol-extracted, ethanol-precipitated, and the pellets washed in 70% ethanol, dried and resuspended in sterile water. Acetyl histone H3 Ab and normal rabbit IgG (17-615) were from Millipore; CREB (sc-186), c-Rel (sc-272), NFkB p65 (sc-372), STAT2 (sc-476), p53 DO1 (sc-126) and AcH4 K16 (sc-8662) were from Santa Cruz Biotechnology, Santa Cruz, CA; and p53 (9282) was from Cell Signaling. Quantitative PCR (QPCR) was used to quantify precipitated NF-κB2 promoter-specific DNA segments. We have performed two sets of QPCR: one with NF-κB2 specific primers and another with GAPDH specific primers. The second set of QPCR (with GAPDH) has been done to normalize the NF-κB2 values as GAPDH expression remains unchanged by p53. Primers used to analyze ChIP samples were: GAPDH F: 5’-GTC AAC GGA TTT GGT CGT ATT-3’ and R: 5’-GAT CTC GCT CCT GGA AGA TGG-3’; NF-κB2 F: 5’-GAG GGA GGA GGG GGC TTA ACC C-3’ and R: 5’-CGG GAG GCC CTC GAC AGT CTA C-3’; MCM6 F: 5’-TTT GCT TAC TGC CGA GGA TT-3’ and R: 5’-GCC GTT CAT TGG TCA GGT-3’; EBAG9 F: 5’-AAA TTT GCG TGA CCT TAC TG-3’ and R: 5’-TGC ACA GAT GAG AAA AAC AG-3’; Cyclin B2 F: 5’-AAC TTC CGC CCA CCC ACT ACA AAC-3’ and R: 5’-CAC TCT CGC ACT CTC ATT GGC TGA A-3’; and Integrin alpha 6 F: 5’-CGC TGT GAT CAT TTT TGA GGG TTG T-3’ and R: 5’-GGA CAG AAT TGT GGT TGC CGA GTA G-3’.
Luciferase reporter assays
H1299 cells were transfected in triplicate with 200ng of the promoter-luciferase reporter constructs and 1μg of vector only (pCMVBam) or p53 expression plasmid for 48 h [44]. After transfection, cells were harvested and luciferase activity measured using the Promega luciferase assay kit (#E1500, Promega, Madison, WI) according to the manufacturer’s instructions. Cell extracts were normalized to each other based on total protein concentration and luciferase activity detected using a Luminometer from Turner Designs.
Western blotting
Immunoblottings were carried out as described [8]. NF-κB2 levels were detected using an antibody from Upstate Biotechnology (#05-361; Charlottesville, VA). Actin levels were detected using the AC-15 antibody (Sigma; St. Louis, MO), p53 was detected using the p53 antibody PAb 1801, and Erk2 was detected using ERK2 (sc-154) antibody from Santa Cruz Biotechnology. Westerns blots were developed by the ECL method (GE Healthcare; Piscataway, NJ). SK-BR-3 blot was developed using Li-COR system as described [43].
Migration assays
Cell migration was carried out using wound-healing (scratch) assays, as previously described [45]. Briefly, cells were trypsinized, plated in quadruplicate in 12-well cell culture plates and incubated at 37°C until cells were completely confluent. At this time, a sterile pipette tip was used to scratch across the surface of the plate, removing the complete layer of cells within the scratch area. Following cell removal, each well was washed once with PBS and then replaced with growth medium. Immediately following, the width of the scratch was measured at six specific points under a 5x objective using a light microscope and AxioVision software (Carl Zeiss Microimaging, Thornwood, NY). Cells were incubated at 37°C from 20-60h depending on the cell line under study, at which time the scratch width was measured at the same position as at time 0.
Apoptosis-DNA Ladder Assay
H1048 cells stably expressing an shRNA against p53 or GFP were used for this assay along with a positive control (U937 cells treated with camptothecin, provided in the kit from Roche cat. # 11835246001). Two million cells were used for preparation of DNA for each sample. Cells were lysed with the binding/lysis buffer, and the lysate applied to filter tubes which contain glass fiber fleece to bind nucleic acids. Impurities are removed by washing and DNA is eluted off of the columns. 2μg of each DNA sample was run in a 1% agarose TBE gel and run at 68V until full separation of the positive control DNA ladder was visible.
Experimental and Results
Reducing p53 levels by RNAi in cancer cells with mutant p53 lowers gain of function activities
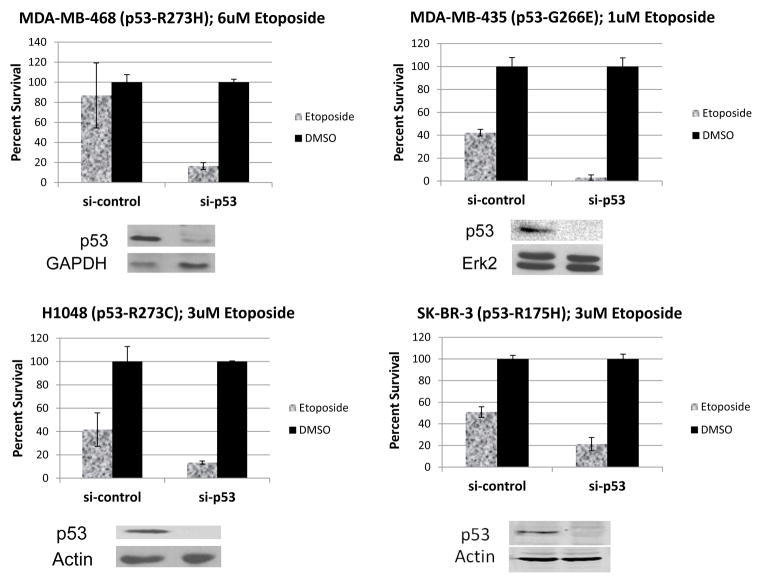
If mutant p53 expression in cancer cells induces GOF phenotypes such as increased tumorigenicity, growth rate, chemoresistance and motility, we hypothesized that by reducing the level of mutant p53 in those cells we should be able to see reduction of these properties. Therefore, we wanted to test whether that principle holds using cancer cell lines expressing mutant p53. Figure 1 shows the results of colony formation assays performed with cell lines MDA-MB-468, MDA-MB-435, H1048 and SK-Br-3, after treatment with etoposide or DMSO. Cells were treated with the drug or DMSO for 48h after being nucleofected with siRNA specific for p53 or control (scrambled or luciferase) as described in Materials and methods. The data demonstrate that these cell lines become more chemosensitive when the mutant p53 level is reduced; thus the p53 mutants in these cells show GOF activity.
Figure 1. Reducing p53 levels in cancer cells with mutant p53 by RNAi lowers chemoresistance.
The indicated cells were transfected with control (si-luc or si-scrambled) or p53-specific siRNA, treated with appropriate concentrations of etoposide for 48h depending on the sensitivity of the cell line to the drug, and colony formation determined (upper panels) as described in Materials and methods. Immunoblots show the efficacy of p53 siRNA treatment. Erk2, GAPDH, or actin was used as a loading control (lower panels). Percent survival is shown in the figures with error bars representing standard deviation from the average number of colonies. The percent survival was calculated by dividing the average number of drug-treated colonies by the average number of DMSO-treated colonies and multiplying by 100. Experiments were done in triplicate and data shown are representative of multiple independent repeats.
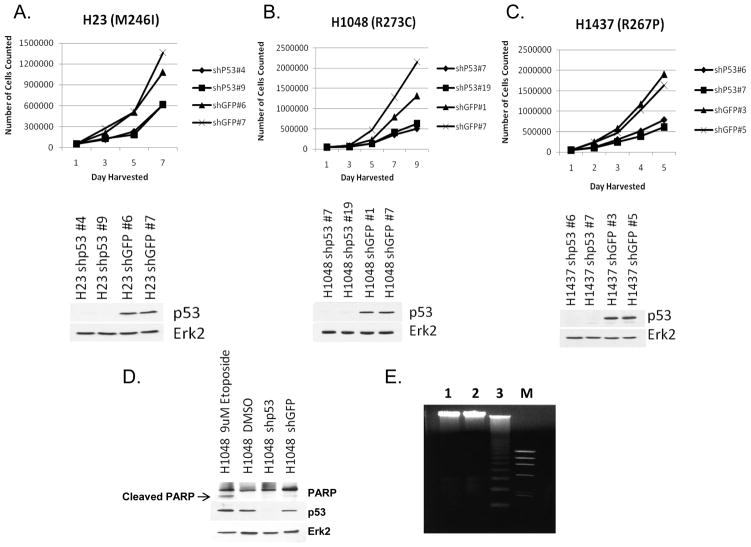
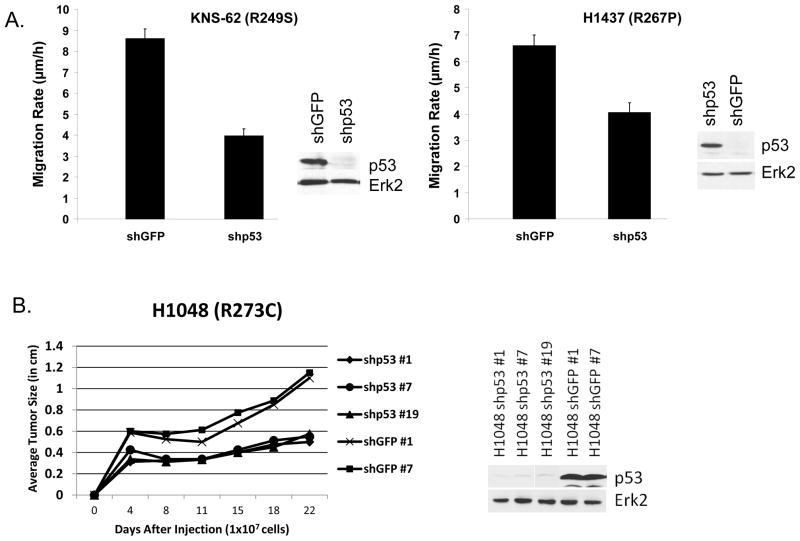
Figure 2 shows growth assay data of cancer lines H23, H1048 and H1437 when their p53 levels were reduced by lentivirus infection expressing shp53, compared to shGFP controls. The data indicate that reduction of mutant p53 level has a significant effect of reducing the growth rate of H23, H1048 and H1437 lung cancer cells. Thus mutant p53 controls the rate of growth of cancer cells expressing GOF mutant p53. We carried out a PARP cleavage assay (to detect apoptosis), which showed no significant differences between the shGFP and shp53 lentivirus infected H1048 cells (Figure 2D) suggesting slower proliferation instead of induction of apoptosis. We have also performed a DNA degradation assay using a kit from Roche (Inc.) using H1048 cells expressing shRNA for GFP or p53 to further detect apoptosis. The data shown in Figure 2E indicated that while in the positive control we could see nice DNA ladder formation, H1048 cells (expressing shRNA for GFP or p53) did not show any indication of apoptosis corroborating our earlier data. This conclusively shows that mutant p53 reduction in H1048 cells does not cause apoptosis. Figure 3A shows motility assays of KNS-62 and H1473 lung cancer cells after their p53 levels were knocked down by lentivirus expressing p53 shRNA, compared to shGFP controls. The data indicate that reduction of mutant p53 level decreases cell motility. Thus, mutant p53 contributes to the increased motility rate of cancer cells expressing GOF mutant p53.
Figure 2. p53 knock down causes decrease in growth rate in cancer cell lines.
A–C Mutant p53 levels were knocked down in three cancer cell lines by lentivirus expressing p53 shRNA as indicated and their growth rates measured in comparison to the corresponding cell lines generated after infecting with lentivirus expressing GFP shRNA as described in Materials and methods. Cells were harvested every other day and cell numbers were determined by Coulter Counter. Error bars represent standard deviation between the average cell counts. Experiments were done in triplicate, and data are representative of multiple independent repeats. Immunoblots show the efficacy of p53 knock down. ERK2 was used as a loading control. D. For the H1048 cells, we performed a PARP cleavage assay to check for caspase activation using PARP antibody from Cell Signaling. E. Apoptosis DNA-Ladder assay of H1048 cells stably transfected with shp53 (1) or shGFP (2). U937 cells after treatment with CAM (3) were used as a positive control for apoptosis.
Figure 3. p53 knock down in cancer cell lines with naturally occurring p53 mutations causes a decrease in rate of motility and tumor growth.
A. The indicated cell lines were cultured to confluence, a scratch made in the monolayer, and the distance measured as described in Methods. p>0.001 for both. Two mutant p53 knocked down clones (confirmed by Western blot, as indicated) were used for each cell line with the average migration between the clones shown. Error bars represent standard deviation from the mean migration rate. B. Mutant p53 levels were knocked down in the lung cancer cell line H1048 (p53-R273C) by lentivirus expressing p53 shRNA or GFP shRNA (used as our control cell line) as indicated and its tumorigenic ability measured after subcutaneous injection into nude mice. For all injections, 1x107 cells/250μl media were used. Mice were injected subcutaneously and tumors allowed to grow to a maximum size of 1cm, measuring periodically as described before (17, 18). Three different clones of H1048 cells with mutant p53 levels knocked down by shRNA were used in comparison to two GFP shRNA control cell lines to eliminate clonal variations. Average tumor size was calculated by taking the average of the width and length of each tumor, then taking the average of all tumors from the particular cell line.
Figure 3B shows tumorigenicity assays of H1048 lung cancer cells when their p53 levels were knocked down by lentivirus expressing p53 shRNA, compared to shGFP controls. The data indicate that inhibiting mutant p53 expression leads to a reduction in tumor growth rate. Thus mutant p53 controls the rate of tumorigenicity of cancer cells expressing GOF mutant p53. We used three shp53 clones and two shGFP clones to rule out clonal variations, and got similar results. Taken together, these studies demonstrate that p53 mutants contribute to multiple GOF phenotypes in a wide range of cancer cell lines with endogenous p53 mutations.
Thus, different cell lines with different endogenous mutant p53 tested show similar loss of GOF activities in reducing the level of mutant p53 (Figures 1–3), suggesting the generality of the conclusion that mutant p53 levels determine GOF phenotypes.
Reducing NF-κB2 levels in cancer cells with mutant p53 by RNAi lowers GOF activity of mutant p53-expressing cells
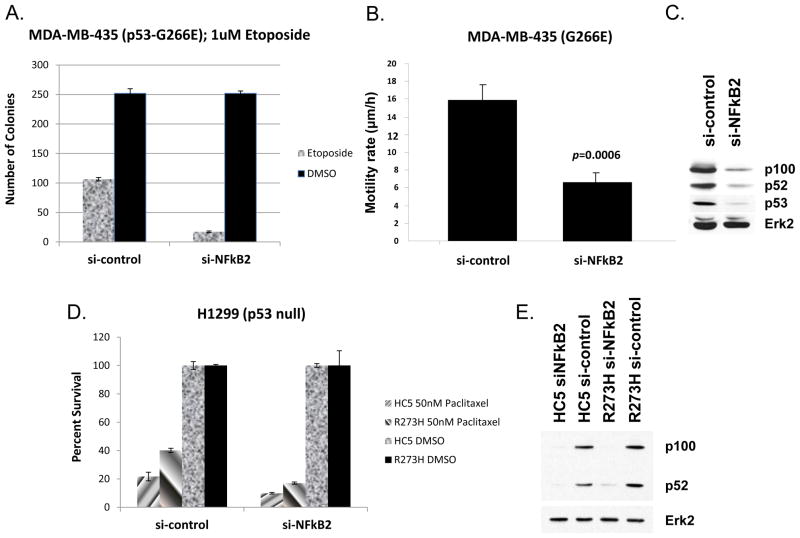
We proposed earlier that mutant p53-induced GOF activity (such as chemoresistance) in cancer cells may be due to induction of NF-κB2 by mutant p53. If that is the case then reducing the level of NF-κB2 in cancer cell lines with mutant p53 should reduce chemoresistance. Since reduction of mutant p53 levels reduces GOF activities in a number of cancer cell lines, we tested the idea whether a similar effect can be seen by reducing the level of NF-κB2. RNAi directed against NF-κB2 using MDA-MB-435 melanoma cells harboring a p53-G266E substitution (one of the cell lines tested in p53 knock down experiments above) was used for this. The data presented in Figure 4A and 4C show that RNAi directed against NF-κB2 reduces the level of expression of the corresponding gene, and reduces resistance to the chemotherapeutic drug etoposide substantially versus DMSO. Furthermore, cell motility – another GOF p53 phenotype – is significantly inhibited by NF-κB2 RNAi (Figure 4B). Lowering of NF-κB2 lowers p53 level a bit also; this is possibly because of the fact that p53 promoter sequences have a prominent NF-κB site and it responds to NF-κB activity [46]. p53 RNAi also inhibited cell migration (not shown), consistent with results with NF-κB2 RNAi. Thus, NF-κB2 at least partially regulates two mutant p53 GOF phenotypes in these cells, suggesting the possibility that mutant p53 may induce its GOF activity via NF-κB2 up-regulation, an observation we reached by expressing mutant p53 in H1299 lung carcinoma cell lines that are normally p53 null [47]. Figures 4D and 4E show reduced resistance to the chemotherapeutic drug paclitaxel after treatment with RNAi against NF-κB2 in H1299 cells expressing either an empty vector (HC5) or the p53 mutant R273H, and Western analysis showing decreased NF-κB2 protein expression. Our data obtained by p53 knock down experiments, using cancer cell lines with naturally occurring mutant p53, and ectopic expression of mutant p53, in p53 null lung cancer cells, are similar, and interchangeable in this respect. Having determined that, we decided to determine the mechanism of up-regulation of NF-κB2 by mutant p53 using H1299 cell systems where mutant p53 was expressed (see below).
Figure 4. Reduction of NF-κB2 causes reduction of drug sensitivity, growth rate and rate of and motility.
A. Chemoresistance of MDA-MB-435 cells depends on the NF-κB2 level. The indicated cells were transfected with control or NF-κB2-specific siRNA, treated with etoposide, and colony formation determined as described in Materials and methods (upper panels). Percent survival is shown in the figure with error bars representing standard deviation from the average number of colonies from triplicate samples. B. Motility of MDA-MB-435 cells depends on the NF-κB2 level. MDA-MB-435 cells were transfected as above. 48h later, standard Transwell migration assays were carried out, and migrating cells stained and counted as described [52]. Bar = +/− 1SD from the mean of 18 samples. C. NF-κB2 and Erk2 levels were determined by immunoblotting for MDA-MB-435 cells used in A and B. D. H1299 cells expressing an empty vector (HC5) or p53-R273H were treated with control (si-scrambled) or RNAi against NF-κB2, treated with paclitaxel, and colony formation determined as described in Materials and methods. E. Western analysis of NF-κB2 knock down by siRNA in H1299 cells expressing an empty vector or p53-R273H.
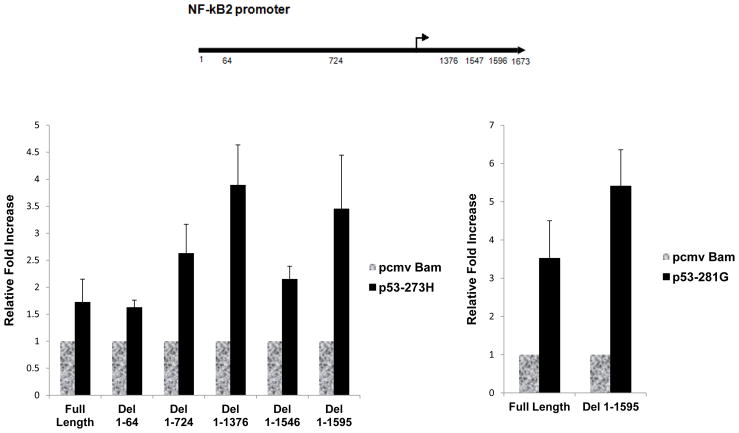
Promoter deletion analysis by transient transcription assays does not indicate a mutant p53 response element
To test whether we could detect a mutant p53 response element on the NF-κB2 promoter, we generated a number of deletion mutants of the NF-κB2 promoter [8] by PCR and subcloned them into the pGL3-basic plasmid vector (Promega). We tested their promoter function by transient transfection in H1299 p53-null lung carcinoma cells as described [8]. After harvesting the cells at 48 h post-transfection, we prepared extracts and determined the luciferase activity as described [8]. Data presented in Figure 5 show that none of the deletion mutants eliminated the transactivation capacity of mutant p53–R273H. Some of the deletions resulted in higher transactivation by mutant p53 suggesting that some sequences of the promoter may act negatively on transactivation; the reason for that needs further investigation. We also tested another mutant, p53-D281G, with the full-length and the largest promoter deletion mutant. Both were transactivated, showing that this sequence independence has generality as far as mutant p53 is concerned. Thus, transient transcriptional analysis failed to provide evidence for a mutant p53 response element on the NF-κB2 promoter. These data are in agreement with those published by us earlier [7] where EGFR promoter deletion did not result in identification of a mutant p53 response element either. Therefore, it is possible that the gene specificity is attained at the chromatin level.
Figure 5. Promoter deletion analysis by transient transcription analysis does not indicate a mutant p53 response element.
A number of NF-κB2 [8] promoter deletion mutants were generated by PCR, and their promoter activity was measured by transient transcriptional analysis using H1299 p53-null lung cancer cell lines transfected with empty vector as control, or p53-R273H (left panel) or p53-D281G (right panel). Forty-eight hours after transfection, cells were harvested and luciferase assays were performed using equal amounts of protein. The figure shows a representative example of multiple experiments. Relative fold of activation in luciferase activity has been plotted compared to that obtained by vector alone. Error bars indicate deviations in luciferase readings relative to vector transfected cells. All experiments were done in triplicate.
ChIP analysis indicates mutant p53 induced histone acetylation on the NF-κB2 promoter
Because mutant p53 can demonstrate GOF activity in different cells, and presumably one of its important targets is NF-κB2, we used H1299 cells in which mutant p53-R273H is expressed as a model to explore the mechanism of up-regulation. We wanted to test whether mutant p53 alters the chromatin structure on the endogenous NF-κB2 promoter in H1299 cells. Therefore, we performed ChIP assays using H1299 cells stably transfected with empty vector alone (HC5) or H1299 cells expressing mutant p53-R273H. The data shown in Figure 6A demonstrate that mutant p53 induces formation of acetylated histone H4 (but not acetylated histone H3) on the NF-κB2 promoter in H1299 cells expressing mutant p53–R273H, suggesting that the up-regulation of NF-κB2 expression by mutant p53 seen in these cells may occur via a transactivation role of mutant p53 executed at the NF-κB2 promoter through chromatin modification.
Figure 6. ChIP analysis indicates acetylation of histones on the NF-κB2 promoter.
ChIP analysis was performed on H1299 cells stably transfected with vector (HC5) or mutant p53-R273H (R273H) to test whether histones are preferentially acetylated on the NF-κB2 promoter, and if different transcription factors and mutant p53 are nucleated on the promoter. As described in Materials and methods, cells were crosslinked with formaldehyde, harvested and DNA sonicated to generate 500 bp-2 Kb fragments. Designated proteins were immunoprecipitated using respective antibodies. Cross-linking was reversed and the DNA isolated. QPCR was performed using gene specific primers. Normalized values represent QPCR values normalized to the GAPDH gene whose expression is not significantly affected by mutant p53. The promoter specific primers are located within 1000 bp of the transcriptional start site. A. ChIP performed with antibodies directed against acetylated histone H3 (acetylated at K9 and K14) and H4 (acetylated at K16). B. ChIP performed with antibodies directed against CREB, NF-κB p65, and STAT. C. ChIP performed with antibodies directed against CBP, cRel and p53 DO1. D. ChIP performed with an antibody against the total p53 protein.
Mutant p53 induces binding of CBP and STAT2 on the NF-κB2 promoter
To extend our studies, we used ChIP analysis to determine whether there is preferential enhancement of interaction of one or more transcription factor(s) on the NF-κB2 promoter in H1299 cells stably transfected with vector alone (HC5) or expressing mutant p53-R273H. The transcription factors CREB, NF-κB p65, STAT2, CBP, c-Rel, and p53 were chosen for ChIP analysis based on transcription factor binding sites located on the NF-κB2 promoter as well as known interactions. The data presented in Figure 6B show that STAT2 nucleation, as determined by ChIP assays, is significantly higher in mutant p53 expressing cells. Thus, our results demonstrate that mutant p53 induces STAT2 interaction on the NF-κB2 promoter, and suggests a STAT2-mutant p53 mechanism. Similarly, when tested by ChIP analysis, CBP, a histone acetyltransferase, is nucleated more on the NF-κB2 promoter in the presence of mutant p53 (Figure 6C). This suggests that CBP nucleation enhances histone acetylation on the promoter.
Figure 6 also shows no enhanced nucleation of mutant p53 on the NF-κB2 promoter as we did not observe any additional promoter fragments immunoprecipitated by p53 antibody DO1 or a p53 antibody recognizing the total protein (Figures 6C and 6D). This suggests that under the conditions of our assay, including cross-linking, we were unable to detect mutant p53 directly on the promoter. It is possible that by changing conditions we may be able to detect mutant p53 on the promoter if it is nucleated via protein-protein interactions.
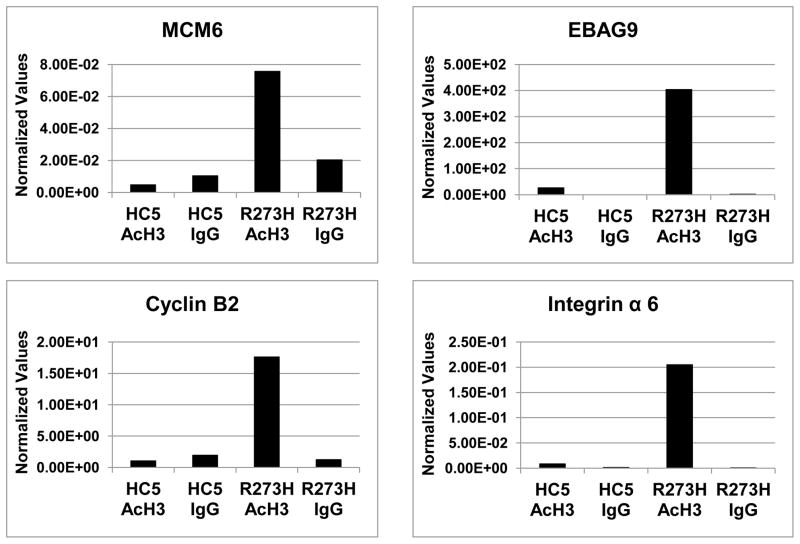
ChIP analysis shows acetylation of histones on mutant p53 target gene promoters, in general
We wanted to test whether the acetylation of histones at the promoters of mutant p53 targets is a general phenomenon, and analyzed a number of mutant p53 targets [8] by ChIP (Figure 7). Thus we performed ChIP assays using H1299 cells stably transfected with empty vector alone (HC5) or cells expressing mutant p53-R273H and targeted primers for a number of genes that we have previously shown to be up-regulated by mutant p53. The data presented in Figure 7 show that all the promoters tested have increased acetylation of histone H3; it is possible that some other promoters may have modification of other histones, such as H4 in the case of the NF-κB2 promoter.
Figure 7. ChIP analysis indicates increased acetylation of histone H3 on multiple mutant p53 target gene promoters.
Promoter regulatory regions of mutant p53 target genes MCM6, EBAG9, Cyclin B2 and Integrin alpha 6 were tested by ChIP assay to determine whether histone H3 becomes preferentially acetylated in the presence of mutant p53, as described in the legend to Figure 6. The primer sequences have been described in Materials and Methods and are based on sequence information obtained at NCBI. The primer sequences are within 1000 bp of the predicted transcriptional start site. Values represent QPCR measurements normalized to the GAPDH gene whose expression is not significantly affected by mutant p53. All these experiments have been performed multiple times on independent biological replicates to verify their authenticity.
Discussion
We and others have shown that expression of tumor-derived mutant p53 in cells leads to increased cell growth rate, cell motility, tumorigenicity, and loss of sensitivity to chemotherapeutic drugs [8, 19, 48] as a manifestation of GOF activity. We have suggested earlier that, in the case of H1299 lung carcinoma cells, the chemoresistance observed by the expression of mutant p53 can be explained in part through mutant p53-dependent transactivation of the NF-κB2 gene [8]. Here, using naturally-occurring cancer cell line models containing GOF p53, we have demonstrated that the presence of p53 mutants leads to GOF phenotypes as illustrated by increased cell growth rate, cell motility, tumorigenicity, and loss of sensitivity to chemotherapeutic drugs (Figures 1–3). We have further shown that the GOF activities are dependent on the level of NF-κB2 (Figure 4), verifying that mutant p53-induced GOF functions use NF-κB2 as a mediator, at least in some instances.
We examined the mechanism of up-regulation of NF-κB2 by mutant p53. Transient transcriptional analysis could not detect any mutant p53 response element (Figure 5). This is not unexpected as our previous studies also failed to detect any [5, 7]. Furthermore, ChIP experiments show that mutant p53 induces chromatin changes through histone acetylation that may cause activation of the NF-κB2 promoter (Figure 6). This histone acetylation perhaps leads to increased interaction of the transcription factor STAT2 to a STAT consensus binding site on the NF-κB2 promoter; this may be the reason for increased NF-κB2 promoter activity in the presence of mutant p53 (Figure 6). Our data also show an interaction of CBP with the NF-κB2 promoter (Figure 6). Since CBP/p300 has histone acetylase activity associated with it, these data indicate that mutant p53 may be enhancing histone acetylation through the use of CBP/p300. Interestingly, interactions of CBP/p300 and STAT, p53 and STAT as well as p53 and CBP/p300 have been reported [49–51]. With that information in hand, we speculate that the STAT1/STAT2 complex on the NF-κB2 promoter might interact with CBP/p300 to open up chromatin and pull down mutant p53. We also speculate that mutant p53 then stimulates further opening up of chromatin via its interaction with CBP/p300 with a positive feed-back loop. At this stage this is conjecture and several steps of this model need to be tested in the future.
The molecular mechanism behind GOF activities by tumor-derived mutant p53 is still unknown. Two possibilities, which again are not mutually exclusive, have evolved. (1) Via protein-protein interactions: Mutant p53 interacts with p53 family members, p73 and p63, and neutralizes their growth suppressive functions leading to the GOF phenotype. In a modification of that hypothesis, mutant p53 may interact with other cellular proteins to gain oncogenic activities. (2) Via mutant p53’s transcriptional activity: Mutant p53 would modulate a number of genes involved in cell growth, survival and oncogenesis. This transcriptional activity is independent of WT p53’s transcriptional functions.
The work presented here shows a possible mechanism of up-regulation of mutant p53 target gene expression with the involvement of chromatin modifications. This chromatin modification is brought into action most likely by CBP which is found to be nucleated on the NF-κB2 promoter. As a result of the histone acetylation we can observe increased STAT interaction on the promoter that might be the reason for enhanced transcription of the mutant p53 target, NF-κB2. What is unknown at this point is how this process is brought into play by mutant p53. Improved techniques are being developed to investigate this. In the future we should be able to elucidate similar mechanisms of modulation of gene expression by mutant p53 in other cases and in other cell lines besides H1299.
Several naturally occurring cancer cell lines with mutant p53 also show gain of function phenotypes
Mutant p53 induces gain of function phenotype in H1299 lung cancer cells by upregulating NF-κB2 transcription
This up-regulation is correlated with increased interaction of CBP and STAT with the NF-κB2 promoter
Acknowledgments
This work was supported by grants from NIH to Sumitra Deb (CA70712 and CA121144) and Swati Palit Deb (CA74172), and Pilot Project Awards from Massey Cancer to Sumitra Deb, Swati Palit Deb, Andrew Yeudall and Brad Windle. We thank Arnold Levine, and Bert Vogelstein, for providing us with cells, plasmids and advice, and Amber Heck and Amanda High for technical assistance.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Prives C, Hall PA. The Journal of pathology. 1999;187:112–126. doi: 10.1002/(SICI)1096-9896(199901)187:1<112::AID-PATH250>3.0.CO;2-3. [DOI] [PubMed] [Google Scholar]
- 2.Vogelstein B, Lane D, Levine AJ. Nature. 2000;408:307–310. doi: 10.1038/35042675. [DOI] [PubMed] [Google Scholar]
- 3.Zhao R, Gish K, Murphy M, Yin Y, Notterman D, Hoffman WH, Tom E, Mack DH, Levine AJ. Genes & development. 2000;14:981–993. [PMC free article] [PubMed] [Google Scholar]
- 4.Chin KVUK, Pastan I, Gottesman MM. Science (New York, NY. 1992;255:459–462. doi: 10.1126/science.1346476. [DOI] [PubMed] [Google Scholar]
- 5.Deb S, Jackson CT, Subler MA, Martin DW. J Virol. 1992;66:6164–6170. doi: 10.1128/jvi.66.10.6164-6170.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cadwell C, Zambetti GP. Gene. 2001;277:15–30. doi: 10.1016/s0378-1119(01)00696-5. [DOI] [PubMed] [Google Scholar]
- 7.Ludes-Meyers JH, Subler MA, Shivakumar CV, Munoz RM, Jiang P, Bigger JE, Brown DR, Deb SP, Deb S. Mol Cell Biol. 1996;16:6009–6019. doi: 10.1128/mcb.16.11.6009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Scian MJ, Stagliano KE, Anderson MA, Hassan S, Bowman M, Miles MF, Deb SP, Deb S. Mol Cell Biol. 2005;25:10097–10110. doi: 10.1128/MCB.25.22.10097-10110.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Scian MJ, Stagliano KE, Ellis MA, Hassan S, Bowman M, Miles MF, Deb SP, Deb S. Cancer Res. 2004;64:7447–7454. doi: 10.1158/0008-5472.CAN-04-1568. [DOI] [PubMed] [Google Scholar]
- 10.Singer S, Ehemann V, Brauckhoff A, Keith M, Vreden S, Schirmacher P, Breuhahn K. Hepatology. 2007;46:759–768. doi: 10.1002/hep.21736. [DOI] [PubMed] [Google Scholar]
- 11.Strano S, Dell'Orso S, Di Agostino S, Fontemaggi G, Sacchi A, Blandino G. Oncogene. 2007;26:2212–2219. doi: 10.1038/sj.onc.1210296. [DOI] [PubMed] [Google Scholar]
- 12.Weisz L, Damalas A, Liontos M, Karakaidos P, Fontemaggi G, Maor-Aloni R, Kalis M, Levrero M, Strano S, Gorgoulis VG, Rotter V, Blandino G, Oren M. Cancer Res. 2007;67:2396–2401. doi: 10.1158/0008-5472.CAN-06-2425. [DOI] [PubMed] [Google Scholar]
- 13.Zalcenstein A, Weisz L, Stambolsky P, Bar J, Rotter V, Oren M. Oncogene. 2006;25:359–369. doi: 10.1038/sj.onc.1209061. [DOI] [PubMed] [Google Scholar]
- 14.Lin J, Teresky AK, Levine AJ. Oncogene. 1995;10:2387–2390. [PubMed] [Google Scholar]
- 15.Lanyi A, Deb D, Seymour RC, Ludes-Meyers JH, Subler MA, Deb S. Oncogene. 1998;16:3169–3176. doi: 10.1038/sj.onc.1201857. [DOI] [PubMed] [Google Scholar]
- 16.Hsiao M, Low J, Dorn E, Ku D, Pattengale P, Yeargin J, Haas M. Am J Pathol. 1994;145:702–714. [PMC free article] [PubMed] [Google Scholar]
- 17.Shi XB, Nesslinger NJ, Deitch AD, Gumerlock PH, deVere White RW. Prostate. 2002;51:59–72. doi: 10.1002/pros.10072. [DOI] [PubMed] [Google Scholar]
- 18.Bristow RG, Peacock J, Jang A, Kim J, Hill RP, Benchimol S. Oncogene. 2003;22:2960–2966. doi: 10.1038/sj.onc.1206405. [DOI] [PubMed] [Google Scholar]
- 19.Blandino G, Levine AJ, Oren M. Oncogene. 1999;18:477–485. doi: 10.1038/sj.onc.1202314. [DOI] [PubMed] [Google Scholar]
- 20.Lee JM, Bernstein A. Proceedings of the National Academy of Sciences of the United States of America. 1993;90:5742–5746. doi: 10.1073/pnas.90.12.5742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wang XJ, Greenhalgh DA, Jiang A, He D, Zhong L, Medina D, Brinkley BR, Roop DR. Oncogene. 1998;17:35–45. doi: 10.1038/sj.onc.1201890. [DOI] [PubMed] [Google Scholar]
- 22.Wang XJ, Greenhalgh DA, Jiang A, He D, Zhong L, Brinkley BR, Roop DR. Mol Carcinog. 1998;23:185–192. doi: 10.1002/(sici)1098-2744(199811)23:3<185::aid-mc7>3.0.co;2-5. [DOI] [PubMed] [Google Scholar]
- 23.Gualberto A, Aldape K, Kozakiewicz K, Tlsty TD. Proceedings of the National Academy of Sciences of the United States of America. 1998;95:5166–5171. doi: 10.1073/pnas.95.9.5166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Albor A, Kaku S, Kulesz-Martin M. Cancer Res. 1998;58:2091–2094. [PubMed] [Google Scholar]
- 25.Murphy KL, Rosen JM. Oncogene. 2000;19:1045–1051. doi: 10.1038/sj.onc.1203274. [DOI] [PubMed] [Google Scholar]
- 26.El-Hizawi S, Lagowski JP, Kulesz-Martin M, Albor A. Cancer Res. 2002;62:3264–3270. [PubMed] [Google Scholar]
- 27.Strano S, Blandino G. Cell cycle (Georgetown, Tex. 2003;2:348–349. [PubMed] [Google Scholar]
- 28.Irwin MS, Kondo K, Marin MC, Cheng LS, Hahn WC, Kaelin WG., Jr Cancer cell. 2003;3:403–410. doi: 10.1016/s1535-6108(03)00078-3. [DOI] [PubMed] [Google Scholar]
- 29.Bohuslav J, Chen LF, Kwon H, Mu Y, Greene WC. The Journal of biological chemistry. 2004;279:26115–26125. doi: 10.1074/jbc.M313509200. [DOI] [PubMed] [Google Scholar]
- 30.Piva R, Belardo G, Santoro MG. Antioxid Redox Signal. 2006;8:478–486. doi: 10.1089/ars.2006.8.478. [DOI] [PubMed] [Google Scholar]
- 31.Xiao G, Fong A, Sun SC. The Journal of biological chemistry. 2004;279:30099–30105. doi: 10.1074/jbc.M401428200. [DOI] [PubMed] [Google Scholar]
- 32.Nadiminty N, Lou W, Lee SO, Lin X, Trump DL, Gao AC. Proceedings of the National Academy of Sciences of the United States of America. 2006;103:7264–7269. doi: 10.1073/pnas.0509808103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Karin M, Cao Y, Greten FR, Li ZW. Nat Rev Cancer. 2002;2:301–310. doi: 10.1038/nrc780. [DOI] [PubMed] [Google Scholar]
- 34.Pham CG, Bubici C, Zazzeroni F, Knabb JR, Papa S, Kuntzen C, Franzoso G. Mol Cell Biol. 2007;27:3920–3935. doi: 10.1128/MCB.01219-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Tamatani T, Azuma M, Ashida Y, Motegi K, Takashima R, Harada K, Kawaguchi S, Sato M. Int J Cancer. 2004;108:912–921. doi: 10.1002/ijc.11640. [DOI] [PubMed] [Google Scholar]
- 36.Liao X, Zhang L, Thrasher JB, Du J, Li B. Molecular cancer therapeutics. 2003;2:1215–1222. [PubMed] [Google Scholar]
- 37.Duan J, Friedman J, Nottingham L, Chen Z, Ara G, Van Waes C. Molecular cancer therapeutics. 2007;6:37–50. doi: 10.1158/1535-7163.MCT-05-0285. [DOI] [PubMed] [Google Scholar]
- 38.You M, Ku PT, Hrdlickova R, Bose HR., Jr Mol Cell Biol. 1997;17:7328–7341. doi: 10.1128/mcb.17.12.7328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Catz SD, Johnson JL. Oncogene. 2001;20:7342–7351. doi: 10.1038/sj.onc.1204926. [DOI] [PubMed] [Google Scholar]
- 40.Wang Z, Zhang B, Yang L, Ding J, Ding HF. The Journal of biological chemistry. 2008;283:10698–10706. doi: 10.1074/jbc.M800806200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Deb SP. Front Biosci. 2002;7:d235–243. doi: 10.2741/A723. [DOI] [PubMed] [Google Scholar]
- 42.Subler MA, Martin DW, Deb S. J Virol. 1994;68:103–110. doi: 10.1128/jvi.68.1.103-110.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Sankala H, Vaughan C, Wang J, Deb S, Graves PR. Archives of biochemistry and biophysics. 512:52–60. doi: 10.1016/j.abb.2011.05.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Deb D, Scian M, Roth KE, Li W, Keiger J, Chakraborti AS, Deb SP, Deb S. Oncogene. 2002;21:176–189. doi: 10.1038/sj.onc.1205035. [DOI] [PubMed] [Google Scholar]
- 45.Wang H, Patel V, Miyazaki H, Gutkind JS, Yeudall WA. Carcinogenesis. 2009;30:165–174. doi: 10.1093/carcin/bgn252. [DOI] [PubMed] [Google Scholar]
- 46.Kirch HC, Flaswinkel S, Rumpf H, Brockmann D, Esche H. Oncogene. 1999;18:2728–2738. doi: 10.1038/sj.onc.1202626. [DOI] [PubMed] [Google Scholar]
- 47.Sirvent JJ, et al. Prognostic value of p53 protein expression and clinicopathological factors in infiltrating ductal carcinoma of the breast. A study of 192 patients. Histol Histopathol. 2001;16(1):99–106. doi: 10.14670/HH-16.99. [DOI] [PubMed] [Google Scholar]
- 48.Koike M, Fujita F, Komori K, Katoh F, Sugimoto T, Sakamoto Y, Matsuda M, Fujita M. Cancer Sci. 2004;95:541–546. doi: 10.1111/j.1349-7006.2004.tb03246.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Livengood JA, Scoggin KE, Van Orden K, McBryant SJ, Edayathumangalam RS, Laybourn PJ, Nyborg JK. The Journal of biological chemistry. 2002;277:9054–9061. doi: 10.1074/jbc.M108870200. [DOI] [PubMed] [Google Scholar]
- 50.Wojciak JM, Martinez-Yamout MA, Dyson HJ, Wright PE. Embo J. 2009;28:948–958. doi: 10.1038/emboj.2009.30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Paulson M, Pisharody S, Pan L, Guadagno S, Mui AL, Levy DE. The Journal of biological chemistry. 1999;274:25343–25349. doi: 10.1074/jbc.274.36.25343. [DOI] [PubMed] [Google Scholar]
- 52.Miyazaki H, Patel V, Wang H, Edmunds RK, Gutkind JS, Yeudall WA. Cancer Res. 2006;66:4279–4284. doi: 10.1158/0008-5472.CAN-05-4398. [DOI] [PubMed] [Google Scholar]