Abstract
Extracellular DNA acts as a cation chelator and induces the expression of antibiotic resistance genes regulated by Mg2+ levels. Here we report the characterization of novel DNA-induced genes in Pseudomonas aeruginosa that are annotated as homologs of the spermidine synthesis genes speD (PA4773) and speE (PA4774). The addition of sublethal concentrations of DNA and membrane-damaging antibiotics induced expression of the genes PA4773 to PA4775, as shown using transcriptional lux fusions and quantitative RT-PCR. Exogenous polyamine addition prevented DNA- and peptide-mediated gene induction. Mutation of PA4774 resulted in an increased outer membrane (OM) susceptibility phenotype upon polymyxin B, CP10A, and gentamicin treatment. When the membrane-localized fluorescent probe C11-BODIPY581/591 was used as an indicator of peroxidation of membrane lipids, the PA4774::lux mutant demonstrated an increased susceptibility to oxidative membrane damage from H2O2 treatment. Addition of exogenous polyamines protected the membranes of the PA4774::lux mutant from polymyxin B and H2O2 treatment. Polyamines from the outer surface were isolated and shown to contain putrescine and spermidine by using high-performance liquid chromatography and mass spectrometry. The PA4774::lux mutant did not produce spermidine on the cell surface, but genetic complementation restored surface spermidine production as well as the antibiotic and oxidative stress resistance phenotypes of the membrane. We have identified new functions for spermidine on the cell surface and propose that polyamines are produced under Mg2+-limiting conditions as an organic polycation to bind lipopolysaccharide (LPS) and to stabilize and protect the outer membrane against antibiotic and oxidative damage.
INTRODUCTION
Pseudomonas aeruginosa is a Gram-negative opportunistic pathogen that is able to cause chronic infections, including wound infections, otitis media, chronic sinusitis, and lung infections in persons with cystic fibrosis (CF) (29, 53, 56). Chronic infections are proposed to be caused by bacterial biofilms, and there is strong evidence to suggest that P. aeruginosa grows as a biofilm in the lungs during chronic infections in CF patients (11, 46, 53). P. aeruginosa is also a major cause of hospital-acquired infections, many of which are due to biofilm colonization of medical implant devices or respirators (14). This opportunistic pathogen possesses high levels of intrinsic antibiotic resistance due to reduced outer membrane permeability, drug inactivation, drug efflux, and mechanisms involving target site mutation (17, 20). However, P. aeruginosa surface-adherent biofilms are 4- to 2,500-fold more resistant to various antimicrobial agents than free-floating planktonic cultures (1, 7, 49, 51). Biofilms are multicellular, surface-associated, microbial communities encased in an extracellular matrix that displays a complex three-dimensional structure and has increased resistance to antimicrobials, environmental stresses, and the host immune response (3, 12, 25). Bacteria growing in a biofilm can evade the host immune response and antibiotic therapy, complicating the treatment process and leading to the development of chronic infections (11, 46, 53).
Gram-negative bacteria possess an outer membrane (OM), which functions as a permeability barrier to extracellular compounds. The asymmetric outer membrane is composed of lipopolysaccharides (LPS) in the outer leaflet and phospholipids in the inner leaflet. LPS is stabilized by the binding of divalent cations such as Mg2+ and Ca2+, which bind neighboring LPS molecules such that displacement of the cations results in destabilization and major disruptions of the cell membrane integrity, ultimately leading to cell death (19). One mechanism of antibiotic resistance involves modifications of LPS that reduce the binding and penetration of cationic antimicrobial peptides. The aminoarabinose LPS modification operon arnBCADTEF-ugd is controlled by the PhoPQ and PmrAB cation-sensing two-component systems and is regulated in response to Mg2+ levels in planktonic cultures (43, 44). Under Mg2+-limiting conditions, this operon is required for antimicrobial peptide resistance (37). Strains with spontaneous polymyxin B resistance mutations in pmrB that constitutively produced aminoarabinose-modified LPS were isolated (47), and in these strains, aminoarabinose addition to the phosphates of the lipid A core masks the negative charges and reduces the binding of cationic antimicrobial peptides to the outer membrane. More recently, polymyxin B-resistant clinical isolates of P. aeruginosa with specific regulatory mutations that demonstrated increased expression of PhoPQ and PmrAB-controlled genes, including the arn operon, were described (59).
In a study to identify biofilm-specific antibiotic resistance determinants in P. aeruginosa, Mah et al. described the ndvB gene that was upregulated in biofilms and was required for the production of periplasmic cyclic glucans, which bind aminoglycosides and prevent their entry into the cytoplasm (41). We recently reported a novel property of DNA as a divalent metal cation chelator, likely due to its highly anionic charge from phosphates in the deoxyribose backbone (49). At high concentrations, DNA is toxic and chelates cations from the OM, leading to major disruptions of both inner and outer membranes and death, similar to the cation-chelating activity of EDTA (49). However, the addition of sublethal levels of DNA to planktonic cells mimics a DNA-rich biofilm and induces the arn operon, resulting in high levels of resistance to antimicrobial peptides and aminoglycosides (49). As extracellular DNA accumulates in the biofilm matrix, we proposed that this outer membrane permeability mechanism of resistance is another example of a biofilm-induced resistance mechanism (49).
In previous studies examining the PhoPQ and PmrAB regulons, we identified the PA4773-PA4775 gene cluster, located immediately upstream of pmrAB, which is induced under Mg2+-limiting conditions (Fig. 1) and is regulated primarily by PmrAB (37, 43, 44). The genes PA4773 and PA4774 show significant similarity to the polyamine synthesis genes speD (52%) and speE (56%) of Escherichia coli. SpeD converts S-adenosylmethionine (SAM) to decarboxylated SAM (dSAM), which is used as a substrate with putrescine to synthesize spermidine by SpeE (40, 62). P. aeruginosa contains speD (PA0654) and speE (PA1687), in addition to their respective homologs PA4773 and PA4774. PA4775 is annotated as a hypothetical protein of unknown function with a PSORT-predicted signal peptide and one predicted transmembrane domain.
Fig 1.
Schematic view of PA4773 to PA4775. PA4773 to PA4775 are located immediately upstream of pmrAB. The locations of the mini-Tn5-lux transposon mutants are indicated (the black triangle indicates a nontranscriptional lux fusion; white triangles indicate transcriptional lux fusions), along with the locations of the promoters and the PmrA binding site, located 67 bp upstream of the start codon of PA4773. Primer pair locations used for qRT-PCR are indicated by the small arrows below the diagram. The putative promoters (P1, P2, and P3) were determined on the basis of transcript length analysis, as shown in Fig. S1 in the supplemental material.
Polyamines are hydrocarbons containing two or more amine groups and are therefore polycationic. In bacteria, spermidine and putrescine are the predominant cytoplasmic polyamines; cadaverine is found in lesser quantities, and the presence of spermine is controversial (60). These molecules are universal, being found in some combination in all cellular life forms (60). Their ubiquitous nature has made defining their exact role within cells difficult. Research is widely available to suggest that they are essential for optimal cell growth and viability, and additionally, they have been linked to microbial pathogenesis, biofilm formation, escape from phagolysosomes, bacteriocin production, toxin activity, and antibiotic resistance (60, 66). Polyamines are frequently associated with the negatively charged nucleic acids (and proteins) in the cytoplasm and have been shown to protect DNA from oxidative damage (16).
Here, we report that PA4773 to PA4775 are induced by the cation-chelating activity of extracellular DNA. We provide evidence that PA4773 and PA4774 constitute an inducible pathway of spermidine synthesis in P. aeruginosa as a replacement for divalent metal cations and have identified novel functions of polyamines in protecting bacterial membranes from antibiotic and oxidative damage.
MATERIALS AND METHODS
Bacterial strains and growth conditions.
All strains and plasmids used are listed in Table 1. P. aeruginosa PAO1 was used as the wild-type strain. The mini-Tn5-lux transposon mutants in PA4773 to PA4775 were previously isolated and shown not to have polar effects on the downstream regulatory pmrAB genes, as previously published (see Fig. S1 in the supplemental material) (37, 43). Unless otherwise stated, cultures were routinely grown at 37°C in BM2 defined minimal medium with either high (2 mM) or low (0.02 mM) MgSO4. BM2 growth medium includes the following components: 0.1 M HEPES (pH 7), 7 mM ammonium sulfate, 20 mM sodium succinate (pH 6.7), 10 μM iron sulfate, 1,600 μM phosphate buffer (pH 7.2), 1.62 μM manganese sulfate, 2.45 μM calcium chloride, 13.91 μM zinc chloride, 4.69 μM boric acid, and 0.67 μM cobalt chloride (48). When added, the source of extracellular DNA was fish sperm DNA (USB).
Table 1.
Bacterial strains used in this studya
| Strain or mutation | Genotype | Reference or source |
|---|---|---|
| Strains | ||
| PAO1 | Wild-type P. aeruginosa | R. E. Hancock |
| PA4773 | PA4773 transposon mutant, nontranscriptional fusion, 98_E1 | 37 |
| PA4773-4::lux | Intergenic PA4773-PA4774 transposon mutant, and transcriptional fusion, 11_A9 | 43 |
| PA4774::lux | PA4774::lux transposon mutant and transcriptional fusion, 38_F9 | 37 |
| PA4775::lux | PA4775::lux transposon mutant and transcriptional fusion, 50_F5 | 37 |
| PA4774::luxCC | PA4774::lux chromosomally complemented with PA4773-PA4775 integrated into attTn7 site | This study |
| PA4773-4::lux phoQ::xylE | Double mutant; transcriptional fusion PA4773-4::lux in a phoQ::xylE mutant background | 43 |
| PA4773-4::lux pmrB::xylE | Double mutant; transcriptional fusion PA4773-4::lux in a pmrB::xylE mutant background | 43 |
| PA3553::lux | PA3553::lux transposon mutant and transcriptional fusion, 53_D10 | 37 |
| Plasmids | ||
| pUCP22 | Ampr, multicopy broad-host-range expression vector | 64a |
| pCR2.1-TOPO | PCR cloning vector, Ampr | Invitrogen |
| pUC18T-mini-Tn7T-Gm | Gmr on mini-Tn7T; mobilizable; for gene insertion in Gms bacteria | 10 |
| pTNS2 | Helper plasmid for integration into the att site | 10 |
| pUC18T-mini-Tn7T-PA4773-PA4775 | pUC18T-mini-Tn7 containing PA4773-PA4775 genes, including native upstream promoter | This study |
Abbreviations: Ampr, ampicillin resistance; Gmr, gentamicin resistance; Gms, gentamicin sensitive.
Gene expression assays.
Gene expression was performed in a high-throughput format using 96-well microplates as previously described (49). Briefly, overnight cultures were grown in BM2 defined medium supplemented with 2 mM or 0.02 mM Mg2+ and extracellular DNA as indicated, diluted 1/1,000 into 150 μl of culture medium in 96-well black plates with a transparent bottom (9520 Costar; Corning Inc.), and overlaid with 50 μl of mineral oil to prevent evaporation. Microplate planktonic cultures were incubated at 37°C in a Wallac Victor3 luminescence plate reader (Perkin-Elmer), and optical density at 600 nm (OD600) (to assess growth) and luminescence (counts per second [CPS]) (to assess gene expression) readings were taken every 20 min throughout 18 h of growth.
Preparation of surface polyamines.
Mid-log-phase cultures (100 ml) were grown under inducing conditions (0.02 mM Mg2+). Cells were collected by centrifugation (10 min, room temperature [RT], 5,000 × g) and normalized to equivalent cell numbers for all strains. Cells were resuspended in 1 ml of 1 M NaCl dissolved in 10 mM HEPES (4-(2-hydroxyethyl)-1-piperazineethanesulfonic acid) buffer (pH 7.4) and incubated for 10 min at 37°C shaking. After incubation, cells were centrifuged three times (10 min, RT, 5,000 × g) to remove whole cells, and the supernatant, which now contained the surface-washed polyamines as previously described (31), was collected.
Measurement of XylE activity.
XylE assays were performed as previously described with minor modifications (43). Twenty-five-milliliter cultures of P. aeruginosa phoQ::xylE (43) were grown to mid-log phase. Cells were collected and subjected to a 1 M NaCl wash, a 50 mM KH2PO4 (pH 7.5) buffer wash, or a 50 mM KH2PO4 (pH 7.5) buffer wash containing 10% acetone followed by sonication, which lysed cells and released the enzyme XylE (catechol 2,3-dioxygenase) from the cytoplasm. XylE assays were performed after the addition of 0.3 mM catechol in 50 mM potassium phosphate buffer to 100 μl of lysates or culture washes. The conversion of catechol to 2-hydroxymuconic semialdehyde was measured every 5 min over the course of 1 h by measuring A405 with a Wallac Victor3 luminescence plate reader (Perkin-Elmer), and the XylE activity was measured as a change in absorbance.
HPLC-UV-mass spectrometry (MS) analysis of polyamines.
Polyamines were analyzed as their dansyl chloride derivatives in reversed-phase high-performance liquid chromatography (HPLC) with modifications of previously established methods (15, 31, 32). The dansylation reaction mixture consisted of 100 μl of NaCl-washed cell surface extract, 100 μl of 0.5 M NaHCO3-Na2CO3 buffer (pH 9.4), and 100 μl of 20 mg/ml dansyl chloride. After incubation for 60 min at 60°C with shaking, methylamine (30 μl of 0.5 mol/liter) was added to the reaction mixture to consume the excess dansyl chloride. The solutions were vortexed, incubated for an additional 30 min at 60°C, centrifuged for 10 min at 13,400 × g, and finally transferred into new vials.
HPLC separation was performed with an Agilent 1200 SL HPLC system with a Luna C18 (2) reverse-phase column (50 by 2 mm) (Phenomenex, Torrance, CA), thermostated at 30°C, with a buffer gradient system composed of 0.1% formic acid in water as mobile phase A and 0.1% formic acid in acetonitrile as mobile phase B at a flow rate of 0.20 ml/min. To 2 μl of sample solution, 9 μl of 0.1% formic acid in water was added, and 10 μl of this solution was injected onto the column. After injection, the column was washed with 95% mobile phase A and 5% mobile phase B for 2 min to effectively remove salts. Elution of the analytes was done by using a linear gradient from 5% to 95% mobile phase B over a period of 25 min. UV absorbance was monitored at 340 nm to detect dansyl-labeled compounds. Mass spectra were acquired in positive-mode ionization using an Agilent 6220 accurate-mass time-of-flight (TOF) HPLC/MS system (Santa Clara, CA) equipped with a dual sprayer electrospray ionization source, with the second sprayer providing a reference mass solution. Mass spectrometric conditions were as follows: drying gas, 10 liters/min at 325°C; nebulizer, 20 lb/in2; mass range, 100 to 1,000 Da; acquisition rate, ∼1.03 spectra/s; fragmentor, 120 V; skimmer, 60 V; capillary, 3,200 V; and instrument state, 2-GHz extended dynamic range. Mass correction was performed for every individual spectrum using peaks at m/z 121.05087 and 922.00979 from the reference solution. Data acquisition was performed using the Mass Hunter software package (version B.02.01.) HPLC-UV-MS data were analyzed by using Agilent Mass Hunter qualitative analysis software (version B.03.01) using a targeted approach.
Construction of chromosomally complemented strain.
Initial attempts to express single genes from pUCP plasmids were unsuccessful due to increased toxicity, as determined by live/dead staining (data not shown), when these genes were expressed individually in high-copy plasmids. Therefore, we cloned PA4773, PA4774, and PA4775 with their native upstream promoter and integrated them into the chromosome at the neutral attTn7 sites using the mini-Tn7 integration system as previously described (9). The genes were PCR amplified using the primers P4773FBamHI and P4775RBamHI and cloned into the TA cloning vector pCR2.1 (Invitrogen). The insert was BamHI excised and subcloned into the BamHI-digested pUC18T-mini-Tn7T-Gm plasmid (9). The P. aeruginosa PA4774::lux mutant strain was transformed simultaneously with pUC18T-mini-Tn7-PA4773-PA4775 and the helper plasmid pTNS2 (10) via electroporation. Transformants were selected on LB agar containing 50 μg/ml gentamicin, and positive clones were confirmed by PCR. The gentamicin resistance cassette was subsequently removed following the protocol by Choi and Schweizer, using Flp-mediated excision (9).
Outer membrane permeability assay.
Outer membrane permeability in response to polymyxin B treatment was assessed as previously described, using a PTI Quantamaster spectrofluorimeter (39). Briefly, P. aeruginosa strains were grown in BM2 with 0.02 mM Mg2+ to mid-log phase (OD600 = 0.5), washed and resuspended in 5 mM HEPES buffer (pH 7.2) containing 5 mM glucose and 0.5 mM carbonyl cyanide m-chlorophenylhydrazone (CCCP), and held at room temperature until testing was completed, which was within 2 h. Twenty microliters of 1-N-phenylnaphthylamine (NPN; 0.5 mM stock) was added to one milliliter of culture (OD600 = 0.5), and baseline NPN fluorescence was recorded for up to 50 s before the addition of 6.4 μg/ml (final concentration) polymyxin B or 20 μg/ml gentamicin. Fluorescence measurements were taken until the counts/s reached a steady state (generally within 150 s). To monitor NPN fluorescence, the excitation/emission wavelengths were set to 350 nm and 420 nm, respectively, both with 5-nm slit widths. To complement the OM permeability phenotypes, control polyamines (Sigma) were exogenously added to the cultures at various concentrations, and cultures were incubated for 10 min prior to challenge with antibiotics.
Quantitative reverse transcription (RT)-PCR.
Bacterial cultures of P. aeruginosa PAO1 and the PA4773, PA4773-4::lux, and PA4774::lux mutants were grown in BM2 medium containing 2 mM or 0.02 mM Mg2+ or 2 mM Mg2+ supplemented with 0.75% DNA to an OD600 of 0.25 to 0.30. Cells were then treated with RNAprotect bacterial reagent (Qiagen) according to the manufacturers' instructions and harvested by centrifugation prior to storage at −80°C. Total RNA was extracted using an RNeasy RNA isolation minikit (Qiagen) and treated with DNase using DNAfree (Ambion), and cDNA was synthesized with a high-capacity cDNA synthesis kit (ABI Biosystems). Prokaryotic gene expression was measured by using iQ SYBR green supermix (Bio-Rad) and bacterial primers specific to PA3553, PA4773, PA4774, PA4775, pmrA, and the proC housekeeping gene (58). Primer sequences are given in Table 2. For quantitative RT-PCR (qRT-PCR), quantification and melting curve analyses were performed with an iQ5 real-time pcr system (Bio-Rad) according to the manufacturer's instructions. Each reaction was done in triplicate, and standard deviations were used to calculate a range of activation (fold) using the 2−ΔΔCT method (38).
Table 2.
Primer sequences
| Primer | 5′–3′ sequencea |
|---|---|
| P4773FBamHI | CCGGATCCGTATCCACCAGCCGTACCTG |
| P4775RBamHI | CCGGATCCTCAGTGATTCCACAATTCCC |
| PA3552rtF | GTAGCGGCATCCATTTCATC |
| PA3552rtR | CATCGACGTTTCTCCAGGAT |
| PA4773rtF | CAGTGGATCGAGGAAAGCAT |
| PA4773rtR | GTACTCCGGCCAGGTATGG |
| PA4774rtF | TTCTACGAGCTGCTGCATTC |
| PA4774rtR | GACCTGGGAGAAGACACTGC |
| PA4775rtF | GTACCGCTGGCCGTCAAC |
| PA4775rtR | GTCTTCGCTCAGCTCGATG |
| pmrArtF | CACCAGGTGACCCTGTCC |
| pmrArtR | CGTAGAGGCTCTGCTCCAGT |
| proCrtF | CAGGCCGGGCAGTTGCTGTC |
| proCrtR | GGTCAGGCGCGAGGCTGTCT |
Restriction sites are underlined.
BODIPY581/591 microscopy.
Overnight cultures were subcultured and grown to mid-log phase in low-Mg2+ (0.02 mM) BM2 medium at 37°C. Cells were then stained with 2 μM C11-BODIPY581/591 for 10 min prior to treatment with 5 mM H2O2 for 10 min. Cells were visualized on 1% agarose beds with a Leica DMI4000 B inverted microscope equipped with an ORCA R2 digital camera and Metamorph software for image acquisition. The excitation and emission filters were 555/25 and 605/52 for red fluorescence and 490/20 and 525/36 for green fluorescence.
Membrane lipid peroxidation assay.
Overnight cultures were subcultured and grown to mid-log phase in low-Mg2+ (0.02 mM) BM2 medium at 37°C. Cells were concentrated to 1 × 109 CFU/ml and stained with 2 μM C11-BODIPY581/591, which was incubated at room temperature to allow membrane incorporation. Cells (1 × 108 CFU/well) were added to 96-well black microplates with transparent bottoms (9520 Costar; Corning Inc.), and an initial fluorescence reading taken using a Wallac Victor3 plate reader (Perkin-Elmer) with emission filters set to 632 nm for red and 535 nm for green. After time zero readings, H2O2 was added (5 to 25 mM), and red and green fluorescence readings were taken every 5 min over the course of 2 h.
Statistical analysis.
Statistical analysis was performed on the data using the unpaired Student's t test to calculate significant differences between PAO1 and mutant strains.
RESULTS
The Mg2+-regulated genes PA4773 to PA4775 are induced by the cation-chelating activity of extracellular DNA.
Previous work has shown PA4773 to PA4775 to be strongly induced by limiting (0.02 mM) Mg2+ and repressed under high (2 mM)-Mg2+ conditions (37, 43, 44). This result was confirmed using the PA4773-4::lux, PA4774::lux, and PA4775::lux strains, which are transcriptional lux fusions and transposon insertion mutants (Fig. 2A). Purified PmrA-His6 has been shown to bind the PA4773 promoter, which contains a PmrA binding site (Fig. 1) (44). PA4773 to PA4775 are also induced by the addition of sublethal doses of antimicrobial peptides under high-Mg2+ conditions, as shown with the PA4774::lux strain (Fig. 2B) (43).
Fig 2.
The Mg2+-regulated genes PA4773 to PA4775 are induced by membrane-damaging antibiotics and extracellular DNA. (A) Expression of PA4773-4::lux, PA4774::lux, and PA4775::lux in low (0.02 mM) and high (2 mM) Mg2+ concentrations. (B) The effects of sub-MIC concentrations of colistin (1.25 μg/ml) and polymyxin B (PxnB) (1.25 μg/ml) on PA4774::lux expression were measured under noninducing conditions (BM2 medium with 2 mM Mg2+). Growth was not affected at these peptide concentrations (data not shown). (C) Effects of increasing concentrations of extracellular DNA (0.5% DNA; 5 mg/ml) added to BM2 medium with 2 mM Mg2+ on PA4773-4::lux, PA4774::lux, and PA4775::lux expression. (D) DNA-mediated induction of PA4774::lux with 1% DNA (10 mg/ml) was neutralized with excess Mg2+ (5 mM), or excess spermidine (Spd) (1.25 mM). Maximum levels of gene expression at 8 h are shown, where gene expression (in counts per second [CPS]) was normalized to growth (OD600). Each experiment was performed at least three times, and representative data are shown. The values shown are the averages from at least three technical replicates with standard deviations. (E) Quantitative RT-PCR was used to measure expression of PA3552, PA4773, and PA4774 in wild-type PAO1 cultured to mid-log phase (OD, 0.25 to 0.3) in BM2 with 0.02 mM Mg2+ or 2 mM Mg2+ with or without 0.75% DNA. The level of gene expression is presented as fold change relative to BM2 with 2 mM Mg2+. Values are means and standard errors of the means from triplicate qPCR experiments on RNA isolated from three independent experiments.
We recently showed that DNA is a cation chelator and induces the expression of the arnBCADTEF-ugd antimicrobial peptide resistance operon (49). To determine if PA4773 to PA4775 are also induced by the presence of extracellular DNA, we measured the expression of lux transcriptional fusions to each gene using a system where extracellular DNA was added to BM2 medium containing high levels of Mg2+ (2 mM). The addition of exogenous DNA to planktonic cultures induced expression of each of the transcriptional reporters PA4773-4::lux, PA4774::lux, and PA4775::lux, in a concentration-dependent manner (Fig. 2C). In this experiment, there are very low levels of expression without exogenous DNA, as the high concentration of Mg2+ (2 mM) represses expression of PA4773 to PA4775 (Fig. 2A). The DNA-mediated induction can be prevented by the addition of excess 5 mM Mg2+, indicating that it is the cation-chelating activity of extracellular DNA that induces expression of these genes (Fig. 2D). DNA-mediated induction can also be prevented by addition of excess spermidine (Fig. 2D).
As the lux reporter strains used to measure promoter activity also had mutations in these genes, quantitative RT-PCR was performed with wild-type PAO1 to show that limiting Mg2+ and exogenous DNA both induced the expression of PA4773 and PA4774. The expression of PA4773 and PA4774 was induced 6- to 17-fold by extracellular DNA and induced up to 800-fold by limiting Mg2+ (Fig. 2E). PA3552 is the first gene in the arn operon and was included as a positive control, as it is known to be both low-Mg2+ and DNA regulated (Fig. 2E) (49). Induction of the arn genes and PA4773 to PA4775 is highest in the presence of 20 μM Mg2+, and the addition of high concentrations of DNA (0.5 to 1%) to medium containing 2 mM Mg2+ cannot mimic the effects of 20 μM Mg2+. The highest level of DNA induction is comparable to gene expression in 500 μM Mg2+, indicating that DNA can chelate up to 75% of the Mg2+ in the medium (data not shown).
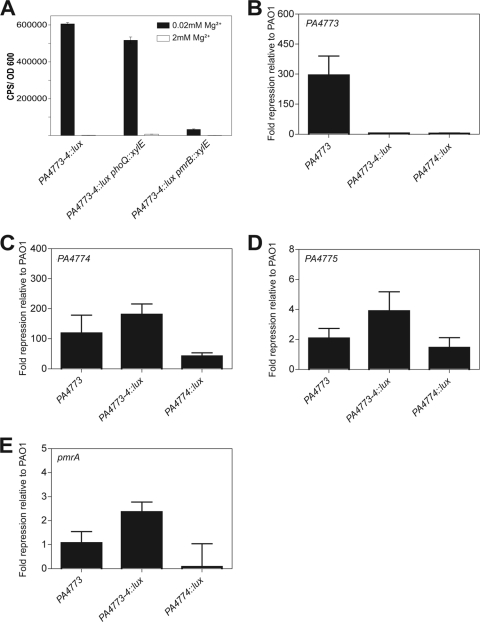
Expression of PA4773-4::lux was monitored under limiting Mg2+ conditions in a single PA4773-4::lux mutant, as well as in a double PA4773-4::lux phoQ::xylE mutant and a double PA4773-4::lux pmrB::xylE mutant. PA4773-4::lux expression was highly induced even when phoQ was inactivated, but no induction was seen when pmrB was inactivated in the double mutant (Fig. 3A). These data indicate that the PmrAB two-component system is required for the expression of PA4773-4::lux under limiting Mg2+ conditions, while the known cation-sensing system PhoPQ is not. As the expression of PA4773-4::lux is Mg2+ regulated, this result suggests that the downstream genes pmrAB are expressed and functional, despite the upstream transposon insertion.
Fig 3.
Transposon insertion into PA4773 and PA4774 does not cause polar effects on the downstream PA4775 or pmrAB genes. (A) Effects of phoQ and pmrB mutation on PA4773-4::lux expression in low (0.02 mM) and high (2 mM) Mg2+ conditions. (B to E) Quantitative RT-PCR analysis of PA4773, PA4774, PA4775, and pmrA gene expression in PA4773, PA4773-4::lux, and PA4774::lux mutants. The level of gene expression is represented as fold change relative to PAO1. All strains were cultured to mid-log phase in BM2 with 0.02 mM Mg2+. Values are means and standard errors of the means from triplicate qPCR experiments on RNA isolated from three independent experiments.
To confirm that transposon insertions in the PA4773, PA4773-4::lux, and PA4774::lux mutants do not cause polar effects on the downstream genes pmrAB, we examined the expression of each of the genes PA4773, PA4774, PA4775, and pmrA in PA4773, PA4773-4::lux, and PA4774::lux mutant backgrounds relative to their expression in PAO1 (Fig. 3B to E). Figure 3B shows that PA4773 is not expressed in a PA4773 mutant, indicated by the ∼300-fold repression in the mutant relative to the wild-type strain, and PA4773 expression is unaffected by downstream insertions. The expression of PA4774 is also repressed in a PA4773 mutant (120-fold) relative to PAO1, indicating that the upstream insertion has a polar effect on the PA4774 gene (Fig. 3C). However, there is a greater loss of expression of PA4773 than of PA4774 in a PA4773 mutant (300-fold versus 120-fold), which suggests that although there is a polar effect, there is residual expression and not a complete loss of PA4774 expression. Expression of PA4774 is also repressed in the PA4773-4::lux (180-fold) and PA4774::lux (40-fold) mutants, as is to be expected (Fig. 3C). The low level of repression observed for PA4775 (1.5- to 3.8-fold) (Fig. 3D) and pmrA (0.3- to 1.7-fold) (Fig. 3E) in PA4773, PA4773-4::lux, and PA4774::lux mutants indicates that the expression of PA4775 and pmrA is unaffected by upstream transposon insertions.
Polyamines are functionally interchangeable with divalent metal cations.
The divalent metal cation Mg2+ binds to LPS in the outer membrane and acts to stabilize LPS (19). The induction of the putative polyamine synthesis genes PA4773 and PA4774 by Mg2+ limitation or by DNA suggests that P. aeruginosa produces its own replacement cation to adapt to the Mg2+ limitation. DNA-mediated induction can be prevented by excess Mg2+ but also by addition of excess spermidine (Fig. 2D). The MIC of spermidine is 32 mM, and there were no effects on growth when spermidine was exogenously supplied to cultures at the concentrations used (data not shown). This result suggests that both divalent metal cations and polyamines can bind and neutralize extracellular DNA.
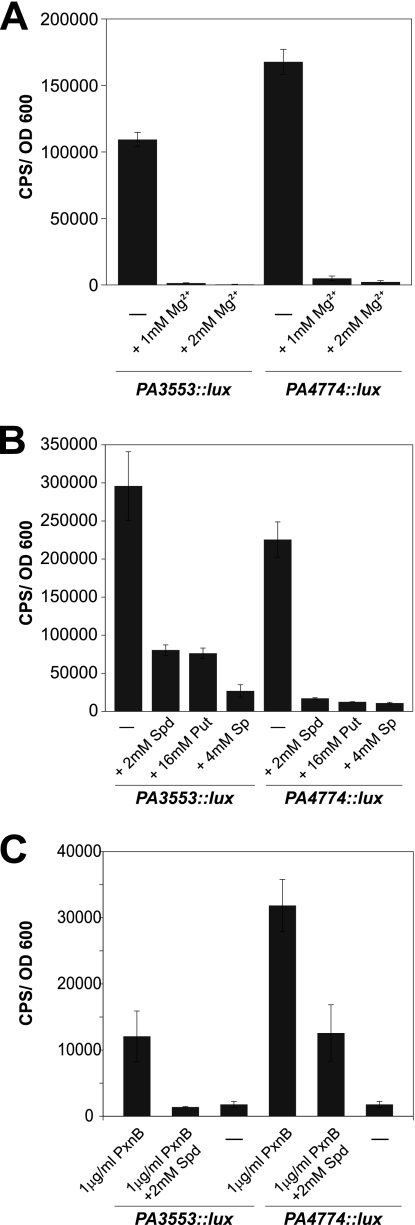
In growth medium with limiting Mg2+, the addition of excess Mg2+ cations (Fig. 4A) or excess polyamines (spermidine, putrescine, and spermine) (Fig. 4B) represses expression of PA4774::lux. Similarly, the polymyxin B-mediated induction of PA4774::lux can also be reduced by the addition of excess 2 mM spermidine (Fig. 4C). These two observations suggest that polyamines are functionally interchangeable and can compete with antimicrobial peptides for cation binding sites in the outer membrane. As an additional control, we showed that expression of the arnBCADTEF-ugd operon (PA3553::lux), a second example of a Mg2+-regulated operon, can also be prevented by the addition of exogenous divalent metal cations (Fig. 4A) or exogenous polyamines (spermidine, putrescine, and spermine) (Fig. 4B). The polymyxin B-mediated induction of PA3553::lux is also repressed by the addition of exogenous spermidine (Fig. 4C). These data support the hypothesis that polyamines can function as organic cations and perform a role similar to that of the divalent metal cations.
Fig 4.
Polyamines are functionally interchangeable with divalent metal cations. Expression of PA3553::lux and PA4774::lux was measured under limiting Mg2+ conditions (0.02 mM) and with the addition of excess 1 mM and 2 mM Mg2+ (A) or the exogenous addition of 2 mM spermidine (Spd), 16 mM putrescine (Put), or 4 mM spermine (Sp) (B). (C) Expression of PA3553::lux and PA4774::lux in BM2 with 2 mM Mg2+ containing polymyxin B (1 μg/ml) with or without 2 mM spermidine or BM2 with 2 mM Mg2+ alone. Maximum gene expression levels at 8 h are shown, where gene expression (in counts per second [CPS]) was normalized to growth (OD600). Each experiment was performed at least three times, and representative data are shown. The values shown are the averages from at least three technical replicates with standard deviations.
Spermidine and putrescine are both localized to the outer membrane of Pseudomonas aeruginosa.
Although polyamines are generally found in the cytoplasm, they have also been identified in the outer membrane and LPS of Salmonella enterica serovar Typhimurium and Escherichia coli (31). While the functions of polyamines in the cytoplasm are well studied, their function in the outer bacterial membrane is poorly understood. We employed the method described by Koski et al. (32) to isolate surface-associated polyamines and labeled them by derivatization with dansyl chloride. P. aeruginosa was grown under limiting Mg2+ conditions to induce expression of PA4773 to PA4775 for surface polyamine isolation and characterization. Cells were washed with 1 M NaCl to remove any noncovalently attached, surface-associated polyamines. Dansyl chloride-labeled polyamines from the NaCl-washed cells were analyzed by HPLC-UV-MS in order to detect all the dansylated compounds.
Figure 5 shows the extracted ion chromatograms (EIC) for the dilabeled putrescine and trilabeled spermidine. Dilabeled putrescine was observed in the mass spectrometer at a retention time of ∼21.7 min as the single-charged species at m/z 555.2085 ± 0.0010 (95% confidence level) (theoretical m/z 555.2094) and the double-charged species at m/z 278.1087 ± 0.0002 (theoretical m/z 278.1083). Trilabeled spermidine was observed in the mass spectrometer at a retention time of ∼25.5 min as the single-charged species at m/z 845.3168 ± 0.0045 (theoretical m/z 845.3183), the double-charged species at m/z 423.1629 ± 0.0014 (theoretical m/z 423.1628), and the triple-charged species at m/z 282.4447 ± 0.0003 (theoretical m/z 282.4443).
Fig 5.
Spermidine and putrescine were detected by HPLC-UV-MS on the outer surfaces of Pseudomonas aeruginosa cells. Extracted ion chromatograms (EIC) are shown for surface-associated polyamines of P. aeruginosa. (A) Di-dansyl chloride putrescine and tri-dansyl chloride spermidine were detected at retention times of ∼21.7 and ∼25.5 min, respectively, from the wild-type P. aeruginosa strain PAO1. (B) The PA4773 mutant contains both putrescine and spermidine on its surface. (C) The PA4774::lux mutant contains only putrescine, with a retention time of 21.7 min. (D) Spermidine production (retention time, 25.5 min) was restored in the surface-associated polyamines of the PA4774::luxCC strain. The EIC shows the m/z signal intensity of putrescine or spermidine (y axis) relative to the HPLC-UV-MS retention time (x axis). (E) XylE activity was measured in the wash fraction of cells treated with buffer (negative control) or washed with 1 M NaCl (NaCl wash), or in the lysate of solvent-treated and sonicated cells (positive control). XylE activity was defined as the change in absorbance over time, after the addition of the substrate catechol.
MS characterization of surface polyamines was performed with wild-type PAO1, the PA4773 and PA4774::lux mutants, and a strain in which PA4774::lux was chromosomally complemented, referred to here as the PA4774::luxCC strain. Unlike the surface fractions from the wild type and the PA4773 mutant, the surface fractions from the PA4774::lux mutant did not produce a UV340 absorbance peak at the 25.5-min retention time of spermidine (Fig. 5C), and only trace amounts of the compound were detected by the mass spectrometer at the limits of detection when a targeted analysis was performed. The production of spermidine was partially restored in the PA4774::luxCC strain (Fig. 5D). For complementation analysis, the genes PA4773 to PA4775 and the 500-bp upstream promoter region of PA4773 were integrated into the neutral attTn7 chromosome site using the mini-Tn7 integration system (9).
We performed the following experiment to ensure the 1 M NaCl wash did not cause cell lysis and the release of cytoplasmic polyamines, which may have subsequently contaminated our surface polyamine preparation. XylE is an enzyme expressed in the cytoplasm. We measured XylE activity in the wash fraction from cells treated with 1 M NaCl and found no release of cytoplasmic XylE (Fig. 5E). A buffer wash served as a negative control, and solvent-treated, sonicated cells served as a positive control (Fig. 5E). This experiment confirms that there was no cell lysis with NaCl treatment, and therefore no contamination of surface polyamines with cytoplasmic polyamines. A similar control experiment was also performed in previous polyamine studies with E. coli and Salmonella and also confirmed that no cytoplasmic contents contaminated the surface polyamines (31).
Spermidine synthesis from the SpeE homolog PA4774 contributes to reduced outer membrane permeability and antimicrobial peptide resistance.
Mechanisms of resistance to antimicrobial peptides involve modifications of teichoic acids and phospholipids in the Gram-positive membrane (33, 55) and modifications to the LPS (47) and phospholipids (22, 30) in Gram-negative bacteria. These membrane modifications reduce the negative surface charge, and we hypothesized that the cationic nature of polyamines would also neutralize the negative surface charge and contribute to antimicrobial peptide resistance. Mutations in PA4773 to PA4775 were previously shown to confer sensitivity to a range of different antimicrobial peptides, implicating this gene cluster in antimicrobial peptide resistance (37, 43). In the MIC assay, the arn mutants were most susceptible to polymyxin B (cyclic peptide), while the polyamine synthesis mutants were most sensitive to the indolicidin-derived peptides (37), suggesting that each modification provides resistance to unique peptides and that together the modifications provide broad-spectrum peptide resistance.
To confirm the protective role of surface polyamines, we utilized the NPN (1-N-phenylnaphthylamine) uptake assay to measure OM permeability in response to antimicrobial peptides (39). The fluorescent probe NPN is hydrophobic and fluoresces weakly within aqueous environments, but it is strongly fluorescent once integrated into the hydrophobic environment of lipid membranes (21, 23, 39, 64). The active transport processes of the inner membrane were inactivated by treatment with a sub-MIC concentration of the proton motive force (PMF) inhibitor carbonyl cyanide m-chlorophenylhydrazone (CCCP) (50). CCCP has previously been shown to inhibit polyamine uptake by intact cells, which is energized by the proton motive force (27).
NPN uptake to both inner and outer membranes after destabilization of the outer membrane by polymyxin B results in a sharp increase in NPN fluorescence (Fig. 6A and B). To determine the relative contribution of the aminoarabinose modification and the effects of polyamines, we compared the NPN uptake profiles of both classes of mutants when they were treated with various peptides. The aminoarabinose modification PA3553::lux mutant was most susceptible to polymyxin B and showed a high degree of NPN integration (Fig. 6A). The polyamine synthesis PA4774::lux mutant had an intermediate susceptibility phenotype compared to PAO1 and PA3553::lux (Fig. 6A). Mutations in PA4773 and PA4775 did not result in outer membrane permeability susceptibility to polymyxin B (data not shown). In addition to measuring the NPN uptake profile, we also calculated the rate of NPN uptake during the first 1,000 ms after polymyxin B treatment. When 6.4 μg/ml of polymyxin B was used, the rate of NPN uptake was 12 times greater in the PA4774::lux (626,660 arbitrary units [AU]/1,000 ms) mutant than in wild-type PAO1 (52,129 AU/1,000 ms). When using 3.2 μg/ml of polymyxin B, the rate of uptake is 6 times greater in the PA4774::lux mutant (138,846 AU/1,000 ms) than in the wild type (23,995 AU/1,000 ms). The chromosomally complemented strain demonstrated a restored polymyxin B resistance phenotype (Fig. 6B).
Fig 6.
Polyamines contribute to antibiotic resistance by protecting the outer membrane from antimicrobial peptide and gentamicin treatment. (A) NPN uptake profiles were measured in the PAO1, PA3553::lux, and PA4774::lux strains from the moment of polymyxin B addition (6.4 μg/ml). (B) NPN uptake profiles of PAO1, the PA4774::lux mutant, and the PA4774::luxCC strain challenged with 6.4 μg/ml polymyxin B. (C) NPN uptake profiles of the PAO1, PA3553::lux, and PA4774::lux strains following 10 μg/ml CP10A treatment. (D) NPN uptake profiles following 20 μg/ml gentamicin treatment for PAO1, the PA4774::lux mutant, and the PA4774::luxCC strain. The time frame includes the point of addition of antibiotic (0 s) and the following 60 s. Ten readings/s were recorded. Each experiment was performed three times, and results of a representative experiment are shown in each panel.
In addition to polymyxin B treatment, the PA4774::lux mutant also demonstrated a strong susceptibility to CP10A and gentamicin (Fig. 6C and D). The PA4774::lux mutant is more susceptible than the PA3553::lux mutant to the cationic antimicrobial peptide CP10A, an improved variant of indolicidin, which is consistent with the MIC phenotypes of these mutants (37). Gentamicin is an aminoglycoside that is also cationic and must cross the OM through the process of self-promoted uptake, similar to antimicrobial peptides, in order for it to reach the cytoplasm, where it acts to inhibit protein synthesis (18, 19, 39). The PA4774::luxCC strain demonstrated a strongly decreased membrane permeability to polymyxin B treatment (Fig. 6B) and a partially restored aminoglycoside resistance phenotype (Fig. 6D). PA4773 and PA4774 are annotated as SpeD and SpeE homologs, catalyzing the first and second steps in spermidine synthesis, respectively. Surprisingly, the PA4773 mutant did not have a peptide sensitivity phenotype as determined by the NPN assay. These data, together with the HPLC-UV-MS surface polyamine analysis (Fig. 5), suggest that only PA4774 and the final step in spermidine synthesis are critical for polymyxin B resistance.
Exogenous polyamines protect the outer membrane from polymyxin B treatment.
To confirm that polyamines act on the surface of the cell and protect the membrane from antimicrobial peptide treatment, we performed NPN uptake assays in the presence of exogenous polyamines. In Fig. 7A, it is shown that adding exogenous spermidine to the PA4774::lux mutant prior to polymyxin B treatment (within 10 min) reduced the initial rate and final level of NPN uptake following polymyxin B treatment. We next compared the abilities of various polyamines added exogenously to protect the OM from polymyxin B treatment. Figure 7B demonstrates that when low concentrations (10 μM) of cadaverine, putrescine, spermine, and spermidine are added, only spermine and spermidine reduce the OM permeability in the wild-type strain. The addition of increasing concentrations of exogenous spermidine caused a concentration-dependent reduction of the OM permeability of wild-type PAO1 (Fig. 7C). The addition of exogenous 25 to 50 μM cadaverine and putrescine also protected the OM from polymyxin B treatment, to a level similar to that seen with 10 μM spermidine (data not shown). It should be noted that this assay is a specific measure of the outer membrane properties, and thus surface polyamines can have an effect only if they bind the surface and alter the ability of polymyxin B to act on the membrane. Although polyamines can be imported into the cell, as they are also known to act as carbon sources, the addition of CCCP inhibits polyamine uptake systems (27).
Fig 7.
Exogenous polyamines protect the outer membrane from polymyxin B treatment. (A) NPN uptake profiles were measured in wild-type PAO1 and the PA4774::lux mutant with and without the exogenous addition of 25 μM spermidine (Spd), prior to challenge with 6.4 μg/ml polymyxin B. (B) NPN uptake profiles of wild-type PAO1 without and with the addition of 10 μM cadaverine (Cad), putrescine (Put), spermine (Sp), and spermidine (Spd), prior to challenge with 6.4 μg/ml polymyxin B. (C) NPN uptake profiles of wild-type PAO1 in the presence of increasing concentrations of spermidine (Spd), prior to challenge with 6.4 μg/ml polymyxin B. The time frame includes the point of addition of antibiotic (1 s) and the following 60 s. Ten readings/s were recorded. Each experiment was performed three times, and results of a representative experiment are shown in each panel.
Spermidine protects membrane lipids from oxidative damage.
Reactive oxygen species (ROS) are produced during the normal course of bacterial respiration and are produced by the host immune response during infection. There is some evidence in the literature that bacterial polyamines can act as antioxidants and protect cells from oxidative stress, but the mechanism is poorly understood (8). Polyamines are found in the cytoplasm and are known to protect DNA from oxidative stress (60, 66). ROS cause damage to DNA, protein, and lipids, and given the membrane localization of spermidine and putrescine in P. aeruginosa (Fig. 5), we wanted to determine if polyamines protect membrane lipids from oxidative damage.
The fluorescent probe C11-BODIPY581/591 has been used as an indicator of lipid peroxidation in eukaryotic cells (13, 52), but to our knowledge this is the first time it has been used in live bacteria. The probe integrates into membranes and undergoes a shift from red to green fluorescence emission upon peroxidation (13). The C11 lipid tail likely causes BODIPY581/591 insertion into the outer membrane, similar to what we have shown for other lipophilic dyes, i.e., FM 4-64 and FM 1-43 (36). We labeled mid-log-phase P. aeruginosa wild-type PAO1 and PA4774::lux cells with C11-BODIPY581/591 and showed that C11-BODIPY581/591 uniformly labels the P. aeruginosa membrane with red fluorescence (Fig. 8). To subject the cells to oxidative stress, P. aeruginosa was first labeled with C11-BODIPY581/591 and then treated with 5 mM H2O2. The conversion of red to green fluorescence in the bacterial membrane upon oxidative stress from H2O2 exposure was observed using fluorescence microscopy (Fig. 8).
Fig 8.
The fluorescent probe C11-BODIPY581/591 localized to the P. aeruginosa membrane as an indicator of lipid peroxidation. Mid-log-phase cultures of PAO1 and the PA4774::lux mutant were stained with 2 μM C11-BODIPY581/591 prior to treatment with 5 mM H2O2. (A) Phase-contrast images; (B) red fluorescence channel; (C) green fluorescence channel; (D) merged images. Images were taken with a 100× objective.
To measure the degree of oxidative damage, we performed a quantitative analysis of lipid peroxidation using a fixed amount of C11-BODIPY581/591 bound to membranes and measured the green fluorescence as an indicator of oxidative damage to lipids. As shown in Fig. 9A, PAO1 cells grown under noninducing conditions for PA4774 expression (2 mM Mg2+) displayed a concentration-dependent increase in peroxidation of membrane lipids upon exposure to increasing amounts of H2O2. Figure 9B illustrates that PAO1 cells grown under inducing conditions for PA4774 expression (0.02 mM Mg2+) showed resistance to oxidative damage by H2O2. To determine if the spermidine synthesis genes PA4773 and PA4774 contribute to this increased oxidative stress resistance, the phenotypes of these mutants were compared to that of wild-type PAO1. The PA4774::lux mutant showed a statistically significant increase in susceptibility to peroxidation membrane damage (Fig. 9C and D), while the PA4773 and PA4775::lux mutants were unaffected (data not shown). The PA4774::luxCC strain demonstrated parental levels of resistance to 25 mM H2O2 treatment (Fig. 9C). The exogenous addition of 1 mM spermidine was also able to restore near-parental levels of resistance to 10 mM H2O2 treatment in the PA4774::lux mutant (Fig. 9D). These data indicate that spermidine production protects membrane lipids from oxidative damage.
Fig 9.
The PA4774::lux mutant is more susceptible to lipid peroxidation than wild-type PAO1. Mid-log-phase cultures were stained with 2 μM C11-BODIPY581/591 and exposed to various concentrations of H2O2, and lipid peroxidation (green fluorescence) was measured every 10 min for 2 h. (A) Lipid peroxidation in PAO1 cultures grown under noninducing conditions with 2 mM Mg2+ and treated with increasing concentrations of H2O2. (B) Effect of growth under low-Mg2+ (0.02 mM) or high-Mg2+ (2 mM) conditions on resistance to oxidative membrane damage in PAO1 following 25 mM H2O2 treatment. (C) To complement the PA4774::lux mutant lipid peroxidation phenotype, the PA4774::luxCC strain was challenged with 25 mM H2O2 treatment (P < 0.01). (D) The addition of 1 mM exogenous spermidine (Spd) was tested for the ability to restore parental levels of lipid peroxidation to the PA4774::lux mutant following 10 mM H2O2 treatment (P < 0.05). (C and D) Green C11-BODIPY581/591 fluorescence was measured after 100 min of H2O2 exposure. Each value shown is the average of three technical replicates with standard deviations, and each experiment was performed three times. The unpaired Student's t test was used to compare the levels of lipid peroxidation among strains.
DISCUSSION
Polyamines are essential molecules for both prokaryotic and eukaryotic cells. Although the cytoplasmic functions of polyamines are well studied, the role of polyamines in the membrane is poorly understood. Polyamines have been previously identified in the outer membrane and LPS of Gram-negative bacteria (31) and linked to surface-associated functions that include spheroplast stabilization and attachment during biofilm formation (26, 54, 62). Here we present evidence for novel membrane functions of polyamines, obtained using approaches that specifically examined the role of polyamines in protecting the bacterial outer membrane from antibiotic treatment and oxidative stress.
In addition to regulation by membrane-damaging antibiotics and Mg2+ levels, the PA4773-PA4774 spermidine synthesis genes are also induced by the cation-chelating activity of extracellular DNA (Fig. 2). Collectively, the expression of these genes in the presence of multiple agents that cause membrane stress suggests a role for polyamines in membrane repair and stabilization. We confirmed the protective role of polyamines against antibiotic treatment by using the NPN uptake assay. The outer membrane of the PA4774::lux mutant had increased susceptibility to polymyxin B, CP10A, and gentamicin treatment (Fig. 6). The NPN assay also showed that exogenous polyamine addition protected the OM of both the wild-type PAO1 and the PA4774::lux strain from polymyxin B damage (Fig. 7). We showed a protective role for exogenous polyamines at low polyamine concentrations (10 to 50 μM), which is less than the intracellular concentration of spermidine (6 mM) and putrescine (20 mM) in E. coli (62). We also provided evidence that polyamines can act as a substitute for inorganic cations (Mg2+ and Ca2+), which are normally found in the outer membrane bound to LPS and function to cross-bridge and stabilize LPS in the outer membrane (19, 21) (Fig. 2D). Polyamines were interchangeable with Mg2+ cations in preventing expression of the spermidine synthesis genes and the arn operon, which is required for aminoarabinose modification of LPS (Fig. 4A and B). Exogenous polyamines also reduced peptide-induced expression of both PA3553::lux and PA4774::lux (Fig. 4C), suggesting that antimicrobial peptides and polyamines can compete for binding sites in the OM, further supporting the idea that polyamines act to stabilize the outer membrane to protect against antimicrobial attack. Spermidine (which has a 3+ charge) is more protective than the other bacterial polyamines cadaverine and putrescine (2+ charge) (Fig. 7), which may be due to the extra positive charge. As such, spermine (4+ charge) would be predicted to be more protective than spermidine. However, this was not the case at low concentrations (Fig. 7B), and at concentrations higher than 10 μM, spermine disrupts the OM permeability and has the lowest MIC of all polyamines (data not shown). The higher toxicity of spermine may help to explain why it is not frequently found in bacteria.
Polyamines were isolated from the P. aeruginosa cell surface and their identity confirmed using HPLC-UV-MS analysis (Fig. 5). This method of isolating surface polyamines has shown that the majority of polyamines isolated are specifically from the LPS in the outer membrane, with no contamination of cytoplasmic polyamines (Fig. 5E) (31). The lack of spermidine in the outer membrane of the PA4774::lux (speE homolog) mutant was the cause of increased susceptibility to polymyxin B, CP10A, and gentamicin treatment. The polyamine protection of the outer membrane likely occurs by binding to the negative surface charges, thus preventing the self-promoted uptake of antimicrobial peptides and aminoglycosides, which compete with cations for binding to LPS in the outer membrane (19). Thus, under limiting Mg2+ conditions or in the presence of extracellular DNA, spermidine production by P. aeruginosa is a novel mechanism that contributes to modification of the outer membrane surface. The noncovalent interactions of polyamines with the bacterial surface may work in addition to the aminoarabinose modification of the phosphates of LPS, both functioning to ensure that the negative surface charges are masked and limit membrane destabilization by cationic antibiotics.
PA4773 is annotated as a SAM decarboxylase, whose end product, dSAM, is a substrate with putrescine for spermidine synthesis by PA4774. Within the P. aeruginosa genome, there are the speE (PA0654) and speD (PA1687) genes, in addition to the homologs studied here. We propose that the genes PA4773 and PA4774 represent an inducible pathway for spermidine production. The PA4773-PA4775 gene cluster is coordinately expressed under limiting Mg2+ conditions (37, 43, 44) and in the presence of extracellular DNA (Fig. 2). However, mutation in the first gene in the cluster (PA4773) did not affect the NPN uptake, lipid peroxidation (data not shown), or spermidine production phenotypes (Fig. 5). One possible explanation is that the production of dSAM by PA4773 was performed by the true SpeD enzyme, implying a possible redundancy in gene function. And although the transposon insertion in PA4773 had a polar effect on downstream PA4774 expression, PA4774 expression was not completely lost (Fig. 3). A PA4774::lux mutant showed increased susceptibility to OM damage by antibiotics and oxidative stress, indicating that the final step in spermidine synthesis is essential for the production of spermidine and resistance to polymyxin B and oxidative stress.
Polyamines have been implicated in antibiotic resistance in P. aeruginosa. Paradoxically, high concentrations of exogenous spermidine (20 mM) can both increase and decrease the susceptibility of P. aeruginosa to various classes of antibiotics (34, 35). Similarly, a recent study showed that under conditions of increased intracellular spermidine production, E. coli, P. aeruginosa, and other bacteria are more resistant to tetracycline but more sensitive to kanamycin (4). The mechanism that accounts for these contrasting effects on antibiotic resistance is not understood. We show here that spermidine can stabilize the outer membrane and contribute to resistance, but the increased susceptibility may be due to the membrane-destabilizing effects of high polycation concentrations on the outer membrane, thus facilitating the entry of certain antibiotics (21).
We also showed that polyamines protect membrane lipids from oxidative damage using a new approach with a fluorescent indicator of lipid peroxidation. Microscopy was used to confirm the membrane localization of C11-BODIPY581/591, which is therefore a targeted indicator of damage to outer membrane lipids. A previous report implicated cadaverine in protecting E. coli from nitrosative stress, although the mechanism of protection was unclear (6). The presence of polyamines on the cell surface may act to scavenge ROS (16), thereby limiting the damage to the lipids buried in the membrane. It appears that polyamines can protect different macromolecules (lipids and DNA) in different cellular compartments (membrane and cytoplasm) from oxidative damage, dependent on their localization.
The identification of polyamines on the outer surface of P. aeruginosa (Fig. 5) indicates that polyamines are transported to the surface, and possibly secreted. There is evidence of putrescine and cadaverine secretion in E. coli using antiporter transport proteins (28, 45). A spermidine excretion protein complex, MdtJI, which belongs to the small multidrug resistance family of drug exporters, has been identified in E. coli by Higashi et al. (24). The function of secreted polyamines is unclear, but it is tempting to speculate secreted polyamines may play a role in cross-linking the anionic biofilm matrix polymers or may provide an attractive conditioning layer. Polyamines have been implicated in biofilm formation (26, 54), and there are examples of secreted proteins that cross-link and reinforce the exopolysaccharide matrix, thus promoting biofilm integrity (5). Extracellular polyamines have also been proposed to play a signaling role in biofilm formation (26). Our future work will attempt to identify the polyamine secretion system by testing the role of the known spermidine transport pathways in P. aeruginosa (40) and putative polyamine antiporters in the transport of polyamines to the cell surface, and possibly the extracellular environment.
We recently described the cation-chelating activity of DNA, which activates the PhoPQ two-component system and expression of the arn antimicrobial peptide resistance operon in P. aeruginosa (49). The spermidine synthesis genes PA4773 and PA4774 are a second example of DNA-induced genes that contribute to antimicrobial peptide resistance. These genes are likely expressed in P. aeruginosa growing in DNA-rich environments, including growth in biofilms, where extracellular DNA accumulates as a matrix polymer (2, 65), or during growth in the CF lung, where neutrophil DNA accumulates to high concentrations (57, 61, 63), possibly due to the formation of neutrophil extracellular traps (42). Herein we describe a dual, protective role for spermidine in resistance to aminoglycoside antibiotics, antimicrobial peptides, and oxidative stress, which can protect P. aeruginosa from antibiotic treatment and possibly from the host immune response.
Supplementary Material
ACKNOWLEDGMENTS
This work was funded by the Westaim Corporation, Alberta Science and Research Authority (ASRA) and Cystic Fibrosis Canada. S.L. holds the Westaim-ASRA chair in Biofilm Research. H.M. is the recipient of a Cystic Fibrosis Canada fellowship.
We thank R. Whittal and B. Reiz of the University of Alberta Mass Spectrometry Facility for their technical assistance in collecting of the HPLC-MS data, with instrument funding from a Western Economic Diversification grant. Additional thanks go to R. Yates of the University of Calgary for use of the spectrofluorimeter and J. B. McPhee for comments on the manuscript.
Footnotes
Published ahead of print 9 December 2011
Supplemental material for this article may be found at http://jb.asm.org/.
REFERENCES
- 1. Aaron SD, et al. 2002. Single and combination antibiotic susceptibilities of planktonic, adherent, and biofilm-grown Pseudomonas aeruginosa isolates cultured from sputa of adults with cystic fibrosis. J. Clin. Microbiol. 40:4172–4179 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Allesen-Holm M, et al. 2006. A characterization of DNA release in Pseudomonas aeruginosa cultures and biofilms. Mol. Microbiol. 59:1114–1128 [DOI] [PubMed] [Google Scholar]
- 3. Anderson GG, O'Toole GA. 2008. Innate and induced resistance mechanisms of bacterial biofilms. Curr. Top. Microbiol. Immunol. 322:85–105 [DOI] [PubMed] [Google Scholar]
- 4. Bernier SP, Letoffe S, Delepierre M, Ghigo JM. 2011. Biogenic ammonia modifies antibiotic resistance at a distance in physically separated bacteria. Mol. Microbiol. 81:705–716 [DOI] [PubMed] [Google Scholar]
- 5. Borlee BR, et al. 2010. Pseudomonas aeruginosa uses a cyclic-di-GMP-regulated adhesin to reinforce the biofilm extracellular matrix. Mol. Microbiol. 75:827–842 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Bower JM, Mulvey MA. 2006. Polyamine-mediated resistance of uropathogenic Escherichia coli to nitrosative stress. J. Bacteriol. 188:928–933 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Ceri H, et al. 1999. The Calgary biofilm device: new technology for rapid determination of antibiotic susceptibilities of bacterial biofilms. J. Clin. Microbiol. 37:1771–1776 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Chattopadhyay MK, Tabor CW, Tabor H. 2003. Polyamines protect Escherichia coli cells from the toxic effect of oxygen. Proc. Natl. Acad. Sci. U. S. A. 100:2261–2265 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Choi KH, Schweizer HP. 2006. Mini-Tn7 insertion in bacteria with single attTn7 sites: example Pseudomonas aeruginosa. Nat. Protoc. 1:153–161 [DOI] [PubMed] [Google Scholar]
- 10. Choi KH, et al. 2005. A Tn7-based broad-range bacterial cloning and expression system. Nat. Methods 2:443–448 [DOI] [PubMed] [Google Scholar]
- 11. Costerton JW, Stewart PS, Greenberg EP. 1999. Bacterial biofilms: a common cause of persistent infections. Science 284:1318–1322 [DOI] [PubMed] [Google Scholar]
- 12. Davies D. 2003. Understanding biofilm resistance to antibacterial agents. Nat. Rev. Drug Discov. 2:114–122 [DOI] [PubMed] [Google Scholar]
- 13. Drummen GP, van Liebergen LC, Op den Kamp JA, Post JA. 2002. C11-BODIPY(581/591), an oxidation-sensitive fluorescent lipid peroxidation probe: (micro)spectroscopic characterization and validation of methodology. Free Radic. Biol. Med. 33:473–490 [DOI] [PubMed] [Google Scholar]
- 14. Gaynes R, Edwards JR. 2005. Overview of nosocomial infections caused by gram-negative bacilli. Clin. Infect. Dis. 41:848–854 [DOI] [PubMed] [Google Scholar]
- 15. Guo K, Li L. 2009. Differential 12C-/13C-isotope dansylation labeling and fast liquid chromatography/mass spectrometry for absolute and relative quantification of the metabolome. Anal. Chem. 81:3919–3932 [DOI] [PubMed] [Google Scholar]
- 16. Ha HC, et al. 1998. The natural polyamine spermine functions directly as a free radical scavenger. Proc. Natl. Acad. Sci. U. S. A. 95:11140–11145 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Hancock RE. 1998. Resistance mechanisms in Pseudomonas aeruginosa and other nonfermentative gram-negative bacteria. Clin. Infect. Dis. 27(Suppl. 1):S93–S99 [DOI] [PubMed] [Google Scholar]
- 18. Hancock RE. 1997. The bacterial outer membrane as a drug barrier. Trends Microbiol. 5:37–42 [DOI] [PubMed] [Google Scholar]
- 19. Hancock RE. 1984. Alterations in outer membrane permeability. Annu. Rev. Microbiol. 38:237–264 [DOI] [PubMed] [Google Scholar]
- 20. Hancock RE, Speert DP. 2000. Antibiotic resistance in Pseudomonas aeruginosa: mechanisms and impact on treatment. Drug Resist Updat. 3:247–255 [DOI] [PubMed] [Google Scholar]
- 21. Hancock RE, Wong PG. 1984. Compounds which increase the permeability of the Pseudomonas aeruginosa outer membrane. Antimicrob. Agents Chemother. 26:48–52 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Hebecker S, et al. 2011. Alanyl-phosphatidylglycerol synthase: mechanism of substrate recognition during tRNA-dependent lipid modification in Pseudomonas aeruginosa. Mol. Microbiol. 80:935–950 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Helander IM, Mattila-Sandholm T. 2000. Fluorometric assessment of gram-negative bacterial permeabilization. J. Appl. Microbiol. 88:213–219 [DOI] [PubMed] [Google Scholar]
- 24. Higashi K, et al. 2008. Identification of a spermidine excretion protein complex (MdtJI) in Escherichia coli. J. Bacteriol. 190:872–878 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Jensen PO, Givskov M, Bjarnsholt T, Moser C. 2010. The immune system vs. Pseudomonas aeruginosa biofilms. FEMS Immunol. Med. Microbiol. 59:292–305 [DOI] [PubMed] [Google Scholar]
- 26. Karatan E, Duncan TR, Watnick PI. 2005. NspS, a predicted polyamine sensor, mediates activation of Vibrio cholerae biofilm formation by norspermidine. J. Bacteriol. 187:7434–7443 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Kashiwagi K, Kobayashi H, Igarashi K. 1986. Apparently unidirectional polyamine transport by proton motive force in polyamine-deficient Escherichia coli. J. Bacteriol. 165:972–977 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Kashiwagi K, Miyamoto S, Suzuki F, Kobayashi H, Igarashi K. 1992. Excretion of putrescine by the putrescine-ornithine antiporter encoded by the potE gene of Escherichia coli. Proc. Natl. Acad. Sci. U. S. A. 89:4529–4533 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Kirketerp-Moller K, et al. 2008. Distribution, organization, and ecology of bacteria in chronic wounds. J. Clin. Microbiol. 46:2717–2722 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Klein S, et al. 2009. Adaptation of Pseudomonas aeruginosa to various conditions includes tRNA-dependent formation of alanyl-phosphatidylglycerol. Mol. Microbiol. 71:551–565 [DOI] [PubMed] [Google Scholar]
- 31. Koski P, Vaara M. 1991. Polyamines as constituents of the outer membranes of Escherichia coli and Salmonella typhimurium. J. Bacteriol. 173:3695–3699 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Koski P, Helander IM, Sarvas M, Vaara M. 1987. Analysis of polyamines as their dabsyl derivatives by reversed-phase high-performance liquid chromatography. Anal. Biochem. 164:261–266 [DOI] [PubMed] [Google Scholar]
- 33. Kristian SA, Durr M, Van Strijp JA, Neumeister B, Peschel A. 2003. MprF-mediated lysinylation of phospholipids in Staphylococcus aureus leads to protection against oxygen-independent neutrophil killing. Infect. Immun. 71:546–549 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Kwon DH, Lu CD. 2006. Polyamines increase antibiotic susceptibility in Pseudomonas aeruginosa. Antimicrob. Agents Chemother. 50:1623–1627 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Kwon DH, Lu CD. 2006. Polyamines induce resistance to cationic peptide, aminoglycoside, and quinolone antibiotics in Pseudomonas aeruginosa PAO1. Antimicrob. Agents Chemother. 50:1615–1622 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Lewenza S, Vidal-Ingigliardi D, Pugsley AP. 2006. Direct visualization of red fluorescent lipoproteins indicates conservation of the membrane sorting rules in the family Enterobacteriaceae. J. Bacteriol. 188:3516–3524 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Lewenza S, et al. 2005. Construction of a mini-Tn5-luxCDABE mutant library in Pseudomonas aeruginosa PAO1: a tool for identifying differentially regulated genes. Genome Res. 15:583–589 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Livak KJ, Schmittgen TD. 2001. Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCT method. Methods 25:402–408 [DOI] [PubMed] [Google Scholar]
- 39. Loh B, Grant C, Hancock RE. 1984. Use of the fluorescent probe 1-N-phenylnaphthylamine to study the interactions of aminoglycoside antibiotics with the outer membrane of Pseudomonas aeruginosa. Antimicrob. Agents Chemother. 26:546–551 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Lu CD, Itoh Y, Nakada Y, Jiang Y. 2002. Functional analysis and regulation of the divergent spuABCDEFGH-spuI operons for polyamine uptake and utilization in Pseudomonas aeruginosa PAO1. J. Bacteriol. 184:3765–3773 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Mah TF, et al. 2003. A genetic basis for Pseudomonas aeruginosa biofilm antibiotic resistance. Nature 426:306–310 [DOI] [PubMed] [Google Scholar]
- 42. Marcos V, et al. 2010. CXCR2 mediates NADPH oxidase-independent neutrophil extracellular trap formation in cystic fibrosis airway inflammation. Nat. Med. 16:1018–1023 [DOI] [PubMed] [Google Scholar]
- 43. McPhee JB, Lewenza S, Hancock RE. 2003. Cationic antimicrobial peptides activate a two-component regulatory system, PmrA-PmrB, that regulates resistance to polymyxin B and cationic antimicrobial peptides in Pseudomonas aeruginosa. Mol. Microbiol. 50:205–217 [DOI] [PubMed] [Google Scholar]
- 44. McPhee JB, et al. 2006. Contribution of the PhoP-PhoQ and PmrA-PmrB two-component regulatory systems to Mg2+-induced gene regulation in Pseudomonas aeruginosa. J. Bacteriol. 188:3995–4006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Meng SY, Bennett GN. 1992. Nucleotide sequence of the Escherichia coli cad operon: a system for neutralization of low extracellular pH. J. Bacteriol. 174:2659–2669 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Moreau-Marquis S, Stanton BA, O'Toole GA. 2008. Pseudomonas aeruginosa biofilm formation in the cystic fibrosis airway. Pulm. Pharmacol. Ther. 21:595–599 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Moskowitz SM, Ernst RK, Miller SI. 2004. PmrAB, a two-component regulatory system of Pseudomonas aeruginosa that modulates resistance to cationic antimicrobial peptides and addition of aminoarabinose to lipid A. J. Bacteriol. 186:575–579 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Mulcahy H, Charron-Mazenod L, Lewenza S. 2010. Pseudomonas aeruginosa produces an extracellular deoxyribonuclease that is required for utilization of DNA as a nutrient source. Environ. Microbiol. 12:1621–1629 [DOI] [PubMed] [Google Scholar]
- 49. Mulcahy H, Charron-Mazenod L, Lewenza S. 2008. Extracellular DNA chelates cations and induces antibiotic resistance in Pseudomonas aeruginosa biofilms. PLoS Pathog. 4:e1000213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Murata T, Tseng W, Guina T, Miller SI, Nikaido H. 2007. PhoPQ-mediated regulation produces a more robust permeability barrier in the outer membrane of Salmonella enterica serovar Typhimurium. J. Bacteriol. 189:7213–7222 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Nickel JC, Ruseska I, Wright JB, Costerton JW. 1985. Tobramycin resistance of Pseudomonas aeruginosa cells growing as a biofilm on urinary catheter material. Antimicrob. Agents Chemother. 27:619–624 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Pap EH, et al. 1999. Ratio-fluorescence microscopy of lipid oxidation in living cells using C11-BODIPY(581/591). FEBS Lett. 453:278–282 [DOI] [PubMed] [Google Scholar]
- 53. Parsek MR, Singh PK. 2003. Bacterial biofilms: an emerging link to disease pathogenesis. Annu. Rev. Microbiol. 57:677–701 [DOI] [PubMed] [Google Scholar]
- 54. Patel CN, et al. 2006. Polyamines are essential for the formation of plague biofilm. J. Bacteriol. 188:2355–2363 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Peschel A, et al. 1999. Inactivation of the dlt operon in Staphylococcus aureus confers sensitivity to defensins, protegrins, and other antimicrobial peptides. J. Biol. Chem. 274:8405–8410 [DOI] [PubMed] [Google Scholar]
- 56. Ramadan HH. 2006. Chronic rhinosinusitis and bacterial biofilms. Curr. Opin. Otolaryngol. Head Neck Surg. 14:183–186 [DOI] [PubMed] [Google Scholar]
- 57. Ranasinha C, et al. 1993. Efficacy and safety of short-term administration of aerosolised recombinant human DNase I in adults with stable stage cystic fibrosis. Lancet 342:199–202 [DOI] [PubMed] [Google Scholar]
- 58. Savli H, et al. 2003. Expression stability of six housekeeping genes: A proposal for resistance gene quantification studies of Pseudomonas aeruginosa by real-time quantitative RT-PCR. J. Med. Microbiol. 52:403–408 [DOI] [PubMed] [Google Scholar]
- 59. Schurek KN, et al. 2009. Involvement of pmrAB and phoPQ in polymyxin B adaptation and inducible resistance in non-cystic fibrosis clinical isolates of Pseudomonas aeruginosa. Antimicrob. Agents Chemother. 53:4345–4351 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Shah P, Swiatlo E. 2008. A multifaceted role for polyamines in bacterial pathogens. Mol. Microbiol. 68:4–16 [DOI] [PubMed] [Google Scholar]
- 61. Shak S, Capon DJ, Hellmiss R, Marsters SA, Baker CL. 1990. Recombinant human DNase I reduces the viscosity of cystic fibrosis sputum. Proc. Natl. Acad. Sci. U. S. A. 87:9188–9192 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Tabor CW, Tabor H. 1985. Polyamines in microorganisms. Microbiol. Rev. 49:81–99 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Ulmer JS, et al. 1996. Engineering actin-resistant human DNase I for treatment of cystic fibrosis. Proc. Natl. Acad. Sci. U. S. A. 93:8225–8229 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Vaara M. 1992. Agents that increase the permeability of the outer membrane. Microbiol. Rev. 56:395–411 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64a. West SE, Schweizer HP, Dall C, Sample AK, Runyen-Janecky LJ. 1994. Construction of improved Escherichia-Pseudomonas shuttle vectors derived from pUC18/19 and sequence of the region required for their replication in Pseudomonas aeruginosa. Gene 148:81–86 [DOI] [PubMed] [Google Scholar]
- 65. Whitchurch CB, Tolker-Nielsen T, Ragas PC, Mattick JS. 2002. Extracellular DNA required for bacterial biofilm formation. Science 295:1487. [DOI] [PubMed] [Google Scholar]
- 66. Wortham BW, Patel CN, Oliveira MA. 2007. Polyamines in bacteria: pleiotropic effects yet specific mechanisms. Adv. Exp. Med. Biol. 603:106–115 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.