Abstract
Gamma secretase modulators (GSMs) have received much attention as potential therapeutic agents for Alzheimer’s disease (AD). GSMs increase the ratio between short and long forms of the amyloid-β (Aβ) polypeptides produced by gamma-secretase and thereby decrease the amount of the toxic amyloid species. However, the mechanism of action of these agents is still poorly understood. One recent paper (Richter et al., PNAS 107, 14597–14602 2010) presented data that were interpreted to support direct binding of the GSM sulindac sulfide to Aβ42, supporting the notion that GSM action is linked to direct binding of these compounds to the Aβ domain of its immediate precursor, the 99 residue C-terminal domain of the amyloid precursor protein (C99, also known as the β-CTF). Here, contrasting results are presented that indicate there is no interaction between monomeric sulindac sulfide and monomeric forms of Aβ42. Instead, it was observed that sulindac sulfide is itself prone to form aggregates that can bind non-specifically to Aβ42 and trigger its aggregation. This observation, combined with data from previous work (Beel et al., Biochemistry 48, 11837-11839), suggests both that the poor behavior of some NSAID-based GSMs in solution may obscure results of binding assays and that NSAID-based GSMs do not function by directly targeting C99. It was also observed that another GSM, flurbiprofen, fails to bind to monomeric Aβ42 or to C99 reconstituted into bilayered lipid vesicles. These results disfavor the hypothesis that these NSAID-based GSMs exert their modulatory effect by directly targeting a site located in the Aβ42 domain of free C99.
Alzheimer’s disease (AD) is one of the most common neurodegenerative disorders, affecting more than 26 million people worldwide - a number that is expected to more than quadruple by the year 2050(1). AD is characterized by cognitive decline induced by a loss of neurons and synapses in the cerebral cortex (2). This degeneration of neural activity is associated with the existence of extracellular amyloid plaques and intracellular neurofibrillary tangles, both of which characterize the pathology of the disease (3). The neural plaques are comprised of insoluble deposits of amyloid-β (Aβ) peptides, which result from sequential proteolytic cleavage reactions of the amyloid precursor protein (APP) involving β- and γ-secretase. The APP substrate is first cleaved by β-secretase (BACE-1) to release its large ectodomain from a 99 residue transmembrane bound C-terminal fragment (C99, also referred to as β-CTF). C99 then serves as a substrate for γ-secretase, cleavage by which results in release of the APP intracellular domain (AICD) and theAβ polypeptide. The Aβ produced is not completely homogeneous, but varies in length, with Aβ42 and Aβ40 being the most prominent products. Aβ42 is believed to be the most neurotoxic of the amyloid-β polypeptides due to its particularly high propensity to form toxic aggregates that go on to form the amyloid plaques that are the hallmark of AD pathology.
The well defined pathology of AD, first described in the mid-1980s (4–7), set the stage for the proposal of the “amyloid hypothesis” by John Hardy (8), which postulates that accumulation of Aβ in the brain is the primary cause of AD pathology. The amyloid hypothesis is strongly supported by the observation that all mutations of APP or the Presenilin component ofγ-secretase observed in early onset Alzheimer’s disease (EOAD) or familial Alzheimer’s disease (FAD) result either in an increase in total Aβ levels or in elevated production of Aβ42 relative to Aβ40 –thus supporting the relationship between the production of Aβ42 and the clinical symptoms of AD(2, 8, 9).
To date, there are only five FDA approved treatments for AD in the United States, all of which treat cognitive decline and symptoms of the disease(2). Therefore, the development of a disease-modifying agent – one that prevents or reverses the pathology of the disease —represents a significant unmet medical need. Two promising targets from a drug discovery perspective are β- and γ-secretase. The development of potential therapeutics targeting these two proteins has been extensively reviewed(10–12). Among the proposed strategies, development of agents that modulate Aβ production has recently received much attention. A γ-secretase modulator (GSM) is defined as a molecule that changes the relative proportions of the Aβ isoforms produced by γ-secretase (particularly Aβ42 vs. the less toxic Aβ40), without altering the overall rate of APP processing(2). The first GSMs originated from non-steroidal anti-inflammatory drugs (NSAIDs), which were reported to reduce the occurrence of AD in patients using these drugs (13–15). The early NSAIDs, including sulindac sulfide, flurbiprofen, and ibuprofen, were shown to reduce the levels of the highly amyloidogenic Aβ42(13, 16, 17). Recent photoaffinity-cross-linking experiments led to the proposal that GSMs bind directly to the transmembrane APP/C99 substrate (18) to form a complex that somehow then modulates γ-secretase cleavage. The site of GSM binding within C99 was proposed to be located in its N-terminal Aβ42 domain. However, recent NMR studies from our labs failed to reveal any binding of the GSMs flurbiprofen and fenofibrate to monomeric or dimeric C99 in micellar model membranes, with the binding being detectable only to C99 aggregates, which was found to be of a non-specific and non-stoichiometric nature(19). More recently, Multhaup et al. have countered our findings based on NMR, SPR, and bacterial reporter assay results that they interpreted as providing proof that the NSAID-based GSM sulindac sulfide binds avidly and specifically to both Aβ42 and C99(20, 21).
We hypothesize that alternative interpretations are merited for some of the key results from Multhaup and co-workers, and also that some of the experiments of those works may have had unrecognized artifacts. Here, we provide additional data to both clarify the previously published data and to provide new data informing on this controversy. The results support our earlier contention(19) that monomeric GSMs either do not bind monomeric or dimeric forms of C99 or Aβ42 at all (in solution, in micelles, or in membranes) or bind in a weak and non-specific manner that is likely to be unrelated to their GSM activity. Moreover, these studies revealed that sulindac sulfide forms colloid-like assemblies at concentrations above 50μM, a phenomenon that may have been a source of experimental artifacts in some previous studies of GSMs.
Materials and Methods
Reagents, Peptides, and Proteins
Aβ40 and Aβ42 peptides in both isotopically unlabeled and uniformly-15N labeled forms were obtained from rPeptide, LLC (Bogart, GA). For all experiments, peptides were first “monomerized” as previously described (22, 23) by dissolving lyophilized material in 98% formic acid and then immediately evaporating the solvent. This “monomerized” material was stored at −20°C and thawed immediately before use. The compounds used in this study, sulindac sulfide, sulindac sulfone, and flurbiprofen, were obtained from Toronto Research Chemicals (North York, ON, CA), MP Biomedicals (Solon, OH), and Sigma-Aldrich (St. Louis, MO).
C99 was recombinantly expressed as described by Beel et al(19). The mammalian C99 vector was cloned into a pET-21a vector and then transformed into the BL21(DE3) E. coli strain. Protein overexpression was induced via the addition of isopropyl thiogalactoside to 1mM when cells reached an optical density of approximately 0.8. Cells were harvested and lysed, resulting in C99 localization to inclusion bodies. The inclusion bodies were solubilized using a 0.2% SDS/8M urea buffer. C99 was purified via IMAC, during which SDS and urea were removed and replaced with 0.05% LMPG, a lyso-phospholipid detergent. C99 was eluted from the IMAC column using a buffer containing 250mM imidazole and 0.05% LMPG at pH 7.8. For all experiments performed on C99 in LMPG micelles, the final buffer concentration was 100mM imidazole, 10% LMPG, and 2mM EDTA at pH 6.5.
Sample Preparation
All Aβ40 and Aβ42 samples were prepared by dissolving the “monomerized” polypeptide in 20mM NaOH at a concentration of 1mg/ml. The resulting solution was the diluted with sample buffer (50mM sodium phosphate, pH 7.0, 10% D2O) to the desired concentrations, typically 100μM.
C99 reconstitution into lipid vesicles began with protein purification as described above, with the only difference being that the final elution buffer consisted of 0.2% SDS in lieu of 0.05% LMPG. Purified C99 in SDS was concentrated using centrifugal ultrafiltration to a final concentration of 1mM. The concentrated C99 solution was then mixed with a SDS/lipid mixture of 400mM SDS/75mM POPC/25mM POPG (400mM SDS:100mM lipid), resulting in a clear solution. The C99/SDS/lipid mixture was then subjected to extensive dialysis to remove all SDS present, during which process C99/POPC/POPG vesicles spontaneously formed. The 4L dialysis buffer (50mM imidazole and 2.25mM EDTA at pH 6.5) was changed three times daily. Completion of dialysis was determined when the C99/lipid solution became cloudy and the surface tension of the dialysate indicated complete removal of detergent. The C99/lipid vesicles solution was then extruded using a 50nM filter to generate unilamellar vesicles, concentrated to a 1mM:100mM C99:lipid ratio, and flash frozen for later experiments. For the NMR studies (GSM titrations), the solution was diluted with buffer to achieve 100μM C99 plus 10mM lipid. For vesicle-only control samples, the same dialysis procedure was carried out in parallel, minus C99.
CD Spectroscopy
Far-UV CD spectra were obtained on an AppliedPhotophysics Chirascan spectropolarimeter at ambient temperature. The peptides were analyzed at a concentration of 0.5-1mg/ml, using a quartz cuvette with a pathlength of 0.02 cm (far UV CD, 180–250 nm); the spectra were corrected for contributions from the buffer. Each spectrum represents an average of 3 scans.
Dynamic Light Scattering
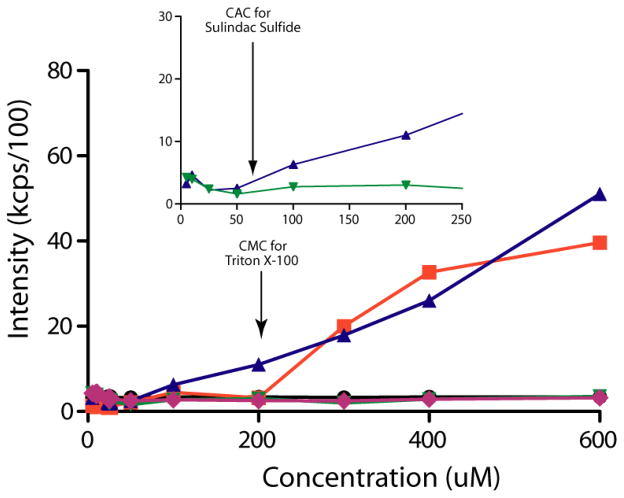
DLS experiments were conducted on a DynaPro Plate Reader WPR-06 (Wyatt Technology Corporation, Santa Barbara, CA) using a laser wavelength of 832.4nm. Briefly, 100uL volumes of solutions of Triton X-100, sulindac sulfide, sulindac sulfone, and flurbiprofen were prepared (from 50mM DMSO stocks) at concentrations of 5, 10, 25, 50, 100, 200, 300, 400, 600, 800, and 1000μM. All solutions were prepared so that the final DMSO concentration was constant at 2% in all samples. Triton X-100 was used as a positive control, and the intensity of the scattered light was measured as a function of drug concentration (Fig 2). All experiments were performed in triplicate at 15°C. Ten acquisitions were performed (10s acquisition time) for each concentration point. Data were processed using the Dynamics 6.10.0.10 software (Wyatt Technology Corporation, Santa Barbara, CA). The average laser light scattering from three experiments were plotted versus concentration to obtain the critical micelle concentration (CMC) or “critical aggregate concentration”, (CAC). The CMC of Triton X-100 was determined to be approximately 200–300μM, a value consistent with that in the literature (24).
Figure 2.
Measurement of the critical aggregation concentration (CAC) by dynamic light scattering (DLS). Scattering intensities were plotted versus concentration, and the CAC was determined as the point when the scattering intensities began to increase. The legend is as follows: buffer only (black circles), Triton X-100 (orange squares), sulindac sulfide (purple triangles), sulindac sulfone (green triangles), and flurbiprofen (green diamonds). Notice that no increase in scattering intensity was observed for buffer, sulindac sulfone, or flurbiprofen. However, a significant increase in scattering intensity was observed for a positive control (Triton X-100) upon micelle formation at 200-300uM and for sulindac sulfide starting above 50 μM, indicating that the latter begins to form aggregates at concentrations above 50μM, which is consistent with NMR data (Figs S3-S5).
NMR Spectroscopy
NMR experiments were performed on Bruker Avance II 500MHz and Avance III 800 MHz NMR spectrometers, both equipped with TCI cryoprobes. 19F NMR measurements were performed on the Avance II 500 MHz system using an SEF cryoprobe. All experiments using the amyloid peptides were performed at 5°C. Relaxation and diffusion measurements were used to verify the oligomeric state of the Aβ peptides used in this study. It is well know that Aβ40 has a lower propensity to form large oligomeric fibril species (25). Thus, this peptide was used to compare the behavior of the Aβ42 species in solution. For 15N relaxation measurements, NMR experiments were performed at 500 MHz. T1 and T2 values were measured as described in Kay et al (26). T1 and T2 values for Aβ40 and Aβ42 were determined from measurements performed on the 500 MHz system using 100 μM solutions of Aβ40 and Aβ42. Peak intensities were measured by integration of the region between 7.5–8.8 ppm. T1 measurements were made using delays of 2, 20, 50, 100, 200, 300, 400, 600, 800, 1000, and 1200 ms. T2 measurements were made using delays of 0, 16, 32, 48, 64, 80, 96, 128, 160, 192, and 240 ms. The intensities were fit to a single exponential function (I (t) = I0e−t/T) using the program GraphPad Prism (GraphPad Software, La Jolla, CA). T1 and T2 values for both Aβ40 and Aβ42 were estimated to be approximately 620ms and 150ms, respectively. The resulting correlation time (τ c) for both molecules was then calculated using the following equation (27):
| (1) |
where T1 and T2 are the respective relaxation times and νn is the spectrometer frequency in Hertz. The resulting calculations yielded correlation times of 3.9 ns for both Aβ40 and Aβ42 which, at 5°C and taking into account the viscosity of water at this temperature (approximately 1.5x that at ambient), corresponds to a protein of approximately 4.9 kDa – a value consistent with the rotational correlation time for a 4.2 kDa protein (as calculated from the Stokes-Einstein equation) (28).
To further classify the oligomeric state of the peptides in solution, pulsed field gradient diffusion measurements were performed at 500 MHz using a stimulated echo experiment (29). Using dioxane in the solution as a reference (as in (30)), solutions of Aβ40 and Aβ42 (both 100 μM) were measured with diffusion gradient strengths varying between 1% and 90% of maximum value. The lengths of the diffusion gradient and stimulated echo were optimized to give a total decay in the protein signal of ~80%. The spectra were acquired with 32K complex points and a spectral window of approximately 6500 Hz. Data was processed using Topspin 2.1 (Bruker Biospin, Billerica, MA). To obtain diffusion decay rates, the dioxane peak and the methyl region of the spectra (0.3-0.7ppm) were integrated at each data point and fit to equation (2) to determine the decay rate:
| (2) |
where the intensities of the protein signals s are plotted as a function of gradient strength, g, to enable determination of the decay rate, d. Decay rates were determined to be 1.6 x 10 −4 s−1 for both peptides. The hydrodynamic radii for Aβ40 and Aβ42 were then calculated as in Wilkins et al (30)to both be approximately 16Å, which as in Wilkins et al. correspond to a polypeptide chain of approximately 42 residues (30). Using this hydrodynamic radius and the viscosity of water at 5 degrees Celsius, the diffusion coefficient, D, was calculated using the Stokes-Einstein relationship:
| (3) |
Where KB is the Boltzmann constant, T is temperature (K), η is solvent viscosity in kg/m.s at 5°C, and r is the hydrodynamic radius in meters.
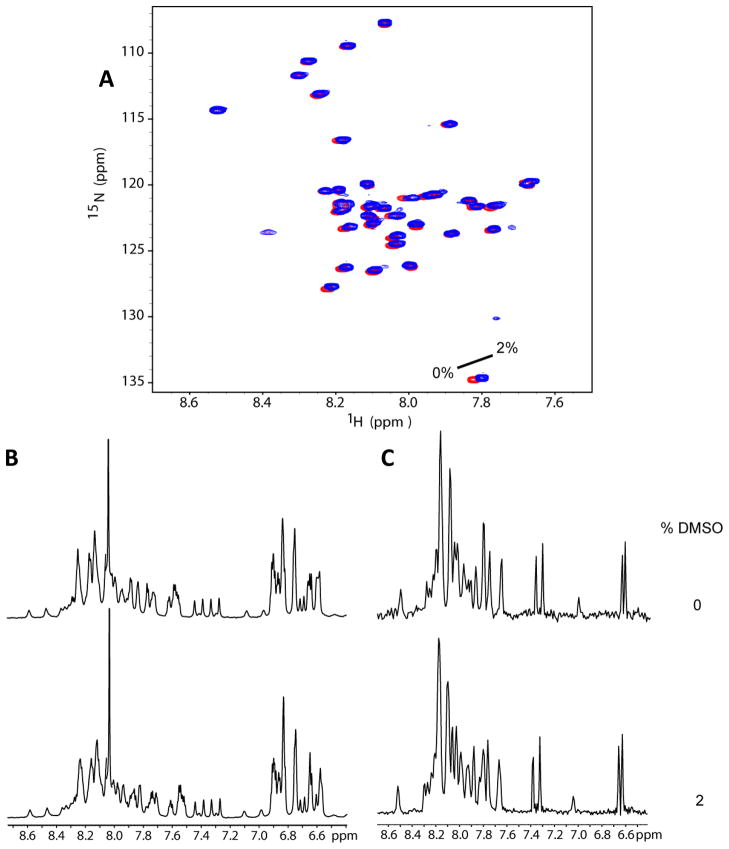
Titrations of 15N-labeled Aβ40 and Aβ42 were performed using an 800 MHz NMR spectrometer at 5°C with 100 μM protein solutions. Titrations were performed with sulindac sulfide, sulindac sulfone, flurbiprofen, and DMSO (as a control) and were carried out in two modes. First, 50mM ligand stocks were prepared in DMSO-d6. Sulindac sulfide and sulindac sulfone were added to 100μM protein solutions at concentrations of 5, 10, 50, 100, 300, and 500μM. Flurbiprofen was added at concentrations of 500μM and 1mM. Control DMSO-only titrations were performed for each series using the final titration point, which contained 2% DMSO by volume. In addition, titrations with sulindac sulfide, sulindac sulfone, and DMSO were performed as in Richter et al (20), where one titration data point was acquired using 100μM Aβ42 and either 300μM sulindac sulfide, 300μM sulindac sulfone, or DMSO-only control. In each case a fresh peptide sample was prepared, the appropriate amount of compound was added (from 50mM DMSO-d6 stock), and spectra were acquired immediately at 5°C. Time course spectra were acquired at t=0, 1hr, and 24hr. When it became evident that the sulindac sulfide was causing aggregation of Aβ42, a separate time course experiment was carried out using identical solutions at t=0, 15min, and 1hr. All two-dimensional 15N-HSQC spectra were acquired using spectral widths of 12,820 Hz and 2432 Hz in the direct and indirect dimensions, respectively. Data were acquired using 1024 X 64 complex data points and 8 scans per increment. Two-dimensional experiments were also accompanied by 1D proton NMR spectra so that the concentrations of the compounds could be monitored throughout the titration (data not shown).
NMR solubility measurements of sulindac sulfide, sulindac sulfone, and flurbiprofen were performed using a 500 MHz NMR spectrometer in experimental buffer (50mM sodium phosphate, pH 7.0, 10% D2O) with 2% DMSO-d6. Briefly, 50mM stock solutions of each compound were prepared in DMSO-d6. For each compound, a 1mM solution was prepared in buffer. Quickly, serial dilutions were made to a final concentration of 5 μM, while keeping the DMSO concentration constant at 2%. 1D proton spectra were measured for each concentration using identical parameters.
To test whether colloidal aggregates of sulindac sulfide can act as promiscuous enzyme inhibitors, β-secretase activity assays were performed in the presence and absence of sulindac sulfide and sulindac sulfone. An NMR-based enzymatic assay was designed using a 19F-labeled BACE-1 substrate peptide — EVNLDAEF(CF3) — where the trifluoromethyl group is at the meta position on the benzyl ring of Phe. BACE-1 cleaves this peptide between the L and D residues, which results in distinct 19F NMR signals for the substrate and product. The assay was conducted in a 96 well plate format, where each well contained 220nM of CHO-expressed BACE-1 prepared in 20mM sodium acetate buffer, pH 5.0, to which were added sulindac sulfide and sulindac sulfone at various concentrations (3.15, 6.25, 12.5, 25, 50, 100, and 200μM) followed by a blank DMSO control and a positive control using an inhibitor with a known Ki value. The reaction was started by addition of 100μM substrate peptide prepared from a 100mM DMSO stock in buffer (20mM sodium acetate, pH 5.0). Cleavage was allowed to proceed for a period of 20 minutes, at which time the reaction was quenched by the addition of 300uL of 8M urea. Samples were then transferred from the 96 well plate to NMR tubes for analysis. 19F NMR spectra showed the presence of both substrate and product, the peak integrals of which were used to calculate the concentration of each species and to assess the degree of inhibition (31).
Surface Plasmon Resonance
SPR experiments were performed using a Biacore S51 instrument and a CM5 sensor chip (GE Healthcare). Monomerized Aβ42 peptide (1mg/ml diluted 1:10 with 10mM sodium acetate, pH 3.4) was immobilized to the sensor chip by standard amine coupling (with ~3000 response units (RU)). Compounds were diluted from DMSO stock solutions in three different running buffers (50mM sodium phosphate, 100mM NaCl, pH 7.0, 2% DMSO (0.2% Tween-20, 0.005% Tween-20, and no detergent). Injections were performed for 55s at a flow rate of 30uL/min and 25 degrees C. Data were analyzed using the Scrubber software (BioLogic Software, Campbell ACT, Australia) and were plotted using GraphPad Prism (GraphPad Software, La Jolla, CA).
Transmission Electron Microscopy
Samples were prepared for transmission electron microscopy (TEM) in an identical manner to the NMR samples. Briefly, “monomerized” peptide was dissolved in 20mM NaOH at a concentration of 1mg/ml. Solutions for TEM were prepared at 100μM. Subsequently, sulindac sulfide and sulindac sulfone were added to final concentrations of 300μM with a DMSO concentration of 2%. Lastly, a drug-free control sample was prepared containing 2% DMSO only. Formvar-coated copper grids were inverted over 50ul sample droplets for 15 minutes. The grids were then briefly rinsed with one drop of ultrapure water, and the excess water was removed by wicking to the side with blotter paper. Samples were then inverted over drops of 2% aqueous uranyl acetate for 15 minutes and the grids were subsequently washed over three drops of ultrapure water. Following air drying, the grids were examined on a Philips CM120 transmission electron microscope (FEI, Inc., Hillsboro, OR) operated at 80kev. Representative images were captured using a Gatan Model 830 SC200 CCD camera (Gatan, Inc., Pleasanton, CA).
Results
Selection of NSAIDs for Study
The finding that certain NSAIDs decreased the production of Aβ42 produced by γ-secretase cleavage led to the observation that these compounds had various effects on the cleavage of APP. For example, compounds such as ibuprofen, fenofibrate, sulindac sulfide, R-flurbiprofen, and indomethacin were shown to decrease the amount of Aβ42 produced. Thus, they were considered Aβ42-lowering NSAIDs (see reviews in (32, 33)). Other compounds, such as celecoxib functioned to increase the amount of Aβ42 and were thus termed Aβ42-increasing NSAIDs. Lastly, several NSAIDs, such as naproxen and sulindac sulfone, were found to have no effect on the production of Aβ42. The sulindacs were chosen because these were the primary focus of the recently published studies(20, 21) that closely concern this paper. Sulindac sulfide acts as a GSM while sulindac sulfone has previously been shown to have no GSM-like effect on the production of Aβ42 and serves as a negative control. The well- characterized GSM R-flurbiprofen was also chosen for a limited number of experiments. It has a much higher aqueous solubility (up to ca. 1 mM) than sulindac sulfide and may therefore be tested as a representative GSM for experiments that were hindered by the relatively low solubility of sulindac sulfide.
Verification of the Oligomeric State of Amyloid Peptides
We sought to reproduce results that were previously interpreted (20) to indicate that GSMs bind specifically and avidly to monomeric Aβ42, results that were previously invoked to support the idea that binding of GSMs to C99 is central to how these compounds modulate amyloid production.
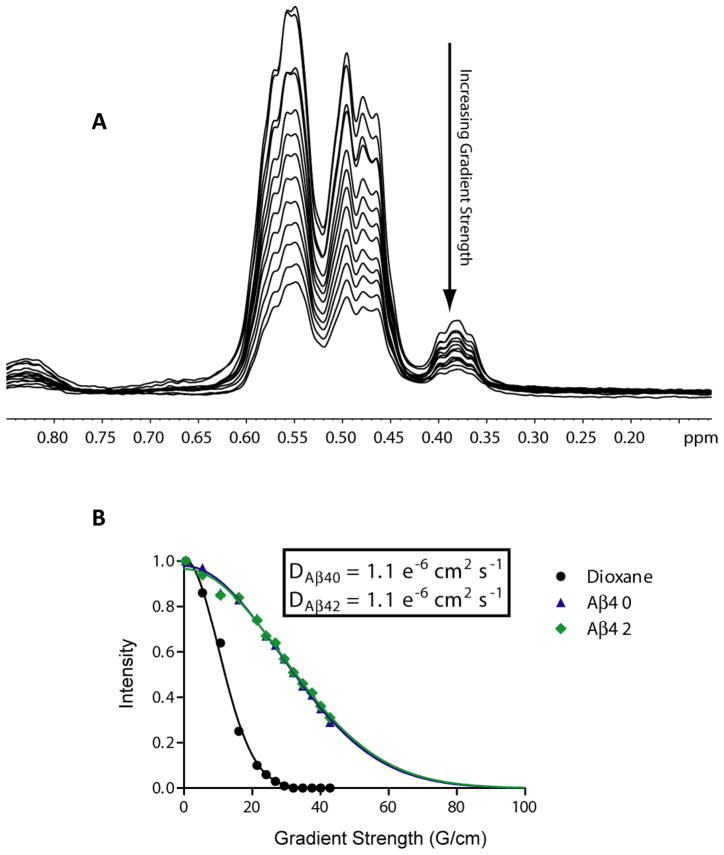
Amyloid peptides, especially Aβ42, are known for their propensity to aggregate and form high molecular weight fibrils. For this reason, we first set out to verify the presence of stable, monomeric polypeptide in our samples. Prior to study, the Aβ40 and Aβ42 peptides were first “monomerized” as previously described (22, 23). CD spectra of Aβ42 were obtained to verify that limited or no β-sheet structure existed in resulting solutions (the presence of which would indicate the formation of fibril-like species). Both peptides exhibited predominantly random coil conformations (Fig S1). The oligomeric states of both the Aβ40 and Aβ42 peptides were then assessed via NMR diffusion measurements. As indicated in Figure 1, the diffusion decay rates of both Aβ40 and Aβ42 were seen to be identical and correspond to a hydrodynamic radius of approximately 16Å, matching that expected for a ~40 residue peptide (Figure S1) (30). These data also enabled calculation of the absolute diffusion coefficients, D, for both peptides (Fig 1B), which also correspond to expected values for a very small protein in an aqueous solution. In addition, we carried out 15N NMR relaxation measurements for both Aβ40 and Aβ42. From these values, a rotational correlation time of 3.9 ns was determined at 5°C (28). This value corresponds to a protein with molecular weight of ~4.9kDa, and confirms that the overwhelming majority of peptides in these samples populated only the monomeric form under the conditions of these experiments (Fig. S2).
Figure 1.
(A) 1H NMR-based translational diffusion data for Aβ42 at Z-gradient strengths varying from 0.5 to 42.3 G/cm. The methyl region of the spectrum between 0.7 and 0.3 ppm was integrated for each point to yield relative intensities that were plotted against gradient strength in (B). The intensities in panel A were measured using dioxane as an internal reference and were fit to a single exponential (see Methods) to determine the hydrodynamic radius and diffusion coefficient, D, as presented in the inset. Data for the dioxane standard is represented by black circles, Aβ40 by blue triangles, and Aβ42 by green diamonds. Curve fits are represented by solid lines of corresponding colors.
Characterization of GSMs
Sulindac sulfide, sulindac sulfone, and flurbiprofen were characterized by dynamic light scattering (DLS) and NMR to determine their solubility and to assess their oligomeric states at the concentrations tested in this and previous work. Using DLS, sulindac sulfide was found to be monomeric below ca. 50 μM. However, between 50 and 100 μM, colloidal aggregates of sulindac sulfide clearly form, indicative of a “critical aggregate concentration” for this compound in the 50–100 μM range (34). At much higher concentrations (starting at 400 μM) sulindac sulfide begins to precipitate, which is the cause of the non-linear increase in laser light scattering above this concentration (Figure 2). Sulindac sulfone and flurbiprofen were found to be monomeric up to concentrations of 1 mM, as the scattering intensity over the entire range of concentrations of these compounds was found to be the same as buffer alone.
As an orthogonal method for measuring compound solubility, 19F NMR experiments were performed at concentrations ranging from 5μM to 1mM in aqueous buffer and with a fixed concentration of 2% DMSO. From the NMR spectra in Figure S3, it is clear that sulindac sulfide begins to form colloidal aggregates at some point between 30 and 62μM as evidenced by the significant broadening and decreasing intensity of the NMR signals at and above 62 μM, demarking the critical aggregation concentration (CAC) of this compound. On the other hand, both sulindac sulfone and flurbiprofen exhibited no significant changes in their NMR spectra (Figs 4–5 and Fig S4 and S5) and appear to remain monodisperse up to concentrations of 1mM. These NMR data confirm and complement the results of DLS and show that sulindac sulfide forms colloidal (water soluble) aggregates with a CAC in the range of 50–60 μM, whereas sulindac sulfone and flurbiprofen remain monomeric up through 500 μM.
Figure 4.
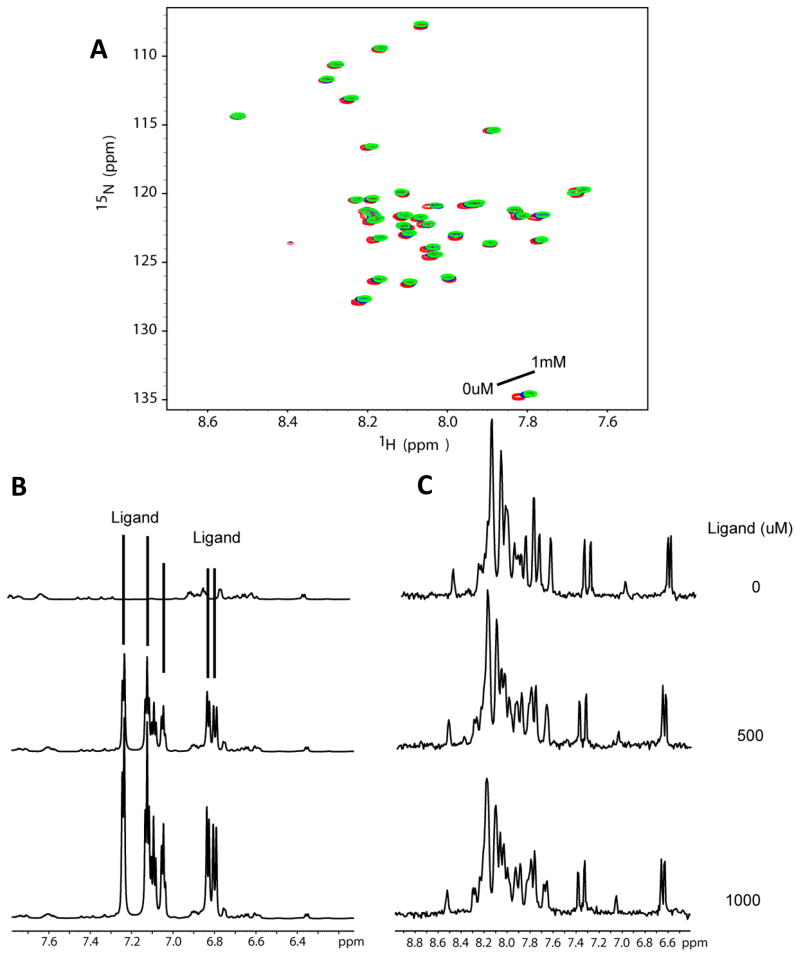
Titration of U-15N-Aβ42 with sulindac sulfone. (A) 15N-HSQC spectra of Aβ42 upon titration of sulindac sulfone at concentrations ranging from 0 to 500 μM. There are no shifts in the peaks of these spectra beyond what is observed for the DMSO-only control titration (see Fig. 6) and peak intensities do not vary. (B) 1H NMR spectra taken at each titration point to allow observation of ligand peaks throughout the titration. It can be seen that the sulindac sulfone peaks remain sharp throughout, reflecting the fact that this compound does not aggregate at concentrations below 500 uM. (C) 1-D 1H NMR projections of the HSQC spectra shown in panel A demonstrate that the solubility of Aβ42 remains unchanged at all points.
Figure 5.
Titration of U-15N-Aβ42 with flurbiprofen. (A) 15N-HSQC spectra of Aβ42 upon titration of flurbiprofen at concentrations of 500uM and 1mM. There are no shifts in the peaks of these spectra beyond what is observed for the DMSO-only control titration (see Fig. 6) and peak intensities do not vary. (B) 1H NMR spectra taken at each titration point to allow observation of ligand peaks throughout the titration. It can be seen that the flurbiprofen peaks remain sharp throughout, reflecting the fact that this compound does not aggregate at concentrations below 1 mM. (C) 1-D 1H NMR projections of the HSQC spectra shown in panel A demonstrate that the solubility of Aβ42 remains unchanged at all titration points.
NMR Titrations of Aβ42 with GSMs
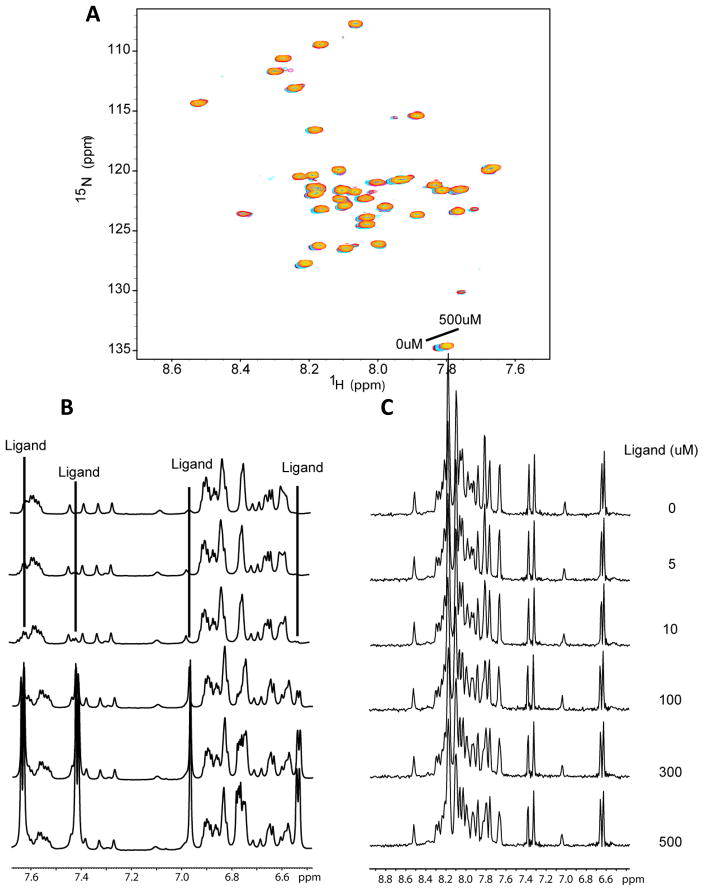
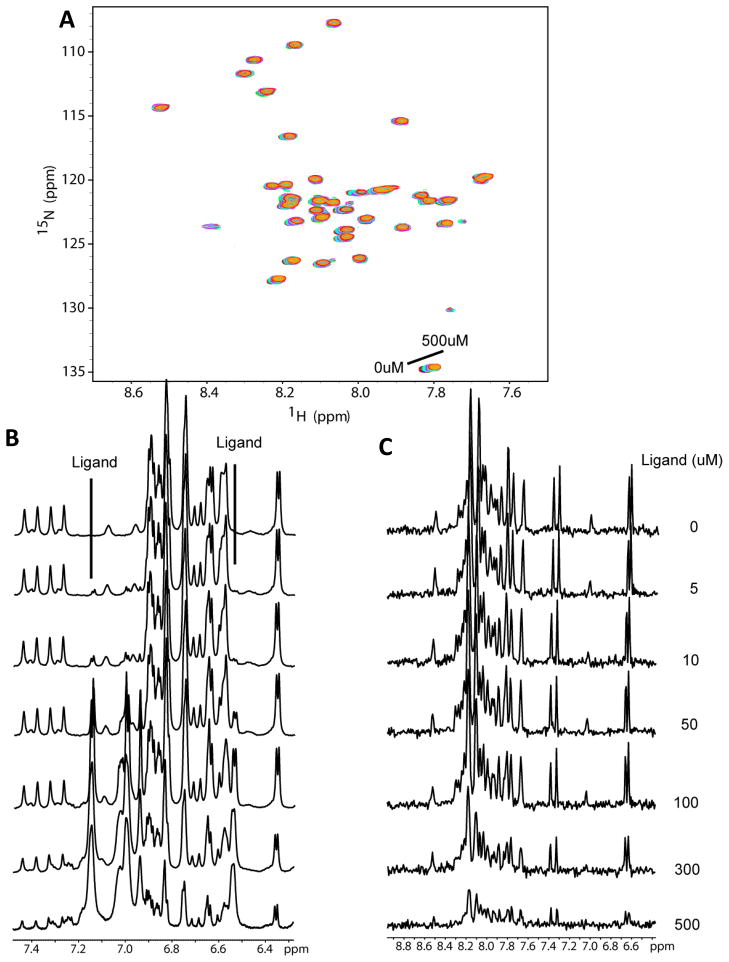
To determine if NMR spectroscopy demonstrates binding of sulindac sulfide to monomeric Aβ42, as claimed in previous work (20) NMR titration experiments were performed with each compound (using 50mM stock solutions of the drugs in DMSO-d6). Control titrations were also performed using DMSO only. Figures 3–6 show 15N- HSQC spectra of Aβ42 titrated with sulindac sulfide and flurbiprofen (both are NSAIDS and GSMs), sulindac sulfone (a NSAID but not a GSM), and DMSO control. It is clear that the titrations for all three NSAIDs led to only very small spectral changes in the HSQC spectrum of Aβ42 (Figures 3A-5A) and that these changes are virtually identical to those observed during the DMSO control titration (Figure 6A). These data provide no evidence for binding of any of the three compounds to monomeric Aβ42. However, in the case of sulindac sulfide, 1-D proton NMR spectra showing resonances both from the GSM and from aromatic sides chains of the peptides (Fig. 3B) reveal that the peaks from monomeric Aβ42 begin to lose intensity at sulindac sulfide concentrations above 50 μM — concentrations at which we have shown this that GSM begins to form colloidal aggregates. This is confirmed by examining the 1-D 1H NMR projections of the 2-D TROSY data (Figure 3C). Such changes were not observed for the flurbiprofen, sulindac sulfone, or for the DMSO control (Figures 4B-C, 5B-C, and 6B-C). These data strongly suggest that colloid formation by sulindac sulfide triggers aggregation of Aβ42.
Figure 3.
Titration of U-15N-Aβ42 with sulindac sulfide. (A) 15N-HSQC spectra of Aβ42 upon titration with sulindac sulfide (from a 50mM stock solution in DMSO) at concentrations ranging from 0 to 500μM. There are no shifts in the peaks of these spectra beyond what is observed for the DMSO-only control titration (see Fig. 6). However, peak intensities decrease at higher sulindac sulfide concentrations. (B) 1H NMR spectra taken at each titration point to allow observation of the ligand peaks throughout the titration. Notice that ligand peaks are observable even at the lowest concentration (5μM) and with a nearly 20-fold excess of protein, but begin to broaden or disappear above 50-100μM, indicating aggregation of the compound. (C) 1-D 1H NMR projections of the HSQC spectra shown in panel A illustrate the decrease in amide 1H signal intensity from the peptide, which demonstrates that Aβ42 begins to aggregate upon addition of sulindac sulfide at concentrations above 50μM.
Figure 6.
DMSO control titration spectra of Aβ42. (A) 15N HSQC spectra of U-15N-Aβ42 upon addition of DMSO-d6 at 0% and 2% (initial and final concentrations in titrations of Figs 3–5). (B) 1-D 1H NMR projections of the HSQC experiments taken in panel A demonstrate that Aβ42 remains soluble and monomeric upon addition of DMSO-d6 to 2%.
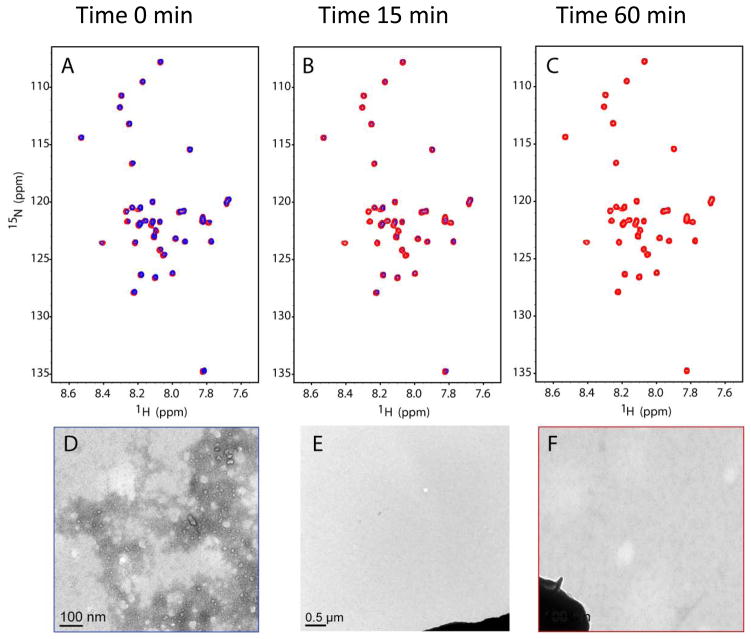
In addition to performing an entire titration series with the NSAIDs, experiments were performed as in Richter et al. (20) where a single point was examined at 1:3 protein:ligand concentration (100μM Aβ42 plus 300μM of either sulindac sulfide, sulindac sulfone, or 2% DMSO control). Upon addition of 300 μM sulindac sulfide, and by the time the sample could be transferred to the spectrometer and the HSQC experiment recorded, some peaks from Aβ42 had begun to disappear (Figure 7). This phenomenon was monitored over the next hour. After 15 minutes, nearly all of the Aβ42 had aggregated and become NMR-invisible. By 1 hour, none of the 15N-HSQC signals were visible in NMR spectrum. This effect was not observed with sulindac sulfone, flurbiprofen, or DMSO (data not shown). To investigate the whereabouts of the Aβ42 peptides, the NMR samples were submitted for transmission electron microscopy (TEM), the results of which are also shown in Fig. 7D-F. Clearly, the addition of sulindac sulfide at concentrations where it forms colloidal aggregates induced the formation of Aβ42 fibrils. On the other hand, addition of DMSO (Fig 7F) or of sulindac sulfone (data not shown) had no effect on the oligomeric state of Aβ42.
Figure 7.
Time course 15N-HSQC spectra of 100 μM U-15N-Aβ42 following addition of sulindac sulfide to 300uM. Panels A, B, and C show spectra taken of 100μM Aβ42 alone (red) and upon addition of sulindac sulfide (blue) at times = 0, 15min, and 1hr, respectively. Notice the decrease in intensity of all the blue peaks as Aβ42 begins to form aggregates. Panels D, E, and F show transmission electron micrographs (66,000x) of 100μM Aβ42 NMR samples fixed to a TEM grid approximately 2hrs after addition of (D) 300uM sulindac sulfide, (E) 300uM sulindac sulfide alone (no protein, 11,600x), and (F) DMSO-only to a final concentration of 2%, matching that in D and E (dark blob in E and F is grid bar included for camera gain). In D, fibrils of Aβ42 are clearly visible.
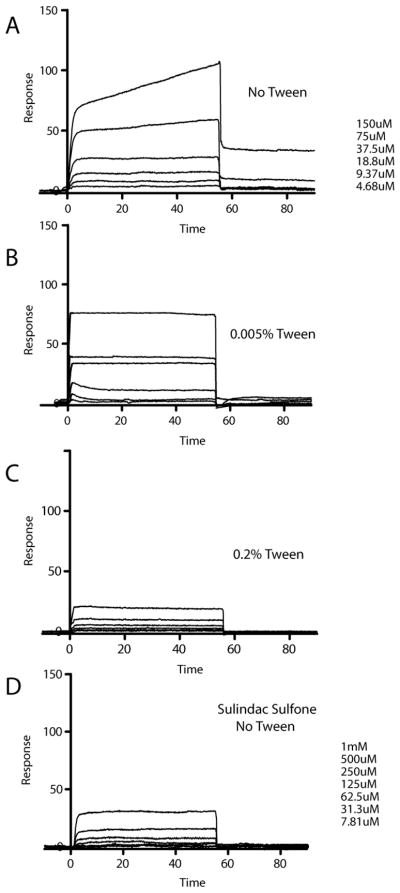
We also looked for direct interaction of three NSAIDs with Aβ42 using surface plasmon resonance (SPR). Previously results from Richter et al (20) suggested that sulindac sulfide binds specifically to Aβ42. However, upon closer inspection of the SPR data presented in the previous work, it can be observed that binding of sulindac sulfide to immobilized Aβ42 was non-saturable over the concentration range tested, suggestive of very weak and/or non-specific binding. To test this hypothesis, we performed SPR experiments in a similar manner as presented in Richter et al. using immobilized Aβ42 peptide. For our studies, we also incorporated varying amounts of Tween-20 in the running buffer— zero detergent (as in the Richter et al.), 0.005% (40 μM, below its CMC of 60μM) and 0.2% (1.6 mM, above CMC) to illuminate whether drug and/or protein aggregation was a factor in the observed SPR response. The results for sulindac sulfone illustrate the patterns expected for the absence of binding (Figure S6). It can be seen that the SPR traces for flurbiprofen (Figure S7), even up to 1 mM, are very similar to those of sulindac sulfone, also indicative of no binding.
In the case of sulindac sulfide (Figure 8), the data is more complex. In the absence of detergent, sulindac sulfide induces a biphasic response suggestive of a rapid binding event followed by a slower second binding event. This second phase is eliminated when the titration is carried out in the presence of a sub-critical micelle concentration (CMC) of detergent, indicating that the slow binding seen in Figure 8A likely represents non-specific association of sulindac sulfide with Aβ42 on the sensor chip — association that can be reduced in the presence of another hydrophobic small molecule (i.e. Tween-20 monomers). When the detergent concentration (Tween-20) is raised still higher to >CMC, it is seen that the SPR response (Figure 8C) is comparable to the negative control SPR response observed at similar concentrations of sulindac sulfone. This indicates that the rapid binding event observed in Figs. 8A-B is of colloidal aggregates of sulindac sulfide to Aβ42. Sub-micellar concentrations of detergents (as in 8B) do not break up those soluble aggregates, but the presence of detergent micelles (as in 8C) effectively dissolves the aggregates, which is seen to eliminate binding.
Figure 8.
SPR analysis of sulindac sulfide with immobilized Aβ42. Overlays of SPR sensorgrams obtained from injections of sulindac sulfide in 50mM sodium phosphate, 50mM NaCl, pH 7 with (A) No detergent, (B) 0.005% Tween-20, and (C) 0.2% Tween-20. Panel D shows corresponding sensorgrams of sulindac sulfone used as a negative control. Aβ42 was immobilized with ~3000 response units (RUs). Compounds at indicated concentration were injected for 55s at a flow rate of 30uL/min at 25 degrees C.
Inhibition of β-Secretase by Sulindac Sulfide
The formation of water soluble colloidal drug aggregates is a commonly encountered phenomenon (33, 35–37). Moreover, such aggregates are known to often have very general and non-specific activities as enzyme inhibitors, sometimes being referred to as “promiscuous inhibitors”(35, 38). To provide additional verification of the nature of the aggregates formed by sulindac sulfide at concentrations above 50 μM we tested to see whether these aggregates have enzyme inhibitory activity. β-secretase (BACE-1) was used as the test enzyme for this experiment. Indeed, we found that sulindac sulfide began to significantly inhibit BACE-1 at concentrations around 50μM, with near complete inhibition being approached at 200μM (Fig S8). No inhibition was observed with sulindac sulfone (Fig S8, panel B), which we showed above does not form aggregates, at least not below 1 mM. We also found that the level of inhibition with sulindac sulfide was significantly reduced by doubling the BACE-1 concentration from 220nM to 440nM (data not shown). Such acute sensitivity to enzyme concentration is a common trait of aggregation-based inhibitors (33, 35–37, 39), which inhibit enzyme action through a non-specific binding mechanism (39). These colloidal aggregates can bind to proteins with high affinity to envelop the protein, preventing substrate access and thus inhibiting protein function (35, 38). Maintaining a constant compound concentration and doubling the enzyme concentration can allow this effect to be at least partially overcome, resulting in decreased inhibition of the enzyme (35). These combined results indicate that not only do aggregates formed by sulindac sulfide trigger fibrillization of Aβ42, but also that these aggregates share properties in common with other “promiscuous inhibitors”.
NMR Titrations of Membrane-Associated C99 with GSMs
In a previous study, we showed that certain GSMs did not bind to C99 monomers and dimers in micellar model membranes(19). Here, we extend this observation to C99 reconstituted into bilayered lipid membranes. Sulindac sulfide, sulindac sulfone, and flurbiprofen each include fluorine atoms, potentiating the use of 19F NMR to monitor binding. 19F NMR chemical shifts are exquisitely sensitive to even very minor changes in local environment. C99 was reconstituted into lipid vesicles with a protein to lipid ration of 1:100 (100μM C99:10mM POPC/POPG). Vesicles were then titrated with sulindac sulfide, sulindac sulfone, and R-flurbiprofen. 19F NMR spectra were acquired for each compound in the presence of protein-free vesicles and in the presence of an identical concentration of vesicles containing reconstituted C99 (100 μM).
Unfortunately, no 19F signal could be detected for sulindac sulfide in both the absence and presence of C99, indicating that this compound binds avidly to the vesicles whether the protein is present or not. Vesicles represent a solids-like environment from an NMR standpoint such that a combination of chemical shift anisotropy and 1H-19F dipolar coupling lead to extensive linebroadening and disappearance of 19F signals when sulindac sulfide binds to the vesicles. However, the results were more clearly interpretable for an alternative GSM, fluribiprofen, and for the negative control, sulindac sulfone. These compounds yield sharp 19F NMR peaks in the presence of protein-free vesicles (Figure 9), which indicates either that these compounds do not bind to lipid bilayers at all or bind only weakly such that exchange between solution and the membrane is rapid on the NMR time scale, such that the free population predominates. When C99 is also present in the vesicles at a C99-to-drug mole-to-mole ratio of 5:1, it can be seen in Figure 8 that there are no changes in the spectra relative to protein-free conditions: chemical shifts, linewidths, and peak intensities are unchanged by the presence of the protein. This indicates that sulindac sulfone and flurbiprofen do not bind to C99 even when the protein is present at a five-fold molar excess over the 20 μM drug concentration.
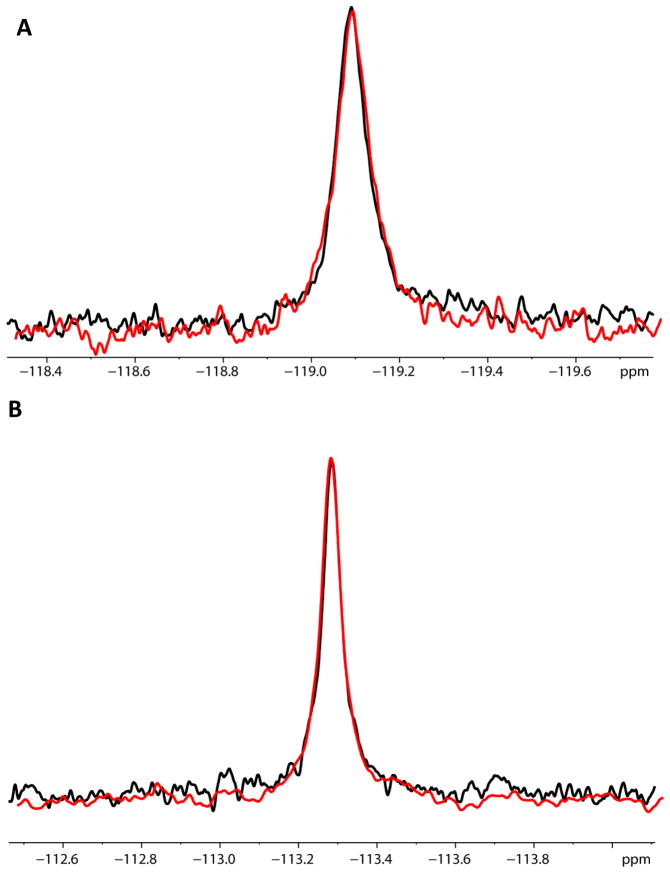
Figure 9.
Comparison of the 19F spectra of flurbiprofen (A) and of sulindac sulfone (B) in the presence of bilayered lipid vesicles in the absence (black) and presence (red) of C99 (A) The samples contained 20μM flurbiprofen both in the absence (black) and in the presence (red) of 100μM C99 incorporated into 10mM POPC/POPG vesicles (1:100 protein:vesicles). (B) The samples contained 20μM sulindac sulfone in both the absence (black) and in the presence (red) of 100μM C99 incorporated into vesicles. All control samples (black) contained only 10mM phospholipid. The lack of change in both sets of spectra indicates that no interaction exists between the compounds and C99.
Discussion
The subject of substrate-targeting GSMs has been a topic of extensive research and discussion over the past several years. Some results have suggested that NSAID-based GSMs directly target the APP substrate (C99) (18, 20, 21), while others, including our previous work(19), disfavor this interpretation (see review in (32)). In addition to showing that non-aggregated C99 in model membranes does not bind GSMs(19), we also presented data suggesting that C99 was very likely to have been in an aggregated form in critical experiments of the original Kukar et al. studies(18). While GSMs do appear to bind to aggregated C99(19), this is unlikely to be relevant to processing of C99 by gamma-secretase in vivo. Moreover, the binding was seen to be non-specific in nature.
Our previous work was disputed in a pair of recent papers by the Multhaup lab (20, 21) which presented data that was interpreted as demonstrating that GSMs, sulindac sulfide in particular, specifically recognize and bind to both membrane-associated C99 and the water soluble monomer form of Aβ42, the latter of which includes the putative GSM binding site proposed in the original work by Kukar et al.Kukar, 2008 #589}. However, we hypothesize that key results and conclusions in the Multhaup papers may have reflected experimental artifacts due to the poor behavior of sulindac sulfide in aqueous solutions. The results of this current work support this hypothesis based on the two primary sets of results, both of which are closely related to the observation that the GSM sulindac sulfide forms colloidal aggregates with a CAC of roughly 50–60 μM. In the first set of results, Richter et al. presented NMR spectra of 100 μM Aβ42 before and after addition of 300 μM sulindac sulfide, which showed a profound drug- induced change in the spectrum of the peptide (Figure 3 in(20)). However, a complete titration series was not carried out, which precludes the possible use of this data to support specific and stoichiometric complex formation between the GSM and Aβ42. Moreover, the NMR spectrum of Aβ42 in the presence of the drug could be interpreted as reflecting the formation of high molecular weight oligomers or aggregates, since many peaks were seen to disappear. The possibility that the GSM might itself be aggregated at 300 μM was not considered, despite the facts that sulindac sulfide is a very hydrophobic compound and that the amyloid-β polypeptides are known to associate non-specifically with small molecule aggregates(33, 37). In the present work, titrations of monomeric Aβ42 by GSMs sulindac sulfide and R-flurbiprofen were followed by NMR spectroscopy, and yielded no evidence for binding of the monomeric drugs to monomeric Aβ42. However, it was found that the colloidal aggregates formed by sulindac sulfide at concentrations above 50-60 μM induced aggregation of Aβ42. Based on this result, we believe that the one point titration presented by the Multhaup lab (20) showing dramatic changes in the NMR spectrum of Aβ42 upon addition of 300 μM sulindac sulfide represents the observation of aggregated Aβ42 formed in response to the presence of colloidal aggregates of sulindac sulfide.
The second set of results involves our repetition of SPR experiments (see Fig 2 in (20)) in which binding of sulindac sulfide to immobilized Aβ42 was tested over a range of drug concentration from 5 to 100 μM. The SPR experiments described by Multhaup et al. (20) demonstrated a linear dose/response, which precludes the conclusion that a specific complex is forming. In addition, the authors noted in the supplementary information that the apparent Kd varied with concentration and that the stoichiometry of the purported Aβ42:Sulindac Sulfide complex varied from 0.2:1 to 2:1. Rather than attributing this to self-association of the compound itself, the authors concluded that the compound bound to multiple binding sites on the Aβ42 peptide. Additionally, these experiments were performed under essentially membrane- and micelle-free conditions. (Tween-20 was present during all these steps, but only at 40 μM, which is below its critical micelle concentration of 60 μM). This is a problem due to the propensity of these proteins to aggregate in the absence of detergent micelles or some other membrane-mimetic medium. Therefore, the immobilized protein present in the SPR experiments (20, 21) was almost certainly in an aggregated form. We reproduced the observation that sulindac sulfide, but not the negative control sulindac sulfone, induces a strong and dose-dependent SPR response. However, when the sulindac sulfide titration was repeated in the presence of Tween-20 micelles, we observed that there was no SPR response beyond what was observed for negative control conditions. This strongly suggests the binding of sulindac sulfide to Aβ42 observed in the earlier work represents non-specific association of these two compounds. Such association is highest when sulindac sulfide is in its colloidal form (at concentrations >50μM) and may also be promoted by the structural properties of sensor chip surface-associated Aβ42, which may itself have aggregate-like properties as a result of being locally concentrated at on the sensor chip surface. Association between the GSM and surface-associated Aβ42 is eliminated by the presence of detergent micelles that can disperse the colloidal drug and can also coat exposed hydrophobic sites on sensor surface-associated Aβ42, making such sites less-susceptible to non-specific hydrophobic interactions with hydrophobic compounds such as sulindac sulfide. We have previously shown that aggregated C99 can bind GSMs in a non-specific fashion (19) and so it is no surprise that this is what was seen by Multhaup and co-workers(21). In the present work, we observed that the GSM R-flurbiprofen exhibits no binding to C99 reconstituted lipid in vesicles. This result extends the conclusions from our earlier observations of the lack of GSM binding to non-aggregated C99 in micellar model membranes to non-aggregated C99 in actual lipid bilayers.
One additional set of experiments from Multhaup and co-workers that yielded support for sulindac sulfide binding to C99 in membranes was a series of ToxR experiments carried out in E. coli (20). In those experiments homodimerization of the transmembrane segment of C99 was assessed following expression into E. coli based on coupling homodimerization of this segment to transcriptional activation of a gene that expresses a colorimetric reporter enzyme. Using this assay, it was seen that sulindac sulfide reduces apparent dimerization of C99 in E. coli in a dose-dependent fashion, consistent with inhibition of dimerization of C99 by GSM binding. These studies were carefully carried out and can indeed be interpreted as being supportive of GSM/C99 binding. However, when conducting in vitro experiments involving GSM drugs, living cells, and an indirect phenotype-based assay the possibility cannot be ruled out that the GSM induces a positive assay response as a result of off-target drug effects that lead to the artifact-based activation of the assay response (i.e., induction of reporter enzyme expression). In light of the biophysical results of this paper, we suggest that this alternative explanation of the ToxR data is very likely applicable.
The experiments and results summarized above lead to the conclusion that the GSM sulindac sulfide does not bind to Aβ42 when both compounds are in monomeric form. On the other hand, the results clearly suggest that two types of non-specific binding occur: those between aggregates of Aβ42 and monomers of NSAID type compounds (sulindac sulfide, as well as flurbiprofen and celecoxib (19)), and those between colloid-type aggregates of sulindac sulfide and monomeric Aβ42. Promiscuous binding of small molecule aggregates to proteins, often accompanied by inhibition of protein function, is a very common occurrence (33, 35–37). Indeed, in this study aggregated sulindac sulfide was found not only to bind to Aβ42, but also to inhibit β-secretase (here used as a representative enzyme). It has previously been shown that Congo red can form colloidal micelle-like aggregates that bind to Aβ and induce its aggregation(37). It has also been observed that a number of drug-like molecules form colloidal aggregates that interact with amyloid-forming yeast prion proteins in a way that inhibits fibril formation(36).
Evidence is accumulating that NSAID-based GSMs do not exert their therapeutic effect by binding to free C99 (review in (32)). Previous publications have suggested that GSMs act by causing conformational changes within Presenilin 1 (PS1)(40–42), or by altering membrane architecture and thereby changing the manner in which γ-secretase cleaves its APP substrate (43). More recent studies have indicated that the action of GSMs may be allosteric in nature. Uemura et al demonstrated that GSM-induced conformational changes in PS1 only occur in the presence of substrate, suggesting that substrate binding to γ-secretase uncovers an allosteric site for GSM binding that is only present in the substrate-enzyme complex (44). Another recent study demonstrated that mutations in the GxxxG motif located in the previously-proposed GSM binding site of C99 still caused an effect on Aβ42 production upon treatment with GSMs (45). The compounds were then shown to display differential or no effects on Aβ42 and Aβ38 levels when PS1 mutants were used, implying disruption of GSM interaction with γ-secretase. These conclusions contradict a free substrate-targeted model of GSM action and instead suggest that these molecules target the γ-secretase enzyme itself or the enzyme-substrate complex (45). The results of this paper rule out binding of GSMs to free C99 in non-aggregated form but do not argue against the possibility that GSMs could interact directly with C99 when is bound to γ-secretase. Taken as a whole, the evidence is becoming overwhelming that GSMs do not specifically target the γ-secretase substrate, at least not prior to its association with the enzyme.
Supplementary Material
Acknowledgments
This work was supported by NIH grant PO1GM080513 (to CRS) and by Alzheimer’s Association grant IIRG-07-59379 (to CRS). Partial support for PB was through NIH T32 GM08320. The authors would like to acknowledge Cynthia Li at Amgen for assistance with CD experiments and Drs. Leszek Poppe, Paul Schnier, and Steve Wood for critical reading of the manuscript.
Abbreviations
- APP
amyloid precursor protein
- β-CTF
99 residue transmembrane C-terminal fragment of the amyloid precursor protein
- BACE-1
β-secretase
- C99
99 residue transmembrane C-terminal domain of the amyloid precursor protein
- NSAID
non-steroidal anti-inflammatory drug
- CAC
critical aggregate concentration
- CD
circular dichroism
- CMC
critical micelle concentration
- E. coli
Escherichia coli
- GSM
gamma-secretase modulator
- MP
membrane protein
- NMR
nuclear magnetic resonance
- POPC
1-palmitoyl-2-oleoyl-phosphatidylcholine
- POPG
1-palmitoyl-2-oleoyl-phosphatidylglycerol
- TM
transmembrane
- TMD
transmembrane domain
- 2-D
two-dimensional
- UV
ultraviolet
Footnotes
This work was supported by NIH grant PO1 GM080513 (to CRS) and by Alzheimer’s Association grant IIRG-07-59379 (to CRS). Partial support for PB was through NIH T32 NIH 5 T32 GM08320.
SUPPORTING INFORMATION AVAILABLE
Eight figures containing further details of the NMR and SPR characterization of the peptides and small molecules used in this study. This material is available free of charge via the Internet at http://pubs.acs.org.
References
- 1.Brookmeyer R, Johnson E, Ziegler-Graham K, Arrighi HM. Forecasting the global burden of Alzheimer's disease. Alzheimers Dement. 2007;3:186–191. doi: 10.1016/j.jalz.2007.04.381. [DOI] [PubMed] [Google Scholar]
- 2.Oehlrich D, Berthelot DJ, Gijsen HJ. Gamma-Secretase Modulators as Potential Disease Modifying Anti-Alzheimer's Drugs. J Med Chem. 2010 doi: 10.1021/jm101168r. [DOI] [PubMed] [Google Scholar]
- 3.Tiraboschi P, Hansen LA, Thal LJ, Corey-Bloom J. The importance of neuritic plaques and tangles to the development and evolution of AD. Neurology. 2004;62:1984–1989. doi: 10.1212/01.wnl.0000129697.01779.0a. [DOI] [PubMed] [Google Scholar]
- 4.Glenner GG, Wong CW. Alzheimer's disease: initial report of the purification and characterization of a novel cerebrovascular amyloid protein. Biochem Biophys Res Commun. 1984;120:885–890. doi: 10.1016/s0006-291x(84)80190-4. [DOI] [PubMed] [Google Scholar]
- 5.Glenner GG, Wong CW, Quaranta V, Eanes ED. The amyloid deposits in Alzheimer's disease: their nature and pathogenesis. Appl Pathol. 1984;2:357–369. [PubMed] [Google Scholar]
- 6.Masters CL, Multhaup G, Simms G, Pottgiesser J, Martins RN, Beyreuther K. Neuronal origin of a cerebral amyloid: neurofibrillary tangles of Alzheimer's disease contain the same protein as the amyloid of plaque cores and blood vessels. EMBO J. 1985;4:2757–2763. doi: 10.1002/j.1460-2075.1985.tb04000.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Masters CL, Simms G, Weinman NA, Multhaup G, McDonald BL, Beyreuther K. Amyloid plaque core protein in Alzheimer disease and Down syndrome. Proc Natl Acad Sci U S A. 1985;82:4245–4249. doi: 10.1073/pnas.82.12.4245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hardy JA, Higgins GA. Alzheimer's disease: the amyloid cascade hypothesis. Science. 1992;256:184–185. doi: 10.1126/science.1566067. [DOI] [PubMed] [Google Scholar]
- 9.Hardy J, Selkoe DJ. The amyloid hypothesis of Alzheimer's disease: progress and problems on the road to therapeutics. Science. 2002;297:353–356. doi: 10.1126/science.1072994. [DOI] [PubMed] [Google Scholar]
- 10.Grill JD, Cummings JL. Current therapeutic targets for the treatment of Alzheimer's disease. Expert Rev Neurother. 10:711–728. doi: 10.1586/ern.10.29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Galimberti D, Scarpini E. Alzheimer's disease: from pathogenesis to disease-modifying approaches. CNS Neurol Disord Drug Targets. 10:163–174. doi: 10.2174/187152711794480438. [DOI] [PubMed] [Google Scholar]
- 12.Aguzzi A, O'Connor T. Protein aggregation diseases: pathogenicity and therapeutic perspectives. Nat Rev Drug Discov. 9:237–248. doi: 10.1038/nrd3050. [DOI] [PubMed] [Google Scholar]
- 13.Weggen S, Rogers M, Eriksen J. NSAIDs: small molecules for prevention of Alzheimer's disease or precursors for future drug development? Trends Pharmacol Sci. 2007;28:536–543. doi: 10.1016/j.tips.2007.09.004. [DOI] [PubMed] [Google Scholar]
- 14.Imbimbo BP. An update on the efficacy of non-steroidal anti-inflammatory drugs in Alzheimer's disease. Expert Opin Investig Drugs. 2009;18:1147–1168. doi: 10.1517/13543780903066780. [DOI] [PubMed] [Google Scholar]
- 15.Imbimbo BP. Why did tarenflurbil fail in Alzheimer's disease? J Alzheimers Dis. 2009;17:757–760. doi: 10.3233/JAD-2009-1092. [DOI] [PubMed] [Google Scholar]
- 16.Weggen S, Eriksen JL, Das P, Sagi SA, Wang R, Pietrzik CU, Findlay KA, Smith TE, Murphy MP, Bulter T, Kang DE, Marquez-Sterling N, Golde TE, Koo EH. A subset of NSAIDs lower amyloidogenic Abeta42 independently of cyclooxygenase activity. Nature. 2001;414:212–216. doi: 10.1038/35102591. [DOI] [PubMed] [Google Scholar]
- 17.Eriksen JL, Sagi SA, Smith TE, Weggen S, Das P, McLendon DC, Ozols VV, Jessing KW, Zavitz KH, Koo EH, Golde TE. NSAIDs and enantiomers of flurbiprofen target gamma-secretase and lower Abeta 42 in vivo. J Clin Invest. 2003;112:440–449. doi: 10.1172/JCI18162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kukar TL, Ladd TB, Bann MA, Fraering PC, Narlawar R, Maharvi GM, Healy B, Chapman R, Welzel AT, Price RW, Moore B, Rangachari V, Cusack B, Eriksen J, Jansen-West K, Verbeeck C, Yager D, Eckman C, Ye W, Sagi S, Cottrell BA, Torpey J, Rosenberry TL, Fauq A, Wolfe MS, Schmidt B, Walsh DM, Koo EH, Golde TE. Substrate-targeting gamma-secretase modulators. Nature. 2008;453:925–929. doi: 10.1038/nature07055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Beel AJ, Barrett P, Schnier PD, Hitchcock SA, Bagal D, Sanders CR, Jordan JB. Nonspecificity of binding of gamma-secretase modulators to the amyloid precursor protein. Biochemistry. 2009;48:11837–11839. doi: 10.1021/bi901839d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Richter L, Munter LM, Ness J, Hildebrand PW, Dasari M, Unterreitmeier S, Bulic B, Beyermann M, Gust R, Reif B, Weggen S, Langosch D, Multhaup G. Amyloid beta 42 peptide (Abeta42)-lowering compounds directly bind to Abeta and interfere with amyloid precursor protein (APP) transmembrane dimerization. Proc Natl Acad Sci U S A. 107:14597–14602. doi: 10.1073/pnas.1003026107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Botev A, Munter LM, Wenzel R, Richter L, Althoff V, Ismer J, Gerling U, Weise C, Koksch B, Hildebrand PW, Bittl R, Multhaup G. The amyloid precursor protein C-terminal fragment C100 occurs in monomeric and dimeric stable conformations and binds gamma-secretase modulators. Biochemistry. 50:828–835. doi: 10.1021/bi1014002. [DOI] [PubMed] [Google Scholar]
- 22.Harmeier A, Wozny C, Rost BR, Munter LM, Hua H, Georgiev O, Beyermann M, Hildebrand PW, Weise C, Schaffner W, Schmitz D, Multhaup G. Role of amyloid-beta glycine 33 in oligomerization, toxicity, and neuronal plasticity. J Neurosci. 2009;29:7582–7590. doi: 10.1523/JNEUROSCI.1336-09.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Schmechel A, Zentgraf H, Scheuermann S, Fritz G, Pipkorn R, Reed J, Beyreuther K, Bayer TA, Multhaup G. Alzheimer beta-amyloid homodimers facilitate A beta fibrillization and the generation of conformational antibodies. J Biol Chem. 2003;278:35317–35324. doi: 10.1074/jbc.M303547200. [DOI] [PubMed] [Google Scholar]
- 24.Ash M, Ash I. The Handbook of Industrial Surfactants. Synapse Information Resources, Inc; Endicott, NY: 2010. [Google Scholar]
- 25.Roher AE, Chaney MO, Kuo YM, Webster SD, Stine WB, Haverkamp LJ, Woods AS, Cotter RJ, Tuohy JM, Krafft GA, Bonnell BS, Emmerling MR. Morphology and toxicity of Abeta-(1-42) dimer derived from neuritic and vascular amyloid deposits of Alzheimer's disease. J Biol Chem. 1996;271:20631–20635. doi: 10.1074/jbc.271.34.20631. [DOI] [PubMed] [Google Scholar]
- 26.Farrow NA, Muhandiram R, Singer AU, Pascal SM, Kay CM, Gish G, Shoelson SE, Pawson T, Forman-Kay JD, Kay LE. Backbone dynamics of a free and phosphopeptide-complexed Src homology 2 domain studied by 15N NMR relaxation. Biochemistry. 1994;33:5984–6003. doi: 10.1021/bi00185a040. [DOI] [PubMed] [Google Scholar]
- 27.Kay LE, Torchia DA, Bax A. Backbone dynamics of proteins as studied by 15N inverse detected heteronuclear NMR spectroscopy: application to staphylococcal nuclease. Biochemistry. 1989;28:8972–8979. doi: 10.1021/bi00449a003. [DOI] [PubMed] [Google Scholar]
- 28.Cavanagh J, Fairbrother WJ, Palmer AG, Skelton NJ. Protein NMR Spectroscopy: Principles and Practice. Academic Press; San Diego, CA: 1996. [Google Scholar]
- 29.Zheng G, Stait-Gardner T, Anil Kumar PG, Torres AM, Price WS. PGSTE-WATERGATE: an STE-based PGSE NMR sequence with excellent solvent suppression. J Magn Reson. 2008;191:159–163. doi: 10.1016/j.jmr.2007.12.001. [DOI] [PubMed] [Google Scholar]
- 30.Wilkins DK, Grimshaw SB, Receveur V, Dobson CM, Jones JA, Smith LJ. Hydrodynamic radii of native and denatured proteins measured by pulse field gradient NMR techniques. Biochemistry. 1999;38:16424–16431. doi: 10.1021/bi991765q. [DOI] [PubMed] [Google Scholar]
- 31.Cheng Y, Judd T, Bartberger M, Chen K, Fremeau R, Hickman D, Hitchcock S, Jordan J, Li V, Lopez P, Louie S, Luo Y, Michelsen K, Nixey T, Powers T, Rattan C, Sickmier E, StJean D, Wahl R, Wen P, Wood S. From Fragment Screening to In Vivo Efficacy: Optimization of a Series of 2-Aminoquinolines as Potent Inhibitors of BACE1. J Med Chem. 2011 doi: 10.1021/jm200544q. [DOI] [PubMed] [Google Scholar]
- 32.Zettl H, Weggen S, Schneider P, Schneider G. Exploring the chemical space of gamma-secretase modulators. Trends Pharmacol Sci. 31:402–410. doi: 10.1016/j.tips.2010.05.007. [DOI] [PubMed] [Google Scholar]
- 33.Coan KE, Shoichet BK. Stoichiometry and physical chemistry of promiscuous aggregate-based inhibitors. J Am Chem Soc. 2008;130:9606–9612. doi: 10.1021/ja802977h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Feng BY, Shoichet BK. Synergy and antagonism of promiscuous inhibition in multiple-compound mixtures. J Med Chem. 2006;49:2151–2154. doi: 10.1021/jm060029z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.McGovern SL, Caselli E, Grigorieff N, Shoichet BK. A common mechanism underlying promiscuous inhibitors from virtual and high-throughput screening. J Med Chem. 2002;45:1712–1722. doi: 10.1021/jm010533y. [DOI] [PubMed] [Google Scholar]
- 36.Feng BY, Toyama BH, Wille H, Colby DW, Collins SR, May BC, Prusiner SB, Weissman J, Shoichet BK. Small-molecule aggregates inhibit amyloid polymerization. Nat Chem Biol. 2008;4:197–199. doi: 10.1038/nchembio.65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lendel C, Bolognesi B, Wahlstrom A, Dobson CM, Graslund A. Detergent-like interaction of Congo red with the amyloid beta peptide. Biochemistry. 49:1358–1360. doi: 10.1021/bi902005t. [DOI] [PubMed] [Google Scholar]
- 38.McGovern SL, Helfand BT, Feng B, Shoichet BK. A specific mechanism of nonspecific inhibition. J Med Chem. 2003;46:4265–4272. doi: 10.1021/jm030266r. [DOI] [PubMed] [Google Scholar]
- 39.Giannetti AM, Koch BD, Browner MF. Surface plasmon resonance based assay for the detection and characterization of promiscuous inhibitors. J Med Chem. 2008;51:574–580. doi: 10.1021/jm700952v. [DOI] [PubMed] [Google Scholar]
- 40.Lleo A, Berezovska O, Herl L, Raju S, Deng A, Bacskai BJ, Frosch MP, Irizarry M, Hyman BT. Nonsteroidal anti-inflammatory drugs lower Abeta42 and change presenilin 1 conformation. Nat Med. 2004;10:1065–1066. doi: 10.1038/nm1112. [DOI] [PubMed] [Google Scholar]
- 41.Berezovska O, Lleo A, Herl LD, Frosch MP, Stern EA, Bacskai BJ, Hyman BT. Familial Alzheimer's disease presenilin 1 mutations cause alterations in the conformation of presenilin and interactions with amyloid precursor protein. J Neurosci. 2005;25:3009–3017. doi: 10.1523/JNEUROSCI.0364-05.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Uemura K, Lill CM, Li X, Peters JA, Ivanov A, Fan Z, DeStrooper B, Bacskai BJ, Hyman BT, Berezovska O. Allosteric modulation of PS1/gamma-secretase conformation correlates with amyloid beta(42/40) ratio. PLoS One. 2009;4:e7893. doi: 10.1371/journal.pone.0007893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Gamerdinger M, Clement AB, Behl C. Effects of sulindac sulfide on the membrane architecture and the activity of gamma-secretase. Neuropharmacology. 2008;54:998–1005. doi: 10.1016/j.neuropharm.2008.02.009. [DOI] [PubMed] [Google Scholar]
- 44.Uemura K, Farner KC, Hashimoto T, Nasser-Ghodsi N, Wolfe MS, Koo EH, Hyman BT, Berezovska O. Substrate docking to gamma-secretase allows access of gamma-secretase modulators to an allosteric site. Nat Commun. 1:130. doi: 10.1038/ncomms1129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Page RM, Gutsmiedl A, Fukumori A, Winkler E, Haass C, Steiner H. Beta-amyloid precursor protein mutants respond to gamma-secretase modulators. J Biol Chem. 285:17798–17810. doi: 10.1074/jbc.M110.103283. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.