Abstract
Helicobacter pylori is a chronic colonizer of the gastric epithelium and plays a major role in the development of gastritis, peptic ulcer disease, and gastric cancer. In its coevolution with humans, the streamlining of the H. pylori genome has resulted in a significant reduction in metabolic pathways, one being purine nucleotide biosynthesis. Bioinformatic analysis has revealed that H. pylori lacks the enzymatic machinery for de novo production of IMP, the first purine nucleotide formed during GTP and ATP biosynthesis. This suggests that H. pylori must rely heavily on salvage of purines from the environment. In this study, we deleted several genes putatively involved in purine salvage and processing. The growth and survival of these mutants were analyzed in both nutrient-rich and minimal media, and the results confirmed the presence of a robust purine salvage pathway in H. pylori. Of the two phosphoribosyltransferase genes found in the H. pylori genome, only gpt appears to be essential, and an Δapt mutant strain was still capable of growth on adenine, suggesting that adenine processing via Apt is not essential. Deletion of the putative nucleoside phosphorylase gene deoD resulted in an inability of H. pylori to grow on purine nucleosides or the purine base adenine. Our results suggest a purine requirement for growth of H. pylori in standard media, indicating that H. pylori possesses the ability to utilize purines and nucleosides from the environment in the absence of a de novo purine nucleotide biosynthesis pathway.
INTRODUCTION
Helicobacter pylori is a Gram-negative, microaerophilic helix-shaped bacterium that colonizes the gastric mucosa of roughly half of the world's population (19, 53). Unlike numerous other pathogenic bacteria capable of existing in diverse environmental niches, H. pylori is only capable of sustained growth within its human host (19). Identified only 27 years ago (45), this bacterium was the first to have two different strains sequenced, allowing for the first genome-wide comparative bioinformatic analysis of a bacterial species (1, 16). The results of this comparative sequencing project (16) as well as subsequent sequencing projects (3) show a relatively small genome with numerous alterations in its metabolic pathways compared with those of other bacterial species. Initial conclusions were that the missing pathway enzymes simply were divergent enough to escape classification by homology screening (16); however, subsequent analysis of the genome has identified 48 potential “dead-end metabolites” (metabolites only consumed or only produced within a metabolic network), indicating missing knowledge about a particular pathway or absence of a fully functional pathway (67). One hypothesis developed to explain these abnormalities was that, having evolved alongside humans for so long, the metabolism of H. pylori was streamlined to coexist in the human niche. Apparent holes in the metabolic pathways of H. pylori suggest the loss of genes no longer required for growth in its relatively stable environment of the human host (67). Opportunistic pathogens generally require a large array of genes to live within multiple and often vastly different environments, leading to much more complex and often redundant metabolic pathways. However, given the human-specific niche and a relatively high genome-wide mutation rate, as well as a high rate of genetic recombination (6, 37), H. pylori has been able to stably and successfully survive within humans such that its metabolic pathways no longer require the redundancy often found in environmental microbes.
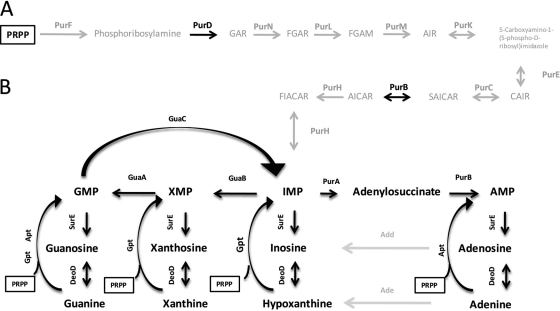
One such redundant pathway found in nature involves the biosynthesis of purine nucleotides. Purine metabolism, quite literally, is the growth-limiting step for all cells, prokaryotic and eukaryotic alike. In terms of bacteria, the rate at which microbes can generate GMP and AMP pools often directly correlates with how fast they can grow (31). A rapid growth rate is often selected for in the presence of numerous competitors, and maximizing access to available purines becomes essential to survival. The predominant method for purine nucleotide generation is through the de novo purine nucleotide biosynthesis pathway, whereby phosphoribosyl pyrophosphate (PRPP) is the initial substrate and, via a series of enzymatic reactions, the purine nucleotide IMP is formed (Fig. 1A). IMP can then be utilized either to generate GMP by the dual reactions of IMP dehydrogenase (IMPDH/GuaB) and GMP synthase (GuaA) or to generate AMP by way of adenylosuccinate synthase (PurA) and adenylosuccinate lyase (PurB). This pathway is initially fed by the metabolism of various amino acids that have been broken down by the organism and, as such, the pathway has a relatively high metabolic cost. The alternate pathway for purine nucleotide formation is the purine salvage pathway (Fig. 1B), so named because instead of generating new purine rings, preexisting purine bases and or nucleosides are taken up from the environment and added directly to PRPP via phosphoribosyltransferases (PRTases), generating nucleotides. The process of purine salvage can be direct, as it is with the generation of GMP from guanine and AMP from adenine, or indirect, as in the generation of IMP or XMP from hypoxanthine and xanthine, respectively. These nucleotides can then be fed into the nucleotide biosynthesis pathway, where they can then be used to generate GMP or AMP. Additionally, based on the established pathway, enzymes such as GMP reductase (GuaC) and adenine/adenosine deaminases (Add/Ade) allow the cycling of nucleotides by creating bypasses in the pathway, thereby allowing for various purine bases to be used for generating both GMP and AMP, instead of only one or the other (46, 55).
Fig 1.
The purine nucleotide biosynthesis pathway homologs in E. coli and H. pylori. (A) E. coli's functioning de novo purine nucleotide biosynthesis pathway, in which a purine ring is formed on PRPP in a stepwise fashion during a series of enzymatic reactions. Homologs for genes required for this de novo purine nucleotide biosynthesis pathway found in the H. pylori genome are presented in black. All enzymes for which no homologs could be found in the H. pylori genome are shown in gray. (B) H. pylori's functioning purine salvage pathway, by which PRPP is affixed to preformed purine rings (obtained from the environment) via PRPTases. E. coli adenosine and adenine deaminase activities without H. pylori annotated homologs are shown in gray. (Figure adapted from Jenkins et al. [35].) Abbreviations: PRPP, 5-phosphoribosyl diphosphate; GAR, glycinamide ribonucleotide; FGAR, N-formylglycinamide ribonucleotide; FGAM, 5′-phosphoribosylformylglycinamidine; AIR, aminoimidazole ribotide; CAIR, 5′-phosphoribosyl-4-carboxy-5-aminoimidazole; SAICAR, 5′-phosphoribosyl-4-(N-succinocarboxamide)-5-aminoimidazole; AICAR, 5-aminoimidazole-4-carboxamide ribotide; FIACAR, 5′-phosphoribosyl-5-formamido-4-imidazolecarboxamide.
Most organisms have both de novo purine nucleotide biosynthesis and purine salvage pathways, allowing generation of DNA and RNA in the presence and absence of external purine sources. Genes comprising either pathway often are found in operons (34–36) and are often regulated by the same regulatory promoter-binding proteins (33, 34) or riboswitches (4, 43), generating feedback loops that allow the organism to alternate between the two synthesis pathways while maintaining adequate purine nucleotide pools. Additionally, the allosteric regulation of nucleotide pools has also been shown in key enzymes in both purine (60, 61) and pyrimidine (59) nucleotide biosynthesis, indicating enzyme activity also may add an additional means of regulation.
Given the importance of purine production and its direct effect on bacterial growth rates, targeting of the enzymes of either the purine de novo synthesis or salvage pathways is currently a heavily investigated field of research. The two currently most examined genes in purine nucleotide biosynthesis, given their importance in both de novo and salvage pathways, are guaB and guaA. Bacterial strains with mutations or deletions in these two genes have been shown to be severely attenuated for growth in Salmonella enterica serotype Typhimurium (23), Yersinia pestis (52), Francisella tularensis (63), Borrelia burgdorferi (36), and Shigella flexneri (40). Studies specific to enzymes found in the de novo purine synthesis pathway have shown that absence of key enzymes in the pathway can directly affect growth rates and virulence in Bacillus anthracis (35), Y. pestis (9), S. Typhimurium (48), and Streptococcus pneumoniae (56). Similarly, the purine salvage pathway has also been investigated in numerous organisms (42, 55). PRTases, the enzymes capable of joining salvaged purine bases with PRPP in order to generate purine nucleotides, have also been studied in numerous pathogens (17, 18, 38, 39).
Purine salvage and utilization have previously been examined in H. pylori (18, 49, 50). Radiolabeling studies have been used to show uptake and incorporation of the purine bases adenine and guanine (and to a lesser extent, hypoxanthine) (50). The presence of adenine, guanine, and hypoxanthine phosphoribosyltransferase activities have been measured from whole-cell lysates (50), and an in-depth look at the enzymatic nature of H. pylori's purified xanthine-guanine phosphoribosyltransferase demonstrated its ability to catalyze the formation of 6-oxopurines (18).
One study has shown that H. pylori can grow in the absence of preformed purines as long as exogenous catalase is present (49). It has been shown that exogenous catalase added to medium can increase the growth of H. pylori by preventing the formation of toxic peroxidation products from unsaturated fatty acids (29) and that bovine serum albumin (BSA) and catalase together can provide the minimal requirements by which H. pylori can grow on blood- or serum-free media (30). Regarding growth in the absence of purines, the hypothesis is that in H. pylori, de novo purine nucleotide biosynthesis requires H. pylori to perform at its metabolic peak, and only when exogenous catalase is present can it achieve that threshold (49). However, since these initial findings, sequence analysis of the genome indicates that H. pylori lacks most of the enzymatic machinery believed to be required for classical de novo purine nucleotide biosynthesis (Fig. 1A) (1, 16) while encoding a robust assortment of salvage pathway-associated genes (Fig. 1B). It has been cautioned (5) that while genomic analysis can be greatly beneficial to understanding bacterial metabolism, the literature is full of instances where measurable enzymatic activities exist in whole-cell lysates despite the apparent absence of a suitable genetic homolog in the H. pylori genome, indicating the potential limitations to a purely bioinformatic approach. However, in defined and standard media, H. pylori requires the presence of at least a single purine base to grow (51, 58, 65, 66), and purine bases were present in trace amounts in the basal medium used in the first H. pylori growth studies that examined the effects of catalase supplementation (30). Other studies have shown that when tissue culture cells are grown in medium lacking purines, H. pylori is able to grow within close proximity to these cells and is also capable of growth in medium that has been previously enriched by tissue cultured cell lines (69), indicating that growth media after 24 and 48 h from uninfected cells can meet H. pylori's purine requirements for growth. These latter findings all support the importance of the purine salvage pathway in H. pylori.
We previously examined H. pylori's purine nucleotide biosynthesis pathway by monitoring the effects of inhibitors of the H. pylori IMPDH enzyme, and we found that the most potent inhibitor tested in this screen also resulted in the reduction of bacterial growth in nutrient-rich medium (26). Further studies have shown that the inhibitory effects of this compound are removed when exogenous guanine is added to minimal, defined medium (32). Given the potential of GuaB as an enzymatic target, we decided to examine the H. pylori purine acquisition pathways more closely in hopes of finding other promising targets for future molecular inhibitors that may prove synergistic when used with IMPDH inhibitors. While activities have already been characterized for other enzymes in this pathway (50), we examined the putative genes encoding these enzymes. The goal of this study was to identify the major enzymatic machinery involved in the H. pylori purine nucleotide biosynthesis pathway and to determine what enzymes are required for the growth and survival of H. pylori on rich and minimal media containing purine bases or nucleosides. Using the Escherichia coli purine nucleotide biosynthesis pathway as a template, we generated mutants of H. pylori G27 with deletions of genes for numerous putative pathway elements: guaA, guaB, guaC, purA, purB, purD, surE, apt, and deoD. In addition, upon failing to insertionally inactivate or delete gpt, we were able to confirm its essentiality by first complementing the mutation with an alternate copy placed elsewhere in the genome and subsequently removing the wild-type gene. Careful examination of the growth of our purine salvage pathway mutants on defined medium revealed that H. pylori possesses a fully functional purine salvage pathway that allows it to grow on any single purine base or nucleoside tested. In the absence of these enzymes, the bacterium exhibits a significantly reduced ability to survive and grow unless provided with various purines. Attempts to grow H. pylori in the absence of purines were unsuccessful, and we conclude that H. pylori strain G27 is incapable of de novo purine nucleotide biosynthesis and relies completely on its salvage pathway for purine nucleotide formation.
MATERIALS AND METHODS
Bacterial strains and growth conditions.
H. pylori strain G27 (3) was generously provided by D. Scott Merrell of the Uniformed Services University of the Health Sciences (Bethesda, MD). Strain Hp1061 (27) was provided by Paul Hoffman of the University of Virginia, and strain 60190 was provided by M. J. Blaser of New York University (13). All bacteria were grown under tissue culture (microaerophilic) conditions, at 37°C with 10% CO2. Bacteria used in all broth culture growth experiments were first grown on blood agar plates (Remel) for 18 h prior to inoculation. All broth cultures were grown in brucella broth (Becton Dickinson), Ham's F-12 defined medium (Gibco), or a derivation of Reynolds and Penn's (RnP) defined medium (58) (without adenine and vitamins D-biotin, folic acid, niacinamide, p-aminobenzoic acid, d-pantothenic acid, pyridoxine hydrochloride, riboflavin, and B12) that was made either purine free or containing single purine bases or nucleosides at a concentration of 0.37 mM. Broth culture supplements included 10% fetal bovine serum (FBS; Gibco) and 1% (vol/vol) amphotericin B (Fungizone; Gibco). Plated bacteria were either grown on blood agar plates, brucella broth-based medium supplemented with 10% FBS with appropriate antibiotics (25 μg/ml chloramphenicol or 25 μg/ml kanamycin), or RnP plates. RnP plates were generated by first making a 3% Bacto agar (Becton, Dickinson) solution in distilled water, autoclaving the solution, bringing it to a temperature of ∼55°C, and then adding it to an equal volume of 0.22-μm filter-sterilized 2× solution of RnP medium (containing single purine bases at a 0.37 mM final concentration).
Generation of purine nucleotide biosynthesis and purine salvage pathway mutants.
Mutant H. pylori strains were generated via a vector-free allelic replacement mutagenesis strategy, as previously described (14). Briefly, the chloramphenicol resistance cassette (cat), initially isolated from Campylobacter coli (70), was PCR amplified. Two pairs of gene-specific primers designated P1/P2 and P3/P4 for every gene to be deleted from the G27 chromosome were used to PCR amplify the up- and downstream regions of each target gene to produce fragments of ∼500 bp flanking the region to be deleted. The P2 and P3 primers contained 5′ sequences upstream of the forward and reverse cat-specific primers. Overlapping, multitemplate PCR was used to first generate P1/P2-cat and cat-P3/P4 products and then once again to combine those products to obtain a P1-cat-P4 construct. PCR products were generated using PfuUltra II Fusion HS DNA polymerase (Stratagene). The resulting product was electrophoresed, gel purified via a QIAquick gel extraction kit (Qiagen), and then naturally transformed directly into H. pylori (70). This vector-free transformation of H. pylori strain G27 was used to generate guaA (HPG27_988/HP0409), guaB (HPG27_788/HP0829), guaC (HPG27_810/HP0854), purA (HPG27_234/HP0255), purB (HPG27_1054/HP1112), purD (HPG27_1163/HP1218), deoD (HPG27_1121/HP1178), surE (HPG27_879/HP0930), and apt (HPG27_531/HP0572) mutants (genetic loci are indicated using the nomenclature from H. pylori G27 and the first sequenced H. pylori strain, 26695, respectively [3, 68]). All genes examined in strains G27 and 26695 are highly homologous and predicted to encode the same putative enzymes (3, 68). Mutant colonies were selected on brucella broth-based medium containing chloramphenicol and confirmed via PCR, using flanking primers ∼100 bp outside the inserted DNA. Internal primers for the target gene were used to confirm the deletion of the wild-type gene, as the knockout mutants should lack the DNA template required for amplification of the PCR product that produces a 500-bp product band seen when wild-type DNA is used as the template. Additional sequencing confirmation of all mutant strains was conducted by ligating the PCR constructs containing mutated genetic loci in addition to ∼100 bp of flanking DNA from chromosomal genomic DNA preparations into the pCR2.1-TOPO vector (Invitrogen) and transforming into either TOP10 (Invitrogen) or DH5α (Invitrogen) E. coli cells for amplification, and then plasmid inserts were sequenced via M13 primer sites. All primers used in this study are listed in Table S1 of the supplemental material.
Complementation of H. pylori.
In order to remove the chromosomal copy of the H. pylori gpt gene, an alternate copy of the gene was placed in the rdxA locus, as previously described (14). Briefly, overlapping, multitemplate PCR was used to generate an alternate copy of the gpt gene to be inserted into the rdxA locus. Overlapping PCR was first used to fuse a kanamycin resistance cassette (aphA3) (24) to the 3′ end of the gpt gene. Next, 500- to 1,000-bp overhanging regions for the 5′ and 3′ ends of the rdxA locus were added to the 5′ and 3′ ends of the PCR construct. Upon transformation into H. pylori, colonies were selected on kanamycin and PCR confirmed, and then an additional round of transformation was conducted using the construct generated to remove the native gpt gene (HPG27_691/HP0735). Colonies were then selected on chloramphenicol and checked via PCR and sequencing.
Due to problems in transformation placement when using the native H. pylori gpt gene (homologous recombination of the PCR construct at the wild-type gpt locus instead of in the rdxA locus), E. coli gpt (gptEC from strain MC4100) was used, as it contained DNA homologous only for the H. pylori rdxA locus and varied significantly enough at the gpt locus that no recombination events occurred at the gpt region. The ∼500-bp promoter of the H. pylori gpt region was first fused to the 5′ end of gptEC, and then the aphA3 and rdxA domains were added, as described above.
Growth and survival assays.
Bacterial growth was measured in 12-well, 2-ml-volume tissue culture-grade polystyrene plates (Fisher). After 24 h, plated bacteria were collected, resuspended in the various media tested, and diluted to the appropriate optical density and/or CFU per ml. Growth and survival were determined by either a change in the optical density at 600 nm (OD600) (DU 530; Beckman Coulter) or from CFU counts conducted 24 or 48 h after inoculation. After OD600 readings were recorded from all cultures, 2 μl of each culture was spotted onto a blood agar plate and allowed to grow for 24 h to confirm the viability of the culture and the absence of contaminants (see Fig. S1 in the supplemental material). Bacterial counts were conducted in duplicate by 10-fold serial dilution on blood agar plates. All growth assays were done in triplicate and repeated at least three independent times. Error bars indicate standard errors of the means and were generated using Prism analysis software.
Catalase growth assays were performed as previously described (49). Briefly, 50-ml flasks containing 10 ml of either RnP medium with 0.1% bovine liver catalase (Sigma) or adenine (Sigma) were inoculated with 18-h, plate-scraped bacteria to an OD600 of approximately 0.1. Cultures were then allowed to grow for 24 h, at 37°C with 10% CO2 and shaking, and the following day, 1 ml from each culture was used to inoculate fresh RnP medium (containing catalase or adenine). One milliliter was used to measure the OD600, and 50 μl was used in 10-fold serial dilutions for CFU counts on blood agar plates. Assays were conducted in triplicate, and CFU counts were conducted in duplicate for each dilution. Assays were conducted in the presence and absence of amphotericin B (Gibco) to ensure the presence of the antifungal had no effect on experimental outcomes.
OD600 absorbance measurements were conducted in 300-ml side-arm flasks (Belco), and OD600 measurements were obtained hourly for 13 h. Thirty milliliters of sterile, RnP medium containing either 0.1% catalase or adenine was added to flasks, which were then placed under normal H. pylori liquid culture conditions (37°C, 10% CO2 with shaking). Samples were collected hourly and in duplicate. The spectrophotometer used for analysis (Spectronic 21D; Milton Roy) was zeroed using phosphate-buffered saline (PBS). Samples were also taken from glass vials containing 5 ml of either 0.1% catalase-containing or adenine-containing medium that were left at room temperature with no shaking as controls. Catalase activity of medium was tested at the start and end of the experiment by pipetting 20 μl of medium onto a coverslip containing a drop of hydrogen peroxide and observing oxygen production.
IMPDH enzymatic assay.
Bacteria were scraped from 24-h blood agar plates and used to inoculate cultures of brucella broth with 10% FBS. Cultures were grown overnight, to an OD600 of ∼0.75. Bacterial pellets were obtained through centrifugation, washed 3 times in PBS, and lysed via French press. Lysates were spun down at ∼6,000 × g to remove nondisrupted cells and then at ∼17,000 × g to remove bacterial membranes. Lysates were assessed for IMPDH activity by measuring the change in absorbance at an optical density of 290 nm, which correlates with XMP production. Assays were conducted in 50 mM Tris (pH 8.0), 100 mM KCl, 1 mM dithiothreitol, 3 mM EDTA, 1 mM IMP, and 1.25 mM NAD at 25°C (protocol generously provided by Lizbeth Hedstrom, Brandeis University). Measurements were taken at 30-s intervals over a 2-hour time period, and each lysate was measured under each condition (plus NAD and IMP, without IMP, and without NAD).
Sequence analysis.
Gene sequences were retrieved from the NCBI Genomic Database and examined using GENtle analysis software. Sequence alignments were conducted using T-Coffee, NCBI's BLAST search engine, or GENtle alignment software. Phylogenetic analysis of Epsilonproteobacteria was conducted via PATRIC, Pathosystems Resource Integration Center (http://patricbrc.vbi.vt.edu/portal/portal/patric/Phylogeny?cType=taxon&cId=209). Metabolic pathway comparisons were conducted using the KEGG Pathway Database.
RESULTS
H. pylori strain G27 requires purine bases for growth.
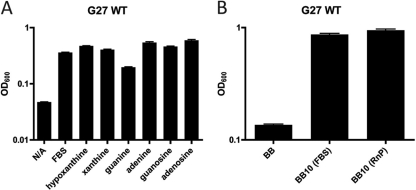
For our model system, we decided to begin our examination of purine utilization by H. pylori strain G27 in a defined medium. RnP defined medium was chosen as a suitable base medium, and we conducted 24-h growth analyses of our wild-type strain using a derivation of this medium containing only single purine bases or nucleosides. We found that strain G27 was capable of growth in RnP media containing any of the purine bases or nucleosides examined (Fig. 2A). When FBS was added to RnP medium lacking any purine bases or nucleosides, robust bacterial growth was also observed; however, RnP medium containing no purine bases or nucleosides and not supplemented with FBS was not sufficient for growth. Additional wild-type H. pylori strains were tested (strains Hp1061 and 60190), and each appeared to possess purine auxotrophy (data not shown). While RnP medium containing purines or FBS appears sufficient for the growth of strain G27, it was noted that OD600 values were significantly higher from the 24-h growth assays in the more nutrient-rich brucella broth medium with either 10% FBS or 10% purine-free RnP medium added (BB10) (Fig. 2B).
Fig 2.
H. pylori strain G27 growth on nutrient-rich (brucella broth) and defined RnP single purine source media. (A) Growth assay in RnP medium containing single purine bases or nucleosides. (B) Growth assays conducted for H. pylori strain G27 in brucella broth either unsupplemented or supplemented with 10% FBS or 10% purine-free RnP medium. Starting OD600 values for RnP medium and brucella broth growth assays were ∼0.03 and 0.1, respectively. Assays were conducted for 24 h. Error bars represent the standard errors of the means. N/A, no additive.
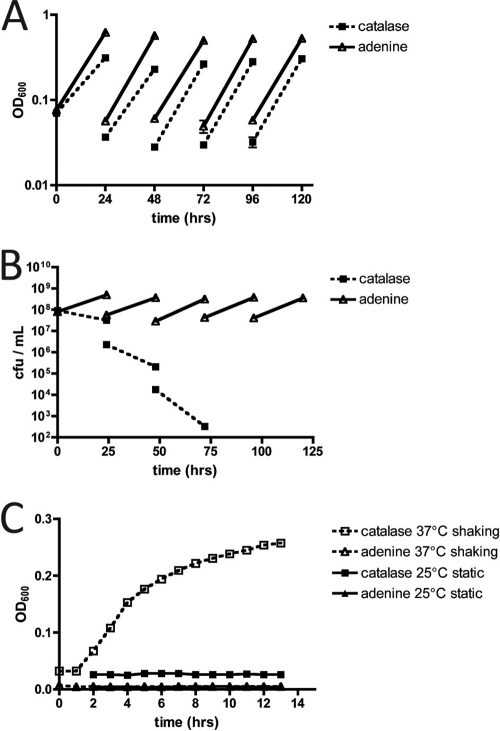
Previous studies have shown that H. pylori is capable of growth in the absence of purine bases when the medium is supplemented with catalase (49). However, since these earlier studies were conducted, sequencing of the H. pylori genome has revealed the apparent absence of most of the genes comprising the classical de novo purine nucleotide biosynthesis pathway (1, 16). In order to examine H. pylori's potentially novel de novo synthesis pathway, we attempted to grow H. pylori in the absence of preformed purines. The protocol, described previously in the literature (49), calls for H. pylori to be serially passaged in purine-free RnP medium in the presence of catalase (0.1% [wt/vol]) or containing only the purine base, adenine. We repeated this experiment, and every 24 h, OD600 values were recorded and CFU per ml were assessed by 10-fold serial dilution. Fresh 10-ml cultures were then inoculated with 1 ml from the previous 24-hour culture, OD600 values were recorded, as were the CFU per ml established prior to 24-hour incubation. Similar to previously reported results (49), absorbance values for both 0.1% catalase and adenine cultures increased after each 24-h passage (Fig. 3A); however, we failed to see a corresponding increase in CFU counts and actually noted a CFU decline after each passage for bacteria grown in catalase compared to those grown in adenine (Fig. 3B). In order to determine the reason for the apparent change in absorbance, hourly OD600 readings of sterile RnP medium (with either catalase or adenine) were taken without adding any bacteria to the cultures. These results showed that medium containing catalase without any bacteria added to it showed an increase in absorbance over 13 h (Fig. 3C). By comparison, RnP medium containing only adenine showed no such increase in absorbance. Plating dilutions from these cultures on blood agar plates resulted in no colony formation from either culture, suggesting that the increase in optical density observed in the presence of catalase was likely due to precipitation under the conditions tested (37°C, 10% CO2, with shaking). RnP medium containing either 0.1% catalase or adenine showed no deviation in absorbance when allowed to sit at room temperature, under static conditions. To ensure the presence of catalase activity, all medium containing 0.1% catalase was tested at the beginning and end of the experiments, based on the production of oxygen when samples of the medium were added to hydrogen peroxide (data not shown). The increase in optical density under normal bacterial growth conditions was not bacteria dependent, and therefore we were unable to confirm the previous results (49). Our studies suggest that H. pylori strain G27 did not grow in RnP medium in the absence of purine bases or nucleosides.
Fig 3.
H. pylori requires purines for growth. (A and B) OD600 values (A) and CFU counts (B) of sequential passages of strain G27. Cultures were grown in defined RnP medium in the presence of either 0.1% bovine liver catalase or adenine. (C) OD600 values were measured hourly for 24 h for culture flasks containing 0.1% bovine liver catalase or adenine with no bacteria added. Error bars represent the standard errors of the means.
H. pylori GMP synthase (ΔguaA) and IMP dehydrogenase (ΔguaB) mutants cannot survive in unsupplemented Ham's F-12 defined medium.
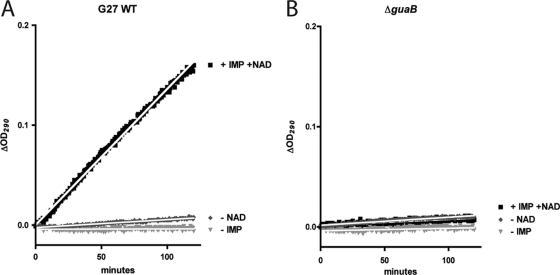
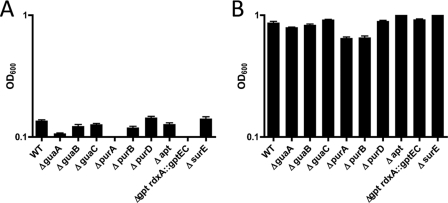
While H. pylori appears to have only a few genes associated with the de novo purine nucleotide biosynthesis pathway, it possesses numerous salvage pathway-associated genes. The presence of the purine salvage pathway in H. pylori had previously been established (50), and we wanted to further explore the limits of this pathway and ascertain the exact set of genes required to utilize given purine bases and nucleosides, first at the level of survival and then for that of sustained growth. Operating under the assumption that H. pylori requires purines salvaged from the environment in order to survive, we generated mutants with deletions of numerous genes in the purine nucleotide biosynthesis pathway (Table 1). To ensure that no other redundant IMPDHs were encoded in the genome, enzyme assays were performed on whole-cell lysates from G27 and the ΔguaB mutant, and the results indicated an absence of enzymatic activity in the deletion mutant (Fig. 4A and B).
Table 1.
Bacterial strains and plasmids used in this study
| Bacterial strain or plasmid | HP locusa in G27 (26695) | Function | Source or reference |
|---|---|---|---|
| H. pylori strains | |||
| G27 WT | 3 | ||
| G27 ΔguaA | HPG27_988 (HP0409) | GMP synthase | This study |
| G27 ΔguaB | HPG27_788 (HP0829) | IMP dehydrogenase | This study |
| G27 ΔguaB rdxA::guaB | HPG27_788 (HP0829) | (Complementation) | This study |
| G27 ΔguaC | HPG27_810 (HP0854) | GMP reductase | This study |
| G27 ΔguaC rdxA::guaC | HPG27_810 (HP0854) | (Complementation) | This study |
| G27 ΔpurA | HPG27_234 (HP0255) | Adenylosuccinate synthase | This study |
| G27 ΔpurB | HPG27_1054 (HP1112) | Adenylosuccinate lyase | This study |
| G27 ΔpurD | HPG27_1163 (HP1218) | Glycinamide ribonucleotide synthetase (putative) | This study |
| G27 Δapt | HPG27_531 (HP0572) | Adenine phosphoribosyltransferase | This study |
| G27 Δapt rdxA::apt | HPG27_531 (HP0572) | (Complementation) | This study |
| G27 Δgpt rdxA::gptEC | HPG27_691 (HP0735) | Guanine phosphoribosyltransferase | This study |
| G27 ΔsurE | HPG27_879 (HP0930) | Stationary-phase survival protein | This study |
| G27 ΔdeoD | HPG27_1121 (HP1178) | Purine nucleoside phosphorylase | This study |
| 60190 WT | 13 | ||
| 1061 WT | 27 | ||
| E. coli strains | |||
| DH5α | Plasmid amplification for sequencing | Lab strain | |
| TOP10 | Plasmid amplification for sequencing | Invitrogen | |
| MC4100 | Cloning of E. coligpt for complementation of H. pylori | Lab strain | |
| Plasmid | |||
| pCR2.1-TOPO | Cloning vector used for sequencing | Invitrogen |
Locus designations for H. pylori strains G27 and 26695, respectively.
Fig 4.
Enzymatic confirmation of the absence of IMPDH activity in ΔguaB mutant lysates. Enzymatic activity was measured for whole-cell lysates from wild-type strain G27 (A) and the ΔguaB deletion mutant (B). Optical density measurements at 290 nm were taken every 30 s for 2 h and used as a measure of XMP production. Enzymatic activity being substrate (IMP) and NAD dependent was shown by conducting the assays in the absence of either IMP or NAD. Lysates were generated twice, assays were each conducted twice, and linear regression lines were calculated for each assay. R2 values for linear regression lines for WT and ΔguaB strains (under NAD addition and IMP addition conditions) were 0.996 and 0.675, respectively, and 1/slope values were 733.8 and 315,600, respectively.
Mutants were first compared to the wild-type strain G27 for growth in brucella broth, either supplemented with 10% FBS (BB10) or unsupplemented (BB), in order to first show that, under rich medium conditions, the mutant strains (i) grew and (ii) survived similarly to the parental, G27 wild-type strain (Fig. 5). Attempts were then made to culture these mutants in commercially available defined medium (Ham's F-12), which contains only a single purine base, hypoxanthine (65, 66). Liquid cultures (2 ml) were inoculated in 12-well tissue culture plates with ∼3.5 × 107 to 4 × 107 CFU/ml. Strain growth levels were compared to wild-type growth for each experiment. H. pylori has previously been shown to only grow to 107 to 108 CFU/ml in F-12 medium not supplemented with FBS (65, 66).
Fig 5.
Nutrient-rich 24-h growth assays for mutant strains used in the study. Deletion mutants were grown overnight in brucella broth (A) or brucella broth supplemented with 10% FBS (B). Starting OD600 values were ∼0.1. Error bars represent the standard errors of the means.
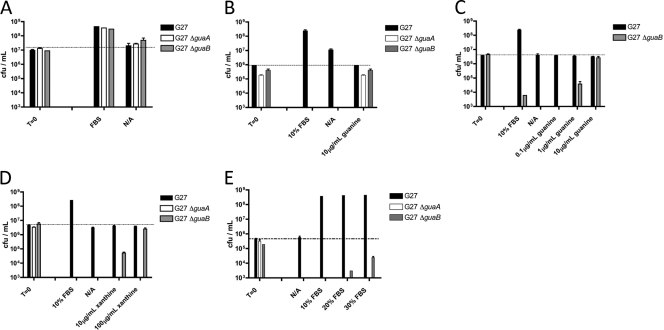
Given the importance of both guaA and guaB in both purine nucleotide de novo synthesis and salvage pathways, we began our observations with deletion mutants for each gene. We found that, without supplemental purines added to F-12 medium, ΔguaA and ΔguaB mutants were incapable of maintaining their initial inoculation levels (∼107 CFU/ml) in liquid culture for 24 h (Fig. 6). GMP reductase (ΔguaC), adenine phosphoribosyltransferase (Δapt), or purine nucleoside phosphorylase (ΔdeoD) mutants showed survival levels similar to that of the wild-type strain, suggesting their ability to still utilize hypoxanthine in the medium (data not shown). For mutants displaying growth defects under these conditions, survival was capable of being restored to the original inoculation levels (for ΔguaA and ΔguaB mutants) when certain purine bases were added to the medium (Fig. 6B to D). The addition of guanine or xanthine was capable of restoring bacterial survival in the ΔguaB mutant, whereas only guanine allowed the ΔguaA mutant to survive at a level comparable with the wild-type strain. Interestingly, 10 times as much xanthine was required to restore the ΔguaB mutant to wild-type levels in the 24-hour survival assay compared to guanine (Fig. 6C and D). When increasing concentrations of FBS were used in the F-12 medium 24-hour survival assay, the ΔguaB mutant had higher CFU counts, whereas the ΔguaA mutant showed no improvement in survival, suggesting the presence of significant levels of xanthine but not guanine in FBS (Fig. 6E).
Fig 6.
guaA, guaB, purA, and purB are essential for bacterial survival in Ham's F-12 medium. (A) CFU counts representing survival of G27 ΔguaA and G27 ΔguaB mutants and their parental strain in nutrient-rich brucella broth supplemented with 10% FBS for 24 h. (B to E) CFU counts representing survival of G27 ΔguaA and G27 ΔguaB mutants and the parental strain in defined medium (Ham's F-12) supplemented with either 10% FBS or increasing concentrations of guanine (B and C), xanthine (D), or increasing concentrations of FBS (E) for 24 h. Starting CFU/ml were between 106 and 107. Dashed lines indicate initial CFU/ml. Error bars represent the standard errors of the means. N/A, no additive.
H. pylori ΔguaA, ΔguaB, ΔguaC, ΔpurA, and ΔpurB mutants exhibit differing capabilities to utilize various purine bases or nucleosides for growth.
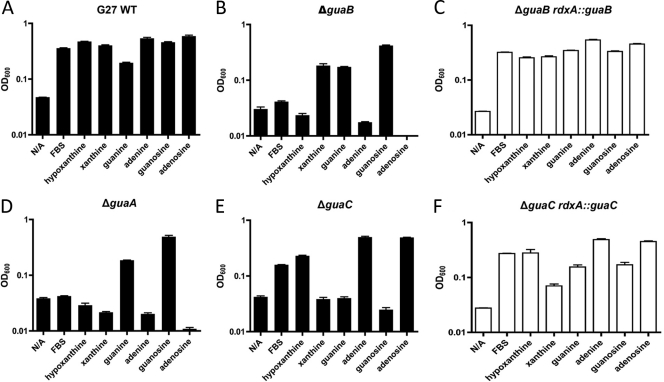
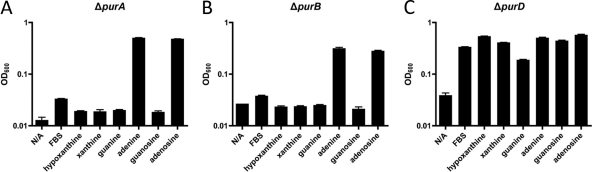
Having identified the requirement for supplemental purines for the survival of H. pylori under laboratory conditions, we next wanted to address the question of whether exogenous purines are required for bacterial growth. Using RnP medium (minus adenine) as a purine-free medium, various purine nucleotide biosynthesis mutants were tested for conditional growth defects in the presence of single purine bases or nucleosides. As stated previously, the parental G27 wild-type strain, as well as all other mutant strains tested, failed to grow in RnP medium lacking all purines. This purine-free RnP medium was then used as a negative control for all subsequent experiments. The ΔguaA mutant was only capable of growth on guanine or guanosine (Fig. 7D). The ΔguaB mutant was only capable of growth in xanthine, guanine, or guanosine (Fig. 7B). The ΔguaC mutant could only grow to wild-type levels in adenine or adenosine and was capable of reduced growth in hypoxanthine (Fig. 7E). When copies of the guaB and guaC genes were inserted into the rdxA locus for ΔguaB and ΔguaC mutants, respectively, the strains were once again capable of growth on all purine bases (Fig. 7C and F). ΔpurA and ΔpurB mutants were only capable of growth on adenine or adenosine (Fig. 8A and B), and notable retardation in growth was seen in both of these mutants in nutrient-rich medium (Fig. 5) as well as on blood agar plates (data not shown). It is worth noting that FBS appears capable of promoting robust growth when added to RnP medium for wild-type G27, as well as some level of growth for the ΔguaC mutant and minimal growth for the ΔguaB mutant. This may be explained by the fact that hypoxanthine is the major purine component found in FBS and that low levels of xanthine are present as well. However, when we repeated these experiments and examined growth levels at both 24 and 48 h, bacteria grown in RnP medium with only FBS added showed a drastic reduction in absorbance and CFU levels between the 24- and 48-h time points, compared to bacteria growing for this same length of time in RnP medium supplemented with purines, which maintained their higher absorbance and CFU levels through 48 h (data not shown). Taken together, these growth analyses indicate an auxotrophic phenotype for purine nucleotide biosynthesis in H. pylori, and they also map out a fully functional purine salvage pathway capable of utilizing any of the four purine bases, in addition to their nucleoside derivatives.
Fig 7.
When grown in RnP defined medium, G27 ΔguaA, ΔguaB, and ΔguaC mutants were capable of growth only in the presence of specific purine bases or nucleosides. OD600 values for 24-h growth assays of H. pylori strain G27 (A) and its ΔguaB (B), ΔguaB rdxA::guaB (C), ΔguaA (D), ΔguaC (E), and ΔguaC rdxA::guaC (F) purine nucleotide biosynthesis mutants in defined RnP medium containing only single purine bases or nucleoside sources. Starting OD600 values were between 0.03 and 0.04. Bars indicate growth of designated mutants (black) and, when applicable, complemented mutant strains (white). Error bars represent the standard errors of the means. N/A, no additive.
Fig 8.
G27 ΔpurA and ΔpurB mutants are capable of growth only in the presence of specific purine bases or nucleosides. OD600 values for 24-h growth assays of H. pylori ΔpurA (A), ΔpurB (B), and ΔpurD (C) strains in defined RnP medium containing only single purine bases or nucleoside sources are shown. Starting OD600 values were between 0.03 and 0.04. Bars indicate growth of designated mutants. Error bars represent the standard errors of the means. N/A, no additive.
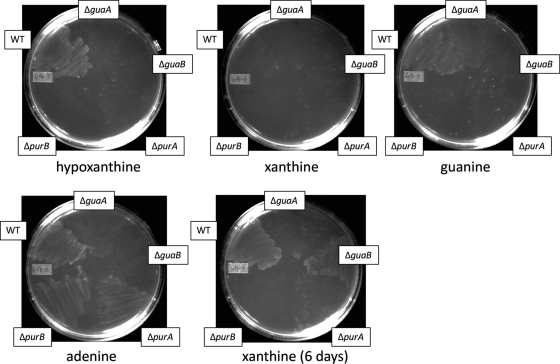
Growth assays were next conducted using agar medium. RnP medium plates containing single purines were used, and mutants were tested on plates containing only hypoxanthine, xanthine, guanine, or adenine (Fig. 9). Bacteria grown for 18 h on blood agar plates were used as the inoculum. The results generally correlated with the broth culture assays, although mutants as well as the wild-type strain appeared to have difficulty growing on xanthine (and to a lesser extent, guanine). This seemed to correlate with what we first noticed in the Ham's F-12 survival assay (Fig. 6): that xanthine, although capable of restoring growth for the ΔguaB mutant, does so inefficiently compared with the purine base guanine.
Fig 9.
Confirmation of mutant growth on plates containing only single purine bases as indicated. Mutants were grown on blood agar plates for 24 h and then streaked onto RnP agar plates containing only single purine bases. The first 4 plates represent 3 days of growth, and the fifth plate is the same xanthine-containing plate after 6 days of growth.
The G27 ΔpurD mutant has no growth defects when grown in the presence of any purine bases or nucleosides.
Phosphoribosylamine-glycine ligase (PurD) catalyzes the second step in de novo purine nucleotide biosynthesis (Fig. 1A), whereby 5-phosphoribosylamine becomes 5′-phosphoribosylglycinamide in the initial generation of the purine ring structure. H. pylori has a purD homolog present in the genome (HP1218) and, given H. pylori's lack of all other genes required for generating purines de novo (Fig. 1A), its function in H. pylori is currently a mystery. The encoded amino acid sequence retains high homology with genes from both Campylobacter jejuni and Helicobacter hepaticus, and an analysis of the catalytic residues indicated that the ribonucleotide- and ATP-binding domains (62) appear intact (see Fig. S2 in the supplemental material).
To ascertain the effects the absence of purD would have on H. pylori growth in our fully defined medium, a knockout mutation in purD was generated and tested for its ability to grow in RnP medium supplemented with single purine bases and nucleosides. We found that the ΔpurD mutant strain showed no difference in its ability to utilize any of the four purine bases or the two purine nucleosides for purine salvage compared with the parental wild-type strain (Fig. 8C), suggesting that PurD does not play any role in purine salvage in H. pylori.
Growth of H. pylori phosphorybosiltransferase mutants in RnP medium indicate a potential for divergent substrate specificities.
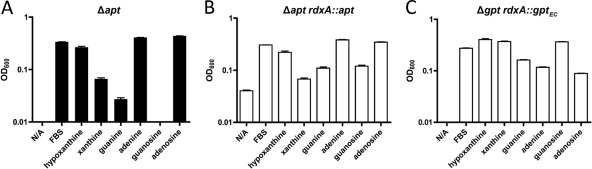
Phosphoribosyltransferases allow PRPP to be added to purine bases to generate nucleotides (Fig. 1) and are required for a functional, purine salvage pathway. H. pylori has two known phosphoribosyltransferases, HP0735 and HP0572, currently believed to encode guanine-phosphoribosyltransferase (Gpt) and adenine-phosphoribosyltransferase (Apt), respectively (1, 3). We were successful in completely deleting the H. pylori apt gene, and interestingly, defined medium growth assays in RnP medium showed a reduced ability of this mutant to utilize guanine or guanosine (and to a lesser extent, xanthine) for growth, while only marginal growth abnormalities were seen when the mutant when grown on adenine (Fig. 10A). This indicates that in H. pylori strain G27, Apt uses guanine as its primary substrate and that the conversion of adenine directly to ATP via Apt potentially is not the major mechanism for ATP generation when H. pylori is grown on adenine. When a copy of apt was inserted into the rdxA locus of the Δapt mutant strain, the ability to grow on guanine was restored (Fig. 10B).
Fig 10.
H. pylori phosphoribosyltransferase mutants show potential substrate specificities. OD600 values for 24-h growth assays of H. pylori strain G27 Δapt (A), Δapt rdxA::apt (B), and Δgpt rdxA::gptEC (C) phosphoribosyltransferase mutants in RnP medium containing only single purine bases or nucleoside sources. Bars indicate growth of designated mutants (black) and, when applicable, complemented mutant strains (white). Starting OD600 values were between 0.03 and 0.04. Error bars represent the standard errors of the means. N/A, no additive.
H. pylori's Gpt has already been shown to be essential under laboratory conditions (11), and we were similarly unable to generate either insertion mutations or complete deletion mutants for this gene (data not shown). To show that gpt is essential in H. pylori and that attempts to delete it were not simply affecting some other downstream essential gene, we attempted to place an alternate copy of the gene elsewhere in the genome (in the rdxA locus) and then remove the native copy. We were unsuccessful in complementing with a copy of the H. pylori gpt, as the complementation construct repeatedly would insert only into the wild-type locus and not the rdxA locus (data not shown). The E. coli gpt gene (gptEC), being divergent enough from the H. pylori homolog, was used for complementation to prevent unwanted transformants at the wild-type locus, and after inserting it into the rdxA locus, we were capable of removing the H. pylori gpt. Subsequent RnP growth assays in this Δgpt rdxA::gptEC mutant showed a slightly reduced ability to grow in the presence of adenine or adenosine (Fig. 10C). Our initial hypothesis was that, due to the presence of a hypoxanthine phosphoribosyltransferase (Hpt) in E. coli, E. coli Gpt may exhibit reduced substrate specificity for hypoxanthine, thus reducing H. pylori's ability to grow on hypoxanthine; however, we saw no growth defects when our mutant was grown on hypoxanthine, suggesting that gptEC is capable of utilizing hypoxanthine at a level adequate for the normal growth of H. pylori. gpt was the only salvage pathway-specific gene that we were unable to delete, indicating that the activity it possesses is essential to purine salvage in H. pylori.
The H. pylori ΔsurE mutant exhibited growth defects when grown in the presence of guanine.
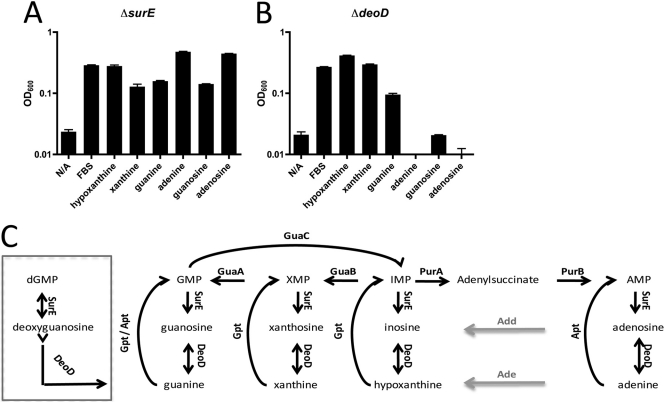
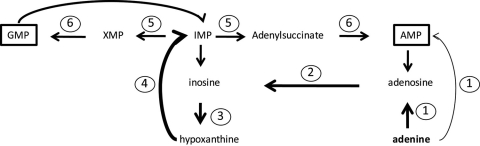
In relation to purine metabolism, surE and deoD, which encode stationary-phase survival protein and purine nucleoside phosphorylase, respectively, are generally studied in the context of purine turnover and degradation (Fig. 11C). Together, they sequentially remove the phosphate and then the sugar moiety from purine nucleotides, leaving the purine base. We tested a ΔsurE mutant in our purine growth assays to determine whether cycling of purine bases affected H. pylori's purine salvage pathway. The deletion mutant showed a slight growth defect when grown on either guanine or guanosine compared to growth on all other purine bases or nucleosides (Fig. 11A). The reasons for this guanine-specific growth defect are not readily apparent; however, one possibility is an alternate route for dGMP synthesis from guanine. The reaction by which SurE removes the phosphate group from nucleotides is believed to be unidirectional, with one exception (Fig. 11C, box): SurE is believed to be able to add or remove the phosphate group from deoxyguanosine and dGMP, respectively. This alternate pathway for dGMP production would be absent due to the removal of surE, and when only guanine is available as a purine source, a diminished ability to generate dGMP could result in growth reduction.
Fig 11.
H. pylori strain G27's purine salvage pathway. OD600 values for 24-hour growth assays of H. pylori strain G27 ΔsurE (A) and ΔdeoD (B) mutants. (C) The putative purine salvage pathway of H. pylori, with nonannotated enzymes adenosine deaminase (Add) and adenine deaminase (Ade) shown by gray lines. An alternate dGMP production pathway utilizing guanine and dependent on SurE is shown in the box. Starting OD600 values were between 0.03 and 0.04. Error bars represent the standard errors of the means. N/A, no additive.
Purine-nucleoside phosphorylase (deoD) is essential for growth on nucleosides, but is also required for growth on the purine base adenine.
Our data thus far suggest that H. pylori is capable of growing on all purine bases and nucleosides tested. We next examined the effects of deleting deoD (encoding purine nucleoside phosphorylase) to see if this would result in the loss of the ability to grow on purine nucleosides. Phosphoribosyltransferases (Gpt and Apt in H. pylori), as a general rule, can only use purine bases as substrates (Fig. 11C), and given that no obvious adenine or guanine kinases are encoded in the genome (1, 16), the only way nucleosides should be able to be utilized in purine salvage in H. pylori is if the sugar moiety attached to the purine base is first removed by nucleoside phosphorylase (DeoD) and then PRPP can be attached to the base to form a nucleotide. In the mutant lacking deoD, the ability to grow in either guanosine or adenosine was severely reduced (Fig. 11B). These results confirm the absence of adenosine kinase and guanosine kinase in H. pylori, as has been previously noted (1, 16, 50). Curiously, we found that the ΔdeoD mutant also was incapable of growth on the purine base adenine. This result hints at the potential existence of an as-yet-unknown adenosine deaminase in H. pylori.
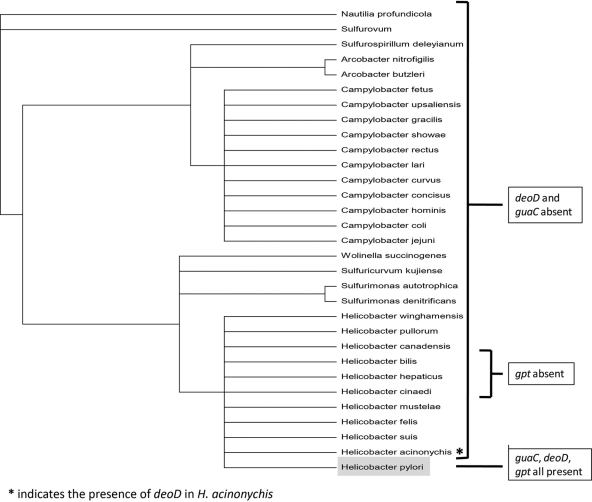
Computational analysis of the purine nucleotide biosynthesis and salvage pathways in Epsilonproteobacteria reveals a correlation between the loss of de novo synthesis pathway genes and the acquisition of purine salvage pathway-associated genes (guaC and deoD).
Microbes adapting to new environments often undergo periods of genomic plasticity, during which they undergo a loss of genes that are no longer essential as well as an acquisition of new environment-specific genes. Regarding the evolutionary biology of H. pylori, bioinformatic analysis strongly indicates such a period of genetic flux existed for H. pylori and that this period appears to coincide with the loss of its de novo purine nucleotide biosynthesis pathway and acquisition of genes that bolster its purine salvage pathway. When three of the genes examined in this study (guaC, deoD, and gpt), which are all associated with H. pylori's purine salvage pathway, were compared against the phylogeny of Epsilonproteobacteria, a remarkable pattern emerged (Fig. 12). Of the Epsilonproteobacteria, H. pylori is the only species to possess all three of these salvage pathway genes. Furthermore, the presence of guaC may be specific to human colonization, as Helicobacter acinonychis strain Sheeba, hypothesized to have successfully jumped hosts from early humans to large felines (21), appears to have lost, or never, acquired a guaC homolog (a more robust cladogram analysis is provided in Fig. S3 of the supplemental material).
Fig 12.
Cladogram of Epsilonproteobacteria, for the order of Campylobacterales (generated via the PATRIC mulitgene phylogentic tree viewer). Species are listed in order of similarity at the genome level. Species are labeled according to the presence or absence of three purine salvage pathway-associated genes: deoD, purine nucleoside phosphorylase; guaC, GMP reductase; gpt, guanine phosphoribosyltransferase. H. pylori, the only bacterium lacking more than 4 of the 10 pur genes required for IMP production and thereby de novo purine nucleotide biosynthesis, is highlighted in gray.
The absence of one or two homologs in certain de novo pathway genes has been established previously in the bacterial literature, and a number of Epsilonproteobacteria, as well as numerous other bacterial species, appear capable of de novo purine nucleotide biosynthesis in the absence of a purK homolog. H. pylori, however, is the only Epsilonproteobacteria to apparently lack homologs to 8 out of the 10 genes that encode enzymes known to be essential for classical de novo purine nucleotide biosynthesis (purF, purT, purN, purL, purM, purK, purE, purC, and purH). Of the two remaining de novo pathway-associated genes, purB also plays a role in the purine salvage pathway, leaving purD as the only “purely de novo” purine nucleotide biosynthesis gene present in H. pylori.
DISCUSSION
H. pylori strain G27 appears capable of growth on all purine bases as well as their nucleoside equivalents. Strains 60190 and 1061 also grow on all purine bases and appear to require exogenous purines for growth. This requirement for purines appears confirmed by bioinformatic analysis of the H. pylori genome, which has indicated the absence of several genes that encode enzymes thought to be essential for de novo purine biosynthesis (1, 3, 16). An illustration of H. pylori's putative de novo purine biosynthesis is present in the literature in the aforementioned report of a 0.1% catalase growth assay that indicated H. pylori is capable of growing in the absence of purines (49). In the course of our study, we were unable to replicate this purine-free growth, despite supplementing RnP medium with 0.1% bovine liver catalase, and we therefore conclude that the presence of catalase is insufficient to allow de novo purine nucleotide biosynthesis to occur.
We were successful in deleting nine genes from H. pylori's purine salvage pathway, and we found multiple genes had unique effects on growth and survival under tested laboratory conditions. Hypoxanthine and adenine both can normally be used by the bacterium to generate ATP, but neither alone can overcome the blockage in the pathway represented in the ΔguaA and ΔguaB mutants in order to generate GTP (Fig. 1). An inability for our ΔguaA mutant to grow on hypoxanthine-rich medium (Ham's F-12) supplemented with 30% FBS combined with the fact that guanine (the only purine base on which our ΔguaA mutant could grow) is found in relatively small amounts in blood and tissue is a good indication that GMP synthase is a promising target for future molecular inhibitors. Our ΔguaB mutant's inability to grow on adenine and hypoxanthine, the two most prominent purine bases present in human tissues and blood, shows IMPDH is also a promising drug target, as has previously been shown (26, 32). We found it interesting that while it was capable of growth on hypoxanthine, our ΔguaC mutant exhibited lower growth levels on this purine source than our wild-type strain. This indicated to us the importance of purine cycling for H. pylori; however, another potential explanation could be that GMP reductase is in fact involved in the generation of GMP from IMP. This reaction has been shown previously (54) and relies on the presence of ammonia. Given the constant production of ammonia via H. pylori's urease, this may explain our results in regard to alterations in growth on hypoxanthine.
The fact that we were able to only generate a deletion mutant for apt and not gpt confirms that GPT is the major phosphoribosyltransferase utilized by H. pylori (11). In addition, the Δapt mutant strain did not appear to have significant growth defects when grown on adenine, compared with the wild-type strain, and its inability to grow in guanine indicated either (i) that H. pylori adenine phosphoribosyltransferase is in fact also its primary guanine-phosphoribosyltransferase, or (ii) that H. pylori Gpt alone is incapable of meeting H. pylori's purine needs when grown only in the presence of guanine. When grown for 48 h, the Δapt mutant was capable of growth in xanthine but never to the levels of growth seen in adenine or hypoxanthine, and it did not grow to the same level as the wild-type strain grown in xanthine (data not shown). Due to the reduced growth observed in H. pylori containing E. coli gpt when grown in adenine and adenosine, we considered the possibility that the H. pylori Gpt might be capable of utilizing adenine as a substrate. Attempts to generate ΔpurA Δapt and ΔpurB Δapt double deletion mutants were unsuccessful, suggesting that H. pylori's GPRTase does not have sufficient substrate specificity to utilize adenine in order to maintain growth solely on adenine.
The inserted gptEC failed to fully complement the deletion of gpt from the H. pylori genome, as was evident from the reduced growth of H. pylori on adenine. As Gpt is not believed to possess APRTase activity, the basis for this observed phenotype is not readily apparent. H. pylori possesses GMP reductase (guaC), which allows it to utilize guanine or xanthine to synthesize either GMP or AMP, as GMP can be reduced to IMP, which is then free to be utilized in AMP synthesis (Fig. 1B). The H. pylori genome does not encode a recognized AMP deaminase, nor does it contain homologs to adenine or adenosine deaminases, which would promote the conversion of adenine/adenosine to hypoxanthine/inosine, thereby allowing AMP/adenosine/adenine to be utilized in the synthesis of GMP. Despite this, the rate at which H. pylori takes up adenine has been shown to be the highest among all the purine bases examined (50), and APRTase activity was found to be the highest of the PRTases examined (50). The question of how H. pylori is capable of growth solely on adenine has been asked previously in the literature, and one study showed adenine deaminase activity from whole-cell lysates (49), indicating the presence of a potentially novel adenine deaminase. The presence of such an enzyme would allow for a pathway by which GMP could be synthesized with only adenine as the initial substrate. It would also explain why H. pylori Gpt is essential under laboratory conditions, as it would be required for the generation of GMP from any of the purine bases (Fig. 11C).
Our genetic evaluation, however, suggests that the activity is instead that of an adenosine deaminase, due to the inability of our ΔdeoD mutant to grow on adenine. We were able to detect what we believed to be adenosine deaminase activity in crude, whole-cell lysates via spectrometry (data not shown); however, due to the limited amounts of enzyme and substrate that could be used via this method, we were unable to measure activity under the conditions in which the original adenine deaminase activity was characterized (49), and a more robust evaluation of this observed activity is under investigation. Assuming the presence of an adenosine deaminase, we present one possible pathway for adenine usage for AMP or GMP production, as outlined in Fig. 13. This model potentially could explain H. pylori's limited growth on adenine when its gpt is removed and replaced with gptEC, as E. coli possesses an additional phosphoribosyltransferase (HPRTase) with high specificity for hypoxanthine (15), and its GPRTase has preferred substrate specificities for guanine and xanthine (15). This model would also explain why Gpt is essential in H. pylori.
Fig 13.
Potential metabolic pathway by which H. pylori is capable of generating GMP and AMP from a single purine source, adenine. The pathway for adenine processing assumes weak APRTase activity and the presence of an adenosine deaminase. (Step 1) Adenine is utilized by APRTase to generate AMP, while simultaneously, purine nucleoside phosphorylase is adding a sugar moiety to adenine (generating adenosine), which allows for recognition of the nucleoside by adenosine deaminase. (2) Adenosine is deaminated to inosine, which is then (3) phosphorlyated to hypoxanthine, allowing for (4) utilization by H. pylori's Gpt. Subsequently, GMP and AMP are produced via the usual fashion, (5) first from IMP to XMP or adenylsuccinate and (6) finally to GMP and AMP.
This model would appear to be thermodynamically favorable, given that nucleoside synthesis is favored over nucleoside phosphorolysis (25) and that the equilibrium for purine base/nucleosides can be tipped in either direction by either PRTase or adenine/adenosine deaminase activities (which use up purine bases/nucleosides, respectively). We found it perplexing that our Δapt mutant maintained its ability to utilize adenine without any significant growth defects, as well as the fact that our ΔpurA and ΔpurB mutants exhibited growth defects even when grown on nutrient-rich media or on adenine. Taken together, there exists the possibility that perhaps very little APRTase activity is present in H. pylori strain G27. If little or no adenine phosphoribosyltransferase activity is present, depleting adenine stores (given that apparently Apt's major substrate in this bacterial strain is in fact guanine), then the accumulation of adenine (in addition to the presence of adenosine deaminase) would further push the equilibrium toward the generation of adenosine. The presence of HPRTase activity would then push the equilibrium in favor of nucleoside phosphorolysis, and increasing amounts of inosine subsequently would be converted to hypoxanthine.
This hypothesis would seemingly conflict with previous findings showing strong APRTase activity in H. pylori lysates (50); however, the presence of adenine deaminase activity (as opposed to adenosine deaminase activity), also noted in the literature (49), would thermodynamically favor strong APRTase activity and would remove the requirement for DeoD for adenine processing (see Fig. S4B in the supplemental material). The presence of apparent adenosine and adenine deaminase activities when only one of the two enzymes is encoded in the genome has been shown previously in other bacteria when a purine nucleoside phosphorylase is also present (20). It has also been shown that the difference between substrate preferences for adenine and adenosine deaminases can be as little as 1 to 4 amino acid substitutions (57), and H. pylori strain variances could explain the presence of either adenosine or adenine deaminase activity. It has been postulated in the literature that H. pylori may possess numerous unidentified genes that, although different from previously described orthologs, are capable of carrying out the same enzymatic functions (16). Strain variability may explain these apparent differences, and further examination of these activities, as well as the identification of the gene encoding the adenine/adenosine deaminase enzyme, must occur prior to further analysis.
Another potential hypothesis is that, due to previous studies using whole-cell lysates to examine APRTase and adenine deaminase activities of H. pylori (using the generation of either AMP or hypoxanthine, respectively, as readouts), single enzymatic activities were assigned to what may have actually been numerous, multistep reactions. In the case of APRTase activity (shown to be almost three times that of HPRTase activity), the generation of AMP results from both the Apt phosphoribosyltransferase as well as the stepwise generation of AMP from adenine described above: via adenine/adenosine deaminase, HPRTase, adenylosuccinate synthase, and lyase. Similarly, adenine deaminase activity as measured by hypoxanthine production could include the stepwise generation of hypoxanthine from adenine via purine nucleoside phosphorylase and an adenosine deaminase. What was originally characterized as purine nucleoside phosphotransferase activity (50) might also have simply been the stepwise removal of the sugar by purine nucleoside phosphorylase (DeoD) activity and subsequent addition of PRPP by the adenine phosphoribosyltransferase (Apt). Identification of the H. pylori gene encoding the adenine/adenosine deaminase enzyme would allow for further exploration of these single and/or stepwise enzymatic activities.
H. pylori has coevolved with humans for hundreds of thousands of years, to the extent that every human migration in recorded history can be traced using the genetic code of their chromosomal, mitochondrial, and Helicobacter DNA (22, 72). Much like intracellular bacterial pathogens, H. pylori has a severely reduced genome, given its limited contact with the “outside” world, and has long since lost its ability to survive outside its human host. As such, its genome is quite streamlined and seemingly well suited for living only within the human gastric mucosa (7). Its metabolic pathways, as a result, have been selected for this relatively stable environment, and superfluous or redundant pathways have long since been removed from the genome (41, 44). H. pylori lives off the nutrients obtained from gastric epithelial cells, either relatively passively in the case of strains lacking the cagA gene and/or possessing the s2/m2 vacA allele, or aggressively in the case of the more pathogenic strains containing cagA and the s1/m1 vacA allelic variant. Adenine, hypoxanthine, and xanthine can be found ubiquitously in mammalian tissues (71), in contrast with guanine, which is found in only low levels in blood and tissue (28). Gastric epithelial cells undergo rapid growth and renewal and require large amounts of purines to sustain their continuous divisions. When these cells undergo apoptosis they exhibit a controlled release of nucleotides via the plasma membrane channel pannexin 1, which is used to recruit phagocytes and regulate selective plasma membrane permeability (12). All of these factors seem to point to a relatively stable, purine-rich environment within the gastric mucosa and in which purine salvage would seem to take precedence over de novo nucleotide biosynthesis. Purine salvage is always energetically favorable compared with de novo synthesis (2, 8), and given the modest purine needs of slow-growing H. pylori, it is quite possible that its salvage pathway is more than adequate for its environmental niche.
In many bacteria, purine nucleotide biosynthesis and salvage genes are directly regulated by the amounts of purines present. Often, numerous purine nucleotide biosynthesis genes are found in the same operons, allowing them to be controlled by a single, regulatory mechanism (34–36). Regulators for purine nucleotide biosynthesis and salvage pathways are abundant; regulatory promoter-binding proteins (33, 34) and riboswitches (4, 43) are utilized for many of these genes in most other bacteria. By comparison, the purine synthesis genes found in H. pylori are spread throughout the genome (44) without any apparent order or regulation. While many are found in operons (64), the other genes in these operons are not believed to be associated with purine nucleotide biosynthesis. We attempted to use bioinformatics to look for the presence of any genetic regulatory schemes in the genes we examined in H. pylori purine salvage and processing pathways, and we were unable to find anything indicating a “classical” regulatory network based on the presence or absence of purines. We did find potential sigma 70 −10 regions for most of these genes, and previous studies have shown that nucleotide biosynthesis-related genes in H. pylori generally have ∼30-nucleotide-long 5′-untranslated regions (UTRs) (compared to much larger and variable 5′-UTRs for most other gene classes, such as protein synthesis or transcription), indicating their apparent optimization for translation (64). The single caveat in regard to the regulation of the purine nucleotide salvage pathway is the apparent upregulation of GMP reductase in the presence of acid, and numerous studies have noted this phenomena (10, 64, 73), although none have yet identified the regulatory mechanism responsible.
In organisms containing both de novo and salvage pathways, regulation of purine pools is generally required in order to switch between the de novo and salvage pathways; however, in the absence of one of these pathways, extensive regulation may simply not be required. An absence of the more classical regulatory elements for most of the genes examined, coupled with H. pylori's inability to survive outside its human host, suggests that H. pylori requires a purine-rich environment. Few classically recognized regulators in general have been identified in H. pylori (1, 5), and while the potential of regulation by antisense or sRNAs has been shown to exist (64), the genes examined in this work do not appear to be regulated even by this newly found means of regulation. The apparent absence of classical regulation at these loci indicates to us that H. pylori potentially exists in a purine-rich environment, and as such, the expression of purine salvage enzymes is “hard-wired,” compared with free-living organisms, which require ranges of regulation due to the dynamic natures of their habitats. Experiments testing this hypothesis are ongoing.
The existence of other bacteria lacking de novo synthesis pathways points out that, given the right microenvironment, purine salvage can be sufficient within a human host (36, 55). As many as 10 years ago, loss of the purine de novo nucleotide biosynthesis pathway was thought to represent a relatively rare evolutionary event. However, due to the plethora of bioinformatic data now available, numerous examples from all manner of genera can be found. Strict intracellular pathogens, such as species from the genera Chlamydia and Rickettsia, the parasitic flagellate protozoan Trypanosoma, Treponema pallidum, Clostridiales family genomospecies strain BVAB3 (isolated from the human vaginal cavity via the Human Microbiome Project), Mycoplasma, Ureaplasma, Mesoplama, Borrelia, and even many lactobacilli, all appear to exist without the presence of the classical de novo purine nucleotide biosynthesis pathway. It is noteworthy that many of these organisms are associated with mucosal epithelial layers, much like H. pylori, and that not all are strict intracellular pathogens. In the end, it is the environment that chooses an organism's genes, and if a microbe can exist for long enough in a niche that does not require the synthesis of purine nucleotides de novo, those genes, no longer under selection, will be lost over time, either passively in the form of point mutations or actively in the form of “genetic black holes” (47).
The evolutionary biology of H. pylori's apparent loss of its de novo pathway appears to also coincide with the acquisition of its purine salvage pathway. Of the Epsilonproteobacteria, H. pylori is the only species known to possess gpt, guaC, and deoD, all three being salvage pathway-associated genes, as well as the only species to lack 8 out of the 10 genes encoding enzymes known to be essential for classical de novo purine nucleotide biosynthesis. When the results of the growth assays conducted in this work are laid against this evolutionary framework, the reasons for this apparent coincidence become clear. Gpt, guanine/xanthine phosphoribosyltransferase, allows purine bases to be salvaged for the generation of purine nucleotides. DeoD allows nucleosides to be utilized for the generation of purine nucleotides and potentially allows for cycling between AMP and GMP via an as-yet-unannotated adenosine deasminase, thereby allowing adenine to be utilized to generate both AMP and GMP. GuaC, GMP reductase, permits cycling between GMP and AMP, allowing salvaged xanthine and guanine to be utilized to generate both GMP and AMP. With these three genes, H. pylori has seemingly gained the ability to utilize any single purine base or nucleoside for the generation of either GMP and/or AMP. Our data as well as previous findings in the literature (49) show that H. pylori also possesses an adenine or adenosine deaminase, which would be yet another example of the presence of a purine salvage pathway enzyme present in H. pylori while very few Epsilonproteobacteria genomes appear to encode adenine or adenosine deaminases. Given a relatively competitor-free, purine-rich environment and H. pylori's slow growth rate, our results suggest that any ancestral de novo purine biosynthesis pathway may not have been required for H. pylori to survive in the human host.
We conclude that H. pylori has developed a purine salvage pathway that allows for the usage of any single purine base or nucleoside to generate both GMP and AMP. Given the lack of de novo synthesis gene homologs in H. pylori, its relatively purine-rich environment, and its lack of growth pressures, we also conclude that, much like other microbes with similar genetic pathways and environments, H. pylori has lost its ability to generate purine nucleotides de novo. Future experiments will be conducted to determine the effects of these various salvage pathway mutants in vivo, thereby potentially hinting at the actual utilization of purine bases by H. pylori during infection. It has already been shown in vitro that targeting the purine uptake and processing machinery in H. pylori has bacteriostatic potential (26, 32), and analogs of purine bases have been shown to have cytotoxic effects (49). Knowledge of what purines are available to the bacterium in its actual gastric niche will help validate the importance of each enzyme in the pathway and will indicate which one enzyme or combination of enzymes offer the best targets to disrupt in this essential metabolic pathway in H. pylori.
Supplementary Material
ACKNOWLEDGMENTS
This research was supported in part by the University of Virginia Cancer Center through the James and Rebecca Craig Research Scholars Award and the NCI Cancer Center Support Grant (P30 CA44579), and G.L. was supported in part by a University of Virginia Cancer Training Grant (5T32 CA 009109).
We give special thanks to Lizbeth Hedstrom, Paul Hoffman, F. Heath Damron, and Laura Porter for their helpful comments in preparing the manuscript.
Footnotes
Published ahead of print 22 December 2011
Supplemental material for this article may be found at http://jb.asm.org/.
REFERENCES
- 1. Alm RA, Trust TJ. 1999. Analysis of the genetic diversity of Helicobacter pylori: the tale of two genomes. J. Mol. Med. 77:834–846 [DOI] [PubMed] [Google Scholar]
- 2. An S, Kumar R, Sheets ED, Benkovic SJ. 2008. Reversible compartmentalization of de novo purine biosynthetic complexes in living cells. Science 320:103–106 [DOI] [PubMed] [Google Scholar]
- 3. Baltrus DA, et al. 2009. The complete genome sequence of Helicobacter pylori strain G27. J. Bacteriol. 191:447–448 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Batey RT, Gilbert SD, Montange RK. 2004. Structure of a natural guanine-responsive riboswitch complexed with the metabolite hypoxanthine. Nature 432:411–415 [DOI] [PubMed] [Google Scholar]
- 5. Berg DE, Hoffman PS, Appelmelk BJ, Kusters JG. 1997. The Helicobacter pylori genome sequence: genetic factors for long life in the gastric mucosa. Trends Microbiol. 5:468–474 [DOI] [PubMed] [Google Scholar]
- 6. Bjorkholm B, et al. 2001. Mutation frequency and biological cost of antibiotic resistance in Helicobacter pylori. Proc. Natl. Acad. Sci. U. S. A. 98:14607–14612 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Blaser MJ. 1997. The versatility of Helicobacter pylori in the adaptation to the human stomach. J. Physiol. Pharmacol. 48:307–314 [PubMed] [Google Scholar]
- 8. Brault JJ, Terjung RL. 2001. Purine salvage to adenine nucleotides in different skeletal muscle fiber types. J. Appl. Physiol. 91:231–238 [DOI] [PubMed] [Google Scholar]
- 9. Brubaker RR. 1970. Interconversion of purine mononucleotides in Pasteurella pestis. Infect. Immun. 1:446–454 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Bury-Mone S, et al. 2004. Responsiveness to acidity via metal ion regulators mediates virulence in the gastric pathogen Helicobacter pylori. Mol. Microbiol. 53:623–638 [DOI] [PubMed] [Google Scholar]
- 11. Chalker AF, et al. 2001. Systematic identification of selective essential genes in Helicobacter pylori by genome prioritization and allelic replacement mutagenesis. J. Bacteriol. 183:1259–1268 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Chekeni FB, et al. 2010. Pannexin 1 channels mediate ‘find-me’ signal release and membrane permeability during apoptosis. Nature 467:863–867 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Cover TL, Tummuru MK, Cao P, Thompson SA, Blaser MJ. 1994. Divergence of genetic sequences for the vacuolating cytotoxin among Helicobacter pylori strains. J. Biol. Chem. 269:10566–10573 [PubMed] [Google Scholar]
- 14. Croxen MA, Sisson G, Melano R, Hoffman PS. 2006. The Helicobacter pylori chemotaxis receptor TlpB (HP0103) is required for pH taxis and for colonization of the gastric mucosa. J. Bacteriol. 188:2656–2665 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Deo SS, Tseng WC, Saini R, Coles RS, Athwal RS. 1985. Purification and characterization of Escherichia coli xanthine-guanine phosphoribosyltransferase produced by plasmid pSV2gpt. Biochim. Biophys. Acta 839:233–239 [DOI] [PubMed] [Google Scholar]
- 16. Doig P, et al. 1999. Helicobacter pylori physiology predicted from genomic comparison of two strains. Microbiol. Mol. Biol. Rev. 63:675–707 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Doyle PS, Kanaani J, Wang CC. 1998. Hypoxanthine, guanine, xanthine phosphoribosyltransferase activity in Cryptosporidium parvum. Exp. Parasitol. 89:9–15 [DOI] [PubMed] [Google Scholar]
- 18. Duckworth M, Menard A, Megraud F, Mendz GL. 2006. Bioinformatic analysis of Helicobacter pylori XGPRTase: a potential therapeutic target. Helicobacter 11:287–295 [DOI] [PubMed] [Google Scholar]
- 19. Dunn BE, Cohen H, Blaser MJ. 1997. Helicobacter pylori. Clin. Microbiol. Rev. 10:720–741 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Endo T, Uratani B, Freese E. 1983. Purine salvage pathways of Bacillus subtilis and effect of guanine on growth of GMP reductase mutants. J. Bacteriol. 155:169–179 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Eppinger M, et al. 2006. Who ate whom? Adaptive Helicobacter genomic changes that accompanied a host jump from early humans to large felines. PLoS Genet. 2:e120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Falush D, et al. 2003. Traces of human migrations in Helicobacter pylori populations. Science 299:1582–1585 [DOI] [PubMed] [Google Scholar]
- 23. Fields PI, Swanson RV, Haidaris CG, Heffron F. 1986. Mutants of Salmonella typhimurium that cannot survive within the macrophage are avirulent. Proc. Natl. Acad. Sci. U. S. A. 83:5189–5193 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Foynes S, et al. 2000. Helicobacter pylori possesses two CheY response regulators and a histidine kinase sensor, CheA, which are essential for chemotaxis and colonization of the gastric mucosa. Infect. Immun. 68:2016–2023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Friedmin M. 1950. Desoxyribose-1-phosphate. II. The isolation of crystalline desoxyribose-1-phosphate. J. Biol. Chem. 184:449–459 [PubMed] [Google Scholar]
- 26. Gollapalli DR, et al. 2010. Structural determinants of inhibitor selectivity in prokaryotic IMP dehydrogenases. Chem. Biol. 17:1084–1091 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Goodwin A, et al. 1998. Metronidazole resistance in Helicobacter pylori is due to null mutations in a gene (rdxA) that encodes an oxygen-insensitive NADPH nitroreductase. Mol. Microbiol. 28:383–393 [DOI] [PubMed] [Google Scholar]
- 28. Hartwick RA, Krstulovic AM, Brown PR. 1979. Identification and quantitation of nucleosides, bases and other UV-absorbing compounds in serum, using reversed-phase high-performance liquid chromatography. II. Evaluation of human sera. J. Chromatogr. 186:659–676 [DOI] [PubMed] [Google Scholar]
- 29. Hazell SL, Graham DY. 1990. Unsaturated fatty acids and viability of Helicobacter (Campylobacter) pylori. J. Clin. Microbiol. 28:1060–1061 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Hazell SL, Markesich DC, Evans DJ, Evans DG, Graham DY. 1989. Influence of media supplements on growth and survival of Campylobacter pylori. Eur. J. Clin. Microbiol. Infect. Dis. 8:597–602 [DOI] [PubMed] [Google Scholar]
- 31. Hedstrom L. 2009. IMP dehydrogenase: structure, mechanism, and inhibition. Chem. Rev. 109:2903–2928 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Hedstrom L, Liechti G, Goldberg JB, Gollapalli DR. 2011. The antibiotic potential of prokaryotic IMP dehydrogenase inhibitors. Curr. Med. Chem. 18:1909–1918 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Husnain SI, Thomas MS. 2008. Downregulation of the Escherichia coli guaB promoter by FIS. Microbiology 154:1729–1738 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Husnain SI, Busby SJ, Thomas MS. 2009. Downregulation of the Escherichia coli guaB promoter by upstream-bound cyclic AMP receptor protein. J. Bacteriol. 191:6094–6104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Jenkins A, et al. 2011. Role of purine biosynthesis in Bacillus anthracis pathogenesis and virulence. Infect. Immun. 79:153–166 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Jewett MW, et al. 2009. GuaA and GuaB are essential for Borrelia burgdorferi survival in the tick-mouse infection cycle. J. Bacteriol. 191:6231–6241 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Kennemann L, et al. 2011. Helicobacter pylori genome evolution during human infection. Proc. Natl. Acad. Sci. U. S. A. 108:5033–5038 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Keough DT, et al. 2009. Inhibition of hypoxanthine-guanine phosphoribosyltransferase by acyclic nucleoside phosphonates: a new class of antimalarial therapeutics. J. Med. Chem. 52:4391–4399 [DOI] [PubMed] [Google Scholar]
- 39. Keough DT, et al. 2010. Plasmodium vivax hypoxanthine-guanine phosphoribosyltransferase: a target for anti-malarial chemotherapy. Mol. Biochem. Parasitol. 173:165–169 [DOI] [PubMed] [Google Scholar]
- 40. Kotloff KL, et al. 2000. Shigella flexneri 2a strain CVD 1207, with specific deletions in virG, sen, set, and guaBA, is highly attenuated in humans. Infect. Immun. 68:1034–1039 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Lara-Ramirez EE, et al. 2011. New implications on genomic adaptation derived from the Helicobacter pylori genome comparison. PLoS One 6:e17300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Lawrence KA, Jewett MW, Rosa PA, Gherardini FC. 2009. Borrelia burgdorferi bb0426 encodes a 2′-deoxyribosyltransferase that plays a central role in purine salvage. Mol. Microbiol. 72:1517–1529 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Mandal M, Boese B, Barrick JE, Winkler WC, Breaker RR. 2003. Riboswitches control fundamental biochemical pathways in Bacillus subtilis and other bacteria. Cell 113:577–586 [DOI] [PubMed] [Google Scholar]
- 44. Marais A, Mendz GL, Hazell SL, Megraud F. 1999. Metabolism and genetics of Helicobacter pylori: the genome era. Microbiol. Mol. Biol. Rev. 63:642–674 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Marshall BJ, Warren JR. 1984. Unidentified curved bacilli in the stomach of patients with gastritis and peptic ulceration. Lancet i:1311–1315 [DOI] [PubMed] [Google Scholar]
- 46. Martinelli LK, et al. 2011. Recombinant Escherichia coli GMP reductase: kinetic, catalytic and chemical mechanisms, and thermodynamics of enzyme-ligand binary complex formation. Mol. Biosyst. 7:1289–1305 [DOI] [PubMed] [Google Scholar]
- 47. Maurelli AT, Fernandez RE, Bloch CA, Rode CK, Fasano A. 1998. “Black holes” and bacterial pathogenicity: a large genomic deletion that enhances the virulence of Shigella spp. and enteroinvasive Escherichia coli. Proc. Natl. Acad. Sci. U. S. A. 95:3943–3948 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. McFarland WC, Stocker BA. 1987. Effect of different purine auxotrophic mutations on mouse-virulence of a Vi-positive strain of Salmonella dublin and of two strains of Salmonella typhimurium. Microb. Pathog. 3:129–141 [DOI] [PubMed] [Google Scholar]
- 49. Mendz GL, Shepley AJ, Hazell SL, Smith MA. 1997. Purine metabolism and the microaerophily of Helicobacter pylori. Arch. Microbiol. 168:448–456 [DOI] [PubMed] [Google Scholar]
- 50. Mendz GL, Jimenez BM, Hazell SL, Gero AM, O'Sullivan WJ. 1994. Salvage synthesis of purine nucleotides by Helicobacter pylori. J. Appl. Bacteriol. 77:674–681 [DOI] [PubMed] [Google Scholar]
- 51. Nedenskov P. 1994. Nutritional requirements for growth of Helicobacter pylori. Appl. Environ. Microbiol. 60:3450–3453 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Oyston PC, et al. 2010. A Yersinia pestis guaBA mutant is attenuated in virulence and provides protection against plague in a mouse model of infection. Microb. Pathog. 48:191–195 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Parsonnet J. 1995. The incidence of Helicobacter pylori infection. Aliment. Pharmacol. Ther. 9(Suppl. 2):45–51 [PubMed] [Google Scholar]
- 54. Patton GC, et al. 2011. Cofactor mobility determines reaction outcome in the IMPDH/GMPR (β/α)8 barrel enzymes. Nat. Chem. Biol. 7:950–958 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Pettersson J, et al. 2007. Purine salvage pathways among Borrelia species. Infect. Immun. 75:3877–3884 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Polissi A, et al. 1998. Large-scale identification of virulence genes from Streptococcus pneumoniae. Infect. Immun. 66:5620–5629 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Pornbanlualap S, Chalopagorn P. 2011. Adenosine deaminase from Streptomyces coelicolor: recombinant expression, purification and characterization. Protein Expr. Purif. 78:167–173 [DOI] [PubMed] [Google Scholar]
- 58. Reynolds DJ, Penn CW. 1994. Characteristics of Helicobacter pylori growth in a defined medium and determination of its amino acid requirements. Microbiology 140:2649–2656 [DOI] [PubMed] [Google Scholar]
- 59. Rodriguez M, Good TA, Wales ME, Hua JP, Wild JR. 2005. Modeling allosteric regulation of de novo pyrimidine biosynthesis in Escherichia coli. J. Theor. Biol. 234:299–310 [DOI] [PubMed] [Google Scholar]
- 60. Ropp PA, Traut TW. 1991. Allosteric regulation of purine nucleoside phosphorylase. Arch. Biochem. Biophys. 288:614–620 [DOI] [PubMed] [Google Scholar]
- 61. Ropp PA, Traut TW. 1991. Purine nucleoside phosphorylase. Allosteric regulation of a dissociating enzyme. J. Biol. Chem. 266:7682–7687 [PubMed] [Google Scholar]
- 62. Sampei G, et al. 2010. Crystal structures of glycinamide ribonucleotide synthetase, PurD, from thermophilic eubacteria. J. Biochem. 148:429–438 [DOI] [PubMed] [Google Scholar]
- 63. Santiago AE, et al. 2009. Characterization of rationally attenuated Francisella tularensis vaccine strains that harbor deletions in the guaA and guaB genes. Vaccine 27:2426–2436 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Sharma CM, et al. 2010. The primary transcriptome of the major human pathogen Helicobacter pylori. Nature 464:250–255 [DOI] [PubMed] [Google Scholar]
- 65. Testerman TL, McGee DJ, Mobley HL. 2001. Helicobacter pylori growth and urease detection in the chemically defined medium Ham's F-12 nutrient mixture. J. Clin. Microbiol. 39:3842–3850 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Testerman TL, Conn PB, Mobley HL, McGee DJ. 2006. Nutritional requirements and antibiotic resistance patterns of Helicobacter species in chemically defined media. J. Clin. Microbiol. 44:1650–1658 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Thiele I, Vo TD, Price ND, Palsson BO. 2005. Expanded metabolic reconstruction of Helicobacter pylori (iIT341 GSM/GPR): an in silico genome-scale characterization of single- and double-deletion mutants. J. Bacteriol. 187:5818–5830 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Tomb JF, et al. 1997. The complete genome sequence of the gastric pathogen Helicobacter pylori. Nature 388:539–547 [DOI] [PubMed] [Google Scholar]
- 69. van Amsterdam K, van der Ende A. 2004. Nutrients released by gastric epithelial cells enhance Helicobacter pylori growth. Helicobacter 9:614–621 [DOI] [PubMed] [Google Scholar]
- 70. Wang Y, Roos KP, Taylor DE. 1993. Transformation of Helicobacter pylori by chromosomal metronidazole resistance and by a plasmid with a selectable chloramphenicol resistance marker. J. Gen. Microbiol. 139:2485–2493 [DOI] [PubMed] [Google Scholar]
- 71. Wishart DS, et al. 2007. HMDB: the Human Metabolome Database. Nucleic Acids Res. 35:D521–D526 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Yamaoka Y. 2009. Helicobacter pylori typing as a tool for tracking human migration. Clin. Microbiol. Infect. 15:829–834 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Zanotti G, Cendron L. 2010. Functional and structural aspects of Helicobacter pylori acidic stress response factors. IUBMB Life 62:715–723 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.