Abstract
Vector-borne pathogens regulate their protein expression profiles, producing factors during host infection that differ from those produced during vector colonization. The Lyme disease agent, Borrelia burgdorferi, produces Erp surface proteins throughout mammalian infection and represses their synthesis during colonization of vector ticks. Known functions of Erp proteins include binding of host laminin, plasmin(ogen), and regulators of complement activation. A DNA region immediately 5′ of erp operons, the erp operator, is required for transcriptional regulation. The B. burgdorferi BpaB and EbfC proteins exhibit high in vitro affinities for erp operator DNA. In the present studies, chromatin immunoprecipitation (ChIP) demonstrated that both proteins bind erp operator DNA in vivo. Additionally, a combination of in vivo and in vitro methods demonstrated that BpaB functions as a repressor of erp transcription, while EbfC functions as an antirepressor.
INTRODUCTION
Successful infection of a host requires that the invading pathogen control its production of virulence determinants. The infectious agent must sense its environment and respond by increasing production of appropriate factors and repressing production of unnecessary ones. These features are especially critical for vector-borne pathogens, which must not only efficiently infect two extremely different host types but also be transmitted back and forth between hosts. Deciphering the regulatory pathways used by pathogens to control production of infection-associated proteins provides significant insight into the infectious nature of those organisms. Moreover, regulatory factors are attractive candidates for development of novel preventative and curative therapies.
The spirochetal bacterium Borrelia burgdorferi, the agent of Lyme disease, is an excellent model organism for the study of gene regulation by a vector-borne pathogen. B. burgdorferi is genetically tractable, and its natural mammal-tick infectious cycle can be replicated in the laboratory. In addition, infection by B. burgdorferi is a significant cause of human morbidity, being the most commonly reported vector-borne disease in the United States and many other parts of the world (51, 55, 56).
B. burgdorferi Erp lipoproteins are produced throughout mammalian infection but are largely repressed during colonization of vector ticks (10, 31, 48, 49). Erp synthesis is greatly enhanced when B. burgdorferi is transmitted from a feeding tick into a warm-blooded host. Regulation of Erp protein production is controlled at the level of transcription (6). Erp proteins are located in the bacterial outer membrane and are exposed to the external environment (25, 32, 41). Known functions of Erp proteins include binding of host plasmin(ogen), laminin, and the complement regulators factor H and factor H-related proteins 1, 2, and 5 (2, 3, 11, 12, 34, 37, 40, 45, 59). These functions indicate roles for Erp proteins in host adherence, dissemination, and resistance to the alternative pathway of complement-mediated killing. Borrelial erp genes are located in mono- or bicistronic operons on extrachromosomal cp32 prophages, most of which replicate autonomously as circular episomes (24, 60, 63, 64, 72). Individual Lyme spirochetes naturally contain numerous different cp32 elements, each with a unique erp locus, and therefore produce multiple, distinct Erp surface proteins. A bacterium simultaneously expresses its entire repertoire of Erp proteins (26).
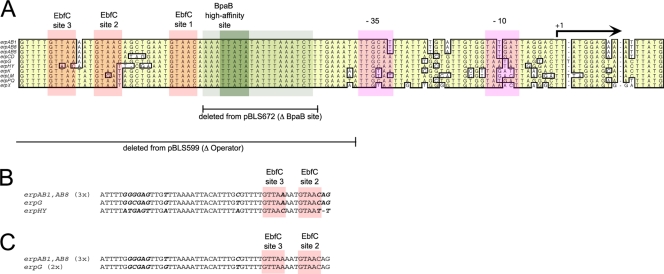
A highly conserved DNA region immediately 5′ of all erp promoters, the erp operator, is required for regulation of erp transcription (see Fig. 1) (6, 10, 64). Two erp operator-binding proteins have been identified, and their binding sites have been characterized: BpaB (borrelial plasmid ParB analogue) and EbfC (erp-binding factor, chromosomal) (4, 13, 52). BpaB binds with high affinity to a 5-bp sequence within the erp operator (13; C. A. Adams, unpublished). Binding of one BpaB protein to that sequence then facilitates binding of additional BpaB molecules along the DNA strand (13). EbfC binds a 4-bp broken palindromic sequence, with all erp operator elements containing 2 to 3 consensus EbfC binding sites adjacent to the BpaB high-affinity site (4, 13, 52). BpaB and EbfC compete with each other for binding to erp operator DNA (13). Like the erp genes, ebfC is poorly expressed in unfed ticks but significantly induced during tick feeding and during mammalian infection (44). For the present work, independent in vivo and in vitro studies were performed to determine the effects of these two proteins on Erp expression. Resulting data indicated that BpaB is a repressor of erp transcription, while EbfC functions as an antirepressor.
Fig 1.
(A) Sequences of the 5′ noncoding DNA of the B. burgdorferi type strain B31, ending with the initiation codon (ATG) of the first erp gene of each locus. Identical nucleotides found in the majority of the 10 loci are boxed and shaded. All of the strain B31 erp loci contain at least 1 consensus EbfC-binding site (GTnAC), plus 1 or 2 additional half-sites (52). Each locus also contains a conserved BpaB-binding region, which consists of an initial binding site (TTATA) and a 19-bp flanking sequence that further stimulates BpaB binding (13; C. A. Adams, unpublished). Regions of noncoding DNA deleted from the mutant erp::gfp fusion constructs pBLS599 and pBLS672 are indicated. (B and C) PCR-amplified portions of DNA sequences bound by EbfC (A) or BpaB (C) in live B. burgdorferi, as assessed by ChIP followed by erp-specific PCR and cloning. Five clones were selected at random, and their inserts were sequenced. Polymorphisms that permit identification of specific erp loci are indicated with boldface italic type.
MATERIALS AND METHODS
Bacterial strains and culture conditions.
All studies used derivatives of the B. burgdorferi type strain, B31, cultured in Barbour-Stoenner-Kelly II medium (73). Chromatin immunoprecipitation (ChIP) was performed using the clonal infectious derivative B31-MI-16, cultured at 34°C (48). For studies requiring B. burgdorferi carrying recombinant plasmids, the readily transformable clonal derivative B31-e2 was used (6, 18). Kanamycin was added to cultures of transformed bacteria at a final concentration of 200 μg/ml. The effects of culture temperature on expression levels of native borrelial proteins were assessed using infectious B31-MI-16, cultured at either 23 or 34°C (17, 29, 48, 62).
Recombinant proteins.
Recombinant BpaB and EbfC were produced as previously described (4, 13). The allele of BpaB carried by the strain B31 lp56 plasmid was used for all studies (13). Recombinant proteins were produced using Escherichia coli strain Rosetta 2 (Novagen, Rockland, MA) and purified from cleared lysates using MagneHis Ni particles (Promega, Madison, WI). Purified proteins were dialyzed with DNA-binding buffer (100 nM dithiothreitol, 50 mM Tris [pH = 7.5], 10% [vol/vol] glycerol, 0.01% [vol/vol] Tween 20, and 0.1% [vol/vol] phenylmethanesulfonyl fluoride). Protein concentrations were determined by the Bradford assay (Bio-Rad, Hercules, CA). Purities were determined by SDS-PAGE and staining with Coomassie brilliant blue. Aliquots were stored at −80°C.
ChIP.
Antiserum directed against BpaB (allele 56) was produced using BALB/c mice, as follows. Mice were injected subcutaneously with 10 μg recombinant BpaB in 80 μl 60% AlOH (m/vol), followed by 2 additional injections 2 weeks apart. One week after the final boost, mice were euthanized, and their blood was pooled and processed into serum. Antiserum was assessed for specificity by immunoblot and enzyme-linked immunosorbent assay (ELISA) with purified recombinant BpaB.
Antiserum directed against EbfC was produced in New Zealand White rabbits by NeoPeptide (Cambridge, MA) using the standard immunization protocol. The vaccinogen was a polypeptide derived from the EbfC sequence, MSSVKSNIDNIKKEM. Antibodies were affinity purified from serum using the vaccinogen polypeptide.
ChIP was performed as previously described (69, 70), with the following modifications. B. burgdorferi B31-MI-16 was cultured at 34°C to mid-exponential phase (approximately 5 × 107 bacteria/ml). Formaldehyde was added to a final concentration of 1%, followed by incubation for 8 min at room temperature while shaking. Cross-linking was stopped by addition of glycine to a final concentration of 0.3 M. Bacteria were pelleted by centrifugation and washed twice with Tris-buffered saline (20 mM Tris [pH 7.5] and 150 mM NaCl). Cell pellets were stored frozen at −80°C. As needed, bacterial pellets were thawed on ice and then resuspended in a 1:4 ratio of lysis buffer to IP buffer (lysis buffer is 10 mM Tris [pH 7.5], 20% sucrose, 50 mM NaCl, and 10 mM EDTA; IP buffer is 50 mM HEPES [pH 7.5], 150 mM NaCl, 1 mM EDTA, 1% Triton X-100, and 0.1% SDS) (69, 70). Lysozyme was added to a final concentration of 5 mg/ml, and the mixture was incubated at 37°C for 30 min. To shear the bacterial DNA, lysates were sonicated using a Branson 102C sonicator (Branson Ultrasonics, Danbury, CT), with 6 pulses of 10 s each at 10% amplitude. Cellular debris was cleared by centrifugation at 12,000 × g for 10 min at 4°C.
Binding of antibodies to resin particles was performed using immunoprecipitation kit-protein A magnetic Dynabeads (Invitrogen, Carlsbad, CA), following the manufacturer's recommended protocol. Antibodies specific for either BpaB or EbfC were utilized. Donkey anti-mouse IgG (Santa Cruz Biotechnology, Santa Cruz, CA) was used as a control for nonspecific antibody binding. Resin beads without added antibody were used in experiments as an additional control.
Borrelial supernatants (800 μl) were incubated with antibody-bead complexes or control beads alone for 20 min at room temperature. The supernatants were poured off, and beads were incubated a second time with a fresh aliquot of lysate. Bead complexes were washed 3 times with IP wash buffer (Invitrogen protein A Dynabead kit) and then resuspended in IP buffer. After transfer to clean microcentrifuge tubes, bead complexes were washed 4 times with IP buffer supplemented with 500 mM NaCl, followed by 2 washes with TE buffer (10 mM Tris and 1 mM EDTA). Beads were resuspended in TE and incubated at 65°C for 18 h. Eluted DNA was purified using DNeasy blood and tissue kits (Qiagen), and antigens were eluted from antibody using elution buffer (immunoprecipitation kit; Invitrogen). Immunoprecipitation was verified by Western blot analysis following antigen elution.
Eluted DNAs were assessed for erp operator DNA by PCR using oligonucleotide primer pairs complementary to conserved sequences flanking erp operators: 164F (5′-TGAGTAGAGCATTTGCAATGGAGAG-3′) and A50R (5′-AAATATATAATTTTGTTACATTTCAG-3′). As a control, oligonucleotide primers which are specific for the B. burgdorferi flaB gene, which does not contain either a BpaB or EbfC site (FLA-1, 5′-CACATATTCAGATGCAGACAGAGG-3′; FLA-2, 5′-CCGGTGCAGCCTGAGCAGTTTGAG-3′) were also used (46, 47). Amplicons were cloned into pCR2.1 (Invitrogen), and the inserts of 5 random clones were sequenced.
Control ChIP reactions that utilized beads alone or beads bound with nonspecific IgG were also subjected to PCR using the primer pairs listed above.
erp::gfp transcriptional fusions and flow cytometry.
A plasmid containing promoterless gfp (pBLS590), an operon fusion between the wild-type erpA operator/promoter and gfp (pBLS591), and a mutant thereof that lacks the entire erp operator (pBLS599) have been described previously (6). Plasmid pBLS672 was created from pBLS591 by removal of the 20-bp high-affinity BpaB-binding site through use of overlap extension PCR (36). Another derivative, pBLJ1, in which all EbfC-binding sites were mutated from the consensus GTnAC to TGATG, was produced. The ability or inability of BpaB and EbfC to bind each DNA construct was assessed by electrophoretic mobility shift assay (EMSA) with each recombinant protein and biotin-labeled DNA probes, as described previously (13, 52). EMSA band intensities were quantified using the Image J software program (http://rsbweb.nih.gov/ij).
All six plasmids were individually introduced into B. burgdorferi B31-e2. Cultures were incubated to mid-exponential phase (approximately 107 bacteria/ml) at either 23 or 34°C. Bacteria were harvested by centrifugation, washed in phosphate-buffered saline (PBS), and resuspended in PBS at approximately 106 cells/ml. Green fluorescent protein (GFP) fluorescence per bacterial cell was analyzed using a FACSCalibur flow cytometer (BD Biosciences, San Jose, CA), with excitation at 488 nm and detection at 530 nm. Each experiment involved measuring a minimum of 50,000 individual bacteria.
In vivo induction of BpaB or EbfC from tetracycline-inducible plasmid constructs.
The previously described pCRW53 replicates autonomously in both B. burgdorferi and E. coli and contains both a constitutively expressed tetR gene and a TetR-repressible promoter, Post, which drives transcription of gfp (71). By use of overlap extension PCR (36), the pCRW53 multiple cloning sites were deleted, and then the gfp gene was removed and replaced with single BamHI and PstI recognition sequences, producing pSZW53-4. The B. burgdorferi strain B31 lp56 bpaB (allele bpaB56) or ebfC gene was amplified by PCR from recombinant plasmid clones and individually ligated into pSZW53-4, producing pBLS705 and pBLS704, respectively. Those two chimeric plasmids and the parental empty vector pSZW53-4 were individually introduced into B. burgdorferi strain B31-e2 by electroporation. Transcription from the Post promoter was induced by addition of anhydrotetracycline (AT), at a final concentration of 0.5 μg/ml, to early-exponential-phase cultures (approximately 105 bacteria/ml). After cultivation to final densities of approximately 107 bacteria/ml, bacteria were harvested by centrifugation and lysed, and proteins were separated by SDS-PAGE. Total proteins were detected by Coomassie brilliant blue staining. Individual proteins were identified by immunoblotting using monospecific antibodies (7, 13, 25, 26, 52) and analyzed densitometrically.
In vitro coupled transcription/translation.
A linear DNA fragment consisting of 471 bp of erp 5′ noncoding DNA fused to gfp was produced by PCR from template pBLS591 (6) using the oligonucleotide primers M13 Forward (5′-GTAAAACGACGGCCAG-3′) and M13 Reverse (5′-CAGGAAACAGCTATGAC-3′). As a control, a similarly sized DNA fragment, which consisted of the B. burgdorferi ospAB promoter and 5′ noncoding DNA fused to gfp, was also produced (16). Bovine serum albumin (BSA) was used for some control experiments. Protein concentrations were determined by Bradford assay (Bio-Rad). Reactions used the cell-free E. coli S30 extract transcription/translation system for linear templates (Promega). Each 75-μl reaction mixture contained 105 nM DNA template, 160 nM (each) protein (alone or together, as well as no added protein), 4 mM NaCl, 4 mM Tris-HCl, 80 μM NaHPO4, and 0.75 nM dithiothreitol (DTT) in the following volumes of kit reagents: 30 μl S30 premix, 22.5 μl E. coli S30 extract, and 7.5 μl 1 mM amino acid mix. To ensure that experimental readouts were due to gfp transcription, rifampin was added to control reactions at a final concentration of 40 μg/ml. Additional control experiments replaced the DNA with 6 μl nuclease-free water. Reactions were lightly mixed and incubated at 37°C for 80 min. Reactions were stopped by incubation on ice for 15 min, and proteins were precipitated with acetone and then resuspended in 85 μl PBS.
For ELISA, 60 μl of resuspended products was added to 380 μl of ELISA coating buffer (50 mM Na2CO3, 500 mM NaHCO3, pH 9.2). GFP product was measured using MACS molecular anti-GFP-horseradish peroxidase conjugate (Miltenyi Biotec, Auburn, CA) and Turbo tetramethyl benzidine (TMB) ELISA (Thermo-Fisher, Pittsburgh, PA). Reactions were stopped with 2 N H2SO4, and absorbance at 450 nm was measured with a Versamax tunable microplate reader. Each experiment was performed with four simultaneous identical reactions, and all experiment was replicated at least three times.
For immunoblotting, reaction products were subjected to SDS-polyacrylamide gel electrophoresis and transferred to nitrocellulose membranes. Membranes were blocked with Sea Block buffer (Thermo-Fisher). GFP was detected by incubation with MACS molecular anti-GFP-horseradish peroxidase conjugate and SuperSignal West Pico chemiluminescent substrate (Thermo-Fisher).
Statistical analyses.
Statistical significance between samples was determined by Student's t test, assuming unequal variance. Graphical representation of ELISA data reflects values that were normalized against the mean absorbance of a reaction which did not contain template DNA.
RESULTS
BpaB and EbfC bind to erp operators in vivo.
Although we earlier found that BpaB and EbfC bind erp operator DNA under in vitro conditions (13, 52), protein-DNA interactions can only be considered biologically relevant if the protein binds DNA when inside the bacterial cell. To that end, we performed chromatin immunoprecipitation (ChIP) of each DNA-binding protein. Following cross-linking treatment of live B. burgdorferi, the cells were lysed, DNA was sheared by sonication, and immunoprecipitation was undertaken with antibodies specific for either BpaB or EbfC. Precipitated DNA fragments were subjected to PCR using oligonucleotide primers specific for erp operator and flanking DNAs. Appropriately sized amplicons were obtained for each experiment and cloned into pCR2.1. For each ChIP, 5 random clones were chosen and their plasmid inserts were sequenced. Control PCRs using oligonucleotide primers specific for the flaB gene, which does not contain either a BpaB or an EbfC site, all failed to generate amplicons, confirming the specificity of these analyses. Control ChIP reactions that used beads alone or beads coated with irrelevant antibody did not generate amplicons with either erp- or flaB-specific primers, further indicating that the BpaB- and EbfC-ChIP results were specific.
Sequencing of anti-BpaB and -EbfC precipitated DNAs revealed that both proteins had bound in vivo to erp operator DNA (Fig. 1B and C). Nucleotide polymorphisms occur among the 5′ noncoding regions of the different erp operons within a single bacterium. EbfC-ChIP yielded 3 clones derived from either erpAB1 or erpAB8 (both are identical in this region) and 1 clone each of erpG and erpHY (Fig. 1B). For BpaB ChIP, the 5 randomly selected PCR fragments consisted of 3 clones derived from erpAB1 or erpAB8 and 2 from erpG (Fig. 1C). Altogether, ChIP results confirmed that BpaB and EbfC bind to multiple erp loci in live B. burgdorferi.
erp operator/promoter-reporter fusions in B. burgdorferi.
B. burgdorferi promoter activity can be assessed in the spirochete using transcriptional fusions between borrelial DNA and gfp (6, 16). Transcription of the reporter gfp gene results in production of GFP, which can be quantified by flow cytometry. This method was utilized to further define the effects of BpaB and EbfC on erp transcription. A promoterless gfp construct served as a negative control. A derived plasmid contains 471 bp of wild-type B. burgdorferi DNA, extending 5′ from the translation initiation codon of erpA, cloned such that the erp promoter drives transcription of gfp. Previous analyses determined that B. burgdorferi carrying this PerpA::gfp fusion plasmid regulated GFP production in response to environmental cues in a manner that directly correlates with native erp transcription and Erp protein production (6). The PerpA::gfp fusion plasmid was mutated as follows: (i) the entire erp operator and 5′ sequences were deleted, (ii) 20 bp that included the high-affinity BpaB-binding site were deleted from the erp operator, or (iii) all EbfC-binding sites were mutated to nonconsensus sequences. EMSAs using labeled probes derived from these constructs confirmed that each protein did not bind DNA that lacked its binding site or contained defective binding sites, while binding of the other protein was not affected (6, 13, 52; data not shown).
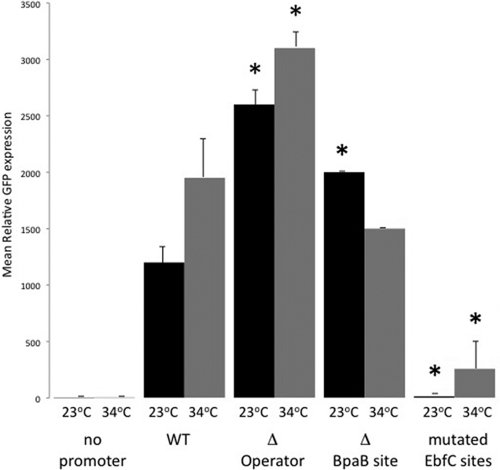
B. burgdorferi carrying the promoterless gfp construct did not produce GFP (Fig. 2) (6). B. burgdorferi produces significantly greater levels of Erp proteins when cultured at 34°C than when cultured at 23°C (58, 62). Likewise, borreliae carrying the wild-type operator-promoter-gfp construct produced greater amounts of GFP when grown at 34° than at 23°C (Fig. 2) (6). Deletion of the entire erp operator resulted in significantly greater expression at both culture temperatures, indicating that the operator is required for transcriptional repression (Fig. 2) (6). Removal of the high-affinity BpaB-binding site from the erp operator resulted in significantly greater production of GFP at 23°C compared with that of the wild-type promoter (Fig. 2). Changing the EbfC-binding sites to nonconsensus sequences significantly reduced GFP production by bacteria cultured at either temperature. Thus, transcription is enhanced if the erp operator cannot bind BpaB and is repressed greatly if EbfC cannot bind.
Fig 2.
GFP production levels of B. burgdorferi containing erp::gfp transcriptional fusions. Constructs contained 471 bp of DNA immediately 5′ of erpA wild type (WT) and mutants thereof, consisting of deletion of all erp DNA 5′ of the promoter −35 sequence (Δ Operator), deletion of 20 bp within the operator that includes the high-affinity BpaB-binding site (Δ BpaB site), and all EbfC-binding sequences mutated to nonconsensus sequences (mutated EbfC sites). GFP was measured by flow cytometry with live B. burgdorferi cultures (6), and levels are reported as mean peak fluorescence values. Results for bacteria containing each mutant construct were compared with those for borreliae containing the WT plasmid. Statistically significant differences from WT results are indicated by asterisks (P < 0.05 by Student's t test).
Overproduction of BpaB and EbfC in B. burgdorferi.
Creation of bpaB and ebfC mutants in B. burgdorferi is proving to be very difficult. Repeated attempts to delete ebfC have failed, suggesting that it is an essential gene (52). Complete deletion of bpaB is complicated in that every known strain of B. burgdorferi contains genes for at least 3 cp32-encoded BpaB proteins, all of which bind to erp operator DNA (13). However, the recent development of an inducible expression system for B. burgdorferi allowed us to take an alternative approach to examine the roles of erp operator-binding proteins in B. burgdorferi. The previously constructed pCRW53 contains tetR, encoding the tet repressor, and a tetracycline-inducible promoter, Post, that functions in B. burgdorferi (71). This plasmid was modified to place a bpaB or ebfC gene such that it is under the transcriptional control of Post. The resulting constructs were transformed into B. burgdorferi, which was cultured in either the absence or presence of the nontoxic inducer molecule anhydrotetracycline (AT), and protein levels were examined by immunoblotting. Addition of AT to cultures of each transformed strain increased BpaB or EbfC to levels well above those of uninduced cultures (Fig. 3).
Fig 3.
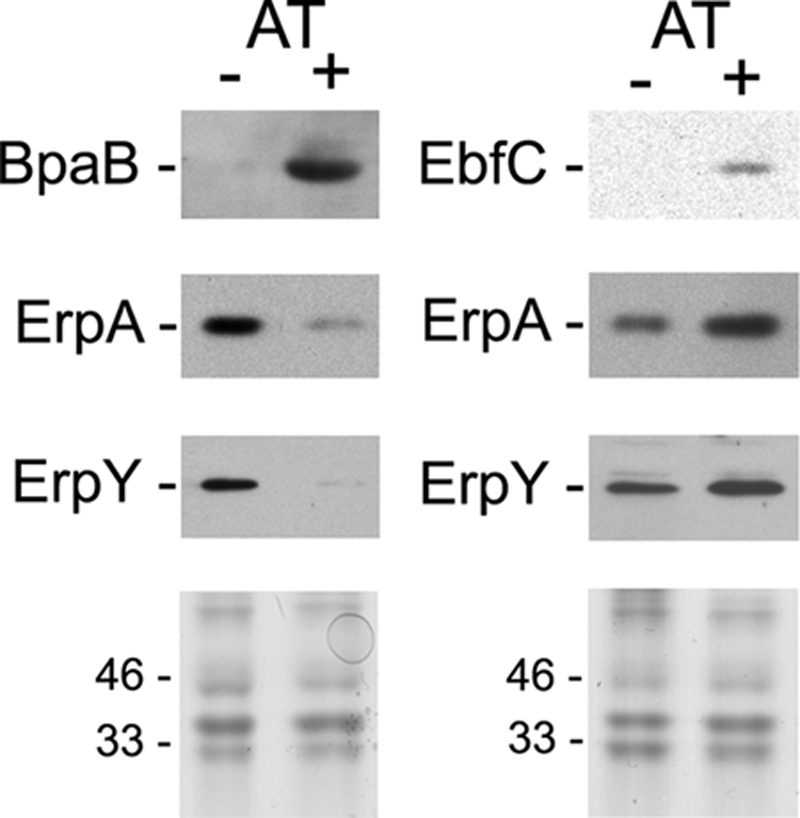
Effects of BpaB or EbfC overproduction by B. burgdorferi, using an anhydrotetracycline (AT)-inducible promoter system. Levels of BpaB, EbfC, ErpA, and ErpY in uninduced (−) and induced (+) bacteria were determined by immunoblotting, using specific antibodies, and densitometry. Lower panels illustrate SDS-PAGE of each bacterial lysate, stained with Coomassie brilliant blue, to confirm equal loading in each lane. Positions of molecular mass standards are shown to the left of each stained gel.
Two unlinked Erp proteins were examined: ErpA, encoded on cp32-1, and ErpY, encoded on cp32-4 (17, 63). Previous work indicated that BpaB proteins from all examined cp32 elements bind the same sequence of all erp operators (13). That was not a surprising conclusion, since the BpaB-binding site sequence of all erp operators is extremely well conserved (Fig. 1A) (1, 6, 10, 43, 60, 64). Further supporting that conclusion, overproduction of lp56-encoded BpaB greatly repressed production of the Erp protein encoded by other cp32 elements (Fig. 3). Those results also indicate that BpaB functions as an erp repressor in trans. Overproduction of EbfC resulted in increased levels of Erp proteins (Fig. 3). Thus, EbfC enhances erp expression in vivo as well as in vitro.
In vitro transcription-translation analyses.
Effects of the erp operator-binding proteins were directly assessed through use of a coupled in vitro transcription-translation system. The utilization of an Escherichia coli S30 extract for these studies conferred several advantages over use of a borrelial extract. Most significantly, E. coli does not produce proteins similar to BpaB, and while those bacteria do produce an EbfC-like protein, it does not specifically bind the same DNA sequence as does the B. burgdorferi protein (19). Thus, the effects of each borrelial protein could be examined in isolation, without complications due to their presence in B. burgdorferi extracts.
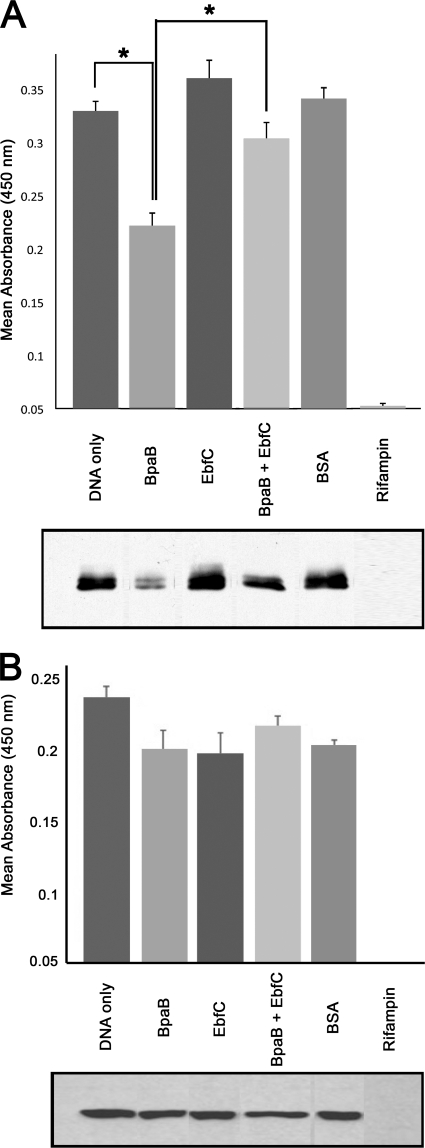
The erpA operator/promoter-gfp construct was utilized for these studies (6). Production of GFP was assayed by both ELISA and immunoblotting. For all these studies, each protein was added to the same final molar concentration. Control studies with added BSA demonstrated that experimental results were not due simply to inclusion of a protein (Fig. 4A and B). Addition of the RNA polymerase inhibitor rifampin completely eliminated readout signals (Fig. 4A and B), confirming that results were dependent on transcription of the Perp::gfp fusion.
Fig 4.
(A) Effects of purified BpaB and/or EbfC proteins on erp expression in a coupled in vitro transcription/translation system. In the upper panel, product levels were quantified by ELISA and are reported as mean absorbances for three independent experiments. Asterisks indicate statistically significant differences (P < 0.001 by Student's t test) between DNA only and BpaB added, or between BpaB alone and BpaB plus EbfC. The lower panel shows anti-GFP immunoblot analyses of one representative series of in vitro transcription/translation reactions. BpaB significantly repressed erp expression, while addition of EbfC counteracted the repressive effect of BpaB. BSA served as a control to confirm that results were specific for each protein. Addition of rifampin completely prevented product formation, demonstrating that results were dependent upon transcription. (B) Control studies using the B. burgdorferi ospAB promoter and 5′ noncoding DNA fused to gfp. The upper panel shows ELISA results. The lower panel shows anti-GFP immunoblotting results. No added protein or combination of proteins had a significant effect upon expression from the ospAB promoter.
Addition of BpaB to the coupled transcription-translation reaction significantly repressed production of GFP (Fig. 4A). EbfC did not exert a significant effect when added by itself. In contrast, simultaneous addition of EbfC and BpaB yielded GFP expression levels that did not differ from those observed when no borrelial protein was present. Those observations are consistent with results of previous studies which determined that EbfC and BpaB compete for binding to erp operator DNA (13). Noting that these studies were performed using nonreplicating template, the data also demonstrate that the effects of BpaB and EbfC on erp expression levels is not due to variations in the cp32 copy number per cell, but the proteins exert their effects directly on erp genes.
As a control, in vitro transcription-translation was also undertaken using a fusion between the B. burgdorferi ospAB promoter and gfp (16). Inclusion of BpaB and/or EbfC did not significantly affect expression from this promoter, indicating that the above-observed effect of BpaB on erp expression was specific to that operon and not a general, nonspecific effect (Fig. 4B).
Effects of growth conditions on BpaB and EbfC.
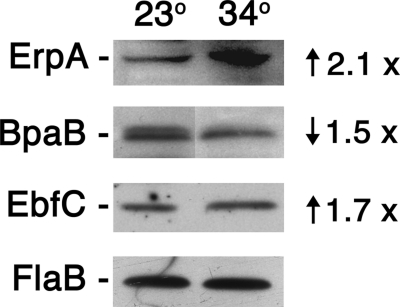
The above-described studies demonstrated that BpaB and EbfC control erp expression. B. burgdorferi produces greater levels of erp transcripts and Erp proteins when cultured at 34°C than when grown at 23°C (58, 62). Therefore, the effects of culture temperature on cellular levels of the erp operator-binding proteins were assessed (Fig. 5). B. burgdorferi cultured at 34°C contained appreciably lower levels of BpaB than did bacteria cultured at 23°C. In contrast, levels of EbfC were higher in bacteria grown at 34°C than they were in bacteria cultured at 23°C. The results for EbfC protein levels correlate with those of previous studies of ebfC mRNA levels (44).
Fig 5.
Representative immunoblots of protein levels in wild-type Borrelia burgdorferi cultured at either 23 or 34°C. B. burgdorferi produce appreciably greater levels of Erp proteins when cultured at 34°C compared with results at 23°C (58, 62). The same bacterial lysates were used for each immunoblot shown. The constitutively expressed FlaB (flagellin) protein was also examined, to serve as a control for equal loading of wells. Comparing results at 23°C versus those at 34°C for these cultures, ErpA levels increased 2.1-fold, BpaB decreased 1.5-fold, and EbfC increased 1.7-fold. This BpaB antiserum, raised against the allele carried by lp56, recognized two of the bacterium's BpaB proteins, both of which were affected by culture temperature in the same manner. The observed differential expression of the EbfC protein corresponds with results of previous studies which found the same effect on ebfC transcript levels (44).
DISCUSSION
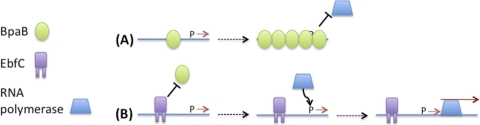
As do other pathogenic bacteria, the Lyme disease spirochete regulates production of numerous proteins during infection processes (51). Research on the borrelial Erp outer surface proteins has revealed multiple properties associated with mammalian infection (2, 3, 11, 12, 34, 37, 40, 45, 59). Given what is known of their functions, it is not surprising that B. burgdorferi Erp proteins are produced during mammalian infection but repressed during colonization of tick vectors (10, 31, 48, 49). The erp operator is required for regulation of erp transcription (6), and the present studies demonstrated that both BpaB and EbfC bind to erp operator DNA in live B. burgdorferi. Results of the studies described in this report, along with previous data, permit the construction of a model of how BpaB and EbfC may control erp expression (Fig. 6). While the model is largely based on studies of cultured B. burgdorferi, such an approach can provide detailed insights into regulatory mechanisms, much as Jacob and Monod's seminal studies of cultured E. coli led to an understanding of lac operon regulation (38, 50).
Fig 6.
A model of the effects of BpaB and EbfC on erp transcription, which incorporates all current data. Both of these DNA-binding proteins serve additional functions in B. burgdorferi, and the borrelial genome contains additional binding sites for each, so effects on erp operons will be dependent upon levels of free BpaB and EbfC proteins in the cell. (A) When cellular levels of free BpaB exceed those of EbfC, a BpaB molecule will bind to the erp operator, which then facilitates binding of additional BpaB proteins to the DNA. BpaB spreading occludes the promoter region, preventing recognition by RNA polymerase and thereby repressing erp expression. (B) When levels of free EbfC exceed those of BpaB, EbfC preferentially binds to the erp operator and competes away BpaB, thereby allowing RNA polymerase to recognize the promoter and transcribe the erp genes.
BpaB initially binds erp operator DNA at a high-affinity site, and deletion of that site significantly reduces binding (13). The initial BpaB-DNA interaction facilitates binding of additional BpaB molecules to the DNA, apparently through protein-protein interactions that stabilize binding to less-desirable DNA sequences (13). Results presented in this report indicate that deletion of the BpaB high-affinity binding site increases erp promoter activity in vivo, and BpaB represses erp transcription in vitro and represses Erp protein levels when overproduced in B. burgdorferi. Altogether, these data indicate that BpaB is a repressor of erp transcription. We hypothesize that repression is due to the spreading of BpaB along erp DNA, which would occlude the promoter elements from recognition by RNA polymerase (Fig. 6A). Such a mechanism of transcriptional repression has been proposed for other bacterial DNA-binding proteins that similarly spread along DNA (8, 20, 28, 39, 42, 53, 54).
The lp56-carried allele of BpaB, expressed in B. burgdorferi from a recombinant plasmid, influenced the erp operons of the bacterium's native cp32-1 and cp32-4. Two important conclusions can be made from that result: (i) BpaB functions as an erp repressor in trans, and (ii) BpaB proteins encoded by one cp32 element will affect erp operons on other cp32s. Lyme spirochetes naturally carry several different cp32 elements, each carrying its own allele of BpaB and having its own erp locus. Our results indicate that the entire cohort of a spirochete's cp32-encoded BpaB proteins controls all of its erp genes. Such cross talk would facilitate the previously observed coexpression of Erp proteins (26, 32).
erp operons contain 2 to 3 consensus EbfC-binding sites adjacent to the BpaB high-affinity site (Fig. 1) (13, 52). EbfC and BpaB compete for binding to erp operator DNA (13). An earlier study observed that B. burgdorferi transcribes significantly higher levels of ebfC mRNA while within feeding ticks or during mammalian infection than while colonizing unfed ticks (44). This parallels the pattern of erp transcription during the borrelial mammal-tick infectious cycle (10, 31, 48, 49). The present studies indicate that EbfC counteracts repression by BpaB in vitro, and overexpression of EbfC in B. burgdorferi increases levels of Erp proteins. Moreover, mutation of the erp operator EbfC-binding sites to nonconsensus sequences resulted in very low transcription levels in B. burgdorferi. Together, these data indicate that EbfC is an antirepressor of erp transcription (Fig. 6B). EbfC by itself did not increase erp expression and therefore does not appear to be a transcriptional activator of erp operons.
Several lines of evidence indicate that both of these DNA-binding proteins carry out additional functions for B. burgdorferi. All relapsing-fever Borrelia species lack erp operons, yet they encode BpaB and EbfC proteins that are nearly identical to those of the Lyme disease spirochetes (61). BpaB plays a role in maintenance of the Erp-encoding cp32 prophages, possibly analogous to the ParB/SopB proteins of other bacterial replicons (9, 21, 65, 66). Related to that point, several such plasmid maintenance proteins also function as transcriptional repressors through mechanisms like that proposed for BpaB and the erp operons (8, 20, 39, 53, 54). Although the spirochete phylum diverged from the rest of the kingdom Eubacteria many millions of years ago, the retention of EbfC homologs by almost all members of the Eubacteria suggests that these proteins perform central, critical functions for many prokaryotes. Several characteristics of EbfC indicate that it is a type of nucleoid-associated (“histone-like”) protein (52; our unpublished results). The ebfC gene is located on the B. burgdorferi chromosome, while erp operons are located on cp32 prophages. These bacteriophages have evidently commandeered a spirochetal protein for their own devices, joining the long list of viruses that utilize host factors as regulators.
It is notable that erp operon transcription is influenced by two distinct proteins, in that erp transcription will be dependent upon relative levels of each DNA-binding protein in the bacterial cell. Moreover, the additional functions of BpaB and EbfC indicate that their effects on erp transcription will be dependent upon cellular levels of free proteins, that is, protein that is not bound elsewhere in the genome. For example, high concentrations of free BpaB in the cytoplasm could result in either erp repression or derepression, depending upon whether free EbfC levels were low or high, respectively.
The conservation of BpaB- and EbfC-binding sites in all known erp operator elements indicates that both proteins will likely exert influence on transcription of all erp operons (1, 5, 6, 33, 43, 60, 64). Yet other factors can also affect levels of erp expression. As is evident from the DNA alignments illustrated in Fig. 1 and elsewhere (1, 5, 6, 33, 43, 60, 64), the −10 and −35 sequences of erp promoters exhibit considerable diversity, which can impact promoter activity and transcript levels (6, 23). Moreover, some erp promoter variants are poorly recognized by the borrelial housekeeping sigma factor RpoD but are better recognized by another sigma factor, RpoS (so called because its sequence resembles that of E. coli RpoS, although the B. burgdorferi RpoS is not associated with stress response) (14, 15, 22, 23, 27). Available data suggest that the B. burgdorferi RpoS-RNA polymerase holoenzyme recognizes the same promoter sequences as does the RpoD-containing enzyme and also variants thereof that are not recognized by RpoD, similar to the E. coli RpoS sigma factor (14, 15, 22, 23, 27, 30, 35, 67). It is not clear whether the variations in erp promoter sequences are due to degenerating transcriptional elements of unnecessary or defective genes, physiological benefits reaped from expressing some operons using both RpoD and RpoS but others using only RpoS, or combinations of those two causes. Very little is known about the mechanistic details of interactions between B. burgdorferi RpoS and erp promoters, since the majority of published studies purporting to be on that topic actually examined the distinct E. coli RpoS sigma factor (22, 23). However, a study of erp promoter-gfp fusions in B. burgdorferi strain 297 found that one of that strain's erp loci, named ospF, was not recognized by borrelial RpoD but was dependent upon borrelial RpoS, while another locus, named ospE, did utilize RpoD (23). Both of those strain 297 operons contain consensus BpaB- and EbfC-binding sites in their operators and are very likely to also be influenced by the two DNA-binding proteins. To argue otherwise requires that one invoke a mechanism by which BpaB and EbfC are excluded from a subset of erp operators, a complexity for which no evidence has been found. Intriguingly, a chimeric promoter consisting of the strain 297 ospF −35 and ospE −10 sequences was not functional in either wild-type or rpoS B. burgdorferi (23), indicating sequence defects in some promoters that, to the best of our knowledge, have yet to be explored. In addition, the metabolite 4,5-dihydroxy-2,3-pentanedione, also known as autoinducer 2 or AI-2, enhances expression of Erp proteins through an unknown mechanism (57, 68). It is worthwhile to note that neither RpoS nor AI-2 has been tested for its ability to control erp expression during actual mammal infection or tick colonization. Yet the effects of those factors on cultured B. burgdorferi strongly suggest that they, and BpaB and EbfC, do perform regulatory roles in nature.
Previous studies and our further ChIP analyses and global RNA sequencing indicate that both BpaB and EbfC bind sites throughout the borrelial genome, in addition to erp operator elements, and control production of numerous other bacterial genes (44; our unpublished results). Defining the mechanisms by which the Lyme disease spirochete controls the relative levels of the BpaB and EbfC DNA-binding proteins will provide further insight into how this pathogen senses and adapts to changes throughout its vertebrate-arthropod infectious cycle. In addition, the near ubiquity of EbfC proteins among diverse bacterial species implies that studies of the borrelial ortholog will have ramifications throughout the kingdom Eubacteria.
ACKNOWLEDGMENTS
This work was funded by National Institutes of Health grants R01-AI044254 to Brian Stevenson and R01-AI063261 to Wolfram Zückert.
We thank Greg Bowman and Jennifer Strange for assistance with flow cytometry analyses, Jay Carroll for providing the PospAB::gfp fusion construct, and Kelly Babb, Gavin Ellis, Michael Fried, Jason Johnston, Jennifer Miller, Heather O'Daniel, and Brett Spear for their help and insightful comments on this work and the manuscript.
Footnotes
Published ahead of print 9 December 2011
REFERENCES
- 1. Akins DR, et al. 1999. Molecular and evolutionary analysis of Borrelia burgdorferi 297 circular plasmid-encoded lipoproteins with OspE- and OspF-like leader peptides. Infect. Immun. 67:1526–1532 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Alitalo A, et al. 2002. Complement inhibitor factor H binding to Lyme disease spirochetes is mediated by inducible expression of multiple plasmid-encoded outer surface protein E paralogs. J. Immunol. 169:3847–3853 [DOI] [PubMed] [Google Scholar]
- 3. Antonara S, Chafel RM, LaFrance M, Coburn J. 2007. Borrelia burgdorferi adhesins identified using in vivo phage display. Mol. Microbiol. 66:262–276 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Babb K, et al. 2006. Borrelia burgdorferi EbfC, a novel, chromosomally-encoded protein, binds specific DNA sequences adjacent to erp loci on the spirochete's resident cp32 prophages. J. Bacteriol. 188:4331–4339 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Babb K, El-Hage N, Miller JC, Carroll JA, Stevenson B. 2001. Distinct regulatory pathways control the synthesis of Borrelia burgdorferi infection-associated OspC and Erp surface proteins. Infect. Immun. 69:4146–4153 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Babb K, McAlister JD, Miller JC, Stevenson B. 2004. Molecular characterization of Borrelia burgdorferi erp promoter/operator elements. J. Bacteriol. 186:2745–2756 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Barbour AG, Hayes SF, Heiland RA, Schrumpf ME, Tessier SL. 1986. A Borrelia-specific monoclonal antibody binds to a flagellar epitope. Infect. Immun. 52:549–554 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Bartosik AA, Lasocki K, Mierzejewska J, Thomas CM, Jagura-Burdzy G. 2004. ParB of Pseudomonas aeruginosa: interactions with its partner ParA and its target parS and specific effects on bacterial growth. J. Bacteriol. 186:6983–6998 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Beaurepaire C, Chaconas G. 2005. Mapping of essential replication functions of the linear plasmid lp17 of B. burgdorferi by targeted deletion walking. Mol. Microbiol. 57:132–142 [DOI] [PubMed] [Google Scholar]
- 10. Brissette CA, et al. 2008. Lyme borreliosis spirochete Erp proteins, their known host ligands, and potential roles in mammalian infection. Int. J. Med. Microbiol. 298(Suppl 1):257–267 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Brissette CA, et al. 2009. Borrelia burgdorferi infection-associated surface proteins ErpP, ErpA, and ErpC bind human plasminogen. Infect. Immun. 77:300–306 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Brissette CA, Verma A, Bowman A, Cooley AE, Stevenson B. 2009. The Borrelia burgdorferi outer-surface protein ErpX binds mammalian laminin. Microbiology 155:863–872 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Burns LH, et al. 2010. BpaB, a novel protein encoded by the Lyme disease spirochete's cp32 prophages, binds to erp Operator 2 DNA. Nucleic Acids Res. 38:5443–5455 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Caimano MJ, Eggers CH, Hazlett KRO, Radolf JD. 2004. RpoS is not central to the general stress response in Borrelia burgdorferi but does control expression of one or more essential virulence determinants. Infect. Immun. 72:6433–6445 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Caimano MJ, et al. 2007. Analysis of the RpoS regulon in Borrelia burgdorferi in response to mammalian host signals provides insight into RpoS function during the enzootic cycle. Mol. Microbiol. 65:1193–1217 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Carroll JA, Stewart PE, Rosa P, Elias AF, Garon CF. 2003. An enhanced GFP reporter system to monitor gene expression in Borrelia burgdorferi. Microbiology 149:1819–1828 [DOI] [PubMed] [Google Scholar]
- 17. Casjens S, et al. 2000. A bacterial genome in flux: the twelve linear and nine circular extrachromosomal DNAs of an infectious isolate of the Lyme disease spirochete Borrelia burgdorferi. Mol. Microbiol. 35:490–516 [DOI] [PubMed] [Google Scholar]
- 18. Casjens S, van Vugt R, Tilly K, Rosa PA, Stevenson B. 1997. Homology throughout the multiple 32-kilobase circular plasmids present in Lyme disease spirochetes. J. Bacteriol. 179:217–227 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Cooley AE, et al. 2009. DNA-binding by Haemophilus influenzae and Escherichia coli YbaB, members of a widely-distributed bacterial protein family. BMC Microbiol. 9:137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Ebersbach G, Gerdes K. 2005. Plasmid segregation mechanisms. Annu. Rev. Genet. 39:453–479 [DOI] [PubMed] [Google Scholar]
- 21. Eggers CH, et al. 2002. Identification of loci critical for replication and compatibility of a Borrelia burgdorferi cp32 plasmid and use of a cp32-based shuttle vector for the expression of fluorescent reporters in the Lyme disease spirochaete. Mol. Microbiol. 43:281–295 [DOI] [PubMed] [Google Scholar]
- 22. Eggers CH, Caimano MJ, Radolf JD. 2004. Analysis of promoter elements involved in the transcription initiation of RpoS-dependent Borrelia burgdorferi genes. J. Bacteriol. 186:7390–7402 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Eggers CH, Caimano MJ, Radolf JD. 2006. Sigma factor selectivity in Borrelia burgdorferi: RpoS recognition of the ospE/ospF/elp promoters is dependent on the sequence of the −10 region. Mol. Microbiol. 59:1859–1875 [DOI] [PubMed] [Google Scholar]
- 24. Eggers CH, Samuels DS. 1999. Molecular evidence for a new bacteriophage of Borrelia burgdorferi. J. Bacteriol. 181:7308–7313 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. El-Hage N, et al. 2001. Surface exposure and protease insensitivity of Borrelia burgdorferi Erp (OspEF-related) lipoproteins. Microbiology 147:821–830 [DOI] [PubMed] [Google Scholar]
- 26. El-Hage N, Stevenson B. 2002. Simultaneous coexpression of Borrelia burgdorferi Erp proteins occurs through a specific, erp locus-directed regulatory mechanism. J. Bacteriol. 184:4536–4543 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Elias AF, et al. 2000. Altered stationary-phase response in a Borrelia burgdorferi rpoS mutant. J. Bacteriol. 182:2909–2918 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Fang FC, Rimsky S. 2008. New insights into transcriptional regulation by H-NS. Curr. Opin. Microbiol. 11:113–120 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Fraser CM, et al. 1997. Genomic sequence of a Lyme disease spirochaete, Borrelia burgdorferi. Nature 390:580–586 [DOI] [PubMed] [Google Scholar]
- 30. Gaal T, et al. 2001. Promoter recognition and discrimination by EσS RNA polymerase. Mol. Microbiol. 42:939–954 [DOI] [PubMed] [Google Scholar]
- 31. Gilmore RD, Jr, Mbow ML, Stevenson B. 2001. Analysis of Borrelia burgdorferi gene expression during life cycle phases of the tick vector Ixodes scapularis. Microbes Infect. 3:799–808 [DOI] [PubMed] [Google Scholar]
- 32. Hefty PS, Jolliff SE, Caimano MJ, Wikel SK, Akins DR. 2002. Changes in the temporal and spatial patterns of outer surface lipoprotein expression generate population heterogeneity and antigenic diversity in the Lyme disease spirochete, Borrelia burgdorferi. Infect. Immun. 70:3468–3478 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Hefty PS, et al. 2001. Regulation of OspE-related, OspF-related, and Elp lipoproteins of Borrelia burgdorferi strain 297 by mammalian host-specific signals. Infect. Immun. 69:3618–3627 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Hellwage J, et al. 2001. The complement regulatory factor H binds to the surface protein OspE of Borrelia burgdorferi. J. Biol. Chem. 276:8427–8435 [DOI] [PubMed] [Google Scholar]
- 35. Hengge-Aronis R. 2002. Stationary phase gene regulation: what makes an Escherichia coli promoter σS-selective? Curr. Opin. Microbiol. 5:591–595 [DOI] [PubMed] [Google Scholar]
- 36. Ho SN, Hunt HD, Horton RM, Pullen JK, Pease LR. 1989. Site-directed mutagensis by overlap extension using polymerase chain reaction. Gene 77:51–59 [DOI] [PubMed] [Google Scholar]
- 37. Hovis KM, et al. 2006. Selective binding of Borrelia burgdorferi OspE paralogs to factor H and serum proteins from diverse animals: possible expansion of the role of OspE in Lyme disease pathogenesis. Infect. Immun. 74:1967–1972 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Jacob F, Monod J. 1961. Genetic regulatory mechanisms in the synthesis of proteins. J. Mol. Biol. 3:318–356 [DOI] [PubMed] [Google Scholar]
- 39. Kalnin K, Stegalkina S, Yarmolinsky M. 2000. pTAR-encoded proteins in plasmid partitioning. J. Bacteriol. 182:1889–1894 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Kenedy MR, Akins DR. 2011. The OspE-related proteins inhibit complement deposition and enhance serum resistance of Borrelia burgdorferi, the Lyme disease agent. Infect. Immun. 79:1451–1457 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Lam TT, et al. 1994. Outer surface proteins E and F of Borrelia burgdorferi, the agent of Lyme disease. Infect. Immun. 62:290–298 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Lang B, et al. 2007. High-affinity DNA binding sites for H-NS provide a molecular basis for selective silencing within proteobacterial genomes. Nucleic Acids Res. 35:6330–6337 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Marconi RT, Sung SY, Hughes CAN, Carlyon JA. 1996. Molecular and evolutionary analyses of a variable series of genes in Borrelia burgdorferi that are related to ospE and ospF, constitute a gene family, and share a common upstream homology box. J. Bacteriol. 178:5615–5626 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Medrano MS, et al. 2007. Regulators of expression of the oligopeptide permease A proteins of Borrelia burgdorferi. J. Bacteriol. 189:2653–2659 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Metts MS, McDowell JV, Theisen M, Hansen PR, Marconi RT. 2003. Analysis of the OspE determinants involved in binding of factor H and OspE-targeting antibodies elicited during Borrelia burgdorferi infection. Infect. Immun. 71:3587–3596 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Miller JC. 2005. Example of real-time quantitative reverse transcription-PCR (Q-RT-PCR) analysis of bacterial gene expression during mammalian infection: Borrelia burgdorferi in mouse tissues. Curr. Protoc. Microbiol. 2005:1D.3 [DOI] [PubMed] [Google Scholar]
- 47. Miller JC, Narayan K, Stevenson B, Pachner AR. 2005. Expression of Borrelia burgdorferi erp genes during infection of non-human primates. Microb. Pathog. 39:27–33 [DOI] [PubMed] [Google Scholar]
- 48. Miller JC, von Lackum K, Babb K, McAlister JD, Stevenson B. 2003. Temporal analysis of Borrelia burgdorferi Erp protein expression throughout the mammal-tick infectious cycle. Infect. Immun. 71:6943–6952 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Miller JC, von Lackum K, Woodman ME, Stevenson B. 2006. Detection of Borrelia burgdorferi gene expression during mammalian infection using transcriptional fusions that produce green fluorescent protein. Microb. Pathog. 41:43–47 [DOI] [PubMed] [Google Scholar]
- 50. Miller JH, Reznikoff WS. 1980. The operon. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY [Google Scholar]
- 51. Radolf JD, Caimano MJ, Stevenson B, Hu LT. Of ticks, mice, and men: understanding the two host lifestyles of Lyme disease spirochetes. Nat. Rev. Microbiol., in press [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Riley SP, et al. 2009. Borrelia burgdorferi EbfC defines a newly-identified, widespread family of bacterial DNA-binding proteins. Nucleic Acids Res. 37:1973–1983 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Rodionov O, Lobocka M, Yarmolinsky M. 1999. Silencing of genes flanking the P1 plasmid centromere. Science 283:546–549 [DOI] [PubMed] [Google Scholar]
- 54. Sawitzke JA, et al. 2002. Transcriptional interference by a complex formed at the centromere-like partition site of plasmid P1. J. Bacteriol. 184:2447–2454 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Stanek G, Strle F. 2003. Lyme borreliosis. Lancet 362:1639–1647 [DOI] [PubMed] [Google Scholar]
- 56. Steere AC, Coburn J, Glickstein L. 2004. The emergence of Lyme disease. J. Clin. Invest. 113:1093–1101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Stevenson B, Babb K. 2002. LuxS-mediated quorum sensing in Borrelia burgdorferi, the Lyme disease spirochete. Infect. Immun. 70:4099–4105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Stevenson B, Bono JL, Schwan TG, Rosa P. 1998. Borrelia burgdorferi Erp proteins are immunogenic in mammals infected by tick bite, and their synthesis is inducible in cultured bacteria. Infect. Immun. 66:2648–2654 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Stevenson B, El-Hage N, Hines MA, Miller JC, Babb K. 2002. Differential binding of host complement inhibitor factor H by Borrelia burgdorferi Erp surface proteins: a possible mechanism underlying the expansive host range of Lyme disease spirochetes. Infect. Immun. 70:491–497 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Stevenson B, Miller JC. 2003. Intra- and interbacterial genetic exchange of Lyme disease spirochete erp genes generates sequence identity amidst diversity. J. Mol. Evol. 57:309–324 [DOI] [PubMed] [Google Scholar]
- 61. Stevenson B, et al. 2000. The relapsing fever spirochete Borrelia hermsii contains multiple, antigen-encoding circular plasmids that are homologous to the cp32 plasmids of Lyme disease spirochetes. Infect. Immun. 68:3900–3908 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Stevenson B, Schwan TG, Rosa PA. 1995. Temperature-related differential expression of antigens in the Lyme disease spirochete, Borrelia burgdorferi. Infect. Immun. 63:4535–4539 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Stevenson B, Tilly K, Rosa PA. 1996. A family of genes located on four separate 32-kilobase circular plasmids in Borrelia burgdorferi B31. J. Bacteriol. 178:3508–3516 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Stevenson B, Zückert WR, Akins DR. 2001. Repetition, conservation, and variation: the multiple cp32 plasmids of Borrelia species, p 87–100 In Saier MH, García-Lara J. (ed), The spirochetes: molecular and cellular biology. Horizon Press, Wymondham, United Kingdom [Google Scholar]
- 65. Stewart PE, Chaconas G, Rosa P. 2003. Conservation of plasmid maintenance functions between linear and circular plasmids in Borrelia burgdorferi. J. Bacteriol. 185:3202–3209 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Stewart PE, Thalken R, Bono JL, Rosa P. 2001. Isolation of a circular plasmid region sufficient for autonomous replication and transformation of infectious Borrelia burgdorferi. Mol. Microbiol. 39:714–721 [DOI] [PubMed] [Google Scholar]
- 67. Typas A, Hengge R. 2006. Role of the spacer between the −35 and −10 regions in sigmas promoter selectivity in Escherichia coli. Mol. Microbiol. 59:1037–1051 [DOI] [PubMed] [Google Scholar]
- 68. von Lackum K, et al. 2006. Functionality of Borrelia burgdorferi LuxS: the Lyme disease spirochete produces and responds to the pheromone autoinducer-2 and lacks a complete activated-methyl cycle. Int. J. Med. Microbiol. 296(Suppl 40):92–102 [DOI] [PubMed] [Google Scholar]
- 69. Wade JT, Reppas NB, Church GM, Struhl K. 2005. Genomic analysis of LexA binding reveals the permissive nature of the Escherichia coli genome and identifies unconventional target sites. Genes Develop. 19:2619–2630 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Wade JT, Struhl K. 2004. Association of RNA polymerase with transcribed regions in Escherichia coli. Proc. Natl. Acad. Sci. U. S. A. 101:17777–17782 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Whetstine CR, Slusser JG, Zückert WR. 2009. Development of a single-plasmid-based regulatable gene expression system for Borrelia burgdorferi. Appl. Environ. Microbiol. 75:6553–6558 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Zhang H, Marconi RT. 2005. Demonstration of cotranscription and 1-methyl-3-nitroso-nitroguanidine induction of a 30-gene operon of Borrelia burgdorferi: evidence that the 32-kilobase circular plasmids are prophages. J. Bacteriol. 187:7985–7995 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Zückert WR. 2007. Laboratory maintenance of Borrelia burgdorferi. Curr. Protoc. Microbiol. 12C:1–10 [DOI] [PubMed] [Google Scholar]