Abstract
Enteric fever is an invasive life-threatening systemic disease caused by the Salmonella enterica human-adapted serovars Typhi and Paratyphi. Increasing incidence of infections with Salmonella enterica serovar Paratyphi A and the spreading of its antibiotic-resistant derivates pose a significant health concern in some areas of the world. Herein, we describe a molecular and phenotypic characterization of an S. Paratyphi A strain accounted for a recent paratyphoid outbreak in Nepal that affected at least 37 travelers. Pulsed-field gel electrophoresis analysis of the outbreak isolates revealed one genetic clone (pulsotype), confirming a single infecting source. Genetic profiling of the outbreak strain demonstrated the contribution of specific bacteriophages as a prime source of genetic diversity among clinical isolates of S. Paratyphi A. Phenotypic characterization in comparison with the S. Paratyphi A ATCC 9150 reference sequenced strain showed differences in flagellar morphology and increased abilities of the outbreak strain with respect to its motility, invasion into nonphagocytic cells, intracellular multiplication, survival within macrophages, and higher induction of interleukin-8 (IL-8) secreted by host cells. Collectively, these differences suggest an enhanced virulence potential of this strain and demonstrate an interesting phenotypic variation among S. Paratyphi A isolates. In vivo profiling of 16 inflammatory cytokines in patients infected with the outbreak strain revealed a common profile of a remarkable gamma interferon (IFN-γ) induction together with elevated concentrations of tumor necrosis factor alpha (TNF-α), IL-6, IL-8, IL-10, and IL-15, but not IL-12, which was previously demonstrated as elevated in nontyphoidal Salmonella infections. This apparent profile implies a distinct immune response to paratyphoid infections.
INTRODUCTION
Salmonella enterica is a Gram-negative, facultative intracellular pathogen posing a major public health concern worldwide. The single species S. enterica consists of over 2,500 closely related serovars, which share 96 to 99% sequence similarity (14). Salmonella, like many other bacterial pathogens, harbors clusters of virulence genes, known as Salmonella pathogenicity islands (SPIs), that have been acquired via horizontal gene transfer. These genomic clusters are considered to be “quantum leaps” in the evolution of Salmonella (18) and play a fundamental role in its pathogenesis (20) and host specificity (1).
Several Salmonella enterica serovars, such as S. Typhimurium and S. Enteritidis, are ubiquitous and “generalist” pathogens that can infect a broad range of animal hosts, causing different clinical manifestations, including gastroenteritis or occasionally septicemia in humans, lethal diarrhea in calves, or a systemic disease in genetically susceptible mice. In contrast, S. Typhi and S. Paratyphi (collectively referred to as typhoidal serovars) are host specific, causing an enteric fever disease in humans (38). Enteric fever is an invasive life-threatening systemic disease with a global annual estimation of over 25 million cases, resulting in more than 200,000 deaths (9). In recent years, for unknown reasons, the incidence of infections with serovar Paratyphi A is increasing, and in some regions of the globe (particularly in south-east Asia), it is accountable for up to 50% of all enteric fever cases (33, 35). Enteric fever is a mostly food- and waterborne disease, and like all Salmonella infections is transmitted by the fecal-oral route. Initial gastrointestinal infection causes brief, often asymptomatic enteritis followed by invasion through the gut mucosa to underlying macrophages and lymphoid tissue. Bacteria can survive and multiply intracellularly within lymphoid follicles, mesenteric lymph nodes, and the mononuclear phagocyte system. Systemic infection with bacteremia and fever develops at 8 to 14 days postinfection, accompanied by bacterial spreading to systemic sites such as the liver, spleen, and bone marrow. Secondary infection of the small bowel can occur via secretion of bacteria through the enterohepatic cycle (reviewed in reference 17).
Due to the lack of a suitable animal model, much of our understanding of typhoidal serovars' pathogenesis is extrapolated from the susceptible mouse strains infection with the nontyphoidal serovar (NTS), which normally does not cause a systemic disease in humans. Although this model has been crucial in understanding many aspects of Salmonella pathogenicity, conclusions regarding the virulence of S. Paratyphi in humans and host response to the infection must be carefully interpreted.
During October 2009, while traveling to Nepal, 38 healthy young Israeli travelers developed enteric fever caused by an S. Paratyphi A strain. Herein we describe molecular and cellular characterization of this outbreak isolate in comparison to the characterized S. Paratyphi A ATCC 9150 strain. In addition, we studied the human immune response to the S. Paratyphi A infection and determined the inflammatory cytokine profile during the acute phase of the disease. Our results highlight patterns of genetic variation among S. Paratyphi A strains, demonstrate differences in virulence capabilities in vitro, and reveal important aspects of the human immune response to a paratyphoid infection in vivo.
MATERIALS AND METHODS
Bacterial strains and growth conditions.
Bacterial strains utilized in this study are listed in Table 1 and include 24 S. Paratyphi A blood isolates obtained from the 2009 outbreak and 13 sporadic S. Paratyphi A isolates obtained during 2003 to 2009 (mostly from returning travelers). Bacterial cultures were routinely maintained in Luria-Bertani (LB; BD Difco) liquid medium at 37°C. LB or xylose lysine deoxycholate (XLD; BD Difco) agar plates were used when appropriate.
Table 1.
Bacterial strains used in this study
| Strain | Genotype and description | Source (reference) |
|---|---|---|
| S. Paratyphi A | ||
| ATCC 9150 | Sequenced reference strain | SGSCa (31) |
| AKU 12601 | Sequenced strain | SGSC (21) |
| S. Typhi CT18 | SGSC (37) | |
| S. Typhimurium | ||
| DT104 96-5227 | Harbors SGI-1 | National Microbiology Laboratory, Public Health Agency of Canada (2) |
| 14028s | SGSC (23) | |
| SARC-13 | Harbors HPI | SGSC (3) |
| E DH5α | F− ϕ80lacZΔM15 Δ(lacZYA-argF)U169 deoR recA1 endA1 hsdR17(rk− mk+) supE44 thi-1 gyrA96 relA1 λ− | Lab collection |
| Clinical isolates of S. Paratyphi A | ||
| 3182 | 2009 Nepal outbreak | Stool |
| 36056/7 | 2007 traveler from Nepal | Blood |
| 45157 | 2009 Nepal outbreak | Blood |
| 45167 | 2009 Nepal outbreak | Blood |
| 45375 | 2009 Nepal outbreak | Blood |
| 45792 | 2009 Nepal outbreak | Blood |
| 45838 | 2009 Nepal outbreak | Blood |
| 45842/7 | 2007 traveler from Nepal | Blood |
| 46162 | 2009 Nepal outbreak | Blood |
| 46167 | 2009 Nepal outbreak | Blood |
| 46185 | 2009 Nepal outbreak | Blood |
| 46236 | 2009 Nepal outbreak | Blood |
| 46562 | 2009 Nepal outbreak | Blood |
| 46900 | 2009 Nepal outbreak | Blood |
| 47412 | 2009 Nepal outbreak | Blood |
| 50661 | 2009 infected lab personnel | Blood |
| 50676 | 2009 Nepal outbreak | Blood |
| 51190 | 2009 traveler from Nepal | Blood |
| 83698 | 2003 traveler from India | Blood |
| 83753 | 2003 traveler from India | Blood |
| 93223 | 2004 traveler from Romania | Blood |
| 105493 | 2006 traveler from Thailand and Nepal | Blood |
| 108003 | 2007 traveler from India | Blood |
| 108599 | 2007 traveler from India | Stool |
| 113498 | 2008 traveler from Sri Lanka | Stool |
| 118239 | 2008 traveler from India | Stool |
| 119989 | 2008 traveler from Thailand and India | Blood |
| 124597 | 2009 traveler from India | Blood |
| 126142 | 2009 Nepal outbreak | Blood |
| 126206 | 2009 Nepal outbreak | Blood |
| 126239 | 2009 Nepal outbreak | Blood |
| 126240 | 2009 Nepal outbreak | Blood |
| 126471 | 2009 Nepal outbreak | Blood |
| 126495 | 2009 Nepal outbreak | Blood |
| 126496 | 2009 Nepal outbreak | Blood |
| 126497 | 2009 Nepal outbreak | Blood |
| 126498 | 2009 Nepal outbreak | Blood |
| 126499 | 2009 Nepal outbreak | Blood |
| 126640 | 2009 Nepal outbreak | Blood |
SGCS, Salmonella Genetic Stock Center, University of Calgary, Calgary, Alberta, Canada.
Genotyping by PFGE.
Pulsed-field gel electrophoresis (PFGE) analysis was carried out according to the standardized Salmonella protocol defined by CDC PulseNet (43), using S. Braenderup H9812 as a molecular standard. XbaI-digested Salmonella DNA embedded in agarose plugs was subjected to PFGE analysis at 14°C in a CHEF DR III system (Bio-Rad Laboratories) using the following protocol: voltage, 6 V/cm for 19 h; initial pulse, 2.2 s; final pulse, 63.8 s; angle, 120°; buffer, 0.5× Tris-borate-EDTA. PFGE-generated DNA profiles were processed by the Bionumerics software V 5.1 (Applied Maths, Sint-Martens Latem, Belgium) using the Dice coefficients with a 1% position tolerance and optimization values. Cluster analysis was performed by the unweighted-pair group mean analysis (UPGMA) method.
Southern blot hybridization.
Primers used in this study are listed in Table 2. DNA primers were purchased from IDT, and PCR was carried out using ReddyMix PCR (Thermo Scientific) or PfuUltra II fusion HS DNA polymerase (Stratagene). For Southern blot analysis, 1 μg of genomic DNA was digested at 37°C for 16 to 18 h with PstI, subjected to electrophoresis in 1.0% agarose gels before being capillary transferred, and cross-linked onto Hybond-N membranes (Amersham Biosciences). Genomic DNA from Escherichia coli DH5α was included as a negative control in all hybridizations. S. Typhi CT18 and S. Typhimurium DT104, 14028s, and SARC-13 were used as positive controls. Southern blots were processed using the digoxigenin (DIG) DNA labeling and detection kit (Roche Applied Sciences), followed by an anti-DIG detection according to the manufacturer's protocol.
Table 2.
Primers used in this study
| Primer name | Sequence | Use |
|---|---|---|
| spi7vex-F | CGACATTTTTCCTGCTTTCG | Southern blot probe for SPI-7 |
| spi7vex-R | ATGCGGCTTTCACCTTAGC | |
| spi8-F | AAATCAGGTAAGGCATCAAAGG | Southern blot probe for SPI-8 |
| spi8-R | CTCAGGTGTTCCATCACTTTCC | |
| spi10sef-F | TTCATTGTCTCTGTTTTTCTGATTG | Southern blot probe for SPI-10 |
| spi10sef-R | TTAAATAGGTATCAACGGGAATG | |
| spi15-F | CTTGCACCTTTATCATCACTGG | Southern blot probe for SPI-15 |
| spi15-R | AGTGTCTCCCTCCTGAACTGC | |
| spi17-F | GCAATAGCGGTTTTATCTGTGG | Southern blot probe for SPI-17 |
| spi17-R | GTTAAATGTCCCCAATCAACG | |
| SGI1-F | TTTCGCCAATCGAATAATCC | Southern blot probe for SGI-1 |
| SGI1-R | ACTTGAACCCAATGCTCTGC | |
| hpi1-F | GACCTGACCTGGCATTTAACC | Southern blot probe for HPI |
| hpi1-R | GCATTGCTTAATGTCTGCATCC | |
| sspH1-F | CCGGTAACTGTCAGATCAGGTC | PCR for sspH1 in Gifsy-3 |
| sspH1-R | TCCCTGTTACACTGAATATTCTCC | |
| ssek3-F | TATCAATCTCAAATCATGG | PCR for ssek3 in ST64B |
| ssek3-R | CGCGTTTATATCATACGTTTGC | |
| AbsentPhages-F | TTAATCGCGCGAAAGAAGC | PCR for the region SPA2552-SPA2626 |
| AbsentPhages-R | TTCAGCGTAATCCGAAGAC | |
| SPA-1-F | GCACAGGGCCAAACTGAAGGATGG | Presence of phage SPA-1 |
| SPA-1-R | GAATTGCCGGAACAGCGGCG | |
| new SPA-2-F | CCTAAGTGATGCCCTAGACACATGC | Presence of phage SPA-2 |
| new SPA-2-R | CAACGGGGAGTTGAAAAATATGCGC | |
| SPA-3-F | GCGCACTTTATCTGCGCCGC | Presence of phage SPA-3 |
| SPA-3-R | ACTTTCGACCACGCCAGCCG |
CGH.
For comparative genomic hybridization (CGH), genomic DNA from S. Typhimurium LT2 and S. Paratyphi A (clone 45157) was extracted from overnight cultures grown in LB using the GenElute kit (Sigma-Aldrich) according to the manufacturer's instructions. DNA labeling and hybridization to the STv7E pan- Salmonella microarray (http://www.sdibr.org/Faculty/mcclelland/mcclelland-lab/mcclelland-protocols) were performed as previously described (41). An Agilent microarray scanner G2505B was used for image acquisition, and signal intensities were quantified with the Spotreader software (Niles Scientific). Data normalization, analysis, and determination of the presence or absence of genes are described elsewhere (41).
Tissue culture conditions and bacterial infection.
The human epithelial cell line HeLa and the murine macrophage-like RAW264.7 cell line were cultured in a high-glucose (4.5 g/liter) Dulbecco's modified Eagle medium (DMEM) supplemented with 10% heat-inactivated fetal bovine serum (FBS), 1 mM pyruvate, and 2 mM l-glutamine. The human colonic adenocarcinoma Caco-2 cell line was grown in DMEM–F-12 medium supplemented with 20% FBS and 2 mM l-glutamine. All cell lines were cultured at 37°C in a humidified atmosphere with 5% CO2. Epithelial cells and RAW264.7 macrophages were seeded at 5 × 104 and 2.5 × 105 cells/ml, respectively, in a 24-well tissue culture dish 18 to 24 h prior to bacterial infection, and experiments were carried out using the gentamicin protection assay as previously described (15). HeLa and Caco-2 cells were infected at a multiplicity of infection (MOI) of ∼1:50 with Salmonella strains that had been subcultured from an overnight culture and grown for 3 h to late logarithmic phase under aerobic conditions or with stationary-phase cultures grown under microaerophilic conditions. RAW264.7 cells were infected at an MOI of 1:10 using overnight stationary-phase-grown cultures. At the desired time points postinfection (p.i.), cells were washed three times with phosphate-buffered saline (PBS) and harvested by addition of lysis buffer (0.1% SDS, 1% Triton X-100 in PBS). Appropriate dilutions were plated on LB plates for bacterial enumeration by CFU count. Salmonella invasion was determined by the number of intracellular Salmonella cells at 2 h p.i. divided by the number of infecting bacteria. Survival and intracellular multiplication (in macrophages and in nonphagocytic cells, respectively) were determined by the number of recovered intracellular salmonellae at 24 h p.i. divided by the number of invading salmonellae at 2 h p.i.
Motility assay.
Ten microliters of overnight Salmonella cultures grown in LB broth were placed onto 0.3% agar LB plates. The plates were incubated for 6 h at 37°C without being inverted.
Transmission electron microscopy.
Salmonella strains grown on soft agar plates were suspended in PBS and adsorbed onto 200-mesh Formvar/carbon-coated copper grids. The grids were negatively stained with 2% aqueous uranyl acetate for 30 s. Images were obtained using a JEOL-1200EX (Jeol, Japan) transmission electron microscope at the Tel-Aviv University Electron Microscopy Facility.
Leukocyte count.
White blood cell (WBC) counts were compared between two groups of patients: 24 patients (14 males and 10 females; average age, 27 ± 7 years) with positive blood cultures for S. Paratyphi A who were hospitalized in the Sheba Medical Center between 2003 and 2010 and 23 patients (14 females, 9 males; average age, 55 ± 18 years) with an E. coli-positive blood culture who were hospitalized between 2010 and 2011. Hematological data were retrieved from the hospital laboratory reports documenting the first blood specimens drawn from the patients with their administration. Data retrieval and analysis were done according to the local ethics committee-approved protocol.
Cytokine analysis.
In vitro IL-8 analysis was performed following Caco-2 cell infection with S. Paratyphi A strains as described above. Secreted IL-8 concentrations were determined 18 h p.i. using a Q-Plex array chemiluminescent IL-8 kit (Quansys Biosciences), according to the manufacturer's protocol. In vivo cytokine analysis was done using blood samples collected from 10 S. Paratyphi A-infected patients. The first blood sample was taken from all patients on admission during bacteremia and prior to antibiotic treatment. A second, matching blood sample was collected from the same patients 12 to 14 weeks after recovery. Serum was separated and kept at −80°C until final analysis. Cytokines were analyzed using the Q-Plex human cytokine screen system (Quansys Biosciences) based on a multiplex enzyme-linked immunosorbent assay (ELISA) approach according to the manufacturer's instructions. Chemiluminescent signal acquisition and quantification of spot intensity were done using the Quansys Q-View imager with Q-View software. The concentrations of cytokines were determined against 5-point standard curves using the Q-View software program. The statistical significance was calculated by the one sample t test, against a theoretical mean of 1, with a two-tailed P value. P < 0.05 was considered to be statistically significant. Informed, written consent was obtained from all subjects. The study was approved by the ethics committee of Sheba Medical Center in accordance with the Helsinki II Declaration.
RESULTS AND DISCUSSION
Molecular fingerprinting of an S. Paratyphi A strain accounted for an outbreak in Nepal.
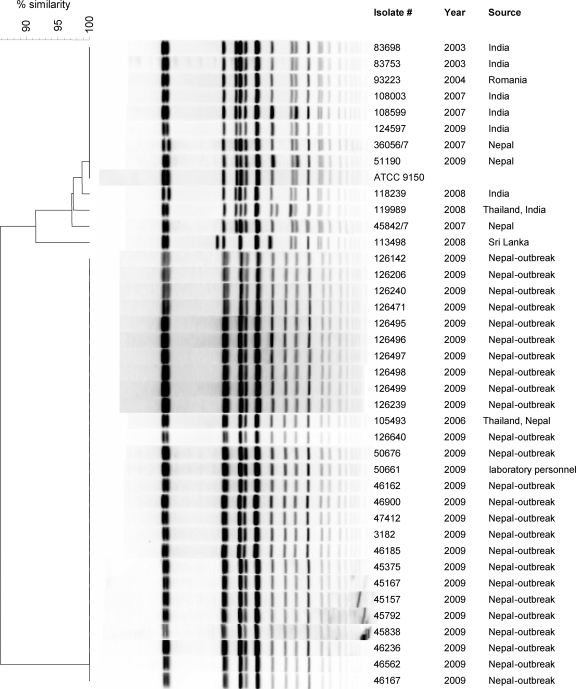
During October 2009, 38 patients (20 males and 18 females; average age, 24.8 ± 4.4 years) were hospitalized in different medical centers in Israel with acute enteric fever and positive blood cultures for S. Paratyphi A. All of the patients were returning travelers from Nepal and reported visiting the city of Pokhara during the same time. To examine a possible contamination from a single infection source, the genetic similarity between these isolates was determined by PFGE. This analysis integrated 40 S. Paratyphi A isolates, including 26 isolates from the 2009 outbreak, 12 sporadic isolates obtained from returning travelers from various places during 2004 to 2008, an isolate from a laboratory technician who developed a paratyphoid fever in November 2009, and the S. Paratyphi A ATCC 9150 reference strain (29). Macrorestriction discriminated the examined isolates into 6 distinct profiles (pulsotypes) as shown in Fig. 1. Twenty-five of 26 of the 2009 outbreak isolates had an identical pulsotype, supporting the assumption of a common infection source for this outbreak. A single isolate (51190), from a patient who returned from Nepal later than the others, showed a pulsotype indistinguishable from ATCC 9150 and a few previous sporadic isolates, suggesting that isolate 51190 was acquired from a different source than the rest of the outbreak isolates. Moreover, the PFGE pattern of the October 2009 outbreak isolates was identical to that of a previous clone (105493), isolated at 2006 from a patient returning from Thailand and Nepal, who most likely got infected with the same strain. The outbreak pulsotype was also identical to the isolate obtained from the laboratory technician (50661) who got infected after handling patients' fecal samples, which might reflect the high infectivity potential of this outbreak strain.
Fig 1.
Molecular fingerprinting of S. Paratyphi A strains. Forty S. Paratyphi A isolates, including 26 clones isolated from the 2009 outbreak, an isolate from infected laboratory personnel (50661), 12 sporadic strains isolated during 2003 to 2008, and the ATCC 9150 reference strain were subjected to PFGE analysis using macrorestriction fingerprinting with XbaI. The isolate number, year of isolation, and the source are indicated adjacent to each pulsotype. Genetic similarity (%) was based on Dice coefficients and is presented by the phylogenetic tree.
Nonetheless, despite the different pulsotypes identified, the overall picture demonstrated a high degree of genetic similarity between all S. Paratyphi A isolates. This observation is in agreement with previous studies that reported low diversity among worldwide S. Paratyphi A isolates and supports the idea of a recent emergence of S. Paratyphi A from a single progenitor (21, 31).
Virulence gene profiling of the outbreak strain.
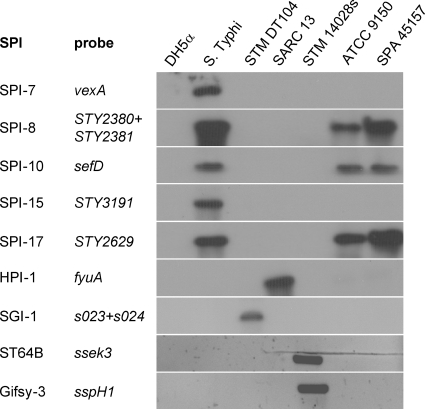
Many of the Salmonella virulence factor genes are organized within the SPIs, genomic islands, and other mobile genetic elements, including lysogenic bacteriophages. Thus far, 21 SPIs have been identified, and in addition to the Salmonella genomic island 1 (SGI-1) and the high-pathogenicity island (HPI), they vary in their distributions among different serovars and even between isolates of the same serovar (reviewed in reference 45). To better characterize the outbreak strain with respect to its virulence gene repertoire, we analyzed the presence of multiple virulence-associated loci in comparison with the ATCC 9150 sequenced strain, using a comparative genome hybridization approach and a pan-Salmonella microarray (41). In addition, to verify some of the microarray results and to explore the presence of elements not represented on the array (such as the Gifsy 3 and ST64B bacteriophages, HPI, and SGI-1), we used Southern blot hybridizations, PCR, and direct sequencing of selected targets. Overall, we found that the outbreak strain consists of an SPI inventory similar to that of ATCC 9150, with 14 SPIs, including SPIs 1 to 6, 8 to 11, and 16 to 18, as well as CS54. Like ATCC 9150, it lacks the SGI-1 and HPI elements, as well as the Gifsy-3 and ST64B prophages (Fig. 2). More detailed information about the presence/absence of more than 150-specific Salmonella virulence-associated genes of the outbreak strain is presented in Table S1 in the supplemental material.
Fig 2.
Molecular typing of the S. Paratyphi A outbreak strain. Southern blot hybridization and PCR were used to determine the presence of the Salmonella pathogenicity islands 7, 8, 10, 15, and 17, HPI, SGI-1, and the virulence-associated prophages ST64B and Gifsy-3 in the genome of the outbreak strain (SPA 45157). E. coli DH5α (DH5α) was used as a negative control for all analyses and S. Typhi CT18 (S. Typhi), S. Typhimurium DT104 (STM DT104), Salmonella Reference Collection C 13 (SARC-13), and S. Typhimurium 14028s (STM 14028s) were used as positive controls. The specific genes used as probes to determine the presence of the above loci are listed. The presence of the prophages ST64B and Gifsy-3 was detected by PCR using primers specific to sseK3 and sspH1, respectively, while the other images show the results of a Southern blot hybridization using the nonradioactive DIG system.
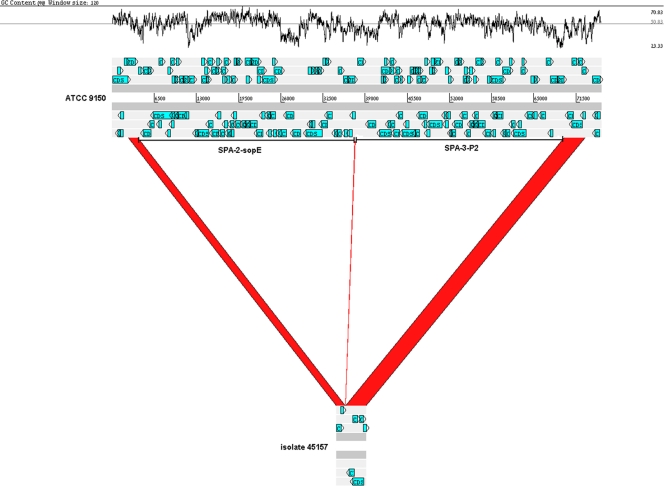
Integrated bacteriophages have been shown to affect the virulence or fitness of Salmonella isolates and often encode virulence factors (4). S. Paratyphi A sequenced strains ATCC 9150 and AKU 12601 harbor three particular phages, designated SPA-1 to -3 (31). Our CGH analysis showed that the 41-kb lambdoid phage SPA-1 (genes SPA2385 to SPA2431) is present in the outbreak strain, excluding three genes (SPA2409, SPA2412, and SPA2417). In contrast, both the 34-kb P2-type phage SPA-2-sopE (SPA2554 to SPA2600), which carries sopE (an invasion-associated gene), and the 25-kb SPA-3-P2 prophage (SPA2601 to SPA2625) are missing from the genome of the outbreak strain. In ATCC 9150, SPA-2-sopE is inserted between two perfect duplicated sequences of 45-bp (ATGTAGGAATTTCGGACGCGGGTTCAACTCCCGCCAGCTCCACCA) located at positions 2657604 and 2691147 that serves as the integration site for this phage. Similarly, SPA-3-P2 is inserted between this sequence located at position 2691147 and a second duplication at position 2723414. PCR analysis using primers flanking SPA-2-sopE and SPA-3-P2 and sequencing of the obtained PCR product confirmed the absence of 65,762 bp from the outbreak clone (Fig. 3). To get a broader perspective of the distribution of these phages, we investigated their presence in the genome of the clinical S. Paratyphi A isolates using PCR. We found that only the outbreak strain and isolate 105493, which shared the same pulsotype, omitted SPA-2-sopE and SPA-3-P2, while the rest of the isolates harbored all three phages (Table 3). These results emphasize the contribution of integrated bacteriophages to the genetic diversity of S. Paratyphi A isolates.
Fig 3.
The outbreak strain lacks a 65.7-kb region of SPA-2-sopE and SPA-3-P2 prophages. A 75,391-bp section of the S. Paratyphi A ATCC 9150 chromosome (positions 2658172 to 2733562) containing SPA-2-sopE and SPA-3-P2 prophages (top scheme) is compared against the sequence of a 4,591-bp PCR fragment amplified from the genome of the outbreak strain (isolate 45157; bottom scheme). The red regions indicate alignment between the two genomes, and the white regions indicate a missing region of 65,762 bp from the outbreak clone, in relation to the reference strain. Open reading frame (ORF) organization is shown by blue arrows, and the locations of SPA-2-sopE and SPA-3-P2 are indicated by the horizontal bars. The GC content (%) is shown in the upper panel. The figure was created using the Artemis Comparison Tool (6) and modified subsequently.
Table 3.
Distribution of phages among reference and clinical isolates of S. Paratyphi A
| Isolate | Origin | Presence or absence of phagea: |
||
|---|---|---|---|---|
| SPA-1 | SPA-2-SopE | SPA-3 | ||
| ATCC 9150 | Salmonella Genetic Stock Center | + | + | + |
| AKU12601 | Salmonella Genetic Stock Center | + | + | + |
| 36056/7 | 2007 traveler from Nepal | + | + | + |
| 45157 | 2009 Nepal outbreak | + | − | − |
| 45842/7 | 2007 traveler from Nepal | + | + | + |
| 51190 | 2009 traveler from Nepal | + | + | + |
| 83698 | 2003 traveler from India | + | + | + |
| 83753 | 2003 traveler from India | + | + | + |
| 93223 | 2004 traveler from Romania | + | + | + |
| 105493 | 2006 traveler from Thailand and Nepal | + | − | − |
| 108003 | 2007 traveler from India | + | + | + |
| 108599 | 2007 traveler from India | + | + | + |
| 113498 | 2008 traveler from Sri Lanka | + | + | + |
| 118239 | 2008 traveler from India | + | + | + |
| 119989 | 2008 traveler from Thailand and India | + | + | + |
| 124597 | 2009 traveler from India | + | + | + |
The presence (+) or absence (−) of SPA-1, SPA-2-SopE, and SPA-3 was determined by PCR using the primers SPA-1-F and SPA-1-R, new SPA-2-F and new SPA-2-R, and SPA-3-F and SPA-3-R, respectively.
Apart from the 75 genes contained within SPA-2-sopE and SPA-3-P2, at least 31 additional genes (mainly metabolic and genes with unknown function) were found to have been deleted from the genome of the outbreak clone (in relation to the ATCC 9150 and/or AKU 12601 genomes; see Table S2 in the supplemental material). Interestingly, quite a few of these genes are actually inactivated (pseudogenes) in ATCC 9150 or in other related genomes and therefore may represent a common pattern of genome degradation in this group of pathogens.
The outbreak strain presents increased virulence in comparison to the ATCC 9150 strain.
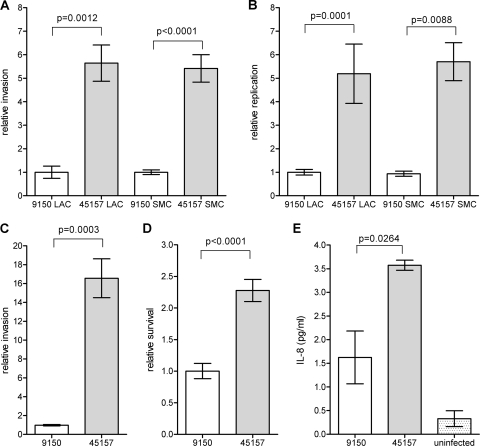
The two hallmarks of Salmonella pathogenicity are its ability to invade nonphagocytic cells and to survive and proliferate within professional phagocytes (reviewed in reference 19). In order to characterize the pathogenic potential of the outbreak strain, we studied these abilities in comparison with the characterized S. Paratyphi A ATCC 9150 strain. Previous studies have shown that Salmonella invasiveness is growth phase as well as oxygen tension dependent (26, 47). Hence, S. Paratyphi A invasion and intracellular replication in a human epithelial cell line were determined under two sets of conditions, including the late logarithmic phase under aerobic conditions (LAC) and the stationary phase under microaerophilic conditions (SMC). Gentamicin protection assays established that the outbreak strain was able to invade HeLa cells 5-fold better than the reference strain under both conditions (Fig. 4A). Intracellular replication of the outbreak strain within nonphagocytic cells was also found to be ∼5-fold higher than that of the reference strain (Fig. 4B). Enhanced invasion of the outbreak strain was also demonstrated in Caco-2 cells (Fig. 4 C), suggesting that its superior invasion is not a cell-line-specific characteristic, but represents a common characteristic of this strain.
Fig 4.
The outbreak strain demonstrates an increased virulence in vitro. The S. Paratyphi A reference strain ATCC 9150 and the outbreak strain (isolate 45157) were grown to either late logarithmic phase under aerobic conditions (LAC) or to a stationary phase under microaerophilic conditions (SMC) and used to infect different host cells. The invasion (A) and intracellular replication (B) of the outbreak strain in relation to the reference strain in HeLa cells are shown, following infection at a multiplicity of infection (MOI) of ∼50:1. (C) S. Paratyphi A strains that were grown to the stationary phase under microaerophilic conditions were used to infect Caco-2 cells at an MOI of ∼50:1. The invasion of the outbreak strain is shown in relation to the invasion of ATCC 9150. (D) RAW264.7 macrophage-like cells were infected with stationary-phase S. Paratyphi A strains at an MOI of ∼10:1. Survival of the outbreak strain (45157) in the macrophages 24 h postinfection, in relation to the reference strain (9150) is presented. (E) Caco-2 cells were infected with S. Paratyphi A strains grown to the stationary phase under microaerophilic conditions. The secreted IL-8 concentration was measured in the tissue culture medium 18 h postinfection using a quantitative ELISA-based chemiluminescent assay (Q-Plex human IL-8 array). Baseline levels of IL-8 were measured in uninfected cells that were included as a control (uninfected). All panels present the mean and the standard error of the mean (SEM; represented by the error bars) of at least 3 independent infections. An unpaired t test with two tails was used to determine the significance of the differences between the compared data.
Infection experiments, using RAW264.7 macrophage-like cells, were consistent with the results above and showed that the survival of the outbreak strain was higher than that of the ATCC 9150 strain (Fig. 4 D).
Salmonella invasion of intestinal epithelial cells leads to induction and secretion of proinflammatory cytokines such as IL-8, which play an important role in the recruitment of inflammatory cells to the site of infection and elicitation of the mucosal inflammatory response (32). Due to the differences found in the invasion abilitiy between the outbreak strain and S. Paratyphi A 9150, we also assessed IL-8 secretion following epithelial cell penetration. Caco-2 cells were infected with S. Paratyphi A strains grown under microaerophilic conditions, and the secreted IL-8 concentration was measured in the medium 18 h postinfection. Correlated with the invasion data, epithelial cells that were infected with the outbreak 45157 strain were found to secrete higher levels of the cytokine IL-8 than cells infected with the 9150 reference strain (Fig. 4E), most likely due to the increased invasion ability of the outbreak strain into host cells.
Collectively, these results suggest that the outbreak strain presents an enhanced virulence potential compared with the ATCC 9150 strain and that different S. Paratyphi A isolates vary in their pathogenicity, at least in vitro.
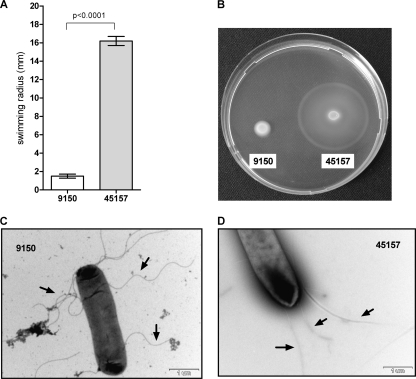
The outbreak strain exhibits enhanced motility and a different flagellar morphology.
Motility is an important virulence trait for Salmonella, facilitating invasion into epithelial cells (25, 28). The observed differences in invasion between the outbreak strain and the reference strain prompted us to assess their motility using the soft agar swimming assay. Comparison between S. Paratyphi A strains grown to the stationary phase in rich LB medium demonstrated significantly higher motility of the outbreak strain (Fig. 5A and B) and provided a possible mechanistic explanation for its increased invasion of host cells. To identify potential differences in the expression or the formation of the flagella between these strains, we applied transmitted electron microscopy. Although both strains were found to be flagellated, the flagellar morphology was found to be different. While ATCC 9150 showed a curly and thicker flagellar structure, the flagella of the outbreak stain were straight and thinner (Fig. 5C and D). Salmonella mutants that produce irregular flagella were described more than 40 years ago and were shown to be impaired in their movement (22). Thus, it is very likely that the structural differences identified in the flagella of both strains affect the degree of their motility.
Fig 5.
The outbreak strain demonstrates enhanced motility and different flagellar morphology. The S. Paratyphi A reference strain (9150) and the outbreak strain (45157) were grown in LB overnight. Ten microliters of each culture was inoculated onto a soft (0.3%) agar plate and incubated at 37°C for 6 h. The ability of the different cultures to move through the soft agar (swim) was measured. The average of five independent experiments with the error bars representing the standard error of the mean is shown (A). One representative experiment was recorded using a Pentax K10D digital camera system (B). S. Paratyphi A strains and surface flagella were negatively stained using 2% uranyl acetate solution and visualized by scanning transmission electron microscopy. Electron micrographs at a ×20,000 magnification of ATCC 9150 (C) and 45157 (D) are presented. Representative flagellar filaments in each strain are indicated by the black arrows.
Together, these experiments demonstrated both functional and phenotypic variations (including in virulence) among different isolates of S. Paratyphi A.
The immune response to an S. Paratyphi A infection in vivo.
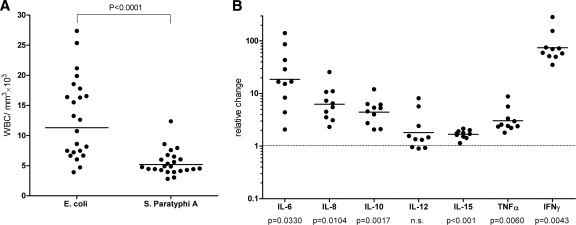
In humans, an acute inflammatory disease, particularly due to bacterial infection, is often associated with a high WBC count (39, 44). An accumulation of circulatory leukocytes has been documented in several Salmonella-infected animals, including monkeys (11), fowl typhoid in chickens caused by S. Gallinarum (16), and in murine typhoid mediated by S. Typhimurium infection (10). To examine whether paratyphoid patients demonstrate increased WBC levels (leukocytosis) during their acute infection, peripheral WBC counts of 24 hospitalized paratyphoid patients were compared to those of a control group of 23 patients with invasive E. coli infections. This comparison showed that while invasive E. coli infections are characterized by increased WBC numbers ([12.9 ± 1.4] × 103/ml), paratyphoid patients' counts ([5.4 ± 0.4] × 103/ml) were within the lower end of the accepted range (4.1 × 103 to 10.9 × 103/ml) (Fig. 6A). These results suggested that leukocytosis does not characterize the immune response to a paratyphoid infection in humans during the acute phase of the disease, as opposed to some other infectious diseases.
Fig 6.
S. Paratyphi A induces a distinct immune response in vivo. (A) The WBC count is shown for 23 patients with invasive infections of E. coli and 24 S. Paratyphi A-infected patients. Each point represents the WBC count of a single patient, with the mean indicated by the horizontal line. An unpaired t test with two tails was used to determine the significance of the difference between the two groups. (B) Serum samples from 10 S. Paratyphi A-infected patients were collected during the acute stage of the disease and 12 to 14 weeks after their convalescence. The serum concentration of 16 inflammatory cytokines was determined by the Q-Plex human cytokine screen kit. The change in the concentrations of 7 cytokines (IL-6, IL-8, IL-10, IL-12, IL-15, TNF-α, and IFN-γ) during the disease versus convalescence is shown, with the calculated P values below. Points represent the fold change (during disease/convalescence) in the serum concentration of specific cytokines as measured in each patient, with the mean indicated by the horizontal line. n.s., not statistically significant.
Different cytokines are known to play a pivotal role in initiating and regulating the innate and adaptive immune responses against Salmonella. A few clinical studies focusing on NTS infections in humans have reported the involvement of several inflammatory pathways, including gamma interferon (IFN-γ) (34, 48), tumor-necrosis factor alpha (TNF-α) (48), IL-6 (27), IL-8 (27), IL-10 (34, 48), IL-12 (34, 48), IL-15 (34), and IL-18 (34). Other studies have examined cytokine concentrations in the serum of S. Typhi-infected patients (5, 24); however, not much is known about the human immune response to a paratyphoid infection. To shed some light on this subject, we analyzed the circulating levels of 16 primary inflammatory cytokines (IL-1α, IL-1β, IL-2, IL-4, IL-5, IL-6, IL-8, IL-10, IL-12p70, IL-13, IL-15, IL-17, IL-23, IFN-γ, TNF-α, and TNF-β) in the serum of 10 paratyphoid patients, all of whom were infected with the outbreak strain. Serum that was taken during the acute phase of the disease from each of these patients was compared to a matching sample obtained from the same patient 12 to 16 weeks after convalescence.
We found in 10/10 patients a dramatic elevation (∼75-fold) of IFN-γ, in addition to a more moderate induction of IL-6 (18.6-fold), IL-8 (6.2-fold), IL-10 (4.4-fold), IL-15 (1.6-fold), and TNF-α (3.0-fold) (Fig. 6B). The serum concentrations of the other 10 tested cytokines (IL-1α, IL-1β, IL-2, IL-4, IL-5, IL-12p70, IL-13, IL-17, IL-23, and TNF-β) were not found to fluctuate between the two time points. To the best of our knowledge, this analysis is the broadest assessment of the cytokine response to a paratyphoid infection in vivo thus far.
IFN-γ, the cytokine which had the most elevated levels among the paratyphoid patients, is the primary cytokine that drives the Th1-type immune response and is involved in the clearance of intracellular pathogens. IFN-γ is produced by T-helper cells, natural killer (NK) cells, and NK-like cells and in synergy with TNF-α, (which was also found to increase in these patients) is required for the initiation of antimicrobial functions of the infected macrophages (7, 12, 50). A remarkable induction of IFN-γ during paratyphoid fever provided evidence that IFN-γ plays a pivotal role in the human response to an S. Paratyphi A infection. Nonetheless, other type 1 cytokines, including IL-17, IL-23, and particularly IL-12, were not increased during the acute stage of the disease. IL-12 is known to activate IFN-γ secretion in Th1 and NK cells and was found to be elevated in NTS infections (34, 48). While we cannot rule out the possibility of an earlier IL-12 induction, its unchanged level during the acute phase of the paratyphoid disease may suggest IFN-γ induction in an IL-12-independent manner. A similar mechanism has been reported in the context of the intracellular parasite Leishmania major infection, demonstrating that the effector function of mature Th1 cells in vivo is independent of IL-12 (8). The lack of IL-12 induction in the presence of a dramatic increase in IFN-γ may indicate the elicitation of a different immune response to a paratyphoid disease rather than the one to an NTS infection. Diverse host-pathogen interplay with S. Paratyphi A versus NTS is intriguing and consistent with accumulating clinical evidence. It has been established that invasive infections caused by NTS, but not by typhoidal serovars, are often associated with immunocompromised adults, in particular in the context of HIV (17). This epidemiological observation implies that certain factors (which are probably malfunctioning in AIDS patients) are required for the immune defense against systemic infection of NTS, but not against typhoidal serovars. Furthermore, recent studies have shown that patients with inherited deficiency of the IL-12/IL-23 system (IL-12p40/IL-12Rβ1) are highly susceptible to invasive extraintestinal NTS infections, but not to S. Typhi nor S. Paratyphi infections, even though some of these patients live in areas where typhoid is endemic (30, 49). Collectively, these observations support the possibility that different inflammatory pathways may be involved in NTS versus typhoidal infections, including a distinct role for the IL-12 pathway.
Additional cytokines found to be induced during paratyphoid infection were IL-6, IL-8, and IL-10. IL-6 is produced in the intestinal mucosa and is particularly important due to its pleiotropic involvement in different pathways (36). Although commonly considered a proinflammatory cytokine, there is also evidence that IL-6 has important anti-inflammatory properties and may exert protective effects in different tissues (51). IL-8 is a potent neutrophil chemotactic factor secreted by intestinal inflammatory cells in response to bacterial invasion (13). IL-8 secretion induced by some Salmonella serovars leads to a massive neutrophil migration and an elicitation of a mucosal inflammatory response (reviewed in reference 19). IL-10 is an anti-inflammatory cytokine, which inhibits antigen presentation to T cells and suppresses phagocytosis and intracellular killing (46). Interestingly, Pie et al. showed that S. Typhimurium induced immunosuppression in mice and caused the production of large amounts of IL-10 (40). As our results indicated a moderate increase in IL-6 and IL-10 together with the lack of leukocytosis, it is tempting to suggest a pathogen-mediated IL-6/IL-10 induction, as a means to prevent T-cell proliferation and to suppress the cellular immune response of the host. Nonetheless, further experimental data are certainly required to support this hypothesis.
The cytokine profile found in the paratyphoid patients is similar to previous cytokine analysis of serum from typhoid fever patients infected with S. Typhi that revealed elevated levels of IL-6, IL-8, TNF-α, and IFN-γ (5, 24). In S. Typhi, the virulence (Vi) capsule is presumed to be central to the host-pathogen interactions and is believed to facilitate evasion of the immune system (42). These results suggest that although S. Paratyphi A lacks the Vi capsule, a significant overlap does exist in the immune response to S. Typhi and S. Paratyphi infections.
Conclusions.
Our study describes genetic and phenotypic characterization of an S. Paratyphi A strain that was identified as the causative agent of a recent paratyphoid outbreak in Nepal. Comparative PFGE with other S. Paratyphi A isolates and microarray-based CGH analysis showed a high degree of genetic homogeneity among different isolates of S. Paratyphi A. This observation is consistent with the notion that S. Paratyphi A strains have evolved recently, on an evolutionary time scale, from a single ancestor (21, 31). Nevertheless, subtle genetic differences between the outbreak and the reference strains were found, including the absence of two prophages (SPA-2-sopE and SPA-3-P2) from the genome of the outbreak strain, as well as sporadically degraded metabolic and other genes. Differences were also established on the phenotypic level. The outbreak strain demonstrated enhanced motility, invasion, and replication in nonphagocytic cells, higher survival in macrophages, and elevated induction of IL-8 secretion by host cells. Accumulatively, these results indicate that despite a very high degree of genetic conservation, different S. Paratyphi A isolates vary in their virulence potential. Cytokine profile analysis during the acute phase of the disease indicated a remarkable induction of IFN-γ in addition to a more moderate increase of IL-6, IL-8, IL-10, IL-15, and TNF-α, but unchanged levels of IL-12. The revealed cytokine profile, together with several previously reported clinical observations, suggests a distinct host response to S. Paratyphi A infection in comparison to salmonellosis caused by NTS strains. Better understanding and a more comprehensive view of the virulence mechanisms and the immune response to the S. Paratyphi infection might facilitate prevention efforts and the development of novel therapeutics against this emerging pathogen.
Supplementary Material
ACKNOWLEDGMENTS
We thank Nati Keller and the staff of the bacteriology laboratory of the Sheba Medical Center for sharing clinical isolates of S. Paratyphi A. We also thank the National Microbiology Laboratory Public Health Agency of Canada for the S. Typhimurium DT104 96-5227 strain.
This work was supported by grant 249241 from the European Community's Seventh Framework Program (PF7/2007-2013).
Footnotes
Published ahead of print 21 December 2011
Supplemental material for this article may be found at http://cvi.asm.org/.
REFERENCES
- 1. Baumler AJ, Tsolis RM, Ficht TA, Adams LG. 1998. Evolution of host adaptation in Salmonella enterica. Infect. Immun. 66:4579–4587 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Boyd DA, Peters GA, Ng L, Mulvey MR. 2000. Partial characterization of a genomic island associated with the multidrug resistance region of Salmonella enterica Typhymurium DT104. FEMS Microbiol. Lett. 189:285–291 [DOI] [PubMed] [Google Scholar]
- 3. Boyd EF, Wang FS, Whittam TS, Selander RK. 1996. Molecular genetic relationships of the salmonellae. Appl. Environ. Microbiol. 62:804–808 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Brussow H, Canchaya C, Hardt WD. 2004. Phages and the evolution of bacterial pathogens: from genomic rearrangements to lysogenic conversion. Microbiol. Mol. Biol. Rev. 68:560–602 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Butler T, Ho M, Acharya G, Tiwari M, Gallati H. 1993. Interleukin-6, gamma interferon, and tumor necrosis factor receptors in typhoid fever related to outcome of antimicrobial therapy. Antimicrob. Agents Chemother. 37:2418–2421 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Carver TJ, et al. 2005. ACT: the Artemis Comparison Tool. Bioinformatics 21:3422–3423 [DOI] [PubMed] [Google Scholar]
- 7. Coburn B, Grassl GA, Finlay BB. 2007. Salmonella, the host and disease: a brief review. Immunol. Cell Biol. 85:112–118 [DOI] [PubMed] [Google Scholar]
- 8. Constantinescu CS, et al. 1998. The role of IL-12 in the maintenance of an established Th1 immune response in experimental leishmaniasis. Eur. J. Immunol. 28:2227–2233 [DOI] [PubMed] [Google Scholar]
- 9. Crump JA, Luby SP, Mintz ED. 2004. The global burden of typhoid fever. Bull. World Health Organ. 82:346–353 [PMC free article] [PubMed] [Google Scholar]
- 10. Dejager L, Pinheiro I, Bogaert P, Huys L, Libert C. 2010. Role for neutrophils in host immune responses and genetic factors that modulate resistance to Salmonella enterica serovar Typhimurium in the inbred mouse strain SPRET/Ei. Infect. Immun. 78:3848–3860 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. DeRubertis FR, Woeber KA. 1973. Accelerated cellular uptake and metabolism of L-thyroxine during acute Salmonella typhimurium sepsis. J. Clin. Invest. 52:78–87 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Eckmann L, Kagnoff MF. 2001. Cytokines in host defense against Salmonella. Microbes Infect. 3:1191–1200 [DOI] [PubMed] [Google Scholar]
- 13. Eckmann L, Kagnoff MF, Fierer J. 1993. Epithelial cells secrete the chemokine interleukin-8 in response to bacterial entry. Infect. Immun. 61:4569–4574 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Edwards RA, Olsen GJ, Maloy SR. 2002. Comparative genomics of closely related salmonellae. Trends Microbiol. 10:94–99 [DOI] [PubMed] [Google Scholar]
- 15. Gal-Mor O, Valdez Y, Finlay BB. 2006. The temperature-sensing protein TlpA is repressed by PhoP and dispensable for virulence of Salmonella enterica serovar Typhimurium in mice. Microbes Infect. 8:2154–2162 [DOI] [PubMed] [Google Scholar]
- 16. Garcia KB-J, Santana A, Freitas-Neto O, Fagliari J. 2009. Experimental infection of commercial layers using a Salmonella enterica serovar Gallinarum strain: leukogram and serum acute-phase protein concentrations. Braz. J. Poult. Sci. 11:263–270 [Google Scholar]
- 17. Gordon MA. 2008. Salmonella infections in immunocompromised adults. J. Infect. 56:413–422 [DOI] [PubMed] [Google Scholar]
- 18. Groisman EA, Ochman H. 1996. Pathogenicity islands: bacterial evolution in quantum leaps. Cell 87:791–794 [DOI] [PubMed] [Google Scholar]
- 19. Haraga A, Ohlson MB, Miller SI. 2008. Salmonellae interplay with host cells. Nat. Rev. Microbiol. 6:53–66 [DOI] [PubMed] [Google Scholar]
- 20. Hensel M. 2004. Evolution of pathogenicity islands of Salmonella enterica. Int. J. Med. Microbiol. 294:95–102 [DOI] [PubMed] [Google Scholar]
- 21. Holt KE, et al. 2009. Pseudogene accumulation in the evolutionary histories of Salmonella enterica serovars Paratyphi A and Typhi. BMC Genomics 10:36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Iino T, Mitani M. 1967. A mutant of Salmonella possessing straight flagella. J. Gen. Microbiol. 49:81–88 [DOI] [PubMed] [Google Scholar]
- 23. Jarvik T, Smillie C, Groisman EA, Ochman H. 2010. Short-term signatures of evolutionary change in the Salmonella enterica serovar Typhimurium 14028 genome. J. Bacteriol. 192:560–567 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Keuter M, et al. 1994. Patterns of proinflammatory cytokines and inhibitors during typhoid fever. J. Infect. Dis. 169:1306–1311 [DOI] [PubMed] [Google Scholar]
- 25. Khoramian-Falsafi T, Harayama S, Kutsukake K, Pechere JC. 1990. Effect of motility and chemotaxis on the invasion of Salmonella typhimurium into HeLa cells. Microb. Pathog. 9:47–53 [DOI] [PubMed] [Google Scholar]
- 26. Lee CA, Falkow S. 1990. The ability of Salmonella to enter mammalian cells is affected by bacterial growth state. Proc. Natl. Acad. Sci. U. S. A. 87:4304–4308 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Lin CH, et al. 2006. The diagnostic value of serum interleukins 6 and 8 in children with acute gastroenteritis. J. Pediatr. Gastroenterol. Nutr. 43:25–29 [DOI] [PubMed] [Google Scholar]
- 28. Liu SL, Ezaki T, Miura H, Matsui K, Yabuuchi E. 1988. Intact motility as a Salmonella typhi invasion-related factor. Infect. Immun. 56:1967–1973 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Liu SL, Sanderson KE. 1995. The chromosome of Salmonella paratyphi A is inverted by recombination between rrnH and rrnG. J. Bacteriol. 177:6585–6592 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. MacLennan C, et al. 2004. Interleukin (IL)-12 and IL-23 are key cytokines for immunity against Salmonella in humans. J. Infect. Dis. 190:1755–1757 [DOI] [PubMed] [Google Scholar]
- 31. McClelland M, et al. 2004. Comparison of genome degradation in Paratyphi A and Typhi, human-restricted serovars of Salmonella enterica that cause typhoid. Nat. Genet. 36:1268–1274 [DOI] [PubMed] [Google Scholar]
- 32. McCormick BA, Colgan SP, Delp-Archer C, Miller SI, Madara JL. 1993. Salmonella typhimurium attachment to human intestinal epithelial monolayers: transcellular signalling to subepithelial neutrophils. J. Cell Biol. 123:895–907 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Meltzer E, Schwartz E. 2010. Enteric fever: a travel medicine oriented view. Curr. Opin. Infect. Dis. 23:432–437 [DOI] [PubMed] [Google Scholar]
- 34. Mizuno Y, et al. 2003. Th1 and Th1-inducing cytokines in Salmonella infection. Clin. Exp. Immunol. 131:111–117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Ochiai RL, et al. 2005. Salmonella paratyphi A rates, Asia. Emerg. Infect. Dis. 11:1764–1766 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Papanicolaou DA, Wilder RL, Manolagas SC, Chrousos GP. 1998. The pathophysiologic roles of interleukin-6 in human disease. Ann. Intern. Med. 128:127–137 [DOI] [PubMed] [Google Scholar]
- 37. Parkhill J, et al. 2001. Complete genome sequence of a multiple drug resistant Salmonella enterica serovar Typhi CT18. Nature 413:848–852 [DOI] [PubMed] [Google Scholar]
- 38. Parry CM, Hien TT, Dougan G, White NJ, Farrar JJ. 2002. Typhoid fever. N. Engl. J. Med. 347:1770–1782 [DOI] [PubMed] [Google Scholar]
- 39. Pfafflin A, Schleicher E. 2009. Inflammation markers in point-of-care testing (POCT). Anal. Bioanal Chem. 393:1473–1480 [DOI] [PubMed] [Google Scholar]
- 40. Pie S, Matsiota-Bernard P, Truffa-Bachi P, Nauciel C. 1996. Gamma interferon and interleukin-10 gene expression in innately susceptible and resistant mice during the early phase of Salmonella Typhimurium infection. Infect. Immun. 64:849–854 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Porwollik S, Wong RM, McClelland M. 2002. Evolutionary genomics of Salmonella: gene acquisitions revealed by microarray analysis. Proc. Natl. Acad. Sci. U. S. A. 99:8956–8961 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Raffatellu M, Wilson RP, Winter SE, Baumler AJ. 2008. Clinical pathogenesis of typhoid fever. J. Infect. Dev. Ctries. 2:260–266 [DOI] [PubMed] [Google Scholar]
- 43. Ribot EM, et al. 2006. Standardization of pulsed-field gel electrophoresis protocols for the subtyping of Escherichia coli O157:H7, Salmonella, and Shigella for PulseNet. Foodborne Pathog. Dis. 3:59–67 [DOI] [PubMed] [Google Scholar]
- 44. Ryan GB, Majno G. 1977. Acute inflammation. A review. Am. J. Pathol. 86:183–276 [PMC free article] [PubMed] [Google Scholar]
- 45. Sabbagh SC, Forest CG, Lepage C, Leclerc JM, Daigle F. 2010. So similar, yet so different: uncovering distinctive features in the genomes of Salmonella enterica serovars Typhimurium and Typhi. FEMS Microbiol. Lett. 305:1–13 [DOI] [PubMed] [Google Scholar]
- 46. Spellberg B, Edwards JE., Jr 2001. Type 1/type 2 immunity in infectious diseases. Clin. Infect. Dis. 32:76–102 [DOI] [PubMed] [Google Scholar]
- 47. Steele-Mortimer O. 2008. Infection of epithelial cells with Salmonella enterica, p 201–211 In DeLeo F, Otto M. (ed), Bacterial pathogenesis: methods and protocols, vol 431 Humana Press, Totowa, NJ: [DOI] [PubMed] [Google Scholar]
- 48. Stoycheva M, Murdjeva M. 2005. Serum levels of interferon-gamma, interleukin-12, tumour necrosis factor-alpha, and interleukin-10, and bacterial clearance in patients with gastroenteric Salmonella infection. Scand. J. Infect. Dis. 37:11–14 [DOI] [PubMed] [Google Scholar]
- 49. van de Vosse E, Ottenhoff TH. 2006. Human host genetic factors in mycobacterial and Salmonella infection: lessons from single gene disorders in IL-12/IL-23-dependent signaling that affect innate and adaptive immunity. Microbes Infect. 8:1167–1173 [DOI] [PubMed] [Google Scholar]
- 50. Vazquez-Torres A, Fang FC. 2001. Salmonella evasion of the NADPH phagocyte oxidase. Microbes Infect. 3:1313–1320 [DOI] [PubMed] [Google Scholar]
- 51. Xing Z, et al. 1998. IL-6 is an antiinflammatory cytokine required for controlling local or systemic acute inflammatory responses. J. Clin. Invest. 101:311–320 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.