Abstract
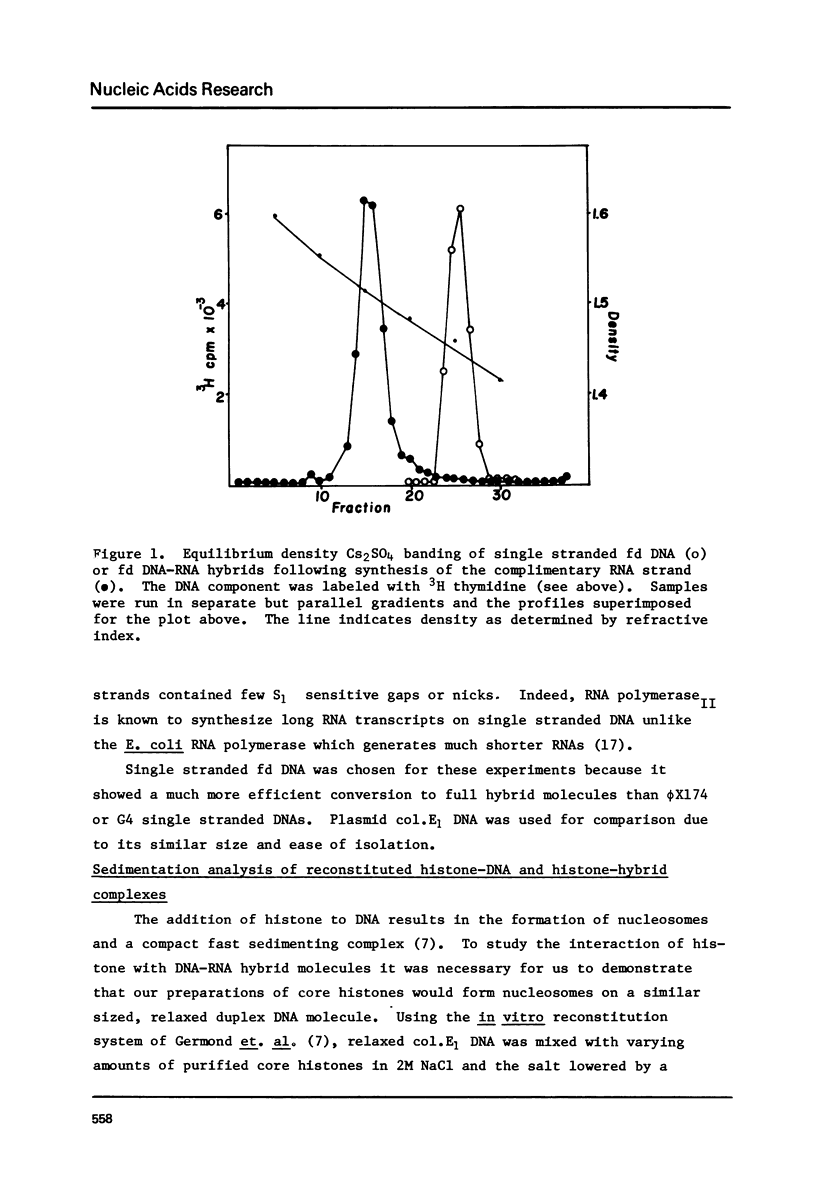
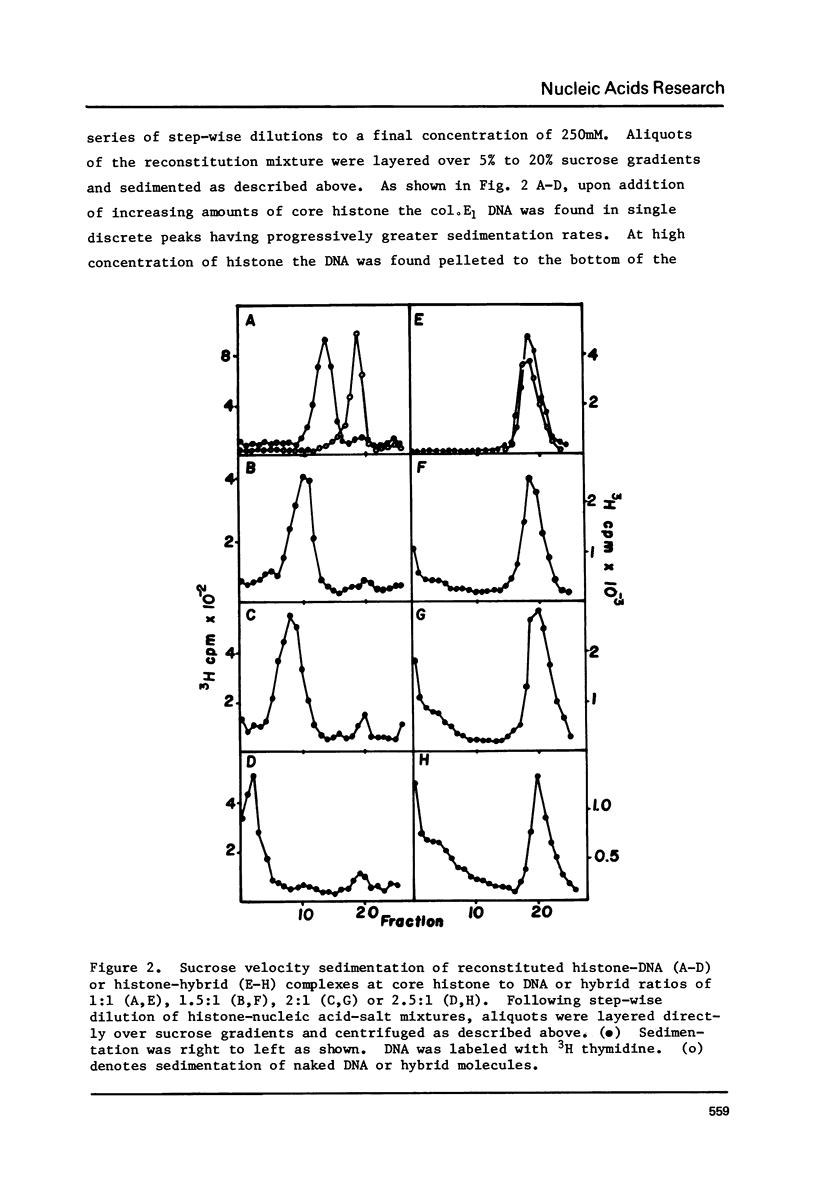
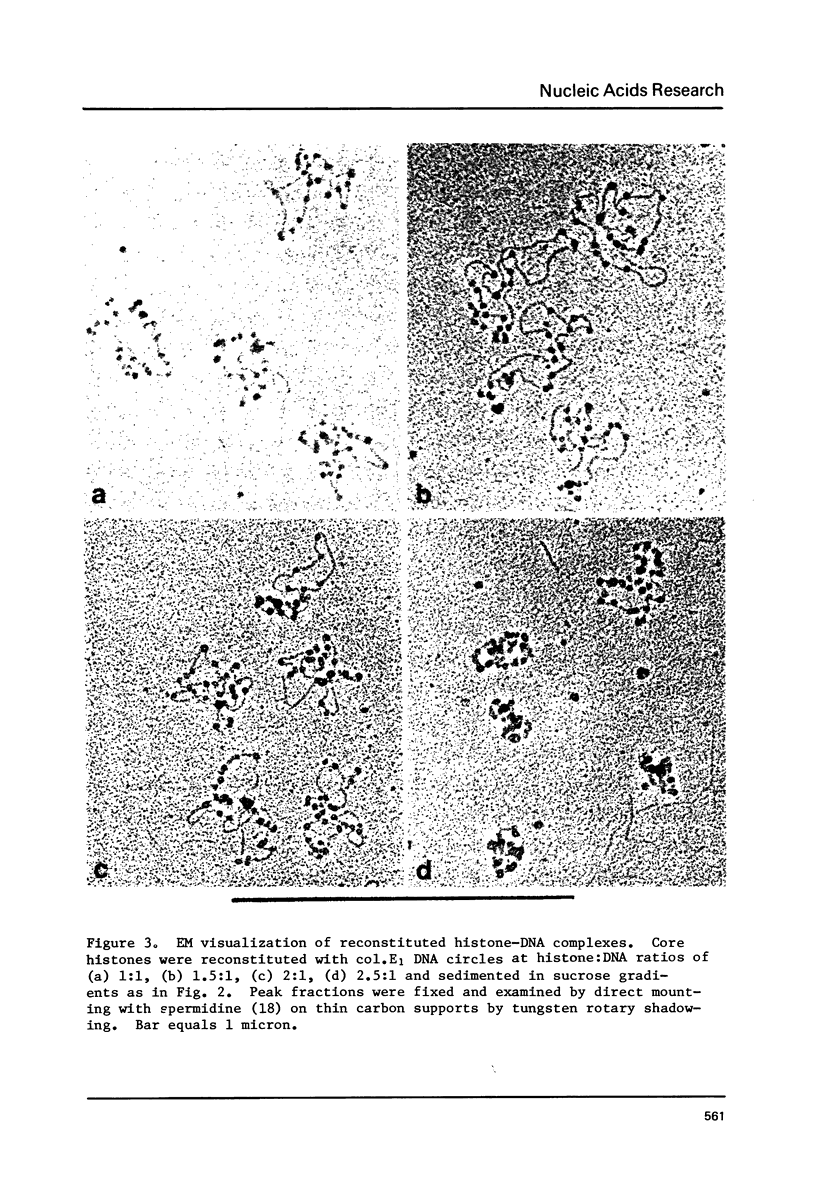
The binding of core histones (H2A, H2B, H3, H4) to a circular plasmid DNA and to a circular DNA-RNA hybrid molecule of similar size has been compared. Circular hybrid molecules were formed from single stranded fd DNA by synthesis of the complimentary strand with ribonucleotides using wheat germ RNA polymerase II. Upon reconstitution of plasmid DNA circles with histone, the sedimentation profiles of the DNA remained sharp by increased several fold in rate. Material from the peak fractions of these sedimentations appeared to be condensed circular loops of nucleosomes when examined by electron microscopy (EM), and the mass ratio of DNA to histone (at the histone concentrations which produced the fastest sedimentations) was typical of native chromatin. In contrast, the sedimentation behavior of DNA-RNA hybrid circles after addition of histone remained unchanged except for a minor fraction which exhibited a broad and faster sedimentation rate. Examination by EM revealed that most of the molecules appeared identical to protein free hybrid circles while the minor, faster sedimenting fraction appeared to be two or more circles bound together by protein aggregates. Finally, a linear molecule consisting of about 3000 base pairs of duplex DNA covalently joined on both ends to 1500 base pairs of RNA-DNA hybrid helix was constructed. Reconstitution of this molecule with core histone showed nucleosome formation only on the central DNA duplex region. Isopycnic banding of fixed hybrid-histone mixtures showed that little or no histone had bound to the bulk of the full hybrid molecules. We suggest that the presence of RNA in a nucleic acid duplex inhibits the condensation of the duplex into a nucleosomal structure by histone.
Full text
PDF



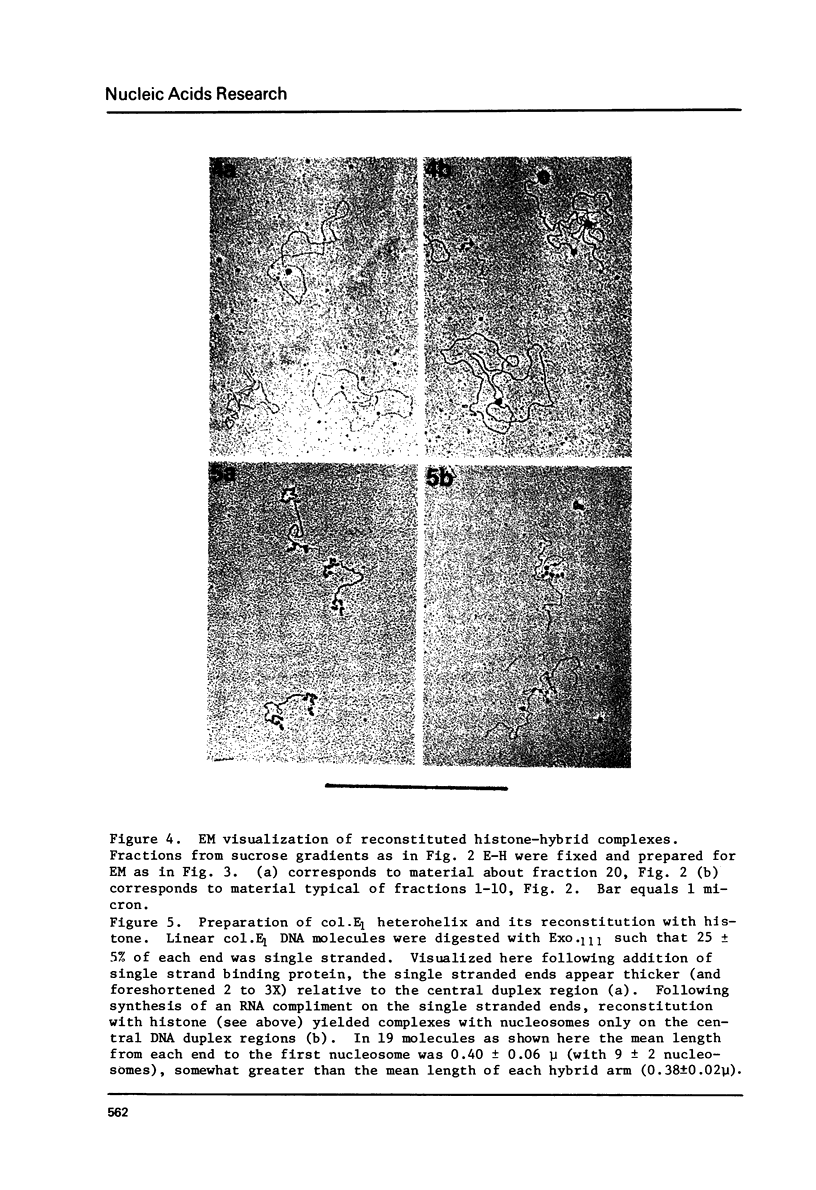

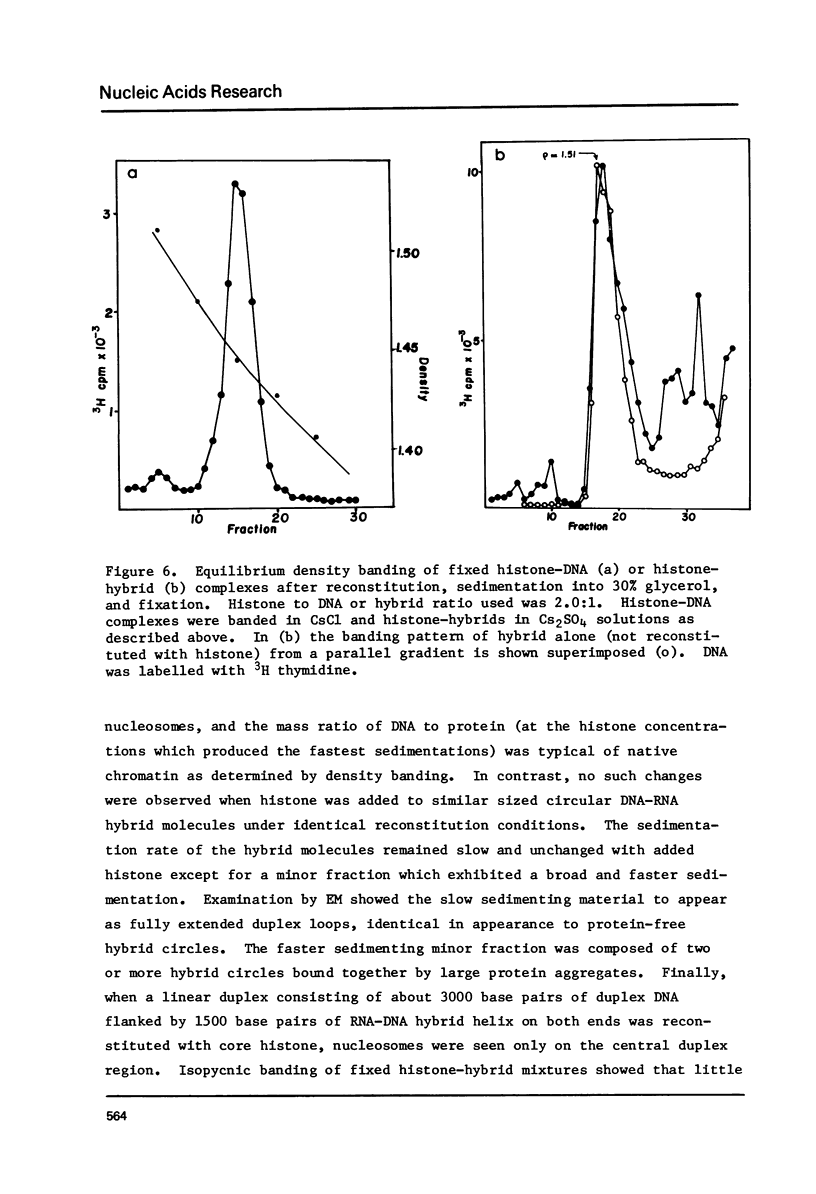


Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- CHAMBERLIN M., BERG P. MECHANISM OF RNA POLYMERASE ACTION: FORMATION OF DNA-RNA HYBRIDS WITH SINGLE-STRANDED TEMPLATES. J Mol Biol. 1964 Feb;8:297–313. doi: 10.1016/s0022-2836(64)80139-x. [DOI] [PubMed] [Google Scholar]
- Carter C. W., Jr Histone packing in the nucleosome core particle of chromatin. Proc Natl Acad Sci U S A. 1978 Aug;75(8):3649–3653. doi: 10.1073/pnas.75.8.3649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christiansen G., Griffith J. Salt and divalent cations affect the flexible nature of the natural beaded chromatin structure. Nucleic Acids Res. 1977 Jun;4(6):1837–1851. doi: 10.1093/nar/4.6.1837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- FULLER W., WILKINS M. H., WILSON H. R., HAMILTON L. D. THE MOLECULAR CONFIGURATION OF DEOXYRIBONUCLEIC ACID. IV. X-RAY DIFFRACTION STUDY OF THE A FORM. J Mol Biol. 1965 May;12:60–76. doi: 10.1016/s0022-2836(65)80282-0. [DOI] [PubMed] [Google Scholar]
- Finch J. T., Lutter L. C., Rhodes D., Brown R. S., Rushton B., Levitt M., Klug A. Structure of nucleosome core particles of chromatin. Nature. 1977 Sep 1;269(5623):29–36. doi: 10.1038/269029a0. [DOI] [PubMed] [Google Scholar]
- Germond J. E., Bellard M., Oudet P., Chambon P. Stability of nucleosomes in native and reconstituted chromatins. Nucleic Acids Res. 1976 Nov;3(11):3173–3192. doi: 10.1093/nar/3.11.3173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greenfield L., Simpson L., Kaplan D. Conversion of closed circular DNA molecules to single-nicked molecules by digestion with DNAase I in the presence of ethidium bromide. Biochim Biophys Acta. 1975 Oct 15;407(3):365–375. doi: 10.1016/0005-2787(75)90104-5. [DOI] [PubMed] [Google Scholar]
- Griffith J. D., Christiansen G. Electron microscope visualization of chromatin and other DNA-protein complexes. Annu Rev Biophys Bioeng. 1978;7:19–35. doi: 10.1146/annurev.bb.07.060178.000315. [DOI] [PubMed] [Google Scholar]
- Griffith J. D. Chromatin structure: deduced from a minichromosome. Science. 1975 Mar 28;187(4182):1202–1203. doi: 10.1126/science.187.4182.1202. [DOI] [PubMed] [Google Scholar]
- Griffith J. D. DNA structure: evidence from electron microscopy. Science. 1978 Aug 11;201(4355):525–527. doi: 10.1126/science.663672. [DOI] [PubMed] [Google Scholar]
- Hancock R. Assembly of new nucleosomal histones and new DNA into chromatin. Proc Natl Acad Sci U S A. 1978 May;75(5):2130–2134. doi: 10.1073/pnas.75.5.2130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laemmli U. K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970 Aug 15;227(5259):680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- Laskey R. A., Mills A. D., Morris N. R. Assembly of SV40 chromatin in a cell-free system from Xenopus eggs. Cell. 1977 Feb;10(2):237–243. doi: 10.1016/0092-8674(77)90217-3. [DOI] [PubMed] [Google Scholar]
- Makino S., Woolford J. L., Jr, Tanford C., Webster R. E. Interaction of deoxycholate and of detergents with the coat protein of bacteriophage f1. J Biol Chem. 1975 Jun 10;250(11):4327–4332. [PubMed] [Google Scholar]
- Modrich P., Zabel D. EcoRI endonuclease. Physical and catalytic properties of the homogenous enzyme. J Biol Chem. 1976 Oct 10;251(19):5866–5874. [PubMed] [Google Scholar]
- Simpson R. T., Künzler P. Cromatin and core particles formed from the inner histones and synthetic polydeoxyribonucleotides of defined sequence. Nucleic Acids Res. 1979 Apr;6(4):1387–1415. doi: 10.1093/nar/6.4.1387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- WILKINS M. H. F., GOSLING R. G., SEEDS W. E. Physical studies of nucleic acid. Nature. 1951 May 12;167(4254):759–760. doi: 10.1038/167759a0. [DOI] [PubMed] [Google Scholar]