Abstract
Subcompartments of the plasma membrane are believed to be critical for lymphocyte responses but few genetic tools exist to test their function. Here we describe a new X-linked B cell deficiency syndrome in mice caused by mutations in Atp11c, a member of the P4 ATPase family thought to serve as flippases concentrating aminophospholipids in the cytoplasmic leaflet of cell membranes. Defective ATP11c decreased the rate of phosphatidylserine translocation in pro-B cells, greatly reduced pre-B and B cell numbers independent of Bcl2-inhibited apoptosis or immunoglobulin gene rearrangement and abolished pre-B cell expansion in response to an Il7 transgene. The only other abnormalities noted were anemia, hyperbilirubinemia and hepatocellular carcinoma. These results identify an intimate connection between phospholipid transport and B lymphocyte function.
Differentiation of B lymphocytes from hematopoietic stem cells is one of the best defined cell differentiation processes in vertebrates. Its analysis by molecular genetics has illuminated fundamental mechanisms for normal immunity, human immunodeficiency diseases, DNA transcription, rearrangement and repair, cell signaling and cancer. B cell differentiation is currently viewed as proceeding in a graded manner, guided by a complex, self-reinforcing network of B cell-specific transcriptional regulators and receptor signaling systems1–4. Understanding how the elements of this network are integrated has only begun to be revealed and hinges upon identifying essential missing steps.
Stepwise differentiation of B cells is demarcated by expression of different cell surface and intracellular proteins5–7. The earliest B lineage biased stage, B lymphoid progenitors (BLPs), expresses the alpha chain of the interleukin 7 receptor (IL-7Rα, CD127), abundant CD43, and Ly6D. It is succeeded by pre-pro B cells (Fraction A cells) that retain these markers but also express B220, and then by pro-B cells (Fraction B), which acquire intermediate expression of CD24 (HSA) and CD19. Development to the pro-B cell stage and beyond requires a feed-forward network of interactions between IL-7 produced by nearby stromal cells, IL-7R signaling activating the transcription factor STAT5, and the B-lineage specific transcription factors Pu-1, EBF-1, E2A and PAX5 (refs. 1,3,4,8). Differentiation to the pro-B cell stage enables recombination of V, D and J elements in the immunoglobulin heavy chain gene, resulting in synthesis of cytoplasmic IgM (cμ) heavy chains that assemble with surrogate light chains and CD79α and β proteins into a pre-B cell receptor (pre-BCR)9. Expression of the pre-BCR intracellularly (cμ+) defines a pre-B cell, enhances the growth response to IL-7 (refs. 10,11), and signals a further increase in CD24, induction of CD25, and downregulates CD43. Pre-B cells switch to light chain gene rearrangement, resulting in assembly of complete IgM molecules with CD79α and β, which are expressed as B cell antigen receptors (BCRs) on the cell surface, and progressively lose IL-7Rα expression. While B cell differentiation depends upon integrated signaling by pre-BCR and IL-7R, much remains to be understood about the initiation and cross-talk between these processes.
A growing body of work suggests that many immunological and cell biological signaling events hinge upon organization of the plasma membrane into specialized substructures, such as lateral concentration of particular lipids into rafts or asymmetrical concentration of specific phospholipids between the exoplasmic and cytoplasmic leaflets12,13. Exploring the function of these membrane specializations in lymphocytes has been limited by the paucity of genetic tools to specifically test their function in vivo, and most experiments depend upon disruptive approaches using detergents and biochemical fractionation. Phosphatidylserine (PS) is an abundant aminophospholipid normally concentrated almost exclusively in the cytoplasmic leaflet13, where it appears to be important for membrane budding and clathrin-coated vesicle formation during post-Golgi transport and endocytosis based on studies in yeast14. PS in the cytoplasmic leaflet also activates protein kinase C beta and provides a docking site for a range of other signaling proteins15. During the early stages of cell death and during certain forms of cell activation such as platelet activation, PS redistributes to the exoplasmic leaflet where it can be recognized by specific PS receptors or blood proteins like clotting factor VIII13. Binding of fluorescent Annexin V to PS in the exoplasmic leaflet is widely used experimentally to measure the initiation of apoptosis. Macrophages bear receptors that recognize exoplasmic PS as “eat-me” signals16,17. Viable cells also display PS during certain conditions independent of apoptosis, notably pre-B cells that have received pre-BCR signals, marginal zone B cells18 and some B cell lymphomas19 but the cause and function of PS display in these circumstances is obscure.
Phospholipids such as PS “flip-flop” very slowly between the inner and outer membrane leaflets in protein-free membranes, with a half-time of hours or days20. Asymmetry of the plasma membrane is actively generated by two groups of ATP-dependent transporters. ABC-type transporters (floppases) are responsible for active transport of specific lipids to the exoplasmic leaflet20. “Flipping” of PS and other aminophospholipids into the inner leaflet against their concentration gradient is greatly accelerated by flippases thought to be members of the P4 ATPase family of 10-transmembrane domain proteins21–23. The functions of the P4 ATPases have mostly been pinpointed by genetic analysis in yeast14 and the role of the fourteen P4 ATPases in humans and other mammals is mostly unexplored. So far only one gene has clearly been implicated in a human disease: ATP8B1 (also known as FIC1) is mutated in two forms of heritable intrahepatic cholestasis24. Here we show that a defect in ATP11c, a ubiquitously expressed P4 ATPase of previously unknown function, decreases B cell PS flippase activity, disrupts B cell development and antibody production, as well as causing hepatocellular carcinoma. This finding reveals a critical role for membrane asymmetry in B lymphocytes and opens up a way to study its function in mammalian cells.
Results
X-linked B cell deficiency and liver disease
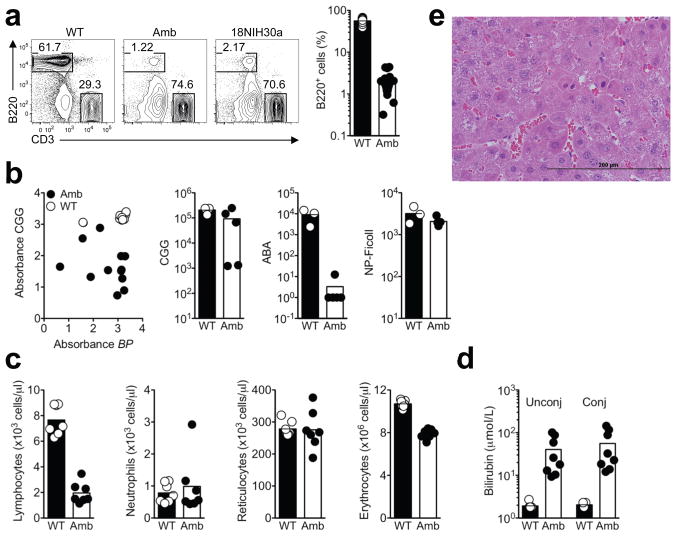
Flow cytometric screening of N-ethyl-N-nitrosourea (ENU)-mutagenized C57BL/6 mouse pedigrees revealed two independent strains, ambrosius and 18NIH30a, with multiple male offspring exhibiting B cell frequencies in blood that were decreased to 3% of controls (Fig. 1a). These animals nevertheless had normal frequencies of T and NK cells (data not shown). B cell deficiency was inherited as a Mendelian recessive, X-linked trait in successive generations. Immunization with inactivated Bordetella pertussis (BP) and alum-precipitated chicken gamma globulin (CGG) coupled to the hapten azo-benzene-arsonate (ABA) showed a variably reduced primary antibody response to both antigens in the mutant animals (Fig. 1b, left). Booster immunization with ABA-CGG 6 weeks later again showed variably decreased antibody response to the CGG protein carrier while antibodies to the ABA hapten - which depends upon antibody hypermutation and selection in germinal centers - was almost absent in all mutant animals (Fig. 1b, middle). In contrast, the mutant mice made a normal IgM antibody response to the T-cell independent antigen, nitrophenyl (NP)-Ficoll (Fig. 1b, right). Thus, the profound decrease in circulating B cells causes a variable humoral immune deficiency.
Figure 1. Identification and initial characterization of strains with reduced B lymphocytes after ENU-induced mutagenesis.
(a) Flow cytometric analysis of the percentage of B220+ B cells in the blood of Atp11c+/0 wild-type (WT), Atp11camb/0 ambrosius (Amb) and 18NIH30a male mice. All data are expressed as a percentage of lymphocytes. Each white circle (WT) and black circle (Amb) represents a single mouse. (b) Primary antibody response 14 days after immunization with inactivated Bordetella pertussis (BP) and alum-precipitated CGG (left). The other panels show antibody to CGG, the hapten ABA, and NP-Ficoll 6 days after booster immunization. Each white circle (WT) and black circle (Amb) represents a single mouse. (c) Number of lymphocytes, neutrophils, reticulocytes and erythrocytes in the blood of Amb and WT littermates. The results are representative of 2 independent experiments with 4–7 mice per group in each experiment. (d) Unconjugated and conjugated bilirubin in the plasma of Amb and control mice. The data are representative of 2 independent experiments with 4 to 7 mice per group and experiment. (e) Typical dysplastic focus on H & E stained liver section from amb mice at 6 months (X400).
In other respects the mutant mice were normal in appearance, size and weight, exhibited no discernable change in gait or behavior, and were fertile. The only other phenotypic abnormalities detected were reduced lymphocytes, normochromic anemia with 30% fewer total erythrocytes but normal reticulocyte numbers, and pronounced yellow colored plasma due to a 30-fold increase in unconjugated and conjugated bilirubin (Fig. 1c,d). Other measures of liver injury, plasma alanine aminotransferase (ALT) and aspartate aminotransferase (AST), were comparable between mutant and control animals (data not shown). Necropsy, however, revealed gross or microscopic liver pathology consistent with hepatocellular carcinoma in 10 out of 10 mutant animals analyzed at 6 months of age and not in normal controls. Liver tumors were well to poorly differentiated hepatocellular carcinomas containing areas of hemorrhage, necrosis and hypervascularity. Tumor development in these mice was likely to have been preceded by the formation of foci of altered hepatocytes (Fig. 1e and Supplementary Fig. 1). Several such foci were present in liver surrounding tumors, exhibiting a striking number of mitotic figures, many of which were multipolar with condensed and asymmetric chromatin aggregation (Supplementary Fig. 1a,b). Other dysplastic features include nuclear anisocytosis, hyperchromasia, pleomorphism and increased nuclear to cytoplasmic ratio (Fig. 1e and Supplementary Fig. 1). Since few experimental mouse models exist for studying hepatocellular carcinoma, the liver pathology in the mutant strain is of considerable interest and will require detailed analysis beyond the scope of the current study.
Next-generation DNA sequencing reveals Atp11c mutations
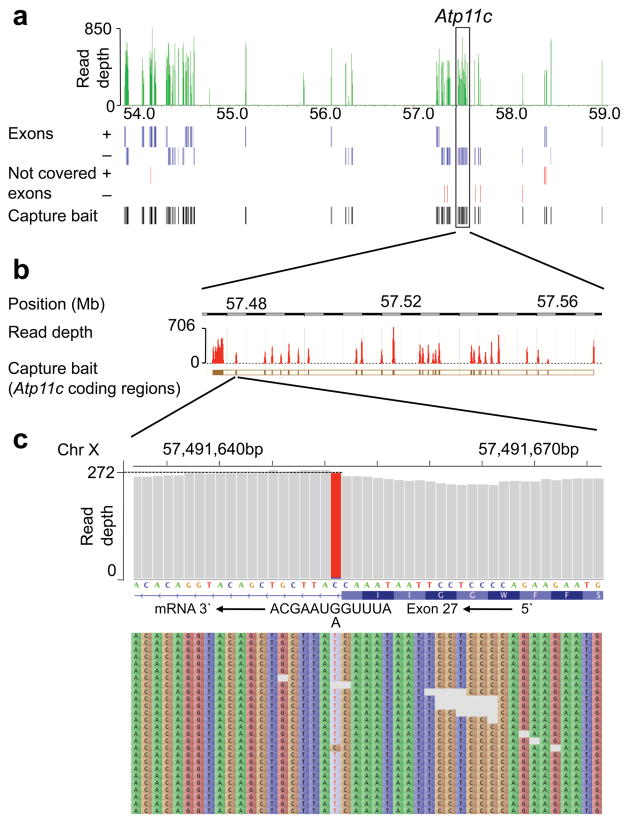
Linkage analysis in (B6xCBA)F2 offspring mapped the mutation to an X-chromosomal region distal to marker rs13483763 at 54,012,901 basepairs (Build NCBI37.1). To capture and sequence Refseq annotated exons in this region of the X-chromosome we designed a custom Agilent Sureselect targeted DNA capture array, from which RNA baits were prepared and hybridized with fragmented genomic DNA from an ambrosius male (Fig. 2a). The enriched DNA was sequenced in paired-end mode for 100 cycles on a single lane of an Ilumina GAIIx sequencer. For the bait-targets, the median read depth from a single lane of data was 138 and 96% of the nucleotides were covered with a read depth of 5 or greater. A median read depth of 137 was obtained across all RefSeq exons in the mutation-containing region and a read depth of 5 or greater for approximately 93% of the nucleotides in these exons. Capture and sequencing was performed in replicate at two facilities, yielding highly reproducible results. These results demonstrate the utility of exon capture and next-generation DNA sequencing to accelerate mutation identification.
Figure 2. Identification of a splice-site mutation in Atp11c by next-generation DNA sequencing.
(a) Read depth from a single GA2x sequencer lane across the tiled region between 54 and 59 Mb on the X-chromosome. Blue lines below denote all annotated exons on the + and − strand, red lines denote exons with a read depth of less than 5, and black lines denote capture baits used to enrich exons from genomic DNA. Atp11c gene is boxed and shown in greater magnification in (b) with read depth and chromosomal coordinates (NCBI build 37.1). (c) View of Atp11c exon 27 splice donor at single nucleotide resolution, read depth across these nucleotides, and sequence of first 30 reads. The C to T change, representing an intronic +1 G to A substitution in the Atp11c sense strand and RNA transcript was the only identified discrepancy between ambrosius DNA and the C57BL/6J mouse reference sequence in two independent capture and sequencing runs, and was confirmed by Sanger sequencing.
Within the interval a single mutation was identified in both capture datasets and confirmed by Sanger sequencing, lying in a previously uncharacterized gene, Atp11c. This was a G to A substitution in the exon 27 splice donor sequence at the +1 position of intron 27 (Fig. 2b,c). The normal +1 G nucleotide is invariant in 5′ splice donor sequences and its substitution would be predicted to abolish splicing, either yielding an in-frame UGA stop codon at the 3′ end of exon 27 (Fig. 2c) or aberrant use of alternative splice donors. PCR-amplification of cDNA from mutant and wild-type bone marrow or spleen with primers located in exon 25 and 29 yielded a shorter product in the mutant animals (Supplementary Fig. 2a), which was shown by sequencing to skip exon 27 and splice exon 26 to exon 28. The resulting deletion of 104 bp introduced a frame-shift after amino acid 1010, abolishing the C-terminal residues encoding the last two transmembrane domains and cytoplasmic tail of the ATP11c protein (Supplementary Figs. 2b and 3). An independent Atp11c mutation was identified by Sanger sequencing in strain 18NIH30a, also disrupting an invariant, essential splice donor nucleotide but in the preceding exon 26 splice donor sequence (intronic +2T to G: TCTGAAGgttagattttt to AACTCTGAAGggtagattttt). Given the severity of these mutations and their similarity to an ATP11c mutation that truncates almost half the protein reported in the accompanying study by Siggs et al25, it is likely that the mutations fully inactivate the protein.
All B cell subsets decreased except marginal zone B cells
The requirement for normal ATP11c in B cells was cell autonomous in chimeric mice reconstituted with an equal mixture of CD45.2 Atp11camb/0 and CD45.1 Atp11c+/0 marrow, so that Atp11camb/0 B cells represented only 1% of B cells (Supplementary Fig. 4). In the same mixed chimeras, Atp11camb/0 T cells and NK cells nevertheless accumulated in approximately equal proportions to their wild-type counterparts, establishing that there is little or no competitive disadvantage of mutant cells in T cells, NK cells or their hematopoietic progenitors.
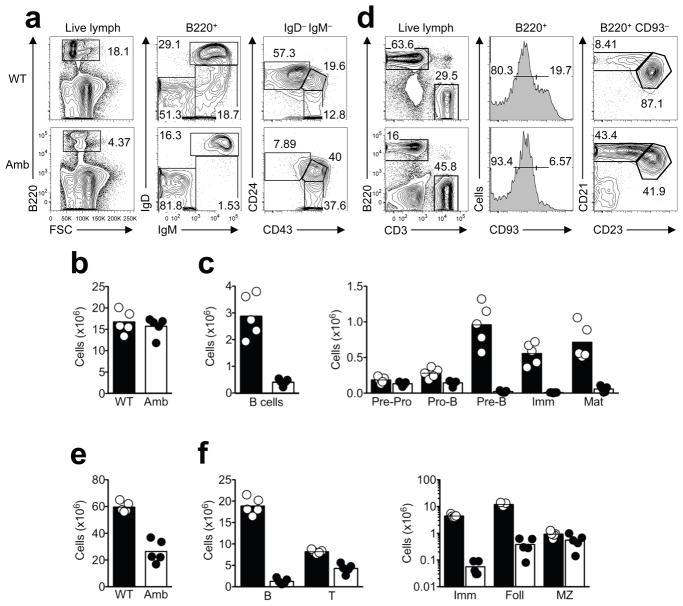
Analysis of hematopoietic progenitors including hematopoietic stem and progenitor cells (HSPCs), multipotent progenitors (MPPs), and common lymphoid progenitors (CLPs) revealed normal or slightly elevated numbers of these populations in the bone marrow of Atp11camb/0 mice compared to their wild-type littermates (Supplementary Fig. 5a,b). A more detailed enumeration of other lineages in the bone marrow showed that Atp11camb/0 mice have normal number of myeloid, erythroid and NK cells and slightly higher number of T cells (Supplementary Fig. 5c,d), indicating that ATP11c is not required for blood cell progenitors and their development into other lineages. A systematic analysis of B cell subsets was performed in Atp11camb/0 and control mice to determine which stages of B cell development were disrupted. In the bone marrow, the absolute number of leukocytes was normal but the percentage and number of B cells were decreased in Atp11camb/0 mice to 15–18% of the numbers in wild-type controls (Fig. 3a–c and Table 1). The number of CD43+ CD24med pro-B cells was 60% of normal, whereas the number of CD43lo CD24hi pre-B cells and IgM+ IgD− immature B cells were only 6% and 1.8% of normal, respectively. IgM+ IgD+ mature recirculating B cells in the bone marrow were 11% of normal numbers, and these expressed much higher densities of IgM compared to wild-type littermates (Fig. 3a–c). When sIgM-negative and lineage marker-negative bone marrow B cell progenitors were sorted by CD43, CD24 and BP-1 expression, there were normal numbers of pre-pro-B cells (Fraction A) in the mutant animals, but progressively reduced numbers of pro-B and pre-B cells in Fractions B, C and C′ (Supplementary Fig. 6). These data establish that ATP11c is required for B cells to differentiate normally past the pro-B cell stage.
Figure 3. ATP11c mutation reduces all B cell subsets except marginal zone B cells.
(a) Representative flow cytometric analysis of bone marrow cells from Atp11c+/0 (WT) or Atp11camb/0 (Amb) male mice. Percentage of B220+ B cells (left); percentage of IgM+ IgD+ mature (Mat) B cells, IgM+ IgD− immature (Imm) B cells, and IgM− IgD− pro- and pre-B cells within the B220+ subset (middle); cells gated on B220+ IgM− IgD− cells, showing the percentage that are CD43− CD24hi pre-B cells, CD43+ CD24med pro-B cells and CD43+ CD24− pre-pro-B cells (right). (b,c) Number of leukocytes and indicated B cell subsets in bone marrow from Atp11c+/0 (mean, black column; individual animals, white circles) or Atp11camb/0 (mean, white columns; individual animals, black circles). (d) Representative flow cytometric analysis of B cell subpopulations in the spleen from Atp11c+/0 (WT) or Atp11camb/0 (Amb) male mice. Percentage of B220+ B cells and CD3+ T cells (left); percentage of B220+-gated cells that are CD93− mature B cells and CD93+ immature B cells (middle); percentage of the CD21hi CD23− marginal zone (MZ) and the CD21med CD23+ follicular (Foll) B cell subset within the B220+ CD93− mature B cells (right). (e,f) Number of leukocytes and lymphocyte subsets in the spleen. Means and individual values shown as in (b,c). Data in panels a-f are representative of at least 5 independent experiments with 2–5 mice per group and experiment.
Table 1. Number of B cells in the spleen and B cell subpopulations in bone marrow.
The numbers are expressed as mean values x105 ± S.E.M in the bone marrow and x106 ± S.E.M in the spleen. B cells (B220+ in live lymphocytes gate) in the spleen. Mature B cell (IgD+ IgM+ in B220+ gate), immature B cells (IgD− IgM+ in B220+ gate), pre-B cell (CD24hi CD43− in IgD− IgM− gate), pro-B cell (CD24int CD43+ in IgD− IgM− gate), pre-pro-B cell (CD24−CD43+ in IgD− IgM− gate) in the bone marrow.
| ATP11c | Transgene | Pre-Pro-B cells | Pro-B cells | Pre-B cells | Immature B cells | Mature B cells | Number of animals (BM) | Spleen B cells | Number of animals (Spleen) |
|---|---|---|---|---|---|---|---|---|---|
| WT | None | 3.99 ± 0.25 | 4.10 ± 0.42 | 28.66 ± 2.04 | 12.50 ± 1.20 | 14.29 ± 0.89 | 14 | 20.06 ± 1.87 | 11 |
| Amb | None | 5.55 ± 0.36 | 2.22 ± 0.42 | 1.74 ± 0.34 | 0.24 ± 0.04 | 1.75 ± 0.25 | 13 | 1.73 ± 0.20 | 10 |
| WT | vavBcl2 | 5.53 ± 0.48 | 6.48 ± 0.65 | 21.48 ± 5.72 | 17.60 ± 1.65 | 69.63 ± 13.55 | 4 | 101.10 ± 11.84 | 4 |
| Amb | vavBcl2 | 6.22 ± 0.49 | 4.78 ± 0.96 | 6.22 ± 1.37 | 3.12 ± 0.26 | 9.98 ± 1.06 | 6 | 6.18 ± 1.37 | 6 |
| WT | IL-7 | 2.36 ± 0.56 | 19.26 ± 2.28 | 262.00 ± 36.5 | 36.36 ± 3.86 | 1.66 ± 0.31 | 5 | 153.30 ± 18.25 | 3 |
| Amb | IL-7 | 5.33 ± 0.21 | 2.33 ± 0.40 | 1.68 ± 0.43 | 0.25 ± 0.03 | 6.30 ± 1.45 | 4 | 7.78 ± 1.94 | 2 |
| WT | MD4 | 2.53 ± 0.29 | 0.50 ± 0.06 | 0.73 ± 0.13 | 15.97 ± 0.89 | 10.07 ± 0.84 | 3 | 10.82 ± 1.64 | 3 |
| Amb | MD4 | 3.28 ± 0.32 | 0.70 ± 0.03 | 0.66 ± 0.03 | 1.98 ± 0.39 | 4.90 ± 1.04 | 5 | 3.00 ± 0.23 | 5 |
In the spleen, the number of B cells in Atp11camb/0 animals was also decreased to 9% of that in wild-type mice (Fig. 3d–f and Table 1). Consistent with the deficits in the bone marrow, immature B cells in the spleen were also decreased to 1% of the numbers in wild-type mice and recirculating follicular B cells decreased to 5% of the normal number. The follicular B cells were located normally based on immunofluorescent staining of spleen sections (not shown) but they displayed an abnormal surface phenotype of higher IgM and CD21 and lower CD23. By contrast, marginal zone B cells were present in normal numbers and normal surface phenotype in Atp11camb/0 (Fig. 3d,f), so that they made up approximately 40% of all splenic B cells. In the peritoneal cavity of Atp11camb/0 mice, the subsets of B1a, B1b and B2 B cells were decreased to less than 10% of normal (Supplementary Fig. 7). These data indicate that marginal zone B cells are unique among B cell subsets by either not requiring or being able to compensate for ATP11c deficiency.
ATP11c deficiency abolishes effects of an Il7 transgene
The transition from pro- to pre-B cells is dependent on signaling through the IL-7 receptor and successful rearrangement of immunoglobulin heavy chain genes10. To test if defects in these processes and subsequent apoptosis caused the developmental block in Atp11camb/0 mice, the mutation was crossed with: (1) Vav-Bcl2 transgenic mice to inhibit apoptosis26; (2) H2Ea-Il7 transgenic mice with greatly increased IL-7 (ref. 27) or (3) MD4 or SWHEL transgenic mice28,29 with Ig heavy and light chain genes already rearranged (Fig. 4 and Table 1).
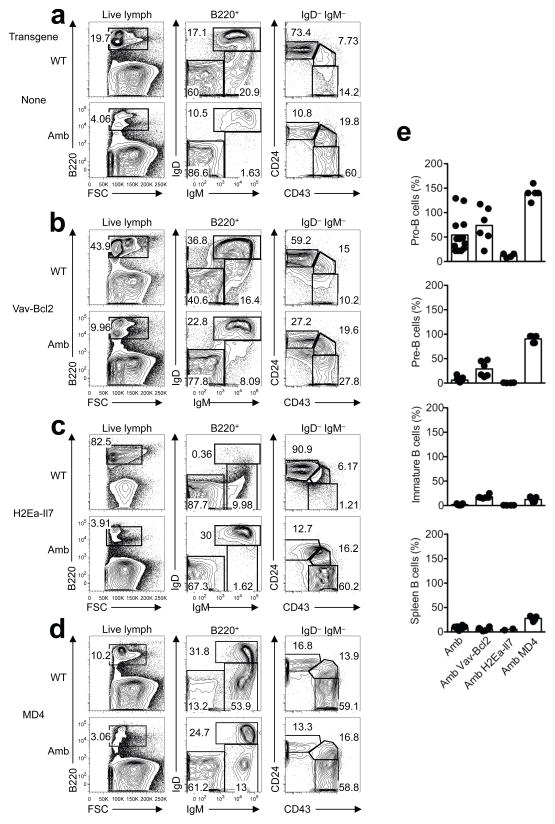
Figure 4. Effects of Bcl-2, IL-7 or BCR transgenes on ATP11c-mutant B cell development.
(a–d) Representative flow cytometric analysis showing the frequency of B cell subpopulations in the bone marrow of (a) non-transgenic mice with wild-type (WT) or mutant (Amb) ATP11c, (b) ATP11c WT or Amb mice with enforced expression of Bcl-2 under the control of the Vav-promoter (Vav-Bcl2), (c) ATP11c WT or Amb mice over-expressing IL-7 under the control of the MHC II Eα gene promoter (H2Ea-Il7), (d) ATP11c WT or Amb mice expressing rearranged Igh and Igk transgenes from the MD4 strain. (e) Graphs show the relative number of pro-B, pre-B, immature B cells in the bone marrow and B cells in the spleen of Atp11camb/0 mutant mice with the indicated transgene, as a percentage the mean number in Atp11c+/0 WT control animals carrying the same transgene. Each circle represents one mouse, columns show means, and data are pooled from multiple experiments with 1–5 mice of each genotype and experiment.
As expected26 Vav-Bcl2 greatly increased the number of mature B cells in control siblings with normal ATP11c. The number of pre-B cells in Atp11camb/0 Vav-Bcl2 mice was 29% of the number in Atp11c+/0 Vav-Bcl2 controls, representing a partial restoration compared to 6% of normal numbers in Atp11camb/0 mutants without Vav-Bcl2 (Fig. 4a,b,e and Table 1). The pre-B cells in Atp11camb/0 Vav-Bcl2 mice nevertheless failed to increase expression of CD24 (Fig. 4b) and CD25 (Supplementary Fig. 8a). Immature B cells were also partly rescued in Atp11camb/0 Vav-Bcl2 animals, although they still represented only 17% of the numbers in Atp11c+/0 Vav-Bcl2 controls (Fig. 4e and Table 1). Likewise spleen B cells remained at 6% of the numbers in Atp11c+/0 Vav-Bcl2 controls and 31% of wild-type mice, and continued to exhibit high IgM. Suppression of apoptosis by enforced Bcl-2 expression was therefore unable to correct the B cell developmental abnormalities caused by ATP11c deficiency.
H2Ea-Il7 transgenic mice with a normal Atp11c gene exhibited a 5-fold increase in bone marrow pro-B cells and 10-fold increase in pre-B cell numbers relative to wild-type mice (Fig. 4c,e and Table 1), consistent with published analyses27. By contrast, there was no effect of the increased IL-7 on the number of pro-B, pre-B, or immature B cells in Atp11camb/0 H2Ea-Il7 mice compared to Atp11camb/0 mice without the IL-7 transgene (Table 1). Increased IL-7 was therefore unable to rescue development of ATP11c-deficient B cells, and instead the mutation abolished the effects of transgenic IL-7 on pro-B and pre-B cells in the bone marrow.
In MD4 transgenic mice, the rearranged Ig heavy and light chain transgenes are normally activated during the pro-B cell stage, bypassing and suppressing RAG-mediated recombination of the endogenous Ig-genes. This lowers the number of pro-B cells and pre-B cells to 12% and 2% of normal numbers, respectively, replacing them with IgM+ IgD− immature B cells that are present in normal numbers in the bone marrow (Table 1). Whereas the Atp11camb mutation decreased the number of pro-B cells in non-transgenic mice, it increased the number of pro-B cells in MD4 transgenic mice to 150% of the numbers in control MD4 animals with normal ATP11c (Fig. 4e and Table 1). As discussed below, the increased numbers of pro-B cells is likely to reflect a developmental delay in activating the rearranged Ig-transgenes within the Atp11camb/0 pro-B cell population. The number of immature B cells in the bone marrow of Atp11camb/0 MD4 animals was nevertheless only partly restored to 11% of the numbers in Atp11c+/0 MD4 mice, whereas spleen and circulating B cells were partly restored to 37% of those in MD4 transgenic mice with normal ATP11c. A similar magnitude of rescue occurred in SWHEL mice with a rearranged Igh gene targeted in its normal location, but only when combined with a rearranged transgenic light chain (Supplementary Fig. 8b). Bypassing the pre-BCR signaling step thus alleviated but did not eliminate the need for ATP11c.
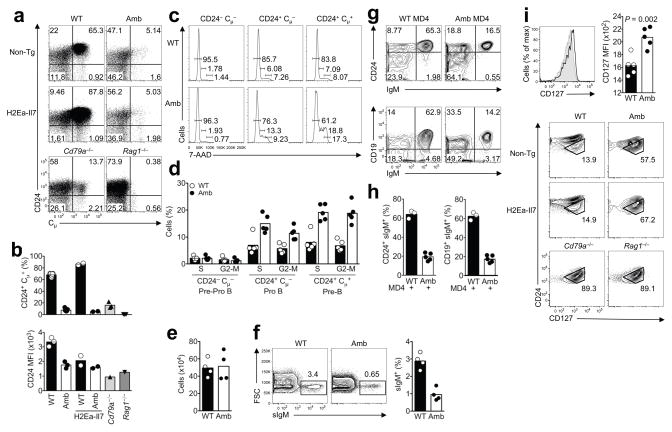
To further characterize the developmental block in bone marrow B cells we performed staining for cytoplasmic μ (cμ) heavy chains in surface IgM− B220lo cells, comparing wild-type or Atp11camb/0 animals with mice homozygous for null mutations eliminating the RAG1 recombinase or the CD79a (Igα) subunit of the pre-BCR and BCR. In wild-type mice, 65% of sIgM− B220lo cells were cμ+ and these were CD24hi due to pre-BCR signaling (Fig. 5a,b). No sIgM− B220lo cells were cμ+ in Rag1−/− mice, consistent with their inability to recombine the heavy chain genes. Fifteen percent of sIgM− B220lo cells were cμ+ in Cd79a−/− marrow, and their low frequency and lower CD24 expression is consistent with the inability of the pre-BCR to assemble and signal. Compared to this baseline frequency of cells with functionally rearranged but unselected IgM heavy chain genes, the frequency of cμ+ cells was even lower in Atp11camb/0 mice, representing only 5% of sIgM− B220lo cells (Fig. 5a,b).
Figure 5. Decreased pro-B cell transition from Ig− to Ig+ cells and absence of response to IL-7 transgene despite increased expression of IL-7Rα.
(a) Representative flow cytometric analysis of the surface expression of CD24 and intracellular expression of IgM (cμ) gated on surface IgM− B220lo 7-AAD− B cells in bone marrow from non-transgenic or IL-7 transgenic mice with wild-type (WT) or mutant (Amb) ATP11c, or from Cd79a−/− and Rag1−/− mice. Numbers indicate percent of gated cells in each quadrant. (b) Percent of intracellular IgM+ CD24+ cells gated as in (a), and geometric mean fluorescence intensity (MFI) of CD24 staining on these cells. The data are representative of 3 different experiments with 1–3 animals per group in each experiment. (c) Representative flow cytometric analysis of the cell cycle of pre-pro-B, pro-B and pre-B cells. The percentages of cycling cells (G0-1, S and G2-M phase) are indicated. (d) The graph shows % of pre-pro-B, pro-B and pre-B cells that are in S and G2-M phase. Data are representative of 2 different experiments with 2–5 mice per group in each experiment. (e) Absolute number of cells after 7 days culture of sorted pro-B cells in the presence of 25 ng/ml IL-7. (f) Representative flow cytometric plots showing the percentage of surface IgM+ cells out of all live B220+ cells after 7 day culture of sorted pro-B cells in the presence of 25 ng/ml IL-7. (g,h) Analysis of bone marrow from mice expressing the rearranged MD4 Igh and Igk transgenes, showing the % of B220low IgD− cells expressing surface IgM and CD24 (left) or CD19 (right). Graphs in (h) are combined from 3 different experiments with 1–3 animals per group in each experiment. (i) Expression of CD127 (IL-7Rα) on CD24med CD19+ IgM− B220lo pro-B cells from Atp11camb/0 (black line) and wild-type (shaded area) mice. The graph shows the mean and standard deviation of CD127 fluorescence on pro-B cells from multiple animals of each genotype, and statistical comparison by student’s t-test. Panels below show representative flow cytometric analysis of CD24 versus CD127 on CD19+ IgM− B220lo 7-AAD− cells in bone marrow from non-transgenic or IL-7 transgenic mice with wild-type (WT) or mutant (Amb) ATP11c, or from Cd79a−/− and Rag1−/− mice. The subset of CD24med pro-B cells is shown in the gate. Data are representative of 3 different experiments with 1–3 animals per group in each experiment.
Since IL-7 is important for pro-B and pre-B cell proliferation we investigated whether their proliferation was diminished by ATP11c deficiency in vivo or in vitro. Flow cytometric staining for DNA content in bone marrow pre-pro-B, pro-B and pre-B cells revealed increased frequencies of mutant pro-B cells and pre-B cells with greater than 2n DNA, indicating that a greater proportion of mutant pro-B and pre-B cells were in S or G2+M phases of cell cycle (Fig. 5c,d). When pro-B cells were flow sorted and cultured with high concentrations of IL-7 (25 ng/ml), mutant and wild-type pro-B cells increased in numbers identically over 7 days, either measured in separate cultures or when their ratios were compared in competitive co-cultures (Fig. 5e and data not shown). However, when the differentiation of these cells was analyzed by measuring the fraction that had become small pre-B and sIgM+ immature B cells, a much lower fraction of the mutant pro-B cells had differentiated (Fig. 5f). These results indicate that ATP11c mutation does not interfere with pro-B cell proliferation but disrupts their differentiation into pre-B and immature B cells.
CD24 was increased on cμ+ cells in Atp11camb/0 marrow, but only to half of the levels on normal cμ+ cells. When the need for Ig-gene rearrangement and pre-BCR signaling was bypassed in MD4 transgenic mice, ATP11c-deficiency still greatly reduced the frequency of B220lo B cells that expressed the Ig genes and instead there was an expanded population of pro-B cells that had not yet activated Ig transgene expression (Fig. 5g,h and Table 1). Collectively, these results indicate that the onset of H-chain expression and the response to pre-BCR assembly are both diminished in the absence of normal ATP11c.
Cμ+ cells normally exhibit a heightened proliferative response to IL-7 as a result of synergy between IL-7R and pre-BCR signaling10,11, so that cμ+ cells among sIgM− B220lo cells are preferentially expanded in H2Ea-Il7 transgenic mice (Fig. 5a,b). Like the earlier data on pro-B and pre-B cell numbers (Table 1), this effect of H2Ea-Il7 was also completely abolished by the Atp11c mutation, so that the low frequency of cμ+ cells in Atp11camb/0 marrow remained unchanged by the Il7 transgene. Expression of IL-7Rα (CD127) on CD19+ CD24med sIgM− B220lo pro-B cells was nevertheless significantly increased in either Atp11camb/0 mice or Atp11camb/0 mice with an H2Ea-Il7 transgene compared to wild-type controls (Fig. 5i), indicating that the absence of an in vivo response to the IL-7 transgene was not explained by failure to express the receptor.
Diminished phosphatidylserine flippase activity
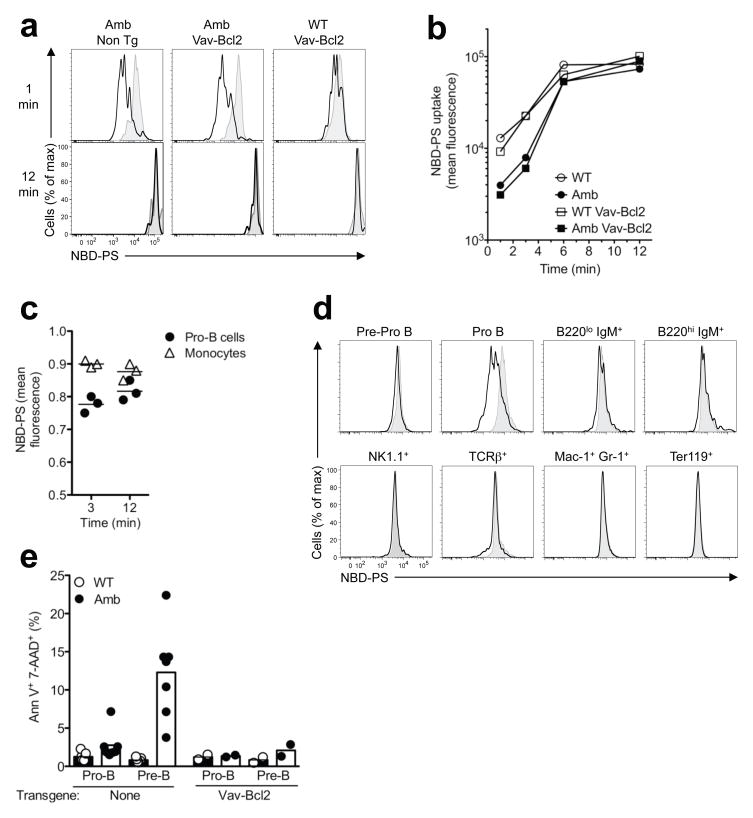
The evidence above that pro-B cells did not differentiate or respond to IL-7 normally focused testing of this cell subset for aminophospholipid flippase activity. Bone marrow from Atp11camb/0 CD45.2 and Atp11c+/0 CD45.1 control animals was mixed and incubated for various amounts of time with the fluorescent phosphatidylserine analogue, NBD-PS. Any dye remaining in the exoplasmic leaflet was then extracted with lipid-free albumin and washed away. The mutant and control pro-B cell subsets were distinguished by antibody staining, and analyzed on a flow cytometer. NBD-PS fluorescence in mutant and wild-type pro-B cells increased rapidly and approached saturation by 12 minutes at 37 C, but was homogeneously less in mutant pro-B cells analyzed at 1 or 3 minutes (Fig. 6a,b). The decreased PS flippase activity in Atp11camb/0 pro-B cells could not be corrected by enforced expression of Bcl-2, and was selective for pro-B cells as it was not as marked in mutant monocytes present in the same samples (Fig. 6a–c). Comparing a broad range of bone marrow cell types revealed that the flippase defect is not only B-lineage specific, but also specific for the pro-B cell stage, indicating a close correlation between flippase activity and cell development (Fig. 6d). The pre-pro-B cells and the immature and mature B cells in the bone marrow, as well as follicular and marginal zone B cells in the spleen of mutant animals, all showed normal flippase activity (Fig. 6d and Supplementary Fig. 9a). Interestingly, the only other blood cell types found with lower flippase activity in ATP11c mutant animals were CD4− CD8− DN and CD4+ CD8+ DP thymocytes (Supplementary Fig. 9b). Similar to pro-B cells, the flippase deficit was stage-specific in the T cell lineage as flippase activity was normal in mature T cells in bone marrow or spleen and in CD4+ or CD8+ SP thymocytes (Fig. 6d and Supplementary Fig. 9a,b). However, the reduction in flippase activity does not cause an obvious defect in DP cell differentiation into SP cells (data not shown). Taken together, these data show that ATP11c is critical for phospholipid flipping in a developmental stage-dependent way and that the B cell lineage is peculiarly sensitive to reduced flippase activity despite a near ubiquitous expression of ATP11c (Supplementary Fig. 10).
Figure 6. ATP11c mutation decreases PS translocation into pro-B cells.
(a) Representative NBD-PS fluorescence profiles after 1 or 12 min incubation in pro-B cells from ATP11c mutant (Amb) or wild-type (WT) mice lacking or carrying the Vav-Bcl2 transgene (black lines), compared to the corresponding CD45.1-marked wild-type pro-B cells in the same tube (shaded grey). (b) NBD-PS geometric mean fluorescence intensity in pro-B cells of the indicated genotypes at different incubation times. (c) Relative NBD-PS fluorescence intensity in bone marrow pro-B cells or monocytes from individual Atp11camb/0 mice, compared to CD45.1-marked wild-type pro-B cells or monocytes stained in the same tube. (d) Representative NBD-PS fluorescence profiles after 1 min incubation in pre/pro-B, pro-B, B220lo IgM+, B220hi IgM+, NK1.1+, TCRβ+, Mac1+ Gr1+ and Ter119+ bone marrow cells from ATP11c mutant (black lines) compared to the corresponding CD45.1-marked wild-type bone marrow cells in the same tube (shaded grey) except for Ter119+ cells which were stained in separate tubes. The data are representative of 3 independent experiments. (e) Percentage of pro- or pre-B cells that are apoptotic as measured by positive staining for Annexin V and 7-AAD, from ATP11c WT or Amb mice with or without the Vav-Bcl2 transgene. The data is from two (non-transgenic) or one (Vav-Bcl2) experiment with 2–5 mice per group and experiment.
To determine if ATP11c mutation leads to a steady-state increase in the concentration of PS in the exoplasmic plasma membrane leaflet of pro-B cells, bone marrow cells were stained with the PS-binding protein Annexin V and the membrane-impermeable DNA dye, 7-aminoactinomycin D (7-AAD). There was no increased Annexin V fluorescence on viable 7-AAD− pro-B or pre-B cells from Atp11camb/0 animals, but the percentage of apoptotic cells staining with both Annexin V and 7-AAD was increased among pre-B cells from Atp11camb/0 mice (Fig. 6e). This increase in apoptotic pre-B cells was suppressed by enforced expression of Bcl-2 (Fig. 6e) indicating that it is an indirect effect of ATP11c deficiency. Thus, the rate of PS translocation into the inner leaflet is diminished in ATP11c-deficient pro-B cells, but remains sufficient to prevent measurable accumulation of PS in the exoplasmic leaflet unless the cells enter apoptosis.
Discussion
This study illuminates a new connection between specialized membrane lipid partitioning, B cell differentiation, and antigen-specific antibody production. A previously unstudied member of the large family of P4-type ATPases, ATP11c, was required for efficient PS flippase activity in developing B-lymphocytes, and was essential for normal B cell differentiation from the pro-B cell stage onwards. As a result of ATP11c mutation, fewer pro-B cells expressed rearranged Igh genes, pro-and pre-B cells appeared unresponsive to supraphysiological concentrations of IL-7 in vivo and differentiated poorly during IL-7 driven proliferation in vitro, there was poor induction of CD24 and CD25 on pre-BCR expressing cells, and a higher percentage of pre-B cells were apoptotic. Little is understood about the relationship between the composition of the inner leaflet of the plasma membrane and cytokine receptor or antigen receptor signaling, and the discovery that ATP11c is critical for B cell differentiation opens up a new avenue to bridge this gap.
With respect to the biochemical function of ATP11c, the simplest interpretation of the data is that it is an essential aminophospholipid flippase in the plasma membrane of pro-B cells, because PS flippase activity is slower in Atp11c mutant cells even when apoptosis is blocked by Bcl-2, and other members of this protein family have been shown to be PS flippases in biochemical reconstitution and yeast genetic studies14,21–23. However, analyses of yeast mutants in P4 ATPases have underscored the potential for effects on protein or organelle trafficking and cell metabolism that could indirectly affect PS flippase activity14,30–32. Future studies will be needed to confirm the PS flippase activity of Atp11c in biochemical reconstitution studies, and to test its relative activity for phosphatidylethanolamine and phosphatidylinositol that are also concentrated on the cytoplasmic leaflet. The observation that PS flippase activity was slower but by no means abolished in Atp11c-mutant pro-B cells implies that other aminophospholipid flippases also act in B cells, consistent with the relatively ubiquitous mRNA expression of many other P4 ATPase family members.
How might the loss of one flippase have such a specific and large effect on pro-B cell differentiation? The data indicate a direct or indirect defect in the response to IL-7R and pre-BCR. In contrast to Il7−/− or Il7ra−/− mice with complete absence of IL-7R signaling33, where B cells fail to develop to the pro-B cell stage, B cells in Atp11c mutant mice develop into pro-B cells, proliferate normally, and display an arrest similar to Il7ra−/− Vav-Bcl2 mice34. Both the in vivo and in vitro defects could all reflect some critical qualitative element of IL-7 signaling, or could be secondary since there is known to be a complex feedforward loop between IL-7 signaling, B cell transcription factors, and pre-BCR signaling1–4. The elevated IL-7R on the surface of mutant pro-B cells may reflect less signaling through the IL-7 receptor, because binding of IL-7 to the IL-7R normally leads to rapid endocytosis of the receptor via clathrin-coated pits necessary for efficient signal transduction35 and IL-7R signaling normally diminishes IL-7R mRNA transcription36,37. The biophysics of antigen receptor or IL-7R signaling is not well understood, but several independent observations provide possible connections with PS concentrations in the cytoplasmic membrane leaflet and in lipid rafts. PS is most concentrated on the inner leaflet of the plasma membrane at endocytic cups and early endosomes13 and in lipid rafts associated with the T cell receptor38, where it serves as a binding site and regulatory factor for proteins with cationic or C2-domains, such as src, ras, rho and PKC family proteins13, and for binding the cationic stretch preceding ITAMs in CD3 subunits39,40. The efficiency of IL-7R signaling has also recently been linked to its association with lipid rafts41,42. Phosphatidylserine has also been found to be externalized and co-capped with the BCR during BCR or pre-BCR signaling43. Thus, it will be important in future studies to test the consequences of Atp11c mutation for accumulation of PS and signal-transducing proteins at subcellular patches where IL-7, pre-BCR or BCR signaling has been initiated.
Why are B cells profoundly compromised by the Atp11c mutation while T cells and most other tissues appear normal? Microarray profiling of Atp11c mRNA shows that it is equally expressed in B and T lymphocytes (data not shown) and most other tissues with highest expression in liver44. PS flippase activity was nevertheless also reduced in DN and DP pro- and pre-T cells, yet the mutation selectively compromised differentiation of pro-B cells. The simplest explanation may be redundancy between ATP11c and other P4 ATPases in other B cell stages and lineages. Alternatively, post-translational regulatory changes in phospholipid flippase, floppase and scramblase activity, and unique membrane events required in bone marrow pro-B and pre-B cells, may explain the stage- and lineage-specific dependence upon ATP11c. The profound requirement for ATP11c in pro-B and pre-B lymphocytes provides a unique experimental avenue for understanding the connections between membrane phospholipid distribution and cell biological functions that are likely to be important for many other cell types, and may provide a new drug target for treating B cell malignancy. The X-chromosomal location of human ATP11c makes it a candidate for inherited B cell deficiency and hepatocellular carcinoma in men.
Methods
Mice and procedures
The ambrosius mouse strain was generated through ENU mutagenesis on a pure C57BL/6 background at the Australian Phenomics Facility, the Australian National University as described previously46. The strain was maintained either by breeding affected males with littermates or with wild-type C57BL/6 females and for mapping purposes crossed to the CBA/H strain. The Vav-Bcl2-Tg26, H2Ea-Il7-Tg27, MD4 strain28 and the Rag1−/− mice have been described previously. The Cd79a−/− mouse strain has a point mutation in Cd79a that leads to a premature stop codon in exon 2 (L. Tze and A. Enders, manuscript in preparation). Except when noted specifically all mice used for this study were between 8–12 weeks old. All experimental mice were maintained on a C57BL/6 background and housed in specific pathogen free conditions at the Australian Phenomics Facility and all animal procedures were approved by the Australian National University Animal Ethics and Experimentation Committee.
Primary immunization with 50 μg ARS-CGG (Biosearch) and 1 x 108 heat- and formalin-inactivated B. pertussis bacteria (Lee Laboratories) and booster immunizations with ARS-CGG and 25 μg NP-Ficoll (Biosearch) and detection of antibodies by enzyme-linked immunosorbent assay were performed as described previously47. Differential absolute blood counts were performed on an ADVIA 2120 Haematology System (Siemens Healthcare Diagnostics)
Determination of serum concentrations of (ALT), (AST) and bilirubin assayed by automated procedures in the Department of Pathology, The Canberra Hospital.
Tumor harvest and histology
Liver tumors, and surrounding liver tissue were removed and fixed in 10% neutral buffered formalin. Liver sections (4 μm) were cut from paraffin-embedded blocks and stained with (H&E) for histological examination.
Sequencing and mutation identification
A SureSelect custom solution array (Agilent) was designed using the online tool, eArray to include Refseq release 43 (NCBI) annotated exons (plus splice donor and acceptor sites) within the 54012901 bp to 60897610 bp chromosome X linkage interval. 100 bp paired end Illumina libraries of the captured regions from a single affected mouse were produced and run in a single lane of an Illumina GAIIx.
Sequence reads were mapped to the NCBIM37 assembly of the reference mouse genome using the bowtie aligner48. Untrimmed reads were aligned allowing a maximum of two sequence mismatches and were discarded where they aligned to the genome more than once. Sequence variants were identified with SAMtools49 and custom perl scripts were subsequently used to identify those which occurred within exons and splice donor-acceptor sites, and were not known or strain-specific variants.
mRNA isolation and amplification from cDNA
Total RNA was isolated with the mirVana Isolation kit (Ambion). cDNA was prepared with the SuperScript First Strand Synthesis System (Invitrogen) using Oligo-dT primers. PCR amplification of parts of the ATP11c gene was performed using. PCR products were amplified by using AccuPrime High Fidelity Taq (Invitrogen) and the following primer sequences: Forward: GGCCCTTCTTACATTGGACA, Reverse: GAAGGTCTGGCGGATAATGA. After gel purification PCR products were sequenced on a ABI 3730 Sequencer (Aplied Biosystems) at the ACRF Biomelecular Resource Facility (JCSMR, ANU) according to the manufacturer’s protocol.
Flow cytometric analysis and cell sorting
Cell suspensions were prepared, counted by trypan blue exclusion and equal number of cells aliquoted into the 96-well round-bottom plates. Cells were then incubated with a primary antibody cocktail containing an appropriate combination of the antibodies each diluted to its optimal concentration in flow cytometry buffer (PBS containing 2% bovine serum and 0.1% NaN3) and incubated for 30 min at 4 °C in the dark. Samples were washed with flow cytometry buffer and resuspended in flow cytometry buffer and analyzed using a LSR II or LSR Fortessa flow cytometer (BD Biosciences).
For intracellular staining cells were fixed and permeabilized with the Foxp3 staining kit from eBioscience. Briefly, cell surface staining of 1 – 2 × 106 cells was performed and cells were washed once with flow cytometry buffer followed by resuspension of the pelleted cells in 50 μl of Fix/Perm solution for 30 min. After one wash with 1x Permeabilization/wash buffer solution, cells were incubated with appropriately conjugated intracellular antibodies. After a final wash step once with the permeabilization/wash buffer and once with flow cytometry buffer, cells were resuspended in flow cytometry buffer and analyzed on a LSR II flow cytometer (BD).
Cell sorting was performed by staining single cell suspensions from the bone marrow of mutant and control mice with appropriate conjugated antibodies in sterile flow cytometry buffer. Pro-B cells were sorted on a Aria I or II flow cytometer (BD Biosciences) and collected in sterile RPMI supplemented with 10 mM HEPES pH 7.2–7.5 (Sigma), 0.1 mM Non-Essential Amino Acid solution (Sigma), 1 mM Sodium Pyruvate (Sigma), 55 μM 2-Merchapthoethanol (GibroBRL), 50 U/ml penicillin-streptomycin (GibroBRL) and 10% heat-inactivated Fetal Calf Serum (FCS) for culturing.
For analysis or sorting the following antibodies were used (Clone names are shown in parenthesis): CD3 (145-2C11), CD4 (RM4-5), CD8 (53-6.7), CD19 (1D3), CD21/35 (7G6), CD23 (B3B4), CD25 (7D4), CD43 (S7), CD45.2 (104), B220 (RA3-6B2), Gr-1 (RB6-8C5), IgM (II/41), NK1.1 (PK136) all from BD; CD5 (53-7.3), CD8 (53-6.7), CD19 (eBio1D3), CD93 (AA4.1), CD93 (C1qRp), CD127 (A7R34), c-Kit (ACK2), B220 (RA3-6B2), IgD (11–26), IgM (II/41), Flt3 (A2F10), Ter119 (TER-119) all from eBiosciences; CD3 (17A2), CD21/35 (7E9), CD23 (B3B4), CD24 (M1/69), CD43 (1B11), CD45 (30-F11), CD45.1 (A20), BP-1 (6C3), IgD (11-26c.2a), Mac-1 (M1/70), TCRβ (H57-597), Sca-1 (D7), Annexin V all from Biolegend; CD24 (91) from Southern Biotechnology, 7-AAD and Streptavidin Qdot 605 from Invitrogen.
For lineage analysis in Supplementary Fig. 5a,b Biotin labeled CD4 (GK1.5), CD5 (53-7.3), CD8, CD11c (HL3), IgM (II/41), B220 (RA3-6B2), Ter119, NK1.1 (PK136), Ly6c (AL21), Gr-1 (RB6-8C5), TCRβ (H57-597), TCRγδ (GL3), Mac-1 (M1/70) all from BD; CD19 (eBio1D3) and IgD (11–26) from eBioscience were used. Streptavidin Qdot 605 from Invitrogen was used as a secondary antibody.
For lineage analysis in Supplementary Fig. 6a,b Biotin labeled CD4 (GK1.5), CD5 (53-7.3), CD8, CD11c (HL3), IgM (II/41), Ter119, NK1.1 (PK136), Ly6c (AL21), Gr-1 (RB6-8C5), TCRβ (H57-597), TCRγδ (GL3) and Mac-1 (M1/70) all from BD were used. Streptavidin Qdot 605 from Invitrogen was used as a secondary antibody.
Generation of Bone Marrow Chimeras
Bone-marrow was extracted from the donor mice by pressurized flow of sterile tissue culture medium through dissected femurs and tibias. Extracted cells were filtered and resuspended in sterile tissue culture medium. Rag1−/− recipient mice were irradiated with a single dose of 450 rads before being intravenously injected with 200 μl of 2 x 106 bone marrow donor cells for each recipient mouse. After reconstitution of the hematopoietic system lymphoid tissues were collected from the recipient mice and single cell suspensions were prepared for analysis on a LSR II flow cytometer (BD Biosciences).
Cell culture
Purified Pro-B cells from bone marrow were counted and then seeded at 5 – 6 x 104 cells per well onto 96-well flat-bottom plates in sterile RPMI supplemented with 10 mM HEPES pH 7.2–7.5 (Sigma), 0.1 mM Non-Essential Amino Acid solution (Sigma), 1 mM Sodium Pyruvate (Sigma), 55 μM 2-Merchapthoethanol (GibroBRL), 50 U/ml of penicillin-streptomycin (GibroBRL) and 10% heat-inactivated Fetal Calf Serum (FCS) and incubated at 37 °C and 5% CO2 for up to 7 days in the presence of 25 ng/ml of IL-7 (R&D Systems). During the culture and at the end of the cultivation period, cells were counted by trypan blue exclusion and stained with appropriate antibodies to cell surface markers and analyzed on a LSR II flow cytometer (BD Biosciences).
Annexin-V staining
Bone marrow and spleen cells from mutant and control animals were isolated and counted by trypan blue exclusion. Equal number of cells from mutant and wild-type was rested for 2 h in the tissue culture medium at 37 °C and 5% CO2. The cells were then stained with appropriate antibodies as detailed above and washed once with flow cytometer buffer and once with Annexin-V binding buffer (Biolegend). Annexin-V and 7-AAD staining were performed in the binding buffer at 20°C for 15 min and washed and resuspended in binding buffer and analyzed on a LSR II flow cytometer (BD Biosciences).
Phospholipid translocase (Flippase) activity assay
Flippase activity assays were performed with mutant and wild-type bone marrow, spleen and thymic cells using the fluorescent analog of phosphatidylserine, NBD-PS (Avanti Polar Lipids, Inc.) as previously reported50 with the following changes. Equal numbers of CD45.1+ marked wild-type cells and CD45.2+ marked mutant cells were mixed and washed with pH 6.0 pre-warmed to 37 °C HBBS (Gibco) supplemented with 5.5mM D-glucose and 20 mM HEPES pH 7.2–7.5. Cells were then incubated with 5 μM NBD-labeled phosphatidylserine in 200 μl of the same solution for the indicated time points at 37 °C. To stop the flipping of NBD-PS cells and to remove the unbound NBD-PS from the cell surface, cells were immediately placed on ice and incubated with HBBS supplemented with 5.5 mM D-glucose, 20 mM HEPES pH 7.2–7.5, 1 % lipid-free BSA for 5 min. After 5 min cells were spun down and washed twice in HBBS supplemented with 5.5 mM D-glucose and 20 mM HEPES pH 7.2–7.5. Cells were then transferred to a 96-well plate and stained with fluorescently labeled antibodies according to standard protocols and analyzed on a LSR II or LSR Fortessa flow cytometer (BD Biosciences).
ATP11c homology model
A profile based alignment generated using Hidden Markov Models as provided by the HHPred server (http://toolkit.tuebingen.mpg.de/). The server identified the Sarcoplasmic reticulum Ca2+-ATPase (1wpg) as the closest relative with a known structure. Based on the alignment the homology model was computed using MODELLER as provided by the server.
Statistical Analysis
The two-tailed student t test was performed. For all statistical analysis, differences were taken to be significant when P < 0.05. All statistical analysis was performed using GraphPad Prism 5 (GraphPad Software).
Supplementary Material
Acknowledgments
We thank the staff of the ANU Bioscience Research Services for excellent animal husbandry, the Australian Phenomics Facility genotyping and mapping team for genetic analysis, D. Howard for help with the bone marrow chimeras, A. Bröer for help with the flippase assay, staff of the Microscopy and flow cytometry resource facility for help with flow cytometry, and the ACRF Biomolecular resource facility and the Australian Genome Research Facility (AGRF) for DNA sequencing. The samples for next generation sequencing were prepared and run in parallel by K. Peng in the ACRF Biomolecular resource facility, JCSMR and the AGRF Next Generation Sequencing and Bioinformatics teams, Brisbane. This study was supported by grants from the National Institutes of Health (C.C.G.), Wellcome Trust (C.C.G.), the Australian Research Council (C.C.G.), the National Health and Medical Research Council (C.C.G.) and the Ramaciotti Foundation (C.C.G. and A.E.). M.Y. was supported by a postgraduate award from the Ministry of National Education, Republic of Turkey and A.E. by the Deutsche Forschungsgemeinschaft (EN 790/1-1).
Footnotes
Author contributions
M.Y. performed and analyzed most of the experiments. C.E.T., S.F., D.L. E.M.K., J.K. and A.E. contributed to experiments; C.M.R. and A.E identified the strain; B.W., D.T.A., Y.Z. and A.E. mapped and identified the mutation. N.C.T. and G.C.F analyzed liver histology and clinical chemistry; S.B. helped with the flippase assay. C.C.G and A.E conceived the research and directed the study. M.Y., C.C.G. and A.E prepared the figures and wrote the paper in consultation with all coauthors.
References
- 1.Malin S, Mcmanus S, Busslinger M. STAT5 in B cell development and leukemia. Curr Opin Immunol. 2010;22:168–176. doi: 10.1016/j.coi.2010.02.004. [DOI] [PubMed] [Google Scholar]
- 2.Ye M, Graf T. Early decisions in lymphoid development. Curr Opin Immunol. 2007;19:123–128. doi: 10.1016/j.coi.2007.02.007. [DOI] [PubMed] [Google Scholar]
- 3.Mandel EM, Grosschedl R. Transcription control of early B cell differentiation. Curr Opin Immunol. 2010;22:161–167. doi: 10.1016/j.coi.2010.01.010. [DOI] [PubMed] [Google Scholar]
- 4.Nutt SL, Kee BL. The transcriptional regulation of B cell lineage commitment. Immunity. 2007;26:715–725. doi: 10.1016/j.immuni.2007.05.010. [DOI] [PubMed] [Google Scholar]
- 5.Welner RS, Pelayo R, Kincade PW. Evolving views on the genealogy of B cells. Nat Rev Immunol. 2008;8:95–106. doi: 10.1038/nri2234. [DOI] [PubMed] [Google Scholar]
- 6.Hardy RR, Kincade PW, Dorshkind K. The protean nature of cells in the B lymphocyte lineage. Immunity. 2007;26:703–714. doi: 10.1016/j.immuni.2007.05.013. [DOI] [PubMed] [Google Scholar]
- 7.Inlay MA, et al. Ly6d marks the earliest stage of B-cell specification and identifies the branchpoint between B-cell and T-cell development. Genes Dev. 2009;23:2376–2381. doi: 10.1101/gad.1836009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wei C, Zeff R, Goldschneider I. Murine pro-B cells require IL-7 and its receptor complex to up-regulate IL-7R alpha, terminal deoxynucleotidyltransferase, and c mu expression. J Immunol. 2000;164:1961–1970. doi: 10.4049/jimmunol.164.4.1961. [DOI] [PubMed] [Google Scholar]
- 9.Melchers F. The pre-B-cell receptor: selector of fitting immunoglobulin heavy chains for the B-cell repertoire. Nat Rev Immunol. 2005;5:578–584. doi: 10.1038/nri1649. [DOI] [PubMed] [Google Scholar]
- 10.Fleming HE, Paige CJ. Pre-B cell receptor signaling mediates selective response to IL-7 at the pro-B to pre-B cell transition via an ERK/MAP kinase-dependent pathway. Immunity. 2001;15:521–531. doi: 10.1016/s1074-7613(01)00216-3. [DOI] [PubMed] [Google Scholar]
- 11.Marshall AJ, Fleming HE, Wu GE, Paige CJ. Modulation of the IL-7 dose-response threshold during pro-B cell differentiation is dependent on pre-B cell receptor expression. J Immunol. 1998;161:6038–6045. [PubMed] [Google Scholar]
- 12.Lingwood D, Simons K. Lipid rafts as a membrane-organizing principle. Science. 2010;327:46–50. doi: 10.1126/science.1174621. [DOI] [PubMed] [Google Scholar]
- 13.Leventis PA, Grinstein S. The distribution and function of phosphatidylserine in cellular membranes. Annu Rev Biophys. 2010;39:407–427. doi: 10.1146/annurev.biophys.093008.131234. [DOI] [PubMed] [Google Scholar]
- 14.Muthusamy BP, Natarajan P, Zhou X, Graham TR. Linking phospholipid flippases to vesicle-mediated protein transport. Biochim Biophys Acta. 2009;1791:612–619. doi: 10.1016/j.bbalip.2009.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Yeung T, et al. Membrane phosphatidylserine regulates surface charge and protein localization. Science. 2008;319:210–213. doi: 10.1126/science.1152066. [DOI] [PubMed] [Google Scholar]
- 16.Park D, et al. BAI1 is an engulfment receptor for apoptotic cells upstream of the ELMO/Dock180/Rac module. Nature. 2007;450:430–434. doi: 10.1038/nature06329. [DOI] [PubMed] [Google Scholar]
- 17.Miyanishi M, et al. Identification of Tim4 as a phosphatidylserine receptor. Nature. 2007;450:435–439. doi: 10.1038/nature06307. [DOI] [PubMed] [Google Scholar]
- 18.Dillon SR, Constantinescu A, Schlissel MS. Annexin V binds to positively selected B cells. J Immunol. 2001;166:58–71. doi: 10.4049/jimmunol.166.1.58. [DOI] [PubMed] [Google Scholar]
- 19.Holder MJ, Barnes NM, Gregory CD, Gordon J. Lymphoma cells protected from apoptosis by dysregulated bcl-2 continue to bind annexin V in response to B-cell receptor engagement: a cautionary tale. Leuk Res. 2006;30:77–80. doi: 10.1016/j.leukres.2005.05.018. [DOI] [PubMed] [Google Scholar]
- 20.Pomorski T, Menon AK. Lipid flippases and their biological functions. Cell Mol Life Sci. 2006;63:2908–2921. doi: 10.1007/s00018-006-6167-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Tang X, Halleck MS, Schlegel RA, Williamson P. A subfamily of P-type ATPases with aminophospholipid transporting activity. Science. 1996;272:1495–1497. doi: 10.1126/science.272.5267.1495. [DOI] [PubMed] [Google Scholar]
- 22.van der Velden LM, van de Graaf SFJ, Klomp LWJ. Biochemical and cellular functions of P4 ATPases. Biochem J. 2010;431:1–11. doi: 10.1042/BJ20100644. [DOI] [PubMed] [Google Scholar]
- 23.Paulusma CC, Elferink RPJO. P4 ATPases--the physiological relevance of lipid flipping transporters. FEBS Lett. 2010;584:2708–2716. doi: 10.1016/j.febslet.2010.04.071. [DOI] [PubMed] [Google Scholar]
- 24.Bull LN, et al. A gene encoding a P-type ATPase mutated in two forms of hereditary cholestasis. Nat Genet. 1998;18:219–224. doi: 10.1038/ng0398-219. [DOI] [PubMed] [Google Scholar]
- 25.Siggs OM, et al. The P4-type ATPase ATP11C is essential for B lymphopoiesis in adult bone marrow. Nat Immunol. doi: 10.1038/ni.2012. this issue. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ogilvy S, et al. Constitutive Bcl-2 expression throughout the hematopoietic compartment affects multiple lineages and enhances progenitor cell survival. Proc Natl Acad Sci USA. 1999;96:14943–14948. doi: 10.1073/pnas.96.26.14943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Mertsching E, Grawunder U, Meyer V, Rolink T, Ceredig R. Phenotypic and functional analysis of B lymphopoiesis in interleukin-7-transgenic mice: expansion of pro/pre-B cell number and persistence of B lymphocyte development in lymph nodes and spleen. Eur J Immunol. 1996;26:28–33. doi: 10.1002/eji.1830260105. [DOI] [PubMed] [Google Scholar]
- 28.Goodnow CC, et al. Altered immunoglobulin expression and functional silencing of self-reactive B lymphocytes in transgenic mice. Nature. 1988;334:676–682. doi: 10.1038/334676a0. [DOI] [PubMed] [Google Scholar]
- 29.Phan TG, et al. B cell receptor-independent stimuli trigger immunoglobulin (Ig) class switch recombination and production of IgG autoantibodies by anergic self-reactive B cells. J Exp Med. 2003;197:845–860. doi: 10.1084/jem.20022144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Gall WE, et al. Drs2p-dependent formation of exocytic clathrin-coated vesicles in vivo. Curr Biol. 2002;12:1623–1627. doi: 10.1016/s0960-9822(02)01148-x. [DOI] [PubMed] [Google Scholar]
- 31.Hua Z, Fatheddin P, Graham TR. An essential subfamily of Drs2p-related P-type ATPases is required for protein trafficking between Golgi complex and endosomal/vacuolar system. Mol Biol Cell. 2002;13:3162–3177. doi: 10.1091/mbc.E02-03-0172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Alder-Baerens N, Lisman Q, Luong L, Pomorski T, Holthuis JCM. Loss of P4 ATPases Drs2p and Dnf3p disrupts aminophospholipid transport and asymmetry in yeast post-Golgi secretory vesicles. Mol Biol Cell. 2006;17:1632–1642. doi: 10.1091/mbc.E05-10-0912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kikuchi K, Lai AY, Hsu CL, Kondo M. IL-7 receptor signaling is necessary for stage transition in adult B cell development through up-regulation of EBF. J Exp Med. 2005;201:1197–1203. doi: 10.1084/jem.20050158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Malin S, et al. Role of STAT5 in controlling cell survival and immunoglobulin gene recombination during pro-B cell development. Nat Immunol. 2010;11:171–179. doi: 10.1038/ni.1827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Henriques CM, Rino J, Nibbs RJ, Graham GJ, Barata JT. IL-7 induces rapid clathrin-mediated internalization and JAK3-dependent degradation of IL-7Ralpha in T cells. Blood. 2010;115:3269–3277. doi: 10.1182/blood-2009-10-246876. [DOI] [PubMed] [Google Scholar]
- 36.Alves NL, van Leeuwen EMM, Derks IAM, van Lier RAW. Differential regulation of human IL-7 receptor alpha expression by IL-7 and TCR signaling. J Immunol. 2008;180:5201–5210. doi: 10.4049/jimmunol.180.8.5201. [DOI] [PubMed] [Google Scholar]
- 37.Park JH, et al. Suppression of IL7Ralpha transcription by IL-7 and other prosurvival cytokines: a novel mechanism for maximizing IL-7-dependent T cell survival. Immunity. 2004;21:289–302. doi: 10.1016/j.immuni.2004.07.016. [DOI] [PubMed] [Google Scholar]
- 38.Zech T, et al. Accumulation of raft lipids in T-cell plasma membrane domains engaged in TCR signalling. EMBO J. 2009;28:466–476. doi: 10.1038/emboj.2009.6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Xu C, et al. Regulation of T cell receptor activation by dynamic membrane binding of the CD3epsilon cytoplasmic tyrosine-based motif. Cell. 2008;135:702–713. doi: 10.1016/j.cell.2008.09.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Aivazian D, Stern LJ. Phosphorylation of T cell receptor zeta is regulated by a lipid dependent folding transition. Nat Struct Biol. 2000;7:1023–1026. doi: 10.1038/80930. [DOI] [PubMed] [Google Scholar]
- 41.Cho JH, Kim HO, Surh CD, Sprent J. T Cell Receptor-Dependent Regulation of Lipid Rafts Controls Naive CD8+ T Cell Homeostasis. Immunity. 2010;32:214–226. doi: 10.1016/j.immuni.2009.11.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Rose T, et al. Interleukin-7 compartmentalizes its receptor signaling complex to initiate CD4 T lymphocyte response. J Biol Chem. 2010;285:14898–14908. doi: 10.1074/jbc.M110.104232. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 43.Dillon SR, Mancini M, Rosen A, Schlissel MS. Annexin V binds to viable B cells and colocalizes with a marker of lipid rafts upon B cell receptor activation. J Immunol. 2000;164:1322–1332. doi: 10.4049/jimmunol.164.3.1322. [DOI] [PubMed] [Google Scholar]
- 44.Wu C, et al. BioGPS: an extensible and customizable portal for querying and organizing gene annotation resources. Genome Biol. 2009;10:R130. doi: 10.1186/gb-2009-10-11-r130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Toyoshima C, Nomura H, Tsuda T. Lumenal gating mechanism revealed in calcium pump crystal structures with phosphate analogues. Nature. 2004;432:361–368. doi: 10.1038/nature02981. [DOI] [PubMed] [Google Scholar]
- 46.Nelms KA, Goodnow CC. Genome-wide ENU mutagenesis to reveal immune regulators. Immunity. 2001;15:409–418. doi: 10.1016/s1074-7613(01)00199-6. [DOI] [PubMed] [Google Scholar]
- 47.Randall KL, et al. Dock8 mutations cripple B cell immunological synapses, germinal centers and long-lived antibody production. Nat Immunol. 2009;10:1283–1291. doi: 10.1038/ni.1820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Langmead B, Trapnell C, Pop M, Salzberg SL. Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol. 2009;10:R25. doi: 10.1186/gb-2009-10-3-r25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Li H, et al. The Sequence Alignment/Map format and SAMtools. Bioinformatics. 2009;25:2078–2079. doi: 10.1093/bioinformatics/btp352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Hanada K, Pagano RE. A Chinese hamster ovary cell mutant defective in the non-endocytic uptake of fluorescent analogs of phosphatidylserine: isolation using a cytosol acidification protocol. J Cell Biol. 1995;128:793–804. doi: 10.1083/jcb.128.5.793. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.