Abstract
Snf1 protein kinase regulates responses to glucose limitation and other stresses. Snf1 activation requires phosphorylation of its T-loop threonine by partially redundant upstream kinases (Sak1, Tos3, and Elm1). Under favorable conditions, Snf1 is turned off by Reg1-Glc7 protein phosphatase. The reg1 mutation causes increased Snf1 activation and slow growth. To identify new components of the Snf1 pathway, we searched for mutations that, like snf1, suppress reg1 for the slow-growth phenotype. In addition to mutations in genes encoding known pathway components (SNF1, SNF4, and SAK1), we recovered “fast” mutations, designated fst1 and fst2. Unusual morphology of the mutants in the Σ1278b strains employed here helped us identify fst1 and fst2 as mutations in the RasGAP genes IRA1 and IRA2. Cells lacking Ira1, Ira2, or Bcy1, the negative regulatory subunit of cyclic AMP (cAMP)-dependent protein kinase A (PKA), exhibited reduced Snf1 pathway activation. Conversely, Snf1 activation was elevated in cells lacking the Gpr1 sugar receptor, which contributes to PKA signaling. We show that the Snf1-activating kinase Sak1 is phosphorylated in vivo on a conserved serine (Ser1074) within an ideal PKA motif. However, this phosphorylation alone appears to play only a modest role in regulation, and Sak1 is not the only relevant target of the PKA pathway. Collectively, our results suggest that PKA, which integrates multiple regulatory inputs, could contribute to Snf1 regulation under various conditions via a complex mechanism. Our results also support the view that, like its mammalian counterpart, AMP-activated protein kinase (AMPK), yeast Snf1 participates in metabolic checkpoint control that coordinates growth with nutrient availability.
INTRODUCTION
The Snf1/AMP-activated protein kinase (AMPK) family is highly conserved in eukaryotes, and its members control responses to metabolic stress (reviewed in references 22, 23, and 26). Mammalian AMPK is activated by increased intracellular AMP-to-ATP ratios under energy-depleting conditions, such as hypoglycemia and hypoxia. AMPK is also regulated by hormones that control whole-body metabolism, including leptin, adiponectin, and ghrelin. Activated AMPK functions to restore energy equilibrium by stimulating ATP-generating pathways and by inhibiting energy-consuming processes. Important functions of AMPK include stimulation of glucose uptake and fatty acid oxidation, as well as downregulation of protein synthesis and cell growth. Accordingly, defects in AMPK signaling have been linked to diseases from diabetes and obesity to cancer, making AMPK a promising target for activation with drugs (reviewed in references 15 and 21).
In the yeast Saccharomyces cerevisiae, Snf1 protein kinase is the ortholog of mammalian AMPK. The most established role of Snf1 is to regulate responses to glucose limitation (for a review, see reference 26). As glucose levels decrease, Snf1 is activated and promotes the use of less preferred (alternative) carbon sources. Activation of Snf1 results from phosphorylation of its conserved T-loop threonine residue (Thr210) by upstream kinases, and cellular levels of phospho-Thr210-Snf1 increase substantially upon glucose limitation (47). S. cerevisiae has three Snf1-activating kinases, Sak1, Tos3, and Elm1, each of which can phosphorylate Thr210 of Snf1 (30, 46, 50, 66). Dephosphorylation and downregulation of Snf1 involves the function of type 1 protein phosphatase Glc7, which is recruited to Snf1 by a targeting protein, Reg1 (47, 75, 76).
Yeast Snf1 has served as a powerful model for studying the principles underlying the structure, function, and regulation of the Snf1/AMPK family members. Studies in yeast characterized the conserved heterotrimeric (αβγ) composition of the kinase complex, with the catalytic α subunit (Snf1), the regulatory/targeting β subunit (Sip1, Sip2, or Gal83), and the stimulatory γ subunit (Snf4) (34, 80). Identification of the Snf1-activating kinases in yeast played a critical role in the identification of mammalian kinases that activate AMPK by phosphorylation of the equivalent activation loop threonine (Thr172) (24, 25, 30–32, 48, 50, 66, 77). At the same time, many aspects of Snf1 regulation in yeast remain unknown.
More recent evidence indicates that, in addition to glucose limitation, Snf1 regulates responses to other unfavorable conditions, including nitrogen limitation, salt stress, alkaline pH, and oxidative stress (29, 47, 52). These findings implicate Snf1 in a broad integration of stress signals and further necessitate the search for the molecular pathways responsible for its regulation, which may have important counterparts in other eukaryotes.
In this study, we explored a new genetic approach to identifying additional snf1-related mutations, based on their genetic interaction with reg1. In addition to mutants with defects in genes encoding known components of the Snf1 pathway (SNF1, SNF4, and SAK1), we isolated mutants with defects in the RasGAP genes IRA1 and IRA2 (68, 69). Cells lacking Ira1, Ira2, or Bcy1, the negative regulatory subunit of cyclic AMP (cAMP)-dependent protein kinase A (PKA) (71, 72), exhibited reduced Snf1 pathway activation in response to glucose limitation. Conversely, Snf1 activation was increased in cells lacking the Gpr1 sugar sensor that contributes to PKA signaling. Our results implicate the Snf1-activating kinase Sak1 as a target for negative regulation by the PKA pathway. PKA not only figures prominently in glucose signaling, but also integrates many other inputs (for reviews, see references 59 and 82) and could contribute to the regulation of Snf1 under various conditions.
MATERIALS AND METHODS
Strains and general methods.
The S. cerevisiae strains used in this study are listed in Table 1. Except for 4011774 and 4013278, all strains were in the Σ1278b genetic background and were descendants of strains MY1384 (MATa; prototroph), MY1401 (MATα ura3Δ leu2Δ his3Δ), and MY1402 (MATa ura3Δ leu2Δ trp1Δ) of the isogenic Sigma2000 series (Microbia, Cambridge, MA). Σ1278b derivatives carrying reg1Δ::URA3, snf1::LEU2, snf1Δ::KanMX6, and sak1Δ::KanMX4 have been described (40, 52, 53); additional derivatives were obtained by genetic crossing and tetrad analysis. To generate Σ1278b derivatives with ira1Δ::KanMX6, ira2Δ::KanMX6, ira2Δ::His3MX6, bcy1Δ::KanMX6, and gpr1Δ::KanMX6, the marker sequences were amplified by PCR with primers flanking the corresponding open reading frames. The mutant alleles were first introduced into wild-type diploids by transformation; all yeast transformations were performed using standard methods (56). The genotypes were confirmed by PCR analysis of genomic DNA. Haploid mutant segregants were recovered from the heterozygous diploids by tetrad analysis. Due to instability and poor viability upon handling and storage, bcy1Δ haploids were generated de novo prior to each experiment from a BCY1/bcy1Δ heterozygous diploid (KY146).
Table 1.
S. cerevisiae strains
| Strain | Genotype | Source |
|---|---|---|
| KY118 | MATaura3Δ leu2Δ his3Δ | This laboratory |
| KY119 | MATaura3Δ leu2Δ trp1Δ | This laboratory |
| KY120 | MATα ura3Δ leu2Δ his3Δ | This laboratory |
| KY121 | MATα ura3Δ leu2Δ trp1Δ | This laboratory |
| KY122 | MATα ura3Δ leu2Δ his3Δ trp1Δ | This laboratory |
| KY123 | MATα ura3Δ leu2Δ his3Δ reg1Δ::URA3 | This study |
| KY124 | MATaura3Δ leu2Δ trp1Δ reg1Δ::URA3 | This study |
| KY125 | MATα ura3Δ leu2Δ trp1Δ snf1Δ::KanMX6 | This study |
| KY126 | MATaura3Δ leu2Δ his3Δ reg1Δ::URA3 snf1::LEU2 | This study |
| KY127 | MATaura3Δ leu2Δ trp1Δ reg1Δ::URA3 sak1-11 | This study |
| KY128 | MATα ura3Δ leu2Δ his3Δ sak1Δ::KanMX4 | This study |
| KY129 | MATaura3Δ leu2Δ his3Δ sak1Δ::KanMX4 | This study |
| KY130 | MATα ura3Δ leu2Δ his3Δ reg1Δ::URA3 fst1-12 | This study |
| KY131 | MATα ura3Δ leu2Δ his3Δ reg1Δ::URA3 fst1-56 | This study |
| KY132 | MATaura3Δ leu2Δ reg1Δ::URA3 fst1-56 | This study |
| KY133 | MATα ura3Δ leu2Δ his3Δ reg1Δ::URA3 fst2-11 | This study |
| KY134 | MATα ura3Δ leu2Δ his3Δ reg1Δ::URA3 fst2-24 | This study |
| KY135 | MATaura3Δ leu2Δ his3Δ reg1Δ::URA3 fst2-24 | This study |
| KY136 | MATaura3Δ leu2Δ trp1Δfst1-56 | This study |
| KY137 | MATaura3Δ leu2Δ trp1Δfst2-11 | This study |
| KY138 | MATaura3Δ leu2Δ trp1Δfst2-24 | This study |
| KY139 | MATα ura3Δ leu2Δ trp1Δ ira1Δ::KanMX6 | This study |
| KY140 | MATα ura3Δ leu2Δ his3Δ trp1Δ ira1Δ::KanMX6 | This study |
| KY141 | MATα ura3Δ leu2Δ his3Δ ira2Δ::KanMX6 | This study |
| KY142 | MATα ura3Δ leu2Δ his3Δ trp1Δ ira2Δ::HisMX6 | This study |
| KY146 | MATa/MATα ura3Δ/ura3Δ leu2Δ/leu2Δ HIS3/his3Δ TRP1/trp1Δ BCY1/bcy1Δ::KanMX6 | This study |
| KY147 | MATaura3Δ leu2Δ his3Δ | This study |
| KY148 | MATaura3Δ leu2Δ his3Δ gpr1Δ::KanMX6 | This study |
| KY149 | MATaura3Δ leu2Δ his3Δ sak1Δ::KanMX4 | This study |
| KY150 | MATaura3Δ leu2Δ his3Δ gpr1Δ::KanMX6 sak1Δ::KanMX4 | This study |
| KY151 | MATα ura3Δ leu2Δ his3Δ reg1Δ::URA3 snf1::LEU2 | This study |
| KY153 | MATaura3Δ leu2Δ his3Δ snf1Δ::KanMX6 | This study |
| 4013278 | MATα ura3Δ leu2Δ his3Δ1 lys2Δ ybr139WΔ::KanMX4 | ATCC |
| 4011774 | MATα ura3Δ leu2Δ his3Δ1 lys2Δ yol083WΔ::KanMX4 | ATCC |
Strains 4011774 and 4013278 were derivatives of BY4742 and were obtained from the American Type Culture Collection (ATCC).
The rich medium was yeast extract-peptone-dextrose (YPD); synthetic complete (SC) medium lacking appropriate supplements was used to select for plasmids (56). Unless otherwise indicated, the media contained abundant (2%) glucose. In all experiments, yeast cells were grown at 30°C.
Isolation of extragenic reg1Δ suppressor mutants.
Cells of strains KY123 and KY124 were grown in liquid YPD medium to late stationary phase. The cells were washed and spread at ∼5 × 107 cells per plate onto 10 plates (five plates per strain) containing synthetic minimal/dextrose medium (SD) containing 4% glucose and appropriate supplements (56). The supplements were uracil (20 mg/liter), leucine (60 mg/liter), histidine (20 mg/liter), and tryptophan (40 mg/liter). Colonies growing at rates exceeding that of the background were collected over the course of 1 week. The isolates were retested for phenotype by streaking on SD.
Plasmids.
Plasmids pIT469 and pRJ217 express LexA-Snf1 and LexA-Snf1-T210A, respectively, from vector pEG202 (17, 39, 42). Plasmid pBM3068 carries a SUC2-lacZ reporter (54), and plasmid pLCLG-Staf carries a STA2-lacZ reporter (36). To construct plasmid pMK9-6, the wild-type SAK1 gene, including 0.72 kb of its upstream regulatory sequence, was amplified by PCR from genomic DNA, cloned into pCRII-TOPO using the TOPO TA cloning kit (Invitrogen), sequenced, and then excised and ligated into the ApaI site of the low-copy-number vector pRS315 (63). pMK11-3 was constructed similarly, except that the genomic DNA used as the template was from strain KY127, which carries a recessive sak1 mutation, designated sak1-11. Sequencing three independent sak1-11 PCR products cloned in pCRII-TOPO indicated that sak1-11 contains a frameshift mutation (deletion of a single nucleotide from the A5 stretch spanning positions 2202 to 2206 of the open reading frame) that truncates the open reading frame after the codon for Lys735. pM31-8 expresses N-terminal triple-hemagglutinin (HA)-tagged Sak1 from the yeast ADH1 promoter of vector pSK134HA (52); the SAK1 sequence was from pM36 (53). pKB2 was derived from pM31-8 by introducing a mutation that changes the Ser1074 codon of SAK1 to an Ala codon, using the QuikChange XL site-directed mutagenesis kit (Stratagene) and a pair of complementary mutagenic primers that also create a silent diagnostic BssHII site overlapping the Ala1074 codon; the construct was confirmed by sequencing. The Ser1139-to-Ala mutation was constructed similarly. Plasmid pKB4 expresses the HA-Sak1-S1074A, S1139A double-mutant protein.
β-Galactosidase assays.
In experiments involving clumpy mutants (fst-ira and bcy1Δ), β-galactosidase activity was assayed in protein extracts (56) and expressed in units defined as follows: 1 unit = optical density at 420 nm (OD420) × 1,000 per min per mg of protein (38). In other experiments, β-galactosidase activity was assayed in permeabilized cells and expressed in Miller units (56).
Immunoblot assays.
Cells were grown under conditions specified in the text. Protein extracts were prepared by the heat inactivation/alkaline treatment method as described previously (51). Briefly, cultures or culture aliquots (3 to 5 ml) were placed in a boiling water bath for 3 min to arrest Snf1 in its culture-specific Thr210 phosphorylation state, followed by cooling, harvesting, mild alkaline treatment, and extraction by boiling in SDS-PAGE loading buffer. Proteins were separated by SDS-PAGE and analyzed by immunoblotting with anti-phospho-Thr172–AMPK (Cell Signaling Technology), which strongly recognizes phospho-Thr210–Snf1; total Snf1 protein levels were determined by using anti-polyhistidine antibody H1029 (Sigma-Aldrich), which strongly recognizes a natural stretch of 13 consecutive histidines (amino acids 18 to 30) present in Snf1 (51). HA-tagged proteins were detected by anti-HA 12CA5 (Sigma-Aldrich). The signals were detected by enhanced chemiluminescence using ECL Plus (Amersham Biosciences) or HyGlo (Denville Scientific).
Immunoprecipitation of HA-tagged Sak1 proteins and analysis of Ser1074 phosphorylation.
Protein extracts were prepared by the heat inactivation/alkaline treatment method as described above, except that a buffer containing 50 mM HEPES, pH 7.3, 5 mM EDTA, 1 mM dithiothreitol (DTT), and 1% SDS was used instead of SDS-PAGE loading buffer for the final extraction stage. After passage through Pierce Detergent Removal Spin Columns (Thermo Scientific), the extracts (200 μg protein) were subjected to immunoprecipitation with anti-HA antibody 12CA5 essentially as described previously (74) in a buffer containing 50 mM HEPES, pH 7.3, 50 mM NaCl, 10% glycerol, 0.1% Triton X-100, 1 mM phenylmethylsulfonyl fluoride (PMSF), and protease inhibitor mixture (Complete Mini; Roche). Unless intended for subsequent phosphatase treatment (see below), the collected material was eluted from protein A beads (25 μl) by boiling in 50 μl of SDS-PAGE loading buffer, and 10-μl samples were analyzed by immunoblotting with custom affinity-purified rabbit anti-phospho-Ser1074–Sak1 antibodies (21st Century Biochemicals). To detect the total levels of HA-tagged Sak1 and Sak1-S1074A, the blots were stripped and reprobed with anti-HA 12CA5. The signals were detected by enhanced chemiluminescence using ECL Plus (Amersham Biosciences) or HyGlo (Denville Scientific).
When phosphatase treatment was involved, the bead-bound immunoprecipitates prepared as described above were resuspended in 200 μl of 1× shrimp alkaline phosphatase (SAP) reaction buffer, split into two halves, and incubated at 37°C for 30 min with or without 10 units of SAP (Promega). The reactions were stopped by adding an equal volume of 2× SDS-PAGE loading buffer and boiling for 5 min. Samples (10 μl) were then analyzed by immunoblotting for Ser1074 phosphorylation and total HA-tagged Sak1 as described above.
RESULTS
Mutant search rationale.
Yeast Snf1 was first identified genetically in straightforward screens for mutants with defects in alternative carbon source utilization (10, 11, 84). With the exception of Snf4, the γ subunit of the Snf1 complex encoded by a unique gene, identification of the components of the Snf1 pathway has been challenging, particularly due to their functional redundancy, precluding the isolation of recessive mutations with robust snf1Δ-like phenotypes. For example, Sak1, Tos3, and Elm1 eluded identification as the Snf1-activating kinases until the discovery of a physical interaction between the Snf1 complex and Sak1 (30, 50, 66). Further studies of the Snf1 pathway could therefore benefit from the use of new, more sensitive genetic screens.
We explored a new genetic approach to identifying additional components of the Snf1 pathway. The approach is based on the Snf1-dependent slow-growth phenotype caused by the reg1Δ mutation. This phenotype is associated with increased phosphorylation of the activation loop Thr210 residue of Snf1 in the absence of Reg1-Glc7 phosphatase (Fig. 1A to C). Our rationale was to identify mutations that restore faster growth by attenuating the hyperactive Snf1 pathway.
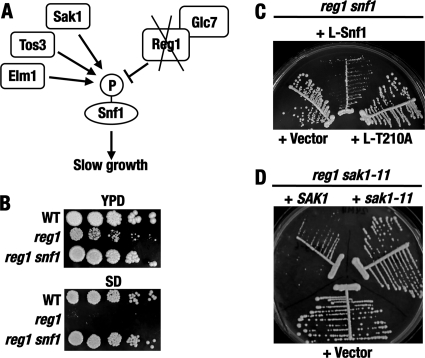
Fig 1.
Rationale of the mutant search. (A) Lack of Reg1 causes Snf1 hyperactivation and slow growth. (B) In addition to causing slow growth on YPD medium, reg1Δ causes a Snf1-dependent growth defect on SD medium. Serial dilutions of cell suspensions were spotted onto plates and grown at 30°C for 3 days. The strains were KY118 (wild-type [WT]), KY124 (reg1), and KY126 (reg1 snf1). (C) The slow growth conferred by the lack of Reg1 depends on the activation loop Thr210 residue of Snf1. Cells of strain KY151 (reg1 snf1) transformed with plasmids expressing LexA-Snf1 (+L-Snf1) or LexA-Snf1-T210A (+L-T210A) or carrying the corresponding vector (+Vector) were streaked on SC medium lacking histidine and grown for 4 days. (D) A fast-growing reg1 suppressor isolate carries a sak1 mutation. Cells of strain KY127 (reg1 sak1-11) transformed with plasmid pMK9-6 (+SAK1) or pMK11-3 (+sak1-11) or with the corresponding vector pRS315 (+Vector) were streaked on SC medium lacking leucine and grown for 4 days.
We further used the fact that reg1Δ mutants grow extremely slowly, if at all, under several conditions less favorable than standard nutrient-rich YPD medium or SC medium. These conditions include the presence of low levels of stressors, such as NaCl, LiCl, or rapamycin (reference 4 and data not shown). We also noted that the reg1Δ mutation causes a significant growth defect on SD medium containing only a limited set of supplements required by the strains used in this study (Fig. 1B). We conducted reg1Δ suppressor searches under several conditions, but the SD-based approach offered the simplest and most productive system. The reg1Δ suppressor mutants central to this paper were obtained by selection on SD.
We isolated 69 spontaneous reg1Δ suppressor mutants. Complementation analyses using knockout tester strains and plasmid-borne genes from our collection suggested that 14 isolates carry mutations in genes encoding known pathway components, namely, Snf1 itself; the γ subunit of the Snf1 complex, Snf4; and, notably, the Snf1-activating kinase Sak1. The snf1 and snf4 isolates were not studied further, but the recovery of sak1 was analyzed in more detail. To confirm genetic linkage, a reg1Δ::URA3 ura3 isolate carrying a putative sak1 allele designated sak1-11 (strain KY127) was crossed to a sak1Δ REG1 ura3 strain (KY128), and the diploid obtained was subjected to tetrad analysis. All reg1Δ::URA3 segregants in 16 tetrads grew at a wild-type rate, consistent with the diploid being homozygous for sak1. Sequencing the sak1-11 allele indicated that it carries a frameshift mutation, so that the last correct codon in sak1-11 is that for Lys735, followed by codons for Thr-Gln-Gln-Arg-Asn and a stop codon (the size of wild-type Sak1 is 1,142 amino acids). Thus, sak1-11 encodes a protein with a significant truncation of the extensive noncatalytic C-terminal domain, which is important for its function (57). Complementing the reg1Δ sak1-11 isolate with plasmid-borne wild-type SAK1 conferred slow growth, whereas introduction of an otherwise identical construct carrying sak1-11 had no discernible effect compared to the corresponding empty vector (Fig. 1D). Collectively, these results demonstrated that the reg1 suppressor approach is sufficiently sensitive to identify mutants with partial defects in the Snf1 pathway.
“Fast” mutations fst1 and fst2.
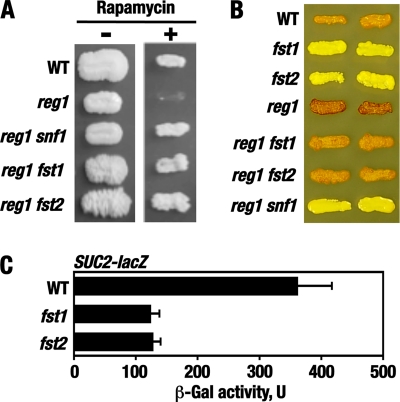
Twenty-four suppressor isolates were distinguished from the rest of the mutants by unusual “cauliflower” colony morphology on YPD medium (Fig. 2A); this morphology was less pronounced on synthetic media, although the mutants still exhibited increased agar adhesion (see Fig. 4). We focused on these mutants; the rest of the mutants remain to be characterized. The “cauliflower” mutations were recessive, and genetic analyses indicated that they all belong to two complementation groups, provisionally dubbed fst1 and fst2, for “fast” growth. Representative fst1 and fst2 alleles exhibited 2:2 segregation in tetrads (Fig. 2B shows results for an fst1 allele), as expected for mutations in single nuclear genes. In addition to improved growth, the fst mutations suppressed reg1Δ for Snf1-dependent rapamycin hypersensitivity (Fig. 3A). The fst mutations conferred significant glycogen accumulation defects, as does snf1 (8, 70), and partially suppressed reg1Δ for Snf1-dependent glycogen hyperaccumulation (Fig. 3B). The fst mutations did not confer detectable defects in growth on nonfermentable carbon sources but caused a reduction in the expression of a SUC2-lacZ reporter (54) under glucose-limiting conditions (Fig. 3C); SUC2 encodes secreted invertase responsible for sucrose and raffinose utilization and is activated in response to glucose limitation in a Snf1-dependent manner (10). Thus, these results suggested that fst1 and fst2 cause partial defects in Snf1 signaling.
Fig 2.
“Fast” reg1 suppressor mutations fst1 and fst2. (A) The fst1 and fst2 mutations suppress the slow growth caused by the lack of Reg1 and confer unusually rough colony morphology. Single cells of strains KY124 (reg1), KY118 (WT), KY127 (reg1 sak1), KY132 (reg1 fst1), and KY135 (reg1 fst2) were micromanipulated on YPD and grown for 4 days. The diameter of the reg1 colony is ∼1 mm. Size comparisons are to scale. (B) Tetrad analysis of fst1. (Top) Tetrad analysis of a control diploid heterozygous for reg1Δ (reg1 × WT). (Bottom) Tetrad analysis of a cross between fast-growing isolate KY130 (reg1 fst1) and wild-type strain KY119 (WT). The cells were grown on YPD medium for 4 days.
Fig 4.
The fst1 and fst2 mutations do not complement ira1Δ and ira2Δ, respectively. Diploids were obtained by mating haploid strains of the indicated genotypes. The strains were KY136 (fst1), KY138 (fst2), KY139 (ira1Δ), and KY141 (ira2Δ). The diploids were grown on SC medium lacking histidine and tryptophan for 4 days (Growth) and tested for glycogen accumulation by staining over iodine crystals (Glycogen). Once the color faded, agar adhesion was tested by rubbing the agar surface with a cell spreader under a stream of water (Adhesion).
Fig 3.
The fst1 and fst2 mutations confer phenotypes consistent with partial defects in the Snf1 pathway. (A) The fst mutations relieve Snf1-dependent rapamycin hypersensitivity caused by the lack of Reg1. (Left) Cells were patched on YPD medium and grown for 3 days (−). (Right) Cells were patched on YPD medium containing 20 ng/ml rapamycin and grown for 5 days (+). The strains were KY120 (WT), KY123 (reg1), KY126 (reg1 snf1), KY130 (reg1 fst1), and KY133 (reg1 fst2). (B) The fst mutations affect glycogen accumulation. Cells were patched on SC medium and grown for 2 days. Glycogen stores were detected by staining over iodine crystals for 1 min (8). Darker color corresponds to more glycogen. The strains were KY119 (WT), KY136 (fst1), KY137 (fst2), KY123 (reg1), KY130 (reg1 fst1), KY133 (reg1 fst2), and KY126 (reg1 snf1). (C) The fst mutations affect SUC2-lacZ expression. Strains KY119 (WT), KY136 (fst1), and KY137 (fst2) transformed with plasmid pBM3068 were grown in liquid SC medium lacking uracil and containing abundant (2%) glucose to mid-log phase and then shifted for 3 h to otherwise identical medium containing limiting (0.05%) glucose. β-Galactosidase (β-Gal) activity was assayed in protein extracts and expressed in units defined in Materials and Methods. The data shown are for glucose-limiting conditions. The values are averages for four or five transformants. In abundant glucose, the values were <10 U. The error bars indicate standard errors.
fst1 and fst2 are mutations in the RasGAP genes IRA1 and IRA2, respectively.
The unusual colony morphology of the fst mutants resembled the morphology caused by the lack of Elm1, one of three Snf1-activating kinases (the morphological effects of the elm1 mutation are unrelated to Elm1's role in Snf1 activation and pertain to its role in the activation of Nim1-related protein kinases involved in morphogenesis checkpoint control [1, 3, 5, 67]). We conducted complementation tests using isogenic elm1Δ tester strains (53), but neither FST1 nor FST2 was found to be allelic to ELM1 (data not shown).
In the course of our experiments, we also noted that the fst1 and fst2 mutants lose viability after prolonged incubation on plates, i.e., after prolonged nutrient limitation. This and other phenotypes conferred by fst1 and fst2 suggested that they cause increased Ras-cAMP signaling and might represent mutations in the IRA1 and IRA2 genes (6, 8, 16, 20, 60, 70). The Ira1 and Ira2 proteins are inhibitors of Ras signaling (RasGAPs) and function by stimulating the intrinsic GTPase activity of the yeast Ras proteins Ras1 and Ras2, thereby promoting their conversion to the nonsignaling GDP-bound form (68, 69).
To determine whether fst1 and fst2 are mutations in IRA1 and IRA2, we performed genetic complementation and linkage analyses. IRA1 and IRA2 are not essential for viability in strains of the Σ1278b genetic background used in this study (19). For complementation tests, ira1Δ and ira2Δ knockout haploids were constructed and crossed to fst1 and fst2 haploids. fst1 did not complement ira1Δ, and fst2 did not complement ira2Δ for defective glycogen accumulation and increased agar adhesion (Fig. 4). The noncomplemented diploids also exhibited sporulation defects, as expected for homozygous ira mutants (68, 69). Due to these sporulation defects, genetic linkage was analyzed using markers located in the vicinity of the IRA1 and IRA2 genes. The marked loci were YBR139W on chromosome II and YOL083W on chromosome XV. Strain KY136 (fst1) was crossed to strain 4013278 (ybr139WΔ::KanMX4), and the resulting diploid was subjected to tetrad analysis. The segregants were scored for glycogen accumulation and kanamycin resistance. The ratio of parental ditype (PD), nonparental ditype (NPD), and tetratype (T) tetrads was as follows: PD/NPD/T = 15:0:0, indicating tight genetic linkage. Tetrad analysis of a cross between strains KY137 (fst2) and 4011774 (yol083WΔ::KanMX4) yielded a PD/NPD/T ratio of 20:0:3, similarly indicating tight linkage; we attribute the incidence of some tetratype tetrads to the 4.1-kb distance between YOL083W and IRA2 and to the possibility that the lesion present in the mutant allele used in this experiment resides toward the distal end of the IRA2 open reading frame, which is very large (9.2 kb). These complementation and linkage analyses provided strong evidence that FST1 is IRA1 and FST2 is IRA2.
The bcy1 mutation affects Snf1 activation.
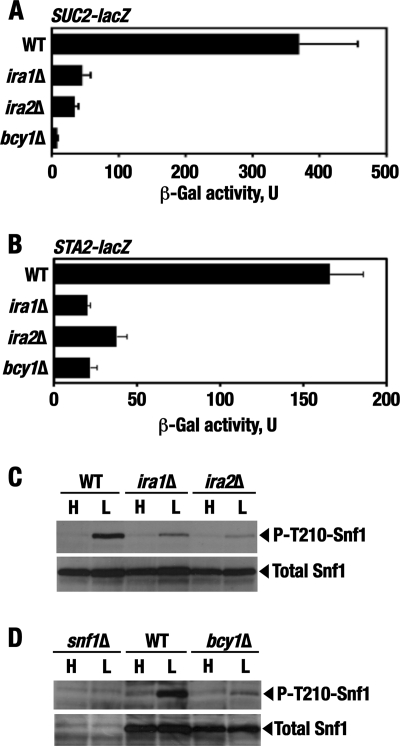
Mutation of IRA1 and IRA2 leads to activation of Ras, whose major cellular role is to stimulate cAMP production by adenylate cyclase and thereby activate cAMP-dependent protein kinase (PKA) (6, 73). The PKA holoenzyme is a tetramer consisting of two negative regulatory subunits (encoded by BCY1) and two catalytic Tpk subunits (encoded by one of three genes, TPK1, TPK2, or TPK3) (9, 71, 72). In the holoenzyme form, PKA is inactive, but cAMP binding to Bcy1 facilitates the release of the active catalytic Tpk subunits. In cells lacking Bcy1, PKA is constitutively activated. We constructed the bcy1Δ mutation and analyzed the expression of the Snf1-dependent SUC2-lacZ reporter as described above. SUC2-lacZ expression in glucose-limited bcy1Δ cells was significantly reduced (Fig. 5A). We also tested a different Snf1-regulated reporter, STA2-lacZ (36). STA2 encodes extracellular glucoamylase, involved in starch utilization, and its expression increases in response to glucose limitation in a Snf1-dependent manner (43). STA2, however, differs from SUC2 in that inhibition of STA2 transcription under glucose-rich conditions involves the Nrg1 and Nrg2 repressors rather than the Mig1 repressor (36, 40). STA2-lacZ expression in glucose-limited bcy1Δ cells was also significantly reduced (Fig. 5B). The negative effects of bcy1Δ on reporter expression were comparable to or exceeded the effects of the ira mutations, suggesting that activation of Ras produces its effects largely via activation of PKA. The similar responses of the two different reporters also suggested that the mutations produce effects upstream of Snf1.
Fig 5.
The ira1Δ, ira2Δ, and bcy1Δ mutations affect Snf1 pathway activation. (A) The ira1Δ, ira2Δ, and bcy1Δ mutations affect SUC2-lacZ expression. Strains transformed with plasmid pBM3068 were grown in liquid SC medium lacking uracil and containing abundant (2%) glucose to mid-log phase and then shifted for 3 h to otherwise identical medium containing limiting (0.05%) glucose. β-Galactosidase activity was assayed in protein extracts and expressed in units defined in Materials and Methods. The data shown are for glucose-limiting conditions. The values are averages for three to seven transformants. In abundant glucose, the values were ≤15 U. The strains were KY122 (WT), KY140 (ira1Δ), KY142 (ira2Δ), and a haploid bcy1Δ derivative (MATa ura3Δ leu2Δ his3Δ trp1Δ bcy1Δ) of KY146. The error bars indicate standard errors. (B) The ira1Δ, ira2Δ, and bcy1Δ mutations affect STA2-lacZ expression. The experiments were performed as for panel C, except that the reporter plasmid used was pLCLG-Staf. The data shown are for glucose-limiting conditions. The values are averages for five transformants. In abundant glucose, the values were ≤8.3 U. (C and D) Cells of the indicated genotypes were grown to mid-log phase in YPD containing 2% glucose (H; high glucose) and then shifted for 1 h to an otherwise identical medium containing 0.05% glucose (L; low glucose). The levels of activated phospho-Thr210–Snf1 (P-T210-Snf1) and total Snf1 protein (Total Snf1) were analyzed by immunoblotting as described in Materials and Methods. The strains were KY121 (WT), KY139 (ira1Δ), and KY142 (ira2Δ) (C) and KY125 (snf1Δ), KY121 (WT), and a haploid bcy1Δ segregant (MATα ura3Δ leu2Δ trp1Δ bcy1Δ) from KY146 (D).
In further support of PKA involvement, the level of phospho-Thr210-Snf1 was reduced in glucose-limited bcy1Δ cells, as it was in ira mutant cells (Fig. 5C and D).
Since the fst/ira mutations suppress reg1Δ for several Snf1-dependent phenotypes, we also used immunoblotting to compare the Snf1 Thr210 phosphorylation status in reg1Δ ira double mutants relative to the reg1Δ single mutant, but the results were inconclusive and are not shown.
Analysis of Sak1 Ser1074 phosphorylation in vivo.
Although Thr210 phosphorylation of Snf1 may not be the only relevant step, we considered the possibility that PKA at least partly affects the Snf1 pathway by affecting a Snf1-activating kinase(s). In this regard, Sak1 appeared to be a particularly attractive candidate. Sak1 makes the largest individual contribution to Snf1 activation (27, 30), and sequence analysis indicates that amino acids 1071 to 1075 (Arg-Arg-Arg-Ser-Ile) in the noncatalytic C terminus of Sak1 (Fig. 6A) conform to the ideal PKA recognition motif, defined as RRXSΦ (37, 61), where X is any amino acid and Φ is a hydrophobic amino acid (Fig. 6B). The site is also flanked by small amino acids (Gly1070, Pro1076, and Ser1077), which is an additional feature favored by PKA (61). Furthermore, we noted that the site is conserved in the Sak1 homologs of other fungi representing several genera (Fig. 6B). Importantly, the noncatalytic C-terminal domain is critical for Sak1 function in vivo (57) and, as such, could contain target sites for positive and/or negative regulation.
Fig 6.
Ser1074 and Ser1139 sites in Sak1. (A) Domain organization of Sak1. KD, kinase domain; NRD, N-terminal regulatory domain; CRD, C-terminal regulatory domain (57). Ser1074 (S1074) and Ser1139 (S1139) are located in the CRD. (B) The Ser1074 PKA motif is embedded in a box of sequence similarity present in Sak1 homologs of fungi representing several genera. Sc, Saccharomyces cerevisiae; Cg, Candida glabrata; Zr, Zygosaccharomyces rouxii; Lt, Lachancea thermotolerans; Ag, Ashbya gossipii. The amino acid positions of the key serine are indicated at the right. (C) Alignment of the Ser1139 site. Sc, S. cerevisiae; Sp, S. paradoxus; Sm, S. mikitae; Cg, C. glabrata. The amino acid positions of the key serine are indicated on the right. In panels B and C, the amino acid identities and similarities to the S. cerevisiae sequence are shaded black and gray, respectively.
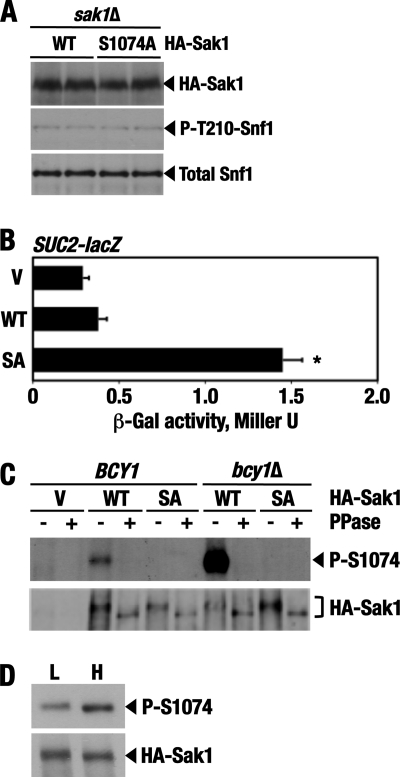
We expressed a triple-HA-tagged Sak1 protein in which Ser1074 is replaced with Ala (HA-Sak1-S1074A). Expression of HA-Sak1-S1074A did not discernibly affect Thr210 phosphorylation of Snf1 (Fig. 7A) but led to a modest (3.4-fold) but consistent increase in the expression of the SUC2-lacZ reporter relative to wild-type HA-Sak1 under high-glucose conditions (Fig. 7B).
Fig 7.
Ser1074 is phosphorylated in vivo. (A) Cells of strain KY129 (sak1Δ) carrying plasmid pM31-8 expressing N-terminal triple-HA-tagged wild-type Sak1 (HA-Sak1; WT) or pKB2 expressing tagged Sak1-S1074A (HA-Sak1; S1074A) were grown to mid-log phase in SC lacking leucine and containing abundant (2%) glucose. The levels of the tagged wild-type and mutant Sak1 proteins (HA-Sak1), phospho-Thr210–Snf1 (P-T210-Snf1), and total Snf1 protein (Total Snf1) were determined by immunoblotting as described in Materials and Methods. (B) Expression of HA-Sak1-S1074A increases SUC2-lacZ expression. Transformants of strain KY129 (sak1Δ) carrying the SUC2-lacZ reporter and expressing HA-Sak1 (WT) or HA-Sak1-S1074A (SA) or carrying the empty vector (V) were grown with plasmid selection in the presence of 2% glucose. β-Galactosidase activity was assayed in permeabilized cells and expressed in Miller units. The values are averages for 10 transformants. The asterisk indicates that the effect is statistically significant (P < 0.01; n = 10). The error bars indicate standard deviations. (C) Wild-type HA-Sak1 (HA-Sak1; WT) and nonphosphorylatable HA-Sak1-S1074A (HA-Sak1; SA) were immunoprecipitated from wild-type cells (BCY1, strain KY118) and bcy1Δ mutant cells (MATa ura3Δ leu2Δ his3Δ bcy1Δ) after growth to mid-log phase in SC lacking leucine and containing 2% glucose. The immunoprecipitates were incubated with (+) or without (−) shrimp alkaline phosphatase (PPase), and Ser1074 phosphorylation was analyzed by immunoblotting with anti-phospho-Ser1074–Sak1 antibodies (P-S1074). The total levels of HA-Sak1 and HA-Sak1-S1074A were determined using anti-HA (HA-Sak1). (D) Wild-type HA-Sak1 was expressed in wild-type strain KY118. Glucose-limited cells were prepared by shifting exponentially growing cells to SC lacking leucine and containing 0.05% glucose for 3 h (L; low glucose). Glucose was then added to the final concentration of 2% for 30 min (H; high glucose). Following immunoprecipitation, Ser1074 phosphorylation (P-S1074) and total levels of HA-Sak1 were analyzed by immunoblotting as described above.
The observed effect was modest, as SUC2-lacZ expression in cells with HA-Sak1-S1074A still increased almost 40-fold (from 1.3 to 49 Miller units) after a 3-h shift from 2% glucose medium to 0.05% glucose medium. In cells with wild-type HA-Sak1, expression increased to a somewhat lower level (31 Miller units), suggesting that Ser1074 is phosphorylated under both high- and low-glucose conditions, as was subsequently confirmed.
We investigated whether Ser1074 is phosphorylated in vivo, using anti-phospho-Ser1074–Sak1 antibodies. Wild-type and bcy1Δ cells expressing HA-Sak1 or HA-Sak1-S1074A were grown in the presence of 2% glucose, and the HA-tagged proteins were immunoprecipitated with anti-HA. In the absence of phosphatase treatment, anti-phospho-Ser1074–Sak1 antibodies detected a phosphorylation signal for HA-Sak1 from wild-type cells and a stronger signal for HA-Sak1 from bcy1Δ cells; no signals were detected for the nonphosphorylatable HA-Sak1-S1074A protein recovered from either wild-type or bcy1Δ cells (Fig. 7C, top). Anti-phospho-Ser1074–Sak1 antibodies did not detect HA-Sak1 in the phosphatase-treated samples. Immunoblotting with anti-HA indicated that the levels of the HA-tagged proteins were comparable in all samples, although there was a noticeable increase in electrophoretic mobility of the phosphatase-treated HA-Sak1 and HA-Sak1-S1074A proteins (Fig. 7C, bottom). Thus, Ser1074 is phosphorylated in vivo, and its increased phosphorylation in bcy1Δ cells is consistent with a role for PKA or a PKA-stimulated kinase, the former being the simpler possibility. The observed phosphatase-induced mobility shift indicates that some kinase or kinases, possibly including Snf1 (44), also phosphorylate Sak1 on a residue(s) other than Ser1074 (and Ser1139 [see below]).
We also tested for a possible stimulatory effect of glucose on Ser1074 phosphorylation. When we added abundant glucose to glucose-limited cells, Ser1074 phosphorylation increased modestly (Fig. 7D). Thus, phosphorylation of Ser1074 alone appears to play only a fine-tuning role in regulation. Despite its modest contribution, however, the underlying mechanism could have long-term evolutionary significance.
Combined effect of Ser1074-to-Ala and Ser1139-to-Ala mutations.
PKA often phosphorylates its targets on multiple sites. Once a protein is established to be a PKA substrate, the probability of its phosphorylation at sites that do not conform to the ideal PKA consensus becomes considerable (61). After Ser1074, the next best match to the PKA consensus in Sak1 is the Ser1139 site (KRRSA) (Fig. 6C). This site seemed compelling for several reasons. First, it has a serine rather than threonine as the putative phosphoacceptor. Second, Ser1139 is preceded by more than 1 arginine residue. Third, the site is well conserved in Saccharomyces species (Fig. 6C). The corresponding C-terminal regions of Sak1 homologs from other genera are also enriched in serine, threonine, arginine, and lysine residues; moreover, a similar PKA motif is present at a similar position in Sak1 of Candida glabrata (KRASW) (Fig. 6C). Finally, Ser1074 and Ser1139 are relatively close to each other (Fig. 6A), suggesting that their phosphorylation could have a combined effect on the same functional aspect of Sak1. We therefore mutated this site (Ser1139 to Ala).
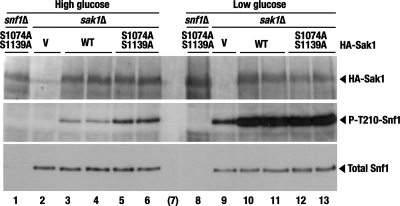
As with HA-Sak1-S1074A, expression of the HA-Sak1-S1139A single-mutant protein did not confer a discernible increase in Snf1 Thr210 phosphorylation in high glucose (data not shown). However, expression of the double mutant HA-Sak1-S1074A, S1139A protein resulted in partially constitutive Thr210 phosphorylation (Fig. 8).
Fig 8.
Expression of double mutant HA-Sak1-S1074A, S1139A confers partially constitutive Thr210 phosphorylation of Snf1. Transformants of strains KY153 (snf1Δ) and KY129 (sak1Δ) expressing HA-Sak1 (WT) or HA-Sak1-S1074A, S1139A, or carrying the empty vector (V) were grown with plasmid selection in the presence of 2% glucose (High glucose) and then shifted to an otherwise identical medium containing 0.05% glucose for 1 h (Low glucose). The levels of wild-type and mutant HA-Sak1 proteins (HA-Sak1), activated phospho-Thr210–Snf1 (P-T210-Snf1), and total Snf1 protein (Total Snf1) were analyzed by immunoblotting. Lane 7 (in parentheses) is empty.
Thus, these results support the idea that Sak1 is a relevant target for negative regulation by the PKA pathway. At the same time, the constitutivity observed for the mutant Sak1 proteins was modest, suggesting several independent possibilities. First, Sak1 could be phosphorylated by PKA on additional sites. Second, Sak1 is not the only relevant target for PKA. Third, PKA is not the only contributor to the negative regulation of Snf1.
Sak1 is not the only relevant target of PKA.
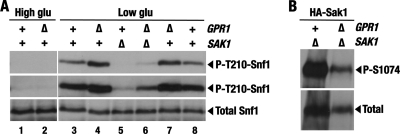
To examine the effects of weakened PKA signaling, we knocked out the GPR1 gene. Gpr1 is a G-protein-coupled receptor involved in glucose sensing and stimulates cAMP signaling in a pathway that is parallel to the Ras-cAMP pathway (45, 55). Since the exact origins of the signals impinging on Snf1 are not completely understood, it seemed particularly interesting to test for a link between Snf1 and Gpr1. It should be noted, however, that the lack of Gpr1 confers a partial defect in PKA signaling and could therefore produce only a partial stimulatory effect on Snf1. While the gpr1Δ mutation did not suffice to confer constitutive Thr210 phosphorylation of Snf1 in high glucose (Fig. 7A, compare lanes 1 and 2), it facilitated better Snf1 activation in response to low glucose (Fig. 9A, compare lanes 3 and 4 or lanes 7 and 8). Thus, Gpr1 partially contributes to the negative control of Snf1.
Fig 9.
Effects of the gpr1 mutation. (A) gpr1Δ facilitates better Thr210 phosphorylation of Snf1 in response to low glucose and partially suppresses sak1Δ. Cells of the indicated genotypes were grown in 2% glucose (High glu) and then shifted to 0.05% glucose (Low glu). The levels of activated phospho-Thr210–Snf1 (P-T210-Snf1) and total Snf1 protein (Total Snf1) were analyzed by immunoblotting. The top two panels represent two different exposures of the same experiment. The strains were KY147 (WT), KY148 (gpr1Δ), KY149 (sak1Δ), and KY150 (sak1Δ gpr1Δ). (B) gpr1Δ does not abolish Ser1074 phosphorylation of Sak1. Cells of strains KY149 (sak1Δ) and KY150 (sak1Δ gpr1Δ) expressing HA-Sak1 from pM31-8 were grown with plasmid selection in the presence of 2% glucose. The levels of phospho-Ser1074–HASak1 (P-S1074) and total HA-Sak1 (Total) were analyzed by immunoblotting.
We envisioned that gpr1Δ and reg1Δ could have combined positive effects on Snf1 activation. We attempted to test this idea, but gpr1Δ reg1Δ double mutants were nonviable.
The lack of Gpr1 did not abolish Ser1074 phosphorylation of Sak1 (Fig. 9B), suggesting that the positive effect of the gpr1Δ mutation on Snf1 activation cannot be mediated exclusively by Sak1. If so, gpr1Δ would be expected to facilitate Snf1 activation even in the absence of Sak1. Indeed, gpr1Δ partially suppressed sak1Δ for the Snf1 activation defect (Fig. 9A, compare lanes 5 and 6), indicating the existence of an additional effector(s). Thus, while these results do not rule out the existence of a Gpr1-PKA-Sak1-Snf1 signaling branch, we conclude that Sak1 is not the only relevant target for negative control by the PKA pathway.
DISCUSSION
Here, we conducted a search for new mutants with possible defects in the Snf1 pathway. We used the fact that the reg1Δ mutation, which confers increased Snf1 activation, also confers very slow growth. We isolated “fast” mutations that, like snf1, relieve the slow-growth phenotype caused by reg1Δ. We recovered recessive mutations, not only in SNF1 and SNF4, but also in SAK1, which encodes one of three partially redundant Snf1-activating kinases. Inactivation of SAK1 alone does not cause growth defects on alternative carbon sources (30, 50), and identification of sak1 in our mutant search suggested that suppression of reg1Δ is a sensitive genetic approach for identifying new components of the Snf1 regulatory network.
We also recovered multiple isolates with mutations in the RasGap genes IRA1 and IRA2, the yeast orthologs of the mammalian NF1 tumor suppressor gene (79). Inactivation of IRA1, IRA2, and BCY1 conferred a reduction in the activation of the Snf1 pathway in response to glucose limitation. These results provided evidence that activation of Ras affects Snf1 activation and that this effect is largely mediated by PKA, although additional contributions by PKA-independent mechanisms cannot be excluded.
The existence of an antagonistic relationship between Ras-PKA and Snf1 was proposed a long time ago, based on the finding that snf1 confers some of the same phenotypes as activation of Ras-PKA (70), but whether Ras-PKA and Snf1 can engage in a hierarchical regulatory relationship remained unclear. More recent evidence indicated that PKA affects the subcellular localization of Sip1 (28), one of three alternate β subunits of the Snf1 complex (Sip1, Sip2, and Gal83). However, Sip1 was reported to be the least abundant of the β subunits (49), and the physiological significance of its PKA-regulated localization is not fully understood. Thus, the present study extends previous findings by implicating PKA in the control of a major aspect of Snf1 function in yeast.
We note that strain-dependent variation in the apparent strength of this PKA-Snf1 connection may be one of the factors that precluded its identification in earlier studies. For example, in our experiments with the dimorphic Σ1278b background, SUC2-lacZ reporter expression was significantly reduced in the ira and bcy1 mutants, whereas expression of constitutive Ras2Val19 had no pronounced effect on SUC2 gene expression in the more commonly used W303 genetic background (81). Σ1278b strains have an unusually high basal level of cAMP signaling (64, 65), and their Snf1 pathway could therefore be more responsive to further increases in cAMP-PKA signaling. Another potential variable to consider is the rate of dephosphorylation of the relevant PKA sites by the cognate phosphatase(s), possibly including Reg1-Glc7 itself.
Our results indicate that the Snf1-activating kinase Sak1 is phosphorylated in a PKA-stimulated manner on Ser1074 within an ideal PKA motif of its C-terminal domain. The C-terminal domain of Sak1 is dispensable for its catalytic activity in vitro (57). A recent study shows that this domain mediates physical interaction with Snf1 and that the segment encompassing amino acids 501 to 740 is required for full Snf1 activation in vivo (44). Ser1074 is located further C-terminal of this segment, in the region that is missing in the product encoded by the inactive sak1-11 allele recovered from our screen (codons 736 to 1142). Ser1074 phosphorylation could make a fine-tuning contribution to modulating the Sak1-Snf1 interaction but could also affect an as-yet-unidentified aspect of Sak1 function. In either case, Ser1074 phosphorylation alone appears to make only a modest contribution to regulation. The underlying mechanism could, however, confer a distinct evolutionary advantage. For example, the Sak1 homolog of the pathogenic fungus C. glabrata, whose Snf1 undergoes regulated T-loop phosphorylation (51), has an ideal PKA recognition motif that directly corresponds to the motif addressed in this work. Moreover, this motif is embedded in a 22-amino-acid region that stands out among the surrounding sequence in sharing more than 80% identity with the region encompassing Ser1074 of S. cerevisiae Sak1, presumably defining a conserved regulatory box. Similar boxes are also found in the Sak1 homologs of fungi representing several genera. Our results also raise the possibility that S. cerevisiae Sak1 is additionally phosphorylated on the nearby Ser1139 residue within another putative PKA recognition motif and that the Ser1074 and Ser1139 phosphorylation events cooperate to negatively control Sak1.
Sak1, however, is not the only relevant target of the PKA pathway. Sequence analysis indicates that another Snf1-activating kinase, Tos3, has an ideal PKA recognition consensus, making it a possible substrate. The third Snf1-activating kinase, Elm1, does not have any immediately compelling PKA motifs but could be regulated by PKA either indirectly or directly by phosphorylation on a near-consensus or nonconsensus site(s). Interestingly, recent evidence indicates that mammalian AMPK is phosphorylated by PKA on a nonconsensus serine that is immediately adjacent to its critical T-loop threonine, and this phosphorylation antagonizes AMPK activation (13). A serine residue is present at the equivalent position in S. cerevisiae Snf1, raising the possibility that PKA could similarly antagonize Snf1 directly by inhibitory T-loop phosphorylation. It is also possible that PKA regulates Thr210 dephosphorylation by Reg1-Glc7, although such a mechanism would not have been detected in our mutant search. In addition, PKA could regulate the Sit4 protein phosphatase, which has been recently reported to contribute to Thr210 dephosphorylation (58). Finally, we cannot exclude the possibility that PKA, whose catalytic isoforms can have opposing physiological functions, plays not only negative but also positive roles in Snf1 regulation. Further experiments will be required to address these and other possibilities in order to fully reconstruct the potentially very complex mechanistic picture.
The molecular mechanisms by which nutrient availability modulates Snf1 activity are not completely understood. PKA figures prominently in glucose signaling and collects its regulatory inputs from multiple effectors, including the Ras, RasGAP, and RasGEF proteins; the Gpr1/Gpa2 G-protein-coupled receptor system; and others (for reviews, see references 59 and 82). The involvement of PKA could therefore account for a portion of the glucose signal affecting Snf1. In addition to glucose signaling, Snf1 participates in responses to other stress conditions, such as nitrogen limitation and exposure to rapamycin (41, 52, 53). PKA is involved in nitrogen and TOR signaling (see reference 82 for a review) and could mediate a relevant signal(s) impinging on Snf1. Consistent with this possibility, we found here that, like snf1, the ira mutations suppress reg1 for rapamycin hypersensitivity.
Our findings lend further support to the value of the yeast Snf1 pathway as a model system for studying cancer-related signaling. The link between AMPK and cancer first transpired from the identification of tumor suppressor LKB1 as an AMPK-activating kinase (30, 78) and tumor suppressor TSC2 as an AMPK target (33). Activated AMPK functions to downregulate mTOR and to stimulate p53-mediated cell cycle arrest (7, 18, 33, 35). AMPK has therefore been proposed to act as a metabolic checkpoint that coordinates cell growth and proliferation with energy availability (35). Our results extend the idea that Snf1 has a similar function in yeast (83). We find it fascinating that the slow-growth phenotype caused by hyperactivation of the Snf1 “tumor suppressor” pathway (reg1Δ) can be so prominently reversed by activation of the Ras “oncogene” pathway in this simple eukaryote. Interestingly, a report using a mouse melanoma model showed that growth factor-activated Ras and oncogenic BRAFV600E downregulate the LKB1-AMPK cascade by a mechanism involving p90RSK-dependent phosphorylation of LKB1 (14); LKB1 is also phosphorylated by PKA (2, 12, 62). We conclude that the Snf1-dependent slow growth of the yeast reg1Δ mutant is not only a useful genetic tool, but also an important phenotype in its own right, reflecting an energy-saving function of Snf1. The exact mechanisms by which activation of Snf1 decelerates growth in yeast are not fully understood but likely involve negative effects on the cell cycle, since the slow growth caused by reg1Δ can be partially rescued by overexpression of the cell cycle progression kinase Cdc28 (our unpublished results). Further analysis of reg1Δ suppressors in yeast could provide new clues to the mechanisms that couple cell growth and proliferation with nutrient availability in eukaryotes.
ACKNOWLEDGMENTS
This work was supported by National Science Foundation grant MCB-0818837 (to S.K.). L.B. was a UWM Advanced Opportunity Program Fellow.
We thank M. Carlson and M. Johnston for reagents and D. Saffarini for helpful comments on the manuscript.
Footnotes
Published ahead of print 2 December 2011
REFERENCES
- 1. Asano S, et al. 2006. Direct phosphorylation and activation of a Nim1-related kinase Gin4 by Elm1 in budding yeast. J. Biol. Chem. 281:27090–27098 [DOI] [PubMed] [Google Scholar]
- 2. Barnes AP, et al. 2007. LKB1 and SAD kinases define a pathway required for the polarization of cortical neurons. Cell 129:549–563 [DOI] [PubMed] [Google Scholar]
- 3. Barral Y, Parra M, Bidlingmaier S, Snyder M. 1999. Nim1-related kinases coordinate cell cycle progression with the organization of the peripheral cytoskeleton in yeast. Genes Dev. 13:176–187 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Bertram PG, et al. 2002. Convergence of TOR-nitrogen and Snf1-glucose signaling pathways onto Gln3. Mol. Cell. Biol. 22:1246–1252 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Blacketer MJ, Koehler CM, Coats SG, Myers AM, Madaule P. 1993. Regulation of dimorphism in Saccharomyces cerevisiae: involvement of the novel protein kinase homolog Elm1p and protein phosphatase 2A. Mol. Cell. Biol. 13:5567–5581 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Broach JR. 1991. RAS genes in Saccharomyces cerevisiae: signal transduction in search of a pathway. Trends Genet. 7:28–33 [DOI] [PubMed] [Google Scholar]
- 7. Buzzai M, et al. 2007. Systemic treatment with the antidiabetic drug metformin selectively impairs p53-deficient tumor cell growth. Cancer Res. 67:6745–6752 [DOI] [PubMed] [Google Scholar]
- 8. Cannon JF, Pringle JR, Fiechter A, Khalil M. 1994. Characterization of glycogen-deficient glc mutants of Saccharomyces cerevisiae. Genetics 136:485–503 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Cannon JF, Tatchell K. 1987. Characterization of Saccharomyces cerevisiae genes encoding subunits of cyclic AMP-dependent protein kinase. Mol. Cell. Biol. 7:2653–2663 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Carlson M, Osmond BC, Botstein D. 1981. Mutants of yeast defective in sucrose utilization. Genetics 98:25–40 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Ciriacy M. 1977. Isolation and characterization of yeast mutants defective in intermediary carbon metabolism and in carbon catabolite derepression. Mol. Gen. Genet. 154:213–220 [DOI] [PubMed] [Google Scholar]
- 12. Collins SP, Reoma JL, Gamm DM, Uhler MD. 2000. LKB1, a novel serine/threonine protein kinase and potential tumour suppressor, is phosphorylated by cAMP-dependent protein kinase (PKA) and prenylated in vivo. Biochem. J. 345:673–680 [PMC free article] [PubMed] [Google Scholar]
- 13. Djouder N, et al. 2010. PKA phosphorylates and inactivates AMPKalpha to promote efficient lipolysis. EMBO J. 29:469–481 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Esteve-Puig R, Canals F, Colome N, Merlino G, Recio JA. 2009. Uncoupling of the LKB1-AMPKalpha energy sensor pathway by growth factors and oncogenic BRAF. PLoS One 4:e4771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Fogarty S, Hardie DG. 2010. Development of protein kinase activators: AMPK as a target in metabolic disorders and cancer. Biochim. Biophys. Acta 1804:581–591 [DOI] [PubMed] [Google Scholar]
- 16. Gil R, Seeling JM. 1999. Characterization of Saccharomyces cerevisiae strains expressing ira1 mutant alleles modeled after disease-causing mutations in NF1. Mol. Cell. Biochem. 202:109–118 [DOI] [PubMed] [Google Scholar]
- 17. Golemis EA, Serebriiskii I, Gjuris J, Brent R. 1997. Current protocols in molecular biology. Wiley, New York, NY [Google Scholar]
- 18. Gwinn DM, et al. 2008. AMPK phosphorylation of raptor mediates a metabolic checkpoint. Mol. Cell 30:214–226 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Halme A, Bumgarner S, Styles C, Fink GR. 2004. Genetic and epigenetic regulation of the FLO gene family generates cell-surface variation in yeast. Cell 116:405–415 [DOI] [PubMed] [Google Scholar]
- 20. Harashima T, Heitman J. 2002. The Galpha protein Gpa2 controls yeast differentiation by interacting with kelch repeat proteins that mimic Gbeta subunits. Mol. Cell 10:163–173 [DOI] [PubMed] [Google Scholar]
- 21. Hardie DG. 2007. AMP-activated protein kinase as a drug target. Annu. Rev. Pharmacol. Toxicol. 47:185–210 [DOI] [PubMed] [Google Scholar]
- 22. Hardie DG. 2007. AMP-activated/SNF1 protein kinases: conserved guardians of cellular energy. Nat. Rev. Mol. Cell Biol. 8:774–785 [DOI] [PubMed] [Google Scholar]
- 23. Hardie DG. 2007. AMPK and SNF1: snuffing out stress. Cell Metab. 6:339–340 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Hawley SA, et al. 2003. Complexes between the LKB1 tumor suppressor, STRADalpha/beta and MO25alpha/beta are upstream kinases in the AMP-activated protein kinase cascade. J. Biol. 2:28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Hawley SA, et al. 2005. Calmodulin-dependent protein kinase kinase-beta is an alternative upstream kinase for AMP-activated protein kinase. Cell Metab. 2:9–19 [DOI] [PubMed] [Google Scholar]
- 26. Hedbacker K, Carlson M. 2008. SNF1/AMPK pathways in yeast. Front. Biosci. 13:2408–2420 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Hedbacker K, Hong SP, Carlson M. 2004. Pak1 protein kinase regulates activation and nuclear localization of Snf1-Gal83 protein kinase. Mol. Cell. Biol. 24:8255–8263 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Hedbacker K, Townley R, Carlson M. 2004. Cyclic AMP-dependent protein kinase regulates the subcellular localization of Snf1-Sip1 protein kinase. Mol. Cell. Biol. 24:1836–1843 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Hong SP, Carlson M. 2007. Regulation of snf1 protein kinase in response to environmental stress. J. Biol. Chem. 282:16838–16845 [DOI] [PubMed] [Google Scholar]
- 30. Hong SP, Leiper FC, Woods A, Carling D, Carlson M. 2003. Activation of yeast Snf1 and mammalian AMP-activated protein kinase by upstream kinases. Proc. Natl. Acad. Sci. U. S. A. 100:8839–8843 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Hong SP, Momcilovic M, Carlson M. 2005. Function of mammalian LKB1 and Ca2+/calmodulin-dependent protein kinase kinase {alpha} as Snf1-activating kinases in yeast. J. Biol. Chem. 280:21804–21809 [DOI] [PubMed] [Google Scholar]
- 32. Hurley RL, et al. 2005. The Ca2+/calmodulin-dependent protein kinase kinases are AMP-activated protein kinase kinases. J. Biol. Chem. 280:29060–29066 [DOI] [PubMed] [Google Scholar]
- 33. Inoki K, Zhu T, Guan KL. 2003. TSC2 mediates cellular energy response to control cell growth and survival. Cell 115:577–590 [DOI] [PubMed] [Google Scholar]
- 34. Jiang R, Carlson M. 1997. The Snf1 protein kinase and its activating subunit, Snf4, interact with distinct domains of the Sip1/Sip2/Gal83 component in the kinase complex. Mol. Cell. Biol. 17:2099–2106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Jones RG, et al. 2005. AMP-activated protein kinase induces a p53-dependent metabolic checkpoint. Mol. Cell 18:283–293 [DOI] [PubMed] [Google Scholar]
- 36. Kartasheva NN, Kuchin SV, Benevolensky SV. 1996. Genetic aspects of carbon catabolite repression of the STA2 glucoamylase gene in Saccharomyces cerevisiae. Yeast 12:1297–1300 [DOI] [PubMed] [Google Scholar]
- 37. Kennelly PJ, Krebs EG. 1991. Consensus sequences as substrate specificity determinants for protein kinases and protein phosphatases. J. Biol. Chem. 266:15555–15558 [PubMed] [Google Scholar]
- 38. Kuchin S, Carlson M. 1998. Functional relationships of Srb10-Srb11 kinase, carboxy-terminal domain kinase CTDK-I, and transcriptional corepressor Ssn6-Tup1. Mol. Cell. Biol. 18:1163–1171 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Kuchin S, Treich I, Carlson M. 2000. A regulatory shortcut between the Snf1 protein kinase and RNA polymerase II holoenzyme. Proc. Natl. Acad. Sci. U. S. A. 97:7916–7920 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Kuchin S, Vyas VK, Carlson M. 2002. Snf1 protein kinase and the repressors Nrg1 and Nrg2 regulate FLO11, haploid invasive growth, and diploid pseudohyphal differentiation. Mol. Cell. Biol. 22:3994–4000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Kuchin S, Vyas VK, Carlson M. 2003. Role of the yeast Snf1 protein kinase in invasive growth. Biochem. Soc. Trans. 31:175–177 [DOI] [PubMed] [Google Scholar]
- 42. Kuchin S, Vyas VK, Kanter E, Hong SP, Carlson M. 2003. Std1p (Msn3p) positively regulates the Snf1 kinase in Saccharomyces cerevisiae. Genetics 163:507–514 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Kuchin SV, Kartasheva NN, Benevolensky SV. 1993. Genes required for derepression of an extracellular glucoamylase gene, STA2, in the yeast Saccharomyces. Yeast 9:533–541 [DOI] [PubMed] [Google Scholar]
- 44. Liu Y, Xu X, Carlson M. 2011. Interaction of SNF1 protein kinase with its activating kinase Sak1. Eukaryot. Cell 10:313–319 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Lorenz MC, et al. 2000. The G protein-coupled receptor gpr1 is a nutrient sensor that regulates pseudohyphal differentiation in Saccharomyces cerevisiae. Genetics 154:609–622 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. McCartney RR, Rubenstein EM, Schmidt MC. 2005. Snf1 kinase complexes with different beta subunits display stress-dependent preferences for the three Snf1-activating kinases. Curr. Genet. 47:335–344 [DOI] [PubMed] [Google Scholar]
- 47. McCartney RR, Schmidt MC. 2001. Regulation of Snf1 kinase. Activation requires phosphorylation of threonine 210 by an upstream kinase as well as a distinct step mediated by the Snf4 subunit. J. Biol. Chem. 276:36460–36466 [DOI] [PubMed] [Google Scholar]
- 48. Momcilovic M, Hong SP, Carlson M. 2006. Mammalian TAK1 activates Snf1 protein kinase in yeast and phosphorylates AMP-activated protein kinase in vitro. J. Biol. Chem. 281:25336–25343 [DOI] [PubMed] [Google Scholar]
- 49. Nath N, McCartney RR, Schmidt MC. 2002. Purification and characterization of Snf1 kinase complexes containing a defined Beta subunit composition. J. Biol. Chem. 277:50403–50408 [DOI] [PubMed] [Google Scholar]
- 50. Nath N, McCartney RR, Schmidt MC. 2003. Yeast Pak1 kinase associates with and activates Snf1. Mol. Cell. Biol. 23:3909–3917 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Orlova M, Barrett L, Kuchin S. 2008. Detection of endogenous Snf1 and its activation state: application to Saccharomyces and Candida species. Yeast 25:745–754 [DOI] [PubMed] [Google Scholar]
- 52. Orlova M, Kanter E, Krakovich D, Kuchin S. 2006. Nitrogen availability and TOR regulate the Snf1 protein kinase in Saccharomyces cerevisiae. Eukaryot. Cell 5:1831–1837 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Orlova M, Ozcetin H, Barrett L, Kuchin S. 2010. Roles of the Snf1-activating kinases during nitrogen limitation and pseudohyphal differentiation in Saccharomyces cerevisiae. Eukaryot. Cell 9:208–214 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Ozcan S, Vallier LG, Flick JS, Carlson M, Johnston M. 1997. Expression of the SUC2 gene of Saccharomyces cerevisiae is induced by low levels of glucose. Yeast 13:127–137 [DOI] [PubMed] [Google Scholar]
- 55. Pan X, Heitman J. 1999. Cyclic AMP-dependent protein kinase regulates pseudohyphal differentiation in Saccharomyces cerevisiae. Mol. Cell. Biol. 19:4874–4887 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Rose MD, Winston F, Hieter P. 1990. Methods in yeast genetics: a laboratory course manual. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY [Google Scholar]
- 57. Rubenstein EM, McCartney RR, Schmidt MC. 2006. Regulatory domains of Snf1-activating kinases determine pathway specificity. Eukaryot. Cell 5:620–627 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Ruiz A, Xu X, Carlson M. 2011. Roles of two protein phosphatases, Reg1-Glc7 and Sit4, and glycogen synthesis in regulation of SNF1 protein kinase. Proc. Natl. Acad. Sci. U. S. A. 108:6349–6354 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Santangelo GM. 2006. Glucose signaling in Saccharomyces cerevisiae. Microbiol. Mol. Biol. Rev. 70:253–282 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Schmelzle T, Beck T, Martin DE, Hall MN. 2004. Activation of the RAS/cyclic AMP pathway suppresses a TOR deficiency in yeast. Mol. Cell. Biol. 24:338–351 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Shabb JB. 2001. Physiological substrates of cAMP-dependent protein kinase. Chem. Rev. 101:2381–2411 [DOI] [PubMed] [Google Scholar]
- 62. Shelly M, Cancedda L, Heilshorn S, Sumbre G, Poo MM. 2007. LKB1/STRAD promotes axon initiation during neuronal polarization. Cell 129:565–577 [DOI] [PubMed] [Google Scholar]
- 63. Sikorski RS, Hieter P. 1989. A system of shuttle vectors and yeast host strains designed for efficient manipulation of DNA in Saccharomyces cerevisiae. Genetics 122:19–27 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Stanhill A, Schick N, Engelberg D. 1999. The yeast ras/cyclic AMP pathway induces invasive growth by suppressing the cellular stress response. Mol. Cell. Biol. 19:7529–7538 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Strudwick N, Brown M, Parmar VM, Schroder M. 2010. Ime1 and Ime2 are required for pseudohyphal growth of Saccharomyces cerevisiae on nonfermentable carbon sources. Mol. Cell. Biol. 30:5514–5530 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Sutherland CM, et al. 2003. Elm1p is one of three upstream kinases for the Saccharomyces cerevisiae SNF1 complex. Curr. Biol. 13:1299–1305 [DOI] [PubMed] [Google Scholar]
- 67. Szkotnicki L, Crutchley JM, Zyla TR, Bardes ES, Lew DJ. 2008. The checkpoint kinase Hsl1p is activated by Elm1p-dependent phosphorylation. Mol. Biol. Cell 19:4675–4686 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Tanaka K, et al. 1990. S. cerevisiae genes IRA1 and IRA2 encode proteins that may be functionally equivalent to mammalian ras GTPase activating protein. Cell 60:803–807 [DOI] [PubMed] [Google Scholar]
- 69. Tanaka K, et al. 1990. IRA2, a second gene of Saccharomyces cerevisiae that encodes a protein with a domain homologous to mammalian ras GTPase-activating protein. Mol. Cell. Biol. 10:4303–4313 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Thompson-Jaeger S, Francois J, Gaughran JP, Tatchell K. 1991. Deletion of SNF1 affects the nutrient response of yeast and resembles mutations which activate the adenylate cyclase pathway. Genetics 129:697–706 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Toda T, et al. 1987. Cloning and characterization of BCY1, a locus encoding a regulatory subunit of the cyclic AMP-dependent protein kinase in Saccharomyces cerevisiae. Mol. Cell. Biol. 7:1371–1377 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Toda T, Cameron S, Sass P, Zoller M, Wigler M. 1987. Three different genes in S. cerevisiae encode the catalytic subunits of the cAMP-dependent protein kinase. Cell 50:277–287 [DOI] [PubMed] [Google Scholar]
- 73. Toda T, et al. 1985. In yeast, RAS proteins are controlling elements of adenylate cyclase. Cell 40:27–36 [DOI] [PubMed] [Google Scholar]
- 74. Treitel MA, Kuchin S, Carlson M. 1998. Snf1 protein kinase regulates phosphorylation of the Mig1 repressor in Saccharomyces cerevisiae. Mol. Cell. Biol. 18:6273–6280 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Tu J, Carlson M. 1994. The GLC7 type 1 protein phosphatase is required for glucose repression in Saccharomyces cerevisiae. Mol. Cell. Biol. 14:6789–6796 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Tu J, Carlson M. 1995. REG1 binds to protein phosphatase type 1 and regulates glucose repression in Saccharomyces cerevisiae. EMBO J. 14:5939–5946 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Woods A, et al. 2005. Ca2+/calmodulin-dependent protein kinase kinase-beta acts upstream of AMP-activated protein kinase in mammalian cells. Cell Metab. 2:21–33 [DOI] [PubMed] [Google Scholar]
- 78. Woods A, et al. 2003. LKB1 is the upstream kinase in the AMP-activated protein kinase cascade. Curr. Biol. 13:2004–2008 [DOI] [PubMed] [Google Scholar]
- 79. Xu GF, et al. 1990. The catalytic domain of the neurofibromatosis type 1 gene product stimulates ras GTPase and complements ira mutants of S. cerevisiae. Cell 63:835–841 [DOI] [PubMed] [Google Scholar]
- 80. Yang X, Jiang R, Carlson M. 1994. A family of proteins containing a conserved domain that mediates interaction with the yeast SNF1 protein kinase complex. EMBO J. 13:5878–5886 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81. Zaman S, Lippman SI, Schneper L, Slonim N, Broach JR. 2009. Glucose regulates transcription in yeast through a network of signaling pathways. Mol. Syst. Biol. 5:245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82. Zaman S, Lippman SI, Zhao X, Broach JR. 2008. How Saccharomyces responds to nutrients. Annu. Rev. Genet. 42:27–81 [DOI] [PubMed] [Google Scholar]
- 83. Zhang J, Vemuri G, Nielsen J. 2010. Systems biology of energy homeostasis in yeast. Curr. Opin. Microbiol. 13:382–388 [DOI] [PubMed] [Google Scholar]
- 84. Zimmermann FK, Kaufmann I, Rasenberger H, Haubetamann P. 1977. Genetics of carbon catabolite repression in Saccharomycess cerevisiae: genes involved in the derepression process. Mol. Gen. Genet. 151:95–103 [DOI] [PubMed] [Google Scholar]