Abstract
l-Proline can be used by Bacillus subtilis as a sole source of carbon or nitrogen. We traced l-proline utilization genetically to the putBCP (ycgMNO) locus. The putBCP gene cluster encodes a high-affinity proline transporter (PutP) and two enzymes, the proline dehydrogenase PutB and the Δ1-pyrroline-5-carboxylate dehydrogenase PutC, which jointly catabolize l-proline to l-glutamate. Northern blotting, primer extension, and putB-treA reporter gene fusion analysis showed that the putBCP locus is transcribed as an l-proline-inducible operon. Its expression was mediated by a SigA-type promoter and was dependent on the proline-responsive PutR activator protein. Induction of putBCP expression was triggered by the presence of submillimolar concentrations of l-proline in the growth medium. However, the very large quantities of l-proline (up to several hundred millimolar) synthesized by B. subtilis as a stress protectant against high osmolarity did not induce putBCP transcription. Induction of putBCP transcription by external l-proline was not dependent on l-proline uptake via the substrate-inducible PutP or the osmotically inducible OpuE transporter. It was also not dependent on the chemoreceptor protein McpC required for chemotaxis toward l-proline. Our findings imply that B. subtilis can distinguish externally supplied l-proline from internal l-proline pools generated through de novo synthesis. The molecular basis of this regulatory phenomenon is not understood. However, it provides the B. subtilis cell with a means to avoid a futile cycle of de novo l-proline synthesis and consumption by not triggering the expression of the putBCP l-proline catabolic genes in response to the osmoadaptive production of the compatible solute l-proline.
INTRODUCTION
The soil-dwelling Gram-positive bacterium Bacillus subtilis lives in a challenging habitat in which the supply of nutrients is often restricted (20, 27, 56). Amino acids are particularly valuable resources for bacteria because they not only can be used as preformed building blocks for protein synthesis but often can also be employed as sole carbon, energy, and nitrogen (or sulfur) sources (22). They enter the habitat of B. subtilis as root exudates (64), as decomposed plant material (27), and as products of lysed or osmotically down-shocked microbial cells (66). B. subtilis can actively seek amino acids as nutrients through chemotaxis (49). Here, we focus on the utilization by B. subtilis of l-proline as a sole carbon and energy source and as a sole nitrogen source.
Many bacteria can employ l-proline as a nutrient, and the catabolism of the amino acid typically involves its enzymatic oxidation to l-glutamate (61, 72), a central metabolite positioned at the intersection of carbon and nitrogen metabolism in many microorganisms (14, 55). Oxidation of proline is catalyzed in a two-step reaction by a flavin-containing proline dehydrogenase (PRODH) (EC 1.5.99.8) to Δ1-pyrroline-5-carboxylate (P5C). This intermediate spontaneously hydrolyzes to γ-glutamate-5-semialdehyde, which is then further oxidized by a NAD-dependent Δ1-pyrroline-5-carboxylate dehydrogenase (P5CDH) (EC 1.5.1.12) to l-glutamate (Fig. 1B). These enzymatic steps can either be carried out by a single bifunctional protein comprising two domains (PRODH-P5CDH) (59) or by monofunctional proteins with separate PRODH and P5CDH activities (32, 68).
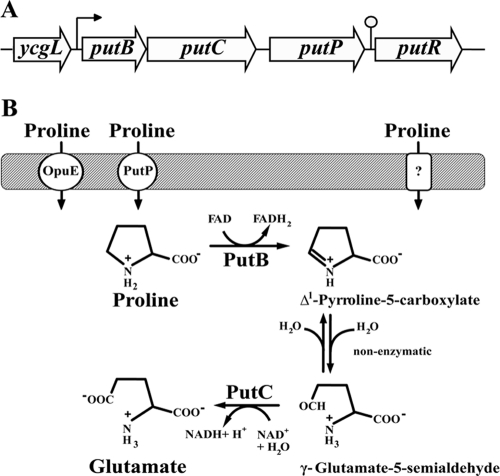
Fig 1.
l-Proline utilization system of B. subtilis. (A) Genetic organization of the putBCP and putR region. The promoter and the predicted Rho-independent terminator for the putBCP gene cluster are indicated by an arrow and a lollipop, respectively. (B) Proposed route for l-proline uptake and catabolism in B. subtilis.
Particularly well-studied examples of proline utilization by microorganisms are the PutPA systems of the enterobacteria Escherichia coli and Salmonella enterica serovar Typhimurium (37, 52, 61, 70, 72). PutP is a high-affinity proline importer and a member of the sodium solute symporter (SSS) (TC2A.21) family, transporters that harness electrochemical Na+ gradients to couple the flow of Na+ ions with the transport of solutes across biological membranes (48). The PutA protein is a trifunctional membrane-associated enzyme comprising both PRODH and P5CDH domains (Fig. 1B) (37, 52) and also contains an N-terminal ribbon-helix-helix DNA-binding domain that endows PutA with the ability to act as a transcriptional repressor (61, 71). Depending on the redox state (52, 70), PutA can switch between its functions as a membrane-associated l-proline-degradative enzyme and a cytoplasmic regulatory protein to repress the expression of the putPA proline utilization gene cluster when no proline is present in the growth medium (41, 46, 61, 71, 72). Transcription of the putPA genes is also regulated via the cyclic AMP (cAMP)-cAMP receptor protein (CRP) complex (13), thereby embedding l-proline utilization into a globally acting regulatory network that prioritizes the use of various carbon sources by microbial cells.
l-Proline utilization has also been investigated in microorganisms other than E. coli and S. enterica serovar Typhimurium. A common denominator of all these microbial proline utilization systems is the induction of put gene expression in response to an external supply of l-proline and the use of the concerted actions of PRODH- and P5CDH-type enzymes to catabolize l-proline to l-glutamate (61). However, the types of regulatory proteins and genetic circuits that control put expression in response to l-proline availability vary considerably in microorganisms (33, 35, 36, 47, 57, 64).
The use of l-proline as a nutrient by B. subtilis is well known (4, 6, 8), but the underlying systems for l-proline uptake and catabolism have so far not been studied in any detail. Recently, the understanding of the genetic control of the use of l-proline as a nutrient by B. subtilis was advanced by the identification of a proline-responsive activator protein (PutR) that controls the expression of the l-proline utilization putBCP (formerly ycgMNO [9]) gene cluster (7, 31). PutR activates putBCP transcription in response to proline availability but can be displaced by an active form of the negatively acting CodY regulatory protein (4, 44, 54) from the putBCP promoter region (7), thereby establishing repression of the gene cluster.
Here, we have focused on physiological aspects of l-proline utilization by B. subtilis and have studied the role of the putBCP-encoded proline transporter PutP and of the PutB (PRODH) and PutC (P5CDH) enzymes. Catabolism of l-proline by B. subtilis poses interesting questions, since l-proline also serves as an important osmostress protectant for the soil bacterium (9, 10). Both the osmotically inducible de novo synthesis of the compatible solute l-proline (11, 67) and its import via the osmotically inducible OpuE transporter (58, 65) confer stress resistance to high-osmolarity challenges.
We discovered that the expression of the catabolic putBCP operon of B. subtilis can be induced in a PutR-dependent fashion by very low concentrations (low μM range) of l-proline present in the growth medium but that the very large quantities of l-proline amassed via de novo synthesis under osmotic stress conditions (several hundred millimolar) do not trigger enhanced putBCP transcription. Physiologically, this allows the B. subtilis cell to avoid a futile cycle of l-proline production and degradation when it faces high-osmolarity surroundings.
MATERIALS AND METHODS
Chemicals.
l-Proline, trans-4-hydroxyproline, thioproline, l-azetidine-2-carboxylic acid (AC), 3,4-dehydro-dl-proline (DHP), the chromogenic substrates for the TreA [phospho-α-(1,1)-glucosidase] enzyme para-nitrophenyl-α-d-glucopyranoside (α-PNPG) and for the ProB enzyme (o-aminobenzaldehyde), and the ninhydrine reagent, as well as the antibiotics chloramphenicol, kanamycin, tetracycline, erythromycin, and spectinomycin, were purchased from Sigma-Aldrich (Steinheim, Germany). Dimethyl-proline (proline betaine) was purchased from Atkins Chemicals (Chengdu, China), and monomethyl-proline (1) was a kind gift from D. Le Rudulier (University of Nice, Nice, France). l-[14C(U)]proline (40 mCi mmol−1) was purchased from DuPont de Nemour GmbH (Neu-Isenburg, Germany).
Bacterial strains.
The E. coli strain DH5α (Invitrogen, Carlsbad, CA) was used for routine cloning purposes and maintenance of cloning vectors and recombinant plasmids. These strains were grown and maintained on Luria-Bertani (LB) agar plates. Solid and liquid media contained, when necessary, antibiotics to select for the presence of plasmids. The B. subtilis wild-type strain JH642 (trpC2 pheA1) (a kind gift of J. Hoch, Scripps Research Institute) and its mutant derivatives were used throughout this study (Table 1).
Table 1.
B. subtilis strains used in this study
| Straina | Relevant genotypeb | Source |
|---|---|---|
| JH642 | trpC2 pheA1 | J. Hoch; BGSCc 1A96 |
| GNB37 | Δ(treA::erm)2 | G. Nau-Wagner |
| MBB1 | Δ(treA::neo)1 | M. Brosius |
| BLOB9 | Δ(opuE::tet)1 | 65 |
| SMB10 | amyE::ϕ(putB′-treA)1 Δ(treA::neo)1 | This study |
| SMB11 | Δ(putP::spc)1 | This study |
| SMB12 | Δ(putP::spc)1 Δ(opuE::tet)1 | This study |
| SMB14 | amyE::ϕ(putB′-treA)1 Δ(putP::spc)1 Δ(treA::neo)1 | This study |
| SMB27 | amyE::ϕ(putB′-treA)1 Δ(putP::spc)1 Δ(opuE::tet)1 Δ(treA::neo)1 | This study |
| SMB28 | amyE::ϕ(putB′-treA)1 Δ(opuE::tet) Δ(treA::neo)1 | This study |
| SMB32 | Δ(putC::neo)2 | This study |
| SMB34 | amyE::ϕ(putB′-treA)1 Δ(putC::neo)2 Δ(treA::erm)2 | This study |
| SMB42 | Δ(putB::spc)3 | This study |
| SMB45 | Δ(putBCP::tet)2 | This study |
| SMB46 | amyE::ϕ(putB′-treA)1 Δ(putBCP::tet)2 Δ(treA::erm)2 | This study |
| SMB49 | amyE::ϕ(putB′-treA)1 Δ(putB::spc)3 Δ(treA::erm)2 | This study |
| TSB2 | amyE::ϕ(putB′-treA)1 Δ(putR::spc) Δ(treA::neo)1 | This study |
| TSB3 | amyE::ϕ(putB′-treA)1 Δ(putC::neo)2 Δ(putR::spc) Δ(treA::erm)2 | This study |
| ACB154 | amyE::ϕ(putB′-treA)1 Δ(treA::kan)2 mcpC::erm | This study |
| BB3330 | SMY Δ(putR::cat) | 7 |
| BB3530 | SMY Δ(putR::spc) | This study |
| OI3280 | trpF7 hisH2 metC mcp::erm | 45 |
All strains except BB3330, BB3530, and OI3280 are derivatives of B. subtilis strain JH642 and therefore carry, in addition to the genetic markers indicated, the trpC2 pheA1 mutations.
The designation amyE::ϕ(putB′-treA)1 indicates that the putB-treA operon gene fusion is stably integrated via a double-recombination event into the chromosomal amyE gene of B. subtilis as a single copy, thereby rendering the fusion strains defective in the extracellular AmyE α-amylase. The ϕ(putB′-treA)1 reporter fusion is linked to a chloramphenicol resistance gene (cat), thereby rendering all strains carrying the amyE::ϕ(putB′-treA)1 construct resistant to the antibiotic chloramphenicol.
BGSC, Bacillus Genetic Stock Center, Ohio State University, Columbus, OH.
Media and growth conditions.
The B. subtilis strains were maintained on LB agar plates; liquid cultures were grown at 37°C in Spizizen's minimal medium (SMM) (26) supplemented with 0.5% (wt/vol) glucose as the carbon source, a solution of trace elements (28), and the amino acids l-tryptophan (20 mg liter−1) and l-phenylalanine (20 mg liter−1) to meet the auxotrophic needs of strain JH642 (trpC2 pheA1) and its derivatives. The medium contained 15 mM NH4Cl as the nitrogen source. When l-proline was used as the sole carbon and energy source, glucose (28 mM) was replaced with 32 mM l-proline to provide the bacterial cells with the same molarity of carbon atoms available to the cells for catabolism. When l-proline was used as the sole nitrogen source, the NH4Cl content (15 mM) of the SMM was replaced with 15 mM l-proline. The antibiotics chloramphenicol, kanamycin, tetracycline, erythromycin, and spectinomycin were used with B. subtilis strains at final concentrations of 5 μg ml−1, 10 μg ml−1, 15 μg ml−1, 1 μg ml−1, and 100 μg ml−1, respectively. Ampicillin and chloramphenicol were used for E. coli cultures at final concentrations of 100 μg ml−1 and 35 μg ml−1, respectively.
Recombinant DNA techniques.
The routine manipulations of plasmid DNA, the construction of recombinant DNA plasmids, the isolation of chromosomal DNA from B. subtilis, and transformation with plasmid or chromosomal DNA were carried out using standard procedures (28). For the detection of homologous sequences by Southern hybridization, we used digoxigenin (DIG)-labeled DNA probes. For the preparation of these hybridization probes and the detection of the hybridization signals with chromosomal DNAs of various B. subtilis strains, we used the DIG DNA Labeling and Detection kit (Roche Diagnostics, Manheim, Germany). The DNA-DNA hybridization conditions used followed the experimental procedures suggested by the manufacturer of the labeling and detection kit. DNA restriction fragments were blotted on a Nytran 13N nylon membrane purchased from Schleicher and Schuell (Dassel, Germany).
Construction of plasmids, putB′-treA reporter strains, and chromosomal gene disruptions.
The construction of a ϕ(putB′-treA)1 transcriptional reporter gene fusion on plasmids and their integration as a single copy into the B. subtilis chromosome at the amyE gene are detailed in the supplemental material. Strains with defects in individual genes of the chromosomal putBCP locus or the entire putBCP gene cluster were constructed by transforming strain JH642 with linearized plasmid DNA carrying the desired gene disruption mutation marked with an antibiotic resistance cassette into strain JH642 and by a subsequent selection for antibiotic-resistant colonies on LB agar plates. Details on the construction of the plasmids used for the generation of these mutant B. subtilis strains can be found in the supplemental material.
Transcription analysis of the putBCP gene cluster by Northern blot analysis.
The transcriptional regulation of the putBCP gene cluster in response to the availability of l-proline in the growth medium and its genetic organization were analyzed by Northern blotting. Total RNA was isolated from B. subtilis strains by the acidic phenol method (30). DIG-labeled single-stranded RNA probes specific for the putB, putC, and putP genes were prepared by in vitro transcription using the DIG RNA-Labeling Kit SP6/T3/T7 (Roche Diagnostics, Mannheim, Germany) according to the procedure described by the manufacturer. Derivatives of the cloning vector pBSK− containing the putB (pSM11), putC (pSM34), or putP (pSM35) gene were used as templates for the T3 RNA polymerase-mediated in vitro transcription reaction. RNA-RNA hybridization and detection of specific put transcripts were performed according to the manufacturer's instructions (Roche Diagnostics, Mannheim, Germany).
Primer extension analysis of the proline-responsive putBCP promoter.
The transcriptional initiation start site of the putBCP mRNA was determined by the primer extension method. Total RNA was isolated by the acidic-phenol method (30) from log-phase cells of the B. subtilis strain JH642 containing pSM13, a plasmid carrying the ′ycgL-putB′ intragenic region that should contain the putBCP promoter (Fig. 1A). A reverse transcription reaction was carried out with 10 μg of total RNA isolated from these cells and 2 μmol of the synthetic oligonucleotide SM28 (5′-CGCCATTTTATTGAGAAAGCCGC-3′, bp 37 to 60 of the putB coding region) labeled at its 5′ end with the infrared dye IRD-800 (Eurofins MWG Operon, Ebersberg, Germany) as described previously (11). The reverse transcription reaction product was analyzed on a 6% DNA-sequencing gel run in a Li-Cor DNA sequencer (type 4000; Eurofins MWG Operon, Ebersberg, Germany). A sequencing ladder produced with the IRD-800-labeled SM28 primer and plasmid pSM13 as the DNA template was run in parallel with the primer extension reaction product on the same DNA-sequencing gel to determine the 5′ end of the putBCP mRNA.
Determination of PutB enzyme activity.
PutB enzyme activity was measured in crude extracts of B. subtilis cells grown in SMM in the absence or presence of 1 mM proline. The assay used followed the method of Dendinger and Brill (17), which monitors the oxidation of proline to Δ1-pyrroline-5-carboxylate (P5C) by determining the formation of the P5C–o-aminobenzaldehyde from the chromogenic substrate o-aminobenzaldehyde in a spectrophotometer. The millimolar extinction coefficient of the P5C–o-aminobenzaldehyde complex is 2.71 mM−1 cm−1 (17). The specific activity of the PutB proline dehydrogenase (also sometimes referred to as proline oxidase) (4) in the crude cell extracts of B. subtilis strains is given as nmol P5C formed per minute and mg protein (U mg protein−1).
TreA enzyme activity assays.
An aliquot (1.5 ml) from cultures of putB-treA B. subtilis fusion strains was harvested by centrifugation for 2 min in an Eppendorf microcentrifuge (15,000 rpm) and resuspended in 0.5 ml Z buffer (42) adjusted to pH 7.0 and containing 1 mg ml−1 lysozyme. After incubation for 10 min at 37°C in an Eppendorf thermomixer, cellular debris was removed by centrifugation (5 min at 12,000 rpm), and the supernatant was then used for TreA activity assays with para-nitrophenyl-α-d-glucopyranoside as the substrate (23). TreA specific activity is expressed in units per mg of protein; protein concentrations were estimated from the optical density of the cell culture (42).
Sensitivity of B. subtilis strains to toxic proline analogues.
The proline analogues AC and DHP are toxic to microorganisms (39, 69). To test the sensitivity of B. subtilis strains to AC and DHP, cultures were grown in SMM with and without the addition of 0.6 M NaCl until they reached an optical density at 578 nm (OD578) of 1.5. A 300-μl aliquot of each culture was then plated on SMM or SMM agar plates with 0.6 M NaCl before a 5-mm paper filter disk, soaked with 10 μl of a 25-mg ml−1 solution of AC or DHP, was placed in the center of each agar plate. The formation of a growth inhibition zone around the filter disk was recorded after incubation of the agar plates at 37°C for 24 to 48 h.
Transport assays with radiolabeled proline.
The kinetic parameters of proline transport via the PutP and OpuE transport systems of B. subtilis were determined in strain SMB11 (PutP+ OpuE−) and BLOB9 (PutP− OpuE+) (Table 1) using l-[14C(U)]proline (40 mCi mmol−1). The strains were cultivated in SMM or SMM containing either 0.4 M or 0.6 M NaCl under putBCP-inducing (growth of the cultures in the presence of 1 mM l-proline) or non-putBCP-induced (growth of the cultures in the absence of l-proline) conditions. Aliquots (2 ml) were removed from the culture when the cells reached the log phase (OD578, about 0.3 to 0.6). Those cultures that were grown in the presence of proline to induce putBCP expression were washed twice with proline-free cultivation medium that had been warmed to 37°C. Various concentrations (1 μM to 40 μM) of l-[14C(U)]proline were added to the cells, and aliquots (0.3 ml) were taken after 40, 80, and 120 s; the cells were then collected by filtration onto a cellulose filter (0.45 μm; Schleicher& Schuell, Dassel, Germany). The filters were washed two times with the proline-free cultivation medium of the cells and subsequently transferred to a scintillation analyzer (Coulter Liquid Scintilliation Analyser 1900CA) with 5 ml scintillation solution. The transport activity of cells is expressed as nmol substrate min−1 mg protein−1. The kinetics of l-[14C(U)]proline uptake was analyzed according to the method of Michaelis-Menten.
Measurements of cellular proline pools.
Cells of the B. subtilis strain SMB10 (Table 1) were cultivated in 20 ml SMM or SMM with 1 M NaCl until they reached an OD578 of 1. One set of the cultures received 5 mM l-proline, and both sets of cultures were then incubated further until they reached an OD578 of 2. Aliquots of the cells were harvested prior to (at an OD578 of 1) and after (at an OD578 of 2) the addition of l-proline, washed with their growth medium (without l-proline), and then assayed for TreA activity to monitor putB-treA expression and proline content. The intracellular content of proline was determined by the method described by Bates et al. (5), which monitors the proline content of samples as a dark-red proline-ninhydrin complex that is measured photometrically at a wavelength of 480 nm. To correlate the colored proline-ninhydrin complex with the proline concentration, a calibration curve was established by treating standard solutions with a known l-proline concentration (0 mM to 10 mM) in the same way as the whole-cell extracts. Intracellular proline concentrations were calculated using a volume for a B. subtilis cell of 0.67 μl per 1 OD578 unit of cell culture (S. Moses, E. P. Bakker, and E. Bremer, unpublished data).
Database searches and alignments of amino acid sequences of proteins related to the PutB, PutC, and PutP proteins.
Proteins that are homologous to the proline catabolic PutB and PutC enzymes, to the proline transporter PutP, and to the proline-responsive PutR activator protein from B. subtilis were searched for via the Web server of the Department of Energy (DOE) Joint Genome Institute (JGI) (http://www.jgi.doe.gov/) or that of the National Center for Biotechnology Information (http://www.ncbi.nlm.nih.gov/) using the BLAST algorithm (2). Protein sequences were aligned and analyzed using ClustalW (63). In silico models of the B. subtilis PutB and PutC proteins were generated with the aid of the SWISS Model server (http://swissmodel.expasy.org) (3).
RESULTS
Predicted functions of the putBCP-encoded proteins for proline utilization.
The ycgMNO gene cluster from B. subtilis encodes two enzymes (YcgM and YcgN) predicted to be involved in l-proline catabolism and a transport protein (YcgO) predicted to mediate l-proline uptake. Since we show below that the ycgMNO-encoded proteins are required for the utilization of l-proline as a nutrient by B. subtilis, we refer here to this gene cluster as putBCP (proline utilization) (Fig. 1A). We avoided the use of putA as a gene designation because the PutA protein in enterobacteria comprises both proline dehydrogenase (PRODH) and Δ1-pyrroline-5-carboxylate dehydrogenase (P5CDH) enzyme activities (37, 52, 59), whereas the PutB and PutC proteins from B. subtilis represent monofunctional PRODH and P5CDH enzymes (see below) (32, 68). The putBCP gene cluster is followed in the same transcriptional orientation by the putR (ycgP) gene, which encodes the PutR protein, the proline-responsive activator of putBCP expression (7, 31). A 155-bp spacer region separates the putBCP gene cluster and the putR gene, and this intergenic region contains a predicted Rho-independent transcriptional terminator sequence (Fig. 1A) and the promoter for the putR gene (7, 31).
Database searches suggest that the B. subtilis PutB protein (303 amino acids) is a monofunctional PRODH (EC 1.5.99.8) (68). PutB exhibits 25% amino acid sequence identity to the PRODH domain (from amino acid 261 to amino acid 612) of the PutA enzyme from E. coli (61). The PutC protein (515 amino acids) from B. subtilis exhibits 37% amino acid sequence identity to the P5CDH domain (from amino acid 650 to amino acid 1130) of the PutA enzyme from E. coli and is predicted to function as a monofunctional NAD-dependent Δ1-pyrroline-5-carboxylate dehydrogenase (P5CDH) (EC 1.5.1.12). It contains the catalytically active residues (Glu-286 and Cys-320) typically found in P5CDH enzymes (32, 61). The crystal structures of the monofunctional PRODH and P5CDH enzymes from Thermus thermophilus were recently reported (32, 68). The amino acid sequence of the PutB protein of B. subtilis is 41% identical to that of the PRODH from T. thermophilus, and the PutC protein sequence is 50% identical to that of the P5CDH from the microorganism, suggesting that the B. subtilis enzymes possess folds similar to those of the PRODH and P5CDH enzymes from T. thermophilus.
The PutP protein (491 amino acids) is predicted to function as a high-affinity proline importer that belongs to the sodium solute symporter family, whose members couple the import of the substrate with the inflow of Na+ ions (48). The B. subtilis PutP transporter exhibits 54% amino acid sequence identity to the well-studied proline importer PutP from E. coli (48). The E. coli PutP protein is predicted to comprise 13 membrane-spanning segments, and those residues known to be involved in l-proline and Na+ binding are conserved in the B. subtilis PutP protein (proline binding, W-85, Y-166, W-270, and Y-274; Na+ binding, Y-15, A-79, M83, A-359, S-362, and T-363) (40, 48). The B. subtilis PutP protein exhibits 61% amino acid sequence identity to the osmotically inducible OpuE transporter that is used by B. subtilis under high-salinity stress conditions to import l-proline as an osmostress protectant (58, 65).
Biochemical and genetic assessment of the putBC-encoded proteins in l-proline catabolism.
The bioinformatic analysis of the predicted enzymatic functions of the PutB and PutC proteins and of the PutP transporter suggests that B. subtilis can import l-proline and oxidize it to l-glutamate via the pathway depicted in Fig. 1B, an l-proline degradation route found in many microorganisms (18, 33, 35, 36, 47, 57, 59, 61, 64, 72). It is known from previous studies that growth of B. subtilis in the presence of l-proline triggers the induction of a proline-catabolizing enzyme (referred to by Atkinson et al. as proline oxidase) that converts l-proline to Δ1-pyrroline-5-carboxylate (4).
To provide biochemical evidence for the suggested l-proline catabolic pathway (Fig. 1B), we assayed proline dehydrogenase (PutB) activity in cleared cell extracts of cultures grown in SMM with glucose or in SMM-grown cultures that received 1 mM l-proline for 80 min prior to cell harvest. The addition of l-proline to B. subtilis cultures grown in SMM increased PRODH activity 5-fold from 0.48 ± 0.05 U (mg protein−1) to 2.69 ± 0.21 U (mg protein−1). This increase in PRODH activity was abolished in a putB mutant that possessed an activity of 0.46 ± 0.02 U (mg protein−1) in cells grown in the absence of l-proline and 0.45 ± 0.02 U (mg protein−1) in cells cultivated in the presence of l-proline.
In addition to these enzymatic studies, proline utilization was also assessed genetically through targeted deletion analysis of the putBCP catabolic genes. The B. subtilis wild-type strain JH642 was able to use l-proline effectively both as a sole carbon and energy source and as a sole nitrogen source (Table 2). Deletion of the entire putBCP locus from the B. subtilis chromosome abolished l-proline utilization (Table 2). Noticeably, a strain with intact putB and putC genes that carries a defect in the l-proline transporter PutP could not use l-proline as a sole carbon and energy source but was proficient in the use of the amino acid as a sole nitrogen source (Table 2). We attribute this finding to reduced, but still significant, import of l-proline in a putP mutant; such a strain is apparently able to import enough l-proline through other transport systems (see Fig. 8) to provide the cells with an adequate level of l-proline for use as a sole nitrogen source.
Table 2.
Use of l-proline as sole carbon and energy source and as sole nitrogen source by B. subtilis
| Strain | Relevant genotype | Growth yield of cultures grown in the presence ofa: |
||
|---|---|---|---|---|
| Glucose and NH4Cl | l-Proline and NH4Cl | Glucose and l-proline | ||
| JH642 | putBCP+ | 3.90 | 4.55 | 4.70 |
| SMB45 | Δ(putBCP::tet)2 | 3.90 | 0.20 | 0.90 |
| SMB11 | Δ(putP::spc)1 | 4.30 | 0.80 | 3.80 |
Cells of the wild-type strain JH642 and its mutant Δ(putBCP::tet)2 and Δ(putP::spc)1 derivatives were cultivated in shake flasks containing (i) SMM with glucose (28 mM) as the carbon and energy source and NH4Cl (15 mM) as the nitrogen source, (ii) glucose (28 mM) as the carbon and energy source and l-proline (15 mM) as the nitrogen source, and (iii) l-proline (32 mM) as the sole carbon and energy source and NH4Cl (15 mM) as the nitrogen source.
PutP- and OpuE-dependent uptake of l-proline.
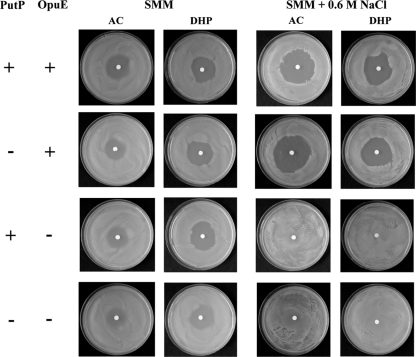
The PutP and OpuE transporters both belong to the sodium solute symporter family (48). The role of OpuE as an l-proline transporter has already been established; OpuE catalyzes the import of l-proline as an osmostress protectant (65), and the transcription of its structural gene (opuE) is induced under high-osmolarity growth conditions (58). We first analyzed the sensitivity of B. subtilis against the toxic proline analogues AC and DHP, which are typically imported into microbial cells through proline transport systems (39, 69). The growth-inhibiting effects of AC and DHP result from the incorporation of these compounds into proteins that then are prone to misfolding. The B. subtilis wild-type strain JH642 is sensitive to both AC and DHP, and the sensitivity to these proline analogues increases under high-salinity growth conditions (Fig. 2). A comparison of the sensitivities to AC and DHP in an isogenic pair of strains expressing either the PutP or OpuE transporter alone showed that AC and DHP sensitivity at high salinity is exclusively dependent on the activity of OpuE (Fig. 2). Thus, either PutP transport activity is inhibited by high salinity or the PutP protein is not present in cells exposed to high-saline growth conditions (Fig. 2). We observed residual AC and DHP sensitivity in a putP opuE double-mutant strain on SMM agar plates (Fig. 2), suggesting that a still uncharacterized l-proline transporter(s) is present in B. subtilis (Fig. 1B). This residual AC and DHP sensitivity is abrogated at high salinity (SMM agar plates containing 0.6 M NaCl) (Fig. 2).
Fig 2.
Sensitivity of B. subtilis and its putP and opuE mutant derivatives to toxic proline analogues. Cells of the wild-type strain JH642 and its mutant derivatives SMB11 (PutP− OpuE+), BLOB9 (PutP+ OpuE−), and SMB12 (PutP− OpuE−) were pregrown in SMM with and without the addition of 0.6 M NaCl. Aliquots (300 μl) of these cultures were then plated on SMM agar plates or SMM agar plates containing 0.6 M NaCl; a paper filter disk soaked with 10 μl of a 25-mg ml−1 solution of the toxic proline analogues AC and DHP was then placed in the middle of each of the agar plates. The agar plates were incubated at 37°C for 24 h (for cells grown on SMM plates) or for 48 h (for cells grown on SMM plates containing 0.6 M NaCl) before the formation of growth inhibition zones around the filter disk was recorded by photography.
l-Proline transport in B. subtilis was then analyzed directly by measuring the kinetic parameters of the PutP and OpuE systems with radiolabeled l-[14C]proline in strains BLOB9 (PutP+ OpuE−) and SMB11 (PutP− OpuE+). The PutP+ strain BLOB9 exhibits high-affinity l-proline transport activity (Km, about 8 μM) with a rather modest capacity (Vmax, about 29 nmol min−1 mg protein−1) in cells that were grown in SMM. However, precultivation of this strain in SMM in the presence of 1 mM l-proline increased l-[14C]proline uptake activity by about 6-fold (Vmax, about 158 nmol min−1 mg protein−1) without influencing the substrate affinity (Km) of the PutP transport system (Table 3). Increases in the external salinity progressively decreased the l-proline transport capacity (Vmax) of the PutP system to the noninduced level, despite the fact that the cells had been cultivated in the presence of l-proline (Table 3).
Table 3.
PutP- and OpuE-mediated uptake of l-[14C]proline by B. subtilis
| Strain | Growth conditionsa | Km (μM) | Vmax (nmol min−1 mg−1) |
|---|---|---|---|
| BLOB9 (PutP+ OpuE−) | SMM | 8 ± 2 | 29 ± 2 |
| SMM + 1 mM Pro | 8 ± 2 | 158 ± 5 | |
| SMM + 0.4 M NaCl | 6 ± 1 | 28 ± 2 | |
| SMM + 0.4 M NaCl + 1 mM Pro | 8 ± 2 | 68 ± 1 | |
| SMM + 0.6 M NaCl + 1 mM Pro | 11 ± 1 | 22 ± 2 | |
| SMB11 (PutP− OpuE+) | SMM | 12 ± 1 | 27 ± 3 |
| SMM + 1 mM Pro | 12 ± 4 | 19 ± 3 | |
| SMM + 0.4 M NaCl | 12 ± 1 | 104 ± 14 | |
| SMM + 0.6 M NaCl | 23 ± 3 | 252 ± 10 |
Cells were grown in SMM either in the absence or presence of the indicated concentrations of NaCl or l-proline to an OD578 of about 0.3 to 0.6. The cells were then harvested by centrifugation and washed twice in prewarmed (37°C) growth medium (SMM or SMM with the indicated salt concentrations) but in the absence of l-proline. For the uptake assays with l-[14C]proline, the substrate concentration was systematically varied between 1 μM and 40 μM; the measured uptake rates were used for the calculation of Km and Vmax values according to Michaelis-Menten kinetics.
The OpuE-mediated l-[14C]proline uptake activity was a mirror image of that of the PutP transporter. There was no induction of OpuE transport activity by l-proline, but growth of the cells in media with increased salinity resulted in progressively increased l-proline uptake activity (Vmax) without strong effects on the affinity (Km) of the OpuE import system (Table 3).
When interpreting the l-[14C]proline transport data summarized in Table 3, one needs to keep in mind that l-proline can still enter a putP opuE double-mutant strain when 1 mM l-proline is provided in the growth medium (data not shown), and such a strain is also somewhat sensitive to AC and DHP (Fig. 2). However, the so-far-uncharacterized third proline importer operating in B. subtilis (Fig. 1A) must be a transport system with a rather modest capacity under the growth conditions we used. This is evident from our finding that there was no l-[14C]proline uptake detectable in strain SMB12 [Δ(putP::spc)1 Δ(opuE::tet)1] at the highest substrate concentration (40 μM) tested in the experiments assessing the kinetic parameters (Table 3) of the PutP and OpuE proline transporters (data not shown).
Northern blot analysis of the transcripts of the putBCP gene cluster.
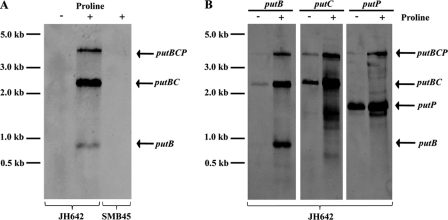
The putB and putC genes are separated by 16 bp, and the intergenic region between putC and putP is 69 bp, suggesting that the putBCP gene cluster is transcribed as an operon. We assessed the transcriptional profile of the putBCP locus by Northern blot analysis using total RNA extracted from cells that were grown in either SMM or SMM in the presence of 1 mM l-proline. Using a single-stranded antisense putB-specific RNA probe, we found that putBCP transcription was inducible by l-proline and that the transcripts detected in the wild-type strain JH642 were absent in strain SMB45 [Δ(putBCP::tet)2] carrying a deletion of the entire putBCP locus (Fig. 3A). The longest detected l-proline-inducible mRNA species, with a measured length of about 3,900 nucleotides, corresponds well to the size of a transcript comprising putBCP, which has a calculated length of 4,018 nucleotides. Hence, the putBCP gene cluster of B. subtilis is expressed as an l-proline-inducible operon. Consistent with the presence of a factor-independent transcriptional terminator in the putP-putR intragenic region (Fig. 1A), the putBCP transcript apparently does not extend into the flanking putR gene (Fig. 1A); in all likelihood, it ends in the vicinity of the predicted Rho-independent transcription terminator (Fig. 1A).
Fig 3.
Northern blot analysis of the putBCP transcript(s). (A) Total RNA was isolated from log-phase cells of the wild-type strain JH642 (lanes 1 and 2) and its putBCP mutant derivative strain SMB45[Δ(putBCP::tet)2] (lane 3) after growth in SMM in the absence (−) or presence (+) of 1 mM l-proline and was then subjected to Northern blot analysis with a single-stranded antisense putB-specific DIG-labeled hybridization probe. (B) Total RNA isolated from the wild-type strain JH642 grown in SMM (−) or SMM with 1 mM l-proline (+) was reacted with single-stranded antisense putB-, putC-, and putP-specific DIG-labeled hybridization probes.
In addition to the full-length putBCP transcript, we observed two l-proline-inducible mRNA species with measured lengths of 2,500 and 800 nucleotides (Fig. 3A). Using putB- and putC-specific probes, we identified these mRNA species as transcripts that comprised either putBC or putB alone (Fig. 3B). The measured lengths of these transcripts correspond closely to the calculated lengths of the putBC (2,476 nucleotides) and putB (912 nucleotides) mRNA species. We cannot distinguish between the possibilities that the putB or putBC mRNA species represents either premature transcription termination products or stable degradation products of the full-length putBCP mRNA. Interestingly, when we used a putP hybridization probe, we also detected an mRNA comprising putP that was present not only in cells cultivated in SMM with l-proline, but also in cells grown in SMM in the absence of l-proline (Fig. 3B). Judging from the length of this constitutively produced mRNA species (estimated length, 1,700 nucleotides), and assuming that it has the same 3′ end as the full-length putBCP transcript, it must be produced from a promoter that is located within the 3′ region of the putC gene, since the length of the calculated putP mRNA segment is only about 1,420 nucleotides. Manual inspection of the corresponding region within the 3′ segment of the putC gene revealed a putative promoter with −35 and −10 regions (TTCAAC-N17-TATCGT) corresponding reasonably well to SigA-type promoter consensus sequences, and this promoter also possessed the TG motif that is frequently found at position −16 in B. subtilis promoters (29). This putative promoter is present 140 bp upstream of the putC stop codon and might therefore direct the constitutive synthesis of the observed 1,700 nucleotide ′putC-putP mRNA species (Fig. 3B).
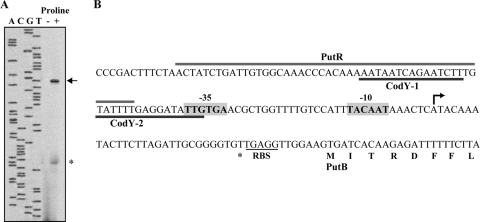
Mapping of the proline-responsive putBCP promoter by primer extension analysis.
To map the 5′ end of the putBCP mRNA, we carried out a primer extension analysis with total RNA isolated from cells of the B. subtilis wild-type strain harboring plasmid pSM13 (′ycgL-putB′) (Fig. 1A) that were grown in either SMM or SMM with 1 mM l-proline. A major l-proline-inducible transcript was detected (Fig. 4A) whose 5′ end corresponds to an A · T base pair located 40 bp upstream of the predicted GTG start codon for the putB reading frame (Fig. 4B), in agreement with other recently reported determinations of the putBCP transcription initiation site (7, 31). Upstream of the transcriptional start site, −10 and −35 elements are present, with features typical of SigA-dependent promoters of B. subtilis (29). The spacing between the −10 and −35 regions of the put promoter is 18 bp, a suboptimal spacing for SigA-type promoters (29).
Fig 4.
Mapping of the l-proline-responsive putBCP promoter by primer extension analysis. (A) Total RNA was isolated from log-phase cells of the B. subtilis strain JH642 [pSM13 (putB′)] cultivated in SMM with glucose as a carbon source or in SMM that contained 1 mM l-proline. A reverse transcription reaction was carried out with this RNA and a synthetic single-stranded DNA oligonucleotide marked with an IRD-800 fluorescent label; the primer used hybridizes to the 5′ region of the putB gene. The same oligonucleotide was used for DNA-sequencing reactions with DNA of plasmid pSM13 to size the 5′ end of the putB mRNA. The arrow marks the full-length reverse transcription product. The mRNA species marked by an asterisk is either a premature termination product of the reverse transcriptase reaction or a degradation product of the full-length reverse transcriptase reaction product. (B) DNA sequence of the putBCP regulatory region. The mapped 5′ end of the putB mRNA is indicated by an arrow; the −10 and −35 elements of the inferred SigA-type putBCP promoter are highlighted. The ribosome-binding site (RBS) and the reading frame of the putB gene are indicated. The marked binding regions for the CodY and PutR regulatory proteins have been mapped through DNA footprinting and mutant analysis (7).
A minor l-proline-inducible mRNA species was also detected in our primer extension experiments (Fig. 4A). However, since its 5′ end is positioned close to the predicted ribosome-binding site of the putB gene (Fig. 4B) and since we did not detect any typical promoter elements in the vicinity of its 5′ end, we interpreted this short mRNA species either as a degradation product of the full-length primer extension reaction product or as a product of a stalling event of the reverse transcriptase.
Use of putB-treA reporter fusions to study induction of gene expression in response to exogenously provided l-proline and proline-related compounds.
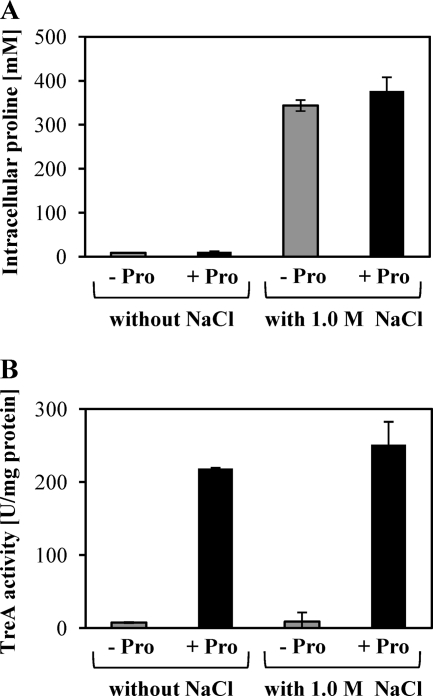
To monitor the expression of the putBCP operon in greater detail, we constructed a putB-treA transcriptional reporter gene fusion and integrated it as a single copy into the amyE locus of the B. subtilis genome. We studied the expression of this fusion in a putBCP+ wild-type genetic background (strain SMB10). First, we analyzed the influence of an exogenous supply of l-proline (1 mM) on the time course of the induction of the putB-treA gene fusion. Expression reached its maximal level about 60 min after the addition of the inducer l-proline to the growth medium; during this time span, we did not observe any enhanced expression of the putB-treA fusion in the control culture that had received no l-proline (Fig. 5A). We then analyzed the dependence of the strength of putB-treA expression on the amount of l-proline that was added to the growth medium. An inducing effect of l-proline was already noticeable when it was present at a concentration of 25 μM in the growth medium (Fig. 5B). It should be noted in this context that the cellular proline pool of B. subtilis cells grown in a minimal medium with glucose as the carbon source has been measured to lie in a range between 16 mM (67) and 10 mM (see Fig. 7). Apparently, this substantial internal l-proline pool does not cause high-level expression of the put gene cluster (Fig. 3A and 5). The very low level of putB-treA expression observed in SMM-grown cells and the enhanced expression of the reporter gene fusion in the presence of an exogenous supply of l-proline were both dependent on the l-proline-responsive PutR regulator (7) (Fig. 5C).
Fig 5.
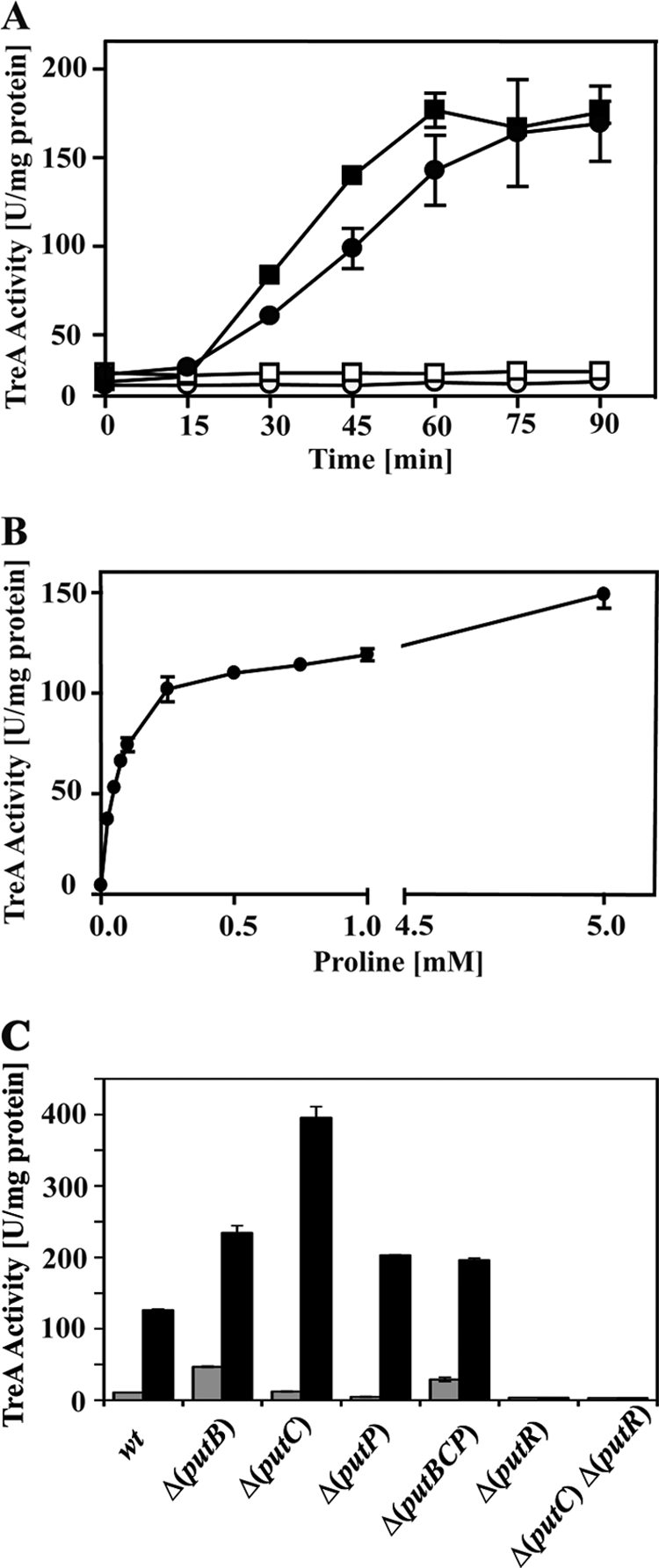
Induction of put expression in response to an external supply of l-proline. Expression of the put genes in response to l-proline in the growth medium was monitored with the aid of a chromosomal putB-treA operon fusion. (A) The reporter strain SMB10 [ϕ(putB-treA)1] was grown in SMM (squares) or SMM with 0.6 M NaCl (circles) to early exponential phase; two cultures (filled symbols) received 1 mM l-proline at time zero, and the induced and noninduced cells were assayed for TreA reporter enzyme activity at the indicated time intervals. (B) The reporter strain SMB10 [ϕ(putB-treA)1] was grown in SMM to early exponential phase, and the cells were assayed for TreA reporter enzyme activity 60 min after they received the indicated amounts of l-proline. (C) ϕ(putB-treA)1 reporter gene fusion activity was measured in cells of various B. subtilis strains that were grown in either the absence (gray bars) or presence (black bars) of 1 mM l-proline; TreA reporter enzyme activity was recorded 60 min after the addition of l-proline to the cultures. The following ϕ(putB-treA)1 fusion strains were used: SMB10 (putBCP+), SMB49 [Δ(putB::spc)3], SMB34 [putB+ Δ(putC::neo)2], SMB14 [putB+ putC+ Δ(putP::spc)1], SMB46 [Δ(putBCP::tet)2], TSB2 [putBCP+ Δ(putR::spc)], and TSB3 [putB+ Δ(putC::neo)2] Δ(putR::spc)]. The values for the TreA activity given represent two independently grown cultures, and for each sample analyzed, the TreA activity was determined twice. wt, wild type. The error bars indicate standard deviations.
We also tested the proline-related compounds thioproline, trans-4-hydroxyproline, monomethyl-proline, and dimethyl-proline (proline betaine) for the ability to induce the expression of the putB-treA reporter fusion; none of these compounds functioned as an inducer for put expression in SMM-grown cells (Fig. 6A). This was also the case for the toxic proline analogues AC and DHP (Fig. 6A). This picture changed, however, when we tested the inducing effects of the above-mentioned proline derivatives and analogues on putB-treA expression in cells cultivated in high-osmolarity medium (SMM containing 0.6 M NaCl). l-Proline still functioned as an inducer under high-salinity growth conditions, but in contrast to SMM-grown cells, both AC and DHP and dimethyl-proline (proline betaine) now functioned as inducers, whereas monomethyl-proline and thioproline still did not function as inducers for putB-treA expression (Fig. 6B). The toxic proline analogues AC and DHP enter the B. subtilis cell under high-salinity growth conditions via the osmotically inducible OpuE (65) transporter (Fig. 2), and their enhancing effects on putBCP expression in osmotically stressed cells (Fig. 6B) can therefore be rationally understood. The fact that they do not induce the expression of the putB-treA reporter fusion in cells cultivated in the absence of salt hint either that these compounds are not substrates for PutP or that the affinity of the PutP transporter for AC and DHP is so low so that they cannot trigger enhanced putB-treA expression or cause increased sensitivity to these toxic compounds (Fig. 2). Notably, the end product of the l-proline degradation pathway, l-glutamate (Fig. 1B), did not trigger significantly enhanced transcription of the putB-treA reporter fusion at either low or high salinity (Fig. 6A and B).
Fig 6.
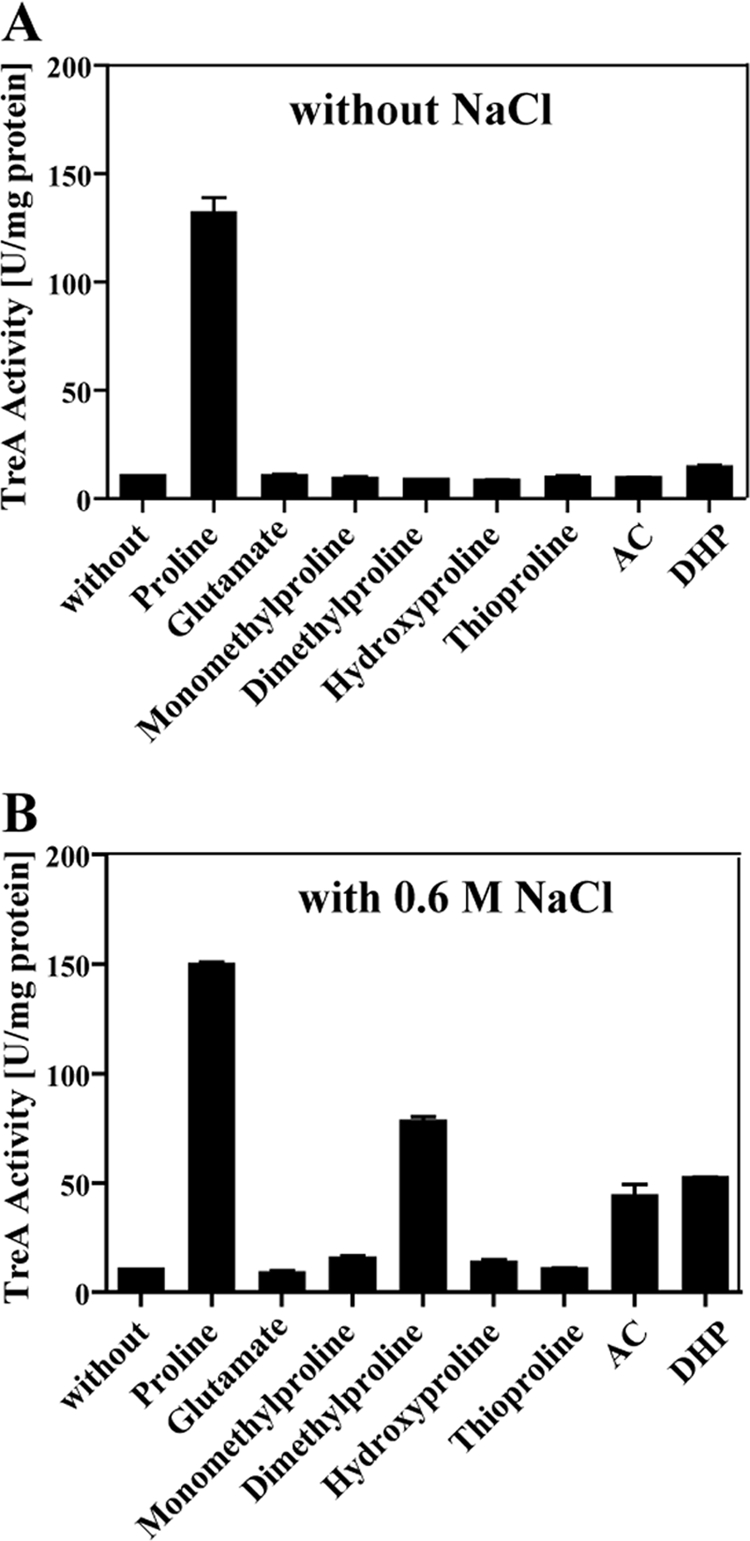
Induction of put expression in response to an external supply of l-proline, proline-derived compounds, and proline analogues. The reporter strain SMB10 [ϕ(putB-treA)1] was pregrown in either SMM (A) or SMM with 0.6 M NaCl (B) overnight, and the cultures were used to inoculate fresh cultures that were then allowed to grow to early exponential phase. To part of the cultures, we added 1 mM l-proline, proline-derived compounds, toxic proline analogues, and glutamate; the cells were then propagated for a further 60 min and were subsequently harvested for TreA reporter enzyme assays. The values for the TreA activity given represent two independently grown cultures, and for each sample analyzed, the TreA activity was determined twice. The error bars indicate standard deviations.
Induction of put expression is not triggered by the large amounts of l-proline synthesized under osmotic stress conditions.
High salinity per se did not trigger putB-treA expression (Fig. 5A and 6B), a rather surprising finding, since it is well known that B. subtilis synthesizes considerable amounts (several hundred millimolar) of the compatible solute l-proline as a cellular defense against high-osmolarity surroundings (11, 67). Induction of the putB-treA reporter fusion by an exogenous supply of l-proline in high-salinity (0.6 M NaCl)-grown cells was still possible, although the kinetics of putB-treA induction was somewhat delayed in comparison to SMM-grown cells (Fig. 5A).
The data presented in Fig. 5A and 6B hint that the osmotically induced l-proline biosynthesis does not trigger putB-treA expression. To investigate this in greater detail, we grew the putB-treA fusion strain SMB10 in either SMM or SMM with 1 M NaCl to an OD578 of 1; the two sets of cultures then received 5 mM l-proline and were further grown until they reached an OD578 of 2. Samples were withdrawn prior to and after the addition of l-proline and assayed for their l-proline contents and TreA reporter enzyme activities. As documented in Fig. 7A, the l-proline pool of the cells grown in SMM with 1 M NaCl rose 34-fold in comparison to that of cells cultivated in SMM; it increased from about 10 mM to about 340 mM. However, the expression of the putB-treA reporter fusion was not triggered by this strong increase in the cellular l-proline content (Fig. 7B). In contrast, the addition of 5 mM l-proline to the growth medium of the cells, regardless of whether they were cultivated in SMM or in SMM with 1 M NaCl, triggered strong expression of the putB-treA reporter fusion (Fig. 7B) but did not lead to a greatly increased cellular proline pool (Fig. 7A). Hence, the B. subtilis cell is “blind” to internally produced l-proline with respect to the induction of putB-treA transcription.
Fig 7.
Synthesis of l-proline by high-salinity-stressed cells does not trigger ϕ(putB-treA)1 expression. Cells of the putB-treA reporter strain SMB10 were grown either in SMM or in SMM with 1 M NaCl to an OD578 of 1, and one part of each culture then received 5 mM l-proline (black bars) while the other part remained untreated (gray bars). The cells were cultivated until they had reached an OD578 of 2, harvested by centrifugation, and assayed for their l-proline contents (A) and TreA reporter enzyme activities (B). The values for the l-proline contents and the TreA activities given represent two independently grown cultures, and for each sample analyzed, the l-proline pool and TreA activity were determined twice. The error bars indicate standard deviations.
l-Proline-mediated induction of putB-treA expression is independent of the putBCP gene products but is dependent on PutR.
The PutA protein from E. coli and S. enterica serovar Typhimurium not only functions as an l-proline-catabolizing bifunctional enzyme (61), it also controls the transcription of the divergently oriented putA and putP genes in response to an external supply of l-proline (72). It possesses an N-terminal ribbon-helix-helix DNA-binding motif (71). This DNA-binding domain is not present in the crystallographically characterized monofunctional PRODH or P5CDH enzyme from T. thermophilus (32, 68), and the PutB and PutC proteins from B. subtilis also lack recognizable DNA-binding motifs.
To test whether any of the putBCP-encoded proteins from B. subtilis would influence the expression of this operon, we introduced the putB-treA reporter gene fusion into an isogenic set of strains that carried various lesions in the putBCP locus. Expression of the reporter gene fusion remained inducible by l-proline in a strain with a deletion of the entire putBCP operon (Fig. 5C). This finding excludes any direct influence of either the proline-catabolizing PutB and PutC enzymes or the proline transporter PutP on putBCP expression. The expression of the putB-treA fusion in a putBCP+ wild-type background was entirely dependent on PutR (Fig. 5C), fully consistent with data recently reported by Belitsky (7) and by Huang et al. (31).
A somewhat higher level of putB-treA induction was noticed in a mutant lacking an intact putB gene (Fig. 5C). Since the PutR activator protein responds directly to l-proline in an in vitro transcription assay system (7), enhanced putB-treA expression can probably be rationalized by the inability of a putB mutant strain to degrade the inducer l-proline. An approximately 4-fold-higher level of putB-treA induction was observed in a strain that has an intact PutB protein but is defective in PutC (Fig. 5C). Such a strain is predicted to accumulate the PRODH reaction product P5C or its spontaneously formed derivative γ-glutamate-5-semialdehyde (Fig. 1B), compounds that might function as effector molecules for PutR. Alternatively, P5C might cause feedback inhibition of the PutB enzyme activity and thus lead to the accumulation of l-proline, the inducer of PutR (7). We note in this context that the Δ(putB::spc)3 and Δ(putC::neo)2 mutants used for this experiment might exert polar effects on downstream-positioned genes in the putBCP operon, and the interpretation of the data obtained with respect to putB-treA induction in the putB and putC mutant strains (Fig. 5C) therefore needs to be viewed with some caution.
Proline-mediated induction of put expression does not depend on PutP- or OpuE-catalyzed proline uptake.
The data documented in Fig. 7 illustrate that B. subtilis can somehow physiologically distinguish between an external supply of l-proline and the l-proline pool amassed through de novo synthesis under osmotic stress conditions (11, 67) to induce put expression. One elegant way by which this could be accomplished would be the monitoring of l-proline import via the PutP transporter and the subsequent communication of this event to the PutR activator protein.
To test this hypothesis, we analyzed the expression of the putB-treA reporter gene fusion in a set of isogenic mutant strains with defects in the proline transporter PutP or the PutP-related OpuE transporter (61% amino acid sequence identity). The expression of the putB-treA reporter gene fusion remained l-proline inducible in both mutant strains, and this was the case even when we tested a putP opuE double mutant (1 mM l-proline was present in the growth medium) (Fig. 8A). Taken together, these data strongly suggest that the above-outlined hypothesis cannot adequately explain the different effects of an external l-proline supply and of an internal l-proline pool on the induction of the putB-treA reporter gene fusion as far as the PutP and OpuE l-proline importers are concerned.
Fig 8.
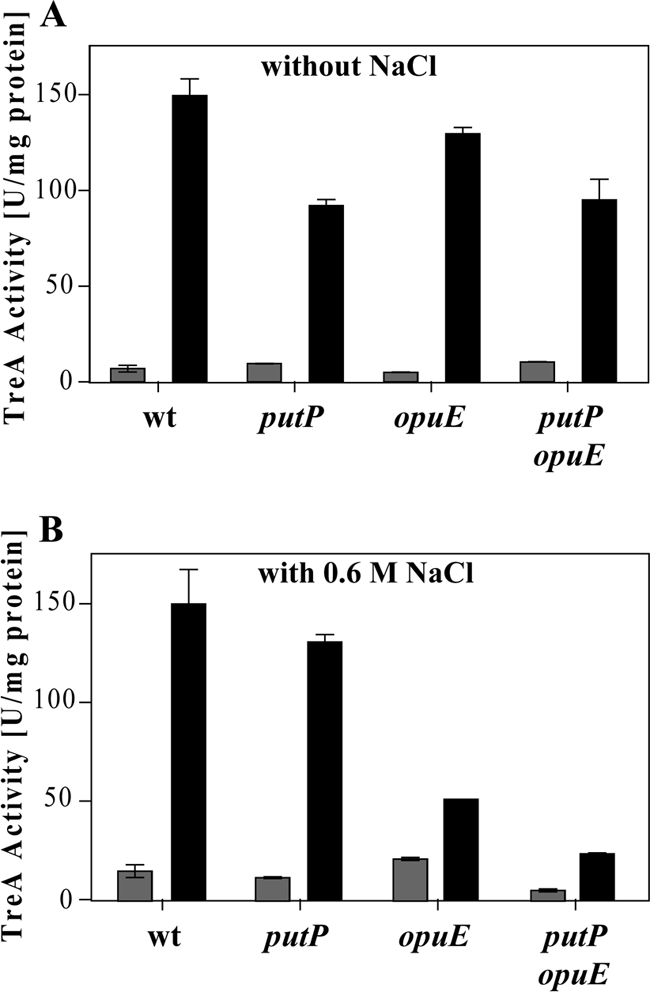
Induction of ϕ(putB-treA)1 reporter gene activity does not depend on intact PutP and OpuE proline transport systems. Cells of the ϕ(putB-treA)1 reporter strains were grown in SMM (A) or SMM with 0.6 M NaCl (B) in the absence (gray bars) or presence (black bars) of 1 mM l-proline. The cells were harvested for TreA reporter enzyme assays 60 min after the cultures received the inducer l-proline. The following ϕ(putB-treA)1 reporter fusion strains were used: SMB10 (putBCP+), SMB14 [Δ(putP::spc)1], SMB28 [Δ(opuE::tet)], and SMB27 [Δ(putP::spc)1 Δ(opuE::tet)]. The values for the TreA activity given represent two independently grown cultures, and for each sample analyzed, the TreA activity was determined twice. The error bars indicate standard deviations.
Consistent with the data on cell growth (Fig. 2) and the influence of high salinity on PutP- and OpuE-mediated l-proline uptake activity (Table 3) obtained with the toxic proline analogues, we observed a significant decrease in the degree of l-proline-mediated induction of the putB-treA reporter gene fusion in strains lacking the osmotically inducible OpuE transporter (58, 65) under high-salinity growth conditions (Fig. 8B).
Proline-mediated induction of put expression does not depend on the McpC chemoreceptor.
Ordal and coworkers have shown that B. subtilis actively seeks l-proline via chemotaxis (49) and that this behavioral response is independent of proline import (51) but depends on the functioning of the membrane-bound methyl-accepting chemotaxis protein McpC (45). We therefore considered the possibility that the induction of putB-treA expression by external l-proline was dependent on the chemoreceptor McpC. To test this hypothesis, we introduced an mcpC::erm gene disruption mutation into the putB-treA reporter strain SMB10 and monitored the l-proline-dependent induction of putB transcription in the resulting strain, ACB154. The data documented in Table 4 conclusively show that the functioning of the McpC chemoreceptor is not required for the induction of put expression in response to an external supply of l-proline.
Table 4.
Influence of the McpC chemoreceptor on putBCP expression
| Straina | Relevant genotypeb |
TreA activity (U mg protein−1) |
||
|---|---|---|---|---|
| putR | mcpC | With l-proline | Without l-proline | |
| SMB10 | + | + | 7 ± 2 | 166 ± 6 |
| TSB2 | − | + | 14 ± 2 | 14 ± 4 |
| ACB154 | + | − | 6 ± 1 | 156 ± 9 |
Each of the B. subtilis strains is derived from the wild-type strain JH642 and carries the same ϕ(putB′-treA)1 reporter gene fusion integrated as a single copy into the chromosomal amyE gene. The strains were pregrown in SMM overnight, and the cultures were used to inoculate fresh cultures that were then allowed to grow to early exponential phase. To part of the cultures, 1 mM l-proline was added, and the cells were then propagated for another 60 min and subsequently harvested for TreA reporter enzyme assays. The values for the TreA activity represent three independently grown cultures, and for each sample analyzed, the TreA activity was determined twice.
+, present; −, absent.
DISCUSSION
The fact that B. subtilis can actively seek l-proline via chemotaxis (45, 49) underscores the function of the amino acid as a nutrient for the soil bacterium (4, 6, 8, 22). We found that the use of l-proline by B. subtilis as a sole carbon and energy or a sole nitrogen source can be traced genetically to the putBCP operon (Table 2). This catabolic gene cluster (Fig. 1A) is linked to the structural gene for the PutR activator protein controlling proline-responsive putBCP expression (7, 31) (Fig. 5C).
The two putBC-encoded catabolic enzymes, the monofunctional PutB PRODH and the monofunctional PutC Δ1-pyrroline-5-carboxylate dehydrogenase (PD5CD), carry out enzymatic reactions that lead to the degradation of l-proline to l-glutamate (Fig. 1B), a proline utilization pathway present in many microorganisms (61, 72). In contrast to the situation found in E. coli and S. enterica serovar Typhimurium (41, 46, 61, 71, 72), our genetic analysis shows that the proline catabolic enzymes PutB and PutC of B. subtilis do not directly participate in the l-proline-mediated regulation of the putBCP gene cluster (Fig. 5C).
The putP-encoded l-proline transporter is a member of the SSS family (48). Its high affinity (Km, about 8 μM) for its substrate and its substantial transport capacity (Vmax, about 158 nmol min−1 mg protein−1 in cells grown in the presence of l-proline) (Table 3) make the PutP uptake system well suited to scavenge this amino acid from scarce environmental sources (27, 66). In addition to the l-proline-inducible full-length putBCP mRNA (Fig. 3A), we detected a putP mRNA species in cells grown in the absence of l-proline (Fig. 3B). This mRNA species is probably produced from a promoter residing in the 3′ region of the putC gene. As a consequence of the translation of the PutP protein from this constitutively synthesized ′putC-putP mRNA species, the B. subtilis cell is predisposed for PutP-dependent scavenging of l-proline from the environment and the ensuing induction of putBCP transcription by the proline-responsive PutR activator protein (7, 31).
Experiments in which we addressed the transcriptional control of the putBCP gene cluster by either Northern blotting (Fig. 3A), primer extension (Fig. 4A), or putB-treA reporter gene studies (Fig. 5 and 6) consistently showed induction of transcription by an exogenous supply of l-proline. In full agreement with recently reported data on put transcription (7, 31), l-proline-mediated induction of the putB-treA reporter fusion used throughout this study was strictly dependent on the PutR activator protein (Fig. 5C). A noticeable effect of the inducer l-proline on putB-treA expression was already recorded in our experiments when 25 μM l-proline was present in the growth medium (Fig. 5B). This low threshold level of the inducer required to trigger enhanced expression of the putBCP operon should permit B. subtilis to use l-proline effectively as a nutrient in natural settings with a predicted low and variable supply of l-proline, e.g., the soil. Indeed, corn root exudates have been shown to contain enough l-proline to induce the expression of the catabolic putAP gene cluster of Pseudomonas putida (64).
in vitro transcription assays with the putBCP regulatory region as the DNA template have shown that PutR responds directly to l-proline to activate putBCP transcription (7). Our in vivo experiments with the putB-treA reporter gene fusion strain suggest possible additional effector molecules for PutR. We found that the toxic proline analogues AC and DHP are also moderately effective in stimulating PutR-dependent putBCP expression (Fig. 6B); these compounds enter the cell under high-salinity growth conditions via the OpuE (65) transporter (Fig. 2). Quite interesting is the inducing effect of dimethyl-proline (proline betaine) on put expression in high-salinity-grown cells (Fig. 6B). This plant-derived compound functions as a compatible solute (25) and is imported by salt-stressed B. subtilis cells through the osmoprotectant uptake systems OpuA, OpuC, and OpuD (9, 53) and not through the proline importer OpuE or PutP (A. Bashir, B. Kempf, and E. Bremer, unpublished results). Since proline betaine, in contrast to l-proline, is metabolically inert in B. subtilis (B. Kempf and E. Bremer, unpublished results), it should function as a gratuitous inducer for the PutR regulatory protein in high-salinity-grown cells.
The most intriguing finding of our study is certainly the observation that enhanced putBCP expression is not triggered by the very large quantities of l-proline (about 340 mM in cells cultivated in the presence of 1 M NaCl) produced via de novo synthesis (11, 67) by osmotically stressed B. subtilis cells (Fig. 7). It thus emerges from our data that the B. subtilis cell can somehow distinguish exogenously provided l-proline from an intracellular l-proline pool built up via de novo synthesis for either anabolic stress (pool size, about 10 to 16 mM) (12, 67) or osmostress (11, 67) protective purposes.
The molecular mechanism(s) responsible for these different effects of external and internal l-proline on the induction of the catabolic putBCP operon is unexplored. One way by which the B. subtilis cell could accomplish this would be to monitor the influx of l-proline through the PutP transporter and then to communicate this information to the PutR activator. Such a regulatory circuit is found, for instance, in E. coli, where the membrane-integrated transcriptional activator CadC, a member of the ToxR family of sensors/regulators, senses lysine availability in the environment indirectly via interactions with the lysine permease LysP (62).
Our data strongly argue against a scenario in which the B. subtilis cell senses the availability of l-proline in its environment by monitoring PutP-mediated l-proline import to upregulate PutR-dependent putBCP expression. Addition of proline (1 mM) to the growth medium triggered putB-treA expression regardless of whether the PutP transporter was intact, and this was the case even when the PutP-related l-proline transporter OpuE was simultaneously missing, as well (Fig. 8A). It is therefore obvious that there is no direct channeling of externally provided proline via the PutP transporter to the PutR regulator, and a direct role of PutP in sensing the presence of the inducer of PutR in the growth medium is clearly ruled out. Our data (Fig. 2 and 8A) show that, in addition to the high-affinity l-proline transporters PutP and OpuE (Table 3), a yet-unidentified third proline transporter is present in B. subtilis. However, this proline import system exhibits rather moderate transport activity under the growth conditions tested, since we did not detect l-[14C]proline uptake in a putP opuE double mutant at a substrate supply of 40 μM (Table 3), but at high proline concentrations (1 mM) it allows enough l-proline import to induce putBCP expression (Fig. 8B). It seems highly unlikely to us that this yet-uncharacterized transport system would be specifically used to monitor the influx of l-proline into the B. subtilis cell in order to regulate the activity of PutR (Fig. 5B).
Another way by which B. subtilis could possibly monitor the presence of l-proline in its environment to induce putBCP expression is the use of the membrane-embedded chemoreceptor protein McpC. l-Proline taxis by B. subtilis (50) is independent of l-proline uptake (51) and strictly requires the functioning of McpC (45). Hence, it seemed possible that B. subtilis would exploit the McpC chemoreceptor to monitor the presence of external l-proline and then, in a deviation from its well-established role in chemotaxis signaling, would communicate this information to the cell to turn on putBCP transcription via the PutR regulator. Our data clearly rule out any role of McpC for the induction of putBCP expression (Table 4).
An alternative scenario worth considering with respect to the noninducing effects of the large amounts of l-proline synthesized by osmotically challenged B. subtilis cells on put expression (Fig. 7) is that the cell actually does not distinguish between externally provided and internally synthesized l-proline. Rather, it seems possible that the cell is somehow blinded to the intracellular proline signal that results from the osmotically instigated high-level proline synthesis (11, 67) either because the PutR activator cannot interact efficiently with the inducer proline or is compartmentalized in such a way that it is not accessible to l-proline or because the PutR protein complexed with proline cannot interact properly with the put regulatory region to induce transcription. However, our finding that an external supply of proline to severely salt-stressed cells still affords put induction (Fig. 5A, 7, and 8) argues that such a scenario is unlikely.
A phenomenon related to but different from the issue we focus on here with respect to the osmoadaptive l-proline biosynthesis and the catabolic use of the amino acid by B. subtilis has been addressed in E. coli and S. enterica serovar Typhimurium (15, 19, 21, 43). These two Gram-negative bacteria can achieve osmoprotection via uptake of the compatible solute l-proline through the osmotically inducible ProP and ProU transporters, but in contrast to B. subtilis (11, 67), they do not synthesize l-proline as an osmoprotectant (16, 38). However, the overproduction of l-proline as a consequence of feedback-resistant ProB variants leads to enhanced osmotic tolerance of S. enterica serovar Typhimurium cells (15). Interestingly, in these strain backgrounds, mutants that are defective in putA possess higher levels of proline than their putA+ counterparts, indicating that part of the newly produced l-proline is catabolized in these artificial proline overproducers (15). On the other hand, Ekena and Maloy (21) reported that the degradation of proline pools accumulated under high salinity is limited due to direct inhibition of the PutA proline degradative enzyme, but this might also be a nonspecific consequence of a more general inhibition of enzyme activity observed in severely osmotically stressed cells (19). In B. subtilis, exogenously provided l-proline is a moderately effective osmoprotectant in direct comparison with the metabolically inert compatible solute glycine betaine (34, 65). This is partially due to putBCP-dependent l-proline degradation (A. Zaprasis, H. Barzantny, T. Hoffmann, and E. Bremer, unpublished data). However, we found that exogenously provided l-proline is, in contrast to the amino acid arginine and the sugar glucose, a carbon source used very inefficiently by osmotically stressed B. subtilis cells (see Fig. S1 in the supplemental material).
It is evident that we do not yet understand an important aspect of the genetic control of the PutBCP-dependent l-proline utilization pathway of B. subtilis with respect to the overall process of cellular adjustment to high-osmolarity environments (24, 60). Whatever the underlying molecular control mechanism might be that prevents strong induction of the catabolic putBCP genes under osmotic stress conditions through newly synthesized l-proline (Fig. 5A and 7), our data highlight the fact that the B. subtilis cell actively prevents the onset of a wasteful and futile cycle of l-proline synthesis and degradation of the newly produced l-proline when it faces high-osmolarity surroundings. Failure to do so would certainly make osmoadaptive l-proline synthesis (11, 67) less effective and thereby in all likelihood render the overall cellular response of B. subtilis to osmotic stress less robust (9, 10).
Supplementary Material
ACKNOWLEDGMENTS
We are very grateful to M. Lippmann and J. Gade for their dedicated and expert technical assistance throughout this study and to V. Koogle for the careful editing of the manuscript. We thank D. Le Rudulier (University of Nice, Nice, France) for the kind gift of monomethyl-proline, and we are grateful to G. W. Ordal, R. Brückner, M. Brosius, M. Jebbar, G. Nau-Wagner, and O. Schmidt-Kittler for generously making various plasmids and mutant strains of B. subtilis available to us. We thank our colleagues L. N. Csonka and J. Wood for inspiring discussions on the ins and outs of proline uptake and utilization.
Financial support for this study was provided by the Deutsche Forschungsgemeinschaft (DFG), by the Fonds der Chemischen Industrie, and by grants from the BMBF via the Bacell-SysMo2 consortium and the LOEWE program of the State of Hessen (via the Centre for Synthetic Microbiology, SynMicro, Marburg) (to E.B.) and by a research grant (GM036718) from the U.S. Public Health Service (to A.L.S.). A. Z. was supported by the International Max Planck Research School for Environmental, Cellular and Molecular Biology (IMPRS MIC-Marburg).
Footnotes
Supplemental material for this article may be found at http://jb.asm.org/.
Published ahead of print 2 December 2011
REFERENCES
- 1. Alloing G, Travers I, Sagot B, Le Rudulier D, Dupont L. 2006. Proline betaine uptake in Sinorhizobium meliloti: characterization of Prb, an Opp-like ABC transporter regulated by both proline betaine and salinity stress. J. Bacteriol. 188:6308–6317 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. 1990. Basic local alignment search tool. J. Mol. Biol. 215:403–410 [DOI] [PubMed] [Google Scholar]
- 3. Arnold K, Bordoli L, Kopp J, Schwede T. 2006. The SWISS-MODEL workspace: a web-based environment for protein structure homology modelling. Bioinformatics 22:195–201 [DOI] [PubMed] [Google Scholar]
- 4. Atkinson MR, Wray LV, Jr, Fisher SH. 1990. Regulation of histidine and proline degradation enzymes by amino acid availability in Bacillus subtilis. J. Bacteriol. 172:4758–4765 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Bates SL, Waldren RP, Teare ID. 1973. Rapid determination of free proline for water-stress studies. Plant Soil 39:205–207 [Google Scholar]
- 6. Baumberg S, Harwood CR. 1979. Carbon and nitrogen repression of arginine catabolic enzymes in Bacillus subtilis. J. Bacteriol. 137:189–196 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Belitsky BR. 2011. Indirect repression by Bacillus subtilis CodY via displacement of the activator of the proline utilization operon. J. Mol. Biol. 413:321–336 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Belitsky BR, Sonenshein AL. 1998. Role and regulation of Bacillus subtilis glutamate dehydrogenase genes. J. Bacteriol. 180:6298–6305 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Bremer E. 2002. Adaptation to changing osmolarity, p 385–391 In Sonenshein AL, Hoch JA, Losick R. (ed), Bacillus subtilis and its closest relatives. ASM Press, Washington, DC [Google Scholar]
- 10. Bremer E, Krämer R. 2000. Coping with osmotic challenges: osmoregulation through accumulation and release of compatible solutes, p 79–97 In Storz G, Hengge-Aronis R. (ed), Bacterial stress responses. ASM Press, Washington, DC [Google Scholar]
- 11. Brill J, Hoffmann T, Bleisteiner M, Bremer E. 2011. Osmotically controlled synthesis of the compatible solute proline is critical for cellular defense of Bacillus subtilis against high osmolarity. J. Bacteriol. 193:5335–5346 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Brill J, Hoffmann T, Putzer H, Bremer E. 2011. T-box-mediated control of the anabolic proline biosynthetic genes of Bacillus subtilis. Microbiology 157:977–987 [DOI] [PubMed] [Google Scholar]
- 13. Chen LM, Maloy S. 1991. Regulation of proline utilization in enteric bacteria: cloning and characterization of the Klebsiella put control region. J. Bacteriol. 173:783–790 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Commichau FM, Gunka K, Landmann JJ, Stülke J. 2008. Glutamate metabolism in Bacillus subtilis: gene expression and enzyme activities evolved to avoid futile cycles and to allow rapid responses to perturbations of the system. J. Bacteriol. 190:3557–3564 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Csonka LN. 1988. Regulation of cytoplasmic proline levels in Salmonella typhimurium: effect of osmotic stress on synthesis, degradation, and cellular retention of proline. J. Bacteriol. 170:2374–2378 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Csonka LN, Hanson AD. 1991. Prokaryotic osmoregulation: genetics and physiology. Annu. Rev. Microbiol. 45:569–606 [DOI] [PubMed] [Google Scholar]
- 17. Dendinger S, Brill WJ. 1970. Regulation of proline degradation in Salmonella typhimurium. J. Bacteriol. 103:144–152 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Deutch CE. 2011. l-Proline nutrition and catabolism in Staphylococcus saprophyticus. Antonie Van Leeuwenhoek 99:781–793 [DOI] [PubMed] [Google Scholar]
- 19. Deutch CE, Hasler JM, Houston RM, Sharma M, Stone VJ. 1989. Nonspecific inhibition of proline dehydrogenase synthesis in Escherichia coli during osmotic stress. Can. J. Microbiol. 35:779–785 [DOI] [PubMed] [Google Scholar]
- 20. Earl AM, Losick R, Kolter R. 2008. Ecology and genomics of Bacillus subtilis. Trends Microbiol. 16:269–275 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Ekena K, Maloy S. 1990. Regulation of proline utilization in Salmonella typhimurium: how do cells avoid a futile cycle? Mol. Gen. Genet. 220:492–494 [DOI] [PubMed] [Google Scholar]
- 22. Fisher SH. 1993. Utilization of amino acids and other nitrogen-containing compounds, p 221–235 In Sonenshein AL, Hoch JA, Losick R. (ed), Bacillus subtilis and other Gram-positive bacteria. ASM Press, Washington, DC [Google Scholar]
- 23. Gotsche S, Dahl MK. 1995. Purification and characterization of the phospho-alpha(1,1)glucosidase (TreA) of Bacillus subtilis 168. J. Bacteriol. 177:2721–2726 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Hahne H, et al. 2010. A comprehensive proteomics and transcriptomics analysis of Bacillus subtilis salt stress adaptation. J. Bacteriol. 192:870–882 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Hanson AD, et al. 1994. Osmoprotective compounds in the Plumbaginaceae: a natural experiment in metabolic engineering of stress tolerance. Proc. Natl. Acad. Sci. U. S. A. 91:306–310 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Harwood CR, Archibald AR. 1990. Growth, maintenance and general techniques, p 1–26 In Harwood CR, Cutting SM. (ed), Molecular biological methods for Bacillus. John Wiley & Sons, Chichester, United Kingdom [Google Scholar]
- 27. Harwood CR, Crawshaw SG, Wipat A. 2001. From genome to function: systematic analysis of the soil bacterium Bacillus subtilis. Comp. Funct. Genomics 2:22–24 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Harwood CR, Cutting SM. 1990. Molecular biological methods for Bacillus. John Wiley & Sons, Chichester, United Kingdom [Google Scholar]
- 29. Helmann JD. 1995. Compilation and analysis of Bacillus subtilis sigma A-dependent promoter sequences: evidence for extended contact between RNA polymerase and upstream promoter DNA. Nucleic Acids Res. 23:2351–2360 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Holtmann G, Bremer E. 2004. Thermoprotection of Bacillus subtilis by exogenously provided glycine betaine and structurally related compatible solutes: involvement of Opu transporters. J. Bacteriol. 186:1683–1693 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Huang SC, Lin TH, Shaw GC. 2011. PrcR, a PucR-type transcriptional activator, is essential for proline utilization and mediates proline-responsive expression of the proline utilization operon putBCP in Bacillus subtilis. Microbiology 157:3370–3377 [DOI] [PubMed] [Google Scholar]
- 32. Inagaki E, et al. 2006. Crystal structure of Thermus thermophilus Δ1-pyrroline-5-carboxylate dehydrogenase. J. Mol. Biol. 362:490–501 [DOI] [PubMed] [Google Scholar]
- 33. Jafri S, Evoy S, Cho K, Craighead HG, Winans SC. 1999. An Lrp-type transcriptional regulator from Agrobacterium tumefaciens condenses more than 100 nucleotides of DNA into globular nucleoprotein complexes. J. Mol. Biol. 288:811–824 [DOI] [PubMed] [Google Scholar]
- 34. Kappes RM, Kempf B, Bremer E. 1996. Three transport systems for the osmoprotectant glycine betaine operate in Bacillus subtilis: characterization of OpuD. J. Bacteriol. 178:5071–5079 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Köcher S, Tausendschön M, Thompson M, Saum SH, Müller V. 2011. Proline metabolism in the moderate halophilic bacterium Halobacillus halophilus: differential regulation of isogenes in proline utilization. Environ. Microbiol. Rep. 3:443–448 [DOI] [PubMed] [Google Scholar]
- 36. Lee JH, Park NY, Lee MH, Choi SH. 2003. Characterization of the Vibrio vulnificus putAP operon, encoding proline dehydrogenase and proline permease, and its differential expression in response to osmotic stress. J. Bacteriol. 185:3842–3852 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Ling M, Allen SW, Wood JM. 1994. Sequence analysis identifies the proline dehydrogenase and Δ1-pyrroline-5-carboxylate dehydrogenase domains of the multifunctional Escherichia coli PutA protein. J. Mol. Biol. 243:950–956 [DOI] [PubMed] [Google Scholar]
- 38. Lucht JM, Bremer E. 1994. Adaptation of Escherichia coli to high osmolarity environments: osmoregulation of the high-affinity glycine betaine transport system ProU. FEMS Microbiol. Rev. 14:3–20 [DOI] [PubMed] [Google Scholar]
- 39. May G, Faatz E, Villarejo M, Bremer E. 1986. Binding protein dependent transport of glycine betaine and its osmotic regulation in Escherichia coli K12. Mol. Gen. Genet. 205:225–233 [DOI] [PubMed] [Google Scholar]
- 40. Mazier S, Quick M, Shi L. 2011. Conserved tyrosine in the first transmembrane segment of solute:sodium symporters is involved in Na+-coupled substrate co-transport. J. Biol. Chem. 286:29347–29355 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Menzel R, Roth J. 1981. Regulation of the genes for proline utilization in Salmonella typhimurium: autogenous repression by the putA gene product. J. Mol. Biol. 148:21–44 [DOI] [PubMed] [Google Scholar]
- 42. Miller JH. 1972. Experiments in molecular genetics. Cold Spring Harbor Laboratory, Cold Spring Harbor, NY [Google Scholar]
- 43. Milner JL, McClellan DJ, Wood JM. 1987. Factors reducing and promoting the effectiveness of proline as an osmoprotectant in Escherichia coli K12. J. Gen. Microbiol. 133:1851–1860 [DOI] [PubMed] [Google Scholar]
- 44. Molle V, et al. 2003. Additional targets of the Bacillus subtilis global regulator CodY identified by chromatin immunoprecipitation and genome-wide transcript analysis. J. Bacteriol. 185:1911–1922 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Müller J, Schiel S, Ordal GW, Saxild HH. 1997. Functional and genetic characterization of mcpC, which encodes a third methyl-accepting chemotaxis protein in Bacillus subtilis. Microbiology 143:3231–3240 [DOI] [PubMed] [Google Scholar]
- 46. Muro-Pastor AM, Ostrovsky P, Maloy S. 1997. Regulation of gene expression by repressor localization: biochemical evidence that membrane and DNA binding by the PutA protein are mutually exclusive. J. Bacteriol. 179:2788–2791 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Nakada Y, Nishijyo T, Itoh Y. 2002. Divergent structure and regulatory mechanism of proline catabolic systems: characterization of the putAP proline catabolic operon of Pseudomonas aeruginosa PAO1 and its regulation by PruR, an AraC/XylS family protein. J. Bacteriol. 184:5633–5640 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Olkhova E, Raba M, Bracher S, Hilger D, Jung H. 2011. Homology model of the Na+/proline transporter PutP of Escherichia coli and its functional implications. J. Mol. Biol. 406:59–74 [DOI] [PubMed] [Google Scholar]
- 49. Ordal GW, Marquez-Magana L, Chamberlin MJ. 1993. Motility and chemotaxis, p 765–784 In Sonenshein AL, Hoch JA, Losick R. (ed), Bacillus subtilis and other Gram-positive bacteria. ASM Press, Washington, DC [Google Scholar]
- 50. Ordal GW, Villani DP, Gibson KJ. 1977. Amino acid chemoreceptors of Bacillus subtilis. J. Bacteriol. 129:156–165 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Ordal GW, Villani DP, Nicholas RA, Hamel FG. 1978. Independence of proline chemotaxis and transport in Bacillus subtilis. J. Biol. Chem. 253:4916–4919 [PubMed] [Google Scholar]
- 52. Ostrovsky de Spicer P, Maloy S. 1993. PutA protein, a membrane-associated flavin dehydrogenase, acts as a redox-dependent transcriptional regulator. Proc. Natl. Acad. Sci. U. S. A. 90:4295–4298 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Smits SH, et al. 2008. The compatible-solute-binding protein OpuAC from Bacillus subtilis: ligand binding, site-directed mutagenesis, and crystallographic studies. J. Bacteriol. 190:5663–5671 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Sonenshein AL. 2005. CodY, a global regulator of stationary phase and virulence in Gram-positive bacteria. Curr. Opin. Microbiol. 8:203–207 [DOI] [PubMed] [Google Scholar]
- 55. Sonenshein AL. 2007. Control of key metabolic intersections in Bacillus subtilis. Nat. Rev. Microbiol. 5:917–927 [DOI] [PubMed] [Google Scholar]
- 56. Sonenshein AL. 2000. Control of sporulation initiation in Bacillus subtilis. Curr. Opin. Microbiol. 3:561–566 [DOI] [PubMed] [Google Scholar]
- 57. Soto MJ, Jimenez-Zurdo JI, van Dillewijn P, Toro N. 2000. Sinorhizobium meliloti putA gene regulation: a new model within the family Rhizobiaceae. J. Bacteriol. 182:1935–1941 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Spiegelhalter F, Bremer E. 1998. Osmoregulation of the opuE proline transport gene from Bacillus subtilis: contributions of the sigma A- and sigma B-dependent stress-responsive promoters. Mol. Microbiol. 29:285–296 [DOI] [PubMed] [Google Scholar]
- 59. Srivastava D, et al. 2010. Crystal structure of the bifunctional proline utilization A flavoenzyme from Bradyrhizobium japonicum. Proc. Natl. Acad. Sci. U. S. A. 107:2878–2883 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Steil L, Hoffmann T, Budde I, Völker U, Bremer E. 2003. Genome-wide transcriptional profiling analysis of adaptation of Bacillus subtilis to high salinity. J. Bacteriol. 185:6358–6370 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Tanner JJ. 2008. Structural biology of proline catabolism. Amino Acids 35:719–730 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Tetsch L, Koller C, Haneburger I, Jung K. 2008. The membrane-integrated transcriptional activator CadC of Escherichia coli senses lysine indirectly via the interaction with the lysine permease LysP. Mol. Microbiol. 67:570–583 [DOI] [PubMed] [Google Scholar]
- 63. Thompson JD, Plewniak F, Thierry JC, Poch O. 2000. DbClustal: rapid and reliable global multiple alignments of protein sequences detected by database searches. Nucleic Acids Res. 28:2919–2926 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Vilchez S, Molina L, Ramos C, Ramos JL. 2000. Proline catabolism by Pseudomonas putida: cloning, characterization, and expression of the put genes in the presence of root exudates. J. Bacteriol. 182:91–99 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. von Blohn C, Kempf B, Kappes RM, Bremer E. 1997. Osmostress response in Bacillus subtilis: characterization of a proline uptake system (OpuE) regulated by high osmolarity and the alternative transcription factor sigma B. Mol. Microbiol. 25:175–187 [DOI] [PubMed] [Google Scholar]
- 66. Welsh DT. 2000. Ecological significance of compatible solute accumulation by micro-organisms: from single cells to global climate. FEMS Microbiol. Rev. 24:263–290 [DOI] [PubMed] [Google Scholar]
- 67. Whatmore AM, Chudek JA, Reed RH. 1990. The effects of osmotic upshock on the intracellular solute pools of Bacillus subtilis. J. Gen. Microbiol. 136:2527–2535 [DOI] [PubMed] [Google Scholar]
- 68. White TA, Krishnan N, Becker DF, Tanner JJ. 2007. Structure and kinetics of monofunctional proline dehydrogenase from Thermus thermophilus. J. Biol. Chem. 282:14316–14327 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Wood JM. 1981. Genetics of L-proline utilization in Escherichia coli. J. Bacteriol. 146:895–901 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Wood JM. 1987. Membrane association of proline dehydrogenase in Escherichia coli is redox dependent. Proc. Natl. Acad. Sci. U. S. A. 84:373–377 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Zhou Y, et al. 2008. Structural basis of the transcriptional regulation of the proline utilization regulon by multifunctional PutA. J. Mol. Biol. 381:174–188 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Zhou Y, Zhu W, Bellur PS, Rewinkel D, Becker DF. 2008. Direct linking of metabolism and gene expression in the proline utilization A protein from Escherichia coli. Amino Acids 35:711–718 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.