Abstract
One of the challenges for developing an H5N1 influenza vaccine is the diversity of antigenically distinct isolates within this subtype. Previously, our group described a novel hemagglutinin (HA) derived from a methodology termed computationally optimized broadly reactive antigen (COBRA). This COBRA HA, when used as an immunogen, elicits a broad antibody response against H5N1 isolates from different clades. In this report, the immune responses elicited by the COBRA HA virus-like particle (VLP) vaccine were compared to responses elicited by a mixture of VLPs expressing representative HA molecules from clade 2.1, 2.2, and 2.3 primary H5N1 isolates (polyvalent). The COBRA HA VLP vaccine elicited higher-titer antibodies to a panel of H5N1 HA proteins than did the other VLPs. Both COBRA and polyvalent vaccines protected vaccinated mice and ferrets from experimental infection with highly lethal H5N1 influenza viruses, but COBRA-vaccinated animals had decreased viral replication, less inflammation in the lungs of mice, and reduced virus recovery in ferret nasal washes. Both vaccines had similar cellular responses postchallenge, indicating that higher-titer serum antibodies likely restrict the duration of viral replication. Furthermore, passively transferred immune serum from the COBRA HA VLP-vaccinated mice protected recipient animals more efficiently than immune serum from polyvalent-vaccinated mice. This is the first report comparing these two vaccine strategies. The single COBRA HA antigen elicited a broader antibody response and reduced morbidity and viral titers more effectively than a polyvalent mixture of primary H5N1 HA antigens.
INTRODUCTION
In addition to yearly epidemics, pandemic outbreaks of influenza have occurred sporadically throughout human history (20, 43). Pandemics occur when a novel pathogenic and transmissible virus emerges into the human population. A critical factor in the emergence of a pandemic virus is that it must be antigenically divergent from the current circulating strains to evade prior immunity in the human population. Therefore, pandemic viruses can potentially emerge from novel subtypes, such as H5N1 or H7N7, or divergent strains of currently circulating subtypes, such as H1N1. Indeed, the influenza pandemic of 2009 was caused by the emergence of a novel, swine-origin H1N1 virus into the human population (8). Avian viruses of the subtypes H5N1, H7N7, and H9N2 have all demonstrated the ability to directly infect humans (51). H5N1 is of particular concern because of the continued cross-species infection and the high pathogenicity of the virus (60% mortality) (54). Although H5N1 has not displayed efficient human-to-human transmission, in vitro studies have established that stable reassortant viruses that retain the pathogenic phenotype of H5N1 can be created with both H3N2 and novel H1N1 viruses (9, 27, 35). Reassortment with transmissible viruses and/or accumulation of mutations could result in the emergence of a highly transmissible H5N1 virus. The genetic compatibility of H5N1 with currently circulating human and swine viruses highlights the need for the development of effective vaccines against H5N1.
Development of prepandemic H5N1 vaccines is complicated by the antigenic diversity within the subtype. Phylogenetic distances of the hemagglutinin (HA) genes of H5N1 viruses distinguish the 10 distinct clades (53). HA-based diversity within clade 2 alone has led to characterization of distinct subclades and sub-subclades. In most human H5N1 influenza infections, the isolates were identified as members of clades 1 or 2, with isolates from clade 2 being detected in over 60 countries and moving westward into the Middle East and Africa (52). Although H5N1 HA proteins display a high degree of similarity (>90% identity), there is little receptor-blocking antibody cross-reactivity between clades. Furthermore, the subclades of clade 2 are antigenically distinct, as determined by the cross-reactivity of receptor-blocking antibodies (53). Despite the risk imposed by highly pathogenic H5N1 influenza, the magnitude of diversity within the subtype complicates vaccine antigen selection for either prepandemic usage or stockpiling. Vaccines that are able to overcome the challenge of antigenic diversity are therefore crucial for effective pandemic preparedness.
Influenza antigenic diversity is not a unique problem for H5N1 vaccine development. Rather, simultaneous circulation of diverse influenza A (H1N1 and H3N2) and influenza B viruses has been a challenge for seasonal influenza vaccine production for over 30 years. The current seasonal vaccine uses a polyvalent formulation to address the issue of distinct viruses circulating simultaneously and therefore is a standard strategy to elicit increased antibody breadth by influenza vaccination. Indeed, multivalent H5N1 vaccines increase the breadth of receptor-blocking antibody responses (12, 37). Another strategy for expanding antibody breadth involves engineering synthetic centralized antigens that are predicted to capture common immune epitopes. Consensus and ancestral sequences have been used for H5N1 vaccine antigens, and these centralized vaccines are a feasible mechanism by which antibody breadth can be expanded (10, 13, 23–25). Our research group has used a consensus-based approach to develop a novel strategy termed computationally optimized broadly reactive antigen (COBRA) (16). Traditional consensus sequences are inherently biased by the input sequences used in alignment and subsequent sequence generation. Because sequencing efforts for H5N1 are reactionary to outbreaks rather than systematic, the available input sequences for generation of any centralized sequence are subject to outbreak dominance and bias. COBRA methodology seeks to overcome this limitation by layering together multiple rounds of consensus sequence generation. This method was specifically applied to the diversity of HA within clade 2 because this clade is the most antigenically diverse, has been implicated in the majority of human infections, and has the widest geographic spread.
Here, we demonstrate that a COBRA HA antigen, based upon HA sequences from human H5N1 isolates, elicits a broader receptor-blocking antibody response than a polyvalent formulation of primary clade 2 HA antigens in mice and ferrets. Both vaccines were able to protect mice and ferrets from highly pathogenic H5N1 challenge, with COBRA-vaccinated animals showing more efficient viral clearance. Additionally, both vaccines protected mice from a divergent clade 1 challenge and immune serum from COBRA-vaccinated animals protected animals more efficiently than serum from polyvalent vaccine recipients.
MATERIALS AND METHODS
Vaccine antigens and preparation.
The design and characterization of the computationally optimized broadly reactive antigen (COBRA) have been described previously (16). Briefly, the COBRA HA antigen was generated by multiple rounds of consensus generation using HA sequences from H5N1 clade 2 human infections collected from 2004 to 2006. Polyvalent vaccine HA antigens were derived via reverse transcription from the following 6:2 reassortant H5N1 viruses: A/Indonesia/5/2005 (clade 2.1; IN/05), A/Whooper Swan/Mongolia/244/2005 (clade 2.2; WS/05), and A/Anhui/1/2005 (clade 2.3; AN/05). All HA antigens were cloned into the pTR600 expression vector (38).
Virus-like particles (VLPs) were generated by transiently transfecting HEK 293T cells with plasmids expressing influenza matrix protein (M1) (A/Puerto Rico/8/1934), neurarninidase (NA) [A/Thailand/1(KAN-1)/2004], and a single H5N1 HA for each preparation. Cells were incubated for 72 h at 37°C, and then supernatants were harvested. Cell debris was cleared by low-speed centrifugation followed by vacuum filtration through a 0.22-μm sterile filter. VLPs were purified by ultracentrifugation (100,000 × g through 20% glycerol, wt/vol) for 4 h at 4°C. Pellets were then resuspended in phosphate-buffered saline (PBS) (pH 7.2) and stored in single-use aliquots at −80°C until use. Total protein concentration was determined by the Micro BCA protein assay reagent kit (Pierce Biotechnology, Rockford, IL). The HA-specific content of each VLP was determined by scanning densitometry as described previously (16). Briefly, purified HA matched to each VLP was electrophoresed with purified VLPs, transferred to a polyvinylidene difluoride (PVDF) membrane, and probed by Western blotting with H5-specific antisera. The relative densities of the HA bands in the purified protein lanes were used to calculate a standard curve, and the densities of the HA bands in the VLP lanes were interpolated. In total, four different VLP preparations were purified and HA content was quantified independently, with each VLP preparation containing one of the three wild-type influenza gene products (IN/05, WS/05, or AN/05) or the COBRA HA.
Mouse studies.
BALB/c mice (Mus musculus, females, 6 to 8 weeks) were purchased from Harlan Sprague Dawley (Indianapolis, IN), housed in microisolator units, allowed free access to food and water, and cared for under USDA guidelines for laboratory animals. Mice were vaccinated with purified COBRA VLPs (3 μg HA) or a polyvalent formulation of VLPs consisting of 1 μg HA each of IN/05, WS/05, and AN/05 (3 μg HA total) via intramuscular injection at week 0 and then boosted at week 3. Vaccines were formulated with Imject alum adjuvant (Imject Alum; Pierce Biotechnology, Rockford, IL) according to the manufacturer's protocol. Fourteen to 21 days after each vaccination, blood was collected from anesthetized mice via the retro-orbital plexus and transferred to a microcentrifuge tube. Tubes were centrifuged, and serum was removed and frozen at −20 ± 5°C.
Three weeks after the final vaccination, mice were challenged intranasally with 5 × 103 PFU of either the highly pathogenic wild-type H5N1 virus A/Whooper Swan/Mongolia/244/2005 (20 mice/group) or the 6:2 reassortant virus with internal genes from the mouse-adapted virus A/Puerto Rico/8/1934 and the surface proteins HA and NA from A/Vietnam/1203/2004 (10 mice/group) in a total volume of 50 μl. Challenge doses for both viruses were established independently and represent approximately 50 50% lethal doses (LD50) (data not shown). Mice were monitored daily for weight loss, disease signs, and death for 14 days after infection. Individual body weights, sickness scores, and death were recorded for each group on each day after inoculation. Sickness scores were determined by evaluating activity (0 = normal, 1 = reduced, 2 = severely reduced), hunched back (0 = absent, 1 = present), and ruffled fur (0 = absent, 1 = present) (46). Experimental endpoints were determined by >20% weight loss or a display of neurological disease, such as hind-limb paralysis. All highly pathogenic wild-type H5N1 influenza virus studies were performed under high-containment biosafety level 3 enhanced conditions (BSL3+). All procedures were in accordance with the National Research Council (NRC) Guide for the Care and Use of Laboratory Animals (33a), the Animal Welfare Act, and the CDC/NIH Biosafety in Microbiological and Biomedical Laboratories guide (7a).
Ferret studies.
Fitch ferrets (Mustela putorius furo, females, 6 to 12 months of age), influenza naïve and descented, were purchased from Marshall Farms (Sayre, PA). Ferrets were housed in pairs in stainless steel cages (Shor-line, Kansas City, KS) containing Sani-chips laboratory animal bedding (P. J. Murphy Forest Products, Montville, NJ). Ferrets were provided with Teklad Global ferret diet (Harlan Teklad, Madison, WI) and fresh water ad libitum. The VLPs were diluted in PBS (pH 7.2) to achieve the final concentration. Ferrets (n = 6) were vaccinated with purified COBRA VLPs (15 μg HA) or a polyvalent formulation of VLPs consisting of 5 μg HA each of IN/05, WS/05, and AN/05 (15 μg HA total) via intramuscular injection at week 0 and then boosted at week 3. Vaccines were formulated with Imject alum adjuvant (Imject Alum; Pierce Biotechnology, Rockford, IL) immediately prior to use according to the manufacturer's protocol. Animals were monitored weekly during the vaccination regimen for adverse events, including weight loss, change in temperature, loss of activity, nasal discharge, sneezing, and diarrhea. Prior to vaccination, animals were confirmed by hemagglutination inhibition (HAI) assay to be seronegative for circulating influenza A (H1N1 and H3N2) and influenza B viruses. Fourteen to 21 days after each vaccination, blood was collected from anesthetized ferrets via the anterior vena cava and transferred to a microcentrifuge tube. Tubes were centrifuged, and serum was removed and frozen at −20 ± 5°C.
Three weeks after the final vaccination, ferrets were challenged intranasally with 1 × 106 PFU of the highly pathogenic H5N1 virus A/Whooper Swan/Mongolia/244/2005 (clade 2.2) in a volume of 0.5 ml in each nostril for a total infection volume of 1 ml. Ferrets were monitored daily for weight loss, disease signs, and death for 14 days after infection. Individual body weights, sickness scores, and death were recorded for each group on each day after inoculation. Sickness scores were determined by evaluating activity (0 = normal, 1 = alert and active after stimulation, 2 = alert but not active after stimulation, 3 = neither active nor alert after stimulation), nasal discharge (0 = absent, 1 = present), sneezing (0 = absent, 1 = present), decreased food intake (0 = absent, 1 = present), diarrhea (0 = absent, 1 = present), dyspnea (0 = absent, 1 = present), and neurological symptoms (0 = absent, 1 = present) as previously described (16). Experimental endpoints were defined as >20% weight loss, development of neurological disease, or an activity score of 3 (neither active nor alert after stimulation). Nasal washes were performed by instilling 3 ml of PBS into the nares of anesthetized ferrets each day for 14 days after inoculation. Washes were collected and stored at −80°C until use. All highly pathogenic wild-type H5N1 influenza virus studies were performed under high-containment biosafety level 3 enhanced conditions (BSL3+). All procedures were in accordance with the NRC Guide for the Care and Use of Laboratory Animals (33a), the Animal Welfare Act, and the CDC/NIH Biosafety in Microbiological and Biomedical Laboratories guide (7a).
ELISA.
An enzyme-linked immunosorbent assay (ELISA) was used to assess total antibody titers to the HA. High-binding, 96-well polystyrene plates (Costar, Lowell, MA) were coated overnight with 50 ng/well of recombinant HA. Coating antigens were derived from the following representative viral isolates: A/Vietnam/1203/2004 (clade 1), A/Indonesia/5/2005 (clade 2.1), A/Whooper Swan/Mongolia/244/2005 (clade 2.2), and A/Anhui/1/2005 (clade 2.3). Plates were blocked with 5% milk diluted in PBS with 0.05% Tween 20. Serum samples were diluted in blocking buffer and added to plates. The serum was 2-fold serially diluted and allowed to incubate for 1 h at room temperature. Plates were washed, and a horseradish peroxidase (HRP)-linked species-specific antibody against IgG was diluted in blocking buffer and added to the plates. Plates were incubated for 1 h at room temperature. Plates were washed, and HRP was developed with 3,3′,5,5′-tetramethylbenzidine (TMB) substrate (Sigma-Aldrich, St. Louis, MO). Plates were incubated in the dark for 15 min, and then the reaction was stopped with 2N H2SO4. Optical densities at a wavelength of 450 nm (OD450s) were read by a spectrophotometer (BioTek, Winooski, VT), and endpoint dilution titers were determined as the reciprocal dilution of the last well which had an OD450 above the mean OD450 plus two standard deviations of naïve animal sera.
HAI assay.
The HAI assay was used to assess functional antibodies to the HA that were able to inhibit agglutination of horse erythrocytes (HRBCs). The protocol was adapted from the CDC laboratory-based influenza surveillance manual (22). To inactivate nonspecific inhibitors, sera were treated with receptor-destroying enzyme (RDE; Denka Seiken, Co., Japan) prior to being tested (4–6, 32, 38). Briefly, three parts RDE was added to one part serum and incubated overnight at 37°C. RDE was inactivated by incubation at 56°C for ∼30 min. RDE-treated serum was 2-fold serially diluted in V-bottom microtiter plates. An equal volume of reassortant virus, adjusted to approximately 8 hemagglutinating units (HAU)/50 μl, was added to each well. The reassortant viruses contained the internal genes from the mouse-adapted strain A/Puerto Rico/8/1934 and the surface proteins HA and NA from the following representative viral isolates: A/Vietnam/1203/2004 (clade 1), A/Indonesia/5/2005 (clade 2.1), A/Whooper Swan/Mongolia/244/2005 (clade 2.2), and A/Anhui/1/2005 (clade 2.3). The plates were covered and incubated at room temperature for 20 min, followed by the addition of 1% horse erythrocytes (Lampire Biologicals, Pipersville, PA) in PBS. Red blood cells (RBCs) were stored at 4°C and used within 72 h of preparation. The plates were mixed by agitation and covered, and the RBCs were allowed to settle for 1 h at room temperature (1). The HAI titer was determined by the reciprocal dilution of the last well which contained nonagglutinated RBCs. Positive and negative serum controls were included for each plate. All mice and ferrets were negative (HAI dilution of ≤1:10) for preexisting antibodies to currently circulating human influenza viruses prior to vaccination.
Plaque assay.
For mouse infections, lung virus titers were evaluated. For ferret infections, nasal wash virus titers were used to assess viral burden. Both lung and nasal wash virus titers were determined using a plaque assay (47, 48). Briefly, lungs from infected mice were harvested postinfection, snap-frozen, and stored at −80°C until use. Samples were thawed and weighed, and single cell suspensions were prepared via passage through a 70-μm mesh (BD Falcon, Bedford, MA) in an appropriate volume of Dulbecco's modified Eagle medium (DMEM) supplemented with penicillin-streptomycin (iDMEM) to achieve a 100 mg/ml final concentration. Cell suspensions were centrifuged at 2,000 rpm for 5 min, and the supernatants were collected.
Madin-Darby canine kidney (MDCK) cells were plated (5 × 105) in each well of a 6-well plate. Samples (lung supernatants for mice and nasal washes for ferrets) were diluted (dilution factors of 1 × 101 to 1 × 106), overlaid onto the cells in 100 μl of iDMEM, and incubated for 1 h. Virus-containing medium was removed and replaced with 2 ml of L-15 medium plus 0.8% agarose (Cambrex, East Rutherford, NJ), and the samples were incubated for 96 h at 37°C with 5% CO2. Agarose was removed and discarded. Cells were fixed with 10% buffered formalin and then stained with 1% crystal violet for 15 min. Following thorough washing in distilled water (dH2O) to remove excess crystal violet, plates were allowed to dry, plaques were counted, and the PFU/g for lung supernatants or PFU/ml for nasal washes were calculated.
Histopathological analysis.
Left lobes of lungs from infected mice were collected 1, 3, and 5 days postinfection and placed into 10% buffered formalin. After fixation, lungs were paraffin embedded and 6-μm sections were prepared for histopathological analysis. Tissue sections were stained with hematoxylin and eosin. For in situ hybridization (ISH), vectors containing 760 bp of the influenza A/California/04/2009 matrix protein were linearized to create antisense and sense templates. 35S-labeled riboprobes were generated using the MAXIscript in vitro transcription kit (Ambion, Austin, TX). ISH was performed as described previously (3). Control riboprobes did not hybridize to lung tissue at any time point postinfection, and noninfected tissue did not show hybridization with viral probes. Hybridized slides were assessed and scored for abundance of foci.
Cellular assays.
The number of anti-influenza-specific cells secreting gamma interferon (IFN-γ) was determined by an enzyme-linked immunosorbent spot (ELISpot) assay (R&D systems, Minneapolis, MN) according to the manufacturer's protocol. Mice were sacrificed at 6 days postinfection (DPI), and spleens and lungs were harvested and prepared in single cell suspensions. Briefly, precoated anti-IFN-γ plates were blocked with RPMI medium plus 10% fetal calf serum (FCS) and antibiotics (cRPMI) for 30 min at room temperature. Medium was removed from the wells, and 105 cells were added to each well. Cells were stimulated with purified recombinant HA from A/Vietnam/1203/2004 (truncated at residue 530; 1 μg/well) or the inactivated 6:2 reassortant virus A/Vietnam/1203/2004 (1:100 dilution of inactivated stock; 100 μl/well) or with the immunodominant H2-Kd CD8+ T cell epitope in H5 HA, HA533 (IYSTVASSL; 1 μg/well) (Pepscan Presto, Leystad, Netherlands). Additional wells were stimulated with phorbol myristate acetate (PMA) (50 ng/well) and ionomycin (500 ng/well) as positive controls or Ova257 (SIINFEKL; 1 μg/well) (Pepscan Presto, Leystad, Netherlands) as a negative control. Additionally, IL-2 (10 U/ml) was added to each well. Plates were incubated at 37°C for 48 h. After incubation, plates were washed four times with R&D wash buffer and were incubated at 4°C overnight with biotinylated anti-mouse IFN-γ. Plates were washed as before and incubated at room temperature for 2 h with streptavidin conjugated to alkaline phosphatase. Plates were washed as before, and spots were developed by incubating at room temperature for 1 h in the dark with 5-bromo-4-chloro-3-indolylphosphate (BCIP)/nitroblue tetrazolium chloride (NBT) chromogen substrate. The plates were washed extensively with deionized (DI) H2O and allowed to dry overnight prior to the spots being counted using an ImmunoSpot ELISpot reader (Cellular Technology Ltd., Cleveland, OH).
The number of anti-HA- and anti-NA-specific antibody-secreting cells (ASC) was determined by a B cell ELISpot assay as previously described (21, 40, 41). Mice were sacrificed at 6 DPI, and spleens and lungs were harvested and prepared in single cell suspensions. Briefly, 0.45-μm PVDF membrane plates (Millipore, Billerica, MA) were coated with either purified recombinant HA from A/Vietnam/1203/2004 or purified recombinant NA from A/Thailand/1(KAN-1)/2004 (250 ng/well) and incubated at 4°C overnight. Plates were washed three times with PBS and blocked with cRPMI at 37°C for 3 to 4 h. Medium was removed from the wells, and 105 cells were added to each well. Plates were incubated at 37°C for 48 h. After incubation, plates were washed as before and incubated at room temperature for 2 h with HRP-conjugated anti-mouse IgG or IgA (Southern Biotech, Birmingham, AL). Plates were washed as before, and spots were developed at room temperature for 1 h in the dark with detection substrate (NovaRED; Vector Labs, Burlingame, CA). The plates were washed extensively with DI H2O and allowed to dry overnight prior to the spots being counted using an ImmunoSpot ELISpot reader (Cellular Technology Ltd., Cleveland, OH).
Passive transfer of sera.
Sera from vaccinated mice were pooled and passively transferred into 9-week-old recipient BALB/c mice (5 mice/group). Equal amounts of serum from each mouse in a particular vaccine group were pooled and heat inactivated for 30 min at 56°C. A total of 200 μl of pooled and inactivated serum was transferred to recipient mice via intraperitoneal (IP) injection. At 24 h posttransfer, mice were infected with the 6:2 reassortant virus with internal genes from the mouse-adapted virus A/Puerto Rico/8/1934 and surface antigens from A/Vietnam/1203/2004 as described above.
Statistical analysis.
Statistical significance of the antibody and cellular immunology data was determined using a two-tailed Student t test to analyze differences between COBRA and polyvalent vaccine groups for each of the different test antigens. Differences in weight loss and sickness scores were analyzed by a two-way analysis of variance (ANOVA) followed by Bonferroni's posttest for each vaccine group at multiple time points (multiparametric). Statistical significance of viral titer data was evaluated using a two-tailed Student t test on log10-transformed values. Significance was defined as a P value of <0.05. Statistical analyses were done using GraphPad Prism software.
RESULTS
VLPs elicit antibody responses in vaccinated mice and ferrets.
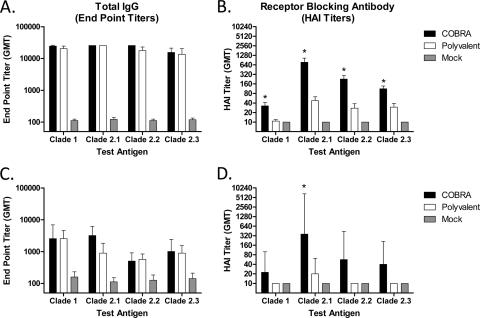
To expand our previous findings comparing COBRA to clade 2.2 VLPs (16), BALB/c mice (n = 30) were vaccinated twice via intramuscular injection with either purified COBRA or polyvalent VLPs, and 2 weeks after the second vaccination, serum was analyzed for antibody responses. All vaccinated mice had high-titer anti-HA antibodies that bound to recombinant HA derived from both clade 1 and various subclades of clade 2 (Fig. 1A). Although the COBRA and polyvalent vaccines elicited similar IgG titers (Fig. 1B), COBRA-vaccinated animals had higher HAI antibody titers for all viruses tested (P < 0.001). In addition to higher HAI titers, COBRA-vaccinated mice had an increased frequency of HAI titers of ≥1:40 for all viruses tested, including those which were components of the polyvalent formulation (Table 1).
Fig 1.
Vaccine-induced serum antibody responses. BALB/c mice (30 mice/group) (A and B) or Fitch ferrets (6 ferrets/group) (C and D) were vaccinated at 0 and 3 weeks, with blood collected 14 to 21 days after each vaccination. (A and C) Total IgG after the second vaccination was determined via ELISA for each vaccine group. (B and D) Receptor-blocking antibody titers after the second vaccination were determined via hemagglutination inhibition (HAI) assays for each vaccine group. Values are the geometric means of the reciprocal dilutions of the last positive wells and the 95% confidence intervals. Significant differences between the COBRA and polyvalent vaccines were determined by a two-tailed Student t test, and a P value of less than 0.05 was considered significant (indicated by an asterisk).
Table 1.
Seroconversion frequency in mice and ferrets after vaccination
| Species | Vaccine antigen | % of animals with seroconversion (no. of animals with seroconversion/total no. of animals of that species) in each cladea |
|||
|---|---|---|---|---|---|
| 1 | 2.1 | 2.2 | 2.3 | ||
| Mouse | COBRA | 60 (18/30) | 100 (30/30) | 100 (30/30) | 100 (30/30) |
| Polyvalent | 3.3 (1/30) | 70 (21/30) | 50 (15/30) | 53 (16/30) | |
| Ferret | COBRA | 33 (2/6) | 67 (4/6) | 50 (3/6) | 50 (3/6) |
| Polyvalent | 0 (0/6) | 33 (2/6) | 0 (0/6) | 0 (0/6) | |
Percent of animals with a HAI titer dilution of ≥1:40 (no. of responders/total no. of animals).
These results were confirmed in the ferret model, where 2 weeks following the final vaccination, all ferrets vaccinated with either the COBRA or polyvalent VLP vaccine had equivalent anti-HA IgG antibody titers against a panel of diverse H5N1 HA molecules (Fig. 1C). COBRA VLP-vaccinated ferrets had HAI titers higher than those of polyvalent VLP-vaccinated animals against all viruses tested (Fig. 1D). However, only the antibodies to the clade 2.1 virus were significantly different (P < 0.05). Furthermore, the frequency of COBRA-vaccinated animals with an HAI titer of ≥1:40 was higher than that of polyvalent VLP-vaccinated ferrets (Table 1).
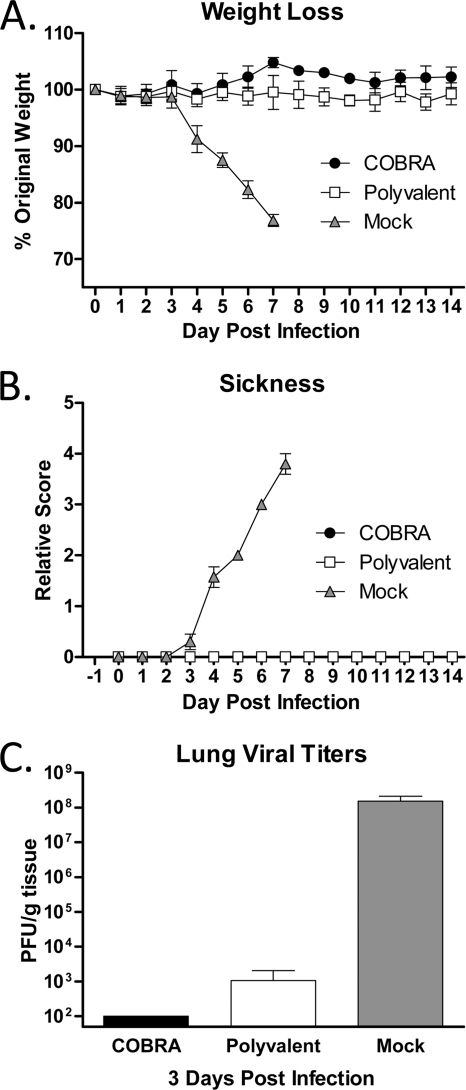
Challenge with highly pathogenic avian influenza (HPAI) H5N1 A/Whooper swan/Mongolia/244/2005 (clade 2.2) virus.
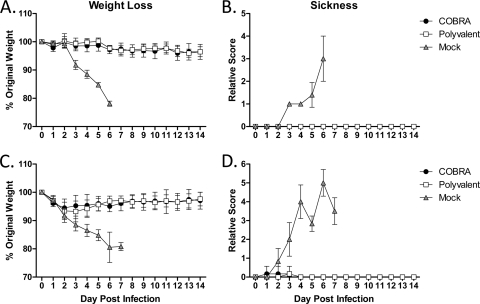
To confirm protective efficacy against highly pathogenic H5N1 infection, vaccinated animals were challenged with a lethal dose (5 × 103 PFU) of the wild-type clade 2.2 isolate A/Whooper Swan/Mongolia/244/2005. All VLP-vaccinated mice were protected from weight loss and death, while mock-vaccinated animals rapidly lost weight and reached experimental endpoints by 6 days postinfection (DPI) (Fig. 2A). Both COBRA and polyvalent VLP-vaccinated mice had mean maximum weight losses of 4% by 12 to 13 DPI. All VLP-vaccinated mice failed to develop any overt signs of disease, while mock-vaccinated mice developed visible illness (Fig. 2B).
Fig 2.
Highly pathogenic clade 2.2 challenge. Vaccinated BALB/c mice (5 mice/group) were infected with 5 × 103 PFU of the highly pathogenic clade 2.2 H5N1 virus A/Whooper Swan/Mongolia/244/2005 (WS/05). (A and B) Mice were monitored daily for weight loss (A) and sickness (B). (C and D) Vaccinated Fitch ferrets (6 ferrets/group) were infected with 1 × 106 PFU of the highly pathogenic clade 2.2 WS/05 virus. Ferrets were monitored daily for weight loss (C) and sickness (D). Values are the means and the standard errors of the mean (SEM) for each group.
Similar to the mice, all VLP-vaccinated ferrets were protected from death following a lethal challenge (1 × 106 PFU). Vaccinated ferrets demonstrated mild weight loss in response to the infection, with COBRA-vaccinated animals having a mean maximum weight loss of 5.5% at 2 DPI and polyvalent-vaccinated animals losing 6.8% by 3 DPI (Fig. 2C). Both groups rapidly recovered weight and failed to develop any significant signs of disease (Fig. 2D). Furthermore, VLP-vaccinated animals had no temperature spikes, while mock-vaccinated animals had temperatures elevated ∼3°C for 1 to 3 DPI (data not shown).
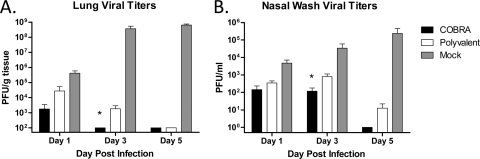
To evaluate vaccine efficacy with a more sensitive output than morbidity and mortality, the viral burden in the respiratory tracts of infected animals was assessed. Both COBRA- and polyvalent-vaccinated mice had reduced lung viral titers as soon as 1 DPI compared to those of mock-vaccinated animals. Furthermore, COBRA-vaccinated mice did not have detectable virus by 3 DPI, while polyvalent-vaccinated mice had prolonged viral replication, with 1.8 × 103 PFU/g detected at 3 DPI (P < 0.05) (Fig. 3A). Additionally, both VLP vaccines prevented extrapulmonary spread of the virus, while mock-vaccinated animals had detectable virus in both kidney and liver by 3 DPI (Table 2). Control of virus replication in ferrets was similar to that observed in mice, although complete clearance of the virus was delayed (Fig. 3B). All VLP-vaccinated animals had decreased recovery of virus in nasal washes compared to that of mock-vaccinated ferrets at every time point tested (P < 0.05). COBRA-vaccinated animals did not have detectable virus by 5 DPI. In contrast, virus replication did not reach undetectable levels until 9 DPI in polyvalent-vaccinated ferrets (data not shown).
Fig 3.
Clade 2.2 viral loads. (A) Vaccinated BALB/c mice (15 mice/group) were infected with 5 × 103 PFU of the highly pathogenic clade 2.2 H5N1 virus A/Whooper Swan/Mongolia/244/2005 (WS/05). Cohorts of mice (5 mice/group) were sacrificed at 1, 3, and 5 days postinfection, lungs were harvested, and viral load was determined by plaque assay. (B) Vaccinated Fitch ferrets (6 ferrets/group) were infected with 1 × 106 PFU of the highly pathogenic WS/05 virus. Nasal washes were collected, and viral load was determined by plaque assay. Values are the mean viral titers and the SEM for each group. Significant differences between COBRA and polyvalent vaccines were determined by a two-tailed Student t test, and a P value of less than 0.05 was considered significant (indicated by an asterisk).
Table 2.
Viral replication in different organs after vaccinations
| Vaccine | Organ | Virus titer (mean ± SD) at each day postinfectiona |
||
|---|---|---|---|---|
| 1 | 3 | 5 | ||
| COBRA | Lung | 2.39 ± 0.86 | <2 | <2 |
| Kidney | <2 | <2 | <2 | |
| Liver | <2 | <2 | <2 | |
| Polyvalent | Lung | 3.05 ± 1.3 | 2.93 ± 0.65 | <2 |
| Kidney | <2 | <2 | <2 | |
| Liver | <2 | <2 | <2 | |
| Mock | Lung | 5.42 ± 0.60 | 8.24 ± 0.678 | 8.79 ± 0.16 |
| Kidney | <2 | 2.57 ± 0.51 | 2.47 ± 0.40 | |
| Liver | <2 | 2.39 ± 0.68 | 2.23 ± 0.40 | |
Log10-transformed viral titers. Titers <2 have no SD listed because values were below the limit of detection of the assay.
Histopathology of infected lungs.
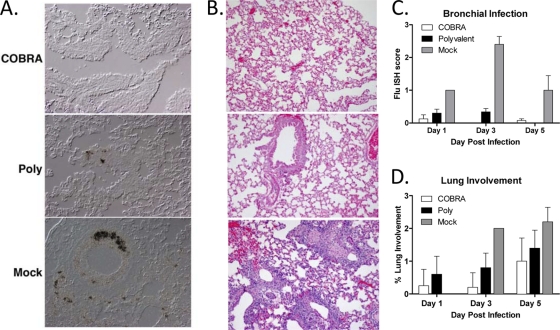
To evaluate the location of influenza viral replication and the severity of influenza-induced inflammation and damage, ISH staining was performed to detect influenza M1 on fixed lung tissues collected at 1, 3, and 5 DPI (Fig. 4). Both COBRA VLP- and polyvalent VLP-vaccinated animals had rare bronchial epithelium infections at 1 DPI (Fig. 4C). At 3 DPI, COBRA VLP-vaccinated animals had no detectable virally infected cells, while animals receiving polyvalent VLP vaccines had detectable bronchial epithelium infections (Fig. 4A and C). This was in contrast to significant bronchial epithelium infection and replication observed in mock-vaccinated animals (Fig. 4A and C).
Fig 4.
Histopathology of infected lungs. Vaccinated BALB/c mice (15 mice/group) were infected with 5 × 103 PFU of the highly pathogenic clade 2.2 H5N1 virus A/Whooper Swan/Mongolia/244/2005 (WS/05). Cohorts of mice (5 mice/group) were sacrificed at 1, 3, and 5 days postinfection. (A and B) IHC for influenza M1 (A) and hematoxylin and eosin staining (B) were performed on sections from paraffin-embedded lung tissue. Representative images are shown from 3 days postinfection. (C) The severity of the influenza ISH foci was assessed in the bronchi at 1, 3, and 5 DPI. The scoring was done as follows: 0, no definitive signal; 1, occasional focus; 2, focus in most fields; and 3, more than one focus per field. (D) The percentage of lung involvement was assessed in lung sections. The scoring was done as follows: 0, <10%; 1, 10 to 24%; 2, 25 to 50%; and 3, >50%.
To determine if vaccination protected animals from lung inflammation, hematoxylin- and eosin-stained sections were evaluated for histopathological changes at 1, 3, and 5 DPI (Fig. 4). COBRA VLP-vaccinated animals had less lung involvement than polyvalent VLP-vaccinated animals at all of the time points examined (Fig. 4B and D). COBRA VLP-vaccinated animals showed minor bronchial inflammation, while polyvalent VLP-vaccinated animals exhibited slight thickening of the bronchial epithelium (Fig. 4B). Both vaccinated groups showed little alveolar involvement compared to that of mock-vaccinated animals. Mock-vaccinated animals demonstrated greater lung involvement than the vaccinated groups and severe alveolar infiltration (Fig. 4B and D).
Challenge of vaccinated mice with A/Vietnam/1203/2004 (clade 1) H5N1 influenza virus.
Having established the clade 2.2 protective profile of both the COBRA and polyvalent VLP vaccines, the efficacy of these vaccines against a more divergent clade 1 challenge was evaluated in mice. COBRA- and polyvalent-vaccinated mice were challenged with the clade 1 reassortant virus A/Vietnam/1203/2004. All VLP-vaccinated animals were protected from weight loss and death, while mock-vaccinated animals rapidly lost weight and reached the experimental endpoint by 7 DPI (Fig. 5A). Furthermore, vaccinated mice also did not develop any signs of disease throughout the course of the study (Fig. 5B). Lungs were harvested at 3 DPI for determination of viral burden (Fig. 5C). COBRA VLP-vaccinated animals did not have detectable virus, while polyvalent VLP-vaccinated animals had 1.1 × 103 PFU/g virus (P = 0.12). Importantly, both vaccines had significantly less recoverable virus than mock-vaccinated animals at 3 DPI (P < 0.01).
Fig 5.
Clade 1 challenge. (A and B) Vaccinated BALB/c mice (4 mice/group) were infected with 5 × 103 PFU of reassortant virus containing the HA and NA genes from the clade 1 H5N1 virus A/Vietnam/1203/2004 (VN/04). Mice were monitored daily for weight loss (A) and sickness (B). Values are the means and standard deviations (SDs) for each group. (C) An additional cohort of vaccinated mice (3 mice/group) was infected, and lungs were harvested 3 days postinfection for analysis of viral burden. Values are the mean viral titers and the SEM for each group.
Analysis of cellular immune responses following challenge.
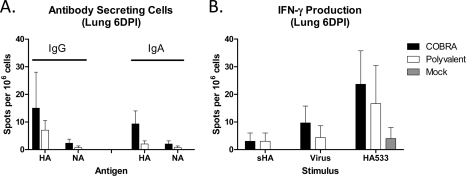
The magnitude of influenza-specific cellular immune responses in the lungs postinfection was evaluated via ELISpot assay for both antibody-secreting cells (ASC) and IFN-γ–producing cells. Vaccinated mice were infected with the clade 1 A/Vietnam/1203/2004 virus, and lungs were harvested at 6 DPI. COBRA- and polyvalent-vaccinated animals had statistically equivalent numbers of both IgG and IgA ASC (P > 0.05) specific for the A/Vietnam/1203/2004 challenge virus HA (Fig. 6A). No ASC were detected in mock-vaccinated animals. Additionally, the majority of the ASC response to infection was specific for HA, as fewer cells were detected for the NA component of either vaccine.
Fig 6.
Postchallenge cellular immune responses. Vaccinated BALB/c mice (3 mice/group) were infected with 5 × 103 PFU of reassortant virus containing the HA and NA genes from the clade 1 H5N1 virus A/Vietnam/1203/2004 (VN/04). Mice were sacrificed 6 days postinfection, lungs were harvested, and the numbers of antibody-secreting cells (A) and IFN-γ-producing cells (B) were determined by ELISpot assay. Values are the mean number of spots and the SEM for each group.
VLP vaccine-primed IFN-γ-secreting cells were also evaluated after infection. IFN-γ-responses were equivalent between the VLP vaccine groups regardless of the stimulating antigen (P > 0.05) (Fig. 6B). As expected, recombinant HA (sHA) and inactivated virus were inefficient stimulators of IFN-γ production compared to the immunodominant HA533 peptide. Overlapping peptide pools spanning the entire HA molecule were also used to stimulate collected cells, and no differences were observed between COBRA and polyvalent VLP vaccines for any of the pools (data not shown). Similar to the ASC data, no influenza-specific IFN-γ responses were detectable above background in mock-vaccinated animals at 6 DPI.
Passive transfer of vaccine-induced immune sera.
The contribution of serum factors to protection from clade 1 challenge was evaluated using a passive-transfer model. Nine-week-old recipient mice were administered pooled sera via IP injection from COBRA-, polyvalent-, and mock-vaccinated mice. Sera from COBRA- and polyvalent-vaccinated mice were confirmed to have equivalent levels of anti-clade 1 binding antibodies prior to transfer (data not shown). The next day, recipient mice were challenged with the clade 1 H5N1 virus. All recipient mice lost weight and had visible signs of morbidity (Fig. 7A and B). Interestingly, recipient mice receiving serum from COBRA VLP-vaccinated mice lost less weight than recipient mice receiving serum from polyvalent VLP-vaccinated mice. Recipient mice receiving COBRA VLP serum had a maximum loss of 5.2% at 6 DPI, and mice receiving polyvalent VLP serum had an 11.8% loss in weight at 7 DPI. Recipient mice that received serum from COBRA VLP-vaccinated mice also began to resolve the clinical symptoms more rapidly than mice receiving serum from polyvalent VLP-vaccinated mice (P < 0.05 at 7 DPI). Although serum from COBRA VLP-vaccinated mice prevented recipient mice from developing illness more efficiently than did serum from polyvalent VLP-vaccinated mice, both COBRA VLP- and polyvalent VLP-elicited sera protected all recipient mice from death. Conversely, all mice receiving serum from mock-vaccinated mice rapidly lost weight, had visible signs of morbidity, and reached the experimental endpoint by 7 DPI.
Fig 7.
Passive-transfer clade 1 challenge. BALB/c mice (10 mice/group) were vaccinated at 0 and 3 weeks, with blood collected 14 to 21 days after each vaccination. Sera collected after the second vaccination were pooled for each vaccine group and administered to naïve recipient mice (5 mice/group). One day after passive transfer, recipient mice were infected with 5 × 103 PFU of reassortant virus containing the HA and NA genes from the clade 1 H5N1 virus A/Vietnam/1203/2004 (VN/04). Mice were monitored daily for weight loss (A) and sickness (B). Values are the means and SDs for each group. Significant differences were determined by a two-way ANOVA with Bonferroni's posttest to evaluate differences between the vaccines at each day. A P value of less than 0.05 was considered significant (indicated by an asterisk).
DISCUSSION
In this study, we compared the immunogenicities and efficacies of two strategies proposed to increase breadth: computationally optimized broadly reactive antigen (COBRA) and a polyvalent mixture of primary antigens. Polyvalent vaccines have been used as a vaccine strategy to increase reactivity for many pathogens, including, but not limited to, influenza (12, 14, 37), monkeypox (19), HIV (2, 39), human papillomavirus (HPV) (15, 17), and pneumococcal disease (45). Polyvalent vaccines consist of a mixture of several antigens and are designed to elicit an immune response that is broader than that elicited by any single component. Seasonal influenza vaccines have traditionally been delivered as polyvalent formulations to address the diversity of currently circulating strains of influenza A (H1N1 and H3N2) and influenza B. Although polyvalent vaccine strategies are undoubtedly effective at expanding breadth, several limitations exist. First, with any polyvalent vaccine, production must be increased to include several different antigens. In the context of H5N1 vaccination, doses required for seroconversion are higher than those required for seasonal vaccines (26, 49), and producing multiple vaccines at higher doses could become a difficult hurdle for vaccine manufacturers. Second, strain selection remains critical to polyvalent vaccine efficacy, as best evidenced by seasonal influenza vaccine escape and continual yearly epidemics. Despite these limitations, the polyvalent strategy remains the standard approach for influenza vaccine design. Therefore, in this study, the breadth of immune responses elicited by COBRA HA antigens was compared to that elicited by a polyvalent mixture of primary H5N1 HA antigens. Previous polyvalent vaccines for H5N1 have focused on inclusion of antigens from different clades (12, 37). The COBRA HA antigen used in these studies is designed specifically to address the diversity present within clade 2 (16), and therefore, selected clade 2 antigens from primary H5N1 isolates were generated for the polyvalent vaccine. Clade 2 is genetically diverse and is divided into distinct subclades, including 2.1, 2.2, 2.3, 2.4, and 2.5, with some subclades being further divided into sub-subclades (53). Furthermore, within clade 2, humans have been infected with isolates representing clades 2.1, 2.2, and 2.3, with the most recent human infections in Egypt identified as clade 2.2 (53). To generate a polyvalent mixture covering the most prevalent subclades of clade 2, representative isolates were selected from clade 2.1 (A/Indonesia/5/2005), clade 2.2 (A/Whooper swan/Mongolia//244/2005), and clade 2.3 (A/Anhui/1/2005).
COBRA and polyvalent VLP vaccines efficiently elicited equivalent levels of broad binding antibodies in mice and ferrets (Fig. 1A and C), but the COBRA VLP vaccine elicited a broader profile of receptor-blocking antibodies (Fig. 1B and D). COBRA-vaccinated ferrets failed to achieve significantly higher levels of receptor-blocking antibodies to test antigens other than those in clade 2.1 (Fig. 1D). This could be due to the reduced number of animals compared to that in the mouse studies or because of the more rigorous nature of the ferret model. Although the ferret receptor-blocking antibody titers did not achieve significance (P > 0.05), the seroconversion frequency was increased compared to that of the polyvalent-vaccinated ferrets (Table 1). The polyvalent VLP vaccine did elicit antibodies to each of the components, confirming the validity of using a polyvalent strategy to increase vaccine breadth (Fig. 1B). Importantly, when any of the polyvalent components were used as a monovalent formulation, the receptor-blocking antibody profile was limited to the homologous test antigen (data not shown). One reason for the decreased receptor-blocking antibody titers in the polyvalent VLP-vaccinated groups could be that each of the components was administered at one-third of the total dose. While this is certainly a possible explanation, the titers of individual components given at a full dose are equivalent to those of the polyvalent vaccine for the homologous test antigen and remain decreased compared to those of the COBRA vaccine (data not shown). Furthermore, prior studies indicated equivalent antibody responses and protection profiles for the different HA doses used in these studies, both the total HA dose and individual-component HA doses (16; unpublished observations).
Both COBRA and polyvalent vaccines protected mice and ferrets from highly pathogenic clade 2 H5N1 viral challenge (Fig. 2). All vaccinated animals were protected from significant weight loss and did not develop overt signs of disease. These findings were consistent with our prior results and confirmed the observation that the COBRA vaccine protects as efficiently as vaccines homologous to the challenge virus (16). Although protection from severe illness and death is certainly critical for pandemic vaccines, decreasing viral replication is also important to limit the potential for both transmission and complications from secondary infections. Efficient human-to-human transmission is an essential factor in pandemic influenza emergence, and to date, H5N1 does not easily spread between people (30, 36, 50). However, if H5N1 were to acquire an efficient human transmission phenotype, the ability of a vaccine to reduce viral titers and thereby limit potential spread in addition to preventing severe disease is an important factor. Animals receiving the COBRA VLP vaccine had decreased levels of replicating virus and returned to baseline more rapidly than polyvalent-vaccinated animals (Fig. 3). Receptor-blocking antibodies are not required to protect experimental animals against severe disease and death induced by highly pathogenic H5N1 influenza infection (18, 28, 29, 34, 42). The findings reported here are consistent with those reports, but higher receptor-blocking antibody titers against the challenge virus did correlate with a reduction in the duration of viral replication (Fig. 1 and 3). Therefore, although high-titer receptor-blocking antibodies are not required for protection from severe disease, the presence of these antibodies may be predictive of a reduced viral burden. Receptor-blocking antibodies may be more effective at preventing nascent virions from infecting new cells and thereby limiting viral replication within the host. Viral replication could provide a more sensitive output for evaluating H5N1 vaccine efficacy.
The goal of these studies was to compare the breadths of two independent broadening strategies: COBRA and polyvalent. Although clade 2 is the most diverse and is spreading westward into the Middle East and Africa, clade 1 is still circulating and causing human disease in Southeast Asia (53). Clade 1 is not represented as a component of our polyvalent mixture and was not part of the COBRA design, and as such, it represents a stringent test to evaluate vaccine efficacy for both broadening strategies. Vaccinated animals had anti-clade 1 binding antibodies at levels equivalent to those of clade 2 test antigens (Fig. 1A and C). Despite these high-titer binding antibodies, both vaccines elicited low to undetectable levels of anti-clade 1 receptor-blocking antibodies (Fig. 1B and D). Consistent with the findings that receptor-blocking antibodies are not predictive of protection from severe disease, all vaccinated animals were protected from weight loss and development of visible illness after challenge with a reassortant virus containing the HA and NA antigens derived from a clade 1 virus (Fig. 5). Furthermore, COBRA-vaccinated animals did not have detectable virus after 3 days postinfection, while polyvalent-vaccinated animals had low levels of virus present. Although the differences were not significant, the viral replication in COBRA-vaccinated animals was below the limit of detection of the assay and animals receiving the polyvalent vaccine had recoverable virus 3 days after clade 1 challenge. One explanation for this finding is the low level of anti-clade 1 receptor-binding antibodies in the COBRA-vaccinated animals (Fig. 1B). These results indicate that even though clade 1 sequences were not included in the design of COBRA, the synthetic antigen serves as an effective vaccine against a divergent virus, even one that is antigenically distinct from the original input sequences. Therefore, we propose that the centralized nature and layered design of the COBRA HA sequence prevent the accumulation of clade-specific, immunodominant, antigenic characteristics present in primary HA sequences. This hypothesis is supported by the phylogenetic location of this COBRA HA sequence and by antigenic modeling that suggests that the COBRA HA retains the most common structure at predicted antigenic regions while primary isolates have clade-specific divergences (16).
In the absence of receptor-blocking antibodies, it is possible that cellular immune responses contribute to the protection from severe disease and death (7, 33, 44). Both COBRA and polyvalent vaccines elicited similar cellular recall responses at 6 days postinfection (Fig. 6). Both IgG and IgA antibody-secreting cells (ASC) specific for the clade 1 HA were recruited to the lungs of vaccinated animals, and this is likely a recall response, as unvaccinated controls did not have any ASC above background at 6 days postinfection (Fig. 6A). ASC in either the spleen or bone marrow were not detectable above the background of the assay in any group before or after challenge (data not shown). Interestingly, the majority of ASC were specific for HA rather than NA. Although the NA content of the vaccines was not standardized, all vaccinated animals had high anti-NA serum IgG prior to challenge (data not shown). It is possible that NA-mediated immunity contributes to protection from challenge in the absence of receptor-blocking antibodies, but the preferential recruitment of anti-HA ASC implies a more critical role for anti-HA binding antibodies. An additional implication for the equivalent numbers of ASC for both vaccine groups is that increased receptor-blocking antibody titers are not directly correlated with increased numbers of ASC (Fig. 1B and 6). This is not a surprising finding, since the titers of binding antibodies were equivalent between the two vaccine groups (Fig. 1A). Importantly, both strategies resulted in recruitment of equivalent numbers of ASC in response to a completely heterologous infection.
One of the proposed advantages for utilizing centralized antigens is expanding the breadth of T cell epitopes (23, 31, 55). Indeed, a consensus-based H5N1 virus elicited IFN-γ–producing cells in response to multiple peptide pools of HA (25). Both the COBRA and polyvalent VLP vaccines elicited IFN-γ responses in the lungs of infected mice by 6 days postinfection, and the numbers of responding cells in the vaccine groups were equivalent, regardless of the stimulating antigen (Fig. 6B). HA533 is the immunodominant, HA-derived CD8+ T cell epitope in BALB/c mice and is conserved in all HA vaccine antigens used in this study (11). The conservation of this epitope in all vaccine strains used in these studies may be responsible for the equivalent responses in the two vaccine groups. Additionally, the COBRA vaccine did not result in expansion of breadth of T cell reactivity across different regions of HA, as measured by stimulation with overlapping peptide pools (data not shown). COBRA failed to expand the breadth of T cell responses in these experiments, which may be a result of the high levels of homology between HA antigens. H5N1 HA protein antigens are >90% identical between strains, and the majority of the diversity is focused on altering antibody binding sites that are usually nonlinear, conformational epitopes. Therefore, any potential T cell epitopes are not changing due to immune pressure and likely remain highly conserved between strains. Indeed, the HA533 BALB/c immunodominant epitope is conserved not only throughout H5N1 strains but also in H1N1 and H9N2.
The presence of B and T cell recall responses in the lungs of vaccinated mice after challenge could be a potential mechanism for protection instead of or in addition to serum antibodies. To evaluate the role of serum factors in protecting mice from a heterologous challenge, we passively transferred immune serum to naïve recipients and challenged with a clade 1 reassortant virus. Both COBRA and polyvalent immune sera protected mice from severe disease and death (Fig. 7). Recipient mice that received immune serum from mice vaccinated with COBRA VLPs lost less weight and recovered from illness more rapidly than recipient mice receiving serum from mice vaccinated with polyvalent VLPs. This result was most likely due to the low levels of anti-clade 1 receptor-blocking antibodies in the serum from COBRA VLP-vaccinated mice (Fig. 1B). In comparison to mice vaccinated directly with VLPs, mice receiving immune serum passively lost more weight and developed more clinical signs following virus challenge. One possibility for this disparity is that for the transferred antibodies to gain access to the site of infection in the airway, there must first be damage to the lung by the virus infection. Transferred antibodies could then function in several ways: direct neutralization of virus, opsonization of viral particles, complement fixation, and/or impairment of viral egress from infected cells. These H5N1 VLP vaccines elicit a mixed antibody isotype profile (IgG1, IgG2a, and IgG2b) that enables these diverse nonneutralizing antibody functions (16). Initial lung damage would then be associated with the observed mild weight loss and development of visible sickness, and the speed of recovery would be related to the quality and function of the transferred antibody. Importantly, this is the first description of passive-transfer-mediated protection by a centralized antigen for H5N1, indicating that serum antibodies are indeed important factors in heterologous protection.
This is the first report comparing the breadth of a centralized H5N1 antigen, COBRA, with the more traditional strategy of a polyvalent mixture. The data presented here indicate that although binding antibodies are sufficient for protection from severe disease and death, receptor-blocking antibodies are essential for reducing viral replication. This observation suggests that in the context of pandemic preparedness, vaccines that elicit receptor-blocking antibodies to a diverse set of viruses would be more effective at limiting viral replication, transmission, and disease impact. Antigenic diversity is a challenge not only for H5N1 influenza but for all influenza subtypes. Polyvalent vaccines are currently utilized to address multiple types and subtypes of influenza simultaneously circulating in the human population. It remains to be determined if COBRA-based vaccine design will be able to overcome diversity between subtypes, as greater levels of diversity and structural limitations are challenges for antigen design. Both COBRA and polyvalent strategies are effective at expanding antibody breadth in the context of influenza vaccination. These two strategies are not mutually exclusive, and an intriguing possibility is a combination thereof: a polyvalent mixture of COBRA antigens. Combining the intrasubtype broadening ability of COBRA with the intersubtype advantages of polyvalent mixtures represents an interesting strategy for expanding breadth within and between subtypes. COBRA-based vaccines are effective at broadening the antibody repertoire against H5N1, and applying this design strategy to other subtypes of influenza warrants further investigation.
ACKNOWLEDGMENTS
We thank R. Sodnomdarjaa and the Mongolian State Central Veterinary Laboratory for providing permission to use the A/Whooper Swan/Mongolia/244/2005 influenza virus.
This work was supported by the National Institutes of Health training grant (T32AI060525 to B.M.G.), National Institutes of Health/National Institute of Allergy and Infectious Diseases awards (K24MH01717 to C.A.W. and U01AI077771 to T.M.R. and C.A.W.), and a collaborative research grant from PATH Vaccine Solutions (to T.M.R.). This work was supported, in part, by a grant from the Pennsylvania Department of Health.
The Pennsylvania Department of Health specifically disclaims responsibility for any analyses, interpretations, or conclusions.
We have no conflict of interest in the results reported in this paper.
Footnotes
Published ahead of print 21 December 2011
REFERENCES
- 1. Askonas B, McMichael A, Webster R. 1982. The immune response to influenza viruses and the problem of protection against infection, p 159–188 In Beare AS. (ed), Basic and applied influenza research. CRC Press, Boca Raton, FL [Google Scholar]
- 2. Barouch DH, et al. 2010. Mosaic HIV-1 vaccines expand the breadth and depth of cellular immune responses in rhesus monkeys. Nat. Med. 16:319–323 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Bissel SJ, et al. 2011. Acute murine H5N1 influenza A encephalitis. Brain Pathol., in press [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Bright RA, et al. 2005. Incidence of adamantane resistance among influenza A (H3N2) viruses isolated worldwide from 1994 to 2005: a cause for concern. Lancet 366:1175–1181 [DOI] [PubMed] [Google Scholar]
- 5. Bright RA, Ross TM, Subbarao K, Robinson HL, Katz JM. 2003. Impact of glycosylation on the immunogenicity of a DNA-based influenza H5 HA vaccine. Virology 308:270–278 [DOI] [PubMed] [Google Scholar]
- 6. Bright RA, Shay DK, Shu B, Cox NJ, Klimov AI. 2006. Adamantane resistance among influenza A viruses isolated early during the 2005–2006 influenza season in the United States. JAMA 295:891–894 [DOI] [PubMed] [Google Scholar]
- 7. Brown DM, Dilzer AM, Meents DL, Swain SL. 2006. CD4 T cell-mediated protection from lethal influenza: perforin and antibody-mediated mechanisms give a one-two punch. J. Immunol. 177:2888–2898 [DOI] [PubMed] [Google Scholar]
- 7a. Centers for Disease Control and Prevention 2009. Biosafety in microbiological and biomedical laboratories, 5th ed Centers for Disease Control and Prevention, Atlanta, GA [Google Scholar]
- 8. Chan M. 11 June 2009, posting date World now at the start of 2009 influenza pandemic. World Health Organization, Geneva, Switzerland: http://www.who.int/mediacentre/news/statements/2009/h1n1_pandemic_phase6_20090611/en/index.html [Google Scholar]
- 9. Chen L-M, Davis CT, Zhou H, Cox NJ, Donis RO. 2008. Genetic compatibility and virulence of reassortants derived from contemporary avian H5N1 and human H3N2 influenza A viruses. PLoS Pathog. 4:e1000072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Chen M-W, et al. 2008. A consensus hemagglutinin-based DNA vaccine that protects mice against divergent H5N1 influenza viruses. Proc. Natl. Acad. Sci. U. S. A. 105:13538–13543 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Chen W, Bennink JR, Morton PA, Yewdell JW. 2002. Mice deficient in perforin, CD4+ T cells, or CD28-mediated signaling maintain the typical immunodominance hierarchies of CD8+ T-cell responses to influenza virus. J. Virol. 76:10332–10337 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Crevar CJ, Ross TM. 2008. Elicitation of protective immune responses using a bivalent H5N1 VLP vaccine. Virol. J. 5:131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Ducatez MF, et al. 2011. Feasibility of reconstructed ancestral H5N1 influenza viruses for cross-clade protective vaccine development. Proc. Natl. Acad. Sci. U. S. A. 108:349–354 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Fiore AE, et al. 2010. Prevention and control of influenza with vaccines. MMWR Morb. Mortal. Wkly. Rep. 59:1–6220075837 [Google Scholar]
- 15. Garland SM, et al. 2007. Quadrivalent vaccine against human papillomavirus to prevent anogenital diseases. N. Engl. J. Med. 356:1928–1943 [DOI] [PubMed] [Google Scholar]
- 16. Giles BM, Ross TM. 2011. A computationally optimized broadly reactive antigen (COBRA) based H5N1 VLP vaccine elicits broadly reactive antibodies in mice and ferrets. Vaccine 29:3043–3054 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Giuliano AR, et al. 2011. Efficacy of quadrivalent HPV vaccine against HPV infection and disease in males. N. Engl. J. Med. 364:401–411 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Govorkova E, Webby R, Humberd J, Seiler J, Webster R. 2006. Immunization with reverse genetics produced H5N1 influenza vaccine protects ferrets against homologous and heterologous challenge. J. Infect. Dis. 194:159–167 [DOI] [PubMed] [Google Scholar]
- 19. Hirao LA, et al. 2011. Multivalent smallpox DNA vaccine delivered by intradermal electroporation drives protective immunity in nonhuman primates against lethal monkeypox challenge. J. Infect. Dis. 203:95–102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Horimoto T, Kawaoka Y. 2005. Influenza: lessons from past pandemics, warnings from current incidents. Nat. Rev. Microbiol. 3:591–600 [DOI] [PubMed] [Google Scholar]
- 21. Joo HM, He Y, Sundararajan A, Huan L, Sangster MY. 2010. Quantitative analysis of influenza virus-specific B cell memory generated by different routes of inactivated virus vaccination. Vaccine 28:2186–2194 [DOI] [PubMed] [Google Scholar]
- 22. Kendal AP, Pereira MS, Skehel JJ. 1982. Concepts and procedures for laboratory-based influenza surveillance. U.S. Department of Health and Human Services, Public Health Service, Centers for Disease Control, Atlanta, GA [Google Scholar]
- 23. Laddy DJ, et al. 2007. Immunogenicity of novel consensus-based DNA vaccines against avian influenza. Vaccine 25:2984–2989 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Laddy DJ, et al. 2009. Electroporation of synthetic DNA antigens offers protection in nonhuman primates challenged with highly pathogenic avian influenza virus. J. Virol. 83:4624–4630 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Laddy DJ, et al. 2008. Heterosubtypic protection against pathogenic human and avian influenza viruses via in vivo electroporation of synthetic consensus DNA antigens. PLoS One 3:e2517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Leroux-Roels I, et al. 2007. Antigen sparing and cross-reactive immunity with an adjuvanted rH5N1 prototype pandemic influenza vaccine: a randomised controlled trial. Lancet 370:580–589 [DOI] [PubMed] [Google Scholar]
- 27. Li C, et al. 2010. Reassortment between avian H5N1 and human H3N2 influenza viruses creates hybrid viruses with substantial virulence. Proc. Natl. Acad. Sci. U. S. A. 107:4687–4692 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Lipatov A, Hoffmann E, Salomon R, Yen H-L, Webster R. 2006. Cross-protectiveness and immunogenicity of influenza A/Duck/Singapore/3/97(H5) vaccines against infection with A/Vietnam/1203/04(H5N1) virus in ferrets. J. Infect. Dis. 194:1040–1043 [DOI] [PubMed] [Google Scholar]
- 29. Lu X, et al. 2006. Cross-protective immunity in mice induced by live-attenuated or inactivated vaccines against highly pathogenic influenza A (H5N1) viruses. Vaccine 24:6588–6593 [DOI] [PubMed] [Google Scholar]
- 30. Maines TR, et al. 2006. Lack of transmission of H5N1 avian-human reassortant influenza viruses in a ferret model. Proc. Natl. Acad. Sci. U. S. A. 103:12121–12126 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. McBurney SP, Ross TM. 2009. Human immunodeficiency virus-like particles with consensus envelopes elicited broader cell-mediated peripheral and mucosal immune responses than polyvalent and monovalent Env vaccines. Vaccine 27:4337–4349 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Mitchell JA, Green TD, Bright RA, Ross TM. 2003. Induction of heterosubtypic immunity to influenza A virus using a DNA vaccine expressing hemagglutinin-C3d fusion proteins. Vaccine 21:902–914 [DOI] [PubMed] [Google Scholar]
- 33. Mozdzanowska K, Maiese K, Gerhard W. 2000. Th cell-deficient mice control influenza virus infection more effectively than Th- and B cell-deficient mice: evidence for a Th-independent contribution by B cells to virus clearance. J. Immunol. 164:2635–2643 [DOI] [PubMed] [Google Scholar]
- 33a. National Research Council 1996. Guide for the care and use of laboratory animals. National Academy Press, Washington, DC [Google Scholar]
- 34. Ninomiya A, Imai M, Tashiro M, Odagiri T. 2007. Inactivated influenza H5N1 whole-virus vaccine with aluminum adjuvant induces homologous and heterologous protective immunities against lethal challenge with highly pathogenic H5N1 avian influenza viruses in a mouse model. Vaccine 25:3554–3560 [DOI] [PubMed] [Google Scholar]
- 35. Octaviani CP, Ozawa M, Yamada S, Goto H, Kawaoka Y. 2010. High level of genetic compatibility between swine-origin H1N1 and highly pathogenic avian H5N1 influenza viruses. J. Virol. 84:10918–10922 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Olsen SJ, et al. 2005. Family clustering of avian influenza A (H5N1). Emerg. Infect. Dis. 11:1799–1801 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Prabakaran M, et al. 2010. Neutralizing epitopes of influenza virus hemagglutinin: target for the development of a universal vaccine against H5N1 lineages. J. Virol. 84:11822–11830 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Ross TM, Xu Y, Bright RA, Robinson HL. 2000. C3d enhancement of antibodies to hemagglutinin accelerates protection against influenza virus challenge. Nat. Immunol. 1:127–131 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Santra S, et al. 2010. Mosaic vaccines elicit CD8+ T lymphocyte responses that confer enhanced immune coverage of diverse HIV strains in monkeys. Nat. Med. 16:324–328 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Sasaki S, et al. 2008. Influence of prior influenza vaccination on antibody and B-cell responses. PLoS One 3:e2975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Sasaki S, et al. 2007. Comparison of the influenza virus-specific effector and memory B-cell responses to immunization of children and adults with live attenuated or inactivated influenza virus vaccines. J. Virol. 81:215–228 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Suguitan AL, et al. 2006. Live, attenuated influenza A H5N1 candidate vaccines provide broad cross-protection in mice and ferrets. PLoS Med. 3:e360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Taubenberger JK, Morens DM. 2009. Pandemic influenza—including a risk assessment of H5N1. Rev. Sci. Tech. 28:187–202 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Teijaro JR, Verhoeven D, Page CA, Turner D, Farber DL. 2010. Memory CD4 T cells direct protective responses to influenza virus in the lungs through helper-independent mechanisms. J. Virol. 84:9217–9226 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Teshale EH, et al. 2008. Effectiveness of 23-valent polysaccharide pneumococcal vaccine on pneumonia in HIV-infected adults in the United States, 1998-2003. Vaccine 26:5830–5834 [DOI] [PubMed] [Google Scholar]
- 46. Toapanta F, Ross T. 2009. Impaired immune responses in the lungs of aged mice following influenza infection. Respir. Res. 10:112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Tobita K. 1975. Permanent canine kidney (MDCK) cells for isolation and plaque assay of influenza B viruses. Med. Microbiol. Immunol. 162:23–27 [DOI] [PubMed] [Google Scholar]
- 48. Tobita K, Sugiura A, Enomote C, Furuyama M. 1975. Plaque assay and primary isolation of influenza A viruses in an established line of canine kidney cells (MDCK) in the presence of trypsin. Med. Microbiol. Immunol. 162:9–14 [DOI] [PubMed] [Google Scholar]
- 49. Treanor JJ, et al. 2001. Safety and immunogenicity of a recombinant hemagglutinin vaccine for H5 influenza in humans. Vaccine 19:1732–1737 [DOI] [PubMed] [Google Scholar]
- 50. Ungchusak K, et al. 2005. Probable person-to-person transmission of avian influenza A (H5N1). N. Engl. J. Med. 352:333–340 [DOI] [PubMed] [Google Scholar]
- 51. Webby RJ, Webster RG. 2003. Are we ready for pandemic influenza? Science 302:1519–1522 [DOI] [PubMed] [Google Scholar]
- 52. Webster R, Govorkova E. 2006. H5N1 influenza: continuing evolution and spread. N. Engl. J. Med. 355:2174–2177 [DOI] [PubMed] [Google Scholar]
- 53. World Health Organization September 2011, posting date Antigenic and genetic characteristics of influenza A(H5N1) and influenza A(H9N2) viruses for the development of candidate vaccine viruses for pandemic preparedness. http://www.who.int/influenza/resources/documents/2011_09_h5_h9_vaccinevirusupdate.pdf [PubMed]
- 54. World Health Organization 15 December 2011, posting date Cumulative number of confirmed human cases of avian influenza A(H5N1) reported to WHO. http://www.who.int/influenza/human_animal_interface/EN_GIP_20111215CumulativeNumberH5N1cases.pdf
- 55. Yan J, et al. 2007. Enhanced cellular immune responses elicited by an engineered HIV-1 subtype B consensus-based envelope DNA vaccine. Mol. Ther. 15:411–421 [DOI] [PubMed] [Google Scholar]