Abstract
Modern technologies have made the sequencing of personal genomes routine. They have revealed thousands of nonsynonymous (amino-acid altering) single nucleotide variants (nSNVs) of protein coding DNA per genome. What do these variants foretell about an individual’s predisposition to diseases? The experimental technologies required to carry out such evaluations at a genomic scale are not yet available. Fortunately, the process of natural selection has lent us an almost infinite set of tests in nature. During the long-term evolution, new mutations and existing variations have been evaluated for their biological consequences in countless species, and outcomes were readily revealed by multispecies genome comparisons. We review studies that have investigated evolutionary characteristics and in silico functional diagnoses of nSNVs found in thousands of disease-associated genes. We conclude that the patterns of long-term evolutionary conservation and permissible divergence are essential and instructive modalities for functional assessment of human genetic variations.
Evolutionary genomic medicine
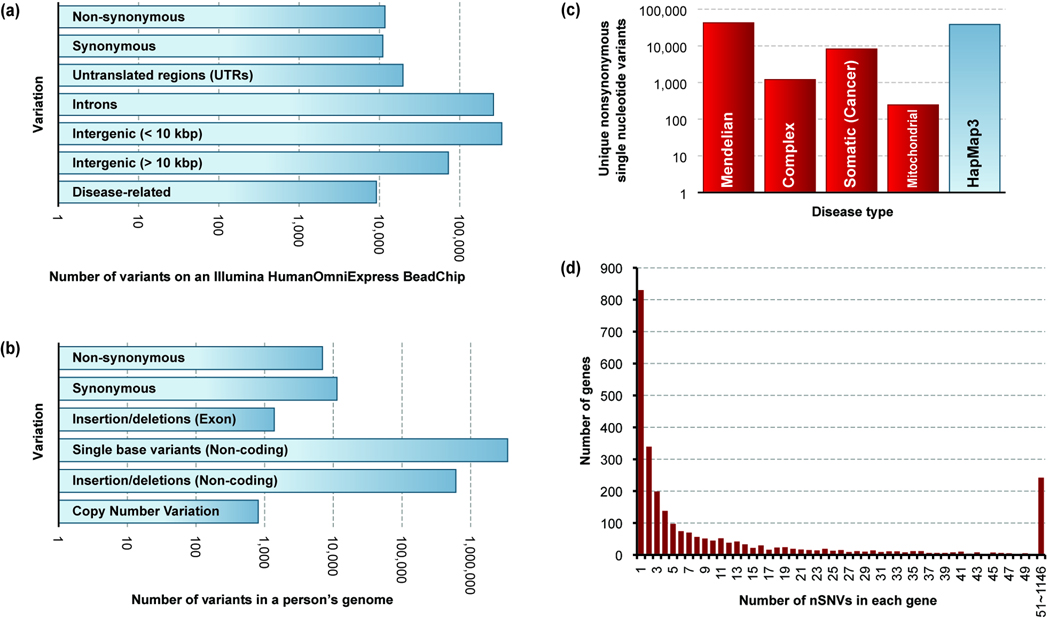
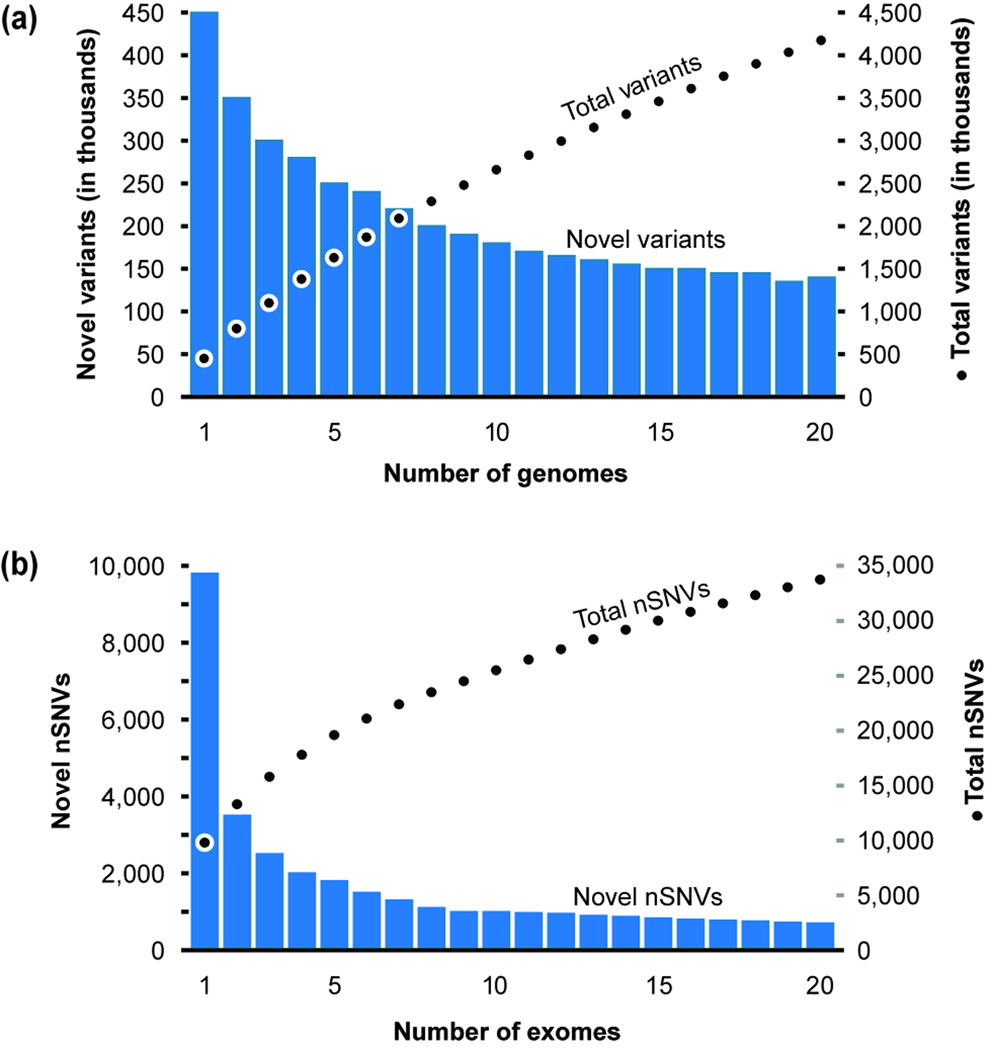
Thousands of individuals in the general public have begun to gain access to their genetic variation profiles by using direct-to-consumer DNA tests available from commercial vendors, which profile hundreds of thousands of genomic markers for a cost of a few hundred dollars (Fig. 1a). Through this genetic profiling, individuals hope to learn about not only their ancestry, but also genetic variations underlying their physical characteristics and predispositions to diseases. In biomedicine, scientists have been profiling variations at genomic markers in healthy and diseased individuals at genome scale in a variety of disease contexts and populations. This has led to the discovery of thousands of disease associated genes and DNA variants [1–6]. Meanwhile, following sharp declines in the per-base cost of sequencing, complete genomic sequencing of individuals and cohorts is underway and expanding [7–11]. Taken together, these efforts have begun to paint a more robust picture of the amount and types of variations found within and between human individuals and populations. Any one personal genome contains more than a million variants, the majority of which are single nucleotide variants (SNVs) (Fig. 1b). With the complete sequencing of each new genome, the number of novel variants discovered is decreasing, but the total number of known variants is growing quickly (Fig. 2a). Our knowledge of the number of disease genes and the total number of known disease-associated SNVs has grown with these advances [12].
Figure 1.
Profiles of personal and population variations. (a) Counts of various types of genetic variants profiled by 23andMe using the Illumina HumanOmniExpress BeadChip. 733,202 SNP identifiers (rsIDs) were retrieved from the Illumina website and mapped to the dbSNP database. Cross-referenced by rsIDs, disease-related variants were determined by using data from HGMD [12] and VARIMED [96] datasets. (b) The numbers of different types of variants found per human genome [97]. (c) The numbers of known non-synonymous single nucleotide variants (nSNVs) in the human nuclear and mitochondrial genomes that are associated with Mendelian diseases, complex diseases, and somatic cancers. Compared to complex diseases and somatic cancers, nSNVs related to Mendelian diseases account for the most variants discovered to date. Data were retrieved from HGMD [12], VARIMED [96], COSMIC [98], MITOMAP [41], and HapMap3 [99] resources. (d) The number of nSNVs in each gene related to Mendelian diseases. The majority of genes have only one or a few mutations, while there are some genes hosting hundreds or even more than 1000 mutations. Data were retrieved from HGMD. The numbers of variants in panels {a–c} are in log10 scale. Information for disease associated variants is shown in red and the personal and population variations are shown in blue.
Figure 2.
Novel SNV discovery with genome and exome sequencing. (a) The number of novel SNVs discovered by sequencing one and more genomes [97]. With increasing numbers of genomes sequenced, the number of novel SNVs decreases (bars), whereas the cumulative count of SNVs increases (filled circles). (b) The number of nSNVs discovered by sequencing one or more exomes [14]. With more exomes sequenced, the number of novel SNVs discovered decreases (bars) and the cumulative count of nSNVs increases (filled circles). Panels a and b are redrawn with permission from [97] and [14], respectively.
Today, a vast majority of the known disease-associated variants are found within protein-coding genes (Fig. 1c) with genome-wide association studies beginning to reveal thousands of non-coding variants. Proteins are encoded in genomic DNA by exon regions, which comprise just ~1% of the genomic sequence (Exome) [11, 13]. It is this part of our genome for which we have the best understanding of how DNA blueprint sequence relates to function, and is arguably the best chance to connect genetic variations with disease pathophysiology. A person’s exome carries about 6,000 – 10,000 amino-acid-altering nonsynonymous SNVs (nSNVs) [2, 7, 9, 10, 14]. These protein point variations are already known to be associated with more than a thousand major diseases [12]. A large number of exome projects are poised to reveal protein mutations of tens of thousands of individuals from disease cohorts and healthy populations for disorders of various complexities [11, 15–17]. With the sequencing of each new exome, we are currently discovering hundreds of new nSNVs, which points to the existence of a large number of different protein alleles in the genomes of humans (Fig. 2b). In addition to the variations arising in the germline, protein-coding regions of somatic cancer cells contain tens of thousands of nonsynonymous mutations of somatic and germline mutational origins (Fig. 1c). Adding to the variation in the nuclear genetic material are thousands of mutations in the mitochondrial genome, many of which are also implicated in diseases (Fig. 1c).
Translating a personal variation profile into useful phenotypic information (e.g., relating to predisposition to disease, differential drug response, and other health concerns) is a grand challenge in the field of genomic medicine. Genomic medicine is concerned with enabling healthcare that is tailored to the individual based on genomic information [18]. This is a daunting task, because common variants derived from large population-based studies typically describe relatively small proportions of disease risk. Additionally, each individual genome carries many private variants that are not typically seen in a limited sampling of the human populations. Although only a small fraction of all personal variations are likely to modulate health, the sheer volume of genomic and exomic variants is far too large to apply traditional laboratory or experimental techniques to aid in their diagnosis. Higher throughput techniques are now becoming available to evaluate the functional consequences of hundreds of specified mutant proteins, or much greater numbers of random mutants. However, these methods are still inadequate to handle the volume of variation information arising from modern sequencing methods in a scalable or economical manner [19–23].
Fortunately, results from the great natural experiment of molecular evolution are recorded in the genomes of humans and other living species. All new mutations and preexisting variations are subjected to the process of natural selection, which eliminates mutants with negative effects on the phenotype. Variations escaping the sieve of natural selection appear in the form of differences among the genomes of humans, great apes, and other species. Through multispecies comparisons of these data, using the models and methods of molecular evolution, it is possible to mine this information and evaluate the severity of each variant computationally (in silico). With the availability of large number genomes from the tree of life, it is becoming clear that evolution can serve as a kind of telescope for exploring the universe of genetic variation. In this evolutionary telescope, the degree of historical conservation of individual position (and regions) and the sets of substitutions permitted among species at individual positions serve as two lenses. This tool has the ability to provide first glimpses into the functional and health consequences of variations that are being discovered by high-throughput sequencing efforts. Consequently, phylomedicine will emerge as an important discipline at the intersection of molecular evolution and genomic medicine with a focus on understanding of human disease and health through the application of long-term molecular evolutionary history. Phylomedicine expands the purview of contemporary evolutionary medicine [24–28] to use evolutionary patterns beyond the short-term history (e.g., populations) by means of multispecies genomics [29, 30].
In the following, we review scientific investigations that have analyzed evolutionary properties of disease-associated nSNVs and predicted function-altering propensities of individual variants in silico using multispecies data. We have primarily focused on variants of exomes, because the function of proteins is currently best understood independent of comparative genomics. Furthermore, protein point mutations are associated with more than 1000 major diseases, and generally with a statistically significant association beyond chance alone. Furthermore, the cost of exome sequencing is declining to the point that the legion of small scientific laboratories and the interested public is now able to economically profile complete exomes [17, 31, 32]. Therefore, the chosen emphasis on exome variations reflects current directions in clinical and research applications of genomic sequencing.
Mendelian (monogenic) diseases
For centuries it has been known that particular diseases run in families, notably in some royal families where there was a degree of inbreeding. Once Mendel’s principles of inheritance became widely known in the early 1900s it became evident from family genealogies that specific heritable diseases fit Mendelian predictions. These are termed Mendelian diseases (reviewed in [33]). Such diseases can have substantial impact on the affected individual but tend to be rare, on the order of one case per several thousand or several tens of thousands of individuals.
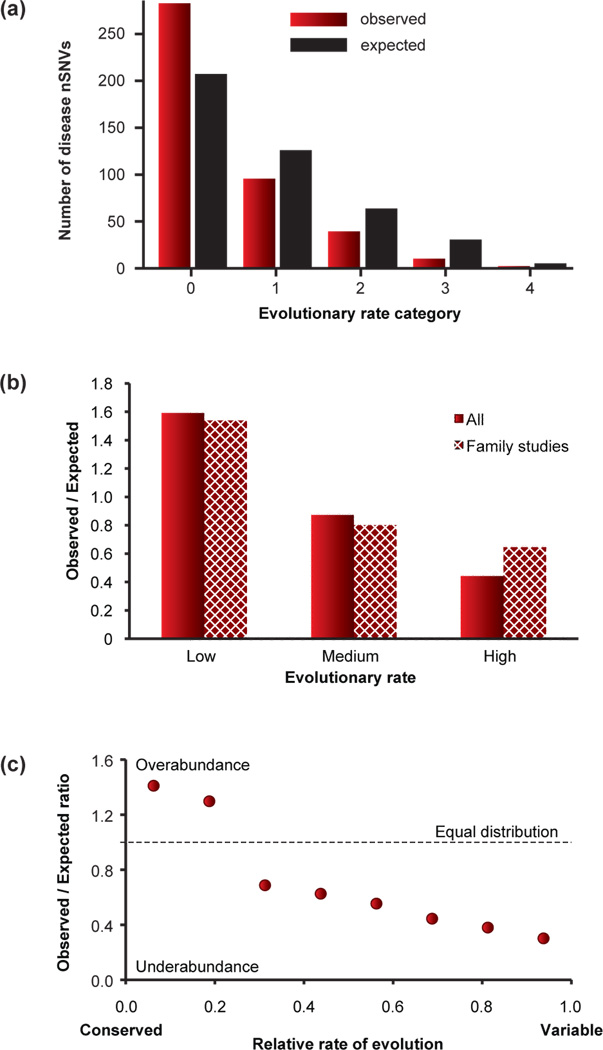
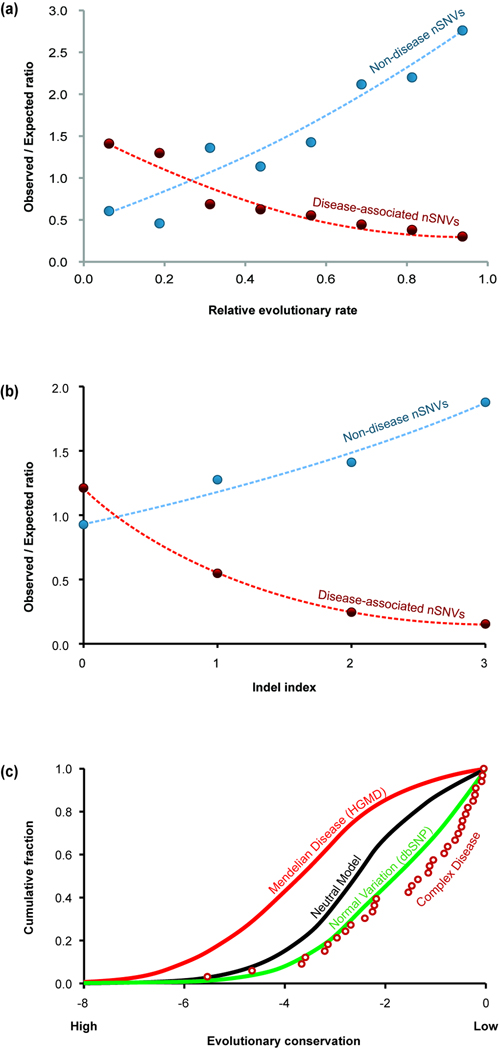
Over the last three decades, mutations in single (candidate) genes in many families have been linked to individual Mendelian diseases (e.g. Box 1). Sometimes more than a hundred SNVs in the same gene have been implicated in a particular disease (Fig. 1d). For example, by the turn of this century, individual patient and family studies revealed over 500 nSNVs in the Cystic fibrosis transmembrane conductance regulator (CFTR) gene for cystic fibrosis (CF). This enabled first efforts to examine evolutionary properties of the positions harboring CFTR nSNVs [29]. The disease-associated nSNVs were found to be overabundant at positions that had permitted only a very small amount of change over evolutionary time [29] (Fig. 3a, b). Soon after, this trend was confirmed at the proteome scale in analyses of thousands of nSNVs from hundreds of genes (Fig. 3c) [34–37]. These patterns were in sharp contrast to the variations seen in non-patients, which are enriched in the fast evolving positions (Fig. 4a) [29, 35]. In population polymorphism data, faster evolving positions also show higher minor allele frequencies than those at slow evolving positions [29, 35], which translates into an enrichment of rare alleles in slow-evolving and functionally important genomic positions [38].
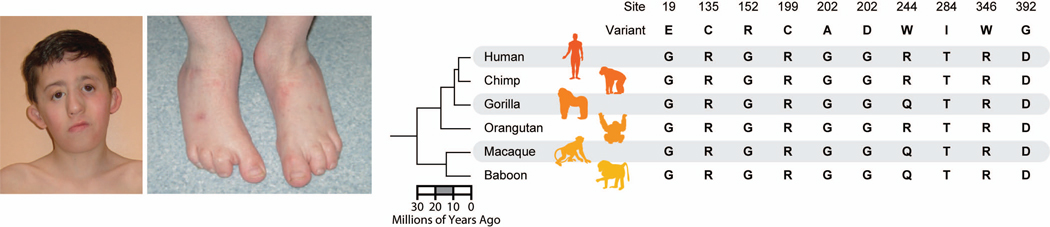
BOX 1. Variation in the dihydroorotate dehydrogenase 1 (DHODH) protein found in individuals suffering from the Miller syndrome.
Miller syndrome is a rare genetic disorder characterized by distinctive craniofacial malformations that occur in association with limb abnormalities (Figure on the left). It is a typical Mendelian disease that is inherited as an autosomal recessive genetic trait. By sequencing the exomes of four affected individuals in three independent kindreds, ten mutations in a single candidate gene, DHODH, were found to be associated with this disease [96]. In the figure on the right, the ten mutations are shown in the context of the DHODH orthologs from six primates (including human) and the timing of their evolutionary relationships (timetree from ref. [57]). They are in slow-evolving sites that are highly conserved not only in primates, but also among distantly related vertebrates. Specifically, 50% of these mutations are found at completely conserved positions among 46 vertebrates, including human. The average evolutionary rate, estimated using methods in ref. [57], for sites containing these disease-related mutations is 0.50 substitutions per billion year, which is ~40% slower than those sites hosting four non-disease-related population polymorphisms of DHODH available in the public databases. Biochemically, the average severity of these ten mutations is more than twice that of the four population polymorphisms, as measured using the Grantham’s [54] index (112 and 55, respectively). PolyPhen-2 [97], a computational program used to predict the propensity of individual amino acid changes at a position to damage protein function, diagnosed all ten mutations to be potentially damaging and the four population polymorphisms to be benign. This case study demonstrated clear patterns of long-term evolutionary conservation for Mendelian disease related variations, and the promising applications of in silico tools in assisting functional diagnosis.
Figure 3.
Evolutionary properties of positions afflicted with disease-associated nonsynonymous single nucleotide variants (nSNVs). (a) The observed and expected numbers of disease associated nSNVs in positions that have evolved with different evolutionary rates in the CFTR protein [29]. The disease associated nSNVs are enriched in positions evolving with the lowest rates, which belong to the rate category 0. (b) The ratio of observed to expected numbers of nSNVs in different rate categories for all CFTR variants (solid pattern; 431 variants) and those reported in publications profiling one or more families (hatched pattern; 59 variants). Data and publications were obtained from HGMD for all variants with deposition date until year 2000. This comparison shows that the initial practice of the use of all available variants, including those reported by clinicians from individual patients (>80% of the variants), did not bias the observed trends. (c) The proteome-scale relationship of the observed/expected ratios of Mendelian disease-associated nSNVs in positions that have evolved with different evolutionary rates. The results are from an analysis of disease associated nSNVs from 2,717 genes (public release of HGMD). Just as for individual diseases, nSNVs are enriched in positions evolving with the lowest rates. Panel a is redrawn with permission from ref. [29].
Figure 4.
The enrichment of disease-associated nSNVs (red) and the deficit of population polymorphisms (blue) in human amino acid positions (a) evolving with different rates and (b) with differ degrees of insertion-deletions [35]. In both cases, smaller numbers on the x axis correspond to more conserved positions. There is an enrichment of disease associated nSNVs and a deficit of population nSNPs in conserved positions. This trend is reversed for the fastest evolving positions. (c) The cumulative distributions of the evolutionary conservation scores for nSNVs associated with Mendelian diseases (solid red line), complex diseases (open red circles), and population polymorphisms (green line). The shift towards the left in Mendelian nSNVs indicates higher position specific evolutionary conservation. Conversely, a shift towards the right in complex disease nSNVs indicates lower evolutionary conservation, which overlaps with normal variations observed in the population. Data for the neutral model (black line) was generated by simulation [37]. Panels a and c are reproduced with permission from refs. [35], [35], and [37], respectively.
Looking at patterns of evolutionary retention at positions, another type of evolutionary conservation, a similar pattern was found: positions preferentially retained over the history of vertebrates were more likely to be involved in Mendelian diseases as compared to the patterns of natural variation (Fig. 4b) [35]. Somatic mutations in a variety of cancers have also been found to occur disproportionately at conserved positions [39, 40]. A similar pattern has emerged for mitochondrial disease-associated nSNVs [41].
The relationship between evolutionary conservation and disease association has been explained by the effect of natural selection [29, 34–37]. There is a high degree of purifying selection on variation at highly conserved positions because of their potential effect on inclusive fitness (fecundity, reproductive success) due to the functional importance of the position [29, 34, 35, 37, 38]. At the faster-evolving positions, many substitutions have been tolerated over evolutionary time in different species. This points to the “neutrality” of some mutations that spread through the population primarily by the process of random genetic drift and appear as fixed differences between species. Therefore, fewer mutations are culled at fast-evolving positions, producing a relative under-abundance of disease mutations at such positions. Of course, the above arguments hold true only when the functional importance of a position has remained unchanged over evolutionary time, an assumption that is expected to be fulfilled for a large fraction of positions in orthologous proteins.
Multigenic (complex) diseases
Despite successes in identifying and mapping genes causing Mendelian diseases, it is now clear that most common diseases with significant genetic components, although they are often seen to cluster in families, do not approximate the simple paradigm of high penetrance based on a dominant/recessive genotype. Instead, common diseases seem to result from a more complex pattern where many genes, and probably other non-genetic factors, contribute in non-additive ways, and individual monogenic factors have a low and inconsistent correlation with the disease phenotype [42–44]. Examples of such diseases include heart disease, asthma, rheumatoid arthritis, and type 2 diabetes [45–49]. These diseases often appear relatively later in life, and the associated SNVs are often present in one or more human populations at substantial frequencies.
An early examination of the evolutionary patterns of the occurrence of a small set of 37 nSNVs associated with complex diseases did not find any tendency for these variations to occur at sites with high conservation (Fig. 4c) [37]. These trends were confirmed with larger datasets containing alleles associated with seven complex diseases [50]. These patterns stand in stark contrast to those seen for Mendelian diseases. At the level of overall rate of protein evolution, genes associated with complex diseases are not under strong purifying selection as compared to proteins implicated in the Mendelian diseases [51]. The rate of nonsynonymous substitutions in complex-disease genes is more than twice that of the Mendelian disease genes [52]. One reason for the lack of evolutionary conservation of positions associated with complex diseases is that their effects appear later in life, which means that these variants are frequently inherited without being acted upon by natural selection and without any impact on fecundity. For this reason, molecular evolutionary analyses are sometimes not deemed to be useful for complex diseases [53].
Evolutionary and biochemical constraints an on disease associated nSNVs
In addition to the evolutionary conservation of the positions in the protein, the biochemical properties of the amino acid change can also provide rich information. Not all changes at a position have an equal effect, because one set of amino acid alternatives could be optimal, another set tolerable, and a third crippling to protein structure and function. Although the actual effect of a mutation is expected to be a complex function of the protein structure and its cellular milieu, many biologists used a simple measure of biochemical difference (Grantham distance [54]) to quantify the severity of amino acid changes. In an analysis of seven genes, it was noted early on that amino acid changes of Mendelian disease associated nSNVs were, on average, 67% more severe than those observed among species in the same proteins [29]. The generality of this trend was confirmed in subsequent analyses of a larger number of Mendelian disease genes [34, 35]. Interestingly, the timing of the onset of a disease also shows correlation with the biochemical severity of an amino acid change: late-onset diseases involve amino acids with smaller biochemical differences [35]. Similarly, the severity of the phenotype also shows a relationship with the biochemical dissimilarity of the variation [e.g., 55]. In addition, the severity of Mendelian nSNVs has been quantified by using the substitution probability of one amino acid into another. These analyses show that disease-associated nSNVs are amino acid changes that are unlike those observed among species proteome-wide [e.g., 29, 34].
A large number of Mendelian disease-associated variations occur at positions that show evolutionary substitutions between species. For example, more than a hundred variants of CFTR protein in CF patients occur at positions that have undergone at least one change (Fig. 3a). In any position, evolutionary differences (substitutions) among species are expected to be neutral in nature, in other words, they are unlikely to have negative fitness effects as long as the protein function has not changed. They constitute a set of evolutionarily permissible alleles (EPAs) at a given position, which are expected to not be involved in diseases at those positions. Indeed, an overwhelming fraction of Mendelian nSNVs (~90%) are not evolutionarily permissible [35, 55, 56]. This is in sharp contrast to population polymorphisms that frequently (59%) appear in the set of EPAs in individual positions [57]. Disease-associated nSNVs in mitochondrial encoded proteins also show similar patterns [58].
Nevertheless, scientists have been interested in investigating why some nSNVs are associated with diseases in humans, but appear as natural alleles in other species [35, 56, 58, 59]. One possibility is that the function of the affected amino acid position has changed either in humans or in other species. In this case, evolutionary differences among species cannot be used to determine permissible amino acids at the affected positions. Another reason for the overlap between the disease nSNVs and evolutionarily permissible alleles is that the amino acid position has undergone compensatory changes. In this case, the negative effects of the mutation(s) at one position of the same or different proteins compensates for the negative effects of the other mutation [35, 56, 59–61]. Such compensation could occur, for example, due to antagonistic pleiotropy [62, 63] or due to protein functional reasons [e.g., 64, 65]. Whatever the reason, the initial mutation needs to escape natural selection for a period of time before it is compensated by another mutation in the same or another protein. This is likely to be possible only for mutations that have very small negative fitness effects, resulting in such mutations occurring at faster evolving positions that are biochemically less radical biochemically [e.g., 35].
Evolutionary diagnosis of function-altering mutations in silico
Over a decade ago, first methods were proposed to predict computationally whether a mutation will negatively affect the structure and function of a human protein [30, 66–68]. These methods, now part of the PolyPhen software package, employed physical properties of the mutational change along with a multispecies alignment as a basis to evaluate mutations. This method showed promise: 69% of mutations associated with human disorders could be correctly diagnosed to be damaging to protein function (true positives) and 66% of known population polymorphisms diagnosed correctly to be non-damaging (true negatives) [67]. Most recently, a true positive rate of 92% was achieved by PolyPhen-2 when only damaging alleles with known effects on the molecular function causing Mendelian diseases were tested [63], which reduced to 73% when all human disease-associated mutations were analyzed. The false positive rate was close to ~20% for PolyPhen-2.
Another early method [sorting intolerant from tolerant (SIFT)] employed multispecies alignments to distinguish between functionally neutral and deleterious amino acid changes [69]. Applications of SIFT and PolyPhen/PolyPhen-2 to predict well-characterized variants in selected sets of genes revealed similar true positive rates for the two programs [70, 71], but these investigations revealed much higher false positive rates (up to 68%). Comparative analyses have also revealed that the prediction accuracy of in silico tools depends on both the algorithm and sequence alignment employed [71–73], with predictions from the PolyPhen-2 showing the least dependence on the alignment employed.
Over the years, these in silico prediction tools have frequently been employed to predict the proportion of benign mutations in newly sequenced human genomes and to prioritize polymorphisms for further experimental research in humans and other species [74–81]. In all of these investigations, the focus has been on diagnosing monogenic disease mutations, because in silico tools based on evolutionary considerations are not expected to be effective for identifying nSNVs associated with complex diseases. The patterns of evolutionarily conservation of known complex disease nSNVs are no different from those of natural polymorphisms found among populations (Fig. 4c).
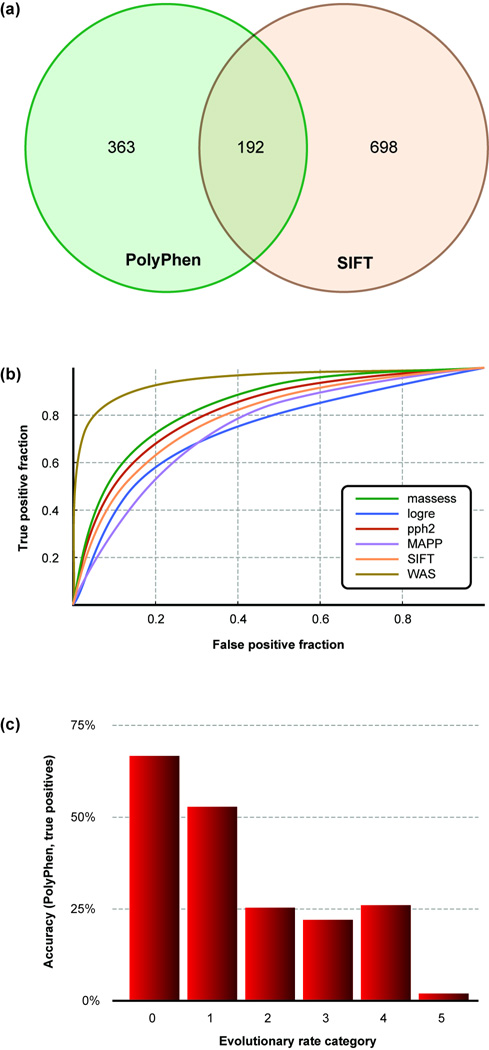
Even for Mendelian disease mutations, in silico diagnosis has been challenging because diagnoses from different programs are not the same for the same variant. For example, PolyPhen and SIFT diagnoses for protein-altering mutations in the Venter genome disagreed more often than they agreed [2] (Fig. 5a). Because of such problems, efforts have gone into the development of composite and ensemble methods that: (i) incorporate increasingly larger numbers of clinical and biological attributes in the decision-making process, and (ii) combine the results from existing tools by using logistic regression, Bayesian neutral networks, decision trees, support vector machines, random forests, and multiple selection rule voting [82–85]. These efforts are beginning to improve prediction accuracy significantly, and one recent method combining many less successful methods into a new composite approach was found to outperform each method used separately (Figure 5b) [85].
Figure 5.
Some applications of evolutionary in silico tools in diagnosing pathogenic variants. (a) The comparison of PolyPhen [100] and SIFT [69] predictions for 7,534 high-quality variants present within the Venter genome [2]. The numbers of variants diagnosed as probably damaging (PolyPhen) and intolerant (SIFT) are shown. The in silico diagnosis of personal variants by different tools produces highly discordant results. (b) ROC (receiver operating characteristic [101]) curves produced by PolyPhen-2 (pph2), SIFT, MAPP, Mutation Assessor (masses), Log R Pfam E-value (logre), and Condel (WAS). Condel used a weighted average of the normalized scores of the other five methods and outperformed each of them [85]. The ROC curve for Condel rises much more quickly, which means that it has a much greater rate of diagnosing damaging variants (true positives) at the expense of much smaller rate of incorrect diagnosis (false positives). (c) The relationship of the accuracy of the PolyPhen prediction for disease-associated nSNVs at positions evolving with different long-term rates (0–5 are categories of slowest to fastest-evolving sites) [57]. This shows that the accuracy of the PolyPhen prediction is the highest for the most slow-evolving positions for disease-associated nSNVs. Panels (a–c) are redrawn with permission from [2], [57], and [85], respectively.
Many evolutionary features used by classical and advanced versions of SIFT and PolyPhen (among others) for diagnosing Mendelian disease variants are also discriminatory for differentiating between driver and passenger mutations [39, 86]. This prompted the development of a hybrid method, CanPredict [86], that integrated gene function information (e.g., gene ontology) to screen somatic mutations (see also [87]). This tool diagnoses mutations found in samples of more than ten patients to be damaging 50% more often than mutations that were seen in only one patient [86]. Driver mutations contribute to cancer progression and have a tendency to be found in many independent samples as compared to passenger mutations that, as the name suggests, hitchhike causing the cells with driver mutations to increase in number by the processes of natural selection and adaptation [39, 40, 88–90]. For mitochondrial DNA (mtDNA), four different tools (including PolyPhen and SIFT) have been combined along with the biochemical features and frequency of variants to evaluate mitochondrial nSNVs [91]. This approach was adopted because only 5% of disease-associated nSNVs in mtDNA were found to be harmful by all four in silico methods, even though each of these SNVs was predicted to be damaging by at least one method [91].
Efforts have been made to identify a priori determinants of the protein position where in silico tools will most probably succeed [57]. This knowledge will empower biologists to quantify the reliability of inference and use the in silico predictions only when they are expected to be reliable. Initial research has revealed a clear-cut relationship between the sensitivity (true positive diagnosis) and specificity (true negative diagnosis) of predictions with the rate at which the given position has evolved over species as diverse as fish and lamprey. The disease-associated nSNVs at slow-evolving positions were more likely to be diagnosed correctly as compared to those at fast-evolving positions (Fig. 5b). This is consistent with earlier findings that the evolutionary rate is overwhelmingly the most important determinant of the accuracy of in silico prediction methods [92, 93]. It is also clear that the accuracy of in silico tools is severely degraded when the observed disease associated variant is found in other species at the same position [57]. Therefore, the in silico diagnosis failures are systematic and probably predictable.
By using evolutionary rates derived from multispecies analyses a priori, it should be possible to develop adaptive classifiers that have a potential to generate more reliable predictions based on the evolutionary context of specific positions. Because high-quality genomic alignments between human and many closely and distantly related species are publicly available, it is possible to enumerate each multi-species aligned position in the human genome to compute position-specific features, such as evolutionary rate of change. These pre-computed evolutionary features could be incorporated into prediction methods to adaptively adjust the classifier thresholds to optimize for the type of nSNVs that are likely to be observed. For example, fast-evolving positions are expected to harbor a higher proportion of neutral nSNVs, so thresholds could therefore be fine-tuned to improve overall accuracy.
Concluding remarks
The cosmic analogy used in the title of this review is intended to convey the enormity of the challenge that researchers in genomic medicine face, as they attempt to decipher functional consequences of the constellation of genomic changes carried in each personal genome. In tackling this challenge, the evolutionary telescope is among a set of initial tools to generate functional predictions. Clearly, the progress made to date prompts enthusiasm, but there is an urgent need to develop better in silico approaches to aid and complement an array of experimental, clinical, and physical tools that must be combined to assay accurately the diversity of the functional effects of the variants present in the human population and of the de novo mutations that continually arise in the natural processes of cell division and population propagation.
Many limits to the use of the evolutionary approaches in genomic medicine are already evident. As mentioned earlier, in silico analysis of nSNVs underlying complex diseases remains a major challenge. Furthermore there are few cases when disease categorization can be seen as a black and white decision: diseases represent a continuum from predominately monogenic to highly polygenic [94]. Some classical monogenic diseases will surely be caused by mutations in multiple genes, whereas some classic polygenic diseases will have a few major effect alleles. This complicates the choice of when to apply evolutionary knowledge in diagnosing the function-altering potential of variants. The distinction between the neutrality and non-neutrality of function alteration is also not straightforward, because it depends on both environmental and genomic contexts (e.g., compensatory mutations) and could well involve fitness trade-offs (e.g., between rapid maturation and risk of disease). Moreover, the extent to which personal variations manifest themselves as health concerns in individuals remains unknown. With an enhanced quantification health and disease, and an improved understanding of genome and disease biology, we will have a better idea of the powers and pitfalls of evolutionary analysis in genomic medicine. At the same time, there is a need to profile exome variants experimentally and connect them with individual health via predictive frameworks. Some cell-based and in vitro assays are already showing promise in deciphering the pathogenic roles of variants in cancers [23, 95], an important step forward towards satisfying the urgent need for the development of higher throughput biological and functional approaches.
Nonetheless, the rapid emergence of clinical genome sequencing has established a pressing need to incorporate evolutionary information into clinical diagnostics. An individual genome contains hundreds of thousands of variants of different antiquities present in an individual genome, and the long-term evolutionary history of genomic positions provides an immediate means to derive and apply predictive and quantitative assessment of the potential functional effect of any given variant observed. Using the evolutionary anatomies of positions, clinicians can be provided ready access to evolutionary-guided in silico diagnostic tools to identify and diagnose observed variants that are most likely to have consequences for the health or clinical course of treatment for a patient.
Textbox 1 Figure I.
Disease-associated genetic variants identified in patients with Miller syndrome.
Acknowledgements
We thank Vanessa Gray and Alicia Varma for literature search, Maxwell Sanderford for mapping 23andMe information to the dbSNP database, and Carol Williams for edits. This work is supported by research grants from National Institutes of Health to S.K.
Glossary
- Complex disease
Refers to any disease having some genetic component of etiology that is characterized as involving the effects of many genes. Complex diseases are typically common in the population, exhibit complex patterns of inheritance, and often involve the interaction of genetic and environmental factors.
- Driver mutation
Somatic mutations implicated as having a causal role in the pathogenesis of cancer.
- Evolutionary retention
A position-specific measure of conservation taking into account the number of times a human amino acid position is missing a homolog in the multiple sequence alignment with other species.
- Exome
The complete collection of (known) exons that ultimately constitute proteins expressed by an individual.
- Genetic drift
The change in the population frequency of alleles due to random sampling of neutral or effectively neutral alleles.
- Mendelian disease
A genetic disease trait exhibiting a Mendelian inheritance pattern for an underlying mutation at a single genetic locus.
- Passenger mutation
Somatic mutations observed in cancer genomes that have not contributed to the cancer’s pathogenesis. Can be seen in high frequencies in tumors if they occur in the same lineage as driver mutations that contribute to the clonal expansion of the cancer cell lineage.
- Purifying selection
A type of directional evolutionary selection that acts to remove deleterious alleles from a population.
- Somatic mutation
A change in the genetic structure that is neither inherited nor passed to offspring.
References
- 1.Li Y, Agarwal P. A Pathway-Based View of Human Diseases and Disease Relationships. PLoS ONE. 2009;4:e4346. doi: 10.1371/journal.pone.0004346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Chun S, Fay JC. Identification of deleterious mutations within three human genomes. Genome Res. 2009;19:1553–1561. doi: 10.1101/gr.092619.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hindorff LA, et al. A Catalog of Published Genome-Wide Association Studies. 2011 www.genome.gov/gwastudies. [Google Scholar]
- 4.Hindorff LA, et al. Potential etiologic and functional implications of genome-wide association loci for human diseases and traits. Proc Natl Acad Sci U S A. 2009;106:9362–9367. doi: 10.1073/pnas.0903103106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.McCarthy MI, et al. Genome-wide association studies for complex traits: consensus, uncertainty and challenges. Nat Rev Genet. 2008;9:356–369. doi: 10.1038/nrg2344. [DOI] [PubMed] [Google Scholar]
- 6.Roberts R, et al. The Genome-Wide Association Study—A New Era for Common Polygenic Disorders. Journal of Cardiovascular Translational Research. 2010;3:173–182. doi: 10.1007/s12265-010-9178-6. [DOI] [PubMed] [Google Scholar]
- 7.Levy S, et al. The diploid genome sequence of an individual human. PLoS Biol. 2007;5:e254. doi: 10.1371/journal.pbio.0050254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bentley DR, et al. Accurate whole human genome sequencing using reversible terminator chemistry. Nature. 2008;456:53–59. doi: 10.1038/nature07517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wang J, et al. The diploid genome sequence of an Asian individual. Nature. 2008;456:60–65. doi: 10.1038/nature07484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wheeler DA, et al. The complete genome of an individual by massively parallel DNA sequencing. Nature. 2008;452:872–876. doi: 10.1038/nature06884. [DOI] [PubMed] [Google Scholar]
- 11.Ng SB, et al. Targeted capture and massively parallel sequencing of 12 human exomes. Nature. 2009;461:272–276. doi: 10.1038/nature08250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Stenson P, et al. The Human Gene Mutation Database: 2008 update. Genome Medicine. 2009;1:1–6. doi: 10.1186/gm13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lander ES, et al. Initial sequencing and analysis of the human genome. Nature. 2001;409:860–921. doi: 10.1038/35057062. [DOI] [PubMed] [Google Scholar]
- 14.Ng SB, et al. Massively parallel sequencing and rare disease. Human Molecular Genetics. 2010;19:R119–R124. doi: 10.1093/hmg/ddq390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ku C-S, et al. Revisiting Mendelian disorders through exome sequencing. Human Genetics. 2011:1–20. doi: 10.1007/s00439-011-0964-2. [DOI] [PubMed] [Google Scholar]
- 16.Choi M, et al. Genetic diagnosis by whole exome capture and massively parallel DNA sequencing. Proceedings of the National Academy of Sciences. 2009;106:19096–19101. doi: 10.1073/pnas.0910672106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Teer JK, Mullikin JC. Exome sequencing: the sweet spot before whole genomes. Human Molecular Genetics. 2010;19:R145–R151. doi: 10.1093/hmg/ddq333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Green ED, Guyer MS. Charting a course for genomic medicine from base pairs to bedside. Nature. 2011;470:204–213. doi: 10.1038/nature09764. [DOI] [PubMed] [Google Scholar]
- 19.Carapito R, et al. Automated high-throughput process for site-directed mutagenesis, production, purification, and kinetic characterization of enzymes. Anal Biochem. 2006;355:110–116. doi: 10.1016/j.ab.2006.04.047. [DOI] [PubMed] [Google Scholar]
- 20.Saboulard D, et al. High-throughput site-directed mutagenesis using oligonucleotides synthesized on DNA chips. Biotechniques. 2005;39:363–368. doi: 10.2144/05393ST04. [DOI] [PubMed] [Google Scholar]
- 21.Sylvestre J. Massive Mutagenesis: high-throughput combinatorial site-directed mutagenesis. Methods Mol Biol. 2010;634:233–238. doi: 10.1007/978-1-60761-652-8_17. [DOI] [PubMed] [Google Scholar]
- 22.van Boxtel R, et al. Systematic generation of in vivo G protein-coupled receptor mutants in the rat. Pharmacogenomics J. 2010 doi: 10.1038/tpj.2010.44. (in press) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Davis EE, et al. TTC21B contributes both causal and modifying alleles across the ciliopathy spectrum. Nat Genet. 2011;43:189–196. doi: 10.1038/ng.756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Williams GC, Nesse RM. The Dawn of Darwinian Medicine. The Quarterly Review of Biology. 1991;66:1–22. doi: 10.1086/417048. [DOI] [PubMed] [Google Scholar]
- 25.Gluckman PD, et al. Principles of evolutionary medicine. Oxford University Press; 2009. [Google Scholar]
- 26.Nesse RM, Williams GC. Why we get sick : the new science of Darwinian medicine. Times Books; 1994. [Google Scholar]
- 27.Harper RMJ. Evolutionary origins of disease. G. Mosdell; 1975. [Google Scholar]
- 28.Stearns SC, Koella JC. Evolution in health and disease. Oxford University Press; 2008. [Google Scholar]
- 29.Miller MP, Kumar S. Understanding human disease mutations through the use of interspecific genetic variation. Hum Mol Genet. 2001;10:2319–2328. doi: 10.1093/hmg/10.21.2319. [DOI] [PubMed] [Google Scholar]
- 30.Sunyaev S, et al. Prediction of deleterious human alleles. Hum Mol Genet. 2001;10:591–597. doi: 10.1093/hmg/10.6.591. [DOI] [PubMed] [Google Scholar]
- 31.Bonetta L. Whole-Genome Sequencing Breaks the Cost Barrier. Cell. 2010;141:917–919. doi: 10.1016/j.cell.2010.05.034. [DOI] [PubMed] [Google Scholar]
- 32.Coffey AJ, et al. The GENCODE exome: sequencing the complete human exome. Eur J Hum Genet. 2011 doi: 10.1038/ejhg.2011.28. (in press) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Harper PS. A short history of medical genetics. Oxford University Press; 2008. [Google Scholar]
- 34.Vitkup D, et al. The amino-acid mutational spectrum of human genetic disease. Genome Biology. 2003;4:R72. doi: 10.1186/gb-2003-4-11-r72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Subramanian S, Kumar S. Evolutionary anatomies of positions and types of disease-associated and neutral amino acid mutations in the human genome. BMC Genomics. 2006;7:306. doi: 10.1186/1471-2164-7-306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kulkarni V, et al. Exhaustive prediction of disease susceptibility to coding base changes in the human genome. BMC Bioinformatics. 2008;9:S3. doi: 10.1186/1471-2105-9-S9-S3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Thomas PD, Kejariwal A. Coding single-nucleotide polymorphisms associated with complex vs. Mendelian disease: evolutionary evidence for differences in molecular effects. Proc Natl Acad Sci U S A. 2004;101:15398–15403. doi: 10.1073/pnas.0404380101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Zhu Q, et al. A genome-wide comparison of the functional properties of rare and common genetic variants in humans. Am J Hum Genet. 2011;88:458–468. doi: 10.1016/j.ajhg.2011.03.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kaminker JS, et al. Distinguishing cancer-associated missense mutations from common polymorphisms. Cancer Res. 2007;67:465–473. doi: 10.1158/0008-5472.CAN-06-1736. [DOI] [PubMed] [Google Scholar]
- 40.Forbes S, et al. COSMIC 2005. Br J Cancer. 2006;94:318–322. doi: 10.1038/sj.bjc.6602928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Montoya J, et al. 20 years of human mtDNA pathologic point mutations: Carefully reading the pathogenicity criteria. Biochimica et Biophysica Acta (BBA) - Bioenergetics. 2009;1787:476–483. doi: 10.1016/j.bbabio.2008.09.003. [DOI] [PubMed] [Google Scholar]
- 42.Lander E, Schork N. Genetic dissection of complex traits. Science. 1994;265:2037–2048. doi: 10.1126/science.8091226. [DOI] [PubMed] [Google Scholar]
- 43.Thomson G, Esposito MS. The genetics of complex diseases. Trends in Genetics. 1999;15:M17–M20. [PubMed] [Google Scholar]
- 44.Botstein D, Risch N. Discovering genotypes underlying human phenotypes: past successes for mendelian disease, future approaches for complex disease. Naure Genetics. 2003:33. doi: 10.1038/ng1090. [DOI] [PubMed] [Google Scholar]
- 45.Fujimura JH. Crafting science : a sociohistory of the quest for the genetics of cancer. Harvard University Press; 1996. [Google Scholar]
- 46.Marenberg ME, et al. Genetic Susceptibility to Death from Coronary Heart Disease in a Study of Twins. New England Journal of Medicine. 1994;330:1041–1046. doi: 10.1056/NEJM199404143301503. [DOI] [PubMed] [Google Scholar]
- 47.Sarafino EP, Goldfedder J. Genetic factors in the presence, severity, and triggers of asthma. Archives of Disease in Childhood. 1995;73:112–116. doi: 10.1136/adc.73.2.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.MacGregor AJ, et al. Characterizing the quantitative genetic contribution to rheumatoid arthritis using data from twins. Arthritis & Rheumatism. 2000;43:30–37. doi: 10.1002/1529-0131(200001)43:1<30::AID-ANR5>3.0.CO;2-B. [DOI] [PubMed] [Google Scholar]
- 49.O'Rahilly S, et al. Genetic Factors in Type 2 Diabetes: The End of the Beginning? Science. 2005;307:370–373. doi: 10.1126/science.1104346. [DOI] [PubMed] [Google Scholar]
- 50.Corona E, et al. Extreme Evolutionary Disparities Seen in Positive Selection across Seven Complex Diseases. PLoS ONE. 2010;5:e12236. doi: 10.1371/journal.pone.0012236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Blekhman R, et al. Natural selection on genes that underlie human disease susceptibility. Curr Biol. 2008;18:883–889. doi: 10.1016/j.cub.2008.04.074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Podder S, Ghosh TC. Exploring the Differences in Evolutionary Rates between Monogenic and Polygenic Disease Genes in Human. Molecular Biology and Evolution. 2010;27:934–941. doi: 10.1093/molbev/msp297. [DOI] [PubMed] [Google Scholar]
- 53.Nathan DG, Orkin SH. Musings on genome medicine: genome wide association studies. Genome Med. 2009;1:3. doi: 10.1186/gm3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Grantham R. Amino Acid Difference Formula to Help Explain Protein Evolution. Science. 1974;185:862–864. doi: 10.1126/science.185.4154.862. [DOI] [PubMed] [Google Scholar]
- 55.Briscoe AD, et al. The spectrum of human rhodopsin disease mutations through the lens of interspecific variation. Gene. 2004;332:107–118. doi: 10.1016/j.gene.2004.02.037. [DOI] [PubMed] [Google Scholar]
- 56.Kondrashov AS, et al. Dobzhansky-Muller incompatibilities in protein evolution. Proc Natl Acad Sci U S A. 2002;99:14878–14883. doi: 10.1073/pnas.232565499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Kumar S, et al. Positional conservation and amino acids shape the correct diagnosis and population frequencies of benign and damaging personal amino acid mutations. Genome Res. 2009;19:1562–1569. doi: 10.1101/gr.091991.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Magalhães J. Human Disease-Associated Mitochondrial Mutations Fixed in Nonhuman Primates. Journal of Molecular Evolution. 2005;61:491–497. doi: 10.1007/s00239-004-0258-6. [DOI] [PubMed] [Google Scholar]
- 59.Gao L, Zhang J. Why are some human disease-associated mutations fixed in mice? Trends Genet. 2003;19:678–681. doi: 10.1016/j.tig.2003.10.002. [DOI] [PubMed] [Google Scholar]
- 60.Kulathinal RJ, et al. Compensated deleterious mutations in insect genomes. Science. 2004;306:1553–1554. doi: 10.1126/science.1100522. [DOI] [PubMed] [Google Scholar]
- 61.Liao BY, Zhang J. Mouse duplicate genes are as essential as singletons. Trends Genet. 2007;23:378–381. doi: 10.1016/j.tig.2007.05.006. [DOI] [PubMed] [Google Scholar]
- 62.Williams GC. Pleiotropy, natural selection, and the evolution of senescence. Evolution. 1957;11:398–411. [Google Scholar]
- 63.He X, Zhang J. Toward a molecular understanding of pleiotropy. Genetics. 2006;173:1885–1891. doi: 10.1534/genetics.106.060269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Lee BC, et al. Analysis of the residue-residue coevolution network and the functionally important residues in proteins. Proteins. 2008;72:863–872. doi: 10.1002/prot.21972. [DOI] [PubMed] [Google Scholar]
- 65.Ferrer-Costa C, et al. Characterization of compensated mutations in terms of structural and physico-chemical properties. J Mol Biol. 2007;365:249–256. doi: 10.1016/j.jmb.2006.09.053. [DOI] [PubMed] [Google Scholar]
- 66.Sunyaev S, et al. Prediction of nonsynonymous single nucleotide polymorphisms in human disease-associated genes. J Mol Med. 1999;77:754–760. doi: 10.1007/s001099900059. [DOI] [PubMed] [Google Scholar]
- 67.Sunyaev S, et al. Towards a structural basis of human non-synonymous single nucleotide polymorphisms. Trends Genet. 2000;16:198–200. doi: 10.1016/s0168-9525(00)01988-0. [DOI] [PubMed] [Google Scholar]
- 68.Sunyaev SR, et al. PSIC: profile extraction from sequence alignments with position-specific counts of independent observations. Protein Eng. 1999;12:387–394. doi: 10.1093/protein/12.5.387. [DOI] [PubMed] [Google Scholar]
- 69.Ng PC, Henikoff S. SIFT: Predicting amino acid changes that affect protein function. Nucleic Acids Res. 2003;31:3812–3814. doi: 10.1093/nar/gkg509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Flanagan SE, et al. Using SIFT and PolyPhen to predict loss-of-function and gain-of-function mutations. Genet Test Mol Biomarkers. 2010;14:533–537. doi: 10.1089/gtmb.2010.0036. [DOI] [PubMed] [Google Scholar]
- 71.Hicks S, et al. Prediction of missense mutation functionality depends on both the algorithm and sequence alignment employed. Hum Mutat. 2011;32:661–668. doi: 10.1002/humu.21490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Reva B, et al. Determinants of protein function revealed by combinatorial entropy optimization. Genome Biol. 2007;8:R232. doi: 10.1186/gb-2007-8-11-r232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Mathe E, et al. Computational approaches for predicting the biological effect of p53 missense mutations: a comparison of three sequence analysis based methods. Nucleic Acids Res. 2006;34:1317–1325. doi: 10.1093/nar/gkj518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Arnett J, et al. Autosomal Dominant Progressive Sensorineural Hearing Loss Due to a Novel Mutation in the KCNQ4 Gene. Arch Otolaryngol Head Neck Surg. 2011;137:54–59. doi: 10.1001/archoto.2010.234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Çalışkan M, et al. Exome sequencing reveals a novel mutation for autosomal recessive non-syndromic mental retardation in the TECR gene on chromosome 19p13. Human Molecular Genetics. 2011;20:1285–1289. doi: 10.1093/hmg/ddq569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Hoefele J, et al. Novel PKD1 and PKD2 mutations in autosomal dominant polycystic kidney disease (ADPKD) Nephrology Dialysis Transplantation. 2010 doi: 10.1093/ndt/gfq720. [DOI] [PubMed] [Google Scholar]
- 77.Saccone SF, et al. SPOT: a web-based tool for using biological databases to prioritize SNPs after a genome-wide association study. Nucleic Acids Research. 2010;38:W201–W209. doi: 10.1093/nar/gkq513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.McGee TL, et al. Novel mutations in the long isoform of the USH2A gene in patients with Usher syndrome type II or non-syndromic retinitis pigmentosa. Journal of Medical Genetics. 2010;47:499–506. doi: 10.1136/jmg.2009.075143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Doherty D, et al. Mutations in 3 genes (MKS3, CC2D2A and RPGRIP1L) cause COACH syndrome (Joubert syndrome with congenital hepatic fibrosis) Journal of Medical Genetics. 2010;47:8–21. doi: 10.1136/jmg.2009.067249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Lee PH, Shatkay H. An integrative scoring system for ranking SNPs by their potential deleterious effects. Bioinformatics. 2009;25:1048–1055. doi: 10.1093/bioinformatics/btp103. [DOI] [PubMed] [Google Scholar]
- 81.Kantaputra PN, et al. Cleft Lip with Cleft Palate, Ankyloglossia, and Hypodontia are Associated with TBX22 Mutations. Journal of Dental Research. 2011;90:450–455. doi: 10.1177/0022034510391052. [DOI] [PubMed] [Google Scholar]
- 82.Huang T, et al. Prediction of Deleterious Non-Synonymous SNPs Based on Protein Interaction Network and Hybrid Properties. PLoS ONE. 2010;5:e11900. doi: 10.1371/journal.pone.0011900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Ng PC, Henikoff S. Predicting the effects of amino acid substitutions on protein function. Annu Rev Genomics Hum Genet. 2006;7:61–80. doi: 10.1146/annurev.genom.7.080505.115630. [DOI] [PubMed] [Google Scholar]
- 84.Mort M, et al. In silico functional profiling of human disease-associated and polymorphic amino acid substitutions. Human Mutation. 2010;31:335–346. doi: 10.1002/humu.21192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Gonzalez-Perez A, Lopez-Bigas N. Improving the assessment of the outcome of nonsynonymous SNVs with a consensus deleteriousness score, Condel. Am J Hum Genet. 2011;88:440–449. doi: 10.1016/j.ajhg.2011.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Kaminker JS, et al. CanPredict: a computational tool for predicting cancer-associated missense mutations. Nucleic Acids Res. 2007;35:W595–W598. doi: 10.1093/nar/gkm405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Carter H, et al. Cancer-Specific High-Throughput Annotation of Somatic Mutations: Computational Prediction of Driver Missense Mutations. Cancer Research. 2009;69:6660–6667. doi: 10.1158/0008-5472.CAN-09-1133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Greenman C, et al. Patterns of somatic mutation in human cancer genomes. Nature. 2007;446:153–158. doi: 10.1038/nature05610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Hon LS, et al. Computational Approaches for Predicting Causal Missense Mutations in Cancer Genome Projects. Current Bioinformatics. 2008;3:46–55. [Google Scholar]
- 90.Bignell GR, et al. Signatures of mutation and selection in the cancer genome. Nature. 2010;463:893–898. doi: 10.1038/nature08768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Bhardwaj A, et al. MtSNP score: a combined evidence approach for assessing cumulative impact of mitochondrial variations in disease. BMC Bioinformatics. 2009;10:S7. doi: 10.1186/1471-2105-10-S8-S7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Tian J, et al. Predicting the phenotypic effects of non-synonymous single nucleotide polymorphisms based on support vector machines. BMC Bioinformatics. 2007;8:450. doi: 10.1186/1471-2105-8-450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Jiang R, et al. Sequence-based prioritization of nonsynonymous single-nucleotide polymorphisms for the study of disease mutations. Am J Hum Genet. 2007;81:346–360. doi: 10.1086/519747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.McKusick VA. Mendelian Inheritance in Man and its online version, OMIM. Am J Hum Genet. 2007;80:588–604. doi: 10.1086/514346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Molatore S, et al. Characterization of a naturally-occurring p27 mutation predisposing to multiple endocrine tumors. Mol Cancer. 2010;9:116. doi: 10.1186/1476-4598-9-116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Chen R, et al. Non-synonymous and synonymous coding SNPs show similar likelihood and effect size of human disease association. PLoS ONE. 2010;5:e13574. doi: 10.1371/journal.pone.0013574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Pelak K, et al. The characterization of twenty sequenced human genomes. PLoS Genet. 2010:6. doi: 10.1371/journal.pgen.1001111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Forbes SA, et al. COSMIC: mining complete cancer genomes in the Catalogue of Somatic Mutations in Cancer. Nucleic Acids Res. 2011;39:D945–D950. doi: 10.1093/nar/gkq929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Altshuler DM, et al. Integrating common and rare genetic variation in diverse human populations. Nature. 2010;467:52–58. doi: 10.1038/nature09298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Ramensky V, et al. Human non-synonymous SNPs: server and survey. Nucleic Acids Res. 2002;30:3894–3900. doi: 10.1093/nar/gkf493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Lasko TA, et al. The use of receiver operating characteristic curves in biomedical informatics. J Biomed Inform. 2005;38:404–415. doi: 10.1016/j.jbi.2005.02.008. [DOI] [PubMed] [Google Scholar]
- 102.Ng SB, et al. Exome sequencing identifies the cause of a mendelian disorder. Nat Genet. 2010;42:30–35. doi: 10.1038/ng.499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Adzhubei IA, et al. A method and server for predicting damaging missense mutations. Nat Meth. 2010;7:248–249. doi: 10.1038/nmeth0410-248. [DOI] [PMC free article] [PubMed] [Google Scholar]