Abstract
Yeast cytosine deaminase catalyzes the hydrolytic deamination of cytosine to uracil as well as the deamination of the prodrug 5-fluorocytosine (5FC) to the anticancer drug 5-fluorouracil. In this study, the role of Glu64 in the activation of the prodrug 5FC was investigated by site-directed mutagenesis, biochemical, NMR, and computational studies. Steady-state kinetics studies showed that the mutation of Glu64 causes a dramatic decrease in kcat and a dramatic increase in Km, indicating Glu64 is important for both binding and catalysis in the activation of 5FC. 19F-NMR experiments showed that binding of the inhibitor 5-fluoro-1H-pyrimidin-2-one (5FPy) to the wild type yCD causes an upfield shift, indicating that the bound inhibitor is in the hydrated form, mimicking the transition state or the tetrahedral intermediate in the activation of 5FC. However, binding of 5FPy to the E64A mutant enzyme causes a downfield shift, indicating that the bound 5FPy remains in an unhydrated form in the complex with the mutant enzyme. 1H and 15N NMR analysis revealed trans-hydrogen-bond D/H isotope effects on the hydrogen of the amide of Glu64, indicating that the carboxylate of Glu64 forms two hydrogen bonds with the hydrated 5FPy. ONIOM calculations showed that the wild type yCD complex with the hydrated form of the inhibitor 1H-pyrimidin-2-one is more stable than the initial binding complex, and in contrast, with the E64A mutant enzyme, the hydrated inhibitor is no longer favored and the conversion has higher activation energy as well. The hydrated inhibitor is stabilized in the wild-type yCD by two hydrogen bonds between it and the carboxylate of Glu64 as revealed by 1H and 15N NMR analysis. To explore the functional role of Glu64 in catalysis, deamination of cytosine catalyzed by the E64A mutant was investigated by ONIOM calculations. The results showed that without the assistance of Glu64, both proton transfers before and after the formation of the tetrahedral reaction intermediate become partially rate-limiting steps. The results of the experimental and computational studies together indicate that Glu64 plays a critical role in both binding and chemical transformation in the conversion of the prodrug 5FC to the anticancer drug 5-fluorouracil.
A major problem in cancer treatment is the toxicity of anticancer drugs to normal cells. Gene directed enzyme prodrug therapy (GDEPT) is aimed at addressing this critical problem by using nontoxic prodrugs, activating them in tumors, and thereby killing cancer cells while minimizing the side effects of anticancer drugs (1–3). One of enzyme-prodrug combinations frequently used in GDEPT is cytosine deaminase (CD) and 5-fluorocytosine (5FC). CD is a metalloenzyme that catalyzes the deamination of cytosine to generate uracil as well as the deamination of 5FC to generate 5-fluorouracil (5FU). 5FU is an anticancer drug used for the treatment of colorectal, breast, stomach, and pancreatic cancers, but, like many anticancer drugs, 5FU is highly toxic, causing side effects such as myelosuppression, mucositis, dermatitis, and diarrhea. 5FC is fairly non-toxic, as CD is absent in human. By activating the prodrug 5FC in the tumor, the CD/5FC-based GDEPT minimizes the side effects of 5FU.
Two CDs have been used as the activating enzyme in the CD/5FC-based GDEPT. One is from E. coli (eCD), and the other from yeast (yCD). The two enzymes have no detectable homology and differ both in size and in tertiary and quaternary structures, arising apparently by convergent evolution. The E. coli enzyme is a homohexameric protein with a molecular weight of 47.5 kDa per subunit and has a TIM-barrel fold (4), whereas the yeast enzyme is a homodimeric protein with a molecular weight of 17.5 kDa per subunit and has a three-layered α-β-α fold (5, 6). Although eCD was used in early GDEPT studies, among wild-type CDs, yCD is preferable for GDEPT, because yCD has a much higher catalytic efficiency for 5FC than eCD (7–9). However, a most recent study showed that a mutant form of eCD is superior to yCD (10).
Because of its biomedical significance, there have been both experimental and computational studies of the catalytic mechanism of yCD. The structure of yCD has been determined by X-ray crystallography at high resolution, including the ligand-free form (5) and the complex form with the inhibitor 1H-pyrimidin-2-one (Py) (5, 6). The bound Py is in the hydrated form 4(R)-hydroxy--3,4-dihydro-1H-pyrimidin-2-one (DHP), mimicking the transition state for the deamination of cytosine. The key elements of the catalytic apparatus of yCD are believed to be a bound zinc ion, which is partially coordinated by bound DHP, and a completely conserved glutamate residue, Glu 64, which has two hydrogen (H) bonds to DHP (Figure 1). The catalytic mechanism of yCD has been investigated by both experimental and computational approaches (11–16). Transient kinetic and NMR studies have shown that product release is rate-limiting in the activation of 5FC by yCD (13). A complete reaction pathway for the deamination of cytosine by yCD has been proposed by two-layered quantum mechanics calculations using the ONIOM method (11) and combined molecular dynamics simulations and ONIOM calculations (14). In the proposed reaction pathway, Glu64 plays a critical role in catalysis, serving as a proton shuttle for both the generation of the tetrahedral reaction intermediate from substrate cytosine and the conversion of the intermediate to the product uracil. However, the role of Glu64 in the enzymatic reaction has not been investigated experimentally. In the present work, we have investigated the role of Glu64 in the activation of 5FC by combined experimental and computational approaches.
Figure 1.
Interactions of the transition state analogue DHP with yCD. The drawing is based on the 1.14 Å-resolution crystal structure of yCD in complex with DHP (PDB code 1P6O) (5). Hydrogen atoms were added using InsightII. The zinc atom (green) is coordinated with His62, Cys91, Cys94 and the 4-hydroxyl group of DHP.
EXPERIMENTAL PROCEDURES
Materials
Restriction enzymes and T4 ligase were purchased from New England Biolabs. Pfu DNA polymerase was purchased from Strategene. [6-3H]-5-fluorocytosine was purchased from Moravek. All other chemicals and biochemicals, including cytosine, 5-fluorocytosine and 5-fluorouracil, were purchased from Sigma-Aldrich.
Chemical synthesis of 5-fluoro-1H-pyrimidin-2-one (5FPy)
5FPy was synthesized via a two-step procedure. 5-fluoro-4-thiouracil (5F4SU) was first prepared from 5FU by thiation with Lawesson’s reagent in 1,4-dioxane according to a published procedure (17). 5FPy was obtained by the Raney-Ni catalyzed desulfurization of 5F4SU (18). The compounds were characterized by spectrophotometry and NMR spectroscopy. All received data were in agreement with literature.
Cloning and Site-Directed Mutagenesis
The plasmid construct for the over-production of yCD with a TEV protease-cleavable N-terminal His tag was made by PCR cloning using the plasmid construct pET17b-yCD (13) as a template. The PCR primers were 5’-GGG ATC CAT ATG GCA AGC AAG TGG GAT CAG-3’ and 5’-GGA ATT CTA CTC ACC AAT ATC TTC AAA CC-3’. The PCR product was digested with NdeI and EcoRI restriction enzymes and ligated with the vector pET-17bHR digested with the same restriction enzymes. The ligation mixture was transformed into E. coli strain DH5α. The correct coding sequence of the cloned yCD gene was verified by DNA sequencing of the entire gene. The resultant plasmid construct (pET17bHR-yCD) was then transformed into E. coli strain BL21(DE3) pLysS for the production of a His-tagged yCD.
The E64A mutation was made by PCR-based site-directed mutagenesis using the plasmid construct pET17b-yCD (13) as a template. The PCR primers were 5’-CC ACA CAT GGT GCG ATC TCC ACT TTG GAA AAC-3’ and 5’-GTT TTC CAA AGT GGA GAT CGC ACC ATG TAG TGT GG-3’. The mutants were selected and verified by DNA sequencing. For the production of a His-tagged E64A mutant enzyme, the mutated gene was excised with NdeI and EcoRI restriction enzymes and cloned into the expression vector pET-17bHR.
The E64D mutation was made by PCR-based site-directed mutagenesis using the plasmid construct pET17bHR-yCD as a template. The PCR primers for the E64D mutagenesis were 5’-CC ACA CTA CAT GGT GAC ATC TCC ACT TTG GAA AAC-3’ and 5’-GTT TTC CAA AGT GGA GAT GTC ACC ATG TAG TGT GG-3’. The mutants were selected and verified again by DNA sequencing.
All the mutant plasmid constructs were transformed into E. coli strain BL21(DE3) pLysS for protein production.
Expression and Purification
All the non-tagged proteins were produced and purified using the same protocol as previously described (13). The His-tagged proteins were purified by chromatography on a Ni-NTA agarose column. The bacterial lysate was loaded onto the column pre-equilibrated with 10 mM imidazole in precooled buffer A (50 mM potassium phosphate buffer, pH 8.0, and 300 mM NaCl). The column was washed with the equilibration buffer until the OD280 of the effluent was <0.05. The column was then eluted with a linear 10–250 mM imidazole gradient in buffer A. Fractions containing the yCD protein were identified by SDS-PAGE and concentrated with an Amicon concentrator using an YM10 membrane. The protein solution was then subjected to TEV protease digestion for 4 h at room temperature in the presence of 0.5 mM EDTA and 1 mM DTT. The digested protein solution was dialyzed against buffer A. The His-tag released by the TEV protease digestion and the undigested protein was removed by loading the dialyzed protein solution onto another Ni-NTA agarose column. The flow-through fraction was collected, concentrated, dialyzed against 2 mM phosphate buffer (pH 8.0), and lyophilized. The protein powder was kept at −80 °C until it was used.
Kinetic Measurements
Steady-state kinetic parameters were measured by a radiometric assay and, for E64A, also by an HPLC assay. For both assays, the buffer contained 50 mM sodium phosphate, pH 7.5, and 50 mM NaCl. The enzyme concentration was 1, 34, and 500 µM for the wild-type yCD, E64D, and E64A, respectively. The 5FC concentration range was 0.08 – 0.68 mM for the wild-type yCD, 6–64 mM for E64D, and 10–100 mM for E64A. A trace amount of [6-3H]-5FC was used to quantify the conversion of 5FC to 5FU. The reaction was initiated and quenched and the product 5FU separated from the substrate 5FC by TLC as previously described (13). The reaction time was 10 s, 8 h, and 30 h for the wild type yCD, E64D, and E64A, respectively. Kinetic parameters were obtained by nonlinear least squares fitting of the data to the standard Michaelis-Menten equation. In the HPLC assay, the reaction was quenched by the addition of an equal volume of 0.1 N HCl. The quenched solutions were injected onto a reverse-phase C18 column (22 × 250 mm). The column was eluted with 20 mM phosphate buffer, pH 2.0 and the elution monitored by UV absorption at 256 nm. The retention times for 5FC and 5FU were 3.6 and 5.8 min, respectively. The peak area of 5FU was integrated and compared with standard samples of known concentrations.
Fluorometric Measurement
The dissociation constant for the binding of 5FPy to the wild-type yCD was measured by fluorometry using a FluoroMax-2 fluorometer. A series of 5FPy solutions of different concentrations were made in 50 mM phosphate buffer, pH 7.5. A yCD stock solution in the same buffer was added into these 5FPy solutions and the mixtures were incubated at room temperature for 10 min in order for them to reach equilibrium. yCD and 5FPy concentrations were 1 µM and 0 – 6.4 µM, respectively. Fluorescence intensities were measured at an emission wavelength of 386 nm with a slit of 4 nm. The excitation wavelength and slit were 320 nm and 4 nm, respectively. A set of controls was obtained in the same manner by adding the buffer instead of the protein stock solution. The Kd value was obtained by nonlinear least-squares fitting of the difference data to the following equation based on an independent one-on-one (one ligand per subunit) binding model:
where ΔF is the fluorescence change upon ligand binding, df is the fluorescence intensity difference per unit concentration between the bound and the free form of 5FPy, Pt and Lt are the total concentrations of the enzyme and 5FPy, respectively, and Kd is the dissociation constant of 5FPy with the yCD enzyme.
Inhibition Assay
The inhibition of the 5FC deamination reaction by 5FPy was measured by the radiometric assay as described earlier. IC50 values were obtained by nonlinear least squares fitting of the inhibition assay data to the equation bellow:
where v is the initial reaction rate, kmin the minimum rate, kmax the maximum rate, and [I] the concentration of the inhibitor. Ki values were calculated by the Cheng-Prusoff equation (19):
where [S] is the concentration of the substrate 5FC.
19F NMR Spectroscopy
NMR samples were prepared by dissolving the wild-type yCD or the E64A mutant enzyme in 100 mM potassium phosphate buffer (pH 7.5) made in D2O. An aliquot of a 5FPy stock solution was added into the protein solutions and the mixtures were incubated for 10 min to reach equilibrium. 19F NMR experiments were performed at 10 or 20 °C on a Varian Inova 600 MHz NMR spectrometer. NMR spectra were acquired with a spectral width of 10,000 Hz, 20,000 complex data points, and 1024 transients and processed with 10 Hz of line broadening. Relaxation delay was 6 s. The 19F chemical shifts were referenced to CF3C6H5, which was set to −63.73 ppm.
1H and 15N NMR Spectroscopy
15N 1H IS-TROSY (20, 21) spectrum was recorded at 25 °C on a Bruker AVANCE 900 MHz NMR spectrometer equipped with a TCI cryoprobe using a sample of 2H/13C/15N-labelled yCD in complex with 5FPy in the 1:1 D2O/H2O buffer. The sample was prepared as previously described (13). Standard jump-return 1H spectra of the apo yCD and the 5FPy complexes of the wild-type and the E64A mutant enzyme were recorded on a Varian INOVA 600 MHz NMR spectrometer with an HCN triple-resonance probehead at either 10 or 25 °C.
ONIOM Calculations
A two-layered ONIOM method (22–29) as implemented in the Gaussian program (Gaussian 03) (30) was used to investigate the effects of the E64A mutation on yCD catalysis. The molecular setup and protocol for the ONIOM calculations were essentially the same as previously described (11). Briefly, the molecular system for the calculations was based on the 1.14 Å-resolution crystal structure of the yCD•DHP complex (PDB code 1P6O) (5). The enzyme was modeled by the catalytic Zn and the active site and neighboring residues, including Ile33, Asn51, Thr60, Leu61, His62, Gly63, Glu64, Ile65, Leu88, Ser89, Pro90, Cys91, Asp92, Met93, Cys94, Thr95, Phe114, Trp152, Phe153, Glu154, Asp155, and Ile156 (11, 12, 14, 15). The system was divided into two layers. The inner layer was composed of the side chains of residue Glu64, His62, Cys91, Cys94, Asn51, Asp155, the zinc ion, the zinc-bound water, and cytosine or Py. The rest of the molecular system constituted the outer layer. Hydrogen atoms were used as link atoms to saturate the dangling bonds. The inner layer was treated with the density functional method B3LYP (31, 32) employing the 6-31G** basis set, and the outer layer by the semi-empirical PM3 method (33, 34). The total ONIOM energy EONIOM is given by (19, 21):
where E(high, model) and E(low, model) are the energies of the inner layer at the high level of theory (B3LYP and the 6-31G** basis set) and at the low level of theory (PM3), respectively, and E(low, real) is the energy of the entire system at the level of PM3.
RESULTS
1. Biochemical Analysis
In order to investigate the role of Glu64 in the activation of the anticancer prodrug 5FC, we replaced Glu64 with alanine and aspartate. Preliminary biochemical characterization of the mutant enzymes without a His tag indicated that the purified mutant enzymes contained a minute amount of eCD, resulting in steady-state kinetic curves requiring two Km’s to fit the data, although the E. coli enzyme is not homologous to the yeast enzyme, different in both size and quaternary structure. In order to get rid of the eCD contamination, both the wild-type yCD and mutants were sub-cloned into a vector for the production of His-tagged proteins. The His-tagged proteins were purified with two Ni-NTA agarose columns. In the first column, proteins bound to Ni-NTA were eluted and collected. The protein solution was then subject to TEV protease digestion to remove the N-terminal His-tag from the tagged yCD. The TEV protease-digested protein solution was loaded to the second Ni-NTA agarose column, in which the flow-through was collected. The second column removed not only undigested yCD but also proteins co-purified with yCD by the first Ni-NTA agarose column. The kinetic behavior of such a yCD mutant enzyme preparation was normal, i.e. requiring only one Km to fit the kinetic data. The protein was purified further by gel filtration (Sephadex G-75) in trial experiments, but the kinetic properties of the protein purified by the three-column procedure were the same as those of the protein purified by the two-column procedure, indicating that the purification procedure with two Ni-NTA columns was effective in removing eCD.
Our analysis was focused on the effects of the E64A mutation, because the mutation completely removed the carboxyl functional group. The steady-state kinetic parameters of the wild-type yCD and the mutant enzymes E64A and E64D are summarized in Table 1. Because of the relatively large errors of the radiometric assay in measuring a small amount of 5FU in the presence of a large amount of 5FC, the kinetic parameters of the E64A mutant enzyme were also determined by an HPLC assay. Both mutations caused a dramatic decrease in kcat and a dramatic increase in Km (Table 1). The E64A mutation caused a decrease in kcat by about 5 orders of magnitude and an increase in Km by about 2 orders of magnitude, resulting in a decrease in kcat/Km by about 8 orders of magnitude. The effects of the E64D mutation were only slightly milder. The kinetic results indicated that Glu64 is involved in both binding and catalysis in the activation of 5FC by yCD, and the longer side-chain is important, as Glu64 cannot be replaced with aspartate with the same functional group but a shorter side-chain.
Table 1.
Kinetic Constants of the Wild-Type yCD and the E64 Mutant Enzymes for the Activation of 5FC
| Protein | Km (mM) | kcat (s−1) | kcat /Km (mM−1s−1) |
|---|---|---|---|
| WT yCD | 0.16 ± 0.01 | 17 ± 0.4 | 106 |
| E64A | 89±30 a | (1.3±0.3)×10−4 a | 1.4 × 10−6 a |
| 33 ± 6 b | (7.3 ± 0.6) × 10−5 b | 2.2 × 10−6 b | |
| E64D | 19±6 | (5.1±1.0)×10−3 | 2.6×10−4 |
Determined by a radiometric assay.
Determined by an HPLC assay.
To further investigate the role of Glu64 in the activation of 5FC, we measured the binding and inhibitory activities of the putative transition state analogue 5FPy. The results are summarized in Table 2. The inhibitory activity of yCD was measured for the E64D mutant enzyme but not for E64A, because the activity of the latter was too low for such an analysis. The E64D mutation caused a decrease in Ki by about 4 orders of magnitude, indicating that the mutant enzyme has a much lower affinity for 5FPy than the wild-type yCD. The Kd value for the binding of 5FPy could be measured by fluorometric titration for the wild-type yCD but not for either of the two mutant enzymes, because the method is not suitable for the weak binding of 5FPy to the mutant enzymes. The Kd value for the binding of 5FPy to the E64A mutant enzyme was estimated by NMR as described below.
Table 2.
Inhibitory and Binding Properties of 5FPy
Measured by fluorometric titration.
Estimated by 19F NMR.
2. 19F NMR Analysis
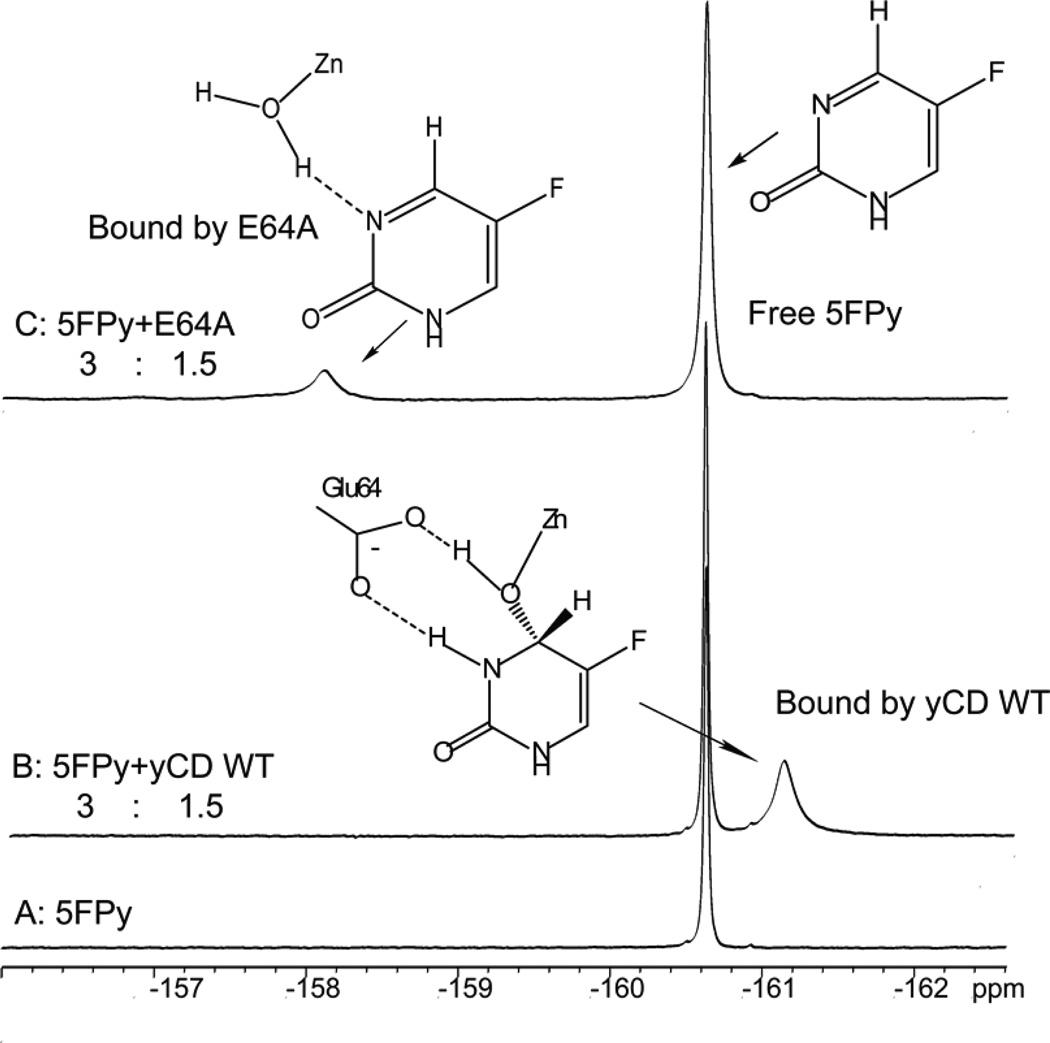
The bound Py in the crystal structure of yCD is in the hydrated form DHP (5, 6). 5FPy differs from Py in having a fluorine atom at position 5 instead of a hydrogen atom. In order to determine whether the bound 5FPy is hydrated like Py and the role of Glu64 in the hydration, 19F-NMR spectra were acquired for 5FPy in the absence or presence of yCD (the wild-type or the E64A mutant enzyme) (Figure 2). In the absence of the enzymes, 5FPy showed a sharp peak at −160.6 ppm. In the presence of a lower concentration of the wild-type yCD, 5FPy showed an additional broad peak due to the bound 5FPy at around −161.1 ppm, upfield of the peak of free 5FPy. In the presence of the same concentration of the E64A mutant enzyme, 5FPy also showed a broad but much smaller peak due to the bound 5FPy at −158.1 ppm, downfield of the peak of free 5FPy. Using model compounds, it has been shown that the 19F NMR signal is a good indicator of the hydration of 5FPy (35). The hydration of 5FPy results in a change in the bonding of C4 from sp2 to sp3, causing a ~1 ppm upfield shift of the 19F NMR signal. This upfield shift of the 19F NMR signal has been used for the determination of the hydration state of 5-fluoropyrimidin-2-one ribonucleoside (35). The dramatic opposite shift of the 19F NMR signal of 5FPy indicated that 5FPy is hydrated when it binds to the wild-type yCD and is not hydrated when it binds to the E64A mutant enzyme. In either case, however, free 5FPy is in slow exchange with the bound 5FPy on the NMR chemical shift timescale. In such an exchange regime, chemical shift does not change with protein or ligand concentration and cannot be used for the estimation of Kd values. However, the relative peak intensities of the NMR signals of the free form and the bound form can be used to estimate the Kd value if the protein and ligand concentrations are in the same order of magnitude of the Kd value. The protein and 5FPy concentrations were suitable for the estimation of the Kd value for the E64A mutant enzyme but not for the wild-type yCD. The Kd value for the binding of 5FPy to the E64A mutant enzyme was estimated to be ~10 mM based on the 19F NMR data, about 5 orders of magnitude higher than that for the binding of 5FPy to the wild-type enzyme. As 5FPy is the hydrated form when it binds to the wild-type yCD and the unhydrated form is favored the hydrated form by a factor of 2.9×103, the Kd for the binding of the hydrated 5FPy, a transition state analogue, to the wild-type CD was estimated to be 2.3 ×10−10 M.
Figure 2.
19F-NMR spectra of the wild-type yCD-5FPy complex and the E64A-5FPy complex. All samples were in a D2O buffer containing 100 mM potassium phosphate, pH 7.5. (A) Spectrum of 5FPy (4.0 mM) in the absence of the wild-type yCD or E64A; (B) spectrum of 5FPy (3.0 mM) in the presence of the wild-type yCD (1.5 mM); (C) spectrum of 5FPy (3.0 mM) in the presence of E64A (1.5 mM).
3. 1H and 15N NMR Analysis
In the crystal structures of the yCD•DHP complex (5), one of the Glu64 side-chain carboxyl oxygen atoms, Oε2, forms a strong H-bond to the O4 H moiety of the hydrated inhibitor with a distance of ~2.5 Å between the two oxygen atoms, and the other carboxyl oxygen, Oε1, forms a normal H-bond with the N3 H imino group of the ligand. Oε1 is also bifurcatedly H-bonded to the backbone amide of the same residue (Figures 1 and 3). Since the bound 5FPy is in the hydrated form (5F-DHP) as indicated by the 19F NMR analysis, we investigated the H-bonding of Glu64 in the yCD•5F-DHP complex by 1H and 15N NMR. Shown in Figure 3 is the close-up view of Glu64 backbone amide resonance in the 15N-1H IS-TROSY (20, 21) spectrum of yCD•5F-DHP. The resonance clearly shows a doublet with a splitting of 17.5 ppb (15.8 Hz at the 900 MHz field for 1H) due to the trans-H-bond D/H isotope effect (36, 37) on the N3 H imino hydrogen of the ligand transmitted via O1, because the corresponding resonance with a similar sample in water leads to a singlet. The large trans-H-bond D/H isotope effect indicates that the Oε1 mediated bifurcated H-bonds are strong (36, 37). Interestingly, a closer inspection on the doublet reveals that it is actually a partial overlap from two sets of doublets with the upper set (upfield in the 15N dimension) having a 5.5 ppb (5.0 Hz at 900 MHz field) upfield shift in the 1H dimension with respect to the lower set (downfield in the 15N dimension). Because no other exchangeable protons are in the vicinity of Glu64 backbone amide, the 5.5 ppb shift difference must be the consequence of the D/H isotope effect on the O4 H hydrogen of the hydrated inhibitor transmitted through the two carboxyl oxygen atoms and two H-bonds. Therefore, the observed long-range trans-H-bond D/H isotope effect convincingly indicates that Oε2⋯H⋯O4 is truly a strong H-bond. This is corroborated by a 16.2-ppm peak in the jump-return 1H spectrum recorded at room temperature, which was assigned to the resonance of the bridging proton, because it disappeared when using either the apo-protein or the E64A mutant enzyme in complex with 5F-DHP (Figure S1). This chemical shift is consistent with those of protons involved in low-barrier H-bonds (LBHBs) (38, 39). There is no any other H-bond with a distance between the two heteroatoms shorter than 2.60 Å in the yCD crystal structures (5). On the other hand, the O4 atom of 5F-DHP is unlikely completely deprotonated by Glu64, otherwise the observed long-range trans-H-bond D/H isotope effect would be much larger. Because the Oε1 mediated H-bonds are strong as indicated by the 17.5 ppb trans-H-bond D/H isotope effect and D/H isotope effects through four covalent bonds can be up to 150 ppb as observed at the active site histidine imidazole ring of the bovine pancreatic enzyme sPLA2 in complex with a phosphonate transition state analogue (40).
Figure 3.
Region of the 15N 1H IS-TROSY spectrum of Glu64 backbone amide resonance. The inset is the schematic diagram of H-bonding between yCD and 5F-DHP derived from the 1.14 Å resolution crystal structure (PDB code: 1P6O) (5). Lengths (Å) of H-bonds are indicated.
4. Computational Analysis
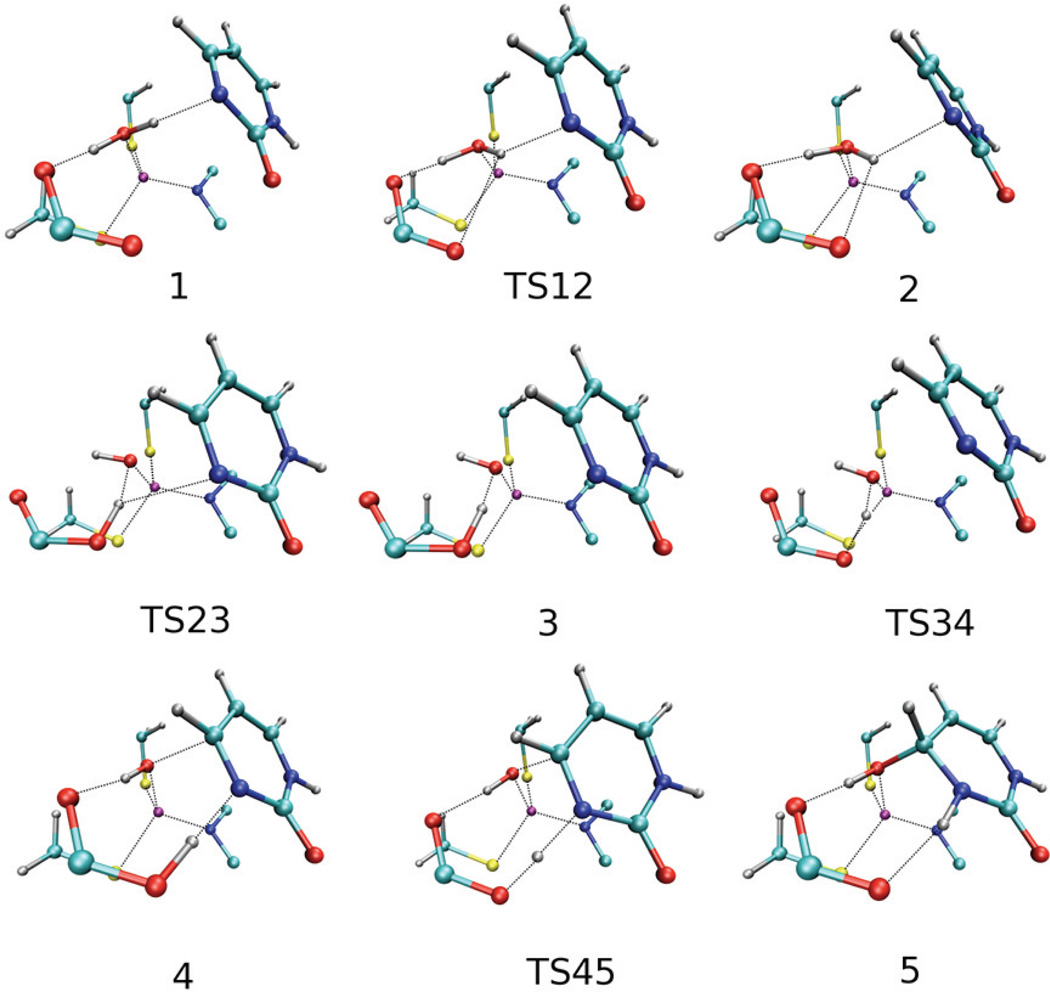
Because 5FPy is hydrated when bound to the wild-type yCD but not hydrated when bound to the E64A mutant enzyme, we investigated the hydration by yCD by ONIOM calculations taking advantage of the availability of the high resolution crystal structure of the enzyme in complex with DHP, the hydrated form of Py. Py was docked in the active site by superposition with DHP in the X-ray crystal structure (5). After optimization, Py shifted slightly from the initial position to give Complex 1 (Figures 4 and 5). Complex 1 is stabilized through a H-bond network and a π-stacking interaction. One new interaction is presented between the zinc-coordinated water OZn and N3 of Py. Glu64 is in the deprotonated form and forms a strong H-bond with the other hydrogen of the zinc-coordinated water. The carbonyl oxygen (O2) of Py is H-bonded to the side chain amide of Asn51 and the backbone amide of Gly63. The amide group at position 1 (N1) of Py is H-bonded to the carboxyl group of Asp155. The latter is also H-bonded to the side chain amide of Asn51. The H-bond between the amide group (Nε2) of His62 and the backbone carbonyl oxygen (Oδ1) of Asp155 helps in maintaining appropriate orientation of the imidazole ring of His62 that stacks with the cytosine ring. Before the hydration occurs, the water is converted to a hydroxide with the assistance of Glu64. First, the H-bond between OZn and N3 is broken and a new H-bond between OZn and Glu64 is formed (not the one already presented). The new conformation is given in Complex 2 (Figures 4 and 5). In this complex, the H-bond OZn…N3 is not completely disrupted. One important feature of this conformation is that the distance between OZn and C4 is shortened because the change of the HOZn position reduces the steric hindrance. Complex 2 is calculated to be less stable than 1 by 2.9 kcal/mol (Figure 6). However, 2 is more stable than 1 by 5.4 kcal/mol at the E(high, model) energy level. The difference of 8.3 kcal/mol indicates the surrounding residues destabilize the complex. The corresponding barrier is 3.2 kcal/mol and there is no barrier found at the E(high, model) energy surface. Second, HOZn is transferred from OZn to Glu64, as seen in Complex 3 (Figures 4 and 5). In this process, the H-bond between OZn and N3 is completely broken. After the proton transfer, a strong H-bond is formed between the Zn-coordinated hydroxide and Glu64-OH. The distance between OZn and C4 is further reduced. Complex 3 is slightly more stable than Complex 2. The energy barrier is calculated to be 5.6 kcal/mol at the ONIOM energy level. Again, there is no barrier found at the E(high, model) level. In the next step, the H-bond between OZn and Glu64-OH is broken and a new H-bond is formed between Glu64-OH and N3, which can be seen in Complex 4 (Figures 4 and 5). The distance between OZn and C4 is shortened to 2.17 Å. The energy barrier is 0.9 kcal/mol at the ONIOM energy level and 1.7 at the E(high, model) energy level. Complex 4 is more stable than the initial Complex 1 by 7.5 kcal/mol at the ONIOM energy level and by 11.8 kcal/mol at the E(high, model) energy level. The Zn-coordinated hydroxide in Complex 4 is well positioned for the nucleophilic attack on C4. The distance between the hydroxide and C4 decreases from 3.01 Å in Complex 1 to 2.66 Å in Complex 2, to 2.44 Å in Complex 3, and 2.17 Å in Complex 4. The nucleophilic attack on C4 is concomitant with the proton transfer from Glu64-OH to N3, resulting in the formation of the tetrahedral product (DHP) as shown in Complex 5 (Figures 4 and 5). The energy barrier is 1.0 kcal/mol at the ONIOM level and 0.7 kcal/mol at the E(high, model) level. The bond lengths, bond angles, and dihedral angles of DHP in complex 5 are in close agreement with those of DHP in the crystal structure (Table 3). The yCD-DHP complex is more stable than the initial yCD•Py complex by 23.2 kcal/mol at the ONIOM level and 30.9 kcal/mol at the E(high, model) level. The difference of 7.7 kcal/mol indicates that the model system is relatively destabilized by the outer layer. 5 is considerably more stable than 1 because of the intrinsic stability of the model system, which suggests that the tight binding of the inhibitor is mainly due to the high affinity of DHP for the wild-type yCD. The results of the calculations using Gaussian 09 on the conversion of Py to DHP and 5FPy to 5F-DHP are similar (data not shown).
Figure 4.
Proposed reaction pathway from Py to DHP catalyzed by the wild type yCD.
Figure 5.
ONIOM optimized structures for the conversion of Py to DHP by the wild type yCD. (1) complex 1, (2) TS12 between complex 1 and 2, (3) complex 2, (4) TS23 between complex 2 and 3, (5) complex 3, (6) TS34 between complex 3 and 4, (7) complex 4, (8) TS45 between complex 4 and 5, (9) complex 5.
Figure 6.
Schematic EONIOM and E(High,model) energy profile for the conversion of Py to DHP catalyzed by the wild-type yCD.
Table 3.
Comparison of the ONIOM Optimized Structure of the Tetrahedral Product 5 with the Crystal Structure of the yCD-Inhibitor Complexa
| Internal coordinate | ONIOM | X-ray structure |
|---|---|---|
| DHP-O4 … Zn | 2.047 | 2.066 |
| His62-Nδ1 ⋯ Zn | 2.007 | 1.991 |
| Cys91-Sγ ⋯ Zn | 2.379 | 2.302 |
| Cys94-Sγ ⋯ Zn | 2.337 | 2.266 |
| DHP-O4 ⋯ C4-DHP | 1.449 | 1.475 |
| DHP-N3 ⋯ C4-DHP | 1.444 | 1.446 |
| ∠Sγ Cys91-Zn-Sγ Cys94 | 115.1 | 117.1 |
| ∠Nδ1 His62-Zn-Sγ Cys91 | 98.4 | 102.0 |
| ∠Nδ1 His62-Zn-Sγ Cys94 | 112.0 | 113.6 |
| ∠ Sγ Cys91-Zn-O4 | 109.5 | 108.3 |
| ∠ Sγ Cys94-Zn-O4 | 104.1 | 103.0 |
| ∠Nδ1 His62-Zn-O4 | 118.2 | 113.1 |
| ∠O4-C4-C5-C6 | 112.6 | 113.1 |
| ∠O4-C4-N3-C2 | 110.8 | 114.4 |
Bond distances are in Å and angles are in degree.
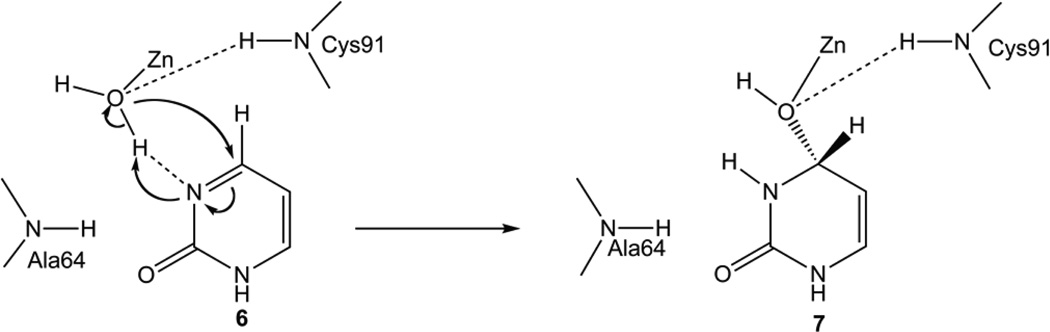
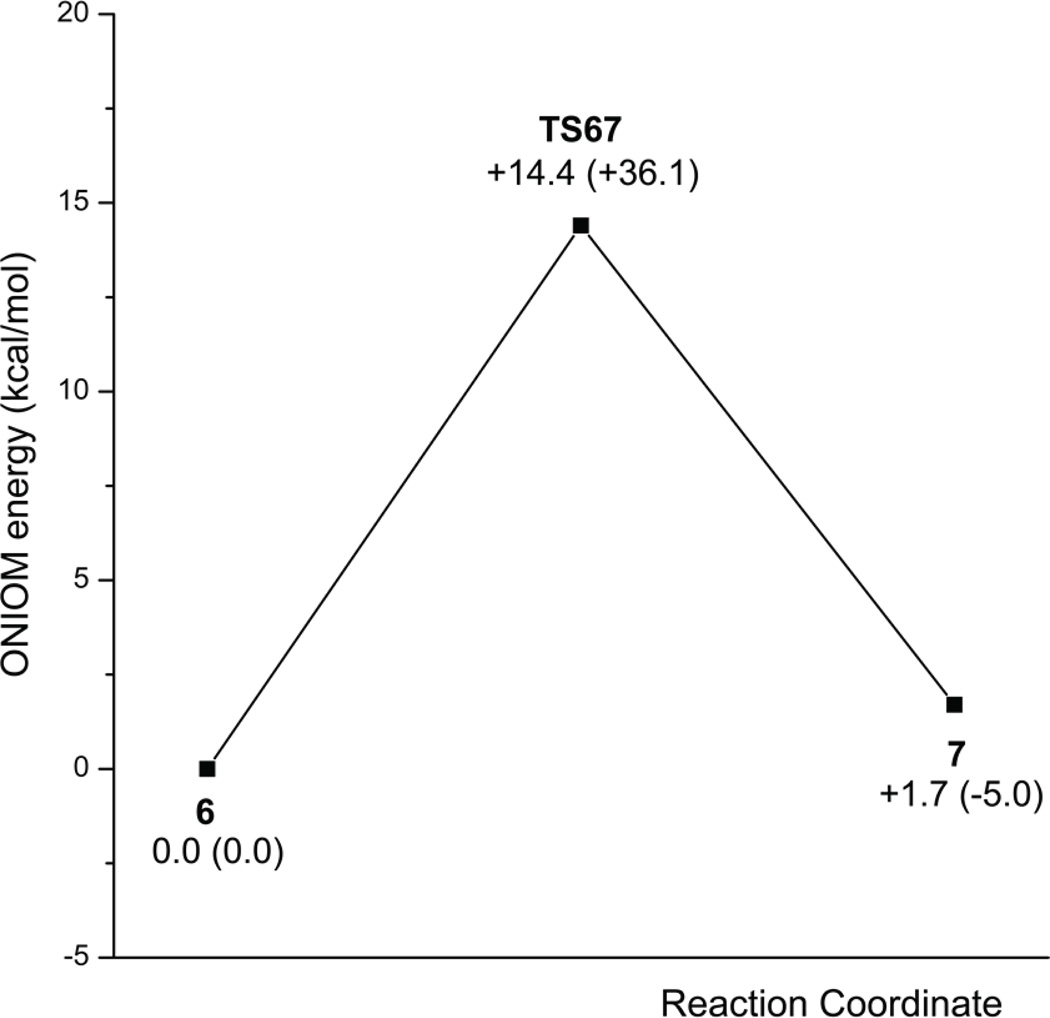
To investigate the effects of the E64A mutation on the hydration, an E64A-Py complex was constructed in the same manner as for the wild-type yCD complex with Py (Complex 6 in Figures 7 and 8), assuming that the structure of the E64A mutant enzyme is the same as that of the wild-type yCD. The conformation of the optimized complex 6 is similar to that of the corresponding wild-type yCD complex. The Zn-coordinated water, however, forms only one H-bond (with N3 of Py), and the other H-bond in the yCD•Py complex is absent, as Glu64 is not present in the mutant enzyme complex. Because of the lack of Glu64, the Zn-coordinated water cannot be converted to hydroxide before the nucleophilic attack on C4. The distance between OZn and C4 is 3.08 Å, longer than that in yCD•Py complex, making it even harder to attack C4. The nucleophilic attack of the Zn-coordinated water on C4 of Py is concomitant with the transfer of one of its protons to N3 of Py to form the tetrahedral product as in Complex 7 (Figures 7 and 8). The approximate geometry of transition state TS67 was found by scanning the HOZn-N3 and OZn-C4 distances from complex 6 to Complex 7. The transition state was found where the OZn-C4 distance is 2.58 Å. The energy barrier is 14.4 kcal/mol at the ONIOM energy level and 36.1 kcal/mol at the E(high, model) level (Figures 7–9), indicating that the protein environment (the outer layer) compensates the barrier by 21.7 kcal/mol. The energy barrier is much higher than the corresponding energy barrier in the hydration catalyzed by the wild-type yCD. It is noted that the proton transfer is transferred slightly ahead of the formation of the OZn-C4 bond. Complex 7 is calculated to be less stable than 6 by 1.7 kcal/mol at the ONIOM level, indicating that the unhydrated form is favored in the active center of the yCD E64A mutant enzyme.
Figure 7.
Proposed reaction pathway from Py to DHP catalyzed by the E64A mutant enzyme.
Figure 8.
ONIOM optimized structures for the conversion of Py to DHP by the E64A mutant enzyme. (1) complex 6, (2) TS67 between complex 6 and 7, (3) complex 7.
Figure 9.
Schematic EONIOM and E(High,model) energy profile for the conversion from Py to DHP catalyzed by the E64A mutant enzyme.
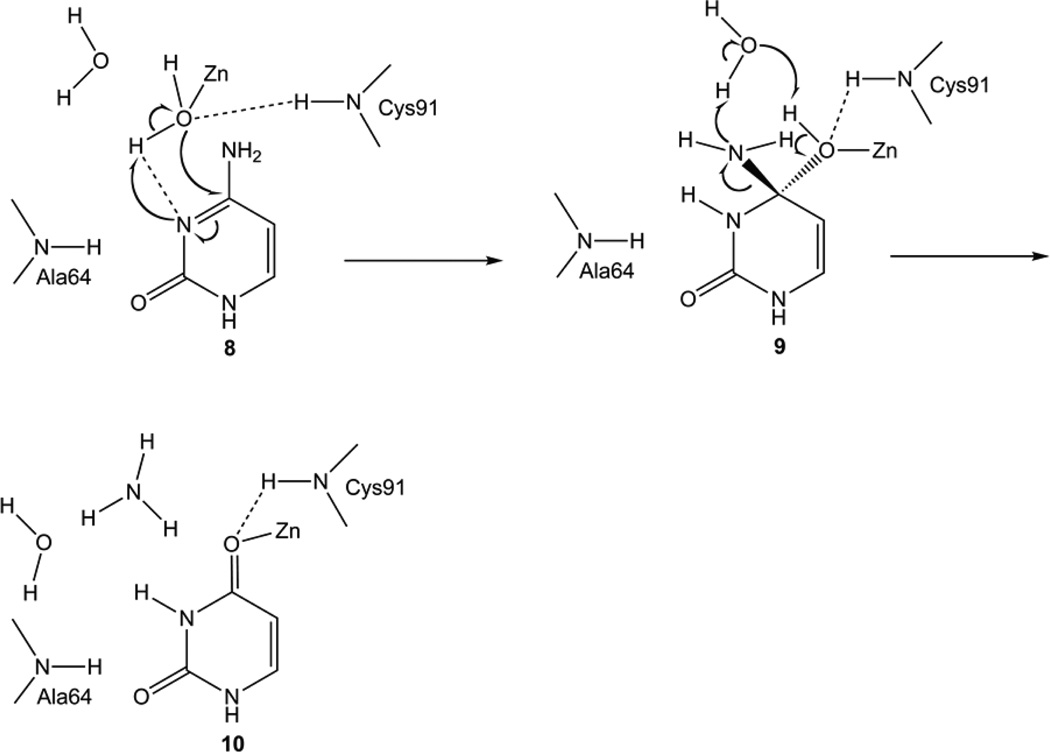
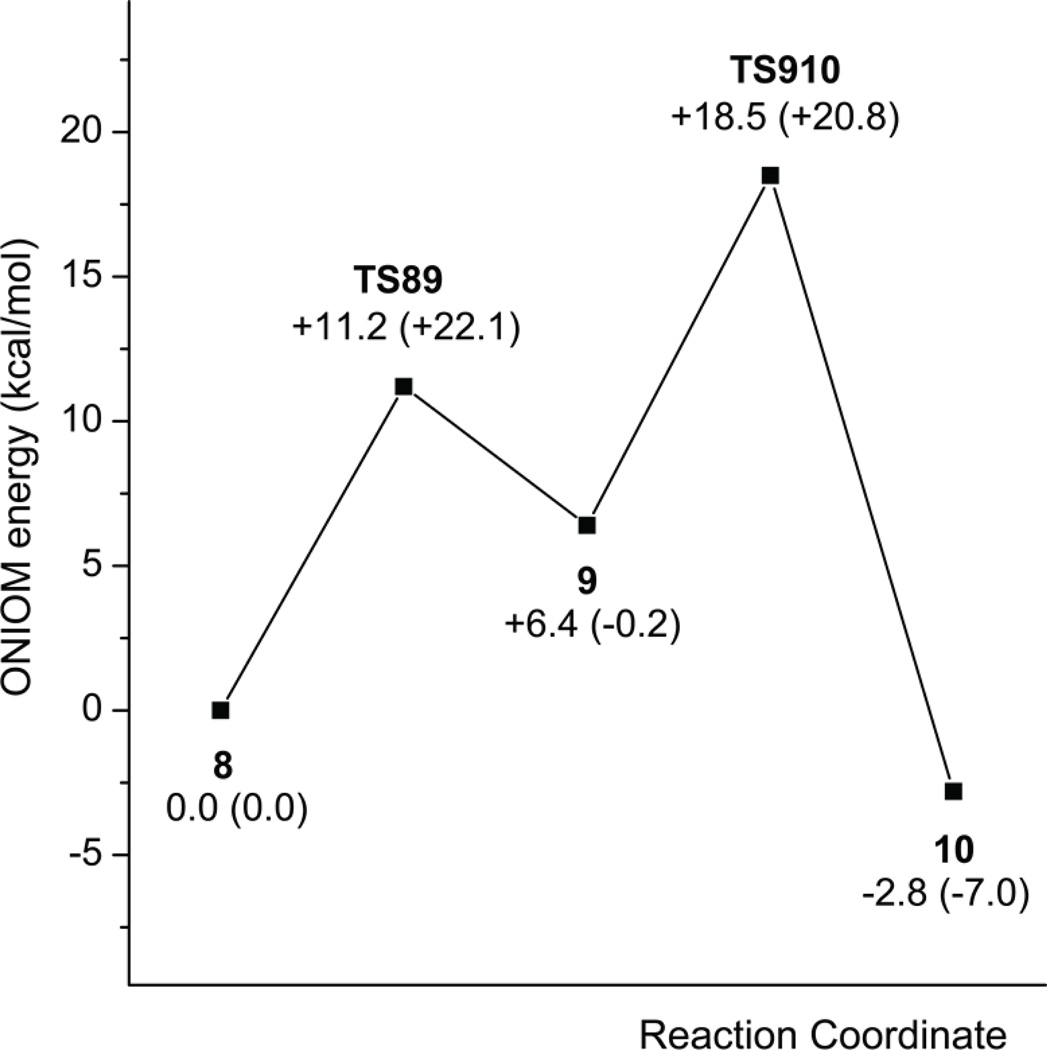
To investigate further the effects of the E64A mutation on the deamination reaction, a complex of the E64A mutant enzyme with the substrate cytosine was constructed as for the construction of the wild-type enzyme-substrate complex (16), assuming again that the structure of the mutant enzyme is the same as that of the wild-type enzyme. A water molecule was added in place of the carboxyl group of Glu64. After optimization, cytosine shifted slightly from the initial position to give Complex 8 (Figures 10 and 11). The interactions between the substrate and enzyme in complex 8 are similar to those in the corresponding wild-type enzyme complex (16) except the interaction involving the Zn-coordinated water and Glu64. In the wild-type enzyme complex, Glu64 forms a H-bond with the Zn-coordinated water and another with the amino group of cytosine. In the mutant enzyme complex, the Zn-coordinated water forms a H-bond with N3 of cytosine, and the amino group of cytosine forms a H-bond with the additional water molecule. As the Zn-coordinated water is H-bonded to N3 and cannot be deprotonated by the additional water molecule, the Zn-coordinated hydroxyl is generated by a direct proton transfer to N3. But the hydroxide is not as well positioned for the nucleophilic attack on C4 as in the corresponding wild-type complex (16). The distance between OZn and C4 is only 2.24 Å in the wild-type enzyme complex but is 2.9 Å in the mutant enzyme complex. The transition state is found when the distance between OZn and C4 is 2.48 Å. The energy barrier for the nucleophilic attack is calculated to be 11.2 kcal/mol at the ONIOM energy level and 22.1 kcal/mol at the E(high, model) level (Figure 12). For the wild-type yCD-catalyzed reaction, the formation of the tetrahedral intermediate is through three steps, in which the highest energy barrier is only 1.9 kcal/mol (11). The stabilization of the tetrahedral intermediate is also effected, because the energy level of the tetrahedral intermediate is 6.4 kcal/mol higher than that of the substrate complex in the mutant enzyme-catalyzed reaction (Figure 12). However the energy level of the same intermediate is 10.8 kcal/mol lower than that of the substrate complex during the wild-type yCD catalyzed reaction (11).
Figure 10.
Conversion of cytosine to the zinc-coordinated uracil catalyzed by the E64A mutant enzyme.
Figure 11.
ONIOM optimized structures for the conversion from cytosine to the zinc-coordinated uracil. (1) complex 8, (2) TS89 between complex 8 and 9, (3) complex 9, (4) TS910 between complex 9 and 10 and (5) complex 10.
Figure 12.
Schematic EONIOM and E(High,model) energy profile for the conversion from cytosine to uracil catalyzed by the E64A mutant enzyme.
Once the tetrahedral intermediate is formed, another proton transfer from OZn to N4 is needed before the C4-N4 bond is cleaved (16). Two possible pathways were explored. In pathway one, the proton is transferred directly to N4 via a four-member ring transition state. In pathway two, the proton transfer is mediated by the additional water molecule. The water donates a proton to N4 and extracts a proton from OZn in a concerted manner. The C4-N4 bond distance increases to 1.64 Å after the proton is transferred to N4. The energy barrier in the second pathway (12.1 kcal/mol at the ONIOM energy level) is dramatically lower than that in the first pathway (22.3 kcal/mol at the ONIOM energy level), indicating that the OZn proton is transferred to N4 via the second pathway. The energy barrier is high when compared with the wild-type yCD catalyzed deamination, in which the energy barrier is only 7.1 kcal/mol at the ONIOM energy level (11).
DISCUSSION
The results of the combined experimental and computational studies reported here indicate that Glu64 is important for both binding and chemical steps in the activation of the anticancer prodrug 5FC. Assuming that the activation of 5FC follows a mechanism similar to that of the deamination of cytosine, it involves the generation of a tetrahedral reaction intermediate from the substrate and the conversion of the intermediate to the product (11). A critical role of Glu64 in the chemical steps is supported by the results of the biochemical analysis of the mutant enzymes. First, the kcat of the E64A mutant enzyme decreases by 5 orders of magnitude relative to that of the wild-type yCD. Our previous transient kinetic and NMR studies showed that product release is rate-limiting in the activation of 5FC by the wild-type yCD (17). Consequently, the kcat of the wild-type enzyme is significantly smaller than the rate constant for the chemical transformation. On the other hand, the rate-limiting step is most likely the chemical transformation in the activation of 5FC by the mutant enzyme, because of its severely impaired catalytic apparatus. Therefore, the kcat of the E64A mutant enzyme should be compared with the corresponding rate constant of the wild-type yCD for the chemical transformation, and thus the E64A mutation slows down the chemical transformation by 6 orders of magnitude. Glu64 contributes to the stabilization of the transition state of the activation of 5FC by ~9 kcal/mol. Second, inhibition and binding studies indicate that the Ki of the putative transition state analogue 5FPy for the inhibition of the E64D mutant enzyme increases by 3 orders of magnitude relative to that for the inhibition of the wild-type yCD and the Kd of 5FPy for the binding of the E64A mutant enzyme increases by 4 orders of magnitude relative to that for the binding of the wild-type yCD, corresponding to ~6 kcal/mol.
A more precise role of Glu64 in the activation of 5FC is revealed by the NMR analysis of the binding of 5FPy to the wild-type and mutant enzymes and the computational analysis of the hydration of Py by the wild-type yCD and the deamination of cytosine by the E64A mutant enzyme. The 19F NMR analysis shows that 5FPy is hydrated when bound to the wild-type yCD but is unhydrated when bound to the E64A mutant enzyme (Figure 2), suggesting that Glu64 plays a critical role in the formation and/or stabilization of the hydrated 5FPy. The ONIOM calculations of the hydration of Py indicate that Glu64 functions as a proton shuttle for the transfer of a proton from the Zn-coordinated water to N3 of Py and the hydration follows a sequential mechanism with N3 protonated first followed by the nucleophilic attack of C4 by the Zn-coordinated hydroxide (Figures 4–6). Furthermore, the complex of the hydrated Py is much more stable than that of the unhydrated Py, based on the dramatic difference in the ONIOM energies of the two complexes (−23.2 kcal/mol). Without the proton shuttle Glu64 in the E64A mutant enzyme, the hydration follows a concerted mechanism with a significantly higher energy barrier (14.4 kcal/mol) and the ONIOM energy of the complex of the hydrated Py is higher than that of the complex of the unhydrated Py (1.7 kcal/mol) (Figures 7–9). The results of the ONIOM calculations suggest that Glu64 both facilitates the hydration of Py and stabilizes the hydrated Py. Since the energy barrier for the hydration of the inhibitor by the E64A mutant enzyme is not insurmountable, the thermodynamic effect is more important. According to the ONIOM calculations, the carboxylate of Glu64 stabilizes the hydrated form of the inhibitor by ~25 kcal/mol. The 15N-1H IS-TROSY analysis (Figure 3) shows that the carboxylate of Glu64 forms two H-bonds with the bound 5F-DHP, just as in the crystal structure of yCD•DHP (5, 6). The two H-bonds must play a major role in the stabilization of the hydrated form of the inhibitor, because 5FPy is predominantly in the unhydrated form when bound to E64A, based on the 19F NMR analysis. As the hydration reaction mimics the formation of the reaction intermediate in the deamination reaction, Glu64 may play a major role in the formation and stabilization of the reaction intermediate in the activation of the anticancer prodrug 5FC.
The ONIOM calculations of the deamination reaction catalyzed by the E64A mutant enzyme show that Glu64 is important for both the formation of the tetrahedral reaction intermediate from the substrate and the conversion of the reaction intermediate to the product (Figures 10–12). Without Glu64 shuttling protons, the barriers for both the formation of the tetrahedral reaction intermediate and its conversion to the product are much higher than those in the wild-type enzyme reaction. Furthermore, the ONIOM energy of the reaction intermediate complex is 10.8 kcal/mol lower than that of the substrate complex in the wild-type enzyme reaction (11) but is 6.4 kcal/mol higher in the mutant enzyme reaction (Figure 12). The combination of these effects makes the mutant enzyme a very sluggish catalyst.
Glu64 is also important for the binding of the anticancer prodrug 5FC. The Km of the E64A mutant enzyme for 5FC increases more than 200-fold in comparison with that of the wild-type enzyme. Because the rate-liming step in the activation of 5FC by the wild-type yCD is the product release, the Km of the enzymatic reaction is a complex parameter, smaller than the Kd for the binding of 5FC (17). On the other hand, the rate-limiting step for the activation of 5FC by the E64A mutant enzyme must be the chemical transformation. Consequently, 5FC is in rapid equilibrium with its complex with the E64A mutant enzyme and thus the Km and Kd values should be similar. Therefore, the Km value of the mutant enzyme is a good measure of the affinity of the mutant enzyme for 5FC and should be compared with the Kd value of the wild-type enzyme for the binding of 5FC, which was determined to be 0.19 mM (13). The ratio of the mutant enzyme’s Km to the wild-type enzyme’s Kd is ~170, indicating that Glu64 contributes to the binding of 5FC by ~3 kcal/mol. The biochemical result is consistent with the complex of the enzyme with cytosine (16) built based on the crystal structure of the complex of yCD with the hydrated Py (10, 11). After energy minimization, Glu64 forms a H-bond with the amino group of cytosine. Disruption of the H-bond by mutagenesis is likely to decrease the affinity of the enzyme for the substrate.
yCD belongs to a family of purine/pyrimidine deaminases with an active center featuring a catalytic zinc coordinated with cysteine and histidine residues and a strictly conserved glutamate. Interestingly, yCD is not homologous to eCD, which has a very different size, a different quaternary structure, and an active center containing a ferrous iron ion coordinated with four histidine residues, an aspartate residue, and a water molecule (4). Among the family of purine/pyrimidine deaminases, yCD is most similar to E. coli cytidine deaminase (CDA), having a catalytic Zn of the same coordination chemistry and a superimposable glutamate (10, 11). However, the functional role of Glu64 of yCD is not the same as that of the corresponding residue Glu104 of E. coli CDA, based on our studies on yCD presented here and the published mutagenesis study on E. coli CDA (23). Both glutamate residues are critical for catalyzing their respective reactions. The E104A mutation of E. coli CDA decreases the kcat of the enzyme by 8 orders of magnitude (23). The effect is more dramatic than that of the E64A mutation of yCD, which decreases the kcat of yCD by 5 orders of magnitude and the rate constant for the chemical transformation by 6 orders of magnitude. Similarly, the fluorinated transition-state analogue 5-fluoropyrimidin-2-one riboside is in the hydrated form when bound to the wild-type E. coli CDA but is in the unhydrated form when bound to the E104A mutant enzyme (22), suggesting that Glu104 also plays a critical role in the generation of the reaction intermediate in the deamination reaction catalyzed by E. coli CDA.
The roles of the two glutamate residues in substrate binding are different. Glu64 of yCD contributes to the stability of the yCD complex with 5FC by ~3 kcal/mol, as discussed earlier. In contrast, Glu104 of E. coli CDA destabilizes the substrate complex of the enzyme E. coli CDA (23). Thus, the Km of the E104A mutant enzyme decreases by a factor of ~30. Interestingly, the Kd for the binding of the fluorinated transition-state analogue 5-fluoropyrimidin-2-one riboside to the E104A mutant enzyme is even lower than that for its binding to the wild-type enzyme, although the fluorinated transition-state analogue is in the unhydrated form when bound to the mutant enzyme, suggesting that Glu104 of E. coli CDA dramatically destabilizes the complex of the unhydrated form of the analogue with the enzyme. It has been proposed that Glu104 of E. coli CDA decreases the energy barrier of the deamination reaction not only by stabilizing the transition state but also by destabilizing the enzyme-substrate complex (22). The destabilization of the enzyme-substrate complex is probably due to the unfavorable interaction between the negatively charged Glu104 and the lone pair of electrons of N3 of cytidine, which becomes a favorable interaction when N3 is protonated during the deamination reaction. E. coli CDA can afford this sacrifice of substrate binding energy for the reduction of the energy barrier of the deamination reaction, because the ribose moiety of cytidine contributes to the binding energy for the formation of the enzyme-substrate complex and consequently the enzyme is still able to maintain its Km in the sub-millimolar range even with the unfavorable interaction between the carboxyl group of Glu104 and N3 of cytidine. On the other hand, without a ribose moiety, cytosine needs a favorable interaction with the carboxyl group of Glu64 of yCD, i.e. a H-bond between the amino group of cytosine and the carboxyl group of Glu64 as described earlier, to keep its Km in the millimolar range. Without this favorable interaction, the Km increases by more than an order of magnitude as shown by this mutagenesis study and a CD with a Km in the tens millimolar range would not be useful for pyrimidine salvage.
H-bonds play critical roles in protein structure, stability, and function. The energy of a typical H-bond is in the range of 1–5 kcal/mol (41), which is particularly suitable for dynamic processes such as molecular recognition and conformational transition. Strong H-bonds, e.g. LBHBs, with energies higher than 10 kcal/mol have also been found in proteins. LBHBs are thought to play an important role in the enormous rate enhancement of enzymatic reactions (42–44), although this proposition is still under debate (45). Despite the importance of H-bonds, no single experimental method is sufficient for the characterization of the full range of H-bonds in proteins. X-ray crystallography is most frequently used for the determination of H-bonds, but the resolutions of the diffraction data of protein crystals are usually insufficient for the placement of H atoms. In contrast, only distances between H atoms are measured in NMR structure determination, whereas heteroatoms are defined by standard covalent geometry. However, H-bonds can be detected directly by NMR through trans-H-bond scalar couplings for small proteins (46) and trans-H-bond deuterium/protium (D/H) isotope effects in favorable situations (36, 37). LBHBs in proteins are characterized generally by a combination of X-ray crystallography and NMR (38, 39), including the distance between the two heteroatoms determined by X-ray crystallography and several parameters measured by NMR, such as proton chemical shift, proton exchange rate protection factor, D/H fractionation factor, and D (or tritium T)/H isotope effects. Unfortunately, most of these parameters are difficult to acquire through direct observation of the bridging proton, in particular for the O⋯H⋯O type of H-bonds, because of the exchange broadening of the proton resonance and the poor NMR properties of 17O, the only NMR active isotope of oxygen. We show that LBHBs can also be identified by a new parameter, trans-H-bond D/H isotope effect (Figure 4). Unlike primary isotope effects (47, 48), secondary isotope effects in proteins can be readily measured by IS-TROSY NMR (20, 21).
In conclusion, the combined experimental and computational studies show that Glu64 plays a critical role in the activation of the anticancer prodrug 5FC. It is important not only for the deamination reaction but also for the binding of the substrate. In the deamination reaction, Glu64 facilitates both the formation of the tetrahedral intermediate and its conversion to the product by shuttling protons. Furthermore, it stabilizes the tetrahedral intermediate and the hydrated form of the inhibitor 5FPy, which mimics the transition state and/or the reaction intermediate. In contrast to Glu104 of E. coli CDA, which destabilizes the enzyme-substrate complex, Glu64 of yCD is also important for substrate binding. Its contribution to substrate binding is of great physiological significance. Without Glu64, the Km of the enzyme would be too high for pyrimidine salvage. The carboxylate of Glu64 forms two strong H-bonds with 5FDHP in the yCD•5F-DHP complex that mimics the transition state or the tetrahedral intermediate of the activation of the prodrug 5FC and such H-bonds can be readily detected by IS-TROSY NMR.
Supplementary Material
Abbreviations used
- CD
cytosine deaminase
- DHP
4(R)-hydroxy-3,4-dihydro-1H-pyrimidin-2-one
- eCD
E. coli cytosine deaminase
- 5F-DHP
4(R)-5-fluoro-hydroxy-3,4-dihydro-1H-pyrimidin-2-one
- 5F4SU
5-fluoro-4-thiouracil
- 5FC
5-fluorocytosine
- 5FPy
5-fluoro-1H-pyrimidin-2-one
- 5FU
5-fluorouracil
- GDEPT
gene directed enzyme prodrug therapy
- IS-TROSY
isotopomer-selective transverse-relaxation-optimized spectroscopy
- ONIOM
our own N-layered integrated molecular orbital + molecular mechanics
- PAGE
polyacrylamide gel electrophoresis
- PCR
polymerase chain reaction
- Py
1H-pyrimidin-2-one
- SDS
sodium dodecyl sulfate
- TLC
thin-layer chromatography
- Tris
2-amino-2-(hydroxymethyl)-1,3-propanediol
- yCD
yeast cytosine deaminase
Footnotes
This work was supported in part by National Institutes of Health of USA (Grant GM51901), the Grant Agency of the Academy of Sciences of the Czech Republic (Projects No. IAA400400812 and IAA400400908), and the Grant Agency of the Czech Republic (Project No. 203/09/1627). This study made use of a Varian INOVA 600 NMR spectrometer at Michigan State University funded in part by National Science Foundation of USA (Grant BIR9512253).
Supporting Information
Downfield region of 1H jump-return spectra of the apo wild-type yCD and the complexes of the wild-type yCD and the E64A mutant enzyme with the inhibitor 5FPy (Figure S1). This material is available free of charge via the Internet at http://pubs.acs.org.
REFERENCES
- 1.Aghi M, Hochberg F, Breakefield XO. Prodrug activation enzymes in cancer gene therapy. J. Gene Med. 2000;2:148–164. doi: 10.1002/(SICI)1521-2254(200005/06)2:3<148::AID-JGM105>3.0.CO;2-Q. [DOI] [PubMed] [Google Scholar]
- 2.Greco O, Dachs GU. Gene directed enzyme/prodrug therapy of cancer: Historical appraisal and future prospectives. J. Cell. Physiol. 2001;187:22–36. doi: 10.1002/1097-4652(2001)9999:9999<::AID-JCP1060>3.0.CO;2-H. [DOI] [PubMed] [Google Scholar]
- 3.Both GW. Recent progress in gene-directed enzyme prodrug therapy: An emerging cancer treatment. Curr. Opin. Mol. Ther. 2009;11:421–432. [PubMed] [Google Scholar]
- 4.Ireton GC, McDermott G, Black ME, Stoddard BL. The structure of Escherichia coli cytosine deaminase. J. Mol. Biol. 2002;315:687–697. doi: 10.1006/jmbi.2001.5277. [DOI] [PubMed] [Google Scholar]
- 5.Ireton GC, Black ME, Stoddard BL. The 1.14 angstrom crystal structure of yeast cytosine deaminase: Evolution of nucleotide salvage enzymes and implications for genetic chemotherapy. Structure. 2003;11:961–972. doi: 10.1016/s0969-2126(03)00153-9. [DOI] [PubMed] [Google Scholar]
- 6.Ko T-P, Lin J-J, Hu C-Y, Hsu Y-H, Wang AH-J, Liaw S-H. Crystal structure of yeast cytosine deaminase. Insights into enzyme mechanism and evolution. J. Biol. Chem. 2003;278:19111–19117. doi: 10.1074/jbc.M300874200. [DOI] [PubMed] [Google Scholar]
- 7.Kievit E, Bershad E, Ng E, Sethna P, Dev I, Lawrence TS, Rehemtulla A. Superiority of yeast over bacterial cytosine deaminase for enzyme/prodrug gene therapy in colon cancer xenografts. Cancer Res. 1999;59:1417–1421. [PubMed] [Google Scholar]
- 8.Kievit E, Nyati MK, Ng E, Stegman LD, Parsels J, Ross BD, Rehemtulla A, Lawrence TS. Yeast cytosine deaminase improves radiosensitization and bystander effect by 5-fluorocytosine of human colorectal cancer xenografts. Cancer Res. 2000;60:6649–6655. [PubMed] [Google Scholar]
- 9.Hamstra DA, Rice DJ, Fahmy S, Ross BD, Rehemtulla A. Enzyme/prodrug therapy for head and neck cancer using a catalytically superior cytosine deaminase. Human Gene Ther. 1999;10:1993–2003. doi: 10.1089/10430349950017356. [DOI] [PubMed] [Google Scholar]
- 10.Johnson AJ, Ardiani A, Sanchez-Bonilla M, Black ME. Comparative analysis of enzyme and pathway engineering strategies for 5FC-mediated suicide gene therapy applications. Cancer Gene Ther. 2011;18:533–542. doi: 10.1038/cgt.2011.6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Sklenak S, Yao LS, Cukier RI, Yan HG. Catalytic mechanism of yeast cytosine deaminase: An ONIOM computational study. J. Am. Chem. Soc. 2004;126:14879–14889. doi: 10.1021/ja046462k. [DOI] [PubMed] [Google Scholar]
- 12.Yao LS, Sklenak S, Yan HG, Cukier RI. A molecular dynamics exploration of the catalytic mechanism of yeast cytosine deaminase. J. Phys. Chem. B. 2005;109:7500–7510. doi: 10.1021/jp044828+. [DOI] [PubMed] [Google Scholar]
- 13.Yao LS, Li Y, Wu Y, Liu AZ, Yan HG. Product release is rate-limiting in the activation of the prodrug 5-fluorocytosine by yeast cytosine deaminase. Biochemistry. 2005;44:5940–5947. doi: 10.1021/bi050095n. [DOI] [PubMed] [Google Scholar]
- 14.Yao LS, Yan HG, Cukier RI. A combined ONIOM quantum chemical-molecular dynamics study of zinc-uracil bond breaking in yeast cytosine deaminase. J. Phys. Chem. B. 2006;110:26320–26326. doi: 10.1021/jp064301s. [DOI] [PubMed] [Google Scholar]
- 15.Yao LS, Yan HG, Cukier RI. A molecular dynamics study of the ligand release path inyeast cytosine deaminase. Biophys. J. 2007;92:2301–2310. doi: 10.1529/biophysj.106.098921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Xu Q, Guo HB, Gorin A, Guo H. Stabilization of a transition-state analogue at the active site of yeast cytosine deaminase: Importance of proton transfers. J. Phys. Chem. B. 2007;111:6501–6506. doi: 10.1021/jp0670743. [DOI] [PubMed] [Google Scholar]
- 17.Felczak K, Bretner M, Kulikowski T, Shugar D. High-yield regioselective thiation of biologically important pyrimidinones, dihydropyrimidinones and their ribo, 2'- deoxyribo and 2',3'-dideoxyribo nucleosides. Nucleos. Nucleot. 1993;12:245–261. [Google Scholar]
- 18.Driscoll JS, Marquez VE, Plowman J, Liu PS, Kelley JA, Barchi JJ., Jr. Antitumor properties of 2(1H)-pyrimidinone riboside (zebularine) and its fluorinated analogs. J. Med. Chem. 1991;34:3280–3284. doi: 10.1021/jm00115a017. [DOI] [PubMed] [Google Scholar]
- 19.Cheng YC, Prussof WH. Relationship between the inhibition constant (ki) and the concentration of inhibitor which causes 50 per cent inhibition (I50) of an enzymatic reaction. Biochem. Pharmacol. 1973;22:3099–3108. doi: 10.1016/0006-2952(73)90196-2. [DOI] [PubMed] [Google Scholar]
- 20.Liu AZ, Li Y, Yao LS, Yan HG. Simultaneous NMR assignment of backbone and side chain amides in large proteins with IS-TROSY. J. Biomol. NMR. 2006;36:205–214. doi: 10.1007/s10858-006-9072-3. [DOI] [PubMed] [Google Scholar]
- 21.Liu AZ, Yao LS, Li Y, Yan HG. TROSY of side-chain amides in large proteins. J. Magn. Reson. 2007;186:319–326. doi: 10.1016/j.jmr.2007.02.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Humbel S, Sieber S, Morokuma K. The IMOMO method: Integration of different levels of molecular orbital approximations for geometry optimization of large systems: Test for n-butane conformation and s(n)2 reaction: Rcl. J. Chem. Phys. 1996;105:1959–1967. [Google Scholar]
- 23.Kuno M, Hannongbua S, Morokuma K. Theoretical investigation on nevirapine and HIV-1 reverse transcriptase binding site interaction, based on ONIOM method. Chem. Phys. Lett. 2003;380:456–463. [Google Scholar]
- 24.Maseras F, Morokuma K. IMOMM - a new integrated ab initio plus molecular mechanics geometry optimization scheme of equilibrium structures and transition-states. J. Comput. Chem. 1995;16:1170–1179. [Google Scholar]
- 25.Svensson M, Humbel S, Froese RDJ, Matsubara T, Sieber S, Morokuma K. ONIOM: A multilayered integrated mo method for geometry optimizations and single point energy predictions. A test for diels-alder reactions and pt(p(t-bu)(3))(2)2 oxidative addition. J. Phys. Chem. 1996;100:19357–19363. [Google Scholar]
- 26.Svensson M, Humbel S, Morokuma K. Energetics using the single point IMOMO (integrated molecular orbital plus molecular orbital) calculations: Choices of computational levels and model system. J. Chem. Phys. 1996;105:3654–3661. [Google Scholar]
- 27.Vreven T, Mennucci B, da Silva CO, Morokuma K, Tomasi J. The ONIOM-PCM method: Combining the hybrid molecular orbital method and the polarizable continuum model for solvation. Application to the geometry and properties of a merocyanine in solution. J. Chem. Phys. 2001;115:62–72. [Google Scholar]
- 28.Vreven T, Morokuma K. On the application of the IMOMO (integrated molecular orbital plus molecular orbital) method. J. Comput. Chem. 2000;21:1419–1432. [Google Scholar]
- 29.Vreven T, Morokuma K, Farkas O, Schlegel HB, Frisch MJ. Geometry optimization with QM/MM, ONIOM, and other combined methods. I. Microiterations and constraints. J. Comput. Chem. 2003;24:760–769. doi: 10.1002/jcc.10156. [DOI] [PubMed] [Google Scholar]
- 30.Frisch MJ, Trucks GW, Schlegel HB, Scuseria GE, Robb MA, Cheeseman JR, Montgomery JJA, Vreven T, Kudin KN, Burant JC, Millam JM, Iyengar SS, Tomasi J, Barone V, Mennucci B, Cossi M, Scalmani G, Rega N, Petersson GA, Nakatsuji H, Hada M, Ehara M, Toyota K, Fukuda R, Hasegawa J, Ishida M, Nakajima T, Honda Y, Kitao O, Nakai H, Klene M, Li X, Knox JE, Hratchian HP, Cross JB, Adamo C, Jaramillo J, Gomperts R, Stratmann RE, Yazyev O, Austin AJ, Cammi R, Pomelli C, Ochterski JW, Ayala PY, Morokuma K, Voth GA, Salvador P, Dannenberg JJ, Zakrzewski VG, Dapprich S, Daniels AD, Strain MC, Farkas O, Malick DK, Rabuck AD, Raghavachari K, Foresman JB, Ortiz JV, Cui Q, Baboul AG, Clifford S, Cioslowski J, Stefanov BB, Liu G, Liashenko A, Piskorz P, Komaromi I, Martin RL, Fox DJ, Keith T, Al-Laham MA, Peng CY, Nanayakkara A, Challacombe M, Gill PMW, Johnson B, Chen W, Wong MW, Gonzalez C, Pople JA. Gaussian 03, revision b.05. Wallingford CT: Gaussian, Inc.; 2003. [Google Scholar]
- 31.Becke AD. Density-functional thermochemistry .3. The role of exact exchange. J. Chem. Phys. 1993;98:5648–5652. [Google Scholar]
- 32.Lee CT, Yang WT, Parr RG. Development of the colle-salvetti correlation-energy formula into a functional of the electron-density. Phys. Rev. B. 1988;37:785–789. doi: 10.1103/physrevb.37.785. [DOI] [PubMed] [Google Scholar]
- 33.Stewart JJP. Optimization of parameters for semiempirical methods .1. Method. J. Comput. Chem. 1989;10:209–220. [Google Scholar]
- 34.Stewart JJP. Optimization of parameters for semiempirical methods .2. Applications. J. Comput. Chem. 1989;10:221–264. [Google Scholar]
- 35.Carlow DC, Short SA, Wolfenden R. Role of glutamate-104 in generating a transition state analogue inhibitor at the active site of cytidine deaminase. Biochemistry. 1996;35:948–954. doi: 10.1021/bi951498y. [DOI] [PubMed] [Google Scholar]
- 36.Liu AZ, Lu ZW, Wang JF, Yao LS, Li Y, Yan HG. NMR detection of bifurcated hydrogen bonds in large proteins. J. Am. Chem. Soc. 2008;130:2428–2429. doi: 10.1021/ja710114r. [DOI] [PubMed] [Google Scholar]
- 37.Liu AZ, Wang JF, Lu ZW, Yao LS, Li Y, Yan HG. Hydrogen-bond detection, configuration assignment and rotamer correction of side-chain amides in large proteins by NMR spectroscopy through protium/deuterium isotope effects. Chembiochem. 2008;9:2860–2871. doi: 10.1002/cbic.200800467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Mildvan AS, Harris TK, Abeygunawardana C. Nuclear magnetic resonance methods for the detection and study of low-barrier hydrogen bonds on enzymes. Methods Enzymol. 1999;308:219–245. doi: 10.1016/s0076-6879(99)08012-x. [DOI] [PubMed] [Google Scholar]
- 39.Bourne CR, Barrow EW, Bunce RA, Bourne PC, Berlin KD, Barrow WW. Inhibition of antibiotic-resistant Staphylococcus aureus by the broad-spectrum dihydrofolate reductase inhibitor rab1. Antimicrob. Agents Chemother. 2010;54:3825–3833. doi: 10.1128/AAC.00361-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Beierlein JM, Karri NG, Anderson AC. Targeted mutations of bacillus anthracis dihydrofolate reductase condense complex structure-activity relationships. J. Med. Chem. 2010;53:7327–7336. doi: 10.1021/jm100727t. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Hibbert F, Emsley J. Hydrogen bonding and chemical reactivity. Adv. Phys. Org. Chem. 1990;26:255–379. [Google Scholar]
- 42.Cleland WW, Frey PA, Gerlt JA. The low barrier hydrogen bond in enzymatic catalysis. J. Biol. Chem. 1998;273:25529–25532. doi: 10.1074/jbc.273.40.25529. [DOI] [PubMed] [Google Scholar]
- 43.Cleland WW, Kreevoy MM. Low-barrier hydrogen-bonds and enzymatic catalysis. Science. 1994;264:1887–1890. doi: 10.1126/science.8009219. [DOI] [PubMed] [Google Scholar]
- 44.Frey PA. Strong hydrogen bonding in molecules and enzymatic complexes. Magn. Reson. Chem. 2001;39:S190–S198. [Google Scholar]
- 45.Schutz CN, Warshel A. The low barrier hydrogen bond (LBHB) proposal revisited: The case of the asp... His pair in serine proteases. Proteins. 2004;55:711–723. doi: 10.1002/prot.20096. [DOI] [PubMed] [Google Scholar]
- 46.Grzesiek S, Cordier F, Jaravine V, Barfield M. Insights into biomolecular hydrogen bonds from hydrogen bond scalar couplings. Prog. Nucl. Magn. Reson. Spectrosc. 2004;45:275–300. [Google Scholar]
- 47.Cassidy CS, Lin J, Frey PA. The deuterium isotope effect on the NMR signal of the low-barrier hydrogen bond in a transition-state analog complex of chymotrypsin. Biochem. Biophys. Res. Commun. 2000;273:789–792. doi: 10.1006/bbrc.2000.2986. [DOI] [PubMed] [Google Scholar]
- 48.Westler WM, Frey PA, Lin J, Wemmer DE, Morimoto H, Williams PG, Markley JL. Evidence for a strong hydrogen bond in the catalytic dyad of transition-state analogue inhibitor complexes of chymotrypsin from proton-triton NMR isotope shifts. J. Am. Chem. Soc. 2002;124:4196–4197. doi: 10.1021/ja017860f. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.