Abstract
Cyanobacteria, including members of the genus Prochlorococcus, contain icosahedral protein microcompartments known as carboxysomes that encapsulate multiple copies of the CO2-fixing enzyme ribulose 1,5-bisphosphate carboxylase/oxygenase (RubisCO) in a thin protein shell that enhances the catalytic performance of the enzyme in part through the action of a shell-associated carbonic anhydrase. However, the exact mechanism by which compartmentation provides a catalytic advantage to the enzyme is not known. Complicating the study of cyanobacterial carboxysomes has been the inability to obtain homogeneous carboxysome preparations. This study describes the first successful purification and characterization of carboxysomes from the marine cyanobacterium Prochlorococcus marinus MED4. Because the isolated P. marinus MED4 carboxysomes were free from contaminating membrane proteins, their protein complement could be assessed. In addition to the expected shell proteins, the CsoS1D protein that is not encoded by the canonical cso gene clusters of α-cyanobacteria was found to be a low-abundance shell component. This finding and supporting comparative genomic evidence have important implications for carboxysome composition, structure, and function. Our study indicates that carboxysome composition is probably more complex than was previously assumed based on the gene complements of the classical cso gene clusters.
INTRODUCTION
Cyanobacteria of the genus Prochlorococcus dominate the world's temperate oceans. Considered to be the most abundant photosynthetic organisms on the planet, Prochlorococcus species are estimated to contribute up to half of the marine biological carbon sequestration and, therefore, are important players in the global carbon cycle (27). Remarkably, members of these tiny phytoplankton species are distributed throughout the water column from the ocean surface to a depth of 200 m and as a genus are able to utilize a broad range of light intensities. High-light-adapted ecotypes like Prochlorococcus marinus MED4 are surface dwellers; low-light-adapted ecotypes like P. marinus MIT9313 are found at greater depths (27, 32). Because of the importance of Prochlorococcus species in the global carbon cycle, the genomes of many different strains have been sequenced.
Prochlorococcus and marine representatives of the genus Synechococcus are classified as α-cyanobacteria based on the arrangement and types of genes that encode their CO2 fixation module, the carboxysome (1, 6). The carboxysome is a protein microcompartment that houses the CO2-fixing enzyme ribulose 1,5-bisphosphate carboxylase/oxygenase (RubisCO) and constitutes the final step of a CO2-concentrating mechanism (CCM) that allows the autotrophic bacteria to grow efficiently at ambient CO2 levels. The first step of the CCM is the active uptake of inorganic carbon into the cell, followed by intracellular accumulation primarily as HCO3−. The bicarbonate is efficiently fixed onto ribulose 1,5-bisphosphate (RubP) and converted to two molecules of 3-phosphoglycerate within the carboxysome by RubisCO, aided by the rapid equilibration of HCO3− with the RubisCO substrate CO2 by the shell-associated carbonic anhydrase CsoSCA (20).
Despite the importance of cyanobacteria in the global carbon cycle, their carboxysomes have not been well characterized; technical difficulties related to cell breakage and contamination with the abundant photosynthetic membranes have prevented purification of the organelle. Intact carboxysomes, all belonging to the α-type, have only been isolated from a few chemoautotrophs. Among these, the sulfur oxidizer Halothiobacillus neapolitanus has emerged as the model organism for direct genetic and biochemical studies because of the ease with which highly purified carboxysomes can be obtained from this species. The polypeptide composition of the H. neapolitanus carboxysome, which also belongs to the α-type, has been assessed quantitatively and qualitatively. The structure and function of most of its protein constituents, as well as the genes encoding the microcompartment, are known (reviewed in reference 44). In contrast, only three individual Prochlorococcus carboxysome proteins have been characterized to any extent: CsoSCA (39), RubisCO (35), and the putative shell protein CsoS1D that is encoded by a gene outside the canonical cso gene cluster (22). We reasoned that the small P. marinus MED4 cells should lend themselves to carboxysome purification because they contain few thylakoid layers and are surrounded by only a thin cell wall (41). Here, we report the first homogeneous preparation of a cyanobacterial carboxysome, its characterization, and evidence for the association of CsoS1D with the carboxysome shell.
MATERIALS AND METHODS
Growth of bacteria.
An axenic culture of Prochlorococcus marinus subsp. pastoris strain CCMP1986 (formerly Prochlorococcus marinus MED4; Provasoli-Guillard National Center for Culture of Marine Phytoplankton) was maintained in 25-ml tissue culture flasks (Falcon) at ambient temperature and a light intensity of 30 μmol · m−2 · s−1. Growth was monitored by measuring fluorescence at 680 nm using a Nanodrop 3300 fluorospectrometer (Thermo). Large-scale cultures for carboxysome isolation were grown in acid-washed 20-liter polycarbonate carboys (Nalgene) until fluorescent measurements peaked, usually between 200 and 250 relative fluorescence units. Small-scale cultures were maintained in Pro99 medium, using Sargasso Seawater as a base (Provasoli-Guillard CCMP); 20-liter cultures were grown in AMP1, using Turk's Island salt mix as a base (24).
Carboxysome isolation.
A 20-liter P. marinus MED4 culture was concentrated to approximately 500 ml by filtration through a Pellicon 2 cassette 0.22-μm GVPP-C unit (Millipore). The cells were pelleted by centrifugation at 10,000 × g for 10 min (JA-25.50 rotor, 4°C) and resuspended in 15 ml cold BEMB buffer (10 mM bicine, 1 mM EDTA, 10 mM MgCl2 · 6H2O, 20 mM NaHCO3, pH 8.0). To prevent proteolysis, the protease inhibitor phenylmethylsulfonyl fluoride was added to a final concentration of 0.6 mM. The cells were sonicated five times with 5-s cycles (Branson Sonifier 450 with microtip, output control set at 6). To reduce the viscosity of the lysate, DNase (100 U) was added and the sample rocked gently at room temperature for 30 min before being subjected to a 10-min clearing spin at 10,000 × g. To the supernatant, 10% Nonidet-P40 (USBioLogic) in BEMB buffer was added to a final concentration of 1%, and the solution was stirred for 30 min before centrifugation at 50,000 × g for 30 min. The resulting opaque white pellet was resuspended in 200 to 400 μl of BEMB buffer and loaded onto a sucrose step gradient (1.1 ml each of 60%, 40%, 20%, and 5% sucrose in BEMB buffer). The gradient was centrifuged at 70,000 × g for 35 min in an L7-65 ultracentrifuge (Beckman). The opaque carboxysome band was harvested with a syringe and diluted with 20 ml BEMB prior to pelleting the carboxysomes by centrifugation at 125,000 × g for 2 h. The final carboxysome pellet was resuspended in 50 to 100 μl BEMB buffer.
TEM.
Photographs of purified P. marinus MED4 carboxysomes were obtained on a Zeiss EM-900 transmission electron microscope (TEM) as described previously (11). An Epson Perfection V700 photo scanner was used to obtain the final images.
Protein analyses.
The protein concentrations of P. marinus MED4 carboxysome samples were determined with the bicinchoninic acid (BCA) assay (Pierce) with bovine serum albumin as the standard. P. marinus MED4 carboxysome peptides were separated by SDS-PAGE in precast 10-to-20% Tris-Tricine Criterion gels (Bio-Rad) and visualized by staining with GelCode Blue (Pierce). Peptides were transferred onto a 0.45-μm nitrocellulose membrane (Bio-Rad) by electroblotting for 2 h at 250 mA. Blots were probed with rabbit polyclonal antibodies raised against various recombinant H. neapolitanus proteins; anti-P. marinus MED4 CsoS1D antibody was provided by Cheryl Kerfeld. Goat anti-rabbit IgG-horseradish peroxidase (HRP) secondary antibody and SuperSignal West Pico chemiluminescent substrate (Thermo) were used to visualize reactive bands. Images were captured and processed in a VersaDoc imaging system (Bio-Rad).
The percent composition of individual carboxysome components was calculated by measuring their relative intensity in stained SDS-PAGE bands, as determined by Quantity One software. Tris-Tricine SDS-PAGE is biased toward low-molecular-weight peptides; therefore, peptide relative intensities were determined from a 10-to-20% gradient Tris-HCl polyacrylamide gel. The ratio of CbbS to CsoS1 observed in Tris-Tricine SDS-PAGE was used to estimate the contribution of each peptide to the single band observed in standard SDS-PAGE. The number of RubisCO molecules per carboxysome was estimated by calculating the volume of an icosahedron with a 90-nm diameter to represent the P. marinus MED4 carboxysome (41) and dividing by the average volume of form IA RubisCO molecules (37). The observed relative intensity of RubisCO (sum of CbbL and CbbS) was divided by the theoretical molecular weight of a P. marinus MED4 RubisCO L8S8 octamer. This ratio was used to calculate moles/mole of RubisCO holoenzyme for the remaining carboxysome components. These numbers were multiplied by the number of estimated RubisCO molecules to determine the copy number of each component.
For copurification experiments, a 100-μl aliquot of purified carboxysomes (650 μg protein) was loaded onto a 600-μl sucrose step gradient in an 8- by 34-mm polycarbonate centrifuge tube (Beckman). The gradient was centrifuged in an MLS-50 rotor (Beckman) at 70,000 × g for 35 min in a Beckman Coulter Optima MAX ultracentrifuge. The gradient was fractionated into 15 aliquots of equal volume using a Brandel fractionator. Undialyzed aliquots were analyzed by SDS-PAGE and immunoblotting as described above.
Carboxysome shells were disrupted by incubation for 3 h at 4°C in a high-salt buffer (100 mM Tris, 10 mM EDTA, 0.5 M NaCl, pH 9.0) that has been shown to disrupt the protein structure of the turnip crinkle virus capsid (12). Incubated samples were centrifuged at 20,000 × g for 30 min at 4°C to generate a shell-enriched pellet fraction and a free-protein-enriched supernatant. The protein content in these fractions was analyzed as described above.
Carboxysome functional assays.
Carbonic anhydrase activity was measured with the stopped-flow changing-indicator method (21). The buffer-indicator mixture used was TAPS [N-tris(hydroxymethyl)methyl-3-aminopropanesulfonic acid]–m-cresol purple (A578) at pH 8.5.
RubisCO kinetic assays were performed as described previously (11) with the following modifications: P. marinus MED4 carboxysomes were not desalted prior to analysis because of the low sample volumes. Consequently, enzyme activation prior to starting the assay was not needed. The appropriate amount of NaHCO3 was added to yield a concentration in the range of 3.4 to 96.4 mM. The concentration of CO2 was calculated using the Henderson-Hasselbalch equation (14). In parallel experiments, the concentration of NaHCO3 was maintained at 60 mM and ribulose 1,5-bisphosphate (RubP) was added to yield a concentration in the range of 50 to 1,600 mM.
Phylogenetic analysis.
Twenty-nine α-cyanobacterial genomes were included in the phylogenetic analysis. Because the genome of Synechococcus sp. strain CB0101 is incomplete and lacks the 16S rRNA sequence, translation initiation factor 2 (IF-2) was used as the protein phylogenetic marker instead (43). CsoS1D was used as the α-carboxysome marker. Protein sequences of IF-2 or CsoS1D were downloaded from IMG-ER (http://img.jgi.doe.gov), aligned with T-Coffee (www.ebi.ac.uk/Tools/msa/tcoffee) (26), trimmed, and then submitted to PhyML (http://www.phylogeny.fr) (10). Maximum-likelihood trees were built with 100 bootstraps and then visualized and edited in Dendroscope (17).
RESULTS
P. marinus MED4 carboxysome purification and composition.
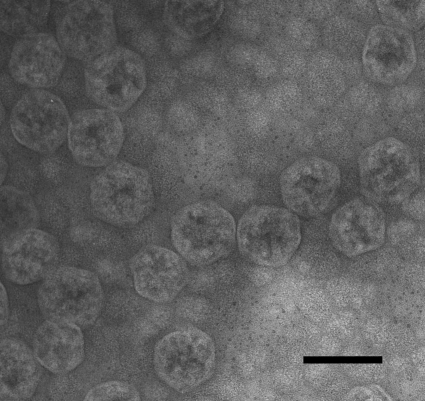
Initial, small-scale carboxysome isolation trials with batch cultures of P. marinus MED4 yielded promising results that prompted the standardization of the growth conditions in 20-liter culture vessels and the development of a robust, large-scale carboxysome purification procedure (detailed in Materials and Methods). Cells harvested when the fluorescence of the culture had reached its peak value yielded pellets of 0.25 to 0.5 g wet weight 20 liter−1, a sufficient amount of starting material for a standard carboxysome purification run. As predicted, the P. marinus MED4 cells lysed readily. A key step proved to be the incubation of the cell lysate with Nonidet P-40 to dissolve the thylakoids. As seen in Fig. 1, the sucrose gradient-purified final carboxysome fraction was free of membrane vesicles and contained apparently intact carboxysomes. The transmission electron micrographs of negatively stained specimens contained many P. marinus MED4 carboxysomes of the expected diameter of approximately 90 nm (41). However, like their counterparts in chemoautotrophs (18), the P. marinus MED4 organelles displayed some size heterogeneity; particles with diameters that range from 70 to 100 nm were also seen in the purified fraction. The purification procedure detailed in Materials and Methods routinely yielded 300 to 400 μg of carboxysome protein per 20 liters of culture.
Fig 1.
Negatively stained TEM image of isolated P. marinus MED4 carboxysomes. The scale bar represents 100 nm.
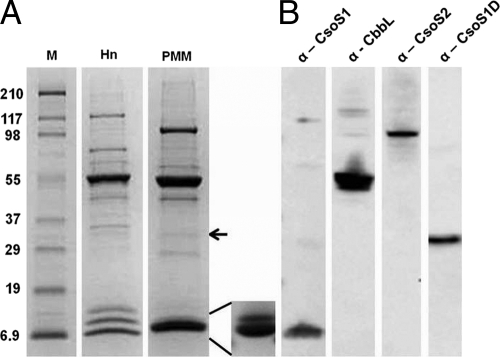
Compared to the well-established polypeptide composition of H. neapolitanus carboxysomes (8), standard SDS-PAGE yielded a much simpler polypeptide pattern for P. marinus MED4 carboxysomes (Fig. 2A). The most abundant band corresponded to the large subunit (CbbL) of RubisCO (Fig. 2B) at an apparent molecular mass of 53.5 kDa. Only one polypeptide of approximately 101 kDa corresponded to the CsoS2 protein (Fig. 2B), whereas in H. neapolitanus carboxysomes, this protein is represented by two polypeptides (CsoS2A and CsoS2B). The faint band above the CbbL band in Fig. 2A is probably the carbonic anhydrase CsoSCA, although its identity was not assessed by immunoblotting because the antibody that recognizes the H. neapolitanus ortholog did not cross-react with the P. marinus MED4 ortholog. Furthermore, the low abundance of this polypeptide prevented identification by mass spectrometry. The single CsoS1 protein and the small subunit of P. marinus MED4 RubisCO (CbbS) migrated as one band of 10.7 kDa but could be separated into separate CbbS and CsoS1 bands (insert in Fig. 2A) by electrophoresis in a 10-to-20% gradient gel Tris-Tricine system. In contrast, the H. neapolitanus CsoS1A and CsoS1C polypeptides migrate as one band together with CsoS4A and CsoS4B, while CbbS and CsoS1B form separate, more slowly migrating bands (Fig. 2A) (8). The P. marinus MED4 CsoS1 band probably also contains the two CsoS4 paralogs that are encoded by the P. marinus MED4 cso gene cluster (22); like CsoSCA, their identity could not be verified because they did not cross-react with the antibody raised against the H. neapolitanus orthologs and their abundance is very low (4). The intensity of the stained bands was used to estimate mass percentages and copy numbers for the P. marinus MED4 carboxysome proteins (Table 1). Assuming that the polypeptides listed in Table 1 include all major protein constituents, the mass of the P. marinus MED4 carboxysome was estimated to be approximately 131 MDa. This value is two-thirds of the H. neapolitanus carboxysome mass calculated in the same manner (15).
Fig 2.
Polypeptide composition of isolated P. marinus MED4 carboxysomes. (A) Stained SDS-polyacrylamide gel of isolated H. neapolitanus (Hn) and P. marinus MED4 (PMM) carboxysomes. Inset: P. marinus MED4 CbbS (upper band) and CsoS1 (lower band) separated in the Tris-Tricine gel system. The arrow marks the CsoS1D polypeptide. M, molecular mass ladder (kDa). (B) Immunoblots of P. marinus MED4 carboxysomes developed with antibodies against P. marinus MED4 CsoS1D, anti-Hn CsoS1, anti-Hn CsoS2, and a commercially available anti-form I RubisCO antibody, respectively.
Table 1.
Protein composition of the PMM carboxysome
| Identity | Molecular mass (kDa) |
% of composition | Copy no. |
||
|---|---|---|---|---|---|
| Theoretical | Observed | Monomer | Oligomer | ||
| CsoS2 | 81.9 | 101 | 12.0 | 163 | |
| CsoSCA | 57.3 | 64 | 2.7 | 58 | 29 |
| CbbL | 52.6 | 53.5 | 44.3 | 1,216 | 152 |
| CsoS1D | 27.5 | 33 | 0.9 | 38 | 12 (6)a |
| CbbS | 12.9 | 12.9 | 13.6 | 1,216 | 152 |
| CsoS1 | 10.4 | 10.7 | 25.4 | 3,232 | 538 |
The number in parentheses indicates the copy number for dimers of stacked trimers (22).
Purified P. marinus MED4 carboxysomes are functional.
The presence of the unique β-carbonic anhydrase CsoSCA is characteristic of α-carboxysomes, and the enzymatic activity of recombinant P. marinus MED4 carboxysomal carbonic anhydrase was established previously (39). Using the stopped-flow changing-indicator assay (21), carbonic anhydrase activity was detected in sucrose gradient-purified P. marinus MED4 carboxysomes (Fig. 3). The turnover number (kcat) of the enzyme for the hydration of CO2 in intact P. marinus MED4 carboxysomes was 0.87 × 104 ± 0.5 × 104 s−1 at pH 8.5. This value is approximately half of the lowest value reported (1.7 × 104 s−1) for other recombinant β-carbonic anhydrases (16, 38) and for the carbonic anhydrase activity of H. neapolitanus carboxysomes (1.8 × 104 s−1) (16).
Fig 3.
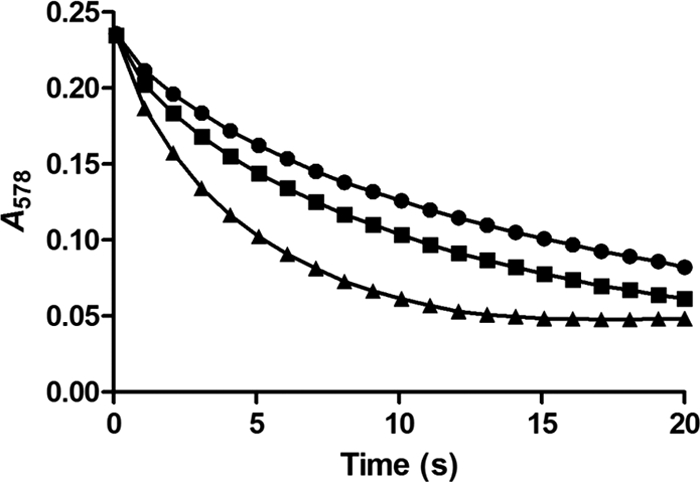
Carbonic anhydrase activity of P. marinus MED4 carboxysomes. The stopped-flow changing-pH-indicator assay (21) was used to measure rates of CO2 hydration in isolated P. marinus MED4 (■) and H. neapolitanus carboxysomes (▲) and compare them with the rates in the uncatalyzed reaction (●). The amounts of P. marinus MED4 and H. neapolitanus carboxysomes used corresponded to a 0.20 μM CsoSCA concentration, as determined by percent composition data (Table 1).
The CO2 fixation ability of purified P. marinus MED4 carboxysomes was assessed with the radiometric RubisCO assay described previously (8). At saturating levels (60 mM) of bicarbonate, a Vmax of 1.28 ± 0.03 μmol · min−1 · mg−1 and an apparent Km for RubP of 169.3 ± 14.6 μM were calculated; saturating levels (0.5 mM) of RubP yielded an apparent KCO2 of 295.1 ± 31.9 μM and a Vmax of 1.27 ± 0.05 μmol · min−1 · mg−1.
CsoS1D is a component of the P. marinus MED4 carboxysome shell.
To determine whether the double-BMC-domain protein CsoS1D, which is encoded by a gene upstream of the canonical cso operon (see Fig. 6C and Table 2), was associated with P. marinus MED4 carboxysomes, sucrose gradient fractions were probed with anti-P. marinus MED4 CsoS1D and with anti-H. neapolitanus CsoS1 antisera. The fractions that yielded CsoS1D and CsoS1 bands of the highest signal intensity clearly coincided (Fig. 4, arrows), suggesting that CsoS1D and CsoS1 copurified during carboxysome purification. Furthermore, blots of purified carboxysomes, when developed with anti-P. marinus MED4 CsoS1D antiserum, identified a very faint polypeptide of approximately 33 kDa in stained SDS-polyacrylamide gels as CsoS1D (Fig. 2A). The relative abundance of CsoS1D was estimated by spot densitometry to be approximately 0.9% of the total carboxysome protein, which corresponds to 38 monomer copies or 6 stacked pseudohexameric trimers (22) per carboxysome.
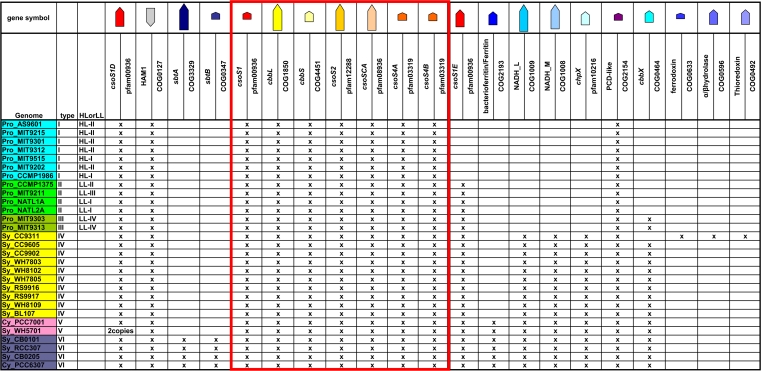
Table 2.
Comparison of cso gene clusters in all sequenced α-cyanobacteriaa
The columns correspond to the order of genes in type VI. The canonical cso gene cluster, from csoS1 to csoS4B, is boxed in red. The relative sizes of the genes are shown by the arrows at the top; warm colors indicate previously known cso genes, and cold colors show proposed putative cso genes. The color coding of the gene symbols in the first column shows types (type I to VI) as indicated in Fig. 6C.
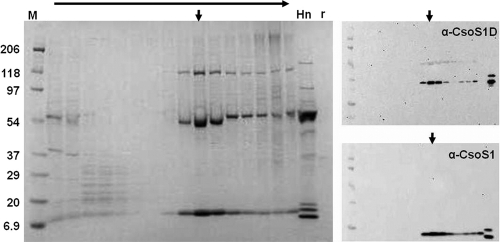
Fig 4.
Copurification of CsoS1D with CsoS1 during sucrose gradient centrifugation of P. marinus MED4 carboxysomes. Aliquots of all gradient fractions (top to bottom indicated by horizontal arrow) were subjected to SDS-PAGE. CsoS1 and CsoS1D were identified by immunoblotting with their respective antibodies. Vertical arrows indicate peak chemiluminescence signals. H. neapolitanus carboxysomes (Hn) and recombinant P. marinus MED4 CsoS1D (r) were included as positive controls. M, molecular mass ladder (kDa).
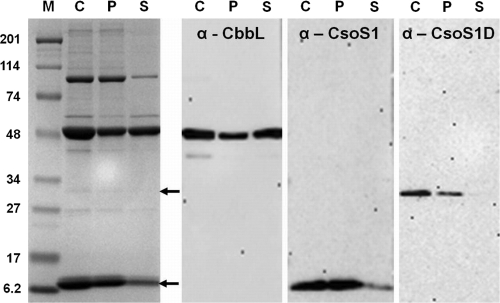
To further ascertain the association of CsoS1D with the shell, P. marinus MED4 carboxysomes were disrupted by incubation in a high-salt buffer (12) and fractionated by centrifugation into a shell-enriched pellet and soluble proteins that had been freed from carboxysomes. Although this procedure did not completely break the P. marinus MED4 organelles (Fig. 5), substantial enrichment of proteins in the pellet and of RubisCO in the supernatant fraction was clearly evident in the stained SDS-polyacrylamide gel and in the immunoblots developed with anti-CbbL and anti-CsoS1 antisera. The fact that CsoS1D protein could only be detected in the shell-enriched pellet fraction firmly established the tight association of the double-domain BMC protein with the P. marinus MED4 carboxysome shell, as was predicted from its structure (22).
Fig 5.
Association of CsoS1D with the P. marinus MED4 carboxysome shell. Carboxysomes (C) were incubated in turnip crinkle virus buffer (see Materials and Methods) and fractionated into a shell-associated pellet (P) and a free-protein-enriched supernatant (S) by centrifugation. CbbL, CsoS1 (lower arrow on stained gel), and CsoS1D (upper arrow on stained gel) polypeptides were identified by immunoblotting as described for Fig. 2 and 4. M, molecular mass ladder (kDa).
Evidence for an expanded carboxysome gene cluster.
The confirmation that CsoS1D is a component of the carboxysome and its position slightly outside the canonical carboxysome gene cluster in many organisms prompted a reconsideration of the inventory of genes that may be carboxysome associated or coregulated with carboxysome-associated proteins.
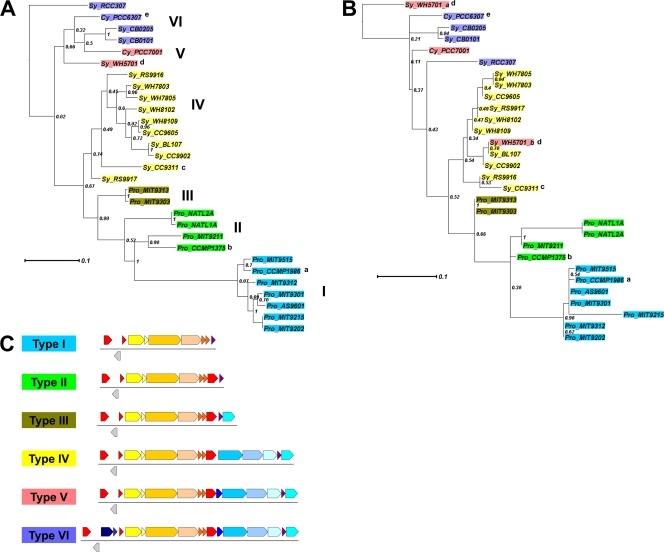
The canonical cso gene cluster (from csoS1 to csoS4B, boxed in red in Table 2), which encodes known components of the α-carboxysome, features the relatively short intergenic distances, conservation across species of gene order and orientation, and functional correlation among clustered genes that are commonly associated with operons (19, 31, 34, 45). The cluster is assumed to be an operon (2, 5, 9), and its genes have been experimentally confirmed to be controlled by a common promoter in Halothiobacillus neapolitanus (3; S. Neidler, F. Cai, and S. Heinhorst, unpublished data). However, a comprehensive survey of the neighborhood of the cso gene cluster in currently available cyanobacterial genomic sequence data revealed additional genes that may be related to carboxysome function and may be coordinately regulated with other known carboxysome genes (Table 2 and Fig. 6). Similar observations have also been made in α-carboxysome-containing chemoautotrophic bacteria (3). In Table 2, cyanobacterial genomes encoding α-carboxysomes are grouped based on their complement of (putative) carboxysome-associated genes and the organization of those genes. Notably, the previously confirmed cso genes (type I in Fig. 6; boxed in red in Table 2) form a conserved core across species. The csoS1D gene is always located upstream from the cso operon, separated by a gene annotated as HAM1 (except in the Synechococcus sp. strain WH5701 genome). Remarkably, the Synechococcus sp. WH5701 genome has two csoS1D genes which fall into different phylogenetic clusters (Fig. 6B). The csoS1D of Sy_WH5701_b groups with Sy_BL107 with a very short branch length and good support (78%), suggesting that this csoS1D copy was gained via a horizontal gene transfer event from a type IV α-cyanobacterium.
Fig 6.
Phylograms of α-cyanobacteria based on IF-2 (A) or CsoS1D (B). Trees are constructed with maximum likelihood (PhyML) with 100 bootstrap replicates. Numbers along the internodes are the percentage of instances each node was supported in 100 bootstrap replicates. Color coding shows types as indicated in panel C. Pro, Prochlorococcus; Sy, Synechococcus; Cy, Cyanobium; a, also known as MED4 or CCMP1378; b, also known as SS120; c, exception—no cbbX and having three additional open reading frames (details in Table 2); d, csoS1D is not present upstream from the cso operon but there are 2 copies of csoS1D genes elsewhere in the genome, marked as “Sy_WH5701_a” and “Sy_WH5701_b” in panel B; e, exception—no cbbX. (C) Schematic diagram of type I to type VI cso gene clusters. The color scheme for genes uses warm colors for previously known cso genes and cold colors for proposed putative cso genes.
Downstream from the canonical cso operon there is an additional, absolutely conserved gene with unknown function (COG2154 in Table 2; purple arrow in Fig. 6). This gene has homology to pterin-4α-carbinolamine dehydratase (PCD), with identities and similarities ranging from 18.5% to 27.4% and 36.2% to 49.4%, respectively, in comparison to the primary structure of PCD from Thermus thermophilus (PDB 1USO). However, in the α-cyanobacterial homolog of PCD, the signature motif for pterin-4α-carbinolamine dehydratase activity (40) is absent. This carboxysome-related PCD-like gene is also present downstream from all known cso gene clusters of chemoautotrophic bacteria (3).
With the exception of the high-light-adapted Prochlorococcus species (type I), all of the marine cyanobacteria contain a gene, csoS1E, encoding an ∼200-amino-acid protein with a single BMC domain, between the canonical cso operon and the PCD-like gene. The correlation of light quality with the presence/absence of this gene suggests that CsoS1E might be important for carboxysome function under low-to-medium light conditions. Due to its high isoelectric point (average pI of 10.4 for all 21 csoS1E genes currently in the database), the translation product of this gene has been difficult to characterize.
DISCUSSION
Cyanobacterial carboxysomes have proven refractory to purification largely because of difficulties encountered with cell breakage and with the separation of intact organelles from other cellular components, notably the abundant thylakoid membranes (13, 29, 33). Consequently, their protein composition is not known and has been inferred mainly from genetic (reviewed in reference 15) and more recent structural genomic evidence (reviewed in reference 7). Because of the unique features of the P. marinus MED4 cso operon and the intriguing possibility that the double-BMC-domain protein CsoS1D, which is encoded by a gene outside the cso operon, is part of the organelle (22), purification and analysis of the P. marinus MED4 carboxysome was of particular interest to us.
Important for our choice of the MED4 strain of Prochlorococcus were the ultrastructural studies by Ting et al. (41), which suggested that the reduced number of photosynthetic membranes, combined with the lack of a prominent peptidoglycan layer, might facilitate the purification of carboxysomes from this cyanobacterium. Indeed, breakage of P. marinus MED4 cells required only light sonication, a treatment that was mild enough to preserve the structural integrity of the carboxysomes. Contaminating thylakoid membranes could be removed by incubation of the cell lysate with the nonionic detergent Nonidet P-40 prior to the final purification step. Carboxysomes in the final fraction were structurally intact, enzymatically active, and remarkably stable compared to other cyanobacterial carboxysomes, which do not withstand sucrose gradient centrifugation (13). It is possible that α-carboxysomes in general are more robust than the larger and less regularly shaped β-carboxysomes found in most cyanobacteria. P. marinus MED4 carboxysomes appear to be particularly resilient, since they were found to be refractory to breakage by the repeated freeze/thaw cycles that easily disrupt H. neapolitanus carboxysomes (39) and instead required prolonged incubation in high-salt buffer at alkaline pH for RubisCO to be released from their interior.
The CO2 fixation activity of purified P. marinus MED4 carboxysomes was lower than that measured with isolated H. neapolitanus organelles. The calculated Vmax of the carboxylation reaction was two-thirds of that reported for H. neapolitanus (11). The Km values of RubisCO for RubP were comparable between the two organisms, but the KCO2 for P. marinus MED4 RubisCO was nearly twice as high as that reported for the H. neapolitanus enzyme (11). These differences in KCO2 values may reflect differences in the intrinsic properties of the two RubisCO species. Scott et al. (35) reported a remarkably high KCO2 for recombinant Prochlorococcus marinus MIT9313 RubisCO expressed in Escherichia coli, suggesting that all members of the genus Prochlorococcus may contain particularly inefficient RubisCO enzymes. It is also possible that the low turnover number of the carboxysome-bound P. marinus MED4 CsoSCA, which rapidly supplies RubisCO inside the carboxysome with its substrate, CO2, limits the catalytic performance of RubisCO and may explain its high apparent KCO2. Distinction between these possibilities requires more extensive comparisons of the catalytic properties exhibited by the carboxy-some-bound enzymes and by RubisCO alone. Because of the limited amounts of purified P. marinus MED4 carboxysomes available, those assays could not be performed in the current study.
In addition to functional characterization of the enzymatic constituents, P. marinus MED4 carboxysome components were identified immunologically using antibodies specific for H. neapolitanus and P. marinus MED4 proteins. Through a combination of both methods, all expected products of the P. marinus MED4 cso operon except the low-abundance CsoS4 proteins were shown to be present in the purified P. marinus MED4 organelles. The estimated 152 RubisCO holoenzyme copies per P. marinus MED4 carboxysome contribute approximately 60% to the total mass, a value that is comparable to that reported for H. neapolitanus (70%) (15). The higher number of RubisCO copies (270) present in carboxysomes from the chemoautotroph reflects differences in average diameter, which translates to an estimated 30 to 40% difference in internal volume between the two carboxysomes (18, 41).
All carboxysome gene clusters, as well as those that encode other bacterial microcompartment types, contain at least two csoS1 (Pfam00936) homologs. In H. neapolitanus, all three csoS1 paralogs are transcribed, and their protein products are abundant components of the carboxysome shell (3). Although the need for multiple paralogs of the major shell protein is not clear, this observation is significant, because the P. marinus MED4 genome is the only one known to contain a cso gene cluster with only one csoS1 gene (22). The CsoS1D protein features two Pfam00936 domains, suggesting that the protein might be a carboxysome component. However, the csoS1D gene is not included in the canonical cso operon that accounts for all currently known α-carboxysome proteins. Our finding that CsoS1D is indeed a component of the P. marinus MED4 carboxysome shell suggests that this protein may be a previously unknown component of all cyanobacterial α-carboxysomes. Because P. marinus MED4 CsoS1D pseudohexamers are able to alternate the conformation of their central pore between the open and closed state, it is believed that the protein may play a role in gated metabolite transport, possibly of the larger RubisCO metabolites RubP and 3-phosphoglycerate, across the carboxysome shell (22). The biochemical evidence provided here for the association of CsoS1D with the P. marinus MED4 carboxysome shell lends credence to a role for of this protein in carboxysome shell function.
Unidentified polypeptide bands have been documented in SDS-polyacrylamide gels of purified H. neapolitanus carboxysomes (8, 11, 15) and are generally assumed to represent low-level contaminants or aggregates of bona fide carboxysome proteins. However, the presence of CsoS1D in the P. marinus MED4 carboxysome and comparative genomics evidence (Table 2) point to the possibility that additional genes not located within the classic cso operon may encode proteins that contribute to carboxysome structure and/or function. In addition to the obvious choices, csoS1E and the PCD-like gene, the CCM-related genes sbtA/B, cbbX, and chpX are also present upstream or downstream from the canonical cso operon in many cyanobacterial genomes. SbtA is a low-CO2-inducible, Na+-dependent bicarbonate transporter that was originally identified in Synechocystis sp. strain PCC6803 (36). Although originally proposed to be a single-component Na+/HCO3− symporter, a small open reading frame (ORF) located directly downstream from sbtA, designated sbtB, is clustered with sbtA and coinduced in Synechocystis sp. PCC6803 under inorganic carbon limitation (42). To date, SbtA has been analyzed genetically and physiologically only in β-cyanobacteria (30, 36); this is the first report that sbtA/B homologues in α-cyanobacteria are clustered with carboxysome genes (Table 1; type VI in Fig. 6C).
The product of the cbbX gene is a form I RubisCO activase (25). The chpX gene encodes a CO2 hydration protein (23). In Synechococcus elongatus PCC7942, the ChpX protein is associated with the NDH-I4 complex, of which NAD(P)H dehydrogenase subunits L and M are two major components. This is consistent with the presence of NADH_L, NADH_M, and chpX in type IV, V, and VI (Table 1) carboxysome gene clusters. In contrast to β-cyanobacteria, in which both ChpX/NDH-I4 (low-affinity/constitutive CO2 uptake activity) and ChpY/NDH-I3 (high-affinity/low-CO2-inducible CO2 uptake activity) complexes have been found (28), α-cyanobacteria only have the genes for the ChpX/NDH-I4 complex or none at all. Synechococcus sp. WH5701 is the only exception; it contains both complexes (28). The activity and affinities of these complexes have yet to be characterized in α-cyanobacteria. Interestingly, a previous study (23) revealed that the CO2 hydration activity of the ChpX/NDH-I4 complex of the β-cyanobacterium Synechococcus elongatus PCC7942 relied on NADPH- and ferredoxin (Fd)-dependent PSI cyclic electron transport; genes encoding ferritin, ferredoxin, and thioredoxin are also found in the neighborhood of canonical cso gene clusters (Table 1).
The grouping of α-cyanobacterial genomes by cso gene cluster organization matches their phylogenetic clustering (Fig. 6). This match is also confirmed by comparing phylograms based on a phylogenetic marker (IF-2) or an α-carboxysome marker (CsoS1D) (Fig. 6A and B). Among the different clusters, type I, represented by P. marinus MED4, seems to have the least complex carboxysome gene cluster, while other types, for example, type VI, might need extra components coordinated with other parts of the CCM to facilitate carboxysome function and form a “superoperon.” Whether these extra genes directly contribute to carboxysome biogenesis and/or function is still an open question. Clearly, our findings have important implications for the structure, function, and regulation of α-carboxysomes and suggest that the protein composition of these important bacterial organelles warrants a closer look beyond what was assumed to be a solved problem.
ACKNOWLEDGMENTS
This work was supported by NSF grants MCB 0818680 (to G.C.C. and S.H.) and MCB 0851070 (to C.A.K., G.C.C., and S.H.).
Footnotes
Published ahead of print 9 December 2011
REFERENCES
- 1. Badger MR, Hanson D, Price GD. 2002. Evolution and diversity of CO2 concentrating mechanisms in cyanobacteria. Funct. Plant Biol. 29:161–173 [DOI] [PubMed] [Google Scholar]
- 2. Beller HR, et al. 2006. The genome sequence of the obligately chemolithoautotrophic, facultatively anaerobic bacterium Thiobacillus denitrificans. J. Bacteriol. 188:1473–1488 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Cai F, Heinhorst S, Shively J, Cannon G. 2008. Transcript analysis of the Halothiobacillus neapolitanus cso operon. Arch. Microbiol. 189:141–150 [DOI] [PubMed] [Google Scholar]
- 4. Cai F, et al. 2009. The pentameric vertex proteins are necessary for the icosahedral carboxysome shell to function as a CO2 leakage barrier. PLoS One 4:e7521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Cannon GC, et al. 2003. Organization of carboxysome genes in the thiobacilli. Curr. Microbiol. 46:115–119 [DOI] [PubMed] [Google Scholar]
- 6. Cannon GC, Heinhorst S, Bradburne CE, Shively JM. 2002. Carboxysome genomics: a status report. Funct. Plant Biol. 29:175–182 [DOI] [PubMed] [Google Scholar]
- 7. Cannon GC, Heinhorst S, Kerfeld CA. 2010. Carboxysomal carbonic anhydrases: structure and role in microbial CO2 fixation. Biochim. Biophys. Acta. 1804:382–392 [DOI] [PubMed] [Google Scholar]
- 8. Cannon GC, Shively JM. 1983. Characterization of a homogenous preparation of carboxysomes from Thiobacillus neapolitanus. Arch. Microbiol. 134:52–59 [Google Scholar]
- 9. Chen X, et al. 2004. Operon prediction by comparative genomics: an application to the Synechococcus sp. WH8102 genome. Nucleic Acids Res. 32:2147–2157 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Dereeper A, et al. 2008. Phylogeny.fr: robust phylogenetic analysis for the non-specialist. Nucleic Acids Res. 36:W465–W469 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Dou Z, et al. 2008. CO2 fixation kinetics of Halothiobacillus neapolitanus mutant carboxysomes lacking carbonic anhydrase suggest the shell acts as a diffusional barrier for CO2. J. Biol. Chem. 283:10377–10384 [DOI] [PubMed] [Google Scholar]
- 12. Golden JS, Harrison SC. 1982. Proteolytic dissection of turnip crinkle virus subunit in solution. Biochemistry 21:3862–3866 [DOI] [PubMed] [Google Scholar]
- 13. Gonzales AD, et al. 2005. Proteomic analysis of the CO2-concentrating mechanism in the open-ocean cyanobacterium Synechococcus WH8102. Can. J. Bot. 83:735–745 [Google Scholar]
- 14. Harpel MR, Lee EH, Hartman FC. 1993. Anion-exchange analysis of ribulose-bisphosphate carboxylase/oxygenase reactions: CO2/O2 specificity determination and identification of side products. Anal. Biochem. 209:367–374 [DOI] [PubMed] [Google Scholar]
- 15. Heinhorst S, Cannon GC, Shively JM. 2006. Carboxysomes and carboxysome-like inclusions, p 141–164 In Shively J. M. (ed), Complex intracellular structures in prokaryotes, vol 2 Springer, Berlin, Germany [Google Scholar]
- 16. Heinhorst S, et al. 2006. Characterization of the carboxysomal carbonic anhydrase CsoSCA from Halothiobacillus neapolitanus. J. Bacteriol. 188:8087–8094 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Huson D, et al. 2007. Dendroscope: an interactive viewer for large phylogenetic trees. BMC Bioinformatics 8:460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Iancu CV, et al. 2010. Organization, structure, and assembly of [alpha]-carboxysomes determined by electron cryotomography of intact cells. J. Mol. Biol. 396:105–117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Jacob E, Sasikumar R, Nair KNR. 2005. A fuzzy guided genetic algorithm for operon prediction. Bioinformatics 21:1403–1407 [DOI] [PubMed] [Google Scholar]
- 20. Kaplan A, Reinhold L. 1999. CO2 concentrating mechanisms in photosynthetic microorganisms. Annu. Rev. Plant Physiol. Plant Mol. Biol. 50:539–570 [DOI] [PubMed] [Google Scholar]
- 21. Khalifah RG. 1971. The carbon dioxide hydration activity of carbonic anhydrase. I. Stop-flow kinetic studies on the native human isoenzymes B and C. J. Biol. Chem. 246:2561–2573 [PubMed] [Google Scholar]
- 22. Klein MG, et al. 2009. Identification and structural analysis of a novel carboxysome shell protein with implications for metabolite transport. J. Mol. Biol. 392:319–333 [DOI] [PubMed] [Google Scholar]
- 23. Maeda S, Badger MR, Price GD. 2002. Novel gene products associated with NdhD3/D4-containing NDH-1 complexes are involved in photosynthetic CO2 hydration in the cyanobacterium, Synechococcus sp. PCC7942. Mol. Microbiol. 43:425–435 [DOI] [PubMed] [Google Scholar]
- 24. Moore LR, et al. 2007. Culturing the marine cyanobacterium Prochlorococcus. Limnol. Oceanogr. Methods 5:353–362 [Google Scholar]
- 25. Mueller-Cajar O, et al. 2011. Structure and function of the AAA+ protein CbbX, a red-type Rubisco activase. Nature 479:194–199 [DOI] [PubMed] [Google Scholar]
- 26. Notredame C, Higgins DG, Jheringa 2000. T-COFFEE: a novel method for fast and accurate multiple sequence alignment. J. Mol. Biol. 302:205–217 [DOI] [PubMed] [Google Scholar]
- 27. Partensky F, Hess WR, Vaulot D. 1999. Prochlorococcus, a marine photosynthetic prokaryote of global significance. Microbiol. Mol. Biol. Rev. 63:106–127 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Price GD, Badger MR, Woodger FJ, Long BM. 2008. Advances in understanding the cyanobacterial CO2-concentrating-mechanism (CCM): functional components, Ci transporters, diversity, genetic regulation and prospects for engineering into plants. J. Exp. Bot. 59:1441–1461 [DOI] [PubMed] [Google Scholar]
- 29. Price GD, Coleman JR, Badger MR. 1992. Association of carbonic anhydrase activity with carboxysomes isolated from the cyanobacterium Synechococcus PCC7942. Plant Physiol. 100:784–793 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Price GD, Woodger FJ, Badger MR, Howitt SM, Tucker L. 2004. Identification of a SulP-type bicarbonate transporter in marine cyanobacteria. Proc. Natl. Acad. Sci. U. S. A. 101:18228–18233 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Price MN, Huang KH, Alm EJ, Arkin AP. 2005. A novel method for accurate operon predictions in all sequenced prokaryotes. Nucleic Acids Res. 33:880–892 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Rocap G, et al. 2003. Genome divergence in two Prochlorococcus ecotypes reflects oceanic niche differentiation. Nature 424:1042–1047 [DOI] [PubMed] [Google Scholar]
- 33. Rodriguez-Buey ML, Marco E, Orus MI. 2005. Isolation of Synechococcus PCC7942 carboxysomes. Ann. Microbiol. 55:81–84 [Google Scholar]
- 34. Salgado H, Moreno-Hagelsieb G, Smith TF, Collado-Vides J. 2000. Operons in Escherichia coli: genomic analyses and predictions. Proc. Natl. Acad. Sci. U. S. A. 97:6652–6657 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Scott KM, et al. 2007. Kinetic isotope effect and biochemical characterization of form IA RubisCO from the marine cyanobacterium Prochlorococcus marinus MIT9313. Limnol. Oceanogr. 52:2199–2204 [Google Scholar]
- 36. Shibata M, et al. 2002. Genes essential to sodium-dependent bicarbonate transport in cyanobacteria: function and phylogenetic analysis. J. Biol. Chem. 277:18658–18664 [DOI] [PubMed] [Google Scholar]
- 37. Shively JM, English RS. 1991. The carboxysome, a prokaryotic organelle: a mini review. Can. J. Bot. 69:957–962 [Google Scholar]
- 38. Smith KS, Ferry JG. 2000. Prokaryotic carbonic anhydrases. FEMS Microbiol. Rev. 24:335–366 [DOI] [PubMed] [Google Scholar]
- 39. So AK-C, et al. 2004. A novel evolutionary lineage of carbonic anhydrase (epsilon class) is a component of the carboxysome shell. J. Bacteriol. 186:623–630 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Suck D, Ficner R. 1996. Structure and function of PCD/DCoH, an enzyme with regulatory properties. FEBS Lett. 389:35–39 [DOI] [PubMed] [Google Scholar]
- 41. Ting CS, Hsieh C, Sundararaman S, Mannella C, Marko M. 2007. Cryo-electron tomography reveals the comparative three-dimensional architecture of Prochlorococcus, a globally important marine cyanobacterium. J. Bacteriol. 189:4485–4493 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Wang H-L, Postier BL, Burnap RL. 2004. Alterations in global patterns of gene expression in Synechocystis sp. PCC 6803 in response to inorganic carbon limitation and the inactivation of ndhR, a LysR family regulator. J. Biol. Chem. 279:5739–5751 [DOI] [PubMed] [Google Scholar]
- 43. Wu M, Eisen J. 2008. A simple, fast, and accurate method of phylogenomic inference. Genome Biol. 9:R151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Yeates TO, Kerfeld CA, Heinhorst S, Cannon GC, Shively JM. 2008. Protein-based organelles in bacteria: carboxysomes and related microcompartments. Nat. Rev. Microbiol. 6:681–691 [DOI] [PubMed] [Google Scholar]
- 45. Zheng Y, Szustakowski JD, Fortnow L, Roberts RJ, Kasif S. 2002. Computational identification of operons in microbial genomes. Genome Res. 12:1221–1230 [DOI] [PMC free article] [PubMed] [Google Scholar]