Abstract
Methanosarcina acetivorans strain C2A is a marine methanogenic archaeon notable for its substrate utilization, genetic tractability, and novel energy conservation mechanisms. To help probe the phenotypic implications of this organism's unique metabolism, we have constructed and manually curated a genome-scale metabolic model of M. acetivorans, iMB745, which accounts for 745 of the 4,540 predicted protein-coding genes (16%) in the M. acetivorans genome. The reconstruction effort has identified key knowledge gaps and differences in peripheral and central metabolism between methanogenic species. Using flux balance analysis, the model quantitatively predicts wild-type phenotypes and is 96% accurate in knockout lethality predictions compared to currently available experimental data. The model was used to probe the mechanisms and energetics of by-product formation and growth on carbon monoxide, as well as the nature of the reaction catalyzed by the soluble heterodisulfide reductase HdrABC in M. acetivorans. The genome-scale model provides quantitative and qualitative hypotheses that can be used to help iteratively guide additional experiments to further the state of knowledge about methanogenesis.
INTRODUCTION
Methanogenic archaea are unique in their ability to grow on low-energy substrates, such as acetic acid, by converting them into methane and other by-products. Methanogens are a critical part of the global carbon cycle, consuming by-products of other natural bioprocesses that would otherwise be recalcitrant in sulfate-poor, anaerobic environments (12). They also play an important role in global warming, since methane is a greenhouse gas 20 times as potent as carbon dioxide (42) and methanogenesis is the primary mechanism for the emission of methane into the atmosphere (2).
Methanosarcina is the only known genus of methanogens with members that can utilize all of the known methanogenic pathways (acetoclastic, methylotrophic, hydrogenotrophic, and methyl reducing) (71). This metabolic diversity makes these species more permissive to metabolic and genetic manipulations than other methanogens. To capitalize on this characteristic, the genomes of three Methanosarcina species have been sequenced (15, 22, 38). In addition, genetic tools have been developed for several of these species, including methods for directed mutagenesis and regulated expression of specific genes (3, 34, 73, 74).
The constraint-based reconstruction and analysis (COBRA) strategy is a powerful paradigm for consolidating large amounts of metabolic knowledge and synthesizing that knowledge into quantitative phenotypic predictions (45, 51). For the performance of constraint-based analysis on an individual organism, its metabolic network is reconstructed from the bottom up, beginning with a sequenced and annotated genome and ending with a network of reactions and reaction-gene associations that directly link genotype and phenotype (68). Many metabolic reconstructions have been curated by hand and have been used to make useful predictions, such as the identification of putative drug targets and the design of novel strains for enhanced biofuel production (45).
In recent years, there have been major advances toward automating much of the reconstruction process (26). Automation is needed to continue the exponential increase in the number of genome-scale metabolic models (43, 45). However, automated reconstructions for methanogens are still particularly problematic for three major reasons: (i) automated predictions tend to be overly homogenized due to their strong reliance on homology-based methods; (ii) reaction and gene databases have more limited coverage of the Archaea than of the other domains of life; and (iii) the energy conservation mechanisms of methanogens are highly specialized (14). Hence, manual curation is necessary in order to obtain reliable predictions from metabolic models of these organisms.
Among methanogens, Methanosarcina acetivorans is notable for its substrate utilization. It can grow and produce methane by using methylated substrates, carbon monoxide, or acetate, but it cannot grow with hydrogen as its primary energy source (58). Also, unlike most methanogens, M. acetivorans is genetically tractable. Therefore, this organism offers opportunities to learn about novel energy conservation mechanisms.
An independent reconstruction for M. acetivorans strain C2A has been reported recently (32). The previously reported reconstruction was curated primarily by using an automated curation pipeline, including the GapFind, GapFill, and GrowMatch algorithms (31, 33). Here we present iMB745, an extensively curated manual reconstruction that differs significantly from the other model published. As many literature sources as were available were integrated to generate a highly accurate list of metabolic reactions, making this reconstruction a valuable knowledge base for this organism. In addition, curation has enabled us to make quantitative phenotypic predictions using constraint-based modeling. We demonstrate the usefulness of this model for probing hypotheses about the workings of incompletely understood parts of the M. acetivorans metabolic network. The analysis thus represents a successful application of the hypothesis-driven modeling approach.
MATERIALS AND METHODS
Model reconstruction.
An initial list of potential reaction-gene associations in Methanosarcina acetivorans strain C2A was generated based on the integration of data from the Kyoto Encyclopedia of Genes and Genomes (KEGG) (30), MetaCyc (13), the Model SEED reconstruction (26), the Transport Protein Analysis Database (TransportDB) (53), and UniProt (70). Reactions from the existing Methanosarcina barkeri strain Fusaro reconstruction (19) and the BiGG database (57) were added if there was sufficient evidence for their inclusion, based on sequence homology and/or literature-based curation, or to fill gaps in the annotation. Some gene suggestions from EFICAZ, which includes evidence from other bioinformatics tools, such as PFAM, were also incorporated (4). Gene associations were verified whenever possible by using bidirectional BLASTP against archaeal protein products with experimentally verified functions (11). In case of conflicts, metabolic functions suggested from the literature were chosen over those suggested in the databases, and inconsistent reactions were removed from the model (see the supplemental material for a comprehensive list). An SBML file containing the iMB745 model may be downloaded at http://price.systemsbiology.net/downloads_tmp.php.
Reaction and metabolite nomenclature consistent with the BiGG database was utilized whenever possible to facilitate comparison with existing manually curated metabolic models (57). Reactions and metabolites without BiGG identifiers were assigned abbreviations in a manner similar to that used for the database (see the supplemental material for a complete list).
Logical gene-protein reaction (GPR) relationships were constructed manually on the basis of literature or database evidence. For example, genes annotated or characterized as separate subunits of a complex were given an “AND” relationship. If there was no evidence of a protein complex catalyzing a reaction with multiple genes, the genes were all assigned an OR relationship.
All intracellular and transport reactions were computationally mass and charge balanced at a pH of 7 based on charges and formulas computed with ACD/Labs software (version 12; Advanced Chemistry Development, Inc.). Charges and formulas are available in the supplemental material.
Construction of the biomass reaction.
The biomass reaction is a sink for essential cell components that represents the consumption of molecular building blocks (such as amino acids and nucleotides) required for cell division. The biomass reaction for Methanosarcina acetivorans strain C2A was modified from that of the closest relative for which a biomass reaction had previously been built, Methanosarcina barkeri strain Fusaro (19). This biomass objective function was first expanded by incorporating more-detailed carbohydrate data from M. barkeri (29) and adding methanofuran-B to the list of required cofactors (40). Then coefficients for lipids were modified on the basis of available data on the unique lipid composition of M. acetivorans (61). Nucleotide and amino acid coefficients specific to M. acetivorans were calculated based on the published genome sequence according to established procedures (68). The coefficients of compounds in the soluble pool were assumed to be the same as those in the M. barkeri biomass equation.
FBA.
Exponential-growth phenotypes were predicted using flux balance analysis (FBA), which has been reviewed previously (49). Briefly, all reactions in the model were represented in a stoichiometric matrix, S, in which each column represented a reaction and each row a metabolite. Hence, the entry (i,j) of S contained the stoichiometric coefficient of metabolite i in reaction j. If metabolite concentrations are assumed to be constant (steady state), conservation of mass requires that Sv be equal to zero, where v is the vector of reaction fluxes (reaction rates). Because there were more reactions than metabolites in the model (as is typical), multiple flux distributions, all of which satisfied the mass balance, were possible.
Reaction fluxes were also constrained by setting minimum and maximum fluxes. In the current study, the reversibility of each reaction was determined based on the literature, database evidence, and thermodynamic calculations. The flux through reversible reactions was unconstrained, while that of irreversible reactions was set to have a minimum v (vmin) of zero. Substrate uptake rates were set to experimentally measured values for purposes of simulations (see the supplemental material for values and references). The reaction rate through the non-growth-associated ATP maintenance reaction (ATPM) was set to 2.5 mmol/g (dry weight)/h to account for upkeep energy costs. This value is somewhat lower than the experimental value of 8.39 mmol/g (dry weight)/h used in the current Escherichia coli model and larger than that in the published M. barkeri model (19). Growth-associated ATP maintenance was included in the biomass equation and was set to 65 mmol/g (dry weight), similar to the values in the M. barkeri and E. coli FBA models (16, 19), to account for energy costs for growth (such as the production of macromolecules from biomass components). Both growth-associated maintenance (GAM) and non-growth-associated maintenance (NGAM) costs were chosen to best match experimentally measured growth and secretion rates. The NGAM is about 1.5 mmol/g (dry weight)/h more than that of the previously published M. barkeri model (19), primarily because ion-pumping inefficiencies for membrane-bound pumps such as Fpo were lumped into the NGAM rather than explicitly stated in the reaction stoichiometry. Detailed calculations and references related to the biomass equation are available in the supplemental material.
Under the assumption that the cell seeks to maximize its growth potential, the specific growth rate was predicted by maximizing the flux through the biomass reaction (max vbiomass) subject to the aforementioned constraints: Sv = 0 and vmin ≤ v ≤ vmax.
Reaction fluxes were predicted in millimoles per gram (dry weight) per hour, and growth rates were predicted per hour. FBA problems were solved using the COBRA toolbox in MATLAB (7) linked to the GLPK linear program solver. Simulations were also repeated using the CPLEX package via the TOMLAB (version 7.0) interface, with identical results. A defined high-salt (HS) medium without a vitamin supplement was used for all simulations. The complete medium composition is listed in the supplemental material; the medium was defined on the basis of the work of Sowers et al. (59).
FVA.
FBA does not necessarily yield a unique flux distribution, although it will yield a unique optimal value for the objective function. Flux variability analysis (FVA) was thus used to calculate the possible range of each flux under optimal growth conditions. Mathematically, the possible range of flux through each reaction i was calculated by maximizing and minimizing its flux (vi) while constraining the objective to be larger than a certain threshold (39). This is performed by minimizing or maximizing vi subject to the following constraints: Sv = 0, vmin ≤ v ≤ vmax, and vbiomass ≥ pct·vbiomass,MAX, where pct is a percentage of maximum biomass production within which the flux range is to be calculated.
All flux variability analyses in this paper were performed with a pct of 100% (the biomass objective function was fixed to its maximum value, within rounding error). Flux variability analysis was performed using the fluxVariability function in the COBRA toolbox (7).
Knockout lethality studies.
For knockout lethality studies, knocked out genes were assigned a value of zero (false) and other genes were assigned a value of 1 (true). The Boolean GPRs were evaluated for every reaction, and reactions with a GPR evaluated as false were removed from the model. After the network was modified in this way, FBA was used to make a growth or no-growth prediction (growth was defined as a predicted vbiomass of >10−5 h−1) for all knockouts of metabolic genes. Lethality predictions were compared to published gene knockout phenotype data (see the supplemental material for references). For substrates with unknown uptake rates (such as monomethylamine), the uptake rate was assumed to be 15 mmol/g (dry weight)/h, similar to the calculated rate for growth on methanol (63), for purposes of FBA simulations.
Calculation of theoretical ATP yield and thermodynamic efficiency.
To calculate the theoretical (maximum possible) ATP yield under various conditions, a flux balance analysis problem was solved, but the flux through the non-growth-associated maintenance (ATPM) reaction was maximized instead of flux through the biomass equation. If necessary, the model was forced to carry flux through a specific ATP-generating pathway (such as acetogenesis) by adding constraints on other ATP-generating pathways (such as methanogenesis). The theoretical ATP yield was calculated by dividing the maximum flux through the ATP maintenance reaction by the CO uptake rate. Thermodynamic efficiency was calculated as the ratio of the theoretical ATP yield predicted by the model and the ATP yield that would be observed if the entire Gibbs energy available from the production of methane and CO2 from a given substrate (under standard conditions) were used to generate ATP (66).
Thermodynamics.
Experimental standard Gibbs free energy changes (ΔG) were unavailable for most of the reactions in the network, but they were available and were included for some methanogenesis reactions (66, 67) and some reactions involved in central metabolism (1) based on experimental Gibbs free energies of formation. When available, the free energy changes were used to help make decisions about reaction reversibility (in combination with direct experimental or modeling evidence).
To estimate the standard Gibbs free energy changes of reactions for which no experimental data were available, Mol files were generated to represent all compounds in the model containing charged structures at a pH of 7. The Mol files contain the structures and locations of charged moieties in each compound in the model. Charges were computed, and charged Mol files were exported, using ACD/Labs software (version 12; Advanced Chemistry Development, Inc.). The Gibbs energy of each compound was estimated using a previously published group contribution method (28). Standard Gibbs free energies of formation and reaction are reported in the supplemental material under the following conditions: temperature, 25°C; pH 7; ionic strength, zero; water in the liquid phase; and all other compounds in the aqueous phase at a concentration of 1 M.
RESULTS
Model reconstruction.
The metabolic network of Methanosarcina acetivorans was curated and validated as described in Materials and Methods. The final network accounts for the activity of 745 metabolic genes and contains 715 intracellular metabolites and 756 reactions (excluding exchange and biomass reactions). The network is considerably larger than that in the manually curated model of M. barkeri (19) and is comparable in size to that found in other genome-scale metabolic reconstructions (17). In addition to reactions included in the model, reactions that were specifically excluded from the model due to literature or modeling evidence were also recorded. Complete lists of reactions included, GPR relationships, and excluded reactions may be found in the supplemental material.
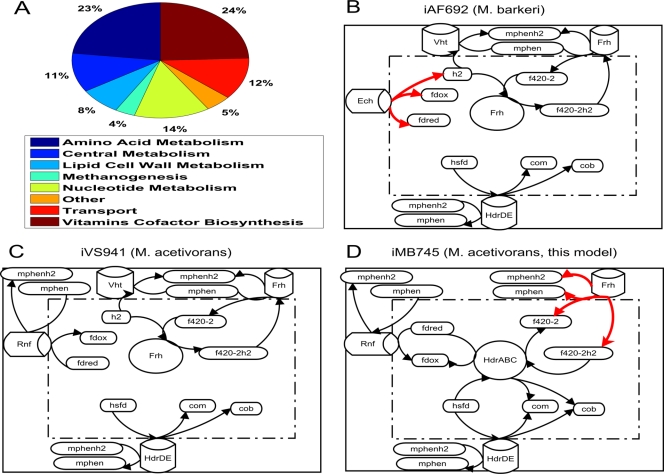
The metabolic network of M. acetivorans consists mostly of reactions required for the synthesis of amino acids, nucleotides, and cofactors (Fig. 1A). This was not surprising given the relatively small number of growth factors required for the growth of this organism. The reconstructed network includes pathways for synthesizing most of the cofactors required for methanogenesis in Methanosarcina species. The exception is methanophenazine, for which, to our knowledge, no complete synthesis pathway has been proposed in any organism. Synthesis pathways were also included for several other cofactors, such as NAD, biotin, flavins, and folate.
Fig 1.
Properties of the Methanosarcina acetivorans model and comparison to other existing Methanosarcina models. (A) After curation, the metabolic model contained reactions related to the synthesis of essential biomass components, cell wall components, and methanogenesis, among others. (B to D) Comparison of the electron transport chains in the three available Methanosarcina models. Reactions indicated by red arrows are reversible in that model. Note that the curated M. acetivorans model includes the Rnf complex and the soluble heterodisulfide reductase (HdrABC) and excludes Frh and Vht, two hydrogenases known to be inactive in M. acetivorans but present and active in M. barkeri. mphen, oxidized methanophenazine; mphenh2, reduced methanophenazine; hsfd, heterodisulfide; com, coenzyme M; cob, coenzyme B.
There are still significant gaps in the knowledge of central metabolic pathways. For example, no homologues to currently known IMP dehydrogenase genes could be found in the genome of M. acetivorans, but the reaction catalyzed by this enzyme is predicted to be essential for nucleic acid synthesis. In addition, several of the methanogenic cofactor synthesis pathways that are completely or partially characterized in Methanocaldococcus jannaschii seem to have diverged in Methanosarcina. Although this is perhaps not surprising given the great evolutionary distance between Methanocaldococcus and Methanosarcina, the identification of these differences could provide a motivation for further investigation into the evolution of these species. A detailed discussion of these differences and other identified gaps in metabolic pathways is provided in the supplemental text.
Comparison to existing genome-scale reconstructions of Methanosarcina species.
The iMB745 model is the third model of members of the genus Methanosarcina to be published. The first was iAF692, a manually curated model of M. barkeri strain Fusaro (19). The iAF692 model was used to estimate the ion-pumping stoichiometry of the Ech hydrogenase and the ATP requirements of nitrogenase in that organism. The second was iVS941, a genome-scale model of M. acetivorans C2A heavily based on automated approaches (32). The iVS941 model was used to study the essentiality of methanogenesis pathways during growth on CO, acetate, and methanol and to predict ways to reconcile simulated knockout lethality predictions with available data.
Many of the differences between iMB745 and each of the previously published models are due to new literature sources for novel metabolic paths unique to the archaea. For example, both of the previously published models include the nonoxidative portion of the pentose phosphate pathway for the synthesis of five-carbon sugars. However, the genes encoding proteins contributing to reactions in that pathway are apparently absent in many methanogens, including both M. acetivorans and M. barkeri. An alternative pathway for the synthesis of ribulose-5-phosphate was recently characterized in Methanocaldococcus jannaschii (23); unlike the pentose phosphate pathway, this pathway generates formaldehyde as a by-product. The genes involved in this pathway had strong homology to genes in M. acetivorans. Therefore, the reactions in the pentose phosphate pathway were excluded from the iMB745 model, and the new pathway was added to the model. Other gaps in the previously existing models were also filled based on recent literature (see the supplemental text for details).
The iMB745 model accounts for key differences in methanogenesis pathways between M. acetivorans and M. barkeri (Fig. 1B to D). Critically, the Fpo and Vht hydrogenases are not functional in M. acetivorans, as has been shown experimentally (24), even though sequence homology suggests that both are present. Due to the strong sequence identity between the M. acetivorans and M. barkeri homologues, the automated reconstruction approach for the previous M. acetivorans reconstruction incorrectly included these reactions in the model. The automated model also did not include the recently characterized soluble heterodisulfide reductase (HdrABC), which plays an important role in methanogenesis during growth on methylated substrates (10).
Other important differences also are found between existing models. For example, M. acetivorans cannot grow on H2 and CO2, and it grows on CO by using a completely different pathway that involves the secretion of acetate, methyl sulfides, and formate (48, 55). M. acetivorans is also able to grow on dimethyl sulfide (DMS), whereas M. barkeri can only perform methanogenesis from that substrate (65). The iMB745 network includes pathways and genes necessary to perform these functions, which are not found in either of the previously existing reconstructions. The iMB745 network also includes novel pathways for the synthesis of methanofuran and cell wall polymers that were newly added to the biomass equation (see Materials and Methods) and accounts for experimentally determined differences in lipid composition from M. barkeri (61). Therefore, the description of essential biochemistry is more complete in this model than in those published previously.
Estimation of Rnf and Mrp ion-pumping stoichiometry.
The stoichiometry of ion pumps in the electron transport chain can have a significant effect on the quantitative predictions of metabolic models (16). In lieu of experimental data, it becomes necessary to estimate the stoichiometry by simulation. In the M. acetivorans model, flux balance analysis was used to estimate the stoichiometry of ion exchange due to the H+/Na+-exchanging complex Mrp and the Rnf complex, two methanogenesis enzymes found in M. acetivorans but not in M. barkeri (54). The Rnf complex is thought to catalyze the reduction of methanophenazine by ferredoxin and to generate either a proton or a sodium motive force (37). The specific ion pumped by Rnf is unknown, but Mrp is strongly upregulated on acetate compared to methylotrophic substrates, suggesting an increased importance of H+/Na+ exchange across the membrane on that substrate (54). Since the ion-pumping activity of Rnf is essential for growth on acetate, and since Mrp and Rnf are coregulated (54), it was assumed for modeling purposes that Rnf pumps sodium ions.
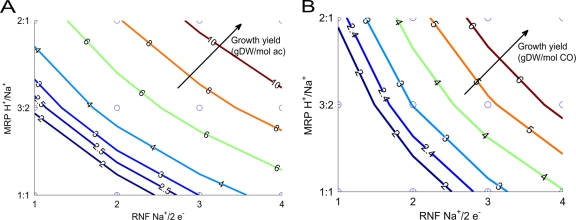
The ATP yield of methanogenesis on substrates that utilize Rnf is strongly dependent on both the number of sodium ions pumped by Rnf and the H+/Na+ exchange ratio of Mrp, neither of which has been determined experimentally. H+/Na+ exchange proteins are known in other organisms that pump protons and sodium ions in 2:1 (64), 3:2 (50), or 1:1 (5) ratios. To find out which combination was most likely in light of experimental growth data, a sensitivity study was performed, varying the number of sodium ions pumped by Rnf as well as the H+/Na+ ratio of Mrp. The closest match between the predicted and experimental growth yields occurred when Rnf was set to pump 3 Na+/2 e− and when Mrp was set to pump 1 H+/Na+ (Fig. 2) (see also the supplemental material). Changes in these ratios had only a minimal effect on predicted product secretion rates (see the supplemental material). Due to the close match with experimental growth yields, these ratios were chosen for all further simulations.
Fig 2.
Analysis of the ion-pumping stoichiometry of Rnf and Mrp during growth on acetate (A) and carbon monoxide (B). Circles represent simulated combinations of ion-pumping stoichiometries for the two pumps. Each colored line is a contour representing a constant predicted growth yield. The experimental growth yields are 2.4 g (dry weight)/mmol during growth on acetate and 2.5 g (dry weight)/mol during growth on CO. Only the combination of a 3 Na/2 e− stoichiometry for Rnf and a 1 Na+/1 H+ stoichiometry for Mrp was consistent with experimental data. See the supplemental material for the analogous simulation of growth on methanol.
The calculated ratios are consistent with thermodynamic data. The Rnf complex utilizes the same electron acceptor (methanophenazine) as the F420 dehydrogenase (Fpo) but uses ferredoxin instead of F420 as the electron donor. The reaction catalyzed by Fpo is coupled to the pumping of two protons across the membrane (6). Since ferredoxin has a lower redox potential than F420, it is reasonable to expect that Rnf can pump more proton equivalents across the membrane than Fpo.
F420 regeneration during growth on carbon monoxide.
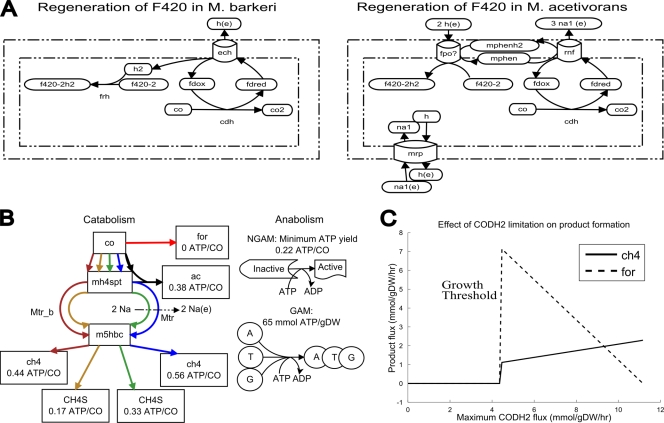
Both M. acetivorans and M. barkeri grow on carbon monoxide by oxidizing it to CO2 and subsequently reducing CO2 to methane (21). In methanogens, the reduction of carbon dioxide to methane requires the oxidation of two equivalents of reduced coenzyme F420. Therefore, in order for a methanogen to grow on carbon monoxide, a mechanism for generating reduced coenzyme F420 must be present in the cell. In CO-grown M. barkeri, reduced F420 is probably regenerated by the generation of molecular hydrogen via the reverse action of Ech hydrogenase, followed by H2-dependent reduction of F420 via the F420-reducing hydrogenase Frh (41, 46). Ech hydrogenase is not present in the M. acetivorans genome, and although an frh operon is present, it does not encode a functional enzyme (24). Thus, the mechanism for F420 regeneration in M. acetivorans during growth on CO remains unknown (21).
Our model suggests that F420 is regenerated by the combined action of F420 dehydrogenase (Fpo) and the Rnf complex (Fig. 3A). In the proposed pathway, Rnf would reduce methanophenazine with ferredoxin, and subsequently, reverse electron transport via Fpo would be used to generate reduced F420. Reverse electron transport by Fpo has not been observed experimentally, but it is thermodynamically feasible in an environment containing excesses of oxidized F420 and reduced methanophenazine. In addition, this hypothesis is consistent with the high levels of Fpo protein and transcript measured during growth on CO (35). Finally, the estimated proton-pumping stoichiometries for Rnf and Fpo (3 and 2 proton equivalents, respectively) suggest that M. acetivorans would conserve one proton for each unit of F420 reduced. This is consistent with the level of conservation in M. barkeri, in which the Ech hydrogenase pumps at least one proton out of the cell (72).
Fig 3.
Analysis of growth of M. acetivorans on carbon monoxide. (A) Regeneration of coenzyme F420 during growth on carbon monoxide for M. barkeri (left) and proposed pathway for M. acetivorans (right). co2, carbon dioxide; na1, Na+. (B) Theoretical ATP yields during growth on CO differed depending on the by-product produced and whether sodium-pumping Mtr or its nonpumping bypass reaction (Mtr_b) was active. Red, formate generation; black, acetogenesis; blue, methanogenesis (with Mtr); green, methyl sulfide production (with Mtr); gold, methyl sulfide production (with Mtr_b); dark red, methanogenesis (with Mtr_b). An ATP/CO ratio of at least 0.22 is required to overcome the non-growth-associated maintenance requirement, and an additional 65 mmol ATP/g (dry weight) is required for growth. m5hbc, co-methyl-co-5-hydroxybenzimidazolylcobamide. (C) FBA predicts that limitation of CO dehydrogenase activity leads to formate production.
As an alternative hypothesis, we also tried to implement a F420-ferredoxin oxidoreductase reaction (FFRED reaction) for the purposes of regenerating coenzyme F420 during growth on CO (10): f420-2(aq) + 2 fdred(aq) + h(aq) → f420-2h2(aq) + 2 fdox(aq), where f420-2 is oxidized F420, f420-2h2 is reduced F420, fdox is ferredoxin, fdred is reduced ferredoxin, and (aq) indicates the aqueous phase. Growth on CO was predicted to be possible if reaction FFRED was added to the model (data not shown). However, the presence of FFRED was also predicted to make a Δrnf mutant viable on acetate, contrary to experimental evidence (N. R. Baun, A. M. Guss, G. Kulkarni, and W. W. Metcalf, unpublished data), and therefore, the reaction was not included in the model. According to the model, a Δrnf mutant growing on acetate could survive with a lower growth rate by reducing coenzyme F420 with FFRED and then generating a proton gradient with Fpo and heterodisulfide reductase (Hdr).
It is possible that an enzyme catalyzing a reaction like FFRED really exists but that the ATP yield of this alternative path is insufficient to make the cell viable for growth on acetate. According to the model, the theoretical maximum wild-type ATP yield of methanogenesis from acetate (utilizing Rnf) is only about 0.75 mmol ATP/mmol acetate, corresponding to ∼65% thermodynamic efficiency (at pH 7) or 30% efficiency (at pH 0) (66). The efficiency is considerably higher than that of methanogenesis from carbon monoxide (0.56 mmol ATP/mmol CO [34%]) or methanol (0.75 mmol ATP/mmol methanol [27%]), possibly because the cell is not viable with less-efficient ATP production from this substrate.
Consistency of the model with knockout lethality data.
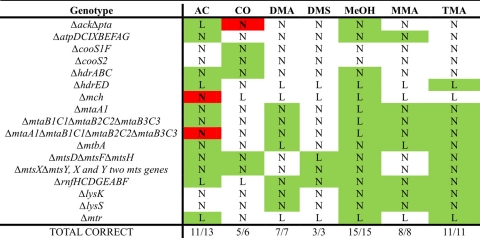
Comparison of knockout lethality predictions with available data indicates that the model correctly predicts the growth/no-growth phenotypes of 60/63 knockout mutants (Table 1). In all cases of incorrect predictions, genes were shown experimentally to be lethal but were predicted to be nonlethal. Two of the incorrect lethality predictions involved acetogenesis during growth on carbon monoxide. The genes encoding Pta and Ack are essential for growth on CO (55). However, flux balance analysis incorrectly predicts that a Δpta Δack mutant can grow by producing methane and carbon dioxide as sole by-products. Although inhibition of Mtr and (if the Mtr bypass indeed exists) another reaction in methanogenesis would cause the Δpta Δack mutation to be lethal, we cannot rule out the possibility that other mechanisms, such as regulatory constraints, are responsible for the essentiality of these genes. There is physiological evidence both supporting (55) and refuting (48) inhibition of methanogenesis reactions by carbon monoxide.
Table 1.
L, lethal; N, nonlethal. AC, acetate; DMA, dimethylamine; DMS, dimethyl sulfide; MeOH, methanol; MMA, monomethylamine; TMA, trimethylamine. The ack gene encodes acetate kinase; the pta gene, phosphotransacetylase; hdr, heterodisulfide reductase; mch, methenyl-tetrahydrosarcinopterin cyclohydrolase; mts, dimethyl sulfide-coenzyme M methyltransferase; rnf, putative ferredoxin-methanophenazine oxidoreductase; mtr, sodium-pumping tetrahydrosarcinopterin-coenzyme M methyltransferase. See the supplemental material for knockout data references.
Green, correct prediction; red, incorrect prediction; no color, no experimental data available for that knockout under those conditions.
A Δmch mutant was predicted to be viable on acetate (Table 1), but this knockout is known to be lethal on that substrate (25). The mch gene was hypothesized to be essential for growth on acetate because it generates reduced F420, which M. acetivorans requires for use in anabolic reactions, such as that involving the F420-dependent glutamate synthase (52). However, during growth on CO, Mch cannot be used to generate reduced F420, because it is required to carry flux in the direction of F420 oxidation. Therefore, for growth on CO, another enzyme (in the model, this is predicted to be Fpo) that is able to reduce F420 must be present. Flux balance analysis predicts that this other enzyme could also be used to reduce F420 during growth on acetate, thus making mch nonessential for growth on acetate. However, the incorrect prediction depended on the ability of M. acetivorans to secrete methyl sulfides during growth on acetate, which is unlikely given that the Mts methyltransferase required for methyl sulfide synthesis is downregulated during growth on acetate (9, 35). Hence, inclusion of the regulatory constraint would fix the phenotype prediction.
Knockout data were useful for refining the model and finding gene annotations in the M. acetivorans genome that are inconsistent with experimental data. For example, M. acetivorans uses Ack and Pta to activate acetate to acetyl coenzyme A (acetyl-CoA) during acetoclastic methanogenesis and cannot grow on acetate without the encoding genes (55). However, the M. acetivorans genome also encodes genes (MA3168 and MA3602) with high sequence identity to the ADP-forming acetyl-CoA synthase of Methanocaldococcus jannaschii, which catalyzes an alternative pathway for activating acetate (44). Inclusion of this reaction would make Pta and Ack nonessential for growth on acetate. On this basis, the reactions in the alternative pathway were excluded from the model.
Consistency of the model with growth phenotype data.
The iMB745 model was used to predict growth phenotypes for wild-type strains of Methanosarcina acetivorans growing on acetate, methanol, and carbon monoxide, the three substrates for which growth and substrate uptake data are available (55, 60, 63). Predicted growth rates were highly dependent on substrate uptake rates, which differed as much as 2-fold depending on the data set used to perform the calculation (see the supplemental material). It was possible to select uptake rates within the experimentally feasible ranges for each substrate that matched the observed growth rates and growth yields within 20% (Table 2).
Table 2.
Growth and secretion rates and yields of M. acetivorans on methanol, acetate, and carbon monoxide using experimentally feasible uptake ratesa
| Substrate | Measured substrate uptake rate (mmol/g [dry wt]/h) | Growth (h−1) |
Growth yield (g [dry wt]/mol substrate) |
CH4 secretion rate (mmol/g [dry wt]/h) |
|||
|---|---|---|---|---|---|---|---|
| Measured | Predicted | Measured | Predicted | Measured | Predicted | ||
| Acetate | 7 | 0.023 | 0.021 | 2.4 | 3.0 | 4.9 | 6.6 |
| Methanol | 20 | 0.098 | 0.086 | 5.2 | 4 | 22 | 13.3 |
| CO | 11.6 | 0.029 | 0.030 | 2.5 | 2.6 | 0.4 | 2.3 |
See references 55, 60, and 63. For simulations on CO, no additional constraints to Hdr or Cdh were assigned. All measured values are averaged across literature sources (see the supplemental material for a complete list of references). Note that due to experimental variability, the measured methanol uptake rate and the secretion of methane on that substrate are inconsistent with mass balance constraints.
Both the growth rates and the secretion rates during growth on acetate closely matched the experimental values. On methanol, the rate of methanogenesis was predicted to be much lower than the experimental value, but the ratios of products are consistent with experimental data. When the ATP yield was maximized during growth on methanol, the predicted ratio of methane to CO2 produced was exactly 3:1, as would be expected to balance redox potentials in the cell (10). When conditions were optimized for growth, the actual ratio of methane to CO2 secreted was predicted in the model to be 3.8:1, because the carbon dioxide-fixing activity of carbon monoxide dehydrogenase (CODH)/acetyl-CoA synthase reduced the net secretion of carbon dioxide. The actual ratio in M. barkeri has been measured as 3.4:1 or 4.4:1 (8).
M. acetivorans produces acetate, formate, methane, and methyl sulfides as by-products when grown on carbon monoxide (in addition to CO2) (48, 55), but the mechanisms of formate and methyl sulfide formation are unclear. Therefore, to make predictions about the conditions necessary to produce these by-products, it was necessary to hypothesize mechanisms for their production.
It is not currently known how or why M. acetivorans generates formate during growth on CO, although it is probably not coupled to methanogenesis (48). It is possible that formate is produced as a by-product of carbon monoxide dehydrogenase during growth on CO to prevent toxic CO accumulation in the cell (48, 56). The CO dehydrogenase enzyme from Rhodospirillum rubrum has been shown to create formate as a by-product, and formate may be formed by a similar mechanism in M. acetivorans (27), although the physiological substrate for the reaction is still unknown (56). In order to investigate formate production, the following reaction was tentatively included in the model: CODH3_SIDERXN: co(g) + h2o(l) → for(aq) + h(aq), where co is carbon monoxide, h2o is water, for is formate, h is H+, (g) indicates the gaseous phase, and (l) indicates the liquid phase. This reaction implies that formate production from CO does not yield ATP, which is likely to be true, since this reaction is endergonic (ΔG0′ = +24 kJ/mol [or ΔG0′ = +6 kJ/mol if CO is treated in the aqueous phase]) under standard conditions (1).
The recently characterized Mts enzymes are necessary for the production of DMS in M. acetivorans, although due to the very low ratio of the dimethyl sulfide production rate to the transcription levels of these enzymes, the in vivo function of these enzymes remains unclear (47). The source of methyl sulfide needed as a substrate for Mts to make dimethyl sulfide is unknown. However, due to the similar structures of sulfide (HS−) and methyl sulfide, one could reasonably hypothesize that methyl sulfide is formed by the same Mts enzyme that produces dimethyl sulfide: m5hbc(aq) + h2s(aq) + h(aq) → ch4s(aq) + 5hbc_red(aq). Here, ch4s is methyl sulfide, h2s is H2S,m5hbc is the methylated form of a cobalamide cofactor utilized in methyltransferases in Methanosarcina (20), and 5hbc_red [5-hydroxybenzimidazolylcob(I)amide] is the unmethylated form of m5hbc.
Despite the inclusion of reactions to make the observed by-products acetate, formate, and methyl sulfides, FBA predicted that only methane and CO2 would be produced as by-products during growth on CO. Consequently, the methane secretion rate was significantly higher than that observed in experiments (Table 2). To investigate the reason for this incorrect prediction, the theoretical ATP yield was calculated for the production of each by-product per mole of CO consumed, as described in Materials and Methods. The theoretical ATP yield from methanogenesis was significantly higher than that from acetogenesis, methyl sulfide production, or formate generation (Fig. 3B). Because most biosynthesis reactions were unconstrained in the direction necessary for biosynthesis, FBA predicted the utilization of pathways with greater ATP production efficiency, because if less substrate is needed to satisfy the ATP requirements of the cell, then more is available to produce biomass. We subsequently examined possible conditions under which these by-products could be produced in an FBA model.
Formate production and regulation of CO levels in M. acetivorans.
M. acetivorans encodes at least two complete CODH operons, and their relative expression during growth on CO may depend on the concentration of CO in the medium (56). Since formate production may be a result of a side reaction of CODH (27), it is tempting to speculate that the level of carbon monoxide in the cell is regulated by the balance of the levels of these proteins, one of which produces formate as a by-product and one of which does not. FBA predicts that if the proposed mechanism for formate production is correct, reduction of the flux through the primary reaction catalyzed by CODH leads to formate production (Fig. 3C). This indicates that CO toxicity could be controlled by balancing the rates of the side reaction and the primary reaction of CODH enzymes.
Inhibition by CO of the methyltransferase Mtr and its possible role in acetogenesis.
The sodium-pumping methyltransferase Mtr catalyzes the reversible transfer of methyl from methyl tetrahydrosarcinopterin to methyl coenzyme M (methyl-CoM) via a cobalamide intermediate. This enzyme is strongly downregulated during growth on CO compared to other substrates (56). It has been hypothesized that methanogenesis is kinetically limited during growth on CO (55), possibly including Mtr, although this hypothesis is controversial (48). Flux balance analysis predicted that if Mtr was notinhibited, the cell could still survive without methanogenesis or acetogenesis by producing methyl sulfides. The theoretical ATP yield of methyl sulfide production, including ATP generation due to the sodium gradient created by Mtr (0.33 ATP/CO), is sufficient to fulfill the ATP maintenance requirement (0.22 ATP/CO) (Fig. 3B). Since acetogenesis is actually essential for growth on carbon monoxide (55) and methyl sulfide production is actually very low, this prediction suggests that either Mtr activity is strongly limited during growth on carbon monoxide or the production of methyl sulfides is kinetically limited.
Despite the downregulation and possible inhibition of Mtr during growth on CO, significant methanogenesis is still observed during growth on this substrate (48, 55). The combination of these observations has inspired the hypothesis that a Mtr bypass reaction exists that performs the same function but does not generate a sodium gradient (21): MTR_BYPASS: mh4spt(aq) + 5hbc_red(aq) + h(aq) → h4spt(aq) + m5hbc(aq), where mh4spt is methyl tetrahydrosarcinopterin. This reaction, when coupled with methyl transfer to coenzyme M, would be strongly thermodynamically favored in the direction of methyl-CoM formation (ΔG0 = −30 kJ/mol) (67). The presence of the bypass reaction could permit tolerance for a wider range of environmental CO concentrations by permitting the cell to balance the increased ATP potential of Mtr with a possibly greater kinetic capacity of the Mtr bypass reaction (21).
To test the effects of the Mtr bypass reaction on metabolism, the bypass reaction was added to the metabolic network, and the sodium-pumping methyltransferase Mtr was removed from the model. The modified model was still predicted to perform only methanogenesis, not acetogenesis, because the theoretical ATP yield of methanogenesis was still higher than that of acetogenesis (0.44 ATP/CO and 0.38 ATP/CO, respectively) (Fig. 3B). In addition, flux variability analysis indicated that no alternative optimal solutions led to acetate secretion. As a result, Mtr inhibition is probably not the sole reason for acetogenesis during growth on CO.
Exploration of an alternate heterodisulfide reductase (HdrABC) on methanol.
HdrABC is a soluble heterodisulfide reductase typically found in methanogens without cytochromes (62). Most methanogens with cytochromes, including Methanosarcina species, use a membrane-bound heterodisulfide reductase, HdrDE, instead of the soluble HdrABC to couple methanogenesis to ATP production (67). Surprisingly, M. acetivorans was found to utilize both types of heterodisulfide reductase during growth on methylotrophic substrates (10).
Since the HdrABC complex is not a sodium or proton pump, it is not clear whether the activity of this complex is coupled to ATP synthesis in M. acetivorans. In Methanothermobacter marburgensis, a methanogen without cytochromes, HdrABC is coupled to ATP synthesis through its interaction with the MvhADG hydrogenase complex (62). The HdrABC-MvhADG complex in M. marburgensis uses an electron bifurcation mechanism, in which the electrons from two equivalents of molecular hydrogen are donated to ferredoxin and to the heterodisulfide (62). M. acetivorans lacks the genes encoding the MvhADG complex, but the similarity of the Hdr enzymes suggests that M. acetivorans HdrABC may also use an electron bifurcation mechanism (Fig. 4A), splitting the electrons of two fully reduced ferredoxins between heterodisulfide and coenzyme F420 (10). Alternatively, HdrABC may simply reduce heterodisulfide with ferredoxin, acting as a sink for excess ferredoxin produced during the oxidation of methanol to CO2.
Fig 4.
Studies on the soluble heterodisulfide reductase HdrABC during growth on methanol. (A) Hypothesized bifurcation mechanism of the soluble heterodisulfide reductase HdrABC during growth on methylotrophic substrates in M. acetivorans. The alternative hypothesis is that the reaction does not involve F420 (dashed lines). formmfr(b), formylmethanofuran(b). (B) Flux balance analysis predicts that during growth on methylotrophic substrates (methanol shown), HdrABC is not used when Rnf is available, but a Δrnf mutant is predicted to carry flux through HdrABC. The growth rate for the Δrnf mutant is predicted to be about 20% less than that for the wild type with bifurcation and 35% less without.
To test the electron bifurcation hypothesis in a genome-scale context, the phenotype of M. acetivorans was simulated with and without electron bifurcation in HdrABC. The addition of two additional constraints was necessary in order to obtain reasonable predictions. Reactions catalyzed by Pta and Ack were disabled so as to prevent acetate secretion, which has not been observed during growth on methanol (36), and the flux through pyruvate-acetyl-CoA oxidoreductase was set to be equal to the wild-type value so as to prevent the secretion of formate and other unobserved by-products during growth on methanol (60). In the presence of Rnf, flux variability analysis did not predict utilization of HdrABC regardless of the mechanism. However, a Δrnf mutant was predicted to utilize HdrABC to oxidize ferredoxin using any optimal flux distribution (Fig. 4B). The Δrnf mutant was predicted to grow 35% more slowly than the wild type without bifurcation and 20% more slowly with bifurcation. A Δrnf mutant actually grew about 25% more slowly on methanol than the wild type (W. Metcalf, unpublished data), so within experimental error it is difficult to tell which mechanism is correct. Further experiments could help elucidate the true mechanism.
DISCUSSION
We have built and manually curated a computable genome-scale model of metabolism in M. acetivorans, only the third methanogen species to be reconstructed (after M. barkeri [19] and Methanocaldococcus jannaschii [69]) and the second in the genus Methanosarcina. We have focused on three approaches to model-guided discovery using the Methanosarcina acetivorans model: (i) identification of knowledge gaps and “missing” reactions in metabolic pathways, (ii) detailed study of metabolic differences between closely related methanogenic species, and (iii) use of constraint-based modeling to study alternative hypotheses about the workings of the metabolic network and the implications of those hypotheses for predicted phenotypes.
The reconstruction endeavor has helped pinpoint gaps in our knowledge of the metabolic networks, both due to unknown differences between different archaeal species and due to differences between the Archaea and other domains of life. Interestingly, even some pathways for the synthesis of specialized methanogenic cofactors, such as those for tetrahydrosarcinopterin and coenzyme M, seem to have diverged from those observed in other methanogens, such as Methanocaldococcus jannaschii. However, the synthesis pathways for other methanogenic cofactors (such as coenzyme B) are conserved across these genera. This observation raises interesting questions about the evolution of these ecologically important organisms and the role of the environment in the selection of metabolic pathways.
Constraint-based modeling proved useful for integrating experimental data from different sources and identifying tensions between data sets. Some of these tensions, such as the disparity between the ability of M. acetivorans to grow on CO and the lethality of a mch knockout, may have been difficult to identify without a genome-scale model and its accompanying predictions. These findings highlight the usefulness of an integrative, genome-scale modeling approach both for validating model predictions and for identifying gaps in our knowledge of methanogen biology.
One of the strengths of genome-scale metabolic modeling is the ability to continually update the model as additional experimental data become available (18). Our investigations of alternative hypotheses for the mechanism of F420 regeneration during growth on carbon monoxide, pathways for synthesis of by-products observed during growth on CO, and the precise reaction catalyzed by the soluble heterodisulfide reductase HdrABC have yielded predictions that can be tested in the laboratory. As additional data become available, improved models may be constructed and used to provide further novel hypotheses in an iterative process that lies at the heart of systems biology.
Supplementary Material
ACKNOWLEDGMENT
We gratefully acknowledge support from the Genomic Sciences Program within the Department of Energy, Office of Biological and Environmental Research (award DE-FG02-10ER64999).
Footnotes
Published ahead of print 2 December 2011
Supplemental material for this article may be found at http://jb.asm.org/.
REFERENCES
- 1. Alberty RA. 2003. Thermodynamics of biochemical reactions. Massachusetts Institute of Technology, Cambridge, MA [Google Scholar]
- 2. Anderson B, et al. April 2010. Methane and nitrous oxide emissions from natural sources. EPA 430-R-10-001. Office of Atmospheric Programs, Environmental Protection Agency, Washington, DC [Google Scholar]
- 3. Apolinario EE, Jackson KM, Sowers KR. 2005. Development of a plasmid-mediated reporter system for in vivo monitoring of gene expression in the archaeon Methanosarcina acetivorans. Appl. Environ. Microbiol. 71:4914–4918 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Arakaki AK, Huang Y, Skolnick J. 2009. EFICAz2: enzyme function inference by a combined approach enhanced by machine learning. BMC Bioinformatics 10:107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Aronson PS. 1985. Kinetic properties of the plasma membrane Na+-H+ exchanger. Annu. Rev. Physiol. 47:545–560 [DOI] [PubMed] [Google Scholar]
- 6. Baumer S, et al. 2000. The F420H2 dehydrogenase from Methanosarcina mazei is a redox-driven proton pump closely related to NADH dehydrogenases. J. Biol. Chem. 275:17968–17973 [DOI] [PubMed] [Google Scholar]
- 7. Becker SA, et al. 2007. Quantitative prediction of cellular metabolism with constraint-based models: the COBRA Toolbox. Nat. Protoc. 2:727–738 [DOI] [PubMed] [Google Scholar]
- 8. Blaut M, Muller V, Fiebig K, Gottschalk G. 1985. Sodium ions and an energized membrane required by Methanosarcina barkeri for the oxidation of methanol to the level of formaldehyde. J. Bacteriol. 164:95–101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Bose A, Kulkarni G, Metcalf WW. 2009. Regulation of putative methyl-sulphide methyltransferases in Methanosarcina acetivorans C2A. Mol. Microbiol. 74:227–238 [DOI] [PubMed] [Google Scholar]
- 10. Buan NR, Metcalf WW. 2010. Methanogenesis by Methanosarcina acetivorans involves two structurally and functionally distinct classes of heterodisulfide reductase. Mol. Microbiol. 75:843–853 [DOI] [PubMed] [Google Scholar]
- 11. Camacho C, et al. 2009. BLAST+: architecture and applications. BMC Bioinformatics 10:421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Capone DG, Kiene RP. 1988. Comparison of microbial dynamics in marine and freshwater sediments: contrasts in anaerobic carbon catabolism. Limnol. Oceanogr. 33:725–749 [Google Scholar]
- 13. Caspi R, et al. 2010. The MetaCyc database of metabolic pathways and enzymes and the BioCyc collection of pathway/genome databases. Nucleic Acids Res. 38:D473–D479 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Deppenmeier U. 2002. The unique biochemistry of methanogenesis. Prog. Nucleic Acid Res. Mol. Biol. 71:223–283 [DOI] [PubMed] [Google Scholar]
- 15. Deppenmeier U, et al. 2002. The genome of Methanosarcina mazei: evidence for lateral gene transfer between bacteria and archaea. J. Mol. Microbiol. Biotechnol. 4:453–461 [PubMed] [Google Scholar]
- 16. Feist AM, et al. 2007. A genome-scale metabolic reconstruction for Escherichia coli K-12 MG1655 that accounts for 1260 ORFs and thermodynamic information. Mol. Syst. Biol. 3:121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Feist AM, Herrgard MJ, Thiele I, Reed JL, Palsson BO. 2009. Reconstruction of biochemical networks in microorganisms. Nat. Rev. Microbiol. 7:129–143 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Feist AM, Palsson BO. 2008. The growing scope of applications of genome-scale metabolic reconstructions using Escherichia coli. Nat. Biotechnol. 26:659–667 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Feist AM, Scholten JCM, Palsson BØ, Brockman FJ, Ideker T. 2006. Modeling methanogenesis with a genome-scale metabolic reconstruction of Methanosarcina barkeri. Mol. Syst. Biol. 2:2006.0004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Ferguson T, Soares JA, Lienard T, Gottschalk G, Krzycki JA. 2009. RamA, a protein required for reductive activation of corrinoid-dependent methylamine methyltransferase reactions in methanogenic archaea. J. Biol. Chem. 284:2285–2295 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Ferry JG. 2010. CO in methanogenesis. Ann. Microbiol. 60:1–12 [Google Scholar]
- 22. Galagan JE, et al. 2002. The genome of M. acetivorans reveals extensive metabolic and physiological diversity. Genome Res. 12:532–542 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Grochowski LL, Xu H, White RH. 2005. Ribose-5-phosphate biosynthesis in Methanocaldococcus jannaschii occurs in the absence of a pentose-phosphate pathway. J. Bacteriol. 187:7382–7389 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Guss AM, Kulkarni G, Metcalf WW. 2009. Differences in hydrogenase gene expression between Methanosarcina acetivorans and Methanosarcina barkeri. J. Bacteriol. 191:2826–2833 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Guss AM, Mukhopadhyay B, Zhang JK, Metcalf WW. 2005. Genetic analysis of mch mutants in two Methanosarcina species demonstrates multiple roles for the methanopterin-dependent C-1 oxidation/reduction pathway and differences in H2 metabolism between closely related species. Mol. Microbiol. 55:1671–1680 [DOI] [PubMed] [Google Scholar]
- 26. Henry CS, et al. 2010. High-throughput generation, optimization and analysis of genome-scale metabolic models. Nat. Biotechnol. 28:977–982 [DOI] [PubMed] [Google Scholar]
- 27. Heo J, Skjeldal L, Staples CR, Ludden PW. 2002. Carbon monoxide dehydrogenase from Rhodospirillum rubrum produces formate. J. Biol. Inorg Chem. 7:810–814 [DOI] [PubMed] [Google Scholar]
- 28. Jankowski MD, Henry CS, Broadbelt LJ, Hatzimanikatis V. 2008. Group contribution method for thermodynamic analysis of complex metabolic networks. Biophys. J. 95:1487–1499 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Kandler O, Hippe H. 1977. Lack of peptidoglycan in the cell walls of Methanosarcina barkeri. Arch. Microbiol. 113:57–60 [DOI] [PubMed] [Google Scholar]
- 30. Kanehisa M, Goto S, Furumichi M, Tanabe M, Hirakawa M. 2010. KEGG for representation and analysis of molecular networks involving diseases and drugs. Nucleic Acids Res. 38:D355–D360 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Kumar VS, Dasika MS, Maranas CD. 2007. Optimization based automated curation of metabolic reconstructions. BMC Bioinformatics 8:212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Kumar VS, Ferry J, Maranas C. 2011. Metabolic reconstruction of the archaeon methanogen Methanosarcina acetivorans. BMC Syst. Biol. 5:28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Kumar VS, Maranas CD. 2009. GrowMatch: an automated method for reconciling in silico/in vivo growth predictions. PLoS Comput. Biol. 5:e1000308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Lessner DJ, Lhu L, Wahal CS, Ferry JG. 2010. An engineered methanogenic pathway derived from the domains Bacteria and Archaea. mBio 1(5):e00243–10 doi:10.1128/mBio.00243-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Lessner DJ, et al. 2006. An unconventional pathway for reduction of CO2 to methane in CO-grown Methanosarcina acetivorans revealed by proteomics. Proc. Natl. Acad. Sci. U. S. A. 103:17921–17926 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Li Q, Li L, Rejtar T, Karger BL, Ferry JG. 2005. Proteome of Methanosarcina acetivorans Part II: comparison of protein levels in acetate- and methanol-grown cells. J. Proteome Res. 4:129–135 [DOI] [PubMed] [Google Scholar]
- 37. Li Q, et al. 2006. Electron transport in the pathway of acetate conversion to methane in the marine archaeon Methanosarcina acetivorans. J. Bacteriol. 188:702–710 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Maeder DL, et al. 2006. The Methanosarcina barkeri genome: comparative analysis with Methanosarcina acetivorans and Methanosarcina mazei reveals extensive rearrangement within methanosarcinal genomes. J. Bacteriol. 188:7922–7931 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Mahadevan R, Schilling CH. 2003. The effects of alternate optimal solutions in constraint-based genome-scale metabolic models. Metab. Eng. 5:264–276 [DOI] [PubMed] [Google Scholar]
- 40. Mahlmann A, Deppenmeier U, Gottschalk G. 1989. Methanofuran B is required for Co2 formation from formaldehyde by Methanosarcina barkeri. FEMS Microbiol. Lett. 61:115–120 [Google Scholar]
- 41. Meuer J, Kuettner HC, Zhang JK, Hedderich R, Metcalf WW. 2002. Genetic analysis of the archaeon Methanosarcina barkeri Fusaro reveals a central role for Ech hydrogenase and ferredoxin in methanogenesis and carbon fixation. Proc. Natl. Acad. Sci. U. S. A. 99:5632–5637 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Milich L. 1999. The role of methane in global warming: where might mitigation strategies be focused? Global Environ. Change 9:179–201 [Google Scholar]
- 43. Milne CB, Kim PJ, Eddy JA, Price ND. 2009. Accomplishments in genome-scale in silico modeling for industrial and medical biotechnology. Biotechnol. J. 4:1653–1670 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Musfeldt M, Schönheit P. 2002. Novel type of ADP-forming acetyl coenzyme A synthetase in hyperthermophilic archaea: heterologous expression and characterization of isoenzymes from the sulfate reducer Archaeoglobus fulgidus and the methanogen Methanococcus jannaschii. J. Bacteriol. 184:636–644 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Oberhardt MA, Palsson BØ, Papin JA. 2009. Applications of genome-scale metabolic reconstructions. Mol. Syst. Biol. 5:320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. O'Brien JM, Wolkin RH, Moench TT, Morgan JB, Zeikus JG. 1984. Association of hydrogen metabolism with unitrophic or mixotrophic growth of Methanosarcina barkeri on carbon monoxide. J. Bacteriol. 158:373–375 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Oelgeschläger E, Rother M. 2009. In vivo role of three fused corrinoid/methyl transfer proteins in Methanosarcina acetivorans. Mol. Microbiol. 72:1260–1272 [DOI] [PubMed] [Google Scholar]
- 48. Oelgeschläger E, Rother M. 2009. Influence of carbon monoxide on metabolite formation in Methanosarcina acetivorans. FEMS Microbiol. Lett. 292:254–260 [DOI] [PubMed] [Google Scholar]
- 49. Orth JD, Thiele I, Palsson BØ. 2010. What is flux balance analysis? Nat. Biotechnol. 28:245–248 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Pinner E, Padan E, Schuldiner S. 1994. Kinetic properties of NhaB, a Na+/H+ antiporter from Escherichia coli. J. Biol. Chem. 269:26274–26279 [PubMed] [Google Scholar]
- 51. Price ND, Reed JL, Palsson BØ. 2004. Genome-scale models of microbial cells: evaluating the consequences of constraints. Nat. Rev. Microbiol. 2:886–897 [DOI] [PubMed] [Google Scholar]
- 52. Raemakers-Franken PC, Brand RJ, Kortstee AJ, Van der Drift C, Vogels GD. 1991. Ammonia assimilation and glutamate incorporation in coenzyme F420 derivatives of Methanosarcina barkeri. Antonie Van Leeuwenhoek 59:243–248 [DOI] [PubMed] [Google Scholar]
- 53. Ren Q, Chen K, Paulsen IT. 2007. TransportDB: a comprehensive database resource for cytoplasmic membrane transport systems and outer membrane channels. Nucleic Acids Res. 35:D274–279 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Rohlin L, Gunsalus RP. 2010. Carbon-dependent control of electron transfer and central carbon pathway genes for methane biosynthesis in the archaean, Methanosarcina acetivorans strain C2A. BMC Microbiol. 10:62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Rother M, Metcalf WW. 2004. Anaerobic growth of Methanosarcina acetivorans C2A on carbon monoxide: an unusual way of life for a methanogenic archaeon. Proc. Natl. Acad. Sci. U. S. A. 101:16929–16934 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Rother M, Oelgeschläger E, Metcalf WM. 2007. Genetic and proteomic analyses of CO utilization by Methanosarcina acetivorans. Arch. Microbiol. 188:463–472 [DOI] [PubMed] [Google Scholar]
- 57. Schellenberger J, Park JO, Conrad TM, Palsson BO. 2010. BiGG: a Biochemical Genetic and Genomic knowledgebase of large scale metabolic reconstructions. BMC Bioinformatics 11:213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Sowers KR, Baron SF, Ferry JG. 1984. Methanosarcina acetivorans sp. nov., an acetotrophic methane-producing bacterium isolated from marine sediments. Appl. Environ. Microbiol. 47:971–978 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Sowers KR, Boone JE, Gunsalus RP. 1993. Disaggregation of Methanosarcina spp. and growth as single cells at elevated osmolarity. Appl. Environ. Microbiol. 59:3832–3839 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Sowers KR, Nelson MJ, Ferry JG. 1984. Growth of acetotrophic, methane-producing bacteria in a pH auxostat. Curr. Microbiol. 11:227–229 [Google Scholar]
- 61. Sprott GD, Dicaire CJ, Choquet CG, Patel GB, Ekiel I. 1993. Hydroxydiether lipid structures in Methanosarcina spp. and Methanococcus voltae. Appl. Environ. Microbiol. 59:912–914 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Stojanowic A, Mander GJ, Duin EC, Hedderich R. 2003. Physiological role of the F420-non-reducing hydrogenase (Mvh) from Methanothermobacter marburgensis. Arch. Microbiol. 180:194–203 [DOI] [PubMed] [Google Scholar]
- 63. Summer H. 2009. Improved approach for transferring and cultivating Methanosarcina acetivorans C2A (DSM 2834). Lett. Appl. Microbiol. 48:786–789 [DOI] [PubMed] [Google Scholar]
- 64. Taglicht D, Padan E, Schuldiner S. 1991. Overproduction and purification of a functional Na+/H+ antiporter coded by nhaA (ant) from Escherichia coli. J. Biol. Chem. 266:11289–11294 [PubMed] [Google Scholar]
- 65. Tallant TC, Krzycki JA. 1997. Methylthiol:coenzyme M methyltransferase from Methanosarcina barkeri, an enzyme of methanogenesis from dimethylsulfide and methylmercaptopropionate. J. Bacteriol. 179:6902–6911 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Thauer RK, Jungermann K, Decker K. 1977. Energy conservation in chemotrophic anaerobic bacteria. Bacteriol. Rev. 41:100–180 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Thauer RK, Kaster AK, Seedorf H, Buckel W, Hedderich R. 2008. Methanogenic archaea: ecologically relevant differences in energy conservation. Nat. Rev. Microbiol. 6:579–591 [DOI] [PubMed] [Google Scholar]
- 68. Thiele I, Palsson BØ. 2010. A protocol for generating a high-quality genome-scale metabolic reconstruction. Nat. Protoc. 5:93–121 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Tsoka S, Simon D, Ouzounis CA. 2004. Automated metabolic reconstruction for Methanococcus jannaschii. Archaea 1:223–229 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. UniProt Consortium 2010. The Universal Protein Resource (UniProt) in 2010. Nucleic Acids Res. 38:D142–D148 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Welander PV, Metcalf WW. 2005. Loss of the mtr operon in Methanosarcina blocks growth on methanol, but not methanogenesis, and reveals an unknown methanogenic pathway. Proc. Natl. Acad. Sci. U. S. A. 102:10664–10669 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Welte C, Krätzer C, Deppenmeier U. 2010. Involvement of Ech hydrogenase in energy conservation of Methanosarcina mazei. FEBS J. 277:3396–3403 [DOI] [PubMed] [Google Scholar]
- 73. Xu H, Aurora R, Rose GD, White RH. 1999. Identifying two ancient enzymes in Archaea using predicted secondary structure alignment. Nat. Struct. Biol. 6:750–754 [DOI] [PubMed] [Google Scholar]
- 74. Zhang JK, White AK, Kuettner HC, Boccazzi P, Metcalf WW. 2002. Directed mutagenesis and plasmid-based complementation in the methanogenic archaeon Methanosarcina acetivorans C2A demonstrated by genetic analysis of proline biosynthesis. J. Bacteriol. 184:1449–1454 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.