Abstract
Aureocin A53 is an antimicrobial peptide produced by Staphylococcus aureus A53. The genetic determinants involved in aureocin A53 production and immunity to its action are organized in at least four transcriptional units encoded by the 10.4-kb plasmid pRJ9. One transcriptional unit carries only the bacteriocin structural gene, aucA. No immunity gene is found downstream of aucA, as part of the same transcriptional unit. Further downstream of aucA is found an operon which contains the three genes aucEFG, whose products seem to associate to form a dedicated ABC transporter. When aucEFG were expressed in RN4220, an aureocin A53-sensitive S. aureus strain, this strain became partially resistant to the bacteriocin. A gene disruption mutant in aucE was defective in aureocin A53 externalization and more sensitive to aureocin A53 than the wild-type strain, showing that aucEFG are involved in immunity to aureocin A53 by active extrusion of the bacteriocin. Full resistance to aureocin A53 was exhibited by transformants carrying, besides aucEFG, the operon formed by two genes, aucIB and aucIA, located between aucA and aucEFG and carried in the opposite strand. AucIA and AucIB share similarities with hypothetical proteins not found in the gene clusters of other bacteriocins. A gene disruption mutant in orf8, located upstream of aucA and whose product exhibits about 50% similarity to a number of hypothetical membrane proteins found in many Gram-positive bacteria, was strongly affected in aureocin A53 externalization but resistant to aureocin A53, suggesting that Orf8 is also involved in aureocin A53 secretion.
INTRODUCTION
Bacteriocins (Bac) are a group of antimicrobial peptides which are produced by bacteria inhabiting diverse environments. They typically inhibit the growth of strains closely related to the producer strain although some bacteriocins have a broad spectrum of activity inhibiting different species of bacteria (21). Bacteriocins produced by Gram-positive bacteria are currently classified into three major classes although a fourth class, composed of cyclic bacteriocins, has already been proposed (40). Class I bacteriocins, the lantibiotics, are small, heat-stable peptides that contain modified amino acids and undergo extensive posttranslational modifications (6). Class II bacteriocins are small, heat-stable peptides which do not contain modified amino acids (29). Class III bacteriocins are heat-labile antimicrobial proteins (5, 21). The majority of the peptide bacteriocins act by permeabilization of the membrane of the sensitive cells, leading to leakage of cellular solutes and, eventually, to cell death (21).
Bacteriocin production is invariably linked to the expression of specific immunity systems which are required for producer self-protection against the toxicity of the bacteriocin (6, 21, 29). For lantibiotics, the protection is mediated by the so-called immunity proteins (LanI or LanH) and/or dedicated ABC transporter proteins (LanEFG). For instance, immunity to the lantibiotic nisin involves a combined action which includes (i) sequestering of the bacteriocin molecules on the bacterial cell membrane by NisI (15) and (ii) removal of the bacteriocin from cells by NisFEG (35). LanH is also a lantibiotic-binding protein which captures the bacteriocin in an energy-independent manner, after which the lantibiotic is transported to the extracellular space by LanEFG in an energy-dependent manner (31). Although LanH presents a function similar to that of LanI (lantibiotic binding), the structures of the two proteins are quite different. For example, NukH, a protein involved in immunity to nukacin ISK-1, is a membrane protein with three transmembrane domains, and its N terminus is located on the cytoplasmic side (31). On the other hand, NisI is a lipoprotein anchored to the extracellular side of the membrane by its N-terminal lipid (39).
In class II bacteriocins, immunity proteins are generally also involved in self-protection. The genetic determinants proposed or confirmed to confer immunity are frequently found closely associated with the bacteriocin structural gene(s), generally as part of the same operon (29). Their products usually have a high pI, are small (ranging from 51 to approximately 150 amino acids), and contain either no or up to four putative transmembrane segments (26). This fact suggests that the immunity proteins can be secreted out of the cell or even be retained in the membrane (14, 16). It has been shown that, for class II bacteriocins that use components of the mannose phosphotransferase system of susceptible cells as targets/receptors, the immunity protein forms a strong complex with the receptor proteins and the bacteriocin and thereby prevents cells from being killed (13).
Another mechanism of immunity has been described for some class II bacteriocins, which relies on the activity of a multidrug transporter protein (19). It seems that these multidrug transporters mediate bacteriocin immunity by removing bacteriocin that enters the cytoplasmic membrane from the outside. Besides conferring immunity, these transporters also function as exit pumps for the bacteriocins, extruding the molecules from their site of production, the cytoplasm, to the extracellular medium (19).
Aureocin A53 is an atypical class II bacteriocin produced by Staphylococcus aureus A53 and encoded by a 10.4-kb plasmid named pRJ9 (20). This bacteriocin is highly active against multidrug-resistant nosocomial staphylococcal strains, including the multidrug-resistant S. aureus (MRSA) clone endemic in most Brazilian hospitals and in other countries (25), Streptococcus agalactiae, and coagulase-positive staphylococcal strains involved in bovine mastitis, a disease of great economic impact (9, 12). It can also inhibit Listeria monocytogenes, an important food-borne pathogen (11). Therefore, aureocin A53 has potential in medical applications and also as a food preservative. However, optimal and rational exploitation of aureocin A53 as an antimicrobial agent requires its production in large scale. This, in turn, necessitates knowledge on all gene products required for its production.
The complete sequence of pRJ9 showed that this plasmid presents 14 putative open reading frames (ORFs) (Fig. 1) (27). One segment of pRJ9 (orf1 to orf6) contains functions related to plasmid mobilization and replication. The second part (orf7 to orf14) appears to carry genes involved in the production of aureocin A53. No gene encoding an immunity protein has been identified in the transcriptional unity which carries the aureocin A53 structural gene, aucA. However, three genes (orf12, orf13, and orf14) were identified in pRJ9, whose products possess similarity to three-component ABC transporters generally involved in immunity to either lantibiotics (6) or cyclic bacteriocins (29, 40). In the present work, the role of these genes and of a putative operon formed by orf11 and orf10, found upstream of orf12, orf13, and orf14, in immunity to aureocin A53 was investigated. During the course of this study another gene, orf8, involved in aureocin A53 secretion was also detected.
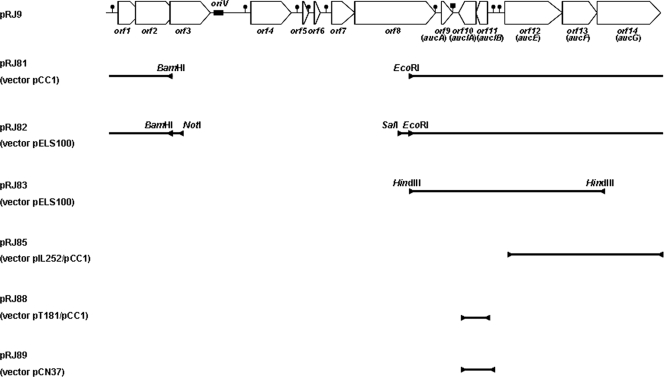
Fig 1.
Genetic map of plasmid pRJ9 (adapted from Fig. 1 of reference 27 with permission of the publisher) showing the inserts cloned into different vectors. The main restriction sites used in the cloning experiments are shown. The inserts found in pRJ85, pRJ88, and pRJ89 were obtained by PCR amplification of the corresponding regions.
MATERIALS AND METHODS
Bacterial strains and growth conditions.
Bacterial strains used in the present study are listed in Table 1. Escherichia coli strains were cultivated at 37°C in Luria-Bertani (LB) broth supplemented with 100 μg ml−1 ampicillin (Ap), 150 μg ml−1 erythromycin (Em), or 12.5 μg ml−1 chloramphenicol (Cm), when appropriate. Lactobacillus species strains were grown in Man-Rogosa-Sharpe (MRS; Oxoid) broth at 30°C. When appropriate, the MRS medium was supplemented with Em at 5 μg ml−1. S. aureus and Micrococcus luteus were grown at 37°C in brain heart infusion (BHI) broth (Difco). S. aureus strains carrying pRJ9 derivatives upon transposon mutagenesis were grown in the presence of 10 μg ml−1 Em. S. aureus strains carrying recombinant plasmids derived from the cloning experiments were grown in the presence of either Em (for derivatives of pCN37 or pIL252) or tetracycline ([Tc] for derivatives of pT181), both at 10 μg ml−1. All media were supplemented with agar at 1.5% or 0.6% (wt/vol), when required.
Table 1.
Plasmids and bacterial strains used in this work
| Plasmid or strain | Relevant feature(s)a | Reference or source |
|---|---|---|
| Plasmids | ||
| pCC1 | 8.1 kb; Cmrori2 oriV | Epicentre |
| pCR2.1-TOPO | 3.9 kb; Kmr | Invitrogen |
| pELS100 | 7.0 kb; shuttle vector E. coli-Lactobacillus; Apr Emr | LMGT |
| pIL252 | 4.6 kb; a LAB vector; Emr | 36 |
| pNO6 | 12.7 kb; shuttle vector E. coli/LAB formed by ligation of pCC1 and pIL252; Cmr Emr | This work |
| pCN37 | 6.3 kb; shuttle vector E. coli-S. aureus; Apr Emr | 8 |
| pT181mcs | 4.8 kb; S. aureus plasmid; Tcr | 2 |
| pNO9 | 12.9 kb; shuttle vector E. coli/S. aureus formed by ligation of pCC1 and pT181mcs; Cmr Tcr | |
| pRJ9 | 10.4 kb; codes for aureocin A53 | 20 |
| pRJ81 | 13.9 kb; Cmr; BamHI/EcoRI-A fragment of pRJ9 cloned into pCC1 | This work |
| pRJ82 | 13.1 kb; BamHI/EcoRI-A fragment of pRJ9 cloned into pELS100 | This work |
| pRJ83 | 10.6 kb; HindIII-A fragment of pRJ9 cloned into pELS100 | This work |
| pRJ84 | 11.0 kb; orf12-orf13-orf14 of pRJ9 cloned into pCC1 | This work |
| pRJ85 | 15.6 kb; orf12-orf13-orf14 cloned into pNO6 | This work |
| pRJ88 | 13.2 kb; orf11-orf10 cloned into pCC1 and ligated to pT181 | This work |
| pRJ89 | 7.0 kb; orf11-orf10 cloned into pCN37 | This work |
| Strains | ||
| E. coli strains | ||
| EPI300 | Epicentre | |
| DH5α | lacZΔM15 hsdR mutant | 33 |
| L. sakei DSM 20017 | Sensitive to aureocin A53 | LMGT |
| M. luteus ATCC 4698 | Sensitive to aureocin A53 | ATCC |
| S. aureus strains | ||
| RN4220 | hsd mutant; sensitive to aureocin A53 | 30 |
| RN9590 | RN4220 carrying pCN37 | 8 |
| A53 | pRJ9; 10.4 kb; Bac+ Imm+ | 20 |
| MB38 | pRJ14 (pRJ9::Tn551); 15.6 kb; Emr | 1 |
| MB139 | pRJ38 (pRJ9::Tn917-lac); 18.9 kb; Emr | 11 |
| MB140 | pRJ39 (pRJ9::Tn917-lac); 18.9 kb; Emr | 11 |
| MB143 | pRJ40 (pRJ9::Tn917-lac); 18.9 kb; Emr | 11 |
| MB145 | pRJ108 (pRJ9::Tn917-lac); 18.9 kb; Emr; low aureocin A53 production | This work |
| MB388 | RN4220 carrying pT181mcs | This work |
| MB420 | RN4220 carrying pRJ88 | This work |
| MB422 | RN4220 carrying pRJ85 and pRJ88 | This work |
| MB425 | RN4220 carrying pNO6 | This work |
| MB426 | RN4220 carrying pRJ85 | This work |
| MB439 | RN4220 carrying pRJ89 | This work |
| MB502 | Plasmidless derivative of strain MB145 | This work |
Imm+, presence of immunity to aureocin A53; hsd mutant, deficient in DNA restriction.
In silico sequence analyses.
Sequence similarity of the predicted amino acid sequences of the proteins encoded by pRJ9 to other proteins was searched using the Basic Local Alignment Search Tool (BLASTn) analysis. Multiple sequence alignments were carried out using BioEdit software. Protein transmembrane domains were predicted by using the TMpred Server (http://www.ch.embnet.org/software/TMPRED_form.html). The pRJ9 DNA sequence was retrieved from GenBank (accession number AF4478813).
DNA techniques.
Plasmids pCC1, pELS100, pIL252, pNO6, pNO9, and pCN37 and their derivatives carrying inserts were isolated from the E. coli host strains by using a QiaPrep Spin Miniprep kit (Qiagen) according to the supplier's instructions. The same kit was used to purify plasmid DNA from either lactic acid bacteria (LAB) or S. aureus. In both cases, the extractions were performed after treatment of the cells, for 30 min at 37°C, with a lysis solution (0 mM Tris-HCl, pH 8.0, 40 mM EDTA, and 25% [wt/vol] sucrose) supplemented with either 5 mg ml−1 lysozyme (Sigma), 15 U ml−1 mutanolysin (Sigma), 100 μg ml−1 RNase A (Sigma), and 2.5 μg ml−1 SDS, for LAB cells, or 1 mg ml−1 lysozyme, 20 μg ml−1 lysostaphin (Sigma), 100 μg ml−1 RNase A, and 2.5 μg ml−1 SDS, for the staphylococcal cells. Restriction enzymes were purchased from Promega, and T4 DNA-ligase was from Invitrogen. The enzymes were used as specified by the suppliers. Agarose gel electrophoreses were performed by standard methods (33).
PCR amplification of the fragment containing the putative orf12-orf13-orf14 operon of pRJ9 was performed using the primers J010A and J011 (Table 2). Each reaction mixture contained the following: 1× Pfx reaction mix, 1.0 mM MgSO4, 10 μM each primer, 1 U of platinum Pfx DNA polymerase (Invitrogen), and 200 ng of template DNA. PCR amplification of the fragment containing the putative orf11-orf10 operon of pRJ9 was performed using the primers ORF10F and ORF11R (Table 2) and Phusion High-Fidelity DNA-Polymerase (Finnzymes Oy). The amplification cycles were performed following the suppliers' recommendations. PCR fragments for cloning experiments were purified by a Qiaex II kit (Qiagen). The nucleotide sequence of the amplicons was verified by dideoxy chain termination sequencing in an automated sequencer (ABI Prism 3100; Perkin Elmer) after the amplicons were cloned into the plasmid vector pCR2.1-TOPO (Invitrogen), according to the manufacturer's instructions.
Table 2.
Primers used in the present study
| Primer | Sequence (5′ → 3′) | Tm (°C)a | Expected amplicon size (bp) |
|---|---|---|---|
| J010A | GGA TCC AAT AAA CAT CAC CCT TAT GT | 55 | 3,041 |
| J011 | GGT GGT GTCGAC GGG ATA TTC CCC AAT CCC CTT | 68 | |
| ORF12-13F | AGA GTG AAG TTA GAG ATG GG | 51 | 381 |
| ORF12-13R | TTT CCT GAC CCA GAA GGC | 55 | |
| ORF13-14F | TAT CCG GTG GCC AAC AAC | 55 | 366 |
| ORF13-14R | GTA ATT ACT GAA GAG ATG CCG | 51 | |
| ORF10F | GTT GAA TAA AGA GCC AAA GGT GCC A | 56 | 735 |
| ORF11R | CAA TTT TAT TCC TCC ATT TCA GAT GA | 52 | |
| ORF10-11F | TTG GAT GCT TGG TTA GAA AA | 50 | 576 |
| ORF10-11R | TTA TCC TTG ATA ATC TTT AGC | 48 | |
| GyrF | GCG TGA ATC ATT TTT AGA TTA TGC G | 53 | 447 |
| GyrR | AAG TTA GGG AAT CGA GCA G | 48 |
Thermal denaturation midpoint temperature.
Plasmid constructions.
All plasmids constructed in this study are also described in Table 1 and shown in Fig. 1. To generate plasmid pRJ81, pCC1 and pRJ9 were cleaved with BamHI and EcoRI. Digestion of pRJ9 generated two fragments of 5.8 kb and 4.6 kb. The 5.8-kb fragment, encompassing the region from the 3′ end of orf8 to the first two thirds of orf2 (Fig. 1), was ligated to linearized pCC1. The resulting plasmid, called pRJ81, was introduced into chemically competent E. coli EPI300.
For construction of pRJ82, plasmids pRJ81 and pELS100, a shuttle vector for E. coli and Lactobacillus spp., were digested with SalI and NotI. The digestion of pRJ81 generated five fragments, and the 6.15-kb fragment (Fig. 1) was isolated and ligated into the linearized pELS100. The new plasmid, called pRJ82, was transferred first to the chemically competent E. coli EPI300 strain and, subsequently, to Lactobacillus sakei DSM20017 by electroporation.
For construction of pRJ83, plasmids pELS100 and pRJ9 were digested with HindIII. These digestions linearized plasmid pELS100 and resulted in four fragments of plasmid pRJ9. The largest one, fragment HindIII-A (3,581 bp; from the 3′ end of orf8 to the 5′ end of orf14) (Fig. 1), was gel purified and ligated to pELS100. pRJ83 was transferred to E. coli EPI300 and, subsequently, to L. sakei DSM20017.
To construct pRJ85, a fragment containing orf12, orf13, and orf14 was amplified by PCR. This amplicon was first ligated to plasmid pCR2.1-TOPO and transferred to chemically competent E. coli DH5α. Five kanamycin-resistant transformants were chosen for plasmid DNA extraction and sequencing to confirm the correct amplicon sequence prior to digestion with EcoRI to liberate the insert. The insert was ligated to pCC1, also digested with EcoRI, generating plasmid pRJ84. Subsequently, pRJ84 was ligated to pIL252, a LAB vector, by the unique BamHI site present on both plasmids, generating plasmid pRJ85. pRJ85 was transferred to E. coli EPI300 and, subsequently, to L. sakei DSM20017.
Plasmid pRJ88, containing an amplicon carrying both orf11 and orf10, was constructed in a manner similar to that described for plasmid pRJ85 but using pT181mcs, a staphylococcal plasmid, instead of pIL252, as a Gram-positive vector. pRJ89 was generated by cloning the amplicon carrying both orf11 and orf10 into the unique EcoRI site of pCN37, an E. coli/S. aureus shuttle vector. Both plasmids were transferred to E. coli DH5α and, subsequently, to S. aureus RN4220 by transformations.
The presence of the correct fragment in all recombinant plasmids was verified by agarose gel electrophoresis, restriction endonuclease analyses, PCR amplification, and DNA sequencing, when appropriate.
Transformations.
Transformation of chemically competent E. coli cells was performed by standard methods (33). LAB and staphylococci were transformed by electroporation according to Aukrust et al. (3) and Schenk and Laddaga (34), respectively.
Plasmid-curing experiments.
Strain MB145, the host strain of plasmid pRJ108, was used for curing experiments. pRJ108 is a pRJ9-derivative tagged with Tn917-lac, which was obtained by the method described by Oliveira et al. (11). MB145 cells were incubated in 5 ml of tryptic soy broth (TSB) medium (Difco) for 24 h at 45°C, after which the culture was diluted 1,000-fold in the same medium and reincubated under the same conditions. This step was repeated until the 10th day, when the culture was serially diluted in 0.85% saline, and dilutions of 10−6 and 10−7 were plated onto TSB agar plates. After incubation at 37°C for 18 h, the colonies were replica plated into Em (10 μg ml−1)-containing medium to test for loss of the erythromycin resistance. Plasmid DNA was isolated from Ems colonies and subjected to electrophoresis in agarose gel to confirm plasmid loss. The sensitivity of the cured derivatives to aureocin A53 was tested by a deferred-antagonism assay on solid BHI medium, as described by Giambiagi-deMarval et al. (20), using them as the indicator and the wild-type strain, A53, as the bacteriocin producer.
Analysis of the toxicity of the inserts cloned into pCC1.
The toxicity of the insert present on pRJ81 was tested in E. coli by induction of the plasmid to a high copy number per cell, according to instructions of the Copy Control PCR cloning kit (Epicentre). Strains EPI300 and EPI300/pCC1 were also included in these experiments as controls. The strains were grown in 5 ml of LB medium at 37°C for 18 h and diluted to an optical density at 600 nm (OD600) of 0.05. Four hundred microliters from each culture was distributed in a Bioscreen honeycomb 100-well plate with addition of the copy control induction agent (Epicentre), according to the manufacturer's instructions. The diluted cultures without addition of the induction agent were used as controls. The growth curves were monitored at 37°C in a Bioscreen C system (Oy Growth Curves AB, Ltd.), which measured kinetically the culture OD600 at time intervals of 30 min, for 24 h. Five replicas of the experiment were performed for each strain. Graphs originated from these measurements were generated using the program Growth Curves, version 2.28, for Microbiology Reader Bioscreen (Labsystems).
Aureocin A53 preparations.
S. aureus A53 was grown in either 100 ml or 1 liter of BHI broth at 37°C, with vigorous shaking for 24 h. The cells were removed by centrifugation at 12,000 × g for 15 min at 4°C, and the supernatant was sterilized by membrane filtration (0.22-μm pores; Sarstedt). Alternatively, aureocin A53 present in the culture supernatant was concentrated by ammonium sulfate precipitation (40%; wt/vol), resuspended in either 10 ml or 100 ml of sterile Milli-Q water, and dialyzed against 0.1 M Na phosphate buffer, pH 6.5.
Antimicrobial activity in all preparations was determined by the dilution method using a microtiter plate assay as described by Ceotto and coworkers (7). Briefly, serial 2-fold dilutions of the bacteriocin preparation were prepared in 100 μl of BHI broth. Each well was seeded with 100 μl of the sensitive strain (106 cells of M. luteus ATCC 4698) in BHI broth. The plates were incubated for 18 h at 37°C. The bacteriocin activity was measured in bacteriocin units (BU) per ml, which represented the reciprocal of the highest supernatant dilution showing at least a 50% inhibition of the bacterial growth in relation to the control with no bacteriocin added, multiplied by 10.
Determination of the aureocin A53 activity kinetics.
An experiment to determine aureocin A53 activity kinetics was performed by reading the OD600 (in a Bioscreen C system) of an aureocin-susceptible culture in the presence of the partially purified bacteriocin at 37°C in intervals of 30 min for a period of 4 h. The partially purified aureocin A53 (640 BU), obtained by ammonium sulfate precipitation, was tested against 109 cells of the indicator strain, M. luteus ATCC 4698, in a 400-μl final volume.
Immunity assays.
The immunity expression to aureocin A53 exhibited by the strains carrying either pRJ9 genes or pRJ9 derivatives tagged with a transposon was tested in microtiter plate assays containing the appropriate broth medium with 2-fold serial dilutions of an aureocin A53 preparation. The bacterial cells under test were collected at the stationary phase of growth, and the OD600 was adjusted to 0.05 by the addition of the appropriate medium. In all assays, 100-μl volumes of these suspensions were added to each well. Controls grown in the absence of the aureocin A53 were also included. The microtiter plates were incubated at the appropriate temperature, according to the bacteria tested, for 24 h, after which the OD620s were measured.
When needed, the immunity to aureocin A53 exhibited by randomly selected transformants was also determined using the Bioscreen C honeycomb 100-well plates as described for the toxicity assays of the inserts cloned into pCC1, with minor modifications. Briefly, 200 μl of 2-fold serial dilutions of an aureocin A53 preparation was distributed in wells of a plate, after which 200 μl from each culture, diluted to an OD600 of 0.05 in the appropriate broth, was added to each well. The growth curves were monitored at either 30°C or 37°C at time intervals of 1 h for a period of 16 to 18 h. In all cases, the assays were performed at least in triplicate. Strains carrying only the vectors used in the cloning experiments, but without inserts, were used as controls.
pRJ9 mutant studies.
Mutants of plasmid pRJ9 were obtained previously by transposon mutagenesis (11). The exact position of the insertion of the transposon into plasmid pRJ9 was determined in the present study by DNA sequencing, using an 18-bp primer (5′-TGT ACC ACT AAT AAC TCA-3′) whose 3′ end is located 101 bp upstream of the left end of the transposon. The presence of aureocin A53 in the culture supernatant and the immunity to aureocin A53 expressed by these mutants were analyzed as described above.
Transcription analyses.
Isolation of total RNA from S. aureus strain A53 was performed with an RNeasy Mini kit (Qiagen), essentially as described by Coelho et al. (10). Briefly, cultures were harvested after 18 h of growth at 37°C. The cells were lysed by incubation with lysozyme (40 mg ml−1; Sigma) and lysostaphin (10 U; Sigma) in Tris-HCl–EDTA (TE) buffer. After this step, the procedure followed the manufacturer's recommendations. RNA preparations were quantified by using a NanoDrop ND1000 Spectrophotometer (Thermo Scientific). DNase I-pretreated RNA (500 ng) was subjected to cDNA synthesis using a Revert Aid H Minus First Strand cDNA Synthesis kit (Fermentas) as described by the manufacturer. Reverse transcription-PCRs (RT-PCRs) were performed with the following primers (Table 2): ORF12-13F and ORF12-13R for cotranscription analysis of aucE and aucF; ORF13-14F and ORF13-14R for cotranscription analysis of aucF and aucG; ORF10-11F and ORF10-11R for cotranscription analysis involving orf11-orf10. An internal control was performed using the primers GyrF and GyrR targeting the housekeeping gene gyrA that codes for the DNA-gyrase subunit A. In addition, a DNA contamination control was performed using this same pair of primers. This control was a PCR using the same conditions of the RT-PCR process without the RT step, and it demonstrated the purity of the RNA preparation. The PCR mixtures consisted of 1× reaction buffer, 1 U of DyNAzyme II DNA-polymerase (Finnzymes Oy), 2.5 mM MgCl2, 10 mM concentration of each deoxynucleoside triphosphate (dNTP; Promega), and 20 pmol of each primer. The PCRs were done essentially as described by Coelho et al. (10), and the amplicons were analyzed on 1% (wt/vol) agarose gels, using 100-bp DNA ladders (from either Fermentas or Promega) as size markers.
RESULTS AND DISCUSSION
Aureocin A53 activity kinetics.
In previous attempts, in spite of the many curing agents tested, we were unable to cure plasmid pRJ9 from strain A53 and, therefore, to prove that pRJ9 carries genetic determinants involved in aureocin A53 immunity (20). In order to investigate the reasons for this failure, the kinetics of aureocin A53 activity was tested against M. luteus in a Bioscreen C system. The OD600 of the susceptible micrococcal culture was drastically reduced by the peptide action: from 1.8 to 1.4 after 15 min, to ∼0.9 after 90 min (50% of reduction), and to ∼0.6 after 4 h, showing that aureocin A53 has lytic activity (data not shown). The lytic activity of aureocin A53 may explain why derivatives of strain A53 devoid of plasmid pRJ9 could never be recovered previously. If pRJ9 carries genes involved in bacteriocin immunity, the plasmid loss is expected to result in cured derivatives which become sensitive to aureocin A53 produced by the remaining Bac-positive (Bac+) cells. Therefore, to avoid the interference of aureocin A53 in the curing experiments, strain MB145, carrying a pRJ9 mutant defective in bacteriocin production, was created by transposon mutagenesis.
Curing of a plasmid pRJ9 derivative.
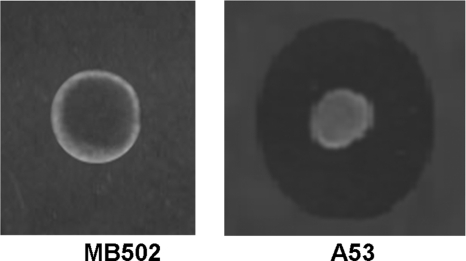
Strain MB145, carrying a pRJ9 derivative tagged with a transposon encoding Em resistance and producing a low level of bacteriocin (40 BU ml−1), was submitted to curing experiments. After 10 days of growth at 45°C, two colonies proved to be Ems. The plasmid loss was confirmed by agarose gel electrophoresis (data not shown). The sensitivity of both Ems colonies to aureocin A53 was also confirmed (Fig. 2; Table 3), indicating that plasmid pRJ9, indeed, carries at least one gene involved in bacteriocin immunity, as found in most bacterial systems (6, 29, 40).
Fig 2.
Sensitivity of strain S. aureus MB502 (a cured derivative of strain MB145 which lost plasmid pRJ108) to aureocin A53. Strains A53 and MB502 were used as bacteriocin producers, and strain MB502 was used as the indicator of aureocin A53 production.
Table 3.
Production of aureocin A53 and immunity to aureocin A53 exhibited by strains either carrying mutations in pRJ9 or cured of pRJ9
| Strain | Position of the transposon insertion (nt)a | Production of aureocin A53 (%) relative to that of strain A53b | Smallest amt (BU) of aureocin A53 with inhibitory activity against the strainc |
|---|---|---|---|
| MB139(pRJ38) | orf6 (4094–4095) | 68 | 1,280 |
| MB38(pRJ14) | orf8 (5131–5132) | 2.5 | 1,280 |
| MB143(pRJ40) | orf8 (5302–5303) | 0.9 | 1,280 |
| MB140(pRJ39) | aucE (7720–7721) | 24 | 80 |
| A53(pRJ9) | Wild-type plasmid | 100 | 1,280 |
| MB502 | Cured of pRJ9 | 0 | 40 |
Determined by DNA sequencing. The numbers in parentheses refer to nucleotide (nt) positions between which the transposon is inserted in each mutant.
Production of aureocin A53 after growth in BHI medium at 37°C for 24 h. The bacteriocin secreted by strain A53 (3,140 BU ml−1) was set as 100%. Data are the means of three independent experiments.
The susceptibility of each strain to aureocin A53 was tested in microtiter plate assays. Data are the means of three independent experiments.
In silico analyses of the products encoded by orf12, orf13, and orf14.
The products of orf12, orf13, and orf14 have 331, 226, and 388 amino acids, respectively. The hydrophobicity profiles of these proteins were determined, and Orf12 and Orf14 were found to have apparently one and four transmembrane domains, respectively (data not shown), suggesting their insertion into the cytoplasmic membrane. On the other hand, Orf13, which does not possess transmembrane domains, shares domains with the consensus sequences of the ABC ATPases found in bacteria and involved in the transport of several bacteriocins (data not shown). These ATP-binding domains, which are responsible for ligation and hydrolysis of ATP, possess a conserved primary sequence whose main motifs, Walker A and Walker B (22, 28), are found in Orf13. Other sequences such as the switch region (which contains a histidine loop), motif D (which contains an invariant Asp), and the signature motif (LSGGQ) are also present in Orf13 (data not shown). Since orf12, orf13, and orf14 products possess similarity to three-component ABC transporters found in some bacteriocin gene clusters and involved in immunity, they will be referred to here as aucE, aucF, and aucG, respectively.
The aucE, aucF, and aucG genes of pRJ9 are transcribed as an operon.
Putative ribosome binding sites (RBS) were found upstream of each auc gene although a canonical one (AGGAGG) was not found preceding any gene. aucE and aucG start with a TTG codon, while aucF starts with an ATG codon (data not shown).
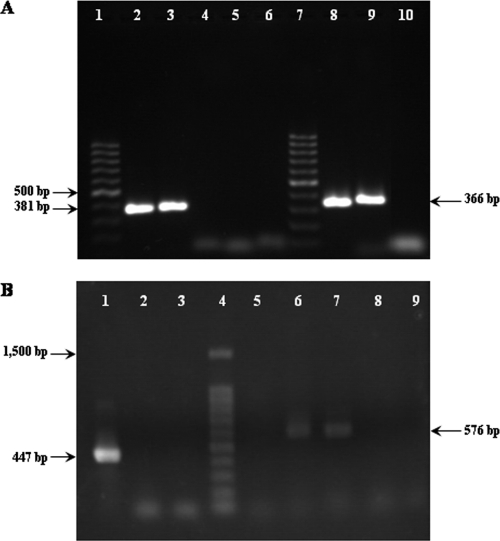
A putative SigA-dependent promoter is located upstream of aucE. A −10 region, with five matches to the consensus sequence (TATAAT), and a −35 region, with four matches to the consensus sequence (TTGACA) (the most conserved bases are shown in boldface), were found 46 bp and 69 bp, respectively, upstream of the start codon TTG, with 17 bp between them. RT-PCR analysis (Fig. 3A) detected amplicons corresponding to cotranscription of either aucE and aucF (lane 3) or aucF and aucG (lane 9), confirming that these genes are coordinately transcribed.
Fig 3.
(A) RT-PCR analysis of the aucEFG genes present on pRJ9. Lanes, 1 and 7, 100-bp DNA ladder (Fermentas Biolabs); lane 2, amplicon from aucE to aucF (381 bp) obtained from pRJ9 as the template; lane 3, amplicon from aucE to aucF obtained from cDNA; lane 4, RNA control; lane 5, blank (no cDNA); lane 6, amplification from aucE to aucF using plasmid pT181 as the template (negative control); lane 8, amplicon from aucF to aucG (366 bp) obtained from pRJ9; lane 9, amplicon from aucE to aucF obtained from cDNA; lane 10, blank (no cDNA). (B) RT-PCR analysis of the orf11-orf10 genes present on pRJ9. Lane 1, internal control (gyrA amplicon; 447 bp) from cDNA obtained from strain A53; lane 2, negative control (gyrA amplification from RNA); lane 3, blank (no cDNA); lane 4, 100-bp DNA ladder (Promega); lane 5, amplification from orf11 to orf10 using plasmid pT181 as the template (negative control); lane 6, amplicon from orf11 to orf10 (576 bp) obtained from pRJ9 as the template; lane 7, amplicon from orf11 to orf10 obtained from cDNA; lane 8, RNA control; lane 9, blank (no cDNA).
Northern blotting experiments detected in strains A53, MB38, and MB143 (the latter two strains carry a transposon insertion into orf8 of pRJ9) a weak hybridization signal, corresponding to an mRNA of ∼2.8 kb (data not shown), a size similar to that expected for a transcript comprising the genes aucEFG. This result confirms that these genes are transcribed as an operon. The ∼2.8-kb signal was not found in strain MB140 because, in this strain, a transposon has inserted into aucE (Table 3), probably disrupting the transcription of the operon. Two additional hybridization signals, corresponding to mRNAs of ∼1.9 and ∼1.3 kb, were also detected in all strains and are consistent with an independent transcription of either aucF and aucG or aucG alone, respectively. These data, together with the analysis of the pRJ9 sequence, suggest the presence of internal promoters in the operon, preceding both aucF and aucG (data not shown).
Cloning of genes involved in immunity to aureocin A53.
Since AucE, AucF, and AucG are part of an ABC transporter which is associated to the membrane, these proteins may be toxic to the host cells under high concentrations. To avoid this problem, cloning experiments were attempted using plasmid pCC1, which contains the F plasmid single-copy origin of replication (ori2) and the inducible high-copy-number oriV. The 5.8-kb BamHI-EcoRI fragment of pRJ9 (Fig. 1) was cloned into plasmid pCC1, resulting in plasmid pRJ81. The toxicity of the insert to E. coli EPI300 cells was then investigated. Plasmid pRJ81 was induced to a high copy number per cell, and the growth of the host strain was analyzed and compared with that of the strains EPI300 and EPI300/pCC1 in a Bioscreen C system. However, no significant difference in growth rates was observed among the three strains tested (data not shown), indicating that the gene products of the 5.8-kb BamHI-EcoRI fragment of pRJ9 were not toxic to E. coli.
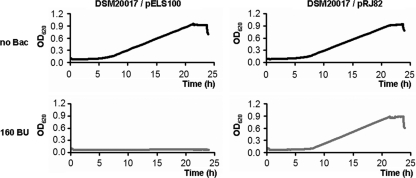
The BamHI-EcoRI insert was subcloned into the shuttle vector pELS100 to generate plasmid pRJ82. The immunity to aureocin A53 exhibited by the lactobacillus transformants carrying pRJ82 was then investigated (Fig. 4). The growth of the control strain L. sakei DSM20017/pELS100 was completely inhibited by 160 BU of aureocin A53. However, strain DSM20017/pRJ82 was not inhibited by any amount of aureocin A53 tested (up to 640 BU), demonstrating full immunity to aureocin A53. Strain DSM20017/pRJ82 was also able to produce aureocin A53 (160 BU ml−1). These results prove that the genetic determinants necessary for expression of immunity to aureocin A53 are located on the 5.8-kb BamHI-EcoRI fragment of pRJ9. The only intact genes found on this fragment are aucA, the bacteriocin structural gene (27), orf10, orf11, aucE, aucF, aucG, and orf1. orf1 is a gene which encodes a protein involved in plasmid mobilization (10a). Therefore, its role in immunity was discarded.
Fig 4.
Growth curves exhibited by lactobacillus strains carrying either pELS100 or pRJ82 (5.8-kb BamHI-EcoRI fragment of pRJ9 cloned into pELS100) in the presence of 160 BU of aureocin A53 (gray curves). The controls (black curves) represent the growth of the strains in the absence of aureocin A53.
Three-component ABC transporters, whose genetic organization is similar to that of the operon aucEFG, have been shown to contribute to bacteriocin immunity by active extrusion of the peptide, reducing the concentration of the bacteriocin in direct contact with the cytoplasmic membrane (4, 23, 35, 41). To investigate if the ABC transporter encoded by aucEFG is involved in immunity to aureocin A53, a deletion of aucG was generated. Plasmid pRJ83 was then constructed by ligation of the HindIII-A fragment of pRJ9 to plasmid pELS100 (Fig. 1). pRJ83 could be transferred to E. coli but not to L. sakei DSM20017, in spite of the many attempts made to transform the latter strain with this recombinant plasmid derived from the same vector used for construction of pRJ82. The only major difference between pRJ83 and pRJ82 is the deletion, in the former plasmid, of orf1 (involved in mobilization) and of about 90% of aucG. As in pRJ82, the aureocin A53 structural gene (aucA) remained intact in pRJ83. These results suggest that AucG, the largest membrane component of the ABC transporter, may play an important role in immunity to aureocin A53, which would explain the lack of cellular viability, in the presence of the bacteriocin, exhibited by the lactobacillus cells harboring this deletion mutant. Similar observations have been described in the literature. According to Rincé et al. (32), who studied genes involved in production of lacticin 481, the immunity to this lantibiotic was lost if any one of the genes lctE, lctF, or lctG was deleted.
To test if aucEFG are the only genes required for immunity to aureocin A53, an amplicon of 3,041 bp, including these three genes and also the putative promoter region, was cloned into the shuttle vector pNO6. The generated plasmid, pRJ85, was transferred to S. aureus RN4220, a strain sensitive to aureocin A53. The resultant transformant showed partial immunity to aureocin A53 (Fig. 5) since it exhibited a much longer lag phase in the presence of aureocin A53 than strain A53, the native host strain of pRJ9. The strain carrying the empty vector (pNO6) was not able to grow in the presence of aureocin A53. Taken together, these results suggest that other gene(s) present on pRJ9 may be required for full expression of immunity to aureocin A53.
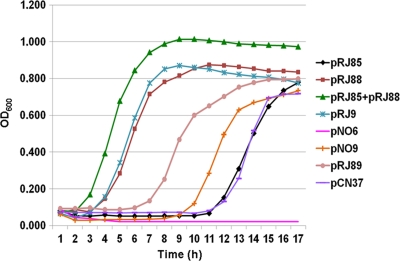
Fig 5.
Growth curves of staphylococcal strains carrying different plasmids in the presence of aureocin A53 (2,048 BU). With the exception of pRJ9, whose host strain is A53, the remaining plasmids are in the genetic background of strain RN4220. pNO6 (pCC1 plus pIL252), pNO9 (pCC1 plus pT181), and pCN37 are the vectors used, and their host strains were employed as controls. pRJ85 represents aucEFG cloned into pNO6; pRJ88 represents aucIB-aucIA cloned into pNO9, and pRJ89 represents aucIB-aucIA cloned into pCN37. The curves were generated with the results representing the means of three independent experiments.
In the insert present on plasmid pRJ82, which conferred full immunity to L. sakei, the products of orf11 and orf10 are the only other candidates to be involved in immunity. orf11 and orf10, whose functions were unknown, are encoded in the opposite strand and in the same reading frame, with no overlapping and no intergenic region between them. RBS could also be found upstream of each orf although a canonical RBS was not found preceding any gene (data not shown). orf11 starts with a TTG codon, while orf10 starts with an ATG codon. A putative SigA-dependent promoter is located upstream of orf11. A −10 region, with five matches to the consensus sequence, and a −35 region, with four matches to the consensus sequence, were found 35 bp and 58 bp, respectively, upstream of the start codon TTG of orf11 (data not shown), with 17 bp between them. RT-PCR analysis (Fig. 3B) detected an amplicon corresponding to cotranscription of both orf11 and orf10 (lane 7), confirming that these genes are coordinately transcribed.
Orf11 and Orf10 share 80% and 61% similarity, respectively, with hypothetical proteins described in Lactococcus lactis that are not found in the gene clusters of other bacteriocins and whose functions are presently unknown. Orf10 shares common characteristics with proteins involved in bacteriocin immunity, especially with those from class II bacteriocins, since it possesses three transmembrane domains (data not shown), high pI (10.1), and 96 amino acids. On the other hand, Orf11 possesses 94 amino acids but does not present any transmembrane domain. Although anionic (pI of 4.9), Orf11 cannot be excluded as part of the immunity system since different mechanisms may operate for different bacteriocins and since aureocin A53 has features distinct from most class II bacteriocins (27). For this reason, a 735-bp amplicon containing both orf11 and orf10, including the putative promoter, was cloned into pCN37, and the recombinant plasmid (pRJ89) (Fig. 1) was also transferred to S. aureus RN4220. However, again, only a partial immunity to aureocin A53 was exhibited by the transformants carrying pRJ89 (Fig. 5). The transformants exhibited a longer lag phase in the presence of aureocin A53 than strain A53 although their lag phases were shorter than the lag phase exhibited by the same host strain carrying the empty vector (pCN37).
To test if the presence of both operons aucEFG and orf11-orf10 would result in a full-immunity phenotype, plasmids pRJ85 and pRJ88 (carrying orf11-orf10 on pNO9) were sequentially introduced into strain RN4220 by transformations. The resultant strain harboring both plasmids exhibited immunity to aureocin A53 at an even higher level than that observed for strain A53, growing faster and reaching a higher cellular density in the presence of the bacteriocin (Fig. 5).
From the data presented in Fig. 5, it can also be observed that plasmid pRJ88 (carrying orf11-orf10 on a pT181 derivative) confers a higher immunity to staphylococcal cells than plasmid pRJ85 (carrying aucEFG on a pIL252 derivative). The immunity conferred by pRJ88 was similar to that conferred by pRJ9, the native staphylococcal plasmid encoding aureocin A53. However, such observations may result from a gene dosage effect due to a much higher copy number of pT181mcs (about 100 copies/cell), a staphylococcal plasmid, than that of pIL252, a LAB plasmid, which appears to be maintained at very low copy number in a staphylococcal background (data not shown). This hypothesis was confirmed by the experiments performed with pRJ89, which carries the operon orf11-orf10 cloned into pCN37, a staphylococcal shuttle plasmid whose copy number (15 to 20 copies/cell) is lower than that of pT181mcs. As shown in Fig. 5, the staphylococcal transformant carrying pRJ89 grew more slowly than the transformant carrying pRJ88 but still faster than the transformant harboring pRJ85. However, here again, these results may be explained by the high copy number of pCN37 in relation to that of pIL252 (data not shown).
To test if cloning of aucEFG into either pT181mcs or pCN37 would result in increased immunity to aureocin A53, attempts were made to construct pT181mcs or pCN37 derivatives carrying this operon. However, recombinant plasmids could never be recovered in four independent experiments performed with each vector. Therefore, these data suggest that under high concentrations the products encoded by these genes may be toxic to the staphylococcal host cells.
Taken together, these data suggest that the operon orf11-orf10 is indeed required for full expression of immunity to aureocin A53, complementing the role played by aucEFG. Therefore, from here, orf11 and orf10 will be referred to as aucIB and aucIA, respectively.
The presence of more than one function involved in immunity to a given bacteriocin has already been described in other systems, such as in some lantibiotics and in cyclic bacteriocins (6, 24, 40). Lantibiotic-producing strains possess either both or just one of the self-protection systems LanFEG and LanI/LanH. LanFEG is observed for mersacidin, epidermin, lacticin 481, and mutacin II, while LanI can be found in the immunity system for epicidin 280, lactocin S, and Pep5 (6). In relation to subtilin and nisin, it has been observed that the expression of either self-protection system confers partial immunity to these lantibiotics; however, the expression of both systems confers full immunity to them (37, 38).
In the case of nukacin ISK-1, a lantibiotic produced by Staphylococcus warneri ISK-1, the self-protection system against this bacteriocin is conferred by NukFEG and NukH (31). Like NukH, AucIA is a membrane protein with three transmembrane domains. However, in contrast to NukH and according to the TMpred program, AucIA could be inserted into the bacterial membrane with its N terminus oriented in the outer interface part of the cell membrane.
The immunity gene of most linear class II bacteriocins is located immediately downstream of, and in the same operon as, the bacteriocin structural gene (21, 29). Therefore, the location of the operon aucIB-aucIA downstream of, but on the opposite strand of, the aureocin A53 structural gene, aucA, is very unusual. However, there are other examples of atypical locations, such as the immunity genes of bacteriocins LsbA and LsbB, enterocin Q, and carnobacteriocin A, which are located next to the structural genes but have the opposite orientations (17, 18, 19).
Analyses of pRJ9 mutants generated upon transposon insertion.
The exact position of the transposon insertion in each strain carrying a pRJ9 mutation was determined by sequencing and is presented in Table 3. Mutants MB139 (pRJ38) and MB140 (pRJ39) have a transposon inserted into orf6 and aucE, respectively. Two other mutant strains, MB38 (pRJ14) and MB143 (pRJ40), have a transposon inserted into orf8.
All mutants showed a reduction in the amount of bacteriocin found in the culture supernatant (Table 3). The mutant affected in orf6 exhibited a 32% reduction in the amount of aureocin A53 secreted. Orf6, a 61-amino-acid polypeptide, shares 100% identity to a hypothetical protein encoded by other S. aureus plasmids not encoding bacteriocin production. Therefore, Orf6 should not be directly involved in aureocin A53 production. Mutants affected in orf8 exhibited the most drastic reduction, which corresponded to less than 3% of the aureocin A53 produced by the wild-type strain, suggesting that orf8 is important for the maintenance of the normal levels of aureocin A53 secretion. Orf8, which seems to form an operon with orf7, shows about 50% similarity to a number of hypothetical membrane proteins found in many Gram-positive bacteria. However, additional studies are needed to investigate its role in aureocin A53 externalization.
Strain MB140, whose transposon is inserted into aucE, also exhibited a low level of bacteriocin secreted into the medium (a 76% reduction), suggesting that AucE also plays a role in the maintenance of the normal levels of aureocin A53 secretion. Since AucE is part of the unique ABC transporter encoded by the aureocin A53 gene cluster, this dedicated ABC transporter may also be involved in bacteriocin externalization, as found in other bacteriocin systems (6, 24, 40).
The mutant affected in aucE (pRJ39) exhibited a 16-fold increase in sensitivity to aureocin A53 compared to strain A53 (Table 3). The remaining mutants did not show differences in the immunity levels to exogenous aureocin A53 compared to the wild-type strain A53. Such results suggest that neither orf6 nor orf8 plays a role in the immunity phenotype and clearly implicate AucE in this phenotype, confirming the data presented above.
In conclusion, aureocin A53 is the first class II linear staphylococcin to have its immunity genes investigated. Our data show that this bacteriocin, differently from most class II bacteriocins and similarly to various lantibiotics and cyclic bacteriocins, presents more than one system involved in the self-protection of its producer strain. One such system involves a dedicated ABC transporter which probably acts by active extrusion of the peptide. The second system involves an operon which encodes a hydrophobic cationic polypeptide, AucIA, which shares characteristics with immunity proteins generally associated to the membrane and found in the gene clusters of many class II bacteriocins, and an anionic protein, AucIB, never described before as part of a bacteriocin immunity system. Moreover, we found evidence of the major role played by a putative membrane protein, Orf8, in aureocin A53 externalization. The individual role played by AucIA and AucIB in immunity to aureocin A53 and the function of Orf8 in aureocin production and/or secretion are under investigation.
ACKNOWLEDGMENTS
J.D.S.N., H.C., M.L.V.C., and L.R.F. were recipients of scholarships from CNPq/Brazil. This study was supported by grants from CNPq, FAPERJ, CAPES, and PRONEX to M.D.C.D.F.B.
Footnotes
Published ahead of print 9 December 2011
REFERENCES
- 1. Araújo SA, Bastos MCF. 1995. Incompatibility and molecular relationships between small bacteriocinogenic plasmids of Staphylococcus aureus. World J. Microbiol. Biotechnol. 11:525–528 [DOI] [PubMed] [Google Scholar]
- 2. Augustin J R, et al. 1992. Genetic analysis of epidermin biosynthetic genes and epidermin-negative mutants of Staphylococcus epidermidis. Eur. J. Biochem. 204:1149–1154 [DOI] [PubMed] [Google Scholar]
- 3. Aukrust TW, Brurberg MB, Nes IF. 1995. Transformation of Lactobacillus by electroporation. Methods Mol. Biol. 47:201–208 [DOI] [PubMed] [Google Scholar]
- 4. Bastos MCF, Ceotto H, Coelho MLV, Nascimento JS. 2009. Staphylococcal antimicrobial peptides: relevant properties and potential biotechnological applications. Curr. Pharm. Biotechnol. 10:38–61 [DOI] [PubMed] [Google Scholar]
- 5. Bastos MCF, Coutinho BG, Coelho MLV. 2010. Lysostaphin: a staphylococcal bacteriolysin with potential clinical applications. Pharmaceuticals 3:1139–1161 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Bierbaum G, Sahl H-G. 2009. Lantibiotics: mode of action, biosynthesis and bioengineering. Curr. Pharm. Biotechnol. 10:2–18 [DOI] [PubMed] [Google Scholar]
- 7. Ceotto H, et al. 2010. Aureocins 4185, bacteriocins produced by Staphylococcus aureus 4185: potential application in food preservation. Foodborne Pathog. Dis. 7:1255–1262 [DOI] [PubMed] [Google Scholar]
- 8. Charpentier E, et al. 2004. Novel cassette-based shuttle vector system for Gram-positive bacteria. Appl. Environ. Microbiol. 70:6076–6085 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Coelho MLV, et al. 2007. Activity of staphylococcal bacteriocins against Staphylococcus aureus and Streptococcus agalactiae involved in bovine mastitis. Res. Microbiol. 158:625–630 [DOI] [PubMed] [Google Scholar]
- 10. Coelho MLV, Ceotto H, Madureira DJ, Nes IF, Bastos MCF. 2009. Mobilization functions of the bacteriocinogenic plasmid pRJ6 of Staphylococcus aureus. J. Microbiol. 47:327–336 [DOI] [PubMed] [Google Scholar]
- 10a. Coutinho BG, Coelho MLV, Ceotto H, Bastos MCF. Revealing the latent mobilization capability of the staphylococcal bacteriocinogenic plasmid pRJ9. J. Mol. Microbiol. Biotechnol., in press [DOI] [PubMed] [Google Scholar]
- 11. De Oliveira SS, et al. 1998. Genetic analysis of the bacteriocin-encoding plasmids pRJ6 and pRJ9 of Staphylococcus aureus by transposon mutagenesis and cloning of genes involved in bacteriocin production. J. Appl. Microbiol. 85:972–984 [DOI] [PubMed] [Google Scholar]
- 12. De Oliveira SS, Abrantes J, Cardoso M, Sordelli D, Bastos MCF. 1998. Staphylococcal strains involved in bovine mastitis are inhibited by Staphylococcus aureus antimicrobial peptides. Lett. Appl. Microbiol. 27:287–291 [DOI] [PubMed] [Google Scholar]
- 13. Diep DB, Skaugen M, Salehian Z, Holo H, Nes IF. 2007. Common mechanisms of target cell recognition and immunity for class II bacteriocins. Proc. Natl. Acad. Sci. U. S. A. 104:2384–2389 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Eijsink VGH, Skeie M, Middelhoven PH, Brurberg MB, Nes IF. 1998. Comparative studies of class IIa bacteriocins of lactic acid bacteria. Appl. Environ. Microbiol. 64:3275–3281 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Engelke G, et al. 1994. Regulation of nisin biosynthesis and immunity in Lactococcus lactis 6F3. Appl. Environ. Microbiol. 60:814–825 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Ennahar S, Sashihara T, Sonomoto K, Ishizaki A. 2000. Class IIa bacteriocins: biosynthesis, structure and activity. FEMS Microbiol. Rev. 24:85–106 [DOI] [PubMed] [Google Scholar]
- 17. Franz CM, et al. 1999. Atypical genetic locus associated with constitutive production of enterocin B by Enterococcus faecium BFE 900. Appl. Environ. Microbiol. 65:2170–2178 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Franz CM, van Belkum MJ, Worobo RW, Vederas JC, Stiles ME. 2000. Characterization of the genetic locus responsible for production and immunity of carnobacteriocin A: the immunity gene confers cross-protection to enterocin B. Microbiology 146:621–631 [DOI] [PubMed] [Google Scholar]
- 19. Gajic O, et al. 2003. Novel mechanism of bacteriocin secretion and immunity carried out by lactococcal multidrug resistance proteins. J. Biol. Chem. 278:34291–34298 [DOI] [PubMed] [Google Scholar]
- 20. Giambiagi-Marval M, Mafra MA, Penido EGC, Bastos MCF. 1990. Distinct groups of plasmids correlated with bacteriocin production in Staphylococcus aureus. J. Gen. Microbiol. 136:1591–1599 [DOI] [PubMed] [Google Scholar]
- 21. Heng NCK, Wescombe PA, Burton JP, Jack RW, Tagg JR. 2007. The diversity of bacteriocins in Gram positive bacteria, p 45–92In M. Riley MA, Chavan MA. (ed), Bacteriocins: ecology and evolution. Springer, Berlin, Germany [Google Scholar]
- 22. Jones PM, George AM. 2002. Mechanism of ABC transporters: A molecular dynamics simulation of a well characterized nucleotide-binding subunit. Proc. Natl. Acad. Sci. U. S. A. 99:12639–12644 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Klein C, Entian K-D. 1994. Genes involved in self-protection against the lantibiotic subtilin produced by Bacillus subtilis ATCC 6633. Appl. Environ. Microbiol. 60:2793–2801 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Maqueda M, et al. 2008. Genetic features of circular bacteriocins produced by Gram-positive bacteria. FEMS Microbiol. Rev. 32:2–22 [DOI] [PubMed] [Google Scholar]
- 25. Nascimento JS, et al. 2006. Bacteriocins as alternative agents for control of multiresistant staphylococcal strains. Lett. Appl. Microbiol. 42:215–221 [DOI] [PubMed] [Google Scholar]
- 26. Nes IF, et al. 1996. Biosynthesis of bacteriocins in lactic acid bacteria. Antonie van Leeuwenhoek 70:113–128 [DOI] [PubMed] [Google Scholar]
- 27. Netz DJ, et al. 2002. Biochemical characterisation and genetic analysis of aureocin A53, a new, atypical bacteriocin from Staphylococcus aureus. J. Mol. Biol. 319:745–756 [DOI] [PubMed] [Google Scholar]
- 28. Nikaido H. 2002. How are the ABC transporters energized? Proc. Natl. Acad. Sci. U. S. A. 99:9609–9610 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Nissen-Meyer J, Rogne P, Oppegard C, Haugen HS, Kristiansen PE. 2009. Structure-function relationships of the non-lanthionine-containing peptide (class II) bacteriocins produced by Gram-positive bacteria. Curr. Pharm. Biotechnol. 10:19–37 [DOI] [PubMed] [Google Scholar]
- 30. Novick RP. 1967. Properties of a cryptic high-frequency transducing phage in Staphylococcus aureus. Virology 33:155–166 [DOI] [PubMed] [Google Scholar]
- 31. Okuda K, Aso Y, Nakayama J, Sonomoto K. 2008. Cooperative transport between NukFEG and NukH in immunity against the lantibiotic nukacin ISK-1 produced by Staphylococcus warneri ISK-1. J. Bacteriol. 190:356–362 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Rincé A, Dufour A, Uguen P, Le Pennec JP, Haras D. 1997. Characterization of the lacticin 481 operon: the Lactococcus lactis genes lctF, lctE, and lctG encode a putative ABC transporter involved in bacteriocin immunity. Appl. Environ. Microbiol. 63:4252–4260 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Sambrook J, Fritsch EF, Maniatis T. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY [Google Scholar]
- 34. Schenk S, Laddaga RA. 1992. Improved method for electroporation of Staphylococcus aureus. FEMS Microbiol. Lett. 73:133–138 [DOI] [PubMed] [Google Scholar]
- 35. Siegers K, Entian K-DK. 1995. Genes involved in immunity to the lantibiotic nisin produced by Lactococcus lactis 6F3. Appl. Environ. Microbiol. 61:1082–1089 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Simon D, Chopin A. 1988. Construction of a vector plasmid family and its use for molecular cloning in Streptococcus lactis. Biochimie 70:559–566 [DOI] [PubMed] [Google Scholar]
- 37. Stein T, Heinzmann S, Solovieva I, Entian K-D. 2003. Function of Lactococcus lactis nisin immunity genes nisI and nisFEG after coordinated expression in the surrogate host Bacillus subtilis. J. Biol. Chem. 278:89–94 [DOI] [PubMed] [Google Scholar]
- 38. Stein T, Heinzmann S, Dusterhus S, Borchert S, Entian K-D. 2005. Expression and functional analysis of the subtilin immunity genes spaIFEG in the subtilin-sensitive host Bacillus subtilis MO1099. J. Bacteriol. 187:822–828 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Takala TM, Saris PE. 2006. C terminus of NisI provides specificity to nisin. Microbiology 152:3543–3549 [DOI] [PubMed] [Google Scholar]
- 40. van Belkum MJ, Martin-Visscher LA, Vederas JC. 2011. Structure and genetics of circular bacteriocins. Trends Microbiol. 19:411–418 [DOI] [PubMed] [Google Scholar]
- 41. Yarmus M, Mett A, Shapira R. 2000. Cloning and expression of the genes involved in the production of and immunity against the bacteriocin lacticin RM. Biochim. Biophys. Acta 1490:279–290 [DOI] [PubMed] [Google Scholar]